Abstract
The tumor microenvironment (TME), controlled by intrinsic mechanisms of carcinogenesis and epigenetic modifications, has, in recent years, become a heavily researched topic. The TME can be described in terms of hypoxia, metabolic dysregulation, immune escape, and chronic inflammation. RNA methylation, an epigenetic modification, has recently been found to have a pivotal role in shaping the TME. The N6-methylation of adenosine (m6A) modification is the most common type of RNA methylation that occurs in the N6-position of adenosine, which is the primary internal modification of eukaryotic mRNA. Compelling evidence has demonstrated that m6A regulates transcriptional and protein expression through splicing, translation, degradation, and export, thereby mediating the biological processes of cancer cells and/or stromal cells and characterizing the TME. The TME also has a crucial role in the complicated regulatory network of m6A modifications and, subsequently, influences tumor initiation, progression, and therapy responses. In this review, we describe the features of the TME and how the m6A modification modulates and interacts with it. We also focus on various factors and pathways involved in m6A methylation. Finally, we discuss potential therapeutic strategies and prognostic biomarkers with respect to the TME and m6A modification.
Keywords: tumor microenvironment, RNA methylation, m6A, hypoxia, metabolic dysregulation, immune escape, chronic inflammation
Graphical abstract
Gu and colleagues demonstrated the characteristics and development of the tumor microenvironment and how RNA methylation modulates and interacts with it.
Introduction
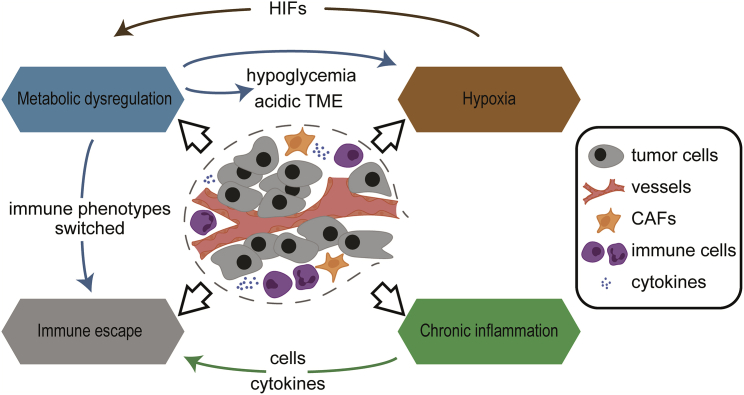
The interaction between malignant cells and their direct environment determines how the tumor progresses through proliferation, differentiation, invasion, metastasis, and even resistance to drug therapy. Referred to as the tumor microenvironment (TME), which acts as the soil of the tumor seed, the TME harbors the extracellular matrix and a mass of heterogeneous stromal cell types and diverse structures, such as neovascularization and immune infiltrates, which are indispensable for the biological activities of neoplasms (Figure 1).1, 2, 3
Figure 1.
Characteristics of TME and their interactions
The characteristics of the tumor microenvironment (TME) comprise hypoxia, metabolism dysregulation, immune escape, and chronic inflammation. The metabolic traits of tumor cells induce hypoxic, hypoglycemic, and acidic TME. Meanwhile, hypoxia-induced HIFs lead to cancer-induced metabolic disorder. Metabolic alterations also switch the function of immune cells from a tumor-active state to a tumor-promoting state and contribute to tumor immune escape. Furthermore, the inflammatory cells and cytokines found in TME tend to facilitate tumor progression and immunosuppression, rather than establish an effective host antitumor response.
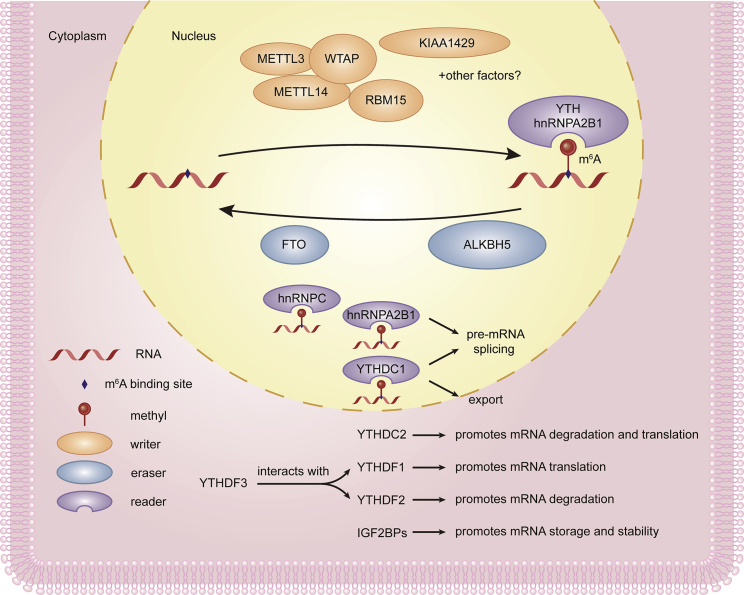
However, the constituent parts of the TME around each tumor are not congenitally the same but are instead widely divergent. These components are affected by a flux of signaling pathways and cytokines. One of these determinants is RNA methylation. Regarded as a type of reversible and dynamic modification, the N6-methylation of adenosine (m6A) is the most prevalent modification of mRNA, accounting for nearly one half of the methylated ribonucleotides and 0.1%–0.4% of adenosine residues. Studies have concluded that RNA methylation, especially the m6A modification, has a nonnegligible role in the diversity and complexity of the TME.4,5 It is acknowledged that methyltransferases, demethylases, and binding proteins, which are compared with writers, erasers, and readers, respectively, are responsible for modulating the expression of m6A modification (Figure 2).6 The methyltransferase-like (METTL)3, METTL14, and WTAP writer proteins act as the catalytic core, structural support, and stabilizer, respectively. The RNA-binding motif protein 15 (RBM15) locates the target sites, whereas the function of KIAA1429 is unknown.7 Erasers, which help to demethylate the m6A modification, consist of fat masses, obesity-associated proteins (FTO), and alkB homolog 5 (ALKBH5).8,9 Readers of the m6A modification on mRNA are the heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1), hnRNPC, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and YT521-B homology (YTH) domain-containing proteins, such as YTHDF1-3 and YTHDC.10, 11, 12 m6A is involved in RNA biogenesis, including in miRNA processing, pre-mRNA splicing, RNA translation, and degradation through the functionally distinct intranuclear and cytoplasmic m6A-binding proteins we mentioned.13, 14, 15, 16, 17, 18, 19 In addition, Zaccara and Jaffrey20 revealed a novel model of DF protein binding and functional capability, in which YTHDF proteins act together to mediate the degradation of m6A-mRNAs, which is contrary to the prevailing model.
Figure 2.
Three kinds of regulators constitute the m6A methylase complex
“Writer” proteins METTL3, METTL14, and WTAP, respectively, act as the catalytic core, structural support, and stabilizer. RBM15 locates the target sites, whereas the function of KIAA1429 is unknown. “Erasers,” which help to demethylate m6A modification, consist of FTO and ALKBH5. “Readers” of m6A modification on mRNA are YTH domain-containing proteins (including YTHDF1-3, YTHDC) and hnRNPA2B1, among others. They recognize m6A sites and then facilitate m6A-RNA biogenesis and functions.
Many studies have discussed DNA and histone methylation in tumor-associated stromal cells.21,22 DNA methylation occurs at the genetic level and frequently acts as a “silencing” epigenetic mark, whereas the posttranslational methylation of histones is known as a type of chromatin modification.23,24 RNA m6A modification is another layer of epigenetic regulation, similar to that of DNA and histone modifications. Nevertheless, the mechanisms through which RNA m6A methylation exerts diverse functions are dramatically different from that of DNA and histone methylation.
In this review, we classify the features of the TME into four categories: hypoxic, metabolic dysregulation, immune escape, and chronic inflammation (Figure 1). We also discuss how RNA m6A methylation participates in the TME, which is necessary for further studies to identify potential diagnostic and therapeutic approaches, to obtain a deeper understanding of carcinogenesis, and to improve prognoses.
m6A modifications interacting with hypoxia in the TME
A hypoxic TEM is a principal feature of most solid tumors. Hypoxia is associated with epigenetics and the expression of downstream genes and, subsequently, regulates glucometabolism, iron metabolism, angiogenesis, proliferation, and apoptosis through the activation of hypoxia inducible factors (HIFs), which are composed of α and β subunits, and through recognition of hypoxia response elements (HREs).25, 26, 27, 28, 29
Several studies have suggested that HIFs significantly influence tumor progression, and these processes cover many factors. HIF-1α expression is linked with P-glycoprotein (P-gp), a multidrug resistance (MDR) gene/transporter. A meta-analysis suggested that the progression of bone tumors, such as differentiation, metastasis, and angiogenesis, is induced by a greater expression of HIF-1α, indicating the prognostic role of HIF-1α.30,31 In hypoxic non-small-cell lung carcinoma (NSCLC) tissue, the upregulation of membrane metallo-endopeptidase (MME) expression in fibroblasts improves invasive and metastatic ability.32 The PEA3 group (known as E1AF and ETV4) can also enhance cancer metastasis by interacting with HIF-1α.33,34 However, HIF-1α inhibits renal cell carcinoma (RCC) tumorigenesis, whereas HIF-2α enhances tumorigenesis by depleting the von Hippel-Lindau (VHL) tumor suppressor and increasing the c-MYC transformation of primary mouse embryo fibroblasts (MEFs).35 Moreover, HIF-2α, rather than HIF-1α, is the main factor of angiogenesis through the upregulation of VEGF.36 In brief, the hypoxic TME is essential for the biological functions of various cancers, and it regulates tumor progression via various relevant factors (Table 1).
