Abstract
Carbapenem-resistant Klebsiella pneumoniae (CRKP) remains a major clinical pathogen and public health threat with few therapeutic options. The mobilome, resistome, methylome, virulome and phylogeography of CRKP in South Africa and globally were characterized. CRKP collected in 2018 were subjected to antimicrobial susceptibility testing, screening by multiplex PCR, genotyping by repetitive element palindromic (REP)-PCR, plasmid size, number, incompatibility and mobility analyses, and PacBio’s SMRT sequencing (n=6). There were 56 multidrug-resistant CRKP, having bla OXA-48-like and bla NDM-1/7 carbapenemases on self-transmissible IncF, A/C, IncL/M and IncX3 plasmids endowed with prophages, traT, resistance islands, and type I and II restriction modification systems (RMS). Plasmids and clades detected in this study were respectively related to globally established/disseminated plasmids clades/clones, evincing transboundary horizontal and vertical dissemination. Reduced susceptibility to colistin occurred in 23 strains. Common clones included ST307, ST607, ST17, ST39 and ST3559. IncFIIk virulent plasmid replicon was present in 56 strains. Whole-genome sequencing of six strains revealed least 41 virulence genes, extensive ompK36 mutations, and four different K- and O-loci types: KL2, KL25, KL27, KL102, O1, O2, O4 and O5. Types I, II and III RMS, conferring m6A (GATC, GATGNNNNNNTTG, CAANNNNNNCATC motifs) and m4C (CCWGG) modifications on chromosomes and plasmids, were found. The nature of plasmid-mediated, clonal and multi-clonal dissemination of blaOXA-48-like and blaNDM-1 mirrors epidemiological trends observed for closely related plasmids and sequence types internationally. Worryingly, the presence of both bla OXA-48 and bla NDM-1 in the same isolates was observed. Plasmid-mediated transmission of RMS, virulome and prophages influence bacterial evolution, epidemiology, pathogenicity and resistance, threatening infection treatment. The influence of RMS on antimicrobial and bacteriophage therapy needs urgent investigation.
Keywords: bacteriophage, carbapenemase, DNA methylation, evolutionary epidemiology, resistance plasmids, restriction modification systems
Data Summary
All data used in this study are found in the supporting Data (S1–S3, available in the online version of this article) and Tables (S1–S6). The genomes of the isolates used in this study have been deposited in DDBJ/ENA/GenBank under BioProject number PRJNA565241 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA565241) and accession numbers VXIW00000000 (KP33), VXIX00000000 (KP32), VXIY00000000 (KP29), VXIZ00000000 (KP15), VXJA00000000 (KP10) and VXJB00000000 (KP8); the versions used in this study are versions VXIW01000000, VXIX01000000, VXIY01000000, VXIZ01000000, VXJA01000000 and VXJB01000000. The accession numbers of complete circular plasmids are VXIX01000005 (pKP32.5_OXA-48), VXIY01000008 (pKP29.9_CTXM-15) and VXIY01000013 (pK29.13_MBELLE), whilst those of partial plasmids are VXJB01000006 (pKP8.6_CTX-M-15), VXJB01000012 (pKP8.12_OXA181), VXJA01000008 (pKP10.8_NDM-1), VXJA01000004 (pKP10.4_OSEI), VXIZ01000004 (pKP15.4_KATLEGO), VXIZ01000012 (pKP15.12_OXA-181), VXIY01000011 (pKP29.11_NDM-7), VXIX01000003 (pKP32.3_CTXM-15), VXIW01000008 (pKP33.8_NDM-1) and VXIW01000004 (pKP10.4_OSEI). All kinetic data (methylation) files have been deposited in GEO under accession number GSE138949 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138949) [GSM4125137 (Kp8); GSM4125138 (Kp10); GSM4125139 (Kp15); GSM4125140 (Kp29); GSM4125141 (Kp32); GSM4588290 (Kp33)].
Impact Statement.
Klebsiella pneumoniae is a major pathogen implicated in numerous nosocomial infections. Worryingly, we show here that K. pneumoniae isolated globally are endowed with rich resistomes and mobilomes that make them almost pandrug-resistant. The isolates in this study contained diverse virulomes and prophages, on both chromosomes and plasmids, with close evolutionary kith or kin to other plasmids identified worldwide. The presence of both bla OXA-48 and bla NDM-1 in the same isolates were observed. Plasmid replicon-specific resistomes, carbapenemases and evolutionary epidemiology are described, showing the resistance gene spectrum and relative prevalence of various plasmid replicons globally. There was a rich diversity of restriction modification systems that could regulate virulence, transcription and plasmid mobility in bacteria, facilitating the epidemiology, resistance, pathogenicity and genomic evolution of the strains, and threatening antimicrobial and bacteriophage therapy.
Introduction
Klebsiella pneumoniae is an encapsulated, non-motile, Gram-negative bacterium first isolated from the lung of a deceased patient with pneumonia in 1882 [1]. These bacteria are known to colonize the human gastrointestinal (GI) tract and oropharynx mucosal surfaces [2]. In addition to K. pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii and MRSA (methicillin-resistant Staphylococcus aureus ) are important hospital pathogens; K. pneumoniae accounts for 3–8 % of several reported infections [3, 4]. These infections are specifically a problem in the elderly, immunocompromised patients and neonates; less frequently, K. pneumoniae infections such as sepsis and pneumonia are community-acquired [5].
Antimicrobial resistance (AMR) in bacteria such as K. pneumoniae has become a major public health concern worldwide, fuelled by misuse and overuse of antibiotics, which increases the evolution of AMR genes (ARGs) and antimicrobial-resistant bacteria [6]. Resistance may be intrinsic, i.e. acquired through mutations, and/or transferred horizontally from one bacterium to another through mobile genetic elements (MGEs) [7]. Among the various AMR mechanisms, acquired resistance through MGEs is most reported [8]. Acquisition of antimicrobial-inactivating enzymes and efflux pump systems are important in the development of multi-drug resistant (MDR) in Enterobacterales , including K. pneumoniae [9, 10], with the resistance-nodulation-division (RND) family of efflux pumps being responsible for ejecting charged and amphiphilic antimicrobials such as aminoglycosides, β-lactams and fluoroquinolones [10, 11]. The use of β-lactams over the years has resulted in widespread escalation of β-lactamases, including carbapenemase-producing K. pneumoniae [12, 13], resulting in increased treatment failure, morbidity and mortality [14]. High-level resistance in K. pneumoniae isolates is typically mediated by the production of carbapenemases and/or porin loss with the production of either extended-spectrum β-lactamase (ESBL) or an AmpC [15]. CTX-M is a plasmid-mediated ESBL that is mostly reported worldwide in Enterobacteriaceae including in K. pneumoniae [16, 17]. The loss of OmpK35 and OmpK36 porins in ESBL-producing K. pneumoniae has been associated with high-level resistance and has been well documented [15, 18].
In Africa and South Africa, K. pneumoniae clones that are commonly reported include ST101, ST152, ST39, ST1414, ST14, ST15 and ST307, with the global ST258 clone being relatively rare [13, 19, 20]. Of the several virulence factors found in K. pneumoniae, K1 and K2 capsular types have been found to proffer hypervirulence characteristics to this species. Hypervirulent K. pneumoniae are highly pathogenic, associated with rmpA, iro, entB, ybtS, mrkD and iucA-D genes. Worryingly, these hypervirulence determinants have been concurrently found on virulence plasmids such as pK2044 and pLVPK, which can also harbour carbapenemases or coexist with carbapenemases in ST11, ST23, ST65 and ST86 clones [21–24]. To date, these hypervirulent strains have been mainly limited to China and South-east Asia, with no reports in Africa.
Carbapenemases are categorized into three classes, class A (e.g. KPC, SME, IMI and GES), class B (e.g. NDM, VIM and IMP) and class D (OXA-48-like) [25, 26]. Carbapenemases are capable of slightly and/or completely hydrolysing β-lactams, including ‘last resort’ carbapenems [27]. Class B carbapenemases, particularly bla NDM genes, have been reported to be more potent than the other groups and cannot be inhibited by commercially available β-lactamase inhibitors such as clavulanic acid, tazobactam or sulbactam [28, 29]. However, metal chelators such as EDTA and mercaptopropionic acid are able to inhibit activity of class B carbapenemases [28].
