Abstract
Circular RNAs (circRNAs) are RNAs with a unique circular structure that is generated from back-splicing processes. These circular molecules were discovered more than 40 years ago but failed to raise scientific interest until lately. Increasing studies have found that these circular RNAs might not just be byproducts of the splicing process but possess important regulatory functions through different cellular events. Most circular RNAs are currently being studied in the field of cancer, and many of them have been confirmed to be involved in the process of tumorigenesis. However, many circular RNAs are implicated in the developmental stages of diseases other than cancer. In this review, we focus on discussing the role of circular RNAs in non-cancer diseases, especially in cardiovascular diseases. Following the summary of the life cycle of circRNAs, we provide input on studying circRNA-protein interactions based on our experience, which modulate protein translocation. Furthermore, we outline the potential of circRNAs to be potent biomarkers, effective therapeutic targets, and potential treatments in cardiovascular diseases as well as other non-cancer fields.
Keywords: circRNAs, circular RNAs, cardiovascular, translocation, biomarker, therapeutic
Graphical abstract

This review focuses on discussing the roles of circRNAs in non-cancer diseases. The authors provide input on studying circRNA-protein interactions, which modulate protein translocations and functions. The therapeutic potentials of circRNAs in cardiovascular diseases and other non-cancer disorders are discussed.
Main text
Viroids were the first circular RNAs (circRNAs) discovered in pathogens. Sanger et al.1 first described viroids in 1976 as “single-stranded and covalently closed circular RNA molecules.” Following this, another study conducted by Hsu and Coca-Prados2 in 1979 described some circRNAs with no free flanking ends. They raised the possibility that the circularity of these RNAs is not dependent on any protein interactions. During that period of time, studies failed to follow up on these unexpected findings due to an explosion of scientific interest in studying the functions of linear RNA, especially after the polymerase chain reaction was invented in 1985. There were some studies in the late 1990s that described transcript products generated from non-canonical splicing. These products were reported to be generated from the deletion in a colon cancer gene3 and human EST-1 gene,4 as well as their isoforms. Later, a few more studies proposed potential biogenesis mechanisms for these circular molecules. A well-known hypothesis proposed by Dubin et al.5 based on the circRNA sry is that complementary intronic sequences drive circularization. Another hypothesis proposed by Pasman et al.6 is that these circular molecules could be produced from nuclear extracts. circRNAs were found to be mostly located in the cytoplasm and expressed in a stage and tissue-specific manner. Up until the beginning of the 21st century, the number of studies on circRNAs increased, but they were still sporadic. People classified circRNAs as non-linear mRNAs,7 RNAs with scrambled exons, or RNAs with exon shuffling.8 circRNAs were generally regarded as products of splicing errors that were rare and held little significance. Although the presence of circRNAs was documented decades ago, very little about their biogenesis and functions were understood until the past decade.
Starting in 2010, advancements in RNA-sequencing technologies along with the buildup of bioinformatics computational pipelines allowed circRNA research to explode. Several bioinformatics studies reported hundreds and thousands of circRNAs that were highly conserved.9,10 In addition to discovering new circRNA products, revised RNA-sequencing techniques allowed for specific detection of circular products without overlapping detection of linear products. These methods include RNase R treatment of the samples before sequencing, which digests linear RNAs without affecting circRNAs, resulting in an enrichment of circRNAs compared to linear RNAs. Since 2017, computational pipelines and databases for circRNA annotation and quantification have been established and developed over time. On top of powerful bioinformatics techniques, novel methods for circRNA verification and validation were also developed.11 Most of these methods were adapted from linear RNA detection techniques. These include quantitative reverse transcriptase PCR, northern blot, and, later on, other quantitative PCRs such as digital PCR were developed. These methods made it possible for wet laboratory scientists to validate data from dry laboratories. Following the identification and validation of circRNAs, methods including vector expression plasmids and siRNA silencing were used to overexpress and knock down circRNAs, respectively, to study their functions. Furthermore, methods such as in situ hybridization and pull-down assays were used to study the underlying molecular mechanisms of circRNAs. With the advent of all of these techniques, circRNA research was able to expand. More and more studies have contributed to the conclusion that circRNAs are not simply byproducts of splicing errors and actually hold critical physiological and pathological functions during biological processes.
In this review, we provide a brief update on the life cycle of circRNAs. We discuss the localization and translocation of circRNAs. We then bring new insights into circRNA functions in the cardiovascular field and discuss the potential of circRNA-based diagnostic and therapeutic strategies in the cardiovascular field as well as other non-cancer diseases.
Life cycle of circRNA: biogenesis and degradation
Biogenesis of circRNA
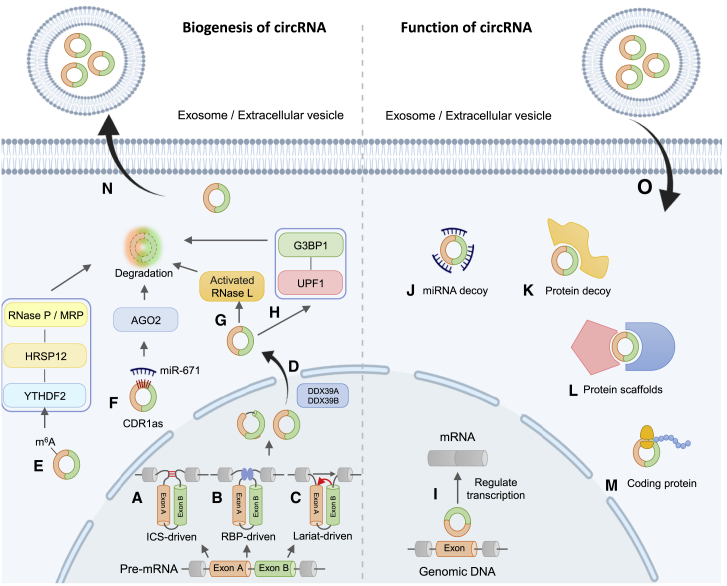
circRNAs have been extensively reported as evolutionarily conserved, stable, and tissue-specific, as well as subcellular location- and developmental stage-specific. circRNAs can be generated through a direct back-splicing process where the 3′ terminal of the RNA is directly joined to the 5′ terminal during the splicing process of precursor mRNA and thereby proceed to form a circular structure. This back-splicing process could be driven by an intronic complementary sequence (Figure 1A) or RNA-binding protein (RBP) (Figure 1B). They can also be generated by a lariat-mediated back-splicing biogenesis model where the RNA sequence that is cut off after the splicing process of the precursor mRNA forms a lariat structure, and such structure is further spliced to form a circular structure (Figure 1C).12 DDX39A and DDX39B are proteins that export the formed circular structure to the cytoplasm from the nucleus (Figure 1D). While DDX39A usually transports short circRNAs, DDX39B assists in forming longer ones.13 Formed circRNAs are ordinarily stable owing to their exclusive circular structure, which defends them from cleavage of the exonuclease. However, circRNAs still undergo degradation.
Figure 1.
Biogenesis and functional mechanisms of circRNA
(A) In the intronic complementary sequence (ICS)-driven back-splicing biogenesis model, the intron regions on both sides of the exons contain complementary sequences, which are paired and tightly connected and promote back-splicing to form a circular structure. (B) In the RNA-binding protein (RBP)-driven back-splicing biogenesis model, under the bridging action of RBP, the splicing sites at both ends of the exon are directly connected and promote back-splicing to form a circular structure. (C) In the lariat-driven back-splicing biogenesis model, the RNA sequence that is cut off during linear RNA splicing forms a lariat structure, and then forms a circular structure through back-splicing. (D) DDX39A and DDX39B proteins transport mature circRNAs from the nucleus to the cytoplasm. (E–H) The degradation process of circRNAs. (E) The YTHDF2 protein recognizes the m6A modification site of circRNA. At the same time, HRSP12 can bridge YTHDF2 and RNase P/MRP to form a complex, thereby initiating the circRNA degradation via RNase P/MRP. (F) MicroRNA miR-671 mediates the binding of CDR1as to AGO2 and triggers the degradation process regulated by PKR activation. (G) Pathological exogenous double-stranded RNA can activate RNase L in cells to degrade circRNAs. (H) The complex of UPF1 and G3BP1 proteins can mediate the degradation of circRNAs. (I) In the process of gene transcription, circRNAs can block exons by binding to genomic DNA, resulting in exon deletion during mRNA splicing. (J) circRNAs can serve as a miRNA decoy, inhibiting miRNA functions by binding to the miRNAs. (K) circRNAs inhibit biological activity of proteins by binding to their functional domain. (L) circRNAs combine with multiple proteins to form complexes and change their biological activity. (M) circRNAs can be translated into polypeptides or proteins. (N) Exosomes or extracellular vesicles can wrap circRNAs and secrete them out of the cell. (O) Under certain conditions, cells can take up exosomes or extracellular vesicles, and the intake circRNAs can function in the recipient cells.
Degradation of circRNA
Due to the unique circular structure of circRNAs, they cannot be eliminated via conventional RNA degradation pathways, and their half-life is longer compared to linear RNA. However, they are still degraded, although their degradation mechanisms are not fully understood. Theoretically, even though the circular structure of circRNAs protects them from exonuclease cleavage, the decay process could still be initiated by endonucleases and completed by a cascade of exonucleases or endonucleases. A recent study indicated that the modification N6-methylation of adenosine (m6A) of circRNA could be recognized by HRSP12, an m6A reader protein, which can interact with the RNase P/MRP endonuclease complex to trigger circRNA degradation14 (Figure 1E). The degradation of circRNAs mediated by synthetic short hairpin RNAs (shRNAs)/small interfering RNAs (siRNAs) is a conventionalmethod used to study the functions of circRNAs. The only example of naturally occurring small RNA-mediated circRNA degradation involves miR-671, which has high complementarity to conserved binding sites on CDR1as and induces AGO2-mediated degradation15 (Figure 1F). Another study found that poly(I:C) stimulation or encephalomyocarditis virus (EMCV) infection introduces pathological exogenous double-stranded RNA (dsRNA) into HeLa cells and activates endoribonuclease RNase L16 (Figure 1G). In addition, there was spontaneous RNase L activation in peripheral blood mononuclear cells (PBMCs) that were derived from patients with systemic lupus erythematosus (SLE).16 Such RNase L-mediated circRNA degradation requires protein kinase R (PKR) activation that is regulated by the formation of intra-dsRNA duplexes, a specific structure formed by many circRNAs. This activated RNase L can cause global degradation of circRNA. For circRNAs with complex secondary structures, the highly structured regions can be recognized by ATP-dependent RNA helicase upstream frameshift 1 (UPF1) as well as endonuclease G3BP1, which subsequently induce circRNA degradation17 (Figure 1H).
In addition to degradation inside the cells, exocytosis or endocytosis might occur to discharge circRNAs from cells or take them up to cells through exosomes18,19 (Figures 1N and 1O). Increasing evidence has shown that circRNAs are present in exosomes.19,20 In some cases, circRNA levels in exosomes could be far more than those in cells.18,21 Exosomal circRNAs can bind to proteins and become more involved in cell-to-cell communication.22 In sum, a lot remains unknown about circRNA degradation and mediation of signal communication, and this awaits to be addressed in future investigations.
Modes of action
Functional mechanisms in diseases
circRNAs have been found to function through some proposed mechanisms: regulating transcription (Figure 1I), acting as microRNA (miRNA) sponges (Figure 1J), regulating other protein activities through binding them or scaffolding them (Figures 1K and 1L), and encoding proteins (Figure 1M). Beyond circRNAs in cancer fields, the functions of circRNAs in other non-cancer diseases, especially cardiovascular diseases, have been reviewed as well.23, 24, 25, 26 In this review, we classify some new and interesting developments based on functional mechanisms of circRNAs in the cardiovascular field (Figure 2).