Table 1.
Functions of TME in tumorigenesis and relevant factors
| TME traits | Factors/types | Downstream factors | Impacts on cancers | References |
|---|---|---|---|---|
| Hypoxia | HIF-1α | upregulates MDR gene (P-gp) | promotes bone tumor progression | 30,31 |
| increases MME expression | improves invasive and metastatic ability of NSCLC | 32 | ||
| upregulates PEA3 group | promotes metastasis | 33,34 | ||
| HIF-2α | increases VHL, decreases c-MYC | promotes RCC tumorigenesis | 35 | |
| upregulates VEGF | induces angiogenesis | 36 | ||
| Metabolic dysregulation | fermentation | induces hypoxia, hypoglycemia, acidic TME | promotes tumor progression | 37 |
| Immune escape | immune system suppression | increases immunosuppressive cells and cytokines | improves capacity of evading antitumor immune responses | 38 |
| immune system ignorance | decreases T cell-inflamed phenotype | |||
| Chronic inflammatory | inflammatory cells, cytokines, chemokines, and exosomes | induces Inflammatory cascades | tumorigenesis promotion or regression | 39,40 |
Abbreviations: NSCLC, non-small-cell lung carcinoma; RCC, renal cell carcinoma
The hypoxic TME activates HIFs and regulates hundreds of genes involved in tumor progression and its biological behaviors. Additionally, many studies have shown that hypoxia also regulates diverse sites of epigenetic mechanisms, including DNA methylation, noncoding RNAs, and histone modifications. Not only can these alterations collaborate with HIFs, they also regulate hypoxic response pathways in a straightforward manner.41 Herein, we will mainly discuss the interaction between hypoxia and RNA methylation.
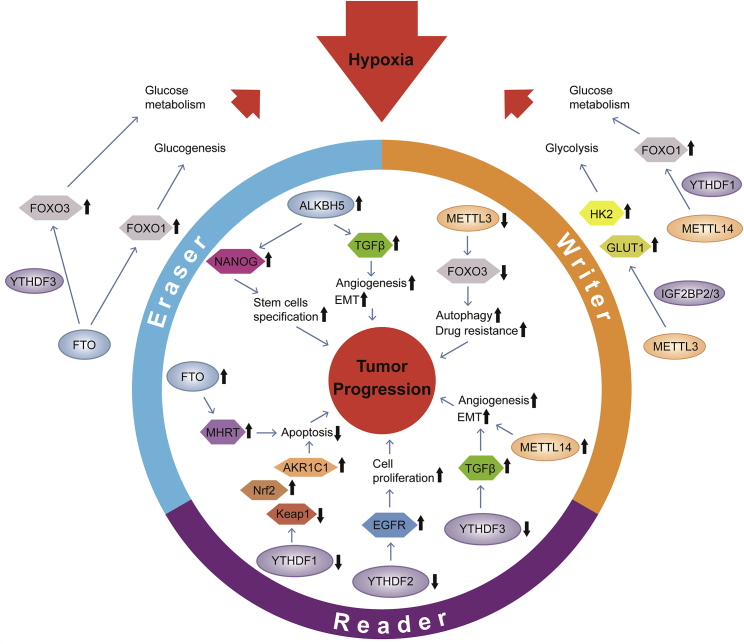
m6A methylation, a posttranscriptional modification, is able to regulate the stability of mRNA during several biological processes, including glucose metabolism, which can modulate the hypoxic condition of solid tumors (Figure 3). Shen and colleagues42 suggested that METTL3 acted at the 5′/3′ UTR regions of HK2 and the 3′ UTR region of SLC2A1 (GLUT1); hence, these two genes, related to the glycolysis pathway, were stabilized by readers, such as IGF2BP2 and IGF2BP3. Glycolysis facilitates the progression of hypoxia and, consequently, promotes tumorigenesis. In addition to promoting glucose uptake and lactate production, METTL3 can also activate mTORC1 signaling.43 Forkhead box O (FOXO) proteins, a family of transcription factors (FOXO1, FOXO3a, FOXO4, and FOXO6 in mammals), are known to bind to the promoters of several target genes and modulate biological behaviors, such as energy production, metabolism, and cellular proliferation. FOXO proteins participate in the regulation of systemic and/or hepatic glucose metabolism to maintain metabolic homeostasis and adapt to nutrient fluctuations.44,45 Notably, the posttranscriptional modifications of FOXO, including m6A methylation, significantly influence the stability of FOXO. Many researchers have identified that methylation regulators modulate the expression of FOXO in several ways. METTL14 binds to FOXO1 mRNA and promotes its translation through the subsequent YTHDF1 recognition of N6-methyladenosine. METTL14 interacts with FOXO1 and acts directly on the promoter regions of target genes.46 Furthermore, crosstalk between METTL3 and FOXO6 triggers the transcription of GPR161 and the regulation of β-defensin expression.47 Researchers have also suggested that FTO induced the mRNA expression of FOXO1 and other genes that were tightly related to glucose metabolism. The FTO-FOXO1 regulatory axis modulates glucogenesis in the liver and lowers fasting blood glucose concentrations, thus, influencing nutrient-rich and hypoxic conditions in solid tumors. FTO also enhances the transcriptional activity of FOXO3 by m6A modification.48, 49, 50 The role of YTHDF3, as a translational promoter of FOXO3 mRNA, has also been uncovered.51 Barretto and colleagues52 illustrated the role of FOXO, which acts as a mediator of hypoxia tolerance. FOXO-mutant animals show disordered glucose metabolism under hypoxic conditions, and low oxygen can induce the upregulation of FOXO. This indicates that FOXO can control homeostasis and cope with hypoxia by regulating physiological processes, such as glucose metabolism (Figure 3). Therefore, m6A regulators can promote or inhibit hypoxia not only through metabolism but also through factors or genes.
Figure 3.
The crosstalk between m6A modification and hypoxia
METTL3 can promote hypoxia through glycolysis regulated by HK2 and GLUT1, whereas hypoxia-downregulated METTL3 can decrease expression of FOXO3 and promote autophagy and drug resistance, thus promoting tumor development. Like METTL3, METTL14 is also regulated by hypoxia when it influences hypoxia conditions through glucose metabolism changed by FOXO1. The expression of METTL14 is upregulated under hypoxia and promotes angiogenesis and EMT through TGF-β. Similarly writers, in an anoxic environment, erasers, and readers promote the upregulation of tumor-related factors and strengthen tumor-related biological processes, so as to promote tumorigenesis and development. In addition, they can help cells adapt to hypoxia or enhance the hypoxic state.