MGEs such as plasmids, transposons, prophages and integrons play a major role in the acquisition and dissemination of ARGs in carbapenem-resistant strains [13, 20, 30, 31]. Among these are large conjugative plasmids that have been associated with horizontal gene transfer (HGT) of carbapenemases between and within Gram-negative bacteria [8]. These plasmids have been reported in K. pneumoniae strains and are associated with multiple replicon groups such as IncF, A/C, L/M, N and X [30]. IncF replicon plasmids are the most predominant and are mainly reported to carry the bla KPC and bla NDM genes in the USA, Canada, Greece, South Africa and Taiwan [13, 20, 30, 32]. The L/M plasmids in K. pneumoniae are more frequently reported in the Czech Republic and Ireland, carrying the bla OXA-48 gene and, more rarely, in Oman carrying the bla NDM gene [33–35]. IncX plasmids are the major vehicles for the bla NDM gene in China and India while IncN plasmids, which are reported rarely, are associated with bla NDM and bla KPC genes in K. pneumoniae strains [20, 36–38].
Besides resistance, plasmids are being increasingly associated with the transmission of virulence factors, resulting in transmission of important pathogenic traits [39–41]. Recently, it is increasingly being realized that restriction modification systems (RMS), consisting of restriction endonucleases (REs) and DNA methylases (MTases), regulate virulence, plasmid mobilization, bacterial immunity, DNA repair, transcription, replication, regulation and resistance in bacteria [42–46]. These suggest the important role of the methylome in bacterial virulence, mobilome and resistome, which ultimately affect the epidemiology of infectious diseases. This study thus seeks to characterize these factors in this important pathogen using isolates from Pretoria (South Africa) and genomes from countries around the world.
Results
Clinical demographics and genome characteristics
The K. pneumoniae isolates (n=56) were isolated from aspirates (n=4), blood cultures (n=17), catheter tips (n=7), swabs (n=11), tissue (n=3) and urine (n=14) (Tables 1 and S1), and were submitted to the referral laboratory from six hospitals and centres including Kalafong hospital (n=10), Mamelodi hospital (n=1), Olievenhoutbosch clinic (n=1), Steve Biko academic hospital (n=36), Tembisa hospital (n=5) and Tshwane rehabilitation centre (n=3). The study population consisted of males (58.9 %) more than females (39.3 %) and results were not available for one participant (Table S1).
Antibiogram-resistome associations
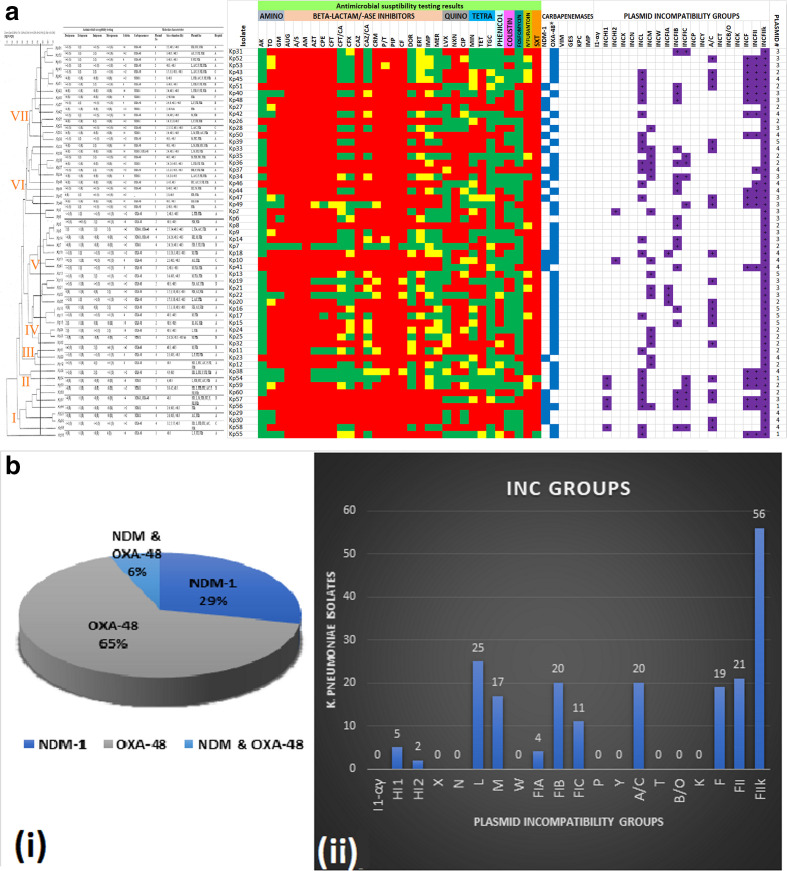
Of the 60 K . pneumoniae with reduced susceptibility to carbapenems as assessed by VITEK, carbapenem resistance could only be confirmed in 56 isolates by the MicroScan system (Table S1). The isolates were MDR, with a substantial number being extensively drug-resistant (KP51, KP27, KP 42 etc.); a few showed pandrug resistance phenomes (e.g. KP56) (Fig. 1a, b(i)). Almost all isolates showed reduced susceptibility to ertapenem (98.2 %), followed by imipenem (66.1 %), doripenem (50 %) and meropenem (48.2 %). Reduced susceptibility to colistin was also observed in 23 (41.1 %) isolates. Among all tested antibiotics, the isolates were most susceptible to amikacin (82.1%), fosfomycin (82.1 %), tigecycline (76.8 %) and levofloxacin (60.7 %) (Fig. 1). The most prevalent carbapenemase detected by PCR was bla OXA-48 (65 %), followed by bla NDM-1 (29 %). No bla GES, bla KPC, bla IMP and/or bla VIM were detected (Fig. 1b(ii)). bla OXA-48-containing isolates were mostly non-resistant to the carbapenems except to ertapenem in selected cases (e.g. KP44, KP40, KP39 and KP2) and to the other carbapenems in a few cases (e.g. KP36, KP49, KP8 and KP55). By contrast, all isolates harbouring bla NDM-1 were resistant to carbapenems. Furthermore, isolates containing both bla OXA-48 and bla NDM-1 genes (e.g. KP10 and KP56) were also resistant to all carbapenems except KP18, which was only resistant to ertapenem and intermediately resistant to the rest (Table S1, Fig. 1).
Fig. 1.
Antibiotic susceptibility patterns, carbapenemase genes and plasmid characteristics of 56 clinical K. pneumoniae isolates arranged according to their clustering patterns on a REP-PCR dendrogram. The isolates clustered into seven main clusters/clades, based on their gel patterns. Antibiotic resistance is shown as red, intermediate resistance is shown as yellow and susceptibility is shown as green. Carbapenemase genes are shown as blue and plasmid replicons are shown as violet/mauve. The number of plasmids is shown in the last column (a). Most of the isolates harboured OXA-48 (65 %) compared to NDM-1 (29 %) whilst a minority had both genes (6 %) (i). IncF plasmid replicons were the commonest plasmid types, being found in all the isolates; IncL, A/C and IncM were also common types (ii) (b).
The resistome of the sequenced isolates were similar and mostly concordant with their respective antibiograms except for fosfomycin to which all isolates were susceptible even though they all harboured the fosA gene. The six isolates carried bla OXA-181 (n=2), bla OXA-48 (n=1), bla NDM-1 (n=2) and bla NDM-7 (n=1) alongside other resistance determinants mediating resistance to aminoglycosides (except amikacin) [aac(3)-lla, aac(6′)-lb-cr, aadA16, aph(3′)-lb, aph(6)-ld, rmtC], quinolones [aac(6′)-lb-cr, oqxA, oqxB, qnrB1, qnrS1], β-lactams (bla OXA-1, bla CTX-M-15, bla SHV, bla TEM-1B), tetracycline (tetA), sulphonamides (sul1, sul2), trimethoprim (dfrA14/27), phenicol (catB3/catA2) and fosfomycin (fosA/A7). KP29 had no tet(A) but was resistant to the tetracyclines; bla SCO-1 was found in only this isolate. The 16S rRNA methyltransferase, rmtC, was found in only KP10 and KP33; however, KP33 was susceptible to amikacin while KP10 was resistant. Chromosomal mutations in parC (S104I) and gyrA (S83I), conferring high-level fluoroquinolone minimum inhibitory concentrations (MICs), were only seen in KP8. Nevertheless, the other isolates, which had no mutations, were also resistant to the fluoroquinolones (KP29 was susceptible to ciprofloxacin and levofloxacin). In addition, extensive mutations and stop codons were identified in the ompK36 genes of the six isolates (Table S1, Data S2).