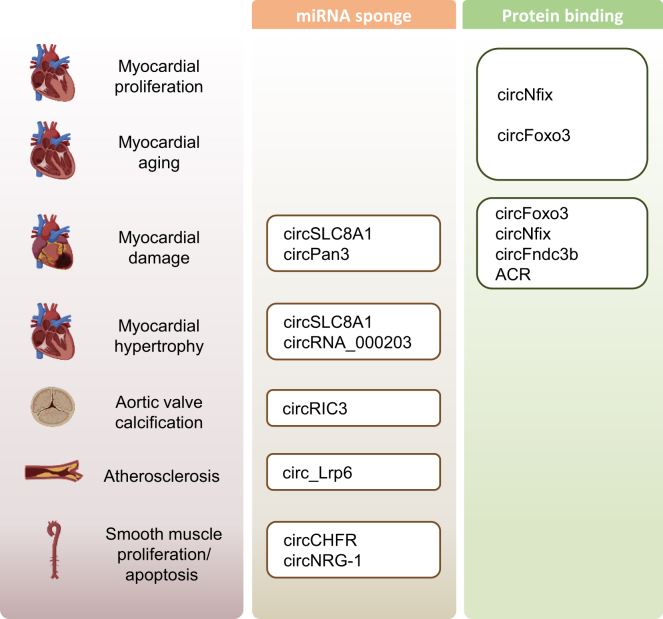
Figure 2.
circRNAs related to cardiovascular diseases and their functional mechanism
The diagram shows relevant circRNA functions in the cardiovascular system. The left column in pink specifies the cardiovascular dysfunction of interest. The middle column in orange indicates circRNAs that could lead to cardiovascular diseases through the miRNA sponge mechanism. The right column in green indicates circRNAs that could lead to cardiovascular diseases through the protein-binding mechanism.
miRNA sponge
The most abundant circRNA in cardiomyocytes is circSLC8A1, which plays a role in myocardial hypertrophy caused by pressure overload27,28 and myocardial damage caused by ischemia-reperfusion29 by sponging miR-133a. In an angiotensin II-induced cardiac hypertrophy mouse model, circRNA_000203 was upregulated in cardiomyocytes and acts as a sponge for miR-26b-5p and miR-140-3p, leading to increased expression of Gata4 and aggravated cardiac hypertrophy.30 A recent study showed that in calcific aortic valve disease (CAVD), circRIC3 is significantly upregulated, leading to osteogenic trans-differentiation of valvular interstitial cells through the circRIC3/miR-204-5p/DPP4 pathway, an indicator of accelerated valvular calcification. It was demonstrated that melatonin inhibits circRIC3 expression, thereby reducing aortic valve calcification.31 Similarly, in vascular smooth muscle cells, overexpression of circLRP6 promotes atherosclerosis development by sponging miR-145.32,33 In aortic smooth muscle, circCHFR promotes the migration and proliferation of the vascular smooth muscle through the miR-370/FOXO1/cyclin D1 pathway;34 meanwhile, circNRG-1 promotes smooth muscle cell apoptosis through miR-193b-5p/NRG-1.35 Not only can circRNA exert regulatory control over miRNA, but miRNA can also regulate circRNA activities. In doxorubicin (Dox)-induced cardiotoxicity, miR-31-5p is upregulated, which suppresses Quaking (QKI), an RBP known to influence circRNA production. As a result, circPAN3 synthesis is suppressed, which leads to cardiomyocyte apoptosis.36
Protein binding
Besides regulating gene expression via the classic miRNA sponging mechanism, circRNAs play roles in cardiac disease via other modes of action. In cardiomyocytes, circFoxo3 expression is significantly higher in older heart tissues compared to younger heart tissues.37 We found that circFoxo3 interacts with ID-1 and E2F1, FAK, and HIF-1α, which are anti-senescence proteins and anti-stress proteins, respectively, preventing their transfer from cytoplasm to nucleus and mitochondria, which blocks their downstream activity. In a Dox-induced mouse model of cardiomyopathy, circFoxo3 promotes cellular senescence and aggravates Dox-induced cardiomyopathy.37 Similarly, the expression of super-enhancer-regulated circNfix is also higher in adult hearts than neonatal hearts. circNfix regulates Gsk3β signaling activity by sponging miR-214, and also binds to Y-box binding protein 1 (Ybx1) and an E3 ubiquitin ligase (Nedd4l) to promote the interaction between them and induce Ybx1 degradation. As a result of these two modes of action, cardiomyocyte proliferation is suppressed. It was shown that inhibiting circNfix led to promotion of cardiomyocyte proliferation and angiogenesis, reduced myocardial dysfunction, and protected the heart after myocardial infarction (MI).38 circFndc3b, whose expression is decreased after MI, has a similar protein binding function. Upregulation of circFndc3b results in the enhancement of angiogenesis, reduction of infarct size, and improvement of post-MI cardiac function by forming a complex with fused in sarcoma (FUS) and vascular endothelial growth factor-A (VEGF-A).39 Lastly, autophagy-related circRNA (ACR) binds to Dnmt3b, which inhibits the methylation of Pink1 gene, thereby promoting Pink1 expression. Pink1 promotes the phosphorylation of FAM65B, which inhibits cell death and autophagy in the heart, ultimately protecting the heart from reperfusion injury.40
Translation
van Heesch et al.’s41 breakthrough discovery from 80 human heart translatomes found that at least 40 circRNAs produced from 39 genes have the ability to encode proteins, including newly detected ribosome-associated circRNAs, such as circCFLAR, circSLC8A1, circMYBPC3, circRYR2, and CDR1as. The authors first identified circRNAs bound to ribosomes to detect translational potential. Then, they compared the nucleic acid sequence of the ribosome binding region with the back-splicing junction region of circRNAs to confirm that the circRNAs bound by ribosomes were in a circular structure. Furthermore, they verified the polypeptide (or microprotein) encoded by the back-splicing junction region of six circRNAs by shotgun mass spectrometry. Translations of 5 out of 40 circRNAs were considered to be m6A-driven, while others were more likely to be IRES-driven.42 These circRNAs may have translation potential in other organs and tissues besides the heart, such as the liver and kidney.41 Although the roles of peptides/microproteins encoded by circRNAs in cardiovascular disease have not been fully uncovered, these findings reveal increased functional diversity of circRNAs, which were previously thought to be non-coding. Elucidating the function of these peptides/microproteins will likely increase our understanding of the physiological and pathological states of the heart.
Subcellular localization and translocation
Even though miRNA sponge mechanisms were applied for loads of circRNA functions in all disease fields, direct binding between circRNA-mRNA or circRNA-protein started to raise interest in their serving as mechanisms underlying circRNA functions. One important consequence of these interactions is the regulation of the localization of proteins.
Cytoplasm localization of proteins can be facilitated by circRNAs. circFoxo3, for instance, a circRNA that is highly expressed in the heart tissues of aged mice and patients, as mentioned before, promoted cellular senescence in vitro and decreased cardiac functions in vivo. circFoxo3 is mainly localized in the cytoplasm, where the interaction between it and ID1 and E2F1 (the anti-senescence proteins), as well as FAK and HIF1α (the anti-stress proteins), led to retention of these proteins in the cytoplasm. As a result, the functions of these affected proteins that act as either transcription factors in the nucleus (ID1, E2F1, HIF1α) or kinase (FAK) in the mitochondria were suppressed, ultimately facilitating senescence of cells37 (Figure 3A).
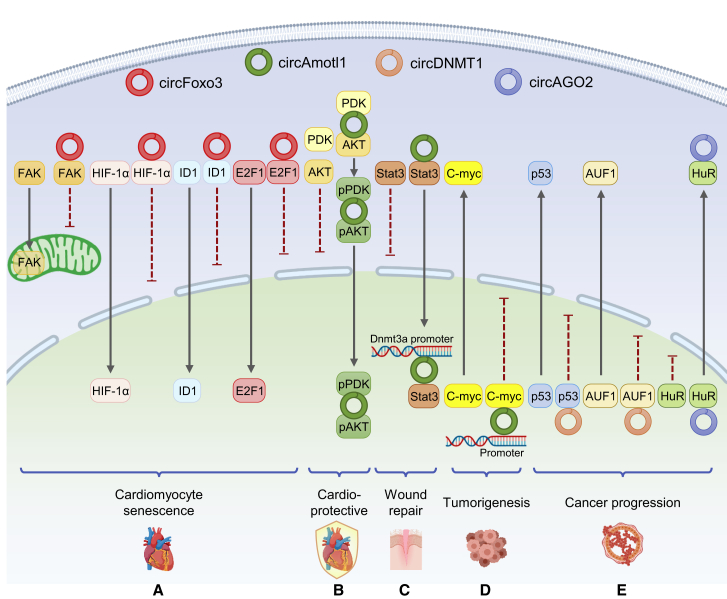
Figure 3.
The role of circRNAs in intracellular protein translocation
(A) circFoxo3 binds to proteins and inhibits their translocation, leading to senescence of cardiomyocytes. circFoxo3 binds to FAK protein and inhibits the translocation of FAK into mitochondria. circFoxo3 binds to HIF-1α, ID1, and E2F1, respectively, and inhibits their nuclear translocation. (B) circAmotl1 can act as a protein scaffold to connect AKT with PDK, leading to their phosphorylation and nuclear translocation, and facilitating cardioprotection. (C) circAmotl1 binds to Stat3 protein and facilitates its nuclear translocation. In the nucleus, circAmotl1 mediates the interaction between the Stat3 and Dnmt3a promoter, thereby promoting Dnmt3a transcription and accelerating wound repair. (D) In the nucleus, circAmotl1 inhibits the translocation of c-myc protein into the cytoplasm by binding to c-myc protein. At the same time, it can also mediate the interaction between c-myc and the promoters of various genes, thereby promoting transcription to promote tumorigenesis. (E) In the nucleus, circDnmt1 inhibits the transfer of p53 and AUF1 protein into the cytoplasm by binding to them, respectively. circAGO2 binds to HuR protein and facilitates its translocation into cytoplasm. Both of these processes can promote cancer progression. The solid black line with an arrow indicates that the protein can translocate across the membrane, and the dotted red line with a horizontal line indicates that the translocation of the protein across the membrane is inhibited.
Other than cytoplasm localization, localization of proteins in the nucleus could be affected by circRNAs as well. Nuclear AKT possesses a protective role in cardiovascular diseases. Previously in our laboratory, we found that circAmotl1 acts as a scaffold for AKT1 and PDK1, promoting AKT1 phosphorylation and nuclear translocation.43 The accumulation of phosphorylated AKT1 in the nucleus resulted in cardioprotective effects in primary cardiomyocytes and in a Dox-induced cardiomyopathy mouse model43 (Figure 3B). circAmotl1 can also accelerate wound repair through binding with STAT3 protein and promote its nuclear translocation.44 Subsequently, STAT3 binds to the promoter of Dnmt3a, which encodes a DNA methyltransferase, thereby activating its transcription. Dnmt3a methylation of the miR-17 promotor suppressed expression of miR-17-5p and increased expression of fibronectin.44 Consequently, these biological cascades led to an increase in cell adhesion, migration, and proliferation, thus accelerating the wound repair process (Figure 3C). Another study from our laboratory showed that circAmotl1 binds to c-myc protein and inhibits its transfer from the nucleus to cytoplasm45 (Figure 3D). The circRNA circAGO2 can bind HuR and facilitate cytoplasmic translocation of HuR, resulting in cancer progression (Figure 3E).46 Alternatively, circDnmt1 can promote tumor progression by binding and inhibiting cytoplasmic translocation of p53 and Auf147 (Figure 3E).
circRNA as biomarkers
Due to their lack of an available 5′ end, circRNAs are not susceptible to digestion by exonucleases,12,48 which makes their half-life up to 19–24 h, about 2.5-fold that of homologous linear RNAs.49 circRNAs can be detected in tissue cells,50 exosomes,18,19,51,52 plasma,53 serum,54 cerebrospinal fluid (CSF),55,56 urine,53 saliva,57,58 and other biological samples. Under pathological conditions, the expression levels of many circRNAs are significantly dysregulated. circRNA levels can be altered under stress to maintain cell homeostasis,59 which usually occurs prior to changes in protein levels. Therefore, circRNAs are ideal biomarkers for diagnosing, establishing prognosis, and monitoring diseases.24,60 Extensive research has been conducted in cardiovascular diseases,23, 24, 25,61, 62, 63, 64 central nervous system (CNS) diseases,65,66 tumors,67,68 and many other diseases.56 Clinical trials have been initiated,69 and multiple patents have already been granted23,70 (Figure 4, second layer; Table 1).