Hypoxia can also affect m6A modification or control the downstream pathway through m6A methylation. Panneerdoss et al.53 demonstrated that the level/activity of writers, erasers, and readers can be regulated by hypoxia. Increased expression of ALKBH5 and METTL14 and decreased expression of YTHDF3 were shown in breast cancer cells under hypoxic conditions, thus contributing to the downregulation of m6A and, consequently, the overexpression of corresponding transcripts, such as transforming growth factor β (TGF-β) and cell cycle proteins in cancer cells. Hypoxia acts as a stimulus that interferes with RNA m6A methylation, followed by tumor progression, angiogenesis, and invasion. In the hypoxic regulation of transcripts encoding human Fe(II) and α-ketoglutarate-dependent JMJD, histone demethylases have been elucidated. HIF-1 acted at the promoter of JMJD-encoding genes and increased their expression, thus inducing histone demethylase activity. Additionally, the JMJD protein products facilitate hypoxic gene induction by remodeling chromatin at other HIF targets. Likewise, TET1, an Fe(II) and α-ketoglutarate-dependent dioxygenase that can convert 5-methylcytosine (5mC) to 5-hydroxy-methylcytosine (5hmC) and induce DNA demethylation activity, is upregulated by HIF-1.54,55 In hypoxic breast cancer cells, Zhang and colleagues56 elucidated that hypoxia-dependent HIF not only compensated for reduced O2 levels but also activated ALKBH5, JMJD2C, and TET1, which modulated the demethylation of mRNA, the demethylation of histone, and the hydroxymethylation of DNA, respectively. As a result, transcriptional and posttranscriptional alterations facilitate the specification and maintenance of breast cancer stem cells (BCSCs). Specifically, exposure to hypoxia upregulates the expression of ALKBH5, which demethylates NANOG mRNA (encoding a pluripotency factor that promotes BCSC specification) by HIF-1α and HIF-2α, whereas the starvation of ALKBH5 expression in human breast cancer cells conversely reduces tumorigenic competence because of the depletion of BCSCs. Hypoxia is tightly related to RNA m6A methylation in the TME through HIF-dependent ALKBH5 expression. Meanwhile, studies are underway to explore whether another m6A demethylase, FTO, is induced by hypoxia in these corresponding cell lines.56 Furthermore, HIF-dependent ZNF217 is a repressive factor of m6A methylation of mRNAs encoding NANOG and KLF4 and is another pluripotency factor regulating BCSC specification. Collectively, hypoxia induces the BCSC phenotype through the variable induction of pluripotency factors and ALKBH5 or ZNF217, which act as negative regulators of RNA methylation.57 RNA m6A methylation has been found to have a physiological role in the hypoxic TME as well as in drug resistance. FOXO3 is described as a critical death-inducing transcription factor downstream of the phosphoinositide 3-kinase (PI3K)-PKB survival signaling pathway. Lin and colleagues58 suggested that METTL3-mediated methylation was critical for m6A-dependent sorafenib sensitivity and that the FOXO3 mRNA 3′ UTR, which is recognized by YTHDF1, acted as a key downstream point. Mechanistically, METTL3 increases the level of m6A modification of FOXO3 so that FOXO3 RNA stability is promoted by YTHDF1 recognition, sustaining m6A-dependent sorafenib sensitivity. Conversely, the METTL3 knockdown-induced downregulation of FOXO3 enhances the autophagy signaling pathway under intratumoral environment-like conditions, such as hypoxia and chemotherapy, consequently leading to the development of drug resistance.58 However, how does hypoxia specifically influence FOXO3 activation? Hypoxic and nutrient-deprived conditions within solid tumors make the “death program” of FOXO3 more complex by influencing the physiological outcome of FOXO3 activation in high-stage neuroblastoma (NB) cells. Hagenbuchner et al.59 found that hypoxia significantly reduced PKB activity and FOXO3 phosphorylation so that FOXO3 activation under hypoxia failed to facilitate cell cycle arrest or cell death. Furthermore, researchers referred to a model as “Janus-faced” when poor activation of FOXO3 drove angiogenesis, whereas the full activation of ectopic FOXO3 induced by hypoxia repressed tumor growth but contributed to tumor cell longevity through the selection of resistant tumor cells. Ultimately, the death program of FOXO3 converts to the “tumor-longevity program.”59 It has also been determined that YTHDF2 suppresses cell proliferation, tumor growth, and activation of MEK and ERK by binding to the N6-methyladenosine sites of the EGFR 3′ UTR and augmenting its degradation. In hepatocellular carcinoma (HCC), hypoxia-induced downregulation of YTHDF2 promotes tumor progression.60 HIF-2α is negatively correlated with YTHDF2, and administration of a HIF-2α antagonist (PT2385) can renovate the YTHDF2-mediated epigenetic machinery, leading to liver cancer suppression.61 Similarly, a deficiency in YTHDF1 has been confirmed in many hypoxic solid tumors. Low expression of YTHDF1 inhibits hypoxia-induced apoptosis and promotes hypoxia adaptation through the Keap1-Nrf2-AKR1C1 axis (in which Keap1 decreases, whereas Nrf2 and AKR1C1 increase).62 There is a positive correlation between FTO and HIF-1α, and HIF-1α can inhibit apoptosis induced by hypoxia by upregulating long noncoding RNA (lncRNA) myosin heavy-chain-associated RNA transcripts (MHRT).63,64 Moreover, hypoxia, lipopolysaccharides and UV light can stabilize mRNA through the coactivator-associated arginine methyltransferase 1 (CARM1)-methylated RNA-binding protein HuR.65
In general, hypoxia, which is the most significant characteristic of the TME and is known to promote tumorigenesis, angiogenesis, invasion, and metastasis, modulates the levels of m6A writers, erasers, and readers, which, in turn, regulate hypoxic conditions and enhance adaptation to hypoxia.
m6A modifications involved in metabolic dysregulation
Compared with healthy cells, tumor cells split glucose to lactic acid in fermentation, even under oxygen-rich conditions, which is known as the Warburg effect. Even though fermentation can only produce two ATPs, whereas oxidative phosphorylation produces 36 ATPs in respiration, in which cells burn organic materials to form carbon dioxide and water, neoplastic cells prefer the fermentation process because it favors tumor growth in several dimensions.66 The metabolic characteristics of tumor cells induce hypoxic, hypoglycemic, and acidic TMEs. Meanwhile, hypoxia-induced HIF significantly enhances angiogenesis and glycolysis, creating a positive feedback loop and leading to a cancer-induced metabolic disorder (Figure 1). Additionally, a lower pH level promotes tumor metastasis, thus, accelerating its progression.37,67 Based on these features, involvement of the most prevalent RNA methylation is nonnegligible.
Alterations in cancer cell metabolism that mediate RNA modifications have been identified, leading to epigenetic and metabolic regulation. Maddocks and colleagues68 found that serine provided single-carbon units and facilitated the methionine cycle and methylation of RNA through de novo ATP synthesis. In contrast, serine depletion significantly decreased the methylation of DNA/RNA and ATP levels, accompanied by lower AMP and AMPK inactivation.68 The metabolic stress conditions we mentioned also increase the expression of FTO through the autophagy and nuclear factor κB (NF-κB) pathways, following the alteration of m6A methylation levels.69 Interestingly, m6A modification can regulate metabolic processes at the level of transcription or translation and influence tumorigenesis and its development. ALKBH3 was found to demethylate 1-meA and 3-meC within intrinsically methylated RNA. N6-methyladenine in mammalian transfer RNA (tRNA) also acts as an ALKBH3 substrate, thus, inducing the protein translation process. In addition, ALKBH3 depletion in human cancer cells subverts proliferation and inhibits protein synthesis accompanied by methylated RNA accumulation.70 m6A modification can not only affect metabolic, infectious, and developmental diseases but also affects metabolism-related cancers by interfering with the metabolic TME of tumor cells and stromal cells. Furthermore, the accumulation of genetic/epigenetic alterations of proto-oncogenes and tumor suppressor genes, which contribute to diseases such as chronic inflammation and metabolic diseases, is suggested to increase tumorigenic competence.71 Studies have validated that m6A modification is a crucial factor in metabolic homeostasis. METTL3 facilitates fatty acid metabolism and regulates the blood glucose balance by alleviating hepatic insulin sensitivity via the N6-methylation of fatty acid synthase (Fasn) mRNA.72 Increased FTO expression causes obesity in mice by the elicitation of a high-fat diet and development of glucose intolerance.73 FTO also mediates splicing of the adipogenic regulatory factor RUNX1T1 by modulating the m6A modification, thereby affecting adipogenesis.74 Meanwhile, RUNX1T1 is an important factor during pancreas development, and a regulatory disorder may activate precancerous cells and extracellular cells, resulting in pancreatic tumors.75 However, how does RNA methylation, particularly m6A modification, regulate tumor cell metabolism? Many studies have indicated that the PI3K-AKT pathway is the most prevalent, activated pathway in human cancers.
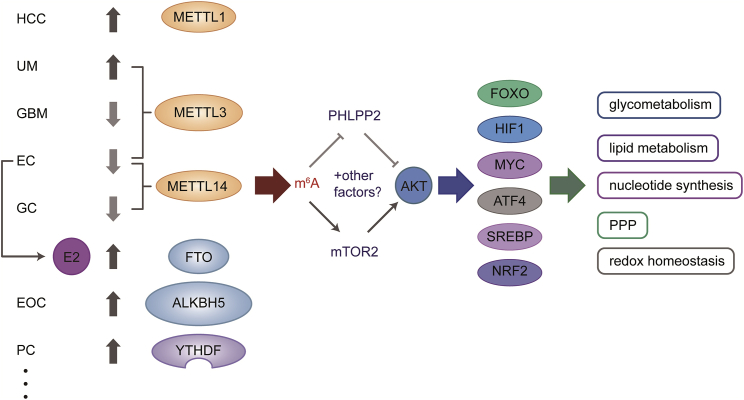
The systemic metabolic homeostasis of individual cells can be maintained through the PI3K-AKT pathway, which can be triggered by insulin, growth factors, and cytokines, leading to the regulation of key metabolic processes, including glucose uptake, glycolysis, nucleotide and lipid synthesis, the pentose phosphate pathway (PPP), and redox homeostasis. AKT directly phosphorylates several metabolic enzymes or regulators of nutrient transport and mediates cellular metabolic reprogramming by activating key downstream effectors, such as mTOR complex 1 (mTORC1), glycogen synthase kinase 3 (GSK3), and members of the forkhead box O (FOXO) family. Intriguingly, in cancer cells, the PI3K-AKT pathway activates nutrient transporters and metabolic enzymes by altering the cellular metabolism program, thus, satisfying the anabolic demands of these neoplasms.76 The PI3K/AKT and mTOR signaling pathways are significantly affected by RNA m6A methylation in most human cancers (Figure 4).77
Figure 4.
AKT pathway, regulated by m6A methylation, mediates the metabolism of diverse cancer cells
The AKT pathway mediates metabolism through several downstream transcription factors, including FOXO, HIF1, MYC, ATF4, SREBP, and NRF2. These factors control the expression of genes that encode metabolic enzymes and modulate the process of glucose uptake, glycolysis, nucleotide and lipid synthesis, pentose phosphate pathway (PPP), and redox homeostasis. The PI3K/AKT and mTOR signaling pathways are significantly affected by m6A RNA methylation regulators in most human cancers, and m6A methylation acts as a regulator of the AKT pathway through decreasing expression of the negative AKT regulator PHLPP2 and by increasing expression of the positive AKT regulator mTORC2.