Mobile colistin resistance determinants (mcr-1 to -10) were not identified, although colistin resistance was recorded in three of the sequenced carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates. In particular, KP10 was susceptible to colistin, had no ccrB gene and had an M66I mutation in pmrA whilst KP15, which was also susceptible to colistin, also had no mutations in the analysed genes. Although KP29 had mutations in ccrB (D189E), kpnE (K112Q) and pmrA (E57G), it was also susceptible. OqxAB, fosA and bla SHV were all found on the chromosomes (Data S2).
REP-PCR phylogenetics
The Repetitive extragenic palindromic (REP)-PCR dendrogram showed seven main clusters, with little or no similarities in the antibiogram, resistome or mobilome of isolates in the same clusters (Fig. 1), i.e. they were not clonally specific. Notwithstanding, isolates from the same hospital/ward had much similar antibiograms, resistomes and mobilomes (Fig. 1, Table S1). Based on the REP-PCR dendrogram and clustering, six representative isolates were selected from three different hospitals and sequenced. Four isolates were from the same hospital but different wards and collection sites including: ward 4, urine (Kp10); vascular surgery ward 4, catheter tip (Kp15); neurology ward, swab (Kp29); and high-care multidiscipline ward, urine (Kp33) (Table S1). Five different sequence types (STs) were identified among the isolates including ST39, ST307, ST607, ST17 and ST3559. Isolates Kp10 and Kp33, carrying bla NDM-1, both belonged to ST39, although they clustered differently on the dendrogram.
Mobilome
Plasmid characterization by gel electrophoresis revealed that most isolates (n=17) carried four plasmids, followed by 16 isolates with two plasmids, 15 isolates with three plasmids, five isolates with five plasmids, and three isolates with only one plasmid. Plasmids sizes ranged from 1.4 to >48.5 kb (Fig. 1, Table S1). Eleven plasmid replicon groups were identified in all the isolates (n=56), which were all positive for IncFIIk (virulent plasmid). The majority of the isolates were positive for IncF (FII, FIB, FIC, FIB), IncL, A/C and IncM plasmids, while only a few isolates were positive for IncHI1 and IncHI2 (Fig. 1b, Table S1). Multi-replicons were reported in 75 % (n=42) of the tested isolates. Two isolates showed the highest multi-replicon combination: one bla OXA-48-producer and one bla NDM-1-producer had six and seven replicon groups, respectively (Fig. 2).
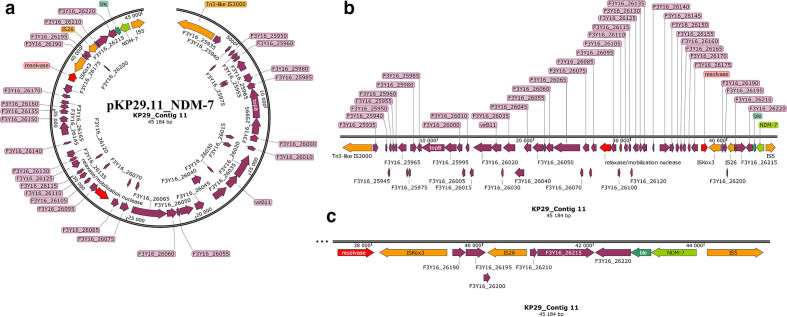
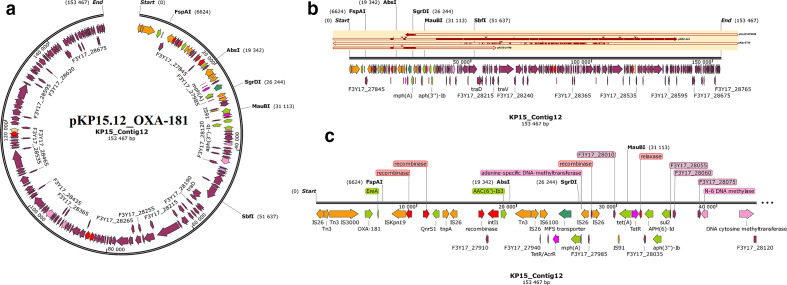
Fig. 2.
Graphical annotation and comparative alignment of pKP29.11_NDM-7. The resistance genes (light green-coloured arrows), methyltransferases/restriction endonucleases (rose-coloured arrows), transposons (orange-coloured arrows), insertion sequences (orange-coloured arrows), integrons (red-coloured arrows), resolvases (red-coloured arrows) and recombinases/integrases (red-coloured arrows) on the plasmid are shown with their orientation (direction of arrow), synteny and immediate environment. Other genes with unknown functions are hidden to make the image less cluttered. A circular version of the plasmid is shown in (a). A linear version of the plasmid and its alignment with other similar plasmids (linear arrows with yellow highlighted background) are shown in (b); regions of alignment are shown as red-filled portions whilst non-aligned areas are shown as empty arrows. An enlarged section of the plasmid focusing on the resistance genes (genomic resistance island) is shown in (c). This plasmid (VXIY01000011) contains NDM-7.
Conjugation experiments were successful on 20 out of the 26 meropenem-resistant K. pneumoniae isolates; 20 donor strains transferred their plasmids to Escherichia coli strain J53-Ar. Among the 20 transferred plasmids, 16 were positive for the bla NDM-1 gene, followed by three bla OXA-48 and one isolate with both bla NDM-1 and bla OXA-48 genes (Fig. 1b).
Sequencing analyses of six isolates showed that five had two major plasmids whilst KP29 had three, which were different from the numbers obtained from the gel electrophoresis analyses, except for KP8 (Figs. 1–6, Table S1); fragments (contigs) of these plasmids were found but had no replicon genes (Table S1). The sizes obtained from the sequencing analyses were also different from that obtained from the gel electrophoresis analyses (Figs. 2–6, S1–S6).
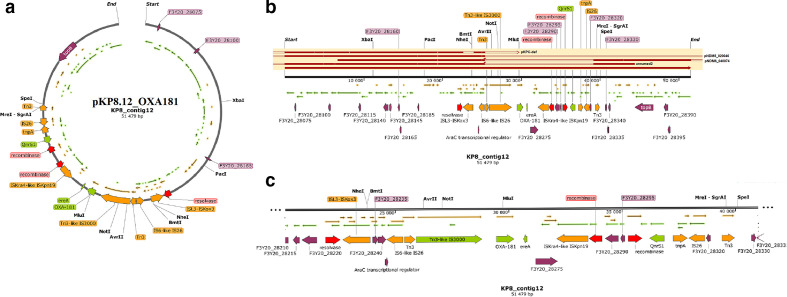
Fig. 3.
Graphical annotation and comparative alignment of pKP8.12_OXA181. The resistance genes (light green-coloured arrows), transposons (orange-coloured arrows), insertion sequences (orange-coloured arrows), integrons (red-coloured arrows), resolvases (red-coloured arrows) and recombinases/integrases (red-coloured arrows) on the plasmid are shown with their orientation (direction of arrow), synteny and immediate environment. Other genes with unknown functions are hidden to make the image less cluttered. A circular version of the plasmid is shown in (a). A linear version of the plasmid and its alignment with other similar plasmids (linear arrows with yellow highlighted background) are shown in (b); regions of alignment are shown as red-filled portions whilst non-aligned areas are shown as empty arrows. An enlarged section of the plasmid focusing on the resistance genes (genomic resistance island) is shown in (c). This plasmid (VXJB01000012) contains OXA-181.