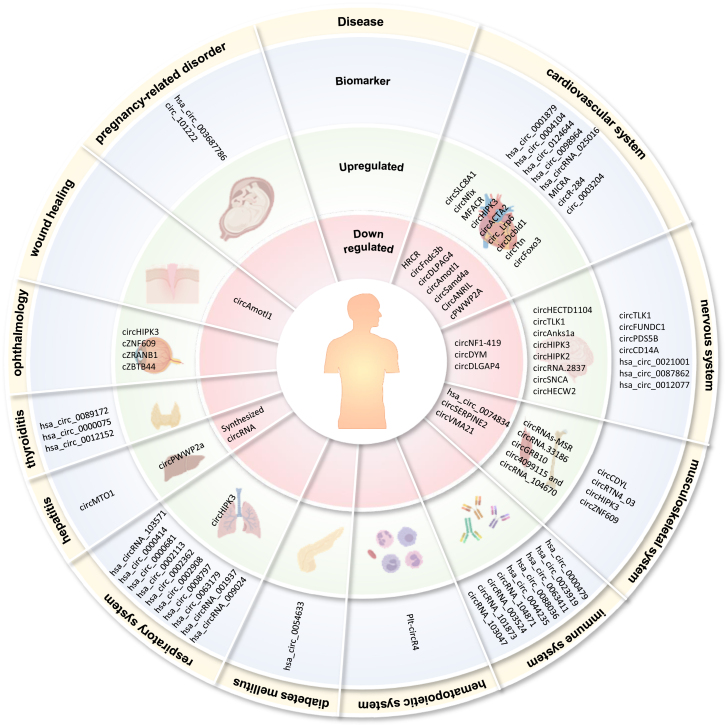
Figure 4.
Implication of circRNAs in various diseases
The outermost layer in beige specifies the diseases of interest. All circRNAs listed have been shown to have their expression levels significantly different between the disease and normal controls in the context of human diseases. The second layer in blue indicates circRNAs that could serve as biomarkers. The third layer in green shows circRNAs that are upregulated in diseases, which might serve as therapeutic targets. In vivo experiments in animal models show that targeted inhibition of their expression levels could obtain a significant therapeutic effect. This innermost layer in pink includes circRNAs that are downregulated in diseases and synthetic circRNAs that can inhibit disease-associated molecules. The dysregulation of circRNAs implies their potential to serve as therapeutic targets. In vivo experiments in animal models suggest that increasing their expression levels may have therapeutic effects.
Table 1.
circRNAs as diagnostic biomarker in diseases
| Disease | circRNA Symbol | circBase ID | Dysregulation | Function | Cohort size | Sample source | ROC |
PMID | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Sensitivity (%) | Specificity (%) | ||||||||
| CAD | – | hsa_circ_0001879 hsa_circ_0004104 |
up | diagnostic biomarker | CAD (n = 412) normal (n = 290) |
PBMCs | hsa_circ_0001879: 0.703 hsa_circ_0004104: 0.700 hsa_circ_0001879+hsa_circ_0004104: 0.742 hsa_circ_0001879+hsa_circ_0004104+risk factors: 0.832 |
83.1 70.7 76.9 66.8 |
54.3 61.4 62.0 89.0 |
31103880 |
| CAD | – | hsa_circ_0124644 hsa_circ_0098964 |
Up | diagnostic biomarker | CAD (n = 137) normal (n = 115) |
blood | hsa_circ_0124644: 0.769 hsa_circ_0124644+risk factors: 0.804 hsa_circ_0124644+ hsa_circ_0098964: 0.811 hsa_circ_0124644+ hsa_circ_00989644+risk factors: 0.843 |
86.1 75.9 82.5 83.2 |
62.6 70.4 73.0 69.6 |
28045102 |
| PoAF | hsa_circRNA_025016 | – | up | independent prognostic indicator | PoAF (n = 365) | plasma | 0.802 | 79.4 | 77.6 | PMC5850143 |
| Cerebral atherosclerosis | circ_0003204 | hsa_circ_0003204 | up | diagnostic biomarker | cerebral atherosclerosis (n = 35) normal (n = 32) |
plasma EVs | circ_0003204: 0.770 circ_0003204+LDL-C: 0.875 |
– | – | 31900142 |
| Atherosclerotic (carotid plaque rupture, stroke) | circR-284 | – | up | diagnostic biomarker | atherosclerotic urgent (n = 41) symptomatic (n = 24) asymptomatic (n = 48) |
serum | circR-284:miR-221: 0.98 circR-284: 0.91 miR-221: 0.47 |
93.0 80.0 49.0 |
97.0 85.0 51.0 |
28779016 |
| AIS | circTLK1 | – | up | diagnostic biomarker | AIS (n = 71) normal (n = 71) |
plasma | 0.868 | 78.9 | 91.5 | 31311824 |
| AIS | circFUNDC1 circPDS5B circCD14A |
– | up | diagnostic biomarker independent prognostic indicator |
AIS (n = 200) normal (n = 100) |
blood | 3 circRNAs: 0.875 (diagnostic biomarker) chang rate of 3 circRNAs: 0.960 (independent prognostic indicator) |
91.0 | 71.5 | 31690252 |
| IMDD | hsa_circ_0087862 hsa_circ_0012077 |
hsa_circ_0087862 hsa_circ_0012077 |
up | diagnostic biomarker | IMDD (n = 10) normal (n = 10) |
CSF exosome | 1.00 | – | – | 31611906 |
| IDD | circRNA_104670 | – | Up | diagnostic biomarker | IDD (n = 10) normal (n = 10) |
tissue | 0.96 | – | – | 30089772 |
| SLE | – | hsa_circ_0000479 | Up | diagnostic biomarker | SLE (n = 64) normal (n = 58) |
PBMCs | 0.730 | 45.3 | 100 | 31608065 |
| RA | – | hsa_circ_0044235 | down | diagnostic biomarker | RA (n = 77) SLE (n = 31) normal (n = 50) |
blood | 0.779 (RA versus normal) 0.721 (RA versus SLE) |
61.04 90.91 |
90.0 45.16 |
30216431 |
| RA | circRNA_104871 circRNA_003524 circRNA_101873 circRNA_103047 |
– | up | diagnostic biomarker | RA (n = 30) normal (n = 25) |
PBMCs | 0.833 0.683 0.676 0.671 |
83.3 83.3 66.7 66.7 |
68.0 60.0 68.0 56.0 |
28618429 |
| ALS | – | hsa_circ_0023919hsa_circ_0063411hsa_circ_0088036 | down up up |
diagnostic biomarker | ALS (n = 60) normal (n = 15) |
PBMCs | 0.952 1.000 0.959 |
90.0 100.0 90.6 |
93.3 100.0 92.3 |
31175544 |
| Pre-diabetes | – | hsa_circ_0054633 | up | diagnostic biomarker | pre-diabetes (n = 63) normal (n = 60) |
blood | hsa_circ_0054633: 0.751 hsa_circ_0054633+risk factors: 0.841 |
90.5 77.8 |
48.3 78.3 |
27878383 |
| T2DM | – | hsa_circ_0054633 | up | diagnostic biomarker | T2DM (n = 64) normal (n = 60) |
blood | hsa_circ_0054633: 0.793 hsa_circ_0054633+risk factors: 0.834 |
71.9 76.6 |
77.8 79.4 |
27878383 |
| GDM | – | hsa_circ_0054633 | up | diagnostic biomarker | 2nd trimester: GDM (n = 40) normal (n = 40) 3rd trimester: GDM (n = 65) normal (n = 65) placenta tissues and neonatal cord blood: GDM (n = 20) normal (n = 20) |
serum | 0.793 (2nd trimester) 0.664 (3rd trimester) 0.747 (placenta tissues) 0.783 (neonatal cord blood) |
57.6 39.1 87.0 87.3 |
90.9 88.7 53.0 60.9 |
30736847 |
| Liver fibrosis | circMTO1 | hsa_circ_0007874 | down | diagnostic biomarker | liver fibrosis (n = 360) normal (n = 360) |
serum | 0.914 (liver fibrosis vs normal) 0.847 (low fibrosis vs normal) 0.934 (medium fibrosis vs normal) 0.962 (high fibrosis vs normal) 0.774 (medium fibrosis vs low fibrosis) 0.880 (high fibrosis vs low fibrosis) 0.762 (high fibrosis vs medium fibrosis) |
75.8 95.0 80.0 90.0 67.5 85.0 52.5 |
90.0 65.0 95.0 97.5 90.0 92.5 92.5 |
31148365 |
| HT | – | hsa_circ_0089172 hsa_circ_0000075 hsa_circ_0012152 |
up | diagnostic biomarker | HT (n = 30) normal (n = 30) |
PBMCs | 0.673 0.715 0.702 |
55.56 55.17 85.19 |
86.21 81.48 59.26 |
31207490 |
| TB | hsa_circRNA_103571 | – | down | diagnostic biomarker | TB (n = 32) normal (n = 29) |
plasma | 0.838 | – | – | 29961269 |
| TB | – | hsa_circ_0000414 hsa_circ_0000681 hsa_circ_0002113 hsa_circ_0002362 hsa_circ_0002908 hsa_circ_0008797 hsa_circ_0063179 |
up | diagnostic biomarker | TB (n = 10) normal (n = 11) |
PBMCs | 0.946 | – | – | 29248507 |
| TB | hsa_circRNA_001937 hsa_circRNA_009024 |
– | Up | diagnostic biomarker | TB (n = 40) normal (n = 40) |
PBMCs | hsa_circRNA_001937: 0.873 hsa_circRNA_009024: 0.810 hsa_circRNA_001937+hsa_circRNA_009024: 0.926 |
85.0 75.0 95.0 |
77.5 80.0 80.0 |
29448254 |
| PE | – | hsa_circ_0036877 | up | diagnostic biomarker | PE (n = 34) normal (n = 110) |
blood | 0.846 | 85.3 | 72.7 | 29643944 |
| PE | circ_101222 | hsa_circ_0029601 | up | diagnostic biomarker | PE (n = 41) normal (n = 41) |
blood | circ_101222: 0.706 ENG: 0.765 circ_101222+ENG: 0.876 |
65.61 68.29 70.73 |
68.54 73.17 80.49 |
26846540 |
ROC, receiver operating characteristic; AUC, area under the curve; PMID, PubMed unique identifier; CAD, coronary artery disease; PBMCs, peripheral blood mononuclear cells; PoAF, postoperative atrial fibrillation; EVs, extracellular vesicles; LDL-C, low-density lipoprotein cholesterol; AIS, acute ischemic stroke; IMDD, immune-mediated demyelinating disease; CSF, cerebrospinal fluid; IDD, intervertebral disc degeneration; SLE, systemic lupus erythematosus; RA, rheumatoid arthritis; ALS, amyotrophic lateral sclerosis; T2DM, type 2 diabetes mellitus; GDM, gestational diabetes mellitus; HT, Hashimoto ’s thyroiditis; TB, tuberculosis; PE, pre-eclampsia; ENG, endoglin.