In HCC, upregulated METTL1 promotes cancer development via the PTEN/AKT signaling pathway. The suppression of AKT activity correspondingly decreases METTL1-mediated malignant phenotypes.78 In uveal melanoma (UM), METTL3 starvation suppresses UM cell proliferation and metastasis through G1 cell cycle arrest by downregulating c-Met, p-AKT, and cell-cycle-related proteins (cyclin and CDK).79 In addition, the lesser expression of METTL3 enhances glioblastoma (GBM) cell proliferation, migration, and invasion by targeting the PI3K/AKT pathway.80 Approximately 70% of endometrial cancers (ECs) show reductions in m6A methylation mediated by either METTL14 mutation or reduced expression of METTL3, which is known as an oncogenic mechanism in EC, leading to increased proliferation and tumorigenesis. Mechanistically, m6A methylation acts as a regulator of the AKT pathway by decreasing the expression of the negative AKT regulator PHLPP2 and increasing the expression of the positive AKT regulator mTORC2 (Figure 4).81 Zhang and colleagues82 revealed that METTL14-depletion-dependent m6A downregulation increased gastric cancer (GC) cell proliferation and invasion through activation of the WNT and PI3K/AKT signaling pathways. Based on the involvement of FTO in metabolic disorders, such as obesity and diabetes, Peng and colleagues50 found that the mRNA of the transcription factor FOXO1 was a direct substrate of FTO and that entacapone acted on the FTO-FOXO1 regulatory axis, thereby reducing body weight and lowering the fasting blood glucose concentrations by regulating gluconeogenesis in the liver and thermogenesis in adipose tissues. Song and colleagues83 elucidated that the growth of pancreatic ductal adenocarcinoma (PDAC) was associated with the miR-21/FOXO1 axis. In EC cells, E2-induced FTO activates the PI3K/AKT and MAPK signaling pathways and enhances tumor cell anabolism and proliferation.84 In epithelial ovarian cancer (EOC), overexpression of ALKBH5 inhibits autophagy, which acts as an important pathway for cell metabolism. Mechanistically, ALKBH5 activates the EGFR-PIK3CA-AKT-mTOR signaling pathway, stabilizing BCL-2 mRNA and facilitating crosstalk between Bcl-2 and Beclin1.85 In prostate cancer (PC), YTHDF2 mediates mRNA degradation by binding to the m6A modification sites of the tumor suppressors LHPP and NKX3-1 with activated phosphorylated AKT. Therefore, tumor progression and the metabolic TEM are regulated.86 In short, RNA m6A methylation is likely to mediate several factors of the AKT pathway and, subsequently, regulate aberrant cell metabolism, including glucose uptake, glycolysis, and anabolic metabolism; hence, a hypoxic, hypoglycemic, and acidic TEM is established.
To conclude, cancer metabolic dysregulation and metabolic reprogramming, including the Warburg effect, contribute to metabolite accumulation, which modulates RNA methylation regulators and subsequent m6A modification. Meanwhile, cancer cells exhibit a number of epigenetic alterations, particularly RNA methylation, that lead to metabolic adaptation and dysregulation through several metabolic pathways, enabling tumor progression. As a result, the crosstalk between m6A modifications and tumor metabolic dysregulation is of clinical value.
m6A modifications participating in tumor immunity
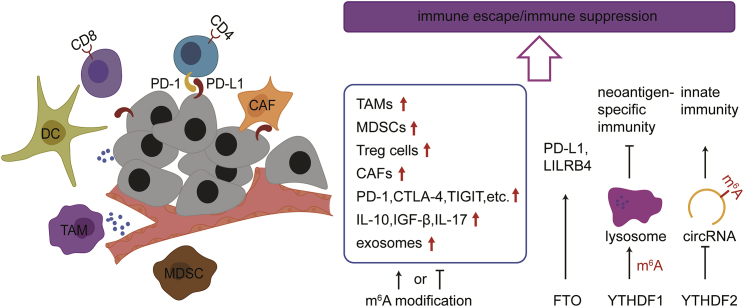
In the TEM, immune cells are different from cancer cells in that immune cells do not yield to Darwinian evolutionary pressure, which would enable them to adapt to progressive tumors but are, instead, often irreversibly subject to local nutrient deprivation. Metabolic alterations, such as tumor-derived lactate and tumor acidity, in the TME switch the phenotype and function of immune cells from a tumor-active state to a tumor-promoting state and lead to tumor immune evasion (Figure 1).87 Neoplasms have the capacity to educate immune cells to evade antitumor immune responses, continuously maintaining their progression. Based on cellular and molecular features of the TEM, several lines of evidence have shown two types of tumor escape. One exhibits a T-cell-inflamed phenotype that comprises infiltrating T cells, an extensive cytokine profile and a type I interferon signature, indicating innate immune activation. These tumors resist immune attack through the immune-system-suppressive pathways. The other phenotype, which lacks a T-cell-inflamed phenotype, resists immune attack by immune system ignorance.38,88 Immunosuppressive cells in the TEM mainly include tumor-associated macrophages (TAMs), regulatory T cells (Tregs), myeloid-deprived suppressor cells (MDSCs), cancer-associated fibroblasts (CAFs), regulatory dendritic cells (DCregs), neutrophils, T helper 17 cells (Th17s), and regulatory B cells (Bregs), whereas immunosuppressive cytokines comprise interleukin-10 (IL-10), TGF-β, IL-17, tumor-derived exosomes, and immune checkpoint molecules, such as programmed cell death-1 (PD-1), programmed cell death-ligand 1 (PD-L1), cytotoxic T lymphocyte antigen 4 (CTLA4), lymphocyte activation gene-3 (LAG-3), T cell immunoglobulin, and immunoreceptor tyrosine-based inhibitory motif (ITIM) domain protein (TIGIT) and T cell immunoglobulin and mucin domain-3 (TIM-3) (Figure 5).89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104
Figure 5.
m6A modification is involved in formation of immune escape or immune suppression
Immunosuppressive cells and cytokines that facilitate immune escape in TME mainly include TAMs, Tregs, MDSCs, CAFs, DCregs, and Bregs, and immunosuppressive cytokines comprise IL-10, TGF-β, IL-17, Th17, tumor-derived exosomes, and immune checkpoint molecule, such as PD-1, CTLA-4, and TIGIT, among others. The m6A score is associated with activation of immunity and PD-1 expression levels to different degrees in various cancers. FTO promotes the expression of immune checkpoint molecules, such as PD-1L and LILRB4. YTHDF1 destructs neoantigen-specific immunity through mediating m6A modification of transcripts that encode lysosomal proteases to promote immune escape. In addition, YTHDF2 isolates m6A-circRNA and acts as an essential factor for suppression of innate immunity.
The methylation of RNA and its regulators, especially m6A, has an indispensable role in tumor biological activities. Chen and colleagues105 established m6A epigenetic module eigengenes by combining hub m6A regulators. The model can reflect the status of the tumor-immune-stromal microenvironment and predict prognosis.105 m6A modifications have a ubiquitous and nonnegligible role in the formation of TME cell-infiltrating characteristics and the three immune phenotypes of tumors, which are immune-excluded, immune-inflamed and immune-desert. Zhang et al. 4 suggested that evaluation of m6A modification patterns could predict stages of tumor inflammation, subtypes, TME stromal activity, genetic variation, and patient prognosis. More specifically, a lower m6A score is associated with an increased mutation burden and activation of immunity, indicating an inflamed TME phenotype, whereas a higher m6A score is characterized by stromal activation and a lack of effective immune infiltration, showing a noninflamed and immune-excluded TME phenotype. Distinct neoantigen loads and responses to anti-PD-1/L1 immunotherapy also existed at different m6A modification levels (Figure 5). Thus, evaluating the m6A modification pattern favors our understanding of the TME infiltration traits and the implementation of immunotherapy strategies.4 m6A modification is not only involved in the oncogenic PI3K-AKT signaling pathway, as mentioned, but is also associated with interferon signaling and immune responses in gastric cancer.82 Next, we turn to experimental evidence for the involvement of m6A in tumor immunity. In head and neck squamous cell carcinoma (HNSCC), PD-L1 expression and tumor-infiltrating immune cells in the TME are significantly associated with m6A-regulator-based signatures and the m6A modification burden, indicating promising therapeutic targets in enhancing immunotherapeutic efficacy.106 In breast cancer, studies have shown that greater RNA m6A methylation characterizes increased expression of HLA-A and more tumor-infiltrating CD8+ T cells, helper T cells, and NK cells but decreased expression of PD-L1, PD-L2, TIM3, and CCR4. Lower RNA m6A modification is linked with the hallmarks of PI3K/AKT signaling in cancer, KRAS signaling, angiogenesis, and shorter overall survival.107 Bioinformatics studies on nasopharyngeal carcinoma (NPC) have indicated that the risk signature for m6A methylation is significantly related to immune infiltration and immune evasion in the TME.108 In GC, the risk score is associated with 16 m6A methylation regulators; hence, modification is regarded as an independent prognostic indicator. The low-risk subgroup shows an upregulation of immune checkpoint molecules, indicating that the m6A signature may be a regulator of immune escape and a potential immunotherapy predictor.109 m6A modification is also involved in the formation of the TME and immune-related pathways in lung adenocarcinoma (LUAD). The leucine-rich pentatricopeptide repeat containing (LRPPRC) gene is negatively correlated with most tumor-infiltrating immune cells. However, m6Sig, a scoring tool, is positively correlated with PD-L1 expression and may reveal TEM features. Additionally, high-m6Sig groups exhibit therapeutic advantages, such as immunotherapy sensitivity.110