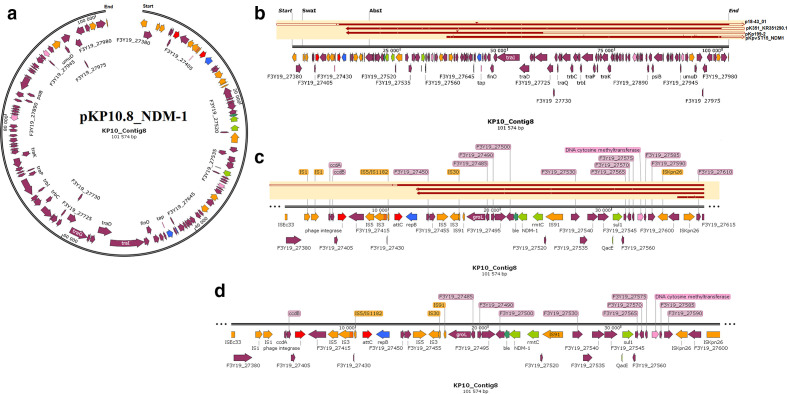
Fig. 4.
Graphical annotation and comparative alignment of pKP10.8_NDM-1. The resistance genes (light green-coloured arrows), replicase genes (blue-coloured arrows), methyltransferases/restriction endonucleases (rose-coloured arrows), transposons (orange-coloured arrows), insertion sequences (orange-coloured arrows), integrons (red-coloured arrows), resolvases (red-coloured arrows) and recombinases/integrases (red-coloured arrows) on the plasmid are shown with their orientation (direction of arrow), synteny and immediate environment. Other genes with unknown functions are hidden to make the image less cluttered. A circular version of the plasmid is shown in (a). A linear version of the plasmid and its alignment with other similar plasmids (linear arrows with yellow highlighted background) are shown in (b); regions of alignment are shown as red-filled portions whilst non-aligned areas are shown as empty arrows. An enlarged section of the plasmid focusing on the resistance genes (genomic resistance island) are shown in (c) and (d). This plasmid (VXJA01000008) contains NDM-1.
Fig. 5.
Graphical annotation and comparative alignment of pKP15.12_OXA-181. The resistance genes (light green-coloured arrows), transporter genes (deep green-coloured arrows), methyltransferases/restriction endonucleases (rose-coloured arrows), transposons (orange-coloured arrows), insertion sequences (orange-coloured arrows), integrons (red-coloured arrows), resolvases (red-coloured arrows) and recombinases/integrases (red-coloured arrows) on the plasmid are shown with their orientation (direction of arrow), synteny and immediate environment. Other genes with unknown functions are hidden to make the image less cluttered. A circular version of the plasmid is shown in (a). A linear version of the plasmid and its alignment with other similar plasmids (linear arrows with yellow highlighted background) are shown in (b); regions of alignment are shown as red-filled portions whilst non-aligned areas are shown as empty arrows. An enlarged section of the plasmid focusing on the resistance genes (genomic resistance island) is shown in (c). This plasmid (VXIZ01000012) contains OXA-181.
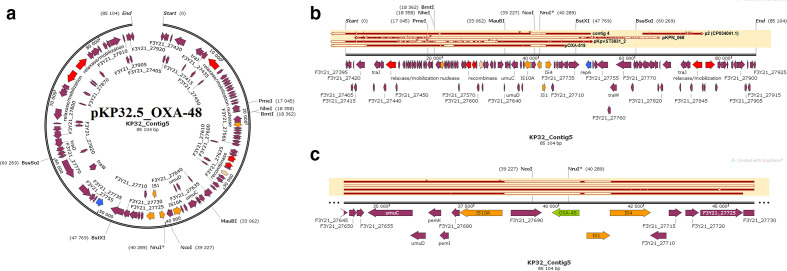
Fig. 6.
Graphical annotation and comparative alignment of pKP32.5_OXA-48. The resistance genes (light green-coloured arrows), replicase genes (blue-coloured arrows), methyltransferases/restriction endonucleases (rose-coloured arrows), transposons (orange-coloured arrows), insertion sequences (orange-coloured arrows), integrons (red-coloured arrows), resolvases (red-coloured arrows) and recombinases/integrases (red-coloured arrows) on the plasmid are shown with their orientation (direction of arrow), synteny and immediate environment. Other genes with unknown functions are hidden to make the image less cluttered. A circular version of the plasmid is shown in (a). A linear version of the plasmid and its alignment with other similar plasmids (linear arrows with yellow highlighted background) are shown in (b), regions of alignment are shown as red-filled portions whilst non-aligned areas are shown as empty arrows. An enlarged section of the plasmid focusing on the resistance genes (genomic resistance island) is shown in (c). This plasmid (VXIX01000005) contains OXA-48.
The three plasmids, pKP32.5_OXA-48, pKP29.9_CTXM-15 and pK29.13_MBELLE, had overlapping ends, making them completely circularized whilst the remaining were partial with no overlapping ends (Figs. 2–6). Bla NDM-1 was present on two plasmids alongside ble, rmtC, sul1 and cytosine MTase within IS1/5/3 and ISKpn26 ISs (insertion sequences) (Fig. 4, S6.1). Bla NDM-7 was found alongside ble within ISs (Fig. 2). Bla OXA-181, together with ereA and QnrS1, was found within composite Tn3-like transposons and ISs; bla OXA-181 was also found with several resistance genes and adenine and cytosine MTases bracketed by Tn3 composite transposons, ISs and IntI1 class 1 integron (Figs 3 and 5). Bla OXA-48 was found on pKP32.5_OXA-48 alone within ISs (Fig. 6) .
Bla CTX-M-15 and bla TEM-1B were found in close synteny on the same plasmids in all the isolates; the bla TEM-1B genetic environment only differed slightly in pKP29.8_CTXM-15 (Figs S1–S6). Adenine and cytosine MTases were also found on the same plasmids as bla CTX-M-15 and bla TEM-1B, within ISs and transposons. Bla OXA-1 and bla SCO-1 (found within ISs, MTase and a recombinase on pKP29.8_CTXM-15) were only found on four plasmids and one plasmid respectively; notably, bla OXA-1 and its rich genetic environment of resistance genes and ISs were on the same plasmids as bla CTX-M-15 and bla TEM-1B, albeit quite distant from the latter, forming a resistance island. Thus, bla CTX-M-15 and bla TEM-1B formed a genomic resistance island with their immediate genetic environment of rich ISs and transposons just as bla OXA-1. Furthermore, other antibiotic and mercury (merABCDETPR) resistance genes, tolA efflux pump, NH2 restriction endonucleases and MTases were also found on ISs, transposons and, mainly, on IntI1 class 1 integrons (Figs S1–S6).
Complete and partial prophage DNAs were identified on both chromosomes and plasmids, including the Klebsi_phiKO2, Cronob_ENT47670, Edward_GF_2, Pectob_ZF40, Phage_Gifsy, and different variants of Salmon (siz), Entero (two) and Escher (three) (Data S3, Figs S7–S11). Complete and incomplete prophages were found in the plasmids of all the isolates. KP15 alone had four complete prophages on two plasmids (pKP15.4_KATLEGO and pKP15.12_OXA-181) whilst the single complete prophages were found on single plasmids in the other isolates. Furthermore, complete and incomplete prophages were also found on chromosomes in these isolates (Figs S7–S11). The same prophages were also found in different isolates (Data S3).
Plasmid evolutionary epidemiology
We undertook a phylogenetic analysis using the plasmids obtained in this study and other closely related plasmids obtained from GenBank. Some of these plasmids, which belonged to different replicon groups and harboured different ARGs, were evolutionary closer to each other, albeit they were from different isolates (Fig. 7). For instance, pKP32.3_CTXM-15 and pKP8.6_CTX-M-15 were of different sizes and from different isolates, but had similar ARGs and replicon types [IncFII(K) and IncFIB(K)] clustered together on the same branch. The same observation was made for pKP10.8_NDM-1 and pKP33.8_NDM-1 as well as for pKP10.4_OSEI and pKP33.4_OSEI, which harboured the same ARGs and replicon types in two different isolates of the same clone (ST39). By contrast, IncX3 bla NDM-7-bearing pKP29.11_NDM-7 plasmid clustered closely with pKP29.9_CTXM-15, an IncFIB(K) and IncFII(K) plasmid, albeit all were from the same isolate. Nevertheless, pKP15.4_KATLEGO and pK29.13_MBELLE, which had IncR and IncFIA(HI1) replicons, clustered separately. IncX3-ColKP3 pKP8.12_OXA181 plasmid was distant from all the other plasmids (Fig. 7). Mauve alignment of these plasmids with other closely related plasmids from GenBank showed similarities in sequence identity and synteny as well as sequence rearrangements (Figs S1–S6).