circRNA as biomarkers in cardiovascular disease
When the heart and blood vessels undergo pathological changes, some disease markers are directly released into the blood circulation and can be easily detected in peripheral blood. For example, hsa_circ_0001879,71 hsa_circ_0004104,71 hsa_circ_0124644,72 and hsa_circ_009896472 are detected in PBMCs of patients with coronary artery disease (CAD). A combination of these circRNAs as biomarkers, CAD risk factors (such as smoking, hypertension, and diabetes mellitus), and conventional markers, including low-density lipoprotein, total cholesterol, and serum creatinine, can be used to diagnose CAD. Some circRNAs can be used to predict the effect of treatment. The expression of hsa_circRNA_025016 in the plasma of patients with coronary artery bypass grafting can predict the occurrence of postoperative atrial fibrillation (PoAF) and inform the need for preventive treatment.73 However, not all diseases involve significant changes in circRNA content in peripheral blood in the early stages. Schulte et al.74 compared the content of circRNA and protein in peripheral blood after MI and concluded that cardiac-related circRNAs have poor detectability in plasma or serum after acute MI. In another study, it was found that MI-associated circRNA (MICRA, mainly transcribed from ZNF609) cannot be used as a biomarker for early diagnosis of acute MI but can accurately predict the risk of left ventricular dysfunction 3–4 months after MI.75
circRNA hsa_circ_0003204 locates predominantly in the cytoplasm of human aorta endothelial cells. This circRNA can inhibit proliferation, migration, as well as tube formation of endothelial cells in atherosclerosis through hsa_circ_0003204/miR-370-3p/transforming growth factor (TGF)-βR2/phosphorylated (phospho-)SMAD3 axis. The content of hsa_circ_0003204 in plasma extracellular vesicles (EVs) of patients with cerebral atherosclerosis is upregulated. The receiver operating characteristic (ROC) curve showed that the area under the curve (AUC) of hsa_circ_0003204 in plasma EVs for cerebral atherosclerosis was predicted to be 0.770. When hsa_circ_0003204 was combined with plasma low-density lipoprotein cholesterol (LDL-C) levels, the AUC increased to 0.875.76
In addition to changes in circRNA levels, the ratio of circRNAs to miRNAs could be used as a biomarker. circR-284 sponges and inhibits miR-221. It was found that the circR-284/miR-221 ratio in the serum of patients with atherosclerosis could be used as an indicator of carotid plaque rupture and stroke.77 The role of circR-284a and miR-221 in these pathological states is not fully understood.77
circRNAs as biomarkers in CNS disease
circRNAs are abundantly expressed in the brain, and their expression profiles are region specific.78 circRNA expression levels and quantitative trait loci in the brain are highly correlated with a variety of complex neurological and psychiatric disorders.79 For example, Alzheimer’s disease (AD) is one kind of neurodegenerative disease typified by a gradual cognitive decrease, with pathological events beginning decades before the presentation of clinical symptoms. Changes in the expression levels of AD-related circRNAs in the parietal cortex can be detected in early, presymptomatic AD.80 Using a whole-transcriptome analysis of an AD model mouse, it was found that competing endogenous RNA networks related to circRNA may be involved in various aspects of AD pathogenesis, including impaired dendritic development and memory. circRNAs, miRNAs, and mRNAs involved in these competing endogenous networks can be used as potential candidates for developing new diagnostic and therapeutic tools for AD.81
In acute ischemic stroke (AIS) patients, circTLK1 in plasma or circFUNDC1, circPDS5B, and circCD14A in whole blood are upregulated.82,83 They can be used as biomarkers for diagnosing AIS. In addition, the changing ratio of circFUNDC1, circPDS5B, and circCD14A levels in blood samples within 7 days after treatment can be used to evaluate the prognosis of AIS treatment.83 Furthermore, hsa_circ_0021001 in peripheral blood of intracranial aneurysm (IA) patients is downregulated, which can be used for the diagnosis of IA. hsa_circ_0021001 levels are associated with aneurysm rupture, Hunt and Hess scale level (a clinical symptom grading system), and timing of surgery. Highly expressed hsa_circ_0021001 is linked to prolonged disease-free survival (DFS) as well as overall survival (OS).84
While some exosomes produced by the CNS can cross the blood-brain barrier (BBB) and enter the blood circulation,18 most of them enter the CSF. These exosomes can immediately reflect the pathological state of the CNS. For example, the increase in hsa_circ_0087862 and hsa_circ_0012077 levels could be detected in the CSF exosomes of patients with immune-mediated demyelinating disease (IMDD). They can be used as biomarkers for early diagnosis of IMDD.55
With regard to mental illness, Mahmoudi et al.85 analyzed more than 90,000 circRNA species in the dorsolateral prefrontal cortex and found that many were depleted in patients with schizophrenia. It was concluded that the biogenesis or stability of circRNAs is reduced in cortical gray matter of patients with schizophrenia. circRNAs could be early diagnosis biomarkers or therapeutic targets of schizophrenia. However, further large-scale clinical research is needed.
circRNAs as biomarkers in musculoskeletal disease
circRNAs are also involved in the physiological and pathological status of skeletal muscle.86,87 Abnormal expression of many circRNAs can be detected in skeletal muscle of myotonic dystrophy type 1 (DM1), such as circCDYL (hsa_circ_0008285), circRTN4_03 (hsa_circ_0001006), circHIPK3, and circZNF609.88 However, a more comprehensive analysis of large-scale clinical samples is needed to determine whether these circRNAs can be used as reliable biomarkers for DM1.89
circRNAs as biomarkers in immune system disease
Some studies have started to focus on circRNAs as biomarkers in autoimmune diseases.90 hsa_circ_0000479 is significantly upregulated in PBMCs of SLE patients.91 Similarly, hsa_circ_0023919, hsa_circ_0063411, and hsa_circ_0088036 levels in PBMCs of amyotrophic lateral sclerosis (ALS) patients could be strong biomarkers for the diagnosis of ALS. The AUC of each circRNA is greater than 0.900, meaning that their specificity and sensitivity are greater than 90%.92 In peripheral blood samples of rheumatoid arthritis (RA) patients, many circRNAs, including hsa_circ_0044235,93 circRNA_104871, circRNA_003524, circRNA_101873, and circRNA_103047,94 are dysregulated and might be potent as biomarkers for the diagnosis of RA.
circRNAs as biomarkers in hematopoietic system disease
circRNAs are widely expressed in human hematopoietic cells. Different types of hematopoietic cells have characteristic circRNA expression profiles at different stages of differentiation.95 The elevated expression of circRNA in enucleated blood cells, such as red blood cells and platelets, suggests their potential important roles in regulating metabolism and responding to environmental factors in enucleated blood cells.95,96 Platelets can release circRNAs through exosomes and participate in intercellular communication. Studies have found that Plt-circR4 is specifically expressed in platelets.20 This suggests that circRNAs can be used as biomarkers for platelet-related diseases.
circRNAs as biomarkers in diabetes mellitus
Many circRNAs are involved in the pathogenesis of diabetes and related vascular complications.97 In peripheral blood samples from pre-diabetes, type 2 diabetes mellitus (T2DM),98 and gestational diabetes mellitus (GDM)99 patients, hsa_circ_0054633 is significantly upregulated. The expression levels of hsa_circ_0054633 in addition to risk factors, such as smoking and hypertension, body mass index, total cholesterol, triglycerides, high-density lipoprotein (HDL), and LDL, can provide important reference values for the diagnosis of diabetes at various stages.98,99
circRNAs as biomarkers in respiratory disease
hsa_circRNA_103571 in the plasma of patients with active tuberculosis (TB) is downregulated,100 while hsa_circ_0000414, hsa_circ_0002113, hsa_circ_0000681, hsa_circ_0002362, hsa_circ_0008797, hsa_circ_0002908, hsa_circ_0063179, hsa_circRNA_001937, hsa_circRNA_009024, and others are upregulated in PBMCs.101,102 These circRNAs could serve as novel biomarkers for the diagnosis of TB.
circRNAs as biomarkers in other diseases
circMTO1 (hsa_circ_0007874) in the serum of chronic hepatitis B (CHB) patients is significantly downregulated, and its expression level is negatively correlated with the degree of severity of liver fibrosis. circMTO1 levels are a good indicator for liver fibrosis and the degree of fibrosis caused by CHB.103 hsa_circ_0089172, hsa_circ_0000075, and hsa_circ_0012152, which are upregulated in PBMCs of Hashimoto’s thyroiditis (HT) patients, have the potential to diagnose HT.104 Upregulation of hsa_circ_0036877105 and circ_101222 (hsa_circ_0029601) was found in the total RNA of PBMCs from pregnant women with pre-eclampsia (PE).106 When combined with the expression of soluble endoglin in plasma, early PE can be effectively predicted.106
circRNA biomarker database
In addition to experimental research, there are network databases, such as BBCancer,107 Circ2Traits,108 CircR2Disease,109 CSCD,110 and miRandola,111 that have recorded disease-related circRNAs based on analysis of high-throughput gene expression profiles that can potentially be used as biomarkers for disease diagnosis.112,113 Most of these candidate circRNAs have not yet been verified by large-scale clinical experiments, so these databases should serve to guide future experiments.
circRNAs as potential therapeutic targets
There is a sophisticated regulatory network consisting of non-coding and coding RNAs, including circRNAs, long non-coding RNAs (lncRNAs), miRNAs, and mRNAs. Under normal conditions, a dynamic balance is maintained.114 Dysregulation of circRNA disrupts this balance, which can significantly contribute to the onset and progression of disease (Figure 4, third layer and innermost layer). Targeting such circRNAs might serve as potential therapeutic approaches.
circRNAs can be delivered to recipient cells via various methods, including lentivirus, adenovirus,115 adeno-associated virus (AAV),116 exosomes,117 and nanoparticles,118,119 to exert their regulatory functions.25 In order to optimize their effects, researchers are attempting to synthesize circRNAs with specific functions.120,121 Synthetic circRNAs can not only possess miRNA sponging abilities similar to endogenous circRNAs,122 but they can also be engineered to have an increased number of miRNA binding sites, thereby enhancing their miRNA sponging capabilities.123, 124, 125 Synthetic circRNAs, up to 5 kb, can also be designed for long-term and large-scale translation.126 By removing the stop codon and circularizing mRNA, that mRNA can be translated into protein.127 The Tornado (Twister-optimized RNA for durable overexpression) expression system can produce circularized RNA aptamers quickly, which can regulate proteins in a stable and efficient manner.128 Despite direct delivery of circRNAs, targeting them using smaller molecules might be a more practical therapeutic method, especially when a circRNA possess a larger size. When a circRNA generates actions by sponging miRNAs, designing siRNAs would be suitable for therapeutic targeting. When a circRNA functions through binding with proteins, designing blocking oligonucleotides that act against binding sites might fit better as a therapeutic approach. When a circRNA regulates downstream activities via translating to peptides or proteins, designing small molecules that target ribosome-binding sites is another possible approach.