Regarding distinct tumors, diverse RNA methylation regulators have different immune effects on tumor progression. Tang and colleagues111 investigated the relationship between the mutation state of m6A-related genes and the immune microenvironment of pancreatic adenocarcinoma (PAAD), suggesting that m6A modification might be involved in immune cell infiltration and that the constitution of the infiltrating immune cells may affect m6A modification. The expression of the eight most-studied m6A-related genes with immune cells in PAAD was involved in their study, and they found that m6A modification could modulate CD8+ T cell aggregation. Specifically, the arm-level gain and deletion of ALKBH5 reduced the infiltration of CD8+ T cells, whereas the arm-level deletions and gains of m6A genes significantly decreased the infiltration of CD4+ T cells. Furthermore, RBM15 is the gene with the highest hazard ratio to PDAC survival, and greater expression of RBM15 stimulates antigen-presenting cells (APCs), enhances inflammation, and upregulates major histocompatibility complex (MHC) class I, Th2s, and Tregs.111 Compiled evidence has illustrated that the high-risk group of lung squamous cell carcinoma (LUSC) has less expression of RNA m6A methylation regulators, including ALKBH5, METTL3, HNRNPC, and KIAA1429, whereas the low-risk LUSC group has higher levels of those regulators. Among the immune cell types estimated, Xu and colleagues112 found that only T follicular helper cells (TFHs) (the CD4+ T cell subpopulation present in B cell follicles) were significantly associated with the pathogenesis of LUSC and the overall survival (OS) of patients with LUSC, indicating that TFHs could independently act as a prognostic LUSC indicator. Furthermore, patients with high-risk LUSC are more sensitive to immunotherapy and chemotherapy.112 Hence, m6A methylation regulators may be associated with TFHs, and they can be used in the development of effective prognostic biomarkers and the exploration of therapeutic targets for patients with LUSC. Li et al.113 found that m6A regulators affected osteosarcoma progression through the humoral immune response. Patients with testicular germ cell tumor (TGCT) and lower expression levels of METTL3 exhibit poorer overall survival and relapse-free survival rates. METTL3 promotes tumor progression by influencing the expression of epithelial-mesenchymal transition (EMT)-related proteins. Additionally, the upregulation of METTL3 activates the tumor immune response (infiltration level of CD8+ and CD4+ T cells and natural killer cells) in TGCTs.114 In mismatch-repair-proficient or microsatellite-instability-low (pMMR-MSI-L) colorectal cancer (CRC) and melanoma, the downregulation of m6A mRNA modification by the starvation of METTL3 and METTL14 elevates cytotoxic tumor-infiltrating CD8+ T cells; promotes the secretion of interferon-gamma (IFN-γ), Cxcl9, and Cxcl10 in the TEM; and even facilitates responses to anti-PD-1 treatment. A mechanical study showed that METTL3/METTL14 depletion enhanced IFN-γ-Stat1-Irf1 by stabilizing Stat1 and Irf1 mRNA via YTHDF, and there was a negative correlation between METTL3/METTL14 and STAT1 in some patients with pMMR-MSI-L CRC tumors. Altogether, METTL3 and METTL14 have been identified as potential therapeutic targets in anticancer immunotherapy.115 ALKBH5 is another potential therapeutic target to promote immunotherapy outcomes in several cancer subtypes. During immune checkpoint blockade (ICB) therapy, such as PD-1, the m6A demethylase ALKBH5 decreases m6A density and splicing events in tumors, and depletion of ALKBH5 sensitizes tumors to cancer immunotherapy by modulating Mct4/Slc16a3 expression, the lactate content of the TEM, and the composition of tumor-infiltrating Tregs and myeloid-derived suppressor cells (MDSCs).116 FTO has an important role in inducing immune evasion (Figure 5). Yang and colleagues69 found that FTO was positively correlated with melanoma tumorigenesis by promoting PD-1L expression. It has been consistently proven that the loss of FTO sensitizes melanoma cells to IFN-γ and anti-PD-1 treatment, which depend on adaptive immunity. Hence, the combination of FTO inhibition with an anti-PD-1 blockade may eliminate resistance to immunotherapy.69 It has also been elucidated that FTO upregulates the expression of leukocyte immunoglobulin-like receptor subfamily B4 (LILRB4), another immune checkpoint molecule in acute myeloid leukemia (AML), by suppressing the YTHDF2-mediated decay of m6A-modified LILRB4 mRNA. In addition, it has been demonstrated that a key m6A reader, YTHDF1, destroys durable neoantigen-specific immunity through interaction with transcripts encoding lysosomal proteases, indicating that mutated m6A modification may promote immune evasion.117 Moreover, the m6A reader YTHDF2 isolates m6A-circular RNA (circRNA) and acts as an essential factor for the suppression of innate immunity. Foreign circRNAs activate antigen-specific T cells, antibody production, and antitumor immunity, but m6A modification prevents immune gene activation and inhibits innate immunity (Figure 5).118
In summary, m6A modification promotes or inhibits tumor progression by regulating the activity of immune cells, the distribution of cytokines, and the role of immune checkpoints in tumor immunity. Importantly, we can predict the immune-infiltration characteristics of the TME and therapeutic efficacy by detecting the abundance of m6A regulators and the m6A level of cancer cells to develop the most effective immunotherapy strategy and obtain the best prognosis for patients.
m6A modification counts in chronic inflammation of the TME
Chronic inflammation is another crucial component of the TME. The inflammatory cells and cytokines found in the TME tend to facilitate tumor progression and immunosuppression, rather than establish an effective host antitumor response (Figure 1). Furthermore, tumors may affect inflammatory cascades, causing tumor promotion or regression, and chemokines and other cytokines have essential roles in these processes.39,40,119
It is well known that inflammatory cytokines, including interleukins, interferons, transforming growth factors, chemokines, and adhesion molecules have key roles in chronic inflammation. Meanwhile, numerous cytokines are modulated by discriminated epigenetic mechanisms, such as DNA, RNA methylation, and histone modifications, and some of these cytokines also act as epigenetic regulators in tumorigenesis and progression.120 In CRC, inflammation and antitumor immunity are key determinants of tumor progression, and cytokine networks are essential for tumor immunology. Cancer mutations and epigenetic adaptations may activate the oncogenic potential of cytokines.121 Chiba et al.122 suggested that chronic inflammation could mediate the expression of microRNAs so that several tumor-related mRNAs or proteins could be changed. These molecular events, regulated by chronic inflammation, subsequently, influence normal cellular function, thus, promoting inflammation-induced tumorigenesis. For instance, the intrinsic regulators of inflammatory responses, such as proinflammatory cytokines, promote genetic mutations and epigenetic alterations, and METTL3/14 and FTO can influence hepatitis C virus biological activities, which may contribute to tumor progression.122
Accumulating evidence has shown that chronic inflammation is significantly associated with HCC. Because G-protein alpha-subunit (GNAS)-activating mutations participate in forming a rare subgroup of inflammatory liver tumors, the roles of GNAS in inflammation-related HCC progression and its underlying mechanism cannot be neglected. In HCC cells, Ding and colleagues123 found that lipopolysaccharide (LPS) stimulation upregulated GNAS expression by enhancing m6A methylation of GNAS mRNA, forming specific inflammatory characteristics of HCC. Moreover, HCC cells show high expression of m6A modification, which is regarded as a result of YTHDF2 reduction. In human HCC cells or mouse hepatocytes, YTHDF2 depletion provokes inflammation, angiogenesis, and metastasis. A mechanical study showed that YTHDF2 promoted the decay of m6A-containing IL-11, which is essential for inflammation-mediated tumorigenesis.61 Additionally, ncRNAs, including miRNAs, lncRNAs, and circRNAs, are involved in cell differentiation, angiogenesis, immune response, inflammatory response, and carcinogenesis. m6A regulators, such as METTL3, ALKBH5, and IGF2BP1, participate in carcinogenesis and inflammatory activities by modifying ncRNAs, which can mediate m6A regulators to influence inflammation and cancer development.124
Therefore, m6A modifications can influence the occurrence and development of chronic inflammation through a variety of inflammatory factors and alter the TEM, especially in HCC. However, a considerable amount of future work is required to identify the inflammatory targets of m6A regulators and to elucidate the functional consequences of such effects in tumors. Additionally, existing crosstalk between m6A modification and chronic inflammation is mainly present in chronic hepatitis, liver cirrhosis, and liver cancer, but the relevant evidence in other diseases is still unknown. Finally, as described in the above four sections, m6A modifications, modulated by diverse m6A regulators, produce multiple biological functions in various TMEs of different cancers (Table 2).
Table 2.