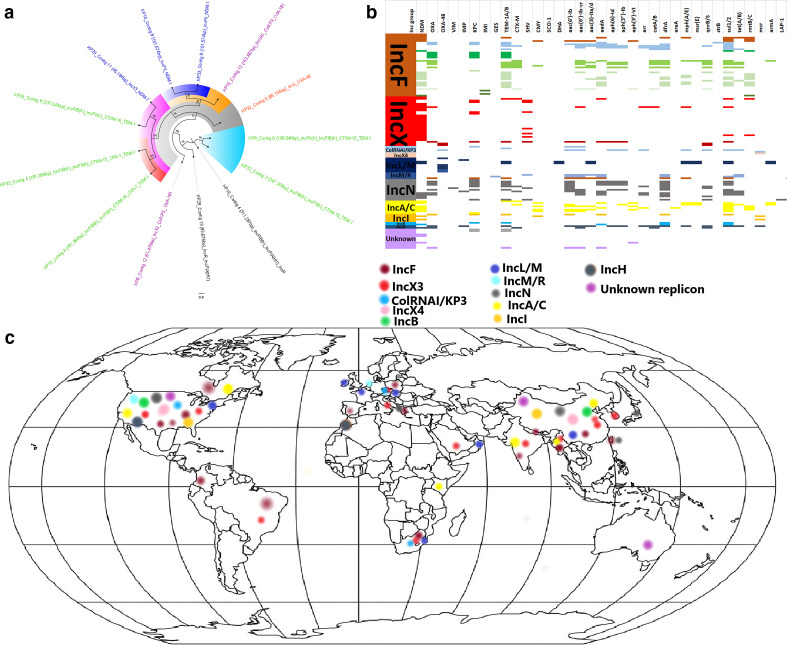
Fig. 7.
Global phylogenetic, resistome and phylogeographical characteristics of carbapenemase-hosting plasmid incompatibility types. The phylogenetic relationships between the various plasmids obtained in this study are shown in (a), with those bearing CTX-M and/or TEM ESBLs labelled with green text whilst those bearing OXA-181, OXA-48 and NDM-1/7 labelled with purple, blue and red text respectively; branches of the same clade have the same highlights. The resistomes of carbapenemase-bearing plasmids isolated worldwide are shown in (b) under various plasmid replicon types; the various plasmid types harbouring carbapenemases and associated resistance genes are also shown. The geographical distribution of the plasmid incompatibility groups is shown in (c), and it is evident that most of these plasmids were isolated from North America, Brazil, Columbia, Europe, South Africa, Kenya, the Middle East, South-East Asia and Australia.
Nucleotide blast analyses and comparative genomics of the carbapenemase-bearing plasmid genomes from this study identified highly similar plasmids with the same replicon types, ARGs and host bacterial species from different countries globally; in a few cases, the replicon types and ARGs differed slightly, albeit the country and bacterial species differed substantially (Fig. 7, S13 and Table S2). In most cases, the plasmids from this study aligned most closely with more than 90 % nucleotide sequence identity and coverage, with plasmids deposited in GenBank, which were from different countries (Table S2, Fig. S13). For instance, pKP8.12_OXA181, a multi-replicon plasmid, was closely aligned with 201 other plasmids of the same or similar replicon group (IncX/ColKP3) from different countries and bacterial hosts but having bla OXA-181 or bla NDM. Similarly, pKP10.8_NDM-1 was closely aligned to 31 other plasmids worldwide of the same replicon with bla NDM. Similar trends were also observed for pKP15.12_OXA-181 (aligned to 33 other IncC/U and ColKP3 plasmids worldwide of the same replicons although they contained few carbapenemases), pKP29.11_NDM-7 (aligned to 231 other IncX3 plasmids worldwide that harboured bla OXA-181, bla KPC or bla NDM) and pKP32.5_OXA-48 (aligned with 59 other IncL plasmids worldwide that also harboured bla OXA-48). Notably, IncX3 plasmids harboured NDM whilst IncX3-ColKP3 plasmids harboured OXA-181 (Fig. S13).
Resistome analyses of the various plasmids showed that IncF, IncN, A/C, ColRNAI/KP3 and IncH had more diverse ARG repertoires compared to IncX, IncL, IncR and IncI (Fig. 7). bla NDM, bla KPC, bla TEM aac(6′)-Ib, aac(3)-II, aadA, dfrA, sul, qnrB/S and rmtB/C were commonly found on all the replicon types except IncX; however, bla NDM, bla KPC and bla SHV were common on IncX plasmids. Indeed, bla NDM was more concentrated on IncX and A/C plasmids than other plasmid types. A phylogenetic analysis of these plasmids could not be undertaken due to their inability to align substantially. Notably, most of these plasmids have been reported from the USA, Europe and South-East Asia, with a few being reported from South America, Africa and Australia. There were more plasmid replicon data from South Africa and Brazil in Africa and South America respectively (Fig. 7, S13).
Virulome
A total of 51 virulence genes were found in the chromosomes of the six genomes, with ccI and traT being found on plasmids. KP8 and KP32 had the lowest set of virulence genes (n=40) whilst the other isolates had all 51 genes. Hypervirulent genes were absent in all the genomes (Table S3, Fig. S14.1-2). The capsule polysaccharide-based typing or the K-loci results showed four different serotypes among the sequenced isolates: KL2 (KP10 and KP33), KL25 (KP15 and KP29), KL27 (KP32) and KL102 (KP8). In addition, the O-loci results also showed four O serotypes: O1v1 (KP10, KP33 and KP15), O2v2 (KP8), O4 (KP32) and O5 (KP29). As shown in Fig. S14.3–8, the K- and O-serotyping was not only clone-specific as different clones shared the same K and O serotypes.
Methylome
Type I, II and III MTases were found in the sequenced isolates (n=6), with type II MTases being the most abundant followed by type I MTases; a single type III MTase (M.Kpn214ORFGP or M.Kpn30104ORFBP) was found chromosomally on all isolates with no known motif. In addition, no motif was identified for the type I RMS in all the isolates and, except for KP15 and KP29, all the type I RMS were found on chromosomes. A complete RMS consisting of REs, MTases and a specificity subunit, representing the hsdRMS operon, were only found on either chromosomes or plasmids (KP15 and KP29) of all but KP32 and KP33; the position of these RMS components on the chromosomes or plasmids shows that they were in very close or overlapping synteny. KP32 had only type I REs with no MTases whilst KP33 had no specificity subunit. Except for KP10 and KP33, all the other isolates had highly unique type I RMS, with R2.KpnLAUORFGP (found on only chromosomes) being the sole common RE in all isolates. Furthermore, the type I REs on the chromosomes of KP15 and KP29 were different from those on the plasmids. Notably, all the isolates had only single type I MTases on either plasmids or chromosomes except KP8, which had two. There were mutations observed in the type I REs (Table S4).
Type II RMS adenine (Dam) and cytosine (Dcm) MTases, which respectively bind to and methylate GATC (all isolates), GATGNNNNNCTG/CAANNNNNNCATC (KP29 only) and CCWGG or CCNGG (all isolates) were identified. Nevertheless, type II MTases with no known motifs were also identified. Notably, all Dams (with GATC motifs) were only found on chromosomes in all isolates whilst most Dcms (with CCWGG or CCNGG motifs) were mainly found on plasmids with a few exceptions: KP8 (M.Kpn10PVDcmP), KP10 and KP33 (M.Kpn0718ORF12365P and M.Sfl2ORFAP), KP15 (M.Kpn3210ORFFP), and KP29 (M.KpnLAUORFBP and M.Kpn3210ORFFP) and KP32 (M.Kpn3210ORFFP, M.KpnNIH30Dcm and M.Sfl2ORFAP) had few chromosomal Dcms. In addition, all the type II REs were on plasmids and were not more than two per isolate compared to several unique MTases on either plasmids or chromosomes. The absence of REs on the chromosomes makes many of these Dcms orphans with no known motifs (Table S4). An interesting observation was the multiple copies of M.Sfl2ORFAP on both plasmids and chromosomes in single strains as well as its common presence in all the strains. No CCWGG motif was found by MotifMaker and GATGNNNNNCTG/CAANNNNNNCATC was only identified by MotifMaker but absent from REBASE. Notably, the type II RE and MTases on either plasmids or chromosomes shared the same CCWGG motif (Table S4).