There is inconclusive evidence for whether circRNAs are immunostimulatory.129 Wesselhoeft et al.130 demonstrated that unmodified exogenous circRNAs are less immunogenic than linear mRNAs. Their circular conformation helps them escape RIG-1 and Toll-like receptor-mediated immune responses and translate into proteins. However, immunogenicity and protein expression stability depend on the purity of the circRNA preparation, as even a small amount of linear RNA can lead to a strong cellular immune response. The N1-methylpseudouridine (m1ψ) and m6A modifications had no significant benefits in reducing the immunogenicity of circRNAs, but they reduced circRNA translation abilities.130 Alternatively, Chen et al.131 reported that exogenous circRNAs trigger an immune response by RIG-1 stimulation. It was found that circRNAs are recognized by RIG-1 and activate an immune response. This immunogenicity is related to the intron sequences involved in circRNA biogenesis. In contrast to circRNAs formed by self-splicing introns (e.g., from phage td introns), circRNAs formed by endogenous intron splicing bind to RBPs that prevent circRNAs from escaping RIG-1 recognition.132 It was found that the m6A modification eliminated the immunogenicity of exogenous circRNA.131 In addition to triggering an immune response in cells, the short dsRNA domain in the secondary structure of circRNAs can bind to dsRNA-activated PKR. It was found that increasing the expression levels of circRNAs in cells inhibited PKR activity, competitively blocked its recognition and binding of pathogen dsRNA, and suppressed a cellular immune response.16,133,134 Overexpression of circRNAs can be used to alleviate abnormal PKR activation and its downstream signaling.16,133,134
Based on the contradictory findings, it is likely that the immunogenicity of circRNAs depends on the type of circRNAs, as well as the mechanism of biogenesis, specific cell type, RNA modifications, and the method of delivery. circRNAs with immunogenicity can potentially be used to trigger or enhance immune activation as a vaccine adjuvant or immune enhancer to induce antiviral or antitumor immunity.131,135, 136, 137 Meanwhile, non-immunogenic circRNAs can be used as potential treatments to exert post-transcriptional regulation in cells.130 Understanding the role of circRNAs in distinct immune processes is relevant to their application in clinical therapy.138
Targeting circRNAs in cardiovascular disease
circRNAs are characteristically expressed at different stages of myocardial development and play key roles in pathways that are specifically active throughout cardiac differentiation.139 Dysregulation of some circRNAs can lead to heart diseases, and targeting these circRNAs or related pathways can serve as a potential therapeutic approach.61 The most abundant circRNA in the heart is circSLC8A1 (also known as circNCX1).140 A shRNA targeting circSLC8A1 in vivo improved myocardial ischemia-reperfusion injury29 and myocardial hypertrophy caused by stress overload.27 circNfix (hsa_circ_0005660) shRNA in the peri-infarcted area in MI model mice38 and a mitochondrial fission and apoptosis-related circRNA (MFACR) siRNA in the coronary arteries141 promoted myocardial cell proliferation and angiogenesis and suppressed myocardial apoptosis and cardiac dysfunction. In vitro experiments showed that circHIPK3 (mmu_circ_0001052) is upregulated in exosomes secreted by hypoxia-treated cardiomyocytes. circHIPK3 can be transferred to cardiac microvascular endothelial cells (CMVECs) through exosomes and attenuate their dysfunction caused by oxidative stress via the circHIPK3/miR-29a/IGF-1 axis.142
In addition, α-smooth muscle actin (α-SMA) is a major component involved in the contraction of vascular smooth muscle cells (VSMCs). Its dysregulation causes a series of pathological manifestations, such as vascular stenosis, hypertension, and atherosclerosis. The NRG-1/circACTA2/miR-548f-5p/α-SMA axis was found to be a critical pathway regulating α-SMA expression in VSMCs.143,144 In addition, circ_Lrp6 (hsa_circ_0009832) plays an important role in regulating VSMC migration, proliferation, and differentiation. Although its expression does not vary significantly under pathological conditions, circ_Lrp6 shRNA inhibited vascular intimal hyperplasia in mouse carotids.32,33 Similarly, circDcbld1 promotes vascular intimal proliferation through the circDcbld1/miR-145-3p/Nrp1 pathway. A chemically modified circDcbld1 siRNA significantly reduced the intimal hyperplasia response after injury.145
Currently, Dox is one of the major cancer drugs in clinical practice. However, some patients can develop congestive heart failure after Dox treatment. Global transcriptome analysis of mouse myocardium exposed to Dox revealed that QKI strongly suppressed the Dox-induced myocardial response. Overexpressing QKI in mice protected the heart from Dox-induced cardiotoxicity by regulating the expression of circRNAs such as circTtn.146 Similarly, a siRNA targeting circFoxo3 also relieved Dox-induced cardiomyopathy.37 circFoxo3 is highly expressed in myocardial tissues of elderly patients, and it promotes myocardial aging. Knockdown of circFoxo3 also reduced senescence of mouse embryonic fibroblasts.37
While the upregulated circRNAs could potentially be targets during disease treatments, overexpressing some downregulated circRNAs could be therapeutic approaches as well. miR-223 promotes cardiac hypertrophy as well as heart failure by affecting activity-regulated cytoskeleton (ARC). Overexpressing heart-related circRNA (HRCR) in a cardiac hypertrophy or heart failure mouse model attenuated disease progression.115 Similarly, circFndc3b overexpression in an MI mouse model resulted in increased circFndc3b binding to FUS protein, increased expression of VEGF, the formation of new blood vessels, and reduced myocardial fibrosis.39 In addition, overexpression of circDLPAG4 in human umbilical vein endothelial cells induced cell migration through the circDLPAG4/miR-143/HECTD1 pathway to reduce ischemia-reperfusion injury.147 Contrary to circFoxo3,37 circAmotl1 is highly expressed in neonatal heart tissue compared to mature heart tissues. It reduced apoptosis and enhanced heart repair by binding to AKT and activating its phosphorylation and nuclear localization, ultimately protecting myocardial cells from Dox-induced myocardial damage.43
In vitro experiments showed that circSamd4a was significantly downregulated in calcified VSMCs. Overexpression of circSamd4a significantly improved intracellular calcium deposition via two pathways: circSamd4a/miR-125a-3p/miR-483-5p/Camsap2 and circSamd4a/miR-125a-3p/Flna.148 In addition, circANRIL was shown to directly bind PES1 and induce nucleolar stress and p53 activation, which promotes apoptosis, inhibits proliferation, and protects vessels from atherosclerosis.149
Crosstalk between vascular endothelial cells and pericytes in microvessels is essential for the stability and remodeling of microvessels. However, diabetes-induced hyperglycemia can interfere with this crosstalk, leading to severe microvascular damage. Overexpression of cPWWP2A (mmu_circ_0000254) in pericytes in a diabetic mouse model protected them from diabetes-induced retinal vascular dysfunction.150
Targeting circRNAs in CNS disease
Many circRNAs participate in neuronal injury mechanisms that occur after ischemic stroke. For example, circHECTD1151 and circTLK182 regulate the expression of TIPARP through miR-142 and miR-335-3p, respectively, and activate autophagy in astrocytes and neurons. shRNA targeting circHECTD1 or circTLK1 in transient middle cerebral artery occlusion (tMCAO) mice significantly reduced neuronal injury as well as neurological deficits following ischemic stroke.82,151
After spinal nerve injury, circAnks1a is significantly upregulated in dorsal horn neurons. circAnks1a regulates neuropathic pain via various modes of action. In the nucleus, circAnks1a ties to the promoter region of Vegfb gene, increasing the recruitment of YBX1 to the Vegfb promoter and ultimately promoting the transcription of Vegfb. In the cytoplasm, circAnks1a enhances the interaction of YBX1 and transportin-1, thereby promoting YBX1 nuclear translocation. Furthermore, circAnks1a can sponge miR-324-3p, promoting the expression of VEGFB protein, which is associated with neuropathic pain. Spinal injection of a siRNA targeting circAnks1a alleviated chronic pain after nerve injury.152 Similarly, the expression of circHIPK3 was upregulated in the dorsal root ganglia of the rat model and in serum of patients with diabetic neuropathic pain. The severity of neuropathic pain was positively correlated with circHIPK3 serum concentration. circHIPK3 shRNA effectively reduced the symptoms of neuropathic pain in rats.153
Many neuropathies involve astrocyte activation. In astrocytes, circHIPK2 promotes autophagy and endoplasmic reticulum activation through the circHIPK2/miR-124-2hg/SIGMAR1/OPRS1 axis to facilitate astrocyte activation.154 circHIPK2 siRNA or shRNA in the hippocampus of a chronic unpredictable stress mouse model inhibited astrocyte dysfunction and improved depressive-like behaviors. However, intravenous administration of siRNA or shRNA did not achieve the same effects, which may be because they cannot cross the BBB. Interestingly, fecal microbiota transplantation from NLRP3-deficient mice allowed microbial metabolites to affect circHIPK2 expression in astrocytes through the gut-brain axis, which significantly improved astrocyte dysfunction and depressive-like behaviors in chronic unpredictable stress mice.155
circRNA.2837 is found to play a role in regulating autophagy in neurons. Its expression is downregulated in the injured sciatic nerve. Microinjection of circRNA.2837 inhibitor into the injured sciatic nerve to further silence circRNA.2837 can protect neurons by inhibiting autophagy in damaged neurons.156 In contrast, pramipexole can effectively inhibit the expression of circSNCA (hsa_circ_0127305) in neuroblastoma cells, reducing cell apoptosis and promoting cell autophagy through the circSNCA/miR-7/SNCA axis, thereby slowing down the progression of Parkinson’s disease.157
In cerebrovascular endothelial cells, circHECW2 can regulate the expression of ATG5 by sponging miR-30d, which promotes endothelial-mesenchymal transition (EMT) and impairs the integrity of the BBB. circHECW2 siRNA in the mouse hippocampus effectively inhibited EMT, which could provide a potential treatment strategy for cerebrovascular damage caused by drug abuse and neuritis.158
circNF1-419 (mmu_circ_0003411) is found to regulate autophagy in senescent astrocytes through the circNF1-419/phosphatidylinositol 3-kinase (PI3K)-1/Akt-AMP-activated protein kinase (AMPK)-mTOR and circNF1-419/PI3K-1/Akt-mTOR signaling pathways. Overexpressing circNF1-419 in the cerebral cortex of mice enhanced autophagy and delayed dementia.159 It has been demonstrated that circDYM regulates microglial activation through ubiquitination of heat shock protein (HSP)90 via the circDYM/miR-9/HECTD1 axis. Individuals with major depressive disorder have significantly lower levels of circDYM. Restoring circDYM levels in the hippocampus of a depression mouse model effectively reduced depression-like behavior.160 In endothelial cells, circDLGAP4 acts as a sponge for miR-143 and inhibits EMT, thus protecting BBB integrity. circDLGAP4 is significantly downregulated in the plasma of AIS patients. Overexpression of circDLGAP4 in tMCAO mice significantly inhibited EMT and reduced the infarcted area, BBB damage, and neurological deficits.161
Targeting circRNA in other diseases
In musculoskeletal system disease, in vitro studies showed that circRNAs-MSR expression in chondrocytes is significantly increased under mechanical stress. circRNAs-MSR may degrade the extracellular matrix of chondrocytes by upregulating tumor necrosis factor (TNF)-α, ultimately contributing to the formation of osteoarthritis (OA).45 In vivo studies have found that circRNA.33186 siRNA injected into the joints of destabilized medial meniscus-induced OA model mice significantly reduced the cartilage destruction and relieved OA.162 Multiple circRNAs were dysregulated in nucleus pulposus (NP) tissues of intervertebral disc degeneration (IDD), such as the downregulation of circGRB10 (hsa_circ_0080210)163 and upregulation of circ4099164 and circRNA_104670.165 In vivo experiments showed that a siRNA AVV targeting circRNA_104670 reduced the severity of IDD lesions.165,166
hsa_circ_0074834 is known to stimulate the osteogenesis-angiogenesis combining process of bone marrow mesenchymal stem cells (BMSCs) through the hsa_circ_0074834/miR-942-5p/ZEB1/VEGF pathway. In BMSCs of bone nonunion patients, hsa_circ_0074834 expression is significantly downregulated. Overexpressing hsa_circ_0074834 in the bone defect model effectively promoted bone regeneration in vivo.167 In addition, circSERPINE2 (hsa_circ_0008365) overexpression in an OA rabbit model significantly improved the surface of cartilage and reduced the formation of osteophytes in joints, degeneration of cartilage matrix, and OA.168 In NP tissues in intervertebral disc degeneration, circVMA21 (hsa_circ_0091702) alleviates cell apoptosis induced by inflammatory cytokines through the circVMA21/miR-200c/XIAP pathway and maintains the balance between extracellular matrix anabolism and catabolism. Overexpressing circVMA21 in the intervertebral disc reduced cell apoptosis. Its overexpression in NP cells inhibited extracellular matrix catabolism and promoted anabolism, thereby reducing intervertebral disc degeneration in rats.169,170
In fibrosis-related disease, TGF-β and LPS can activate circPWWP2a (hsa_circ_0074837) in hepatic stellate cells through the circPWWP2a/miR-203/Fstl1 and circPWWP2a/miR-223/TLR4 pathways, which promote liver fibrosis. circPWWP2a shRNA in a liver fibrosis mouse model reduced the severity of liver fibrosis.171 Fibroblast-to-myofibroblast transition occurs in pulmonary fibrosis, which produces a large amount of collagen. Intratracheal injection of circHIPK3 (hsa_circ_0000284) shRNA effectively reduced pulmonary fibrosis and improved fibroblast dysfunction.172 In skin fibroblasts, ectopic delivery of circAmotl1 increases expression levels of Stat3 and Dnmt3a proteins and facilitates nuclear translocation of Stat3.44 The cascade significantly increases the expression level of fibronectin and thus accelerates wound healing. Gold nanoparticles carrying circAmotl1 in the wound area of mice promoted wound healing.44
In ophthalmology disease, it was demonstrated in retinal endothelial cells of diabetic proliferative retinopathy patients that circHIPK3 (hsa_circ_0000284) sponges miR-30a-3p, resulting in increased expression of VEGF-C, FZD4, and WNT2. Intravitreal injection of circHIPK3 shRNA reduced retinal vascular dysfunction caused by diabetes, including retinal acellular capillaries, vascular leakage, and inflammation.173 cZNF609 (mmu_circ_0001797) is abundantly expressed in endothelial cells.174 It is significantly upregulated in retinal vascular disease175 and neurodegeneration.176 cZNF609 functions as a sponge for miR-615. Intravitreal injection of cZNF609 shRNA reduced retinal vascular damage and inhibited pathological angiogenesis in a mouse model of diabetic retinopathy175 and reduced glial cell proliferation and protected retinal ganglion cells in a glaucoma rat model.176 Knockdown of cZRANB1 (hsa_circ_0000268) in glaucoma models had similar effects, as it affects retinal Müller cell proliferation through the cZRANB1/miR-217/RUNX2 pathway.177 cZBTB44 shRNA in the vitreous of a neovascular. age-related macular degeneration (nAMD) mouse model reduced endothelial cell proliferation, migration, tube formation, and pathological choroidal neovascularization.178
In hepatitis, miR-122 is indispensable in the life cycle of hepatitis C virus (HCV). A synthetic circRNA with eight binding sites for miR-122 was engineered to be an effective miRNA sponge, ultimately inhibiting the production of HCV viral proteins.123
Discussion
Since circRNAs can regulate protein translocation, they are able to modulate protein functions in between nuclear and cytoplasm. Nuclear proteins and cytoplasmic proteins play distinct regulatory roles. While nuclear proteins can affect the first half of the central dogma, including DNA replication and transcription or even change the structure of chromosomes, cytoplasmic proteins can affect the second half of the central dogma, including translation and protein functions. Nuclear translocation is usually mediated by nuclear transport receptors called importins, which transport proteins containing a nuclear localization signal (NLS). circRNAs bring new insights to nuclear translocation processes, as they may not function through the typical NLS-based pathway.