Molecular mechanism and biological functions of m6A in distinct TME of diverse cancers
| TME | Regulators | Cancers | Mechanism | Biological functions | References |
|---|---|---|---|---|---|
| Hypoxia | METTL3 | HCC | methylates FOXO3 | promotes sorafenib sensitivity | 58 |
| ALKBH5 | BC | demethylates NANOG | promotes specification and maintenance of BCSCs | 56 | |
| YTHDF2 | HCC | interacts with HIF-2α | suppresses cancer growth | 65 | |
| Metabolic dysregulation | METTL1 | HCC | upregulates PTEN/AKT signaling pathway | promotes malignant phenotype | 78 |
| METTL3 | UM | upregulates c-MET, p-AKT, cyclin, and CDK | promotes proliferation and metastasis | 79 | |
| GBM | targets PI3K/AKT signaling pathway | suppresses proliferation, migration, and invasiveness | 80 | ||
| METTL14 | EC | regulates expression of PHLPP2 and mTOR2 | suppresses proliferation and tumorigenesis | 81 | |
| GC | mediates WNT and PI3K/AKT signaling pathway | suppresses proliferation and invasiveness | 82 | ||
| ALKBH5 | EOC | demethylates tRNA | promotes protein synthesis | 70 | |
| activates EGFR-PIK3CA-AKT-mTOR signaling pathway | promotes tumor progression | 85 | |||
| FTO | PAAD | demethylates RUNX1T1 | promotes adipogenesis | 74,75 | |
| demethylates FOXO1 | modulates gluconeogenesis and thermogenesis | 50,83 | |||
| EC | activates PI3K/AKT and MAPK signaling pathway | promotes tumor cell anabolism and proliferation | 84 | ||
| YTHDF2 | PC | bounds to LHPP and NKX3-1 | promotes tumor progression | 86 | |
| Immune escape | METTL3 | TGCT | influences EMT and infiltration of T and NK cells | promotes tumor progression | 114 |
| METTL3/14 | CRC | modulates CD8+ T cells, IFN-γ, CXCL9/10 and PD-1 | mediates response to immunotherapy | 115 | |
| RBM15 | PDAC | upregulates APC, MHC I, Th2 cells and Tregs | influences immune responses | 111 | |
| ALKBH5 | PAAD | modulates CD8+ and CD4+ T cells aggregation | influences immune responses | 87 | |
| LUSC | regulates TFH | influences immunotherapy sensitivity | 112 | ||
| modulates Mct4/Slc16a3 expression, lactate content, Tregs and MDSCs | promotes immune escape | 116 | |||
| FTO | MM | promotes PD-1L expression | promotes immune escape | 69 | |
| AML | upregulates LILRB4 expression | regulates immune escape | 117 | ||
| YTHDF1 | AML | interacts with transcripts of lysosomal proteases | suppresses neoantigen-specific immunity | 117 | |
| YTHDF2 | AML | mediates decay of LILRB4 | regulates immune escape | 117 | |
| isolates m6A-circRNA | suppresses innate immunity | 118 | |||
| Chronic inflammatory | METTL3 ALKBH5 |
modifies ncRNAs | influences carcinogenesis and inflammatory activities | 124 | |
| YTHDF2 | HCC | promotes decay of IL11 | suppresses inflammation, angiogenesis, and metastasis | 61 |
Abbreviations: HCC, human hepatocellular carcinoma; BC, breast cancer; UM, uveal melanoma; GBM, glioblastoma; EC, endometrial cancer; GC, gastric cancer; EOC, epithelial ovarian cancer; PAAD, pancreatic adenocarcinoma; PC, prostate cancer; TGCT, Testicular germ cell tumors; CRC, colorectal cancer; PDAC, pancreatic ductal adenocarcinoma; LUSC, lung squamous cell carcinoma; MM, malignant melanoma; AML, acute myeloid leukemia
Discussion
Recently, there has been substantial interest in the TEM and its regulatory network. At the same time, many studies have explored the effects of epigenetic alterations, including RNA methylation, on tumorigenesis and the microenvironment. Unfortunately, previous studies have not generally discussed the relationship between the TEM and m6A methylation, which is the most prevalent modification of mRNA. In this review, we have attempted to introduce some concepts to summarize the features of the TME that have been previously acknowledged: hypoxia, metabolic dysregulation, immune escape, and chronic inflammation. Fortunately, multiple functions of RNA m6A methylation have been identified, which makes it possible to correlate m6A modifications and carcinogenesis. In the meantime, these four features of the TME are induced by tumorigenesis, and reciprocally, they facilitate tumor growth, showing a positive feedback loop. Based on these two truths, we decided to probe the interaction between the TME and RNA methylation, particularly m6A modification. Intriguingly, several lines of research have illustrated that factors of the TME influence m6A modification, which is positively or negatively correlated with the TME. By combining that evidence, we found remarkable interplay among these four characteristics of the TME, especially between hypoxia and metabolic disorders, which are closely related and promote each other. Tumor metabolic features can induce the transformation of the immunophenotype of the TME, thus facilitating tumor immune escape. The role of m6A methylation in the TME is nonnegligible. In the pathogenesis of tumors, the m6A methylation status regulates posttranscriptional modification and enhances or weakens the stability of mRNA, augmenting the expression of proto-oncogenes or impeding that of tumor suppressor genes, thus inducing tumorigenesis. Moreover, m6A methylation can enable the formation of the TME and cell adaptation. On the other hand, the TME has noticeable effects on the expression of m6A methylation regulators through various factors, such as HIFs, which alters the abundance of m6A. Altogether, in the occurrence and development of tumors, m6A methylation and the TME are mutually induced and regulated and can be synergistic or antagonistic.
However, our review is limited by a lack of understanding of the direct connection and internal details between the TEM and m6A methylation in some microenvironment characteristics. For instance, in metabolic dysregulation, whether methylation-related enzymes affect the AKT pathway through nonmethylation pathways and whether the regulation of methylation-related enzymes on the AKT pathway and its downstream molecules directly influence tumor metabolism, deserve further exploration. In addition, because of the scarcity of investigations on the relationship between m6A methylation and chronic inflammation, the pathogenesis and therapeutic targets of tumors with poor prognosis are unknown.
Our study may provide a window of opportunity to introduce TME-targeted therapies that act on RNA methylation, and much remains to be verified before we determine a comprehensive network map between the TME and RNA methylation. We hope that increasing the sensitivity of immunotherapy through appropriate m6A regulators will become available as a third-line and later treatment, and better patient care can be achieved through our further exploration.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81802381), the National Key Research and Development program: The Key Technology of Palliative Care and Nursing for Cancer Patients (2017YFC1309201), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801).
Author contributions
P.M, Y.S., and Y.G. designed the study. Y.G., J.Z., X.W., and Y.F. drafted the manuscript. P.M. and Y.P. critically revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yongqian Shu, Email: yongqian_shu@163.com.
Pei Ma, Email: mapei@njmu.edu.cn.
References
- 1.Riera-Domingo C., Audigé A., Granja S., Cheng W.C., Ho P.C., Baltazar F., Stockmann C., Mazzone M. Immunity, hypoxia, and metabolism-the ménage à trois of cancer: implications for immunotherapy. Physiol. Rev. 2020;100:1–102. doi: 10.1152/physrev.00018.2019. [DOI] [PubMed] [Google Scholar]
- 2.Lin P., Wen D.Y., Chen G., Dang Y.W., He Y., Yang H. Predictive value of hypoxia, metabolism and immune factors for prognosis in hepatocellular carcinoma: a retrospective analysis and multicenter validation study. J. Cancer. 2020;11:4145–4156. doi: 10.7150/jca.41983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denton A.E., Roberts E.W., Fearon D.T. Stromal cells in the tumor microenvironment. Adv. Exp. Med. Biol. 2018;1060:99–114. doi: 10.1007/978-3-319-78127-3_6. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B., Wu Q., Li B., Wang D., Wang L., Zhou Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ear J., Lin S. RNA methylation regulates hematopoietic stem and progenitor cell development. J. Genet. Genomics. 2017;44:473–474. doi: 10.1016/j.jgg.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vågbø C.B., Shi Y., Wang W.L., Song S.H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 12.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y., Dong L., Liu X.M., Guo J., Ma H., Shen B., Qian S.B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019;10:5332. doi: 10.1038/s41467-019-13317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T., Wei Q., Jin J., Luo Q., Liu Y., Yang Y., Cheng C., Li L., Pi J., Si Y. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 19.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 Is a Mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaccara S., Jaffrey S.R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell. 2020;181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M.W., Fujiwara K., Che X., Zheng S., Zheng L. DNA methylation in the tumor microenvironment. J. Zhejiang Univ. Sci. B. 2017;18:365–372. doi: 10.1631/jzus.B1600579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks D.L., Olson R.L., Fernandez-Zapico M.E. Epigenetic control of the tumor microenvironment. Epigenomics. 2016;8:1671–1687. doi: 10.2217/epi-2016-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg M.V.C., Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 25.Subarsky P., Hill R.P. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis. 2003;20:237–250. doi: 10.1023/a:1022939318102. [DOI] [PubMed] [Google Scholar]
- 26.Bristow R.G., Hill R.P. Hypoxia and metabolism: hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Cao Z.L., Han F., Gao Z.C., He Q.Y. Chronic intermittent hypoxia from pedo-stage decreases glucose transporter 4 expression in adipose tissue and causes insulin resistance. Chin. Med. J. (Engl.) 2010;123:463–470. [PubMed] [Google Scholar]