All isolates, including both KP15 and KP29, had mostly m6A modifications (methylated adenines), i.e. resulted in N6-methyladenine (6mA); more than 92 % of GATC and 84 % of GATGNNNNNCTG motifs were found in KP15 and KP29, respectively. Moreover, m4C modification types (methylated cytosines), i.e. that resulted in N4-methylcytosine (4mC), were also seen in all the isolates, albeit fewer than 6mA modifications (Table S4). Analyses of all the K. pneumoniae genomes (from Africa and globally) used in this study showed a higher prevalence of Dams than Dcms in their genomes (data not shown).
Global phylogenomics, phylogeography and resistome
The genomes of the six isolates ranged from 5.6 to 5.9 Mb, with five to 13 contigs, a GC content of 57% and coding sequences ranging from 5461 to 5629. N50 values varied widely between the isolates although the L50 was only between 1 and 2 (Tables S1 and S5).
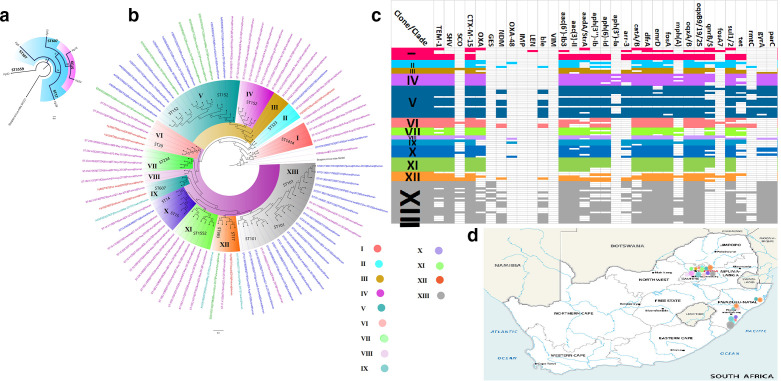
The six isolates belonged to five different clones that clustered differently, with KP10 and KP33 falling within ST39 (Fig. 8a). The isolates also clustered with other K. pneumoniae isolates from Durban, Ozwatini, Pretoria and Pietermaritzburg (clades II, VI, VIII, IX and XII) out of 82 isolates and 13 clades. The same clones/clades were seen circulating within and between Pretoria (Gauteng Province), Ozwatini, Pietermaritzburg and Durban (KwaZulu-Natal Province). K. pneumoniae ST101 is the most prevalent clone in South Africa, followed by ST152; ST101 was most prevalent in Durban, followed by Pretoria. Notably, all the K. pneumoniae strains from South Africa had multiple ARGs, with ST101 and ST152 having the most abundant and diverse repertoire of ARGs, including NDM, GES-5 and CTX-M-15 (Fig. 8c, Tables S5 and S6).
Fig. 8.
Phylogeography and resistome dynamics of K. pneumoniae isolates from South Africa. Each strain is expressed with species name, strain, sequence type, date and country of isolation, and host. The phylogenetic relationship between the six isolates are shown in (a) whilst the phylogenetic relationship of the isolates with other isolates from South Africa is shown in (b). Isolates from this study are labelled red, those from Pretoria are labelled in mauve/purple text, those from Durban are labelled in blue text, those from Pietermaritzburg are labelled in green text, and those from Ozwatini are labelled in turquoise; members of the same clade (labelled I to XIII) are highlighted with the same colour on the branches. The resistomes of the isolates are shown in (c) under the various clades, with members of the same clade having the same colour as the highlights in the phylogenetic tree in (b); blanks refer to absence of resistance genes and filled sections refer to presence of resistance genes. The phylogeography of the various clades is shown in (d); the isolates were mostly from Pretoria and Durban, with some being from Pietermaritzburg and Ozwatini. The evolutionary trajectory of the strains, as shown on the tree (Fig. 8b), shows that some of the clades emerged from other clades or from a common ancestor: for instance, clades IV and V share the same ancestor as clade III, albeit clades IV and V were mainly of ST152. Evidently, ST14 and ST15 as well as ST983 and ST17 are of very close evolutionary distance (Fig. 8b).
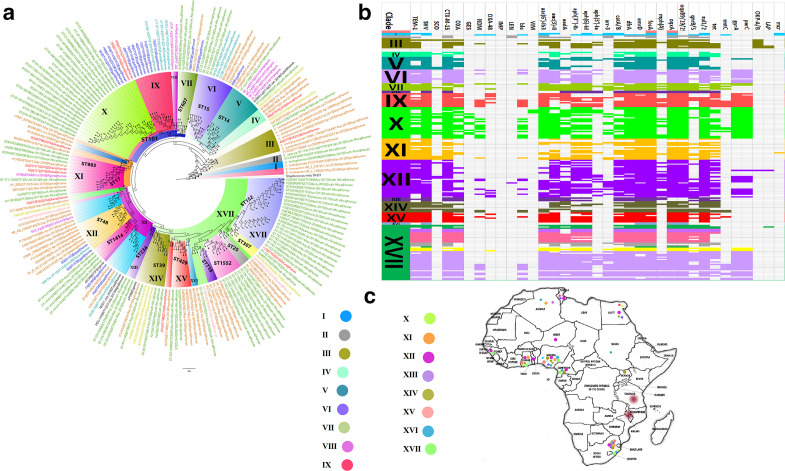
Genomes of K. pneumoniae isolates from Africa were mainly obtained from Algeria, Cameroon, Egypt, Ghana, Guinea, Malawi, Nigeria, Niger, South Africa, Sudan, Tanzania, Togo, Tunisia and Uganda. These clustered in 17 clades and were not restricted to a single country except clades II, VIII, X, XIII and XVI, indicating the distribution of the same clades across several African countries. ST101, which clustered into two clades, was the commonest clone in Africa (South Africa, Egypt, Tunisia, and Ghana) whilst ST152 was also common, but was mainly found in South Africa (Fig. 9). Individual clones were mostly found on the same branches, although different clones were also found within the same clade; in particular, ST716 and ST1552 as well as ST25, ST307 and ST152 clustered on the same branches despite their different clonalities. Multiple ARGs were present in the genomes of all these K. pneumoniae genomes from Africa; notwithstanding this, genes encoding NDM, GES-5 and OXA-48 were only found in a few clades, namely IX, X, XII and XVII (Fig. 9).
Fig. 9.
Phylogeography and resistome dynamics of K. pneumoniae isolates from Africa. Each strain is expressed by species name, strain, sequence type, date and country of isolation, and host on the phylogenetic tree shown in (a) under different label colours representative of the country of origin: green (South Africa), red (Ghana), brown (Nigeria), purple (Algeria), blue (Egypt), turquoise (Tunisia), gold (Cameroon) and black (Sudan, Uganda, unknown country). Members of the same clade (labelled I to XVII) are highlighted with the same colour on the branches. The resistomes of the isolates are shown in (b) under the various clades, with members of the same clade having the same colour as the highlights in the phylogenetic tree in (a); blanks refer to absence of resistance genes and filled sections refer to presence of resistance genes. The phylogeography of the various clades is shown in (c) and most of the clades are concentrated in South Africa, Nigeria, Ghana, Tanzania, Malawi, Algeria, Tunisia and Egypt.