Since proteins are synthesized in the cytoplasm, proteins that function in the nucleus must be transported from the cytoplasm to the nucleus. If circRNAs inhibit or promote the nuclear translocation of proteins, such as histones, transcriptases, and DNA synthases, which make up the structure of chromatin nucleosomes, this will significantly impact subcellular biological processes. While further studies on how circRNAs affect protein nuclear translocation are needed, there are some possibilities. (1) The combination of circRNA and protein changes the tertiary structure of the protein, resulting in conformational changes that disrupt their ability to deform the cell nucleus. (2) When a circRNA and a protein (protein 1) are combined, the binding sites of this protein with its partner protein (protein 2), an intermediary form for protein 1 transmembrane transport, are blocked. (3) Similarly, if a circRNA binds to protein 2, protein 1 cannot enter the nucleus. (4) As seen in the case of circAmotl1 binding to AKT1 and PDK1, the interaction between circRNA and protein results in the modification of the protein (e.g., methylation, phosphorylation), preventing its entry into the nucleus. (5) A circRNA might interact with multiple proteins to form a complex that is too large to enter the nucleus. This may not only prevent the protein from entering the nucleus, but also from entering the mitochondria, lysosomes, and other organelles, or exosomes, or even the cytoskeleton. In all of these cases, circRNAs could also affect the process in the opposite manner to promote the entry of proteins into the nucleus or other organelles.
Due to their circular structure, circRNAs are much more stable than the linear forms and can be used as a suitable biomarker. A large number of studies have proved that they have excellent application value in the diagnosis, prognosis, and monitoring of diseases. Studies show that most circRNAs are involved in many regulatory processes. Some circRNAs have been shown to contribute to a disease through multiple pathways. These circRNAs would be especially effective therapeutic targets, as many mechanisms promoting disease would be simultaneously suppressed. In addition, circRNAs can be artificially synthesized or modified to enhance their therapeutic efficiency as drugs. For example, a photocleavable sequence was added to a siRNA fragment with a circular structure, allowing external stimuli such as light to influence the abundance of a circRNA, which can reduce off-target effects of siRNA.179,180 In a recent review on circRNAs in cancer, we proposed some challenges in targeting circRNAs as treatment, such as how to increase the specificity of treatment and reduce the side effects of off-target effects on other cells and tissues, and how to continuously and accurately regulate the in vivo expression level of circRNAs.181 These are also essential issues that need to be considered in non-tumor disease treatment research. Therefore, we think that, in the near future, circRNAs can first be used as biomarkers for diagnosis. With further research and optimization, circRNAs can be used as new solutions for targeted therapy or local therapy, for some diseases involving multiple gene disorders, and achieve a therapeutic effect of multi-gene coordinated regulation. The mechanisms of circRNA functions have not been fully elucidated. At first, it was thought that many circRNAs functioned as miRNA sponges. Later, it was discovered that circRNAs can also directly bind, sort, store, and also sequester proteins to specific subcellular locations. circRNAs can also act as dynamic scaffolders that regulate protein-protein interactions. Several recent studies found that circRNAs can encode proteins or peptides,42,182 which opens a new area of research for circRNA functioning and has instigated extensive discussions.183, 184, 185, 186 Even though the current circRNA studies could be classified into functional mechanisms such as sponging, protein binding, mRNA regulating, and peptide encoding, whether these functions generated from circRNAs actually act through these specific mechanisms awaits further investigation. More factors need to be considered, including the copy number ratio between circRNAs and their regulatory partners (miRNAs in the sponging mechanism, proteins in the binding mechanism). How the downstream pathway regulated by a circRNA that negatively regulates the function of the circRNA requires more investigation as well. In the research of applying circRNAs for disease treatment, the most studied function of circRNAs is the miRNA sponge mechanism. However, most circRNAs are involved in more than one miRNA regulatory pathway, and each miRNA can cascade to regulate the expression of multiple mRNAs. Therefore, we presume that using circRNAs that function via a miRNA sponge mechanism for disease treatment is prone to some uncontrollable side effects. circRNAs that function in the form of protein binding or translation could directly affect the expression level of proteins, and most of these circRNAs have a single function, so we consider that they may perform well in the process of treatment. In addition, for some circRNAs that can affect gene promoter regions, due to their involvement in the direct regulation of mRNA transcription levels, we think that they have great potential for therapeutic applications.
Currently, research performed on most of the functional mechanisms of a circRNA is based on a specific time point, ignoring the potential changes in circRNA expression and functions over time. Expression differences across developmental stages need to be considered. Furthermore, a circRNA may show different functions during physiological and pathological states. For example, it is possible that some circRNAs play a stress protection role at the beginning of the pathological process, but as they continuously accumulate and fail to be metabolized efficiently, they may be harmful in the later stages of the pathology. Therefore, it is important to examine the expression and function of a circRNA over a period of time.
Perspectives
There are still many unanswered questions. For example, how do circRNAs affect protein entry to and exit from the nucleus? How do circRNAs influence the translocation of a protein by interacting with it? Does the circRNA-protein interaction change the microenvironment and cellular structures? Does nuclear polarity change and thus lead to protein translocation? These questions await further investigation.
In addition to increased stability, what are other essential advantages of a circular structure? Many circRNAs have not yet been found to hold functions; it is important to ask whether they are actively produced by cells on demand or are just byproducts of abnormal splicing. If they are generated actively, what advantages does the circular form offer? If they are mere byproducts, why is there an abundance of stable circRNAs in the body, especially in the brain?
Inspired by the endosymbiotic theory for the origin of mitochondria, we ask: could some circRNAs have originated from microorganisms with circular genetic material (such as viruses) engulfed by eukaryotic cells and integrated into their genome after a long evolutionary process, where these circRNAs continue to perform their biological functions? The immunogenicity of exogenous circRNAs in cells131,132 may lend support to this hypothesis to some degree. Although there is no concrete evidence, we predict that if this hypothesis is proven, it will have a great impact on the research direction of circRNAs and their functional roles. Finally, circRNA nomenclature is inconsistent across studies. The same circRNA appears under different names in different papers. Creating a uniform naming and coding system for circRNAs would be more conducive to sharing findings.
Acknowledgments
This work supported by grants from the Canadian Institutes of Health Research (PJT-153105, PJT-155962, and PJT-166107 to B.B.Y.). Q.Y. and F.L. acknowledge support from the China Scholarship Council (nos. 201906175051 and 201807980003).
Declaration of interests
The authors declare no conflicts of interest.
References
- 1.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 3.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerelle C., Daubersies P., Majérus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubin R.A., Kazmi M.A., Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245–248. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 6.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon R.J., Eperon I.C., Hall L., Samani N.J. A genome-wide survey demonstrates widespread non-linear mRNA in expressed sequences from multiple species. Nucleic Acids Res. 2005;33:5904–5913. doi: 10.1093/nar/gki893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Balool H.H., Weber D., Liu Y., Wade M., Guleria K., Nam P.L., Clayton J., Rowe W., Coxhead J., Irving J. Post-transcriptional exon shuffling events in humans can be evolutionarily conserved and abundant. Genome Res. 2011;21:1788–1799. doi: 10.1101/gr.116442.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang C., Liang D., Tatomer D.C., Wilusz J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 15.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 17.Fischer J.W., Busa V.F., Shao Y., Leung A.K.L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 2020;78:70–84.e6. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preußer C., Hung L.H., Schneider T., Schreiner S., Hardt M., Moebus A., Santoso S., Bindereif A. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles. 2018;7:1424473. doi: 10.1080/20013078.2018.1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M., Prasad N., Levy S., Coffey R.J., Patton J.G., Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao C., Lyu D., Huang S. Circular RNA expands its territory. Mol. Cell. Oncol. 2015;3:e1084443. doi: 10.1080/23723556.2015.1084443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim T.B., Lavenniah A., Foo R.S. Circles in the heart and cardiovascular system. Cardiovasc. Res. 2020;116:269–278. doi: 10.1093/cvr/cvz227. [DOI] [PubMed] [Google Scholar]
- 24.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 25.Lu D., Thum T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. 2019;16:661–674. doi: 10.1038/s41569-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 26.Gomes C.P.C., Schroen B., Kuster G.M., Robinson E.L., Ford K., Squire I.B., Heymans S., Martelli F., Emanueli C., Devaux Y., EU-CardioRNA COST Action (CA17129) Regulatory RNAs in heart failure. Circulation. 2020;141:313–328. doi: 10.1161/CIRCULATIONAHA.119.042474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., Sian S., Ackers-Johnson M.A., Foo R.S. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019;115:1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 28.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 29.Li M., Ding W., Tariq M.A., Chang W., Zhang X., Xu W., Hou L., Wang Y., Wang J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Xu J.D., Fang X.H., Zhu J.N., Yang J., Pan R., Yuan S.J., Zeng N., Yang Z.Z., Yang H. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 2020;116:1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Han D., Zhou T., Zhang J., Liu C., Cao F., Dong N. Melatonin ameliorates aortic valve calcification via the regulation of circular RNA CircRIC3/miR-204-5p/DPP4 signaling in valvular interstitial cells. J. Pineal Res. 2020;69:e12666. doi: 10.1111/jpi.12666. [DOI] [PubMed] [Google Scholar]
- 32.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 33.Heumüller A.W., Dimmeler S. Circular RNA control of vascular smooth muscle cell functions. Circ. Res. 2019;124:456–458. doi: 10.1161/CIRCRESAHA.118.314521. [DOI] [PubMed] [Google Scholar]