- 28.Trisciuoglio D., Gabellini C., Desideri M., Ragazzoni Y., De Luca T., Ziparo E., Del Bufalo D. Involvement of BH4 domain of bcl-2 in the regulation of HIF-1-mediated VEGF expression in hypoxic tumor cells. Cell Death Differ. 2011;18:1024–1035. doi: 10.1038/cdd.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M.M., Wang J., Xie J.X. Regulation of iron metabolism by hypoxia-inducible factors. Sheng Li Xue Bao. 2017;69:598–610. [PubMed] [Google Scholar]
- 30.Ding Z., Yang L., Xie X., Xie F., Pan F., Li J., He J., Liang H. Expression and significance of hypoxia-inducible factor-1 alpha and MDR1/P-glycoprotein in human colon carcinoma tissue and cells. J. Cancer Res. Clin. Oncol. 2010;136:1697–1707. doi: 10.1007/s00432-010-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo D., Ren H., Zhang W., Xian H., Lian K., Liu H. Clinicopathological and prognostic value of hypoxia-inducible factor-1α in patients with bone tumor: a systematic review and meta-analysis. J. Orthop. Surg. Res. 2019;14:56. doi: 10.1186/s13018-019-1101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leithner K., Wohlkoenig C., Stacher E., Lindenmann J., Hofmann N.A., Gallé B., Guelly C., Quehenberger F., Stiegler P., Smolle-Jüttner F.M. Hypoxia increases membrane metallo-endopeptidase expression in a novel lung cancer ex vivo model - role of tumor stroma cells. BMC Cancer. 2014;14:40. doi: 10.1186/1471-2407-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Launoit Y., Baert J.L., Chotteau-Lelievre A., Monte D., Coutte L., Mauen S., Firlej V., Degerny C., Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Wollenick K., Hu J., Kristiansen G., Schraml P., Rehrauer H., Berchner-Pfannschmidt U., Fandrey J., Wenger R.H., Stiehl D.P. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res. 2012;40:1928–1943. doi: 10.1093/nar/gkr978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordan J.D., Bertout J.A., Hu C.J., Diehl J.A., Simon M.C. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rankin E.B., Rha J., Unger T.L., Wu C.H., Shutt H.P., Johnson R.S., Simon M.C., Keith B., Haase V.H. Hypoxia-inducible factor-2 regulates vascular tumorigenesis in mice. Oncogene. 2008;27:5354–5358. doi: 10.1038/onc.2008.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T., Suo C., Zheng C., Zhang H. Hypoxia and metabolism in metastasis. Adv. Exp. Med. Biol. 2019;1136:87–95. doi: 10.1007/978-3-030-12734-3_6. [DOI] [PubMed] [Google Scholar]
- 38.Beatty G.L., Gladney W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015;21:687–692. doi: 10.1158/1078-0432.CCR-14-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 40.Atretkhany K.N., Drutskaya M.S., Nedospasov S.A., Grivennikov S.I., Kuprash D.V. Chemokines, cytokines and exosomes help tumors to shape inflammatory microenvironment. Pharmacol. Ther. 2016;168:98–112. doi: 10.1016/j.pharmthera.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Geismann C., Arlt A. Coming in the air: hypoxia meets epigenetics in pancreatic cancer. Cells. 2020;9:E2353. doi: 10.3390/cells9112353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., Zhang X., Cao Y., Ma D., Zhu X. m6A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H., Gao S., Liu W., Wong C.C., Wu J., Wu J. RNA N6-methyladenosine methyltransferase METTL3 facilitates colorectal cancer by activating the m6A-GLUT1-mTORC1 axis and is a therapeutic target. Gastroenterology. 2021;160:1284–1300.e16. doi: 10.1053/j.gastro.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Murtaza G., Khan A.K., Rashid R., Muneer S., Hasan S.M.F., Chen J. FOXO transcriptional factors and long-term living. Oxid. Med. Cell. Longev. 2017;2017:3494289. doi: 10.1155/2017/3494289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Link W. Introduction to FOXO biology. Methods Mol. Biol. 2019;1890:1–9. doi: 10.1007/978-1-4939-8900-3_1. [DOI] [PubMed] [Google Scholar]
- 46.Jian D., Wang Y., Jian L., Tang H., Rao L., Chen K., Jia Z., Zhang W., Liu Y., Chen X. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics. 2020;10:8939–8956. doi: 10.7150/thno.45178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zong X., Wang H., Xiao X., Zhang Y., Hu Y., Wang F., Wang Y., Lu Z. Enterotoxigenic Escherichia coli infection promotes enteric defensin expression via FOXO6-METTL3-m(6)A-GPR161 signalling axis. RNA Biol. 2021;18:576–586. doi: 10.1080/15476286.2020.1820193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Shen F., Huang W., Qin S., Huang J.T., Sergi C., Yuan B.F., Liu S.M. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2019;104:665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 49.Tao B., Huang X., Shi J., Liu J., Li S., Xu C., Zhong J., Wan L., Feng B., Li B. FTO interacts with FOXO3a to enhance its transcriptional activity and inhibits aggression in gliomas. Signal Transduct. Target. Ther. 2020;5:130. doi: 10.1038/s41392-020-00234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng S., Xiao W., Ju D., Sun B., Hou N., Liu Q., Wang Y., Zhao H., Gao C., Zhang S. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019;11:eaau7116. doi: 10.1126/scitranslmed.aau7116. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., Cao X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. USA. 2019;116:976–981. doi: 10.1073/pnas.1812536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barretto E.C., Polan D.M., Beevor-Potts A.N., Lee B., Grewal S.S. Tolerance to hypoxia is promoted by FOXO regulation of the innate immunity transcription factor NF-κB/relish in Drosophila. Genetics. 2020;215:1013–1025. doi: 10.1534/genetics.120.303219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panneerdoss S., Eedunuri V.K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., Abdelfattah N., Onyeagucha B.C., Cui X., Lai Z. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci. Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mariani C.J., Vasanthakumar A., Madzo J., Yesilkanal A., Bhagat T., Yu Y., Bhattacharyya S., Wenger R.H., Cohn S.L., Nanduri J. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014;7:1343–1352. doi: 10.1016/j.celrep.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard P.J., Loenarz C., Mole D.R., McDonough M.A., Gleadle J.M., Schofield C.J., Ratcliffe P.J. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem. J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 56.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., Semenza G.L. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Z., Niu Y., Wan A., Chen D., Liang H., Chen X., Sun L., Zhan S., Chen L., Cheng C. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagenbuchner J., Rupp M., Salvador C., Meister B., Kiechl-Kohlendorfer U., Müller T., Geiger K., Sergi C., Obexer P., Ausserlechner M.J. Nuclear FOXO3 predicts adverse clinical outcome and promotes tumor angiogenesis in neuroblastoma. Oncotarget. 2016;7:77591–77606. doi: 10.18632/oncotarget.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong L., Liao D., Zhang M., Zeng C., Li X., Zhang R., Ma H., Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., Hu B., Zhou J., Zhao Z., Feng M. Correction to: YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer. 2020;19:137. doi: 10.1186/s12943-020-01257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y., Fan S., Wu M., Zuo Z., Li X., Jiang L., Shen Q., Xu P., Zeng L., Zhou Y. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lappalainen T., Kolehmainen M., Schwab U., Pulkkinen L., de Mello V.D., Vaittinen M., Laaksonen D.E., Poutanen K., Uusitupa M., Gylling H. Gene expression of FTO in human subcutaneous adipose tissue, peripheral blood mononuclear cells and adipocyte cell line. J. Nutrigenet. Nutrigenomics. 2010;3:37–45. doi: 10.1159/000320732. [DOI] [PubMed] [Google Scholar]
- 64.Shen W., Li H., Su H., Chen K., Yan J. FTO overexpression inhibits apoptosis of hypoxia/reoxygenation-treated myocardial cells by regulating m6A modification of Mhrt. Mol. Cell. Biochem. 2021 doi: 10.1007/s11010-021-04069-6. Published online February 6, 2021. [DOI] [PubMed] [Google Scholar]
- 65.Li H., Park S., Kilburn B., Jelinek M.A., Henschen-Edman A., Aswad D.W., Stallcup M.R., Laird-Offringa I.A. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 66.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu L., Cui R., Liu H., Wang F. Emodin and rhein decrease levels of hypoxia-inducible factor-1α in human pancreatic cancer cells and attenuate cancer cachexia in athymic mice carrying these cells. Oncotarget. 2017;8:88008–88020. doi: 10.18632/oncotarget.21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maddocks O.D., Labuschagne C.F., Adams P.D., Vousden K.H. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol. Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang S., Wei J., Cui Y.H., Park G., Shah P., Deng Y., Aplin A.E., Lu Z., Hwang S., He C., He Y.Y. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ueda Y., Ooshio I., Fusamae Y., Kitae K., Kawaguchi M., Jingushi K., Hase H., Harada K., Hirata K., Tsujikawa K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017;7:42271. doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie W., Ma L.L., Xu Y.Q., Wang B.H., Li S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019;518:120–126. doi: 10.1016/j.bbrc.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Church C., Moir L., McMurray F., Girard C., Banks G.T., Teboul L., Wells S., Brüning J.C., Nolan P.M., Ashcroft F.M., Cox R.D. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W., Hao Y.J., Ping X.L., Chen Y.S., Wang W.J. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benitez C.M., Qu K., Sugiyama T., Pauerstein P.T., Liu Y., Tsai J., Gu X., Ghodasara A., Arda H.E., Zhang J. An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet. 2014;10:e1004645. doi: 10.1371/journal.pgen.1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Q., Zhao Y., Hu W., Zhang Y., Wu X., Lu J., Li M., Li W., Wu W., Wang J. m6A RNA modification modulates PI3K/Akt/mTOR signal pathway in gastrointestinal cancer. Theranostics. 2020;10:9528–9543. doi: 10.7150/thno.42971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian Q.H., Zhang M.F., Zeng J.S., Luo R.G., Wen Y., Chen J., Gan L.G., Xiong J.P. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J. Mol. Med. (Berl.) 2019;97:1535–1545. doi: 10.1007/s00109-019-01830-9. [DOI] [PubMed] [Google Scholar]