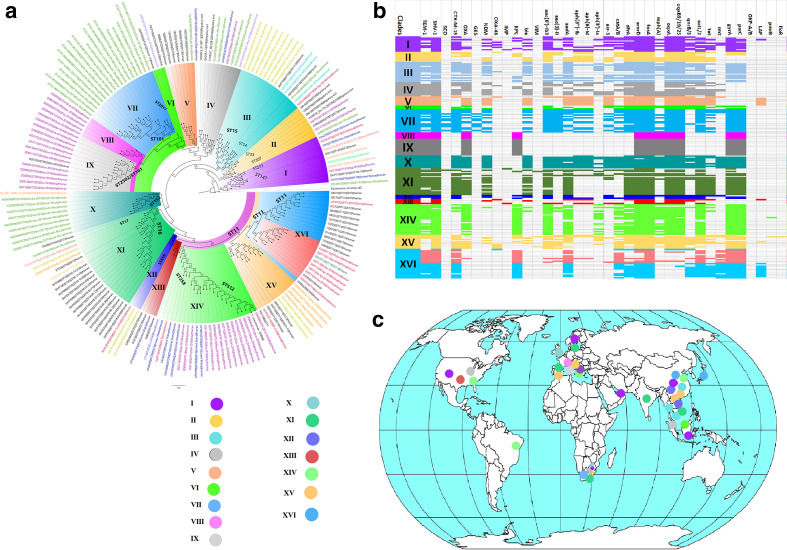
Globally, K. pneumoniae strains producing carbapenemases clustered into 15 main clades, with intercountry transmission being identified in most clades. Italy had numerous ST101, ST2502 and ST512 clones suggestive of local outbreaks. Vietnam had substantial ST15 clones whilst South Africa had ST101 and ST152 clones, suggestive of local clonal transmissions. Worryingly, multiple resistance determinants were seen in all clades (Fig. 10, Table S6).
Fig. 10.
Global phylogeography and resistome dynamics of carbapenemase-producing K. pneumoniae isolates. Each strain is expressed by species name, strain, sequence type, date and country of isolation, and host on the phylogenetic tree shown in (a) under different label colours representative of the country of origin: purple (Italy), green (South Africa), red (China), blue (USA, lemon green (Singapore), orange (UAE), pink (Sweden), turquoise (Thailand), gold (Vietnam), mauve (Malaysia), orange (UK, India or Spain). Members of the same clade (labelled I to XVI) are highlighted with the same colour on the branches. The resistomes of the isolates are shown in (b) under the various clades, with members of the same clade having the same colour as the highlights in the phylogenetic tree in (a); blanks refer to absence of resistance genes and filled sections refer to presence of resistance genes. The phylogeography of the various clades are shown in (c) and most of the clades are concentrated in the USA, Brazil, Europe, UAE, South Africa and South-East Asia.
Discussion
K. pneumoniae is becoming a common cause of fatal and untreatable nosocomial infections worldwide with perennial local outbreaks. Herein, we comprehensively characterized the genomic and epigenomic determinants mediating the MDR, virulence and evolutionary epidemiology of this species. We show that K. pneumoniae strains circulating in South Africa, Africa and globally are bearers of multiple resistance and virulence determinants through clonal, multiclonal and plasmid-mediated dissemination. We also identified and characterized plasmids, transposons, integrons and ISs mobilizing ARGs, virulence genes and RMS within and between K. pneumoniae strains, evinced by a global plasmid evolution and resistome analyses that showed the global distribution of similar plasmid types. More revealing is the RMS and methylation signatures in these strains, which play a crucial role in their virulence, transposition, transcription, replication, DNA repair and resistance regulation [43, 44, 46].
A striking but worrying feature of the isolates used in this study is their MDR phenomes, including resistance to last-resort antibiotics such as colistin and carbapenems, and their widespread distribution in six major healthcare centres in Pretoria (Tshwane municipality) (Table S1). Indeed, the precarious nature of this situation is shown by the recent outbreaks of CRKP infection in hospitals in Pretoria and Johannesburg that resulted in the death of several infants [47, 48]. Previous studies have reported such fatal outbreaks in other public and private sector hospitals in other provinces in South Africa [13, 20, 49]. Hence, this a national more than a local problem necessitating urgent public health interventions to stem its spread [50].
Herein, the CRKP isolates mainly harboured bla OXA-48 and bla NDM-1, which differs from a South African report by the National Institute for Communicable Diseases (NICD) in 2015 in which a high prevalence of bla NDM-1-producing K. pneumoniae in the Gauteng and Kwazulu-Natal provinces and a few reports of bla OXA-48-producing K. pneumoniae in Gauteng and Eastern Cape provinces were described [50]. Similar results were also reported in 2016 by Perovic and colleagues, where a high prevalence of bla NDM-1 was observed in Gauteng province [51]. However, a recent study (2019) reported on an exponential increase in bla OXA-48-like-producing K. pneumoniae strains [19], which agrees with this study and suggests a change in carbapenem resistance determinants in K. pneumoniae in Gauteng. Evidently, NDM-positive strains had higher carbapenem resistance than OXA-48/181 strains (Table S1) [30]. Although isolates with both carbapenemases were also highly resistant, they did not have a higher MIC than those with only NDM (Table S1), which could be due to the possible lack of expression and/or synergy of both genes. The absence of mcr in the isolates suggests chromosomal mutation-mediated colistin resistance.
The REP-PCR, albeit of a poorer resolution than whole-genome-based typing, revealed a major strain that was reported in majority of the K. pneumoniae isolates in this study, mainly carrying the bla OXA-48 gene. One of the isolates in this group was sequenced and we identified bla OXA-181 and ST307, which has been associated with hospital outbreaks, and multiple ARGs such as bla CTX-M-15, bla NDM-1, bla KPC, bla OXA-48 and mcr-1 genes [52–56]. In South Africa, OXA-181-producing K. pneumoniae ST307 isolates were reported previously in private sector hospitals in six provinces, including Gauteng [19]. In this study, the K. pneumoniae ST307 isolates were collected from government sector hospitals in the Tshwane area.
Another strain detected by REP-PCR was found to contain bla OXA-181 and belonged to ST607, a rare clone reported in China and in a neonatal intensive care unit in France [57]. The bla OXA-48-producing K. pneumoniae strain, ST3559, has been recently reported as a novel clone among CRKP isolates collected from hospital wastewater, influent wastewater, river water and riverbed sediments in South Africa [58]. These K. pneumoniae isolates shared the same molecular and MDR characteristics with isolates from the present study (Data S2), suggesting that the same strain is now circulating in hospitals in the Tshwane area. Bla NDM-7, the first to be found in K. pneumoniae ST17 in South Africa, was reported in K. pneumoniae ST147 and ST273 involved in nosocomial cases in Canada, Gabon, Philippines, USA and India [59–63].
Moreover, the phylogeography and resistome analyses of K. pneumoniae genomes from South Africa, Africa, and global carbapenemase-producing K. pneumoniae (CPKP) strains shows the rich resistance repertoire of this species. Notably, similar K. pneumoniae clones are circulating within mainly Gauteng (Pretoria) and Kwazulu-Natal (Durban, Pietermaritzburg and Ozwatini) provinces in South Africa (Fig. 8), suggesting the exchange of carriers of these strains between these distant provinces. Indeed, other non-human factors could be involved in this interprovincial transmission, and the earlier these carriers are identified the better. Furthermore, the presence of the same clades/clones, particularly ST101, ST14, ST15 and ST17, between South Africa, West Africa and North Africa, bearing multiple clinically important ARGs is revealing. Globally, CPKP is mainly concentrated in the USA, South Africa, Europe and South-East Asia, with relatively fewer reports from the Arabian peninsula and Brazil, and further evidence that AMR transcends borders and requires epidemiological investigations to detect and break the transmission chain.
Worryingly, almost all these K. pneumoniae and CPKP isolates co-harboured numerous other ARGs alongside extensive deletions, insertions and mutations in their ompK36 genes (n=6 isolates), which makes them potentially multi-, extensively and pan-drug-resistant, an observation already made in this (Fig. 1, Table S1) and other studies [13, 20]. Coupled with this is the rich virulence genes and capsule repertoire of K. pneumoniae that enable them to escape host immune forces [64]. As shown previously, the virulome was not affected by the isolation source [13]. Worryingly, traT genes, encoding an outer membrane protein that mediates resistance to complement proteins, were found on plasmids in all the isolates (Table S3). Gordon et al. recently found O1 and O2 capsule serotypes, also found in this study, to be most associated with MDR; they also found the K2 serotype, present in KP10 and KP33, to be most common among global K. pneumoniae isolates [64, 65].
Almost 79 capsule polysaccharide types based on the K-loci of K. pneumoniae have been described, and of these, only K1 and K2 serotypes are associated with hypervirulent strains whilst the others are associated with classical strains of K. pneumoniae [66, 67]. Our results showed that two of the sequenced isolates (Kp10 and Kp33), which are highly resistant to carbapenems and harboured the bla NDM-1 gene, were KL2 serotypes. This might indicate that these isolates are K2-hypervirulent K. pneumoniae (K2-hvKP) strains. K2-hvKP strains were not given attention until the report of MDR K2-hvKP strain harbouring bla KPC-2 and bla IMP-4 in China [23]. Following this report, multiple studies, including this one, have reported on carbapenemase production in highly virulent stains of the K2 serotype [68, 69]. Another study in China reported on an ST11 bla KPC-2-producing strain (CR-HvKP1), which is closely related to strains in this study (Data S2); this CR-HvKP1 harboured a virulence plasmid (pLVPK-like) and showed a highly resistant profile [70].
The plasmid evolutionary epidemiology of the plasmids identified in this study, as well as their resistomes, provide deeper insights into the role of plasmids in the dissemination of ARGs in Enterobacterales . Notably, the close evolutionary alignment/distance of plasmids bearing he same or different ARGs, but belonging to different incompatibilities in the same isolate (Fig. 7a), indicates the genetic exchanges (recombinations and rearrangements) that occur between plasmids and between plasmids and chromosomes during replication [71, 72]. Evidently, the very close sequence and resistome similarity between this study’s plasmids and those obtained from different and the same species in other studies worldwide shows the global dissemination of IncF, IncX, A/C, IncN and IncI plasmids and their role in dissemination of ARGs among bacteria.
Furthermore, it shows that not all plasmid types harbour rich resistomes as IncX was mainly limited to only three ARGs. We show that specific plasmid replicons are mostly associated with specific ARGs. Specifically, IncF plasmids harbour more diverse ARGs, including KPC and NDM carbapenemases, whilst IncX harboured less diverse ARGs and was mostly associated with NDM. In addition, OXA carbapenemases were found on mainly L/M and IncX replicons. Interestingly, IncN and IncA/C plasmids also harboured more diverse ARGs, including NDM carbapenemases. Evidently, IncF and IncX appear to be more common replicons than the rest (Fig. 7, Table S2). The relatively lower diversity and abundance of ARGs on these plasmids (Fig. 7) than the resistomes observed in the K. pneumoniae strains (Figs 8–10) is because the K. pneumoniae strains contain multiple plasmids alongside chromosomes, which bear additional ARGs.
Previous studies identified bla NDM-1 on IncF, IncL/M, IncN, A/C and IncX plasmids [30]. Herein, bla NDM-1-producing K. pneumoniae were mostly associated with IncF (FII, F, FIB, FIC), followed by IncL and A/C plasmid replicons (Table S1), which agrees with reports from Nepal, Taiwan, Oman, Myanmar, Canada and South Africa [20, 38, 73, 74]. However, in China, Gabon, India and Japan, NDM-1/7 variants in K. pneumoniae were on IncX3/4 plasmids [69, 75]. The bla OXA-181-producing isolates were on ColKP3, IncX3 and IncF plasmids, similar to previous studies from the Czech Republic, Denmark, São Tomé and Príncipe, and South Africa [19, 33, 76, 77]. In South Africa, IncX3 was found in K. pneumoniae collected during a hospital outbreak [19]. An earlier study reported the significant role that IncL/M plasmids play in the dissemination of bla OXA-48 genes in K. pneumoniae strains worldwide [33, 34, 78–81]. Our findings also demonstrate that the bla OXA-48 gene is usually located on conjugative IncL/M plasmids. The IncHI1B plasmid replicon was responsible for the carriage of bla NDM-1 in strain KP33_1 [82], different from bla NDM-1-producers in this study. A strain reported in the USA (CN1) which harboured an IncFII/FIB multi-replicon showed similar molecular characteristics with bla NDM-1-producers in this study, but it belonged to ST392. Thus, dissemination f ARGs may occur in diverse clones via different plasmid replicon types.
There were discrepancies in plasmid numbers and sizes between the gel-based plasmid characterization and sequencing analyses, which is expected. This could be due to the break-up of the plasmids during the extraction process, leading to smaller sizes and higher plasmid numbers. The PCR-based plasmid typing scheme used in this study was unable to detect IncX3 plasmids, which were revealed with whole-genome sequencing. This is a limitation in areas where this typing scheme is solely used as it needs to be modified with new primers targeting all subtypes of the replicon groups.
No single plasmid harboured both ESBL and carbapenemase genes together and the plasmids harbouring the ESBL genes were of larger sizes with richer resistomes that clustered together on resistance islands, further supporting the fewer resistomes observed on carbapenemase plasmids worldwide (Fig. 7). The genetic environment of all the ARGs and mercuric resistance operons on these self-conjugative plasmids were surrounded by MGEs, which undoubtedly will facilitate their horizontal transmission. Notably, the genetic support of these ARGs, particularly of the ESBLs, were similar to those already reported in other studies: bla CTX-M-15 was always next to ISEc9, bla TEM was always next to an integrase/recombinase in close synteny to bla CTX-M-15, and bla OXA was always surrounded by cat and aac(3′)-II genes. In addition, bla NDM-1/7, bla OXA-181 and bla OXA-48 were also found within several MGEs (Figs 2–6), supporting their horizontal transposition.
Prophages (bacteriophages), which are involved in transduction of genetic material horizontally, were abundant on plasmids and chromosomes, with several partial prophage DNAs, suggesting transduction activity (Figs S7–S12). Indeed, the presence of the same prophages in different isolates and on both plasmids and chromosomes indicates their horizontal movement and importance in influencing MDR and genomic plasticity or evolution. Evidently, their presence on these self-transmissible plasmids suggests that they can be also shared during conjugation. Yet, the presence of prophage DNA and RMS on plasmids and chromosomes is intriguing as RMS identify and destroy foreign DNA, including prophages [43, 44, 83]. However, prophages found on plasmids with the same methylation signature as the host bacterium may escape destruction by REs and CRISPR-Cas complexes. Notwithstanding this, the interplay between phages and RMS on the same plasmids and chromosomes will need further investigation to understand their co-existence and co-evolution.
The isolates were remarkably endowed with rich RMS comprising types I, II and III, with type II RMS being more abundant, as previously reported [44, 46, 83]. Huang et al. recently showed that type I RMS were scarce in CPKP, clearly due to the destruction of carbapenemase-bearing plasmids by the type I RMS; the exception was in isolates that had the type I RMS on plasmids to protect them from REs [84]. It is thus not surprising that plasmids harbouring ESBLs and carbapenemases in this study’s isolates also had RMS that shared the same Dam and Dcm motifs with those on the chromosomes, and CCWGG motifs of type II Dcms were virtually absent on chromosomes but ubiquitous on plasmids (Table S4). Indeed, these RMS were found within MGEs, as already shown [83], which evidently facilitates their movement between plasmids, chromosomes and bacteria. Specifically, the multiple copies of M.Sfl2ORFAP on both plasmids and chromosomes in several isolates suggest a transposition event [83]. Due to the destruction of plasmids or DNA without the same methylation signatures by REs, the presence of the same RMS on plasmids and chromosomes facilitates their safe entry into host bacteria, enhancing dissemination of virulence and resistance plasmids between different species.
The ubiquitous presence of GATC motifs on only chromosomes and their absence on plasmids indicates the conservation of these motifs and their associated Dams in prokaryotes. Notably, the preponderance of type II Dcms with CCWGG motifs on plasmids suggests that their relatively limited presence on chromosomes is due to transposition from plasmids to chromosomes. Recently, environmental bacteria were found to be abundant in Dams with GATC and ATGNNNNNNGCT motifs [43], which were mostly found on chromosomes of this study’s isolates. GATC