- 34.Yang L., Yang F., Zhao H., Wang M., Zhang Y. Circular RNA circCHFR facilitates the proliferation and migration of vascular smooth muscle via miR-370/FOXO1/cyclin D1 pathway. Mol. Ther. Nucleic Acids. 2019;16:434–441. doi: 10.1016/j.omtn.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Zhang S., Yue M., Li Y., Bi J., Liu H. Angiotensin II inhibits apoptosis of mouse aortic smooth muscle cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell Death Dis. 2019;10:362. doi: 10.1038/s41419-019-1590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji X., Ding W., Xu T., Zheng X., Zhang J., Liu M., Liu G., Wang J. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J. Mol. Cell. Cardiol. 2020;140:56–67. doi: 10.1016/j.yjmcc.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 38.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C. Circular RNA circFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z.G., Awan F.M., Du W.W., Zeng Y., Lyu J., Wu D., Gupta S., Yang W., Yang B.B. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du W.W., Yang W., Li X., Awan F.M., Yang Z., Fang L., Lyu J., Li F., Peng C., Krylov S.N. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 48.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maass P.G., Glažar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl.) 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi X., Wang B., Feng X., Xu Y., Lu K., Sun M. circRNAs and exosomes: a mysterious frontier for human cancer. Mol. Ther. Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 53.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen R.X., Chen X., Xia L.P., Zhang J.X., Pan Z.Z., Ma X.D., Han K., Chen J.W., Judde J.G., Deas O. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He J., Ren M., Li H., Yang L., Wang X., Yang Q. Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front. Genet. 2019;10:860. doi: 10.3389/fgene.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Yang T., Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S.Y., Wang J., Ouyang S.B., Huang Z.K., Liao L. Salivary circular RNAs hsa_circ_0001874 and hsa_circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell. Physiol. Biochem. 2018;47:2511–2521. doi: 10.1159/000491624. [DOI] [PubMed] [Google Scholar]
- 59.Fischer J.W., Leung A.K. circRNAs: a regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 2017;52:220–233. doi: 10.1080/10409238.2016.1276882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui X., Wang J., Guo Z., Li M., Li M., Liu S., Liu H., Li W., Yin X., Tao J., Xu W. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol. Cancer. 2018;17:123. doi: 10.1186/s12943-018-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santer L., Bär C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devaux Y., Creemers E.E., Boon R.A., Werfel S., Thum T., Engelhardt S., Dimmeler S., Squire I., Cardiolinc network Circular RNAs in heart failure. Eur. J. Heart Fail. 2017;19:701–709. doi: 10.1002/ejhf.801. [DOI] [PubMed] [Google Scholar]
- 63.Bei Y., Yang T., Wang L., Holvoet P., Das S., Sluijter J.P.G., Monteiro M.C., Liu Y., Zhou Q., Xiao J. Circular RNAs as potential theranostics in the cardiovascular system. Mol. Ther. Nucleic Acids. 2018;13:407–418. doi: 10.1016/j.omtn.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou M.Y., Yang J.M., Xiong X.D. The emerging landscape of circular RNA in cardiovascular diseases. J. Mol. Cell. Cardiol. 2018;122:134–139. doi: 10.1016/j.yjmcc.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 65.Kondo M.A., Mohan A., Mather K.A. Going around in circles: deciphering the role of circular RNAs in neurodegenerative disease. Curr. Opin. Psychiatry. 2020;33:141–147. doi: 10.1097/YCO.0000000000000582. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X., Hamblin M.H., Yin K.J. Noncoding RNAs and stroke. Neuroscientist. 2019;25:22–26. doi: 10.1177/1073858418769556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su M., Xiao Y., Ma J., Tang Y., Tian B., Zhang Y., Li X., Wu Z., Yang D., Zhou Y. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer. 2019;18:90. doi: 10.1186/s12943-019-1002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnaiz E., Sole C., Manterola L., Iparraguirre L., Otaegui D., Lawrie C.H. circRNAs and cancer: biomarkers and master regulators. Semin. Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 69.Braicu C., Zimta A.A., Gulei D., Olariu A., Berindan-Neagoe I. Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell. Mol. Life Sci. 2019;76:1559–1577. doi: 10.1007/s00018-019-03016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bayoumi A.S., Aonuma T., Teoh J.P., Tang Y.L., Kim I.M. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol. Sin. 2018;39:1100–1109. doi: 10.1038/aps.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L., Shen C., Wang Y., Zou T., Zhu H., Lu X., Li L., Yang B., Chen J., Chen S. Identification of circular RNA hsa_circ_0001879 and hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88–96. doi: 10.1016/j.atherosclerosis.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J., Xu Y., Xu S., Liu Y., Yu L., Li Z., Xue X., Wang H. Plasma circular RNAs, hsa_circRNA_025016, predict postoperative atrial fibrillation after isolated off-pump coronary artery bypass grafting. J. Am. Heart Assoc. 2018;7:e006642. [Google Scholar]
- 74.Schulte C., Barwari T., Joshi A., Theofilatos K., Zampetaki A., Barallobre-Barreiro J., Singh B., Sörensen N.A., Neumann J.T., Zeller T. Comparative analysis of circulating noncoding RNAs versus protein biomarkers in the detection of myocardial injury. Circ. Res. 2019;125:328–340. doi: 10.1161/CIRCRESAHA.119.314937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., Burkhardt R., Thiery J., Wagner D.R., Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S., Song G., Yuan J., Qiao S., Xu S., Si Z., Yang Y., Xu X., Wang A. Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFβR2/phosph-SMAD3 axis. J. Biomed. Sci. 2020;27:11. doi: 10.1186/s12929-019-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Bazan H.A., Hatfield S.A., Brug A., Brooks A.J., Lightell D.J., Jr., Woods T.C. Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ. Cardiovasc. Genet. 2017;10:e001720. doi: 10.1161/CIRCGENETICS.117.001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gokool A., Anwar F., Voineagu I. The landscape of circular RNA expression in the human brain. Biol. Psychiatry. 2020;87:294–304. doi: 10.1016/j.biopsych.2019.07.029. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z., Ran Y., Tao C., Li S., Chen J., Yang E. Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Biol. 2019;20:99. doi: 10.1186/s13059-019-1701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dube U., Del-Aguila J.L., Li Z., Budde J.P., Jiang S., Hsu S., Ibanez L., Fernandez M.V., Farias F., Norton J., Dominantly Inherited Alzheimer Network (DIAN) An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma N., Pan J., Ye X., Yu B., Zhang W., Wan J. Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol. Ther. Nucleic Acids. 2019;18:1049–1062. doi: 10.1016/j.omtn.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu F., Han B., Wu S., Yang L., Leng S., Li M., Liao J., Wang G., Ye Q., Zhang Y. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J. Neurosci. 2019;39:7369–7393. doi: 10.1523/JNEUROSCI.0299-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuo L., Zhang L., Zu J., Wang Z., Han B., Chen B., Cheng M., Ju M., Li M., Shu G. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke. 2020;51:319–323. doi: 10.1161/STROKEAHA.119.027348. [DOI] [PubMed] [Google Scholar]
- 84.Teng L., Chen Y., Chen H., He X., Wang J., Peng Y., Duan H., Li H., Lin D., Shao B. Circular RNA hsa_circ_0021001 in peripheral blood: a potential novel biomarker in the screening of intracranial aneurysm. Oncotarget. 2017;8:107125–107133. doi: 10.18632/oncotarget.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahmoudi E., Fitzsimmons C., Geaghan M.P., Shannon Weickert C., Atkins J.R., Wang X., Cairns M.J. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology. 2019;44:1043–1054. doi: 10.1038/s41386-019-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang P., Chao Z., Zhang R., Ding R., Wang Y., Wu W., Han Q., Li C., Xu H., Wang L., Xu Y. Circular RNA regulation of myogenesis. Cells. 2019;8:885. doi: 10.3390/cells8080885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao Y., Chen M., Lian D., Li Y., Li Y., Wang J., Deng S., Yu K., Lian Z. Non-coding RNA regulates the myogenesis of skeletal muscle satellite cells, injury repair and diseases. Cells. 2019;8:988. doi: 10.3390/cells8090988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voellenkle C., Perfetti A., Carrara M., Fuschi P., Renna L.V., Longo M., Sain S.B., Cardani R., Valaperta R., Silvestri G. Dysregulation of circular RNAs in myotonic dystrophy type 1. Int. J. Mol. Sci. 2019;20:1938. doi: 10.3390/ijms20081938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Czubak K., Sedehizadeh S., Kozlowski P., Wojciechowska M. An overview of circular RNAs and their implications in myotonic dystrophy. Int. J. Mol. Sci. 2019;20:4385. doi: 10.3390/ijms20184385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen W., Liu D., Li Q.Z., Zhu H. The function of ncRNAs in rheumatic diseases. Epigenomics. 2019;11:821–833. doi: 10.2217/epi-2018-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo G., Wang H., Ye L., Shi X., Yan K., Lin K., Huang Q., Li B., Lin Q., Zhu L. hsa_circ_0000479 as a novel diagnostic biomarker of systemic lupus erythematosus. Front. Immunol. 2019;10:2281. doi: 10.3389/fimmu.2019.02281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dolinar A., Koritnik B., Glavač D., Ravnik-Glavač M. Circular RNAs as potential blood biomarkers in amyotrophic lateral sclerosis. Mol. Neurobiol. 2019;56:8052–8062. doi: 10.1007/s12035-019-1627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo Q., Zhang L., Li X., Fu B., Deng Z., Qing C., Su R., Xu J., Guo Y., Huang Z., Li J. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin. Exp. Immunol. 2018;194:118–124. doi: 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ouyang Q., Wu J., Jiang Z., Zhao J., Wang R., Lou A., Zhu D., Shi G.P., Yang M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell. Physiol. Biochem. 2017;42:651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 95.Nicolet B.P., Engels S., Aglialoro F., van den Akker E., von Lindern M., Wolkers M.C. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46:8168–8180. doi: 10.1093/nar/gky721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alhasan A.A., Izuogu O.G., Al-Balool H.H., Steyn J.S., Evans A., Colzani M., Ghevaert C., Mountford J.C., Marenah L., Elliott D.J. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zaiou M. circRNAs signature as potential diagnostic and prognostic biomarker for diabetes mellitus and related cardiovascular complications. Cells. 2020;9:659. doi: 10.3390/cells9030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu H., Wu S., Zhu Y., Ye M., Shen J., Liu Y., Zhang Y., Bu S. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin. Epigenetics. 2019;11:22. doi: 10.1186/s13148-019-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yi Z., Gao K., Li R., Fu Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J. Cell. Mol. Med. 2018;22:4076–4084. doi: 10.1111/jcmm.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian Z., Liu H., Li M., Shi J., Li N., Zhang Y., Zhang X., Lv J., Xie X., Bai Y. Potential Diagnostic Power of Blood Circular RNA Expression in Active Pulmonary Tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang Z.K., Yao F.Y., Xu J.Q., Deng Z., Su R.G., Peng Y.P., Luo Q., Li J.M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell. Physiol. Biochem. 2018;45:1230–1240. doi: 10.1159/000487454. [DOI] [PubMed] [Google Scholar]
- 103.Wang W., Dong R., Guo Y., He J., Shao C., Yi P., Yu F., Gu D., Zheng J. circMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell. Mol. Med. 2019;23:5486–5496. doi: 10.1111/jcmm.14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiong S., Peng H., Ding X., Wang X., Wang L., Wu C., Wang S., Xu H., Liu Y. Circular RNA expression profiling and the potential role of hsa_circ_0089172 in Hashimoto’s thyroiditis via sponging miR125a-3p. Mol. Ther. Nucleic Acids. 2019;17:38–48. doi: 10.1016/j.omtn.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu X., Ao J., Li X., Zhang H., Wu J., Cheng W. Competing endogenous RNA expression profiling in pre-eclampsia identifies hsa_circ_0036877 as a potential novel blood biomarker for early pre-eclampsia. Clin. Epigenetics. 2018;10:48. doi: 10.1186/s13148-018-0482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y.G., Yang H.L., Long Y., Li W.L. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123:2113–2118. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 107.Zuo Z., Hu H., Xu Q., Luo X., Peng D., Zhu K., Zhao Q., Xie Y., Ren J. BBCancer: an expression atlas of blood-based biomarkers in the early diagnosis of cancers. Nucleic Acids Res. 2020;48(D1):D789–D796. doi: 10.1093/nar/gkz942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan C., Lei X., Fang Z., Jiang Q., Wu F.X. CircR2Disease: a manually curated database for experimentally supported circular RNAs associated with various diseases. Database (Oxford) 2018;2018:bay044. doi: 10.1093/database/bay044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F., Jin Y., Gao Y., Xia L., Chang H. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Russo F., Di Bella S., Vannini F., Berti G., Scoyni F., Cook H.V., Santos A., Nigita G., Bonnici V., Laganà A. miRandola 2017: a curated knowledge base of non-invasive biomarkers. Nucleic Acids Res. 2018;46(D1):D354–D359. doi: 10.1093/nar/gkx854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shang B.Q., Li M.L., Quan H.Y., Hou P.F., Li Z.W., Chu S.F., Zheng J.N., Bai J. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol. Cancer. 2019;18:138. doi: 10.1186/s12943-019-1071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shan C., Zhang Y., Hao X., Gao J., Chen X., Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 116.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bai H., Lei K., Huang F., Jiang Z., Zhou X. exo-circRNAs: a new paradigm for anticancer therapy. Mol. Cancer. 2019;18:56. doi: 10.1186/s12943-019-0986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Du W.W., Zhang C., Yang W., Yong T., Awan F.M., Yang B.B. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Petkovic S., Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Costello A., Lao N.T., Barron N., Clynes M. Reinventing the wheel: synthetic circular RNAs for mammalian cell engineering. Trends Biotechnol. 2020;38:217–230. doi: 10.1016/j.tibtech.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 122.Liu X., Abraham J.M., Cheng Y., Wang Z., Wang Z., Zhang G., Ashktorab H., Smoot D.T., Cole R.N., Boronina T.N. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O. Functional sequestration of microRNA-122 from hepatitis C virus by circular RNA sponges. RNA Biol. 2018;15:1032–1039. doi: 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rossbach O. Artificial circular RNA sponges targeting microRNAs as a novel tool in molecular biology. Mol. Ther. Nucleic Acids. 2019;17:452–454. doi: 10.1016/j.omtn.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shu Y., Wu K., Zeng Z., Huang S., Ji X., Yuan C., Zhang L., Liu W., Huang B., Feng Y. A simplified system to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol. Ther. Nucleic Acids. 2018;13:556–567. doi: 10.1016/j.omtn.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Costello A., Lao N.T., Barron N., Clynes M. Continuous translation of circularized mRNA improves recombinant protein titer. Metab. Eng. 2019;52:284–292. doi: 10.1016/j.ymben.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 128.Litke J.L., Jaffrey S.R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 2019;37:667–675. doi: 10.1038/s41587-019-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Basavappa M.G., Cherry S. Going in circles: the black box of circular RNA immunogenicity. Mol. Cell. 2019;76:3–5. doi: 10.1016/j.molcel.2019.08.027. [DOI] [PubMed] [Google Scholar]
- 130.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wilusz J.E. Circle the wagons: circular RNAs control innate immunity. Cell. 2019;177:797–799. doi: 10.1016/j.cell.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Flemming A. The enigma of circular RNA. Nat. Rev. Immunol. 2019;19:351. doi: 10.1038/s41577-019-0173-0. [DOI] [PubMed] [Google Scholar]
- 135.Cadena C., Hur S. Antiviral immunity and circular RNA: no end in sight. Mol. Cell. 2017;67:163–164. doi: 10.1016/j.molcel.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li X., Liu C.X., Xue W., Zhang Y., Jiang S., Yin Q.F., Wei J., Yao R.W., Yang L., Chen L.L. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67:214–227.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 137.Wang M., Yu F., Wu W., Zhang Y., Chang W., Ponnusamy M., Wang K., Li P. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017;13:1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen X., Yang T., Wang W., Xi W., Zhang T., Li Q., Yang A., Wang T. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y., Zhang J., Huo C., Ding N., Li J., Xiao J., Lin X., Cai B., Zhang Y., Xu J. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. 2017;24:137–146. doi: 10.1016/j.ebiom.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tan W.L., Lim B.T., Anene-Nzelu C.G., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 141.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Y., Zhao R., Liu W., Wang Z., Rong J., Long X., Liu Z., Ge J., Shi B. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid. Med. Cell. Longev. 2019;2019:7954657. doi: 10.1155/2019/7954657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sun Y., Yang Z., Zheng B., Zhang X.H., Zhang M.L., Zhao X.S., Zhao H.Y., Suzuki T., Wen J.K. A novel regulatory mechanism of smooth muscle α-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ. Res. 2017;121:628–635. doi: 10.1161/CIRCRESAHA.117.311441. [DOI] [PubMed] [Google Scholar]
- 144.Weiser-Evans M.C.M. Smooth muscle differentiation control comes full circle: the circular noncoding RNA, circActa2, functions as a miRNA sponge to fine-tune α-SMA expression. Circ. Res. 2017;121:591–593. doi: 10.1161/CIRCRESAHA.117.311722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rong Z.H., Chang N.B., Yao Q.P., Li T., Zhu X.L., Cao Y., Jiang M.J., Cheng Y.S., Jiang R., Jiang J. Suppression of circDcbld1 alleviates intimal hyperplasia in rat carotid artery by targeting miR-145-3p/neuropilin-1. Mol. Ther. Nucleic Acids. 2019;18:999–1008. doi: 10.1016/j.omtn.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gupta S.K., Garg A., Bär C., Chatterjee S., Foinquinos A., Milting H., Streckfuß-Bömeke K., Fiedler J., Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen L., Luo W., Zhang W., Chu H., Wang J., Dai X., Cheng Y., Zhu T., Chao J. circDLPAG4/HECTD1 mediates ischaemia/reperfusion injury in endothelial cells via ER stress. RNA Biol. 2020;17:240–253. doi: 10.1080/15476286.2019.1676114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ryu J., Kwon D.H., Choe N., Shin S., Jeong G., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Characterization of circular RNAs in vascular smooth muscle cells with vascular calcification. Mol. Ther. Nucleic Acids. 2020;19:31–41. doi: 10.1016/j.omtn.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu C., Ge H.M., Liu B.H., Dong R., Shan K., Chen X., Yao M.D., Li X.M., Yao J., Zhou R.M. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc. Natl. Acad. Sci. USA. 2019;116:7455–7464. doi: 10.1073/pnas.1814874116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R., Wu F., Leng S., Chao J., Zhang J.H. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang S.B., Lin S.Y., Liu M., Liu C.C., Ding H.H., Sun Y., Ma C., Guo R.X., Lv Y.Y., Wu S.L. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat. Commun. 2019;10:4119. doi: 10.1038/s41467-019-12049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang L., Luo T., Bao Z., Li Y., Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem. Biophys. Res. Commun. 2018;505:644–650. doi: 10.1016/j.bbrc.2018.09.158. [DOI] [PubMed] [Google Scholar]
- 154.Huang R., Zhang Y., Han B., Bai Y., Zhou R., Gan G., Chao J., Hu G., Yao H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hu M., Wei X., Li M., Tao L., Wei L., Zhang M., Cheng H., Yuan Y. Circular RNA expression profiles of persistent atrial fibrillation in patients with rheumatic heart disease. Anatol. J. Cardiol. 2019;21:2–10. doi: 10.14744/AnatolJCardiol.2018.35902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhou Z.B., Niu Y.L., Huang G.X., Lu J.J., Chen A., Zhu L. Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol. Ther. Nucleic Acids. 2018;12:718–729. doi: 10.1016/j.omtn.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sang Q., Liu X., Wang L., Qi L., Sun W., Wang W., Sun Y., Zhang H. circSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging (Albany NY) 2018;10:1281–1293. doi: 10.18632/aging.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang L., Han B., Zhang Y., Bai Y., Chao J., Hu G., Yao H. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14:404–418. doi: 10.1080/15548627.2017.1414755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Diling C., Yinrui G., Longkai Q., Xiaocui T., Yadi L., Xin Y., Guoyan H., Ou S., Tianqiao Y., Dongdong W. Circular RNA NF1-419 enhances autophagy to ameliorate senile dementia by binding Dynamin-1 and adaptor protein 2 B1 in AD-like mice. Aging (Albany NY) 2019;11:12002–12031. doi: 10.18632/aging.102529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 160.Zhang Y., Du L., Bai Y., Han B., He C., Gong L., Huang R., Shen L., Chao J., Liu P. circDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry. 2020;25:1175–1190. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bai Y., Zhang Y., Han B., Yang L., Chen X., Huang R., Wu F., Chao J., Liu P., Hu G. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J. Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou Z.B., Huang G.X., Fu Q., Han B., Lu J.J., Chen A.M., Zhu L. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol. Ther. 2019;27:531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo W., Zhang B., Mu K., Feng S.Q., Dong Z.Y., Ning G.Z., Li H.R., Liu S., Zhao L., Li Y. Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 2018;9:319. doi: 10.1038/s41419-017-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang H., He P., Pan H., Long J., Wang J., Li Z., Liu H., Jiang W., Zheng Z. Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 2018;50:27. doi: 10.1038/s12276-018-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Song J., Wang H.L., Song K.H., Ding Z.W., Wang H.L., Ma X.S., Lu F.Z., Xia X.L., Wang Y.W., Fei-Zou, Jiang J.Y. CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp. Mol. Med. 2018;50:94. doi: 10.1038/s12276-018-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Z., Chen X., Xu D., Li S., Chan M.T.V., Wu W.K.K. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52:e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ouyang Z., Tan T., Zhang X., Wan J., Zhou Y., Jiang G., Yang D., Guo X., Liu T. circRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019;10:932. doi: 10.1038/s41419-019-2161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shen S., Wu Y., Chen J., Xie Z., Huang K., Wang G., Yang Y., Ni W., Chen Z., Shi P. circSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 2019;78:826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Collison J. Degenerative disc disease: circular RNA reduces cell death in IVD disease. Nat. Rev. Rheumatol. 2018;14:123. doi: 10.1038/nrrheum.2018.13. [DOI] [PubMed] [Google Scholar]
- 171.Liu W., Feng R., Li X., Li D., Zhai W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging (Albany NY) 2019;11:9569–9580. doi: 10.18632/aging.102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang J.X., Lu J., Xie H., Wang D.P., Ni H.E., Zhu Y., Ren L.H., Meng X.X., Wang R.L. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10:182. doi: 10.1038/s41419-019-1430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 174.Boeckel J.N., Jaé N., Heumüller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 175.Liu C., Yao M.D., Li C.P., Shan K., Yang H., Wang J.J., Liu B., Li X.M., Yao J., Jiang Q., Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang J.J., Liu C., Shan K., Liu B.H., Li X.M., Zhang S.J., Zhou R.M., Dong R., Yan B., Sun X.H. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018;8:3408–3415. doi: 10.7150/thno.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang J.J., Shan K., Liu B.H., Liu C., Zhou R.M., Li X.M., Dong R., Zhang S.J., Zhang S.H., Wu J.H., Yan B. Targeting circular RNA-ZRANB1 for therapeutic intervention in retinal neurodegeneration. Cell Death Dis. 2018;9:540. doi: 10.1038/s41419-018-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou R.M., Shi L.J., Shan K., Sun Y.N., Wang S.S., Zhang S.J., Li X.M., Jiang Q., Yan B., Zhao C. Circular RNA-ZBTB44 regulates the development of choroidal neovascularization. Theranostics. 2020;10:3293–3307. doi: 10.7150/thno.39488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang L., Liang D., Chen C., Wang Y., Amu G., Yang J., Yu L., Dmochowski I.J., Tang X. Circular siRNAs for reducing off-target effects and enhancing long-term gene silencing in cells and mice. Mol. Ther. Nucleic Acids. 2018;10:237–244. doi: 10.1016/j.omtn.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li F., Yang Q., He A.T., Yang B.B. Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin. Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.10.002. Published online October 6, 2020. [DOI] [PubMed] [Google Scholar]
- 182.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schneider T., Bindereif A. Circular RNAs: coding or noncoding? Cell Res. 2017;27:724–725. doi: 10.1038/cr.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Koch L. RNA: translated circular RNAs. Nat. Rev. Genet. 2017;18:272–273. doi: 10.1038/nrg.2017.27. [DOI] [PubMed] [Google Scholar]
- 185.Tatomer D.C., Wilusz J.E. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol. Cell. 2017;66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 186.Servick K. Circular RNAs hint at new realm of genetics. Science. 2017;355:1363. doi: 10.1126/science.355.6332.1363. [DOI] [PubMed] [Google Scholar]