- 79.Luo G., Xu W., Zhao Y., Jin S., Wang S., Liu Q., Chen X., Wang J., Dong F., Hu D.N. RNA m6 A methylation regulates uveal melanoma cell proliferation, migration, and invasion by targeting c-Met. J. Cell. Physiol. 2020;235:7107–7119. doi: 10.1002/jcp.29608. [DOI] [PubMed] [Google Scholar]
- 80.Ji J.W., Zhang Y.D., Lai Y.J., Huang C.G. Mettl3 regulates the proliferation, migration and invasion of glioma cells by inhibiting PI3K/Akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3818–3828. doi: 10.26355/eurrev_202004_20848. [DOI] [PubMed] [Google Scholar]
- 81.Liu J., Eckert M.A., Harada B.T., Liu S.M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang C., Zhang M., Ge S., Huang W., Lin X., Gao J., Gong J., Shen L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766–4781. doi: 10.1002/cam4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song W., Li Q., Wang L., Wang L. Modulation of FoxO1 expression by miR-21 to promote growth of pancreatic ductal adenocarcinoma. Cell. Physiol. Biochem. 2015;35:184–190. doi: 10.1159/000369686. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Z., Zhou D., Lai Y., Liu Y., Tao X., Wang Q., Zhao G., Gu H., Liao H., Zhu Y. Estrogen induces endometrial cancer cell proliferation and invasion by regulating the fat mass and obesity-associated gene via PI3K/AKT and MAPK signaling pathways. Cancer Lett. 2012;319:89–97. doi: 10.1016/j.canlet.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 85.Zhu H., Gan X., Jiang X., Diao S., Wu H., Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J., Xie H., Ying Y., Chen H., Yan H., He L., Xu M., Xu X., Liang Z., Liu B. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer. 2020;19:152. doi: 10.1186/s12943-020-01267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cassim S., Pouyssegur J. Tumor microenvironment: a metabolic player that shapes the immune response. Int. J. Mol. Sci. 2019;21:E157. doi: 10.3390/ijms21010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghiringhelli F., Ménard C., Martin F., Zitvogel L. The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol. Rev. 2006;214:229–238. doi: 10.1111/j.1600-065X.2006.00445.x. [DOI] [PubMed] [Google Scholar]
- 90.Sinha P., Clements V.K., Bunt S.K., Albelda S.M., Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol. 2007;179:977–983. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 91.Dudás J., Fullár A., Bitsche M., Schartinger V., Kovalszky I., Sprinzl G.M., Riechelmann H. Tumor-produced, active interleukin-1β regulates gene expression in carcinoma-associated fibroblasts. Exp. Cell Res. 2011;317:2222–2229. doi: 10.1016/j.yexcr.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmieder A., Michel J., Schönhaar K., Goerdt S., Schledzewski K. Differentiation and gene expression profile of tumor-associated macrophages. Semin. Cancer Biol. 2012;22:289–297. doi: 10.1016/j.semcancer.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y., Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J. Mol. Med. (Berl.) 2016;94:509–522. doi: 10.1007/s00109-015-1376-x. [DOI] [PubMed] [Google Scholar]
- 94.Del Prete G., De Carli M., Almerigogna F., Giudizi M.G., Biagiotti R., Romagnani S. Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J. Immunol. 1993;150:353–360. [PubMed] [Google Scholar]
- 95.Costa N.L., Valadares M.C., Souza P.P., Mendonça E.F., Oliveira J.C., Silva T.A., Batista A.C. Tumor-associated macrophages and the profile of inflammatory cytokines in oral squamous cell carcinoma. Oral Oncol. 2013;49:216–223. doi: 10.1016/j.oraloncology.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 96.Jin W., Dong C. IL-17 cytokines in immunity and inflammation. Emerg. Microbes Infect. 2013;2:e60. doi: 10.1038/emi.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whiteside T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016;126:1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stanczak M.A., Siddiqui S.S., Trefny M.P., Thommen D.S., Boligan K.F., von Gunten S., Tzankov A., Tietze L., Lardinois D., Heinzelmann-Schwarz V. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Invest. 2018;128:4912–4923. doi: 10.1172/JCI120612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vesely M.D., Kershaw M.H., Schreiber R.D., Smyth M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 100.Anderson A.C., Joller N., Kuchroo V.K. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andrews L.P., Marciscano A.E., Drake C.G., Vignali D.A. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017;276:80–96. doi: 10.1111/imr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Manieri N.A., Chiang E.Y., Grogan J.L. TIGIT: a key inhibitor of the cancer immunity cycle. Trends Immunol. 2017;38:20–28. doi: 10.1016/j.it.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 103.Wolf Y., Anderson A.C., Kuchroo V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020;20:173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naidoo J., Dykema A., D’Alessio F. An adapted anti-CTLA4 therapeutic aimed at mitigating the toxicities of checkpoint inhibition. J. Clin. Invest. 2019;129:75–77. doi: 10.1172/JCI125800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen Y.T., Shen J.Y., Chen D.P., Wu C.F., Guo R., Zhang P.P., Lv J.W., Li W.F., Wang Z.X., Chen Y.P. Identification of cross-talk between m6A and 5mC regulators associated with onco-immunogenic features and prognosis across 33 cancer types. J. Hematol. Oncol. 2020;13:22. doi: 10.1186/s13045-020-00854-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yi L., Wu G., Guo L., Zou X., Huang P. Comprehensive analysis of the PD-L1 and immune infiltrates of m6A RNA methylation regulators in head and neck squamous cell carcinoma. Mol. Ther. Nucleic Acids. 2020;21:299–314. doi: 10.1016/j.omtn.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He X., Tan L., Ni J., Shen G. Expression pattern of m6A regulators is significantly correlated with malignancy and antitumor immune response of breast cancer. Cancer Gene Ther. 2020 doi: 10.1038/s41417-020-00208-1. Published online August 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu S., Yu Z., Xiao Z., Zhang Y. Gene signatures and prognostic values of m6A genes in nasopharyngeal carcinoma. Front. Oncol. 2020;10:875. doi: 10.3389/fonc.2020.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mo P., Xie S., Cai W., Ruan J., Du Q., Ye J., Mao J. N6-methyladenosine (m6A) RNA methylation signature as a predictor of stomach adenocarcinoma outcomes and its association with immune checkpoint molecules. J. Int. Med. Res. 2020;48 doi: 10.1177/0300060520951405. 300060520951405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y., Gu J., Xu F., Zhu Q., Chen Y., Ge D., Lu C. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief. Bioinform. 2020:bbaa225. doi: 10.1093/bib/bbaa225. [DOI] [PubMed] [Google Scholar]
- 111.Tang R., Zhang Y., Liang C., Xu J., Meng Q., Hua J., Liu J., Zhang B., Yu X., Shi S. The role of m6A-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ. 2020;8:e9602. doi: 10.7717/peerj.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu F., Zhang H., Chen J., Lin L., Chen Y. Immune signature of T follicular helper cells predicts clinical prognostic and therapeutic impact in lung squamous cell carcinoma. Int. Immunopharmacol. 2020;81:105932. doi: 10.1016/j.intimp.2019.105932. [DOI] [PubMed] [Google Scholar]
- 113.Li J., Rao B., Yang J., Liu L., Huang M., Liu X., Cui G., Li C., Han Q., Yang H. Dysregulated m6A-related regulators are associated with tumor metastasis and poor prognosis in osteosarcoma. Front. Oncol. 2020;10:769. doi: 10.3389/fonc.2020.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo Y., Sun Y., Li L., Mao Y. METTL3 may regulate testicular germ cell tumors through EMT and immune pathways. Cell Transplant. 2020;29 doi: 10.1177/0963689720946653. 963689720946653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L., Hui H., Agrawal K., Kang Y., Li N., Tang R., Yuan J., Rana T.M. m6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020;39:e104514. doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li N., Kang Y., Wang L., Huff S., Tang R., Hui H., Agrawal K., Gonzalez G.M., Wang Y., Patel S.P., Rana T.M. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA. 2020;117:20159–20170. doi: 10.1073/pnas.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Su R., Dong L., Li Y., Gao M., Han L., Wunderlich M., Deng X., Li H., Huang Y., Gao L. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Kempen L.C., Ruiter D.J., van Muijen G.N., Coussens L.M. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur. J. Cell Biol. 2003;82:539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- 120.Yasmin R., Siraj S., Hassan A., Khan A.R., Abbasi R., Ahmad N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediators Inflamm. 2015;2015:201703. doi: 10.1155/2015/201703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.West N.R., McCuaig S., Franchini F., Powrie F. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015;15:615–629. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 122.Chiba T., Marusawa H., Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 123.Ding H., Zhang X., Su Y., Jia C., Dai C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell. Mol. Biol. Lett. 2020;25:8. doi: 10.1186/s11658-020-00204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yi Y.C., Chen X.Y., Zhang J., Zhu J.S. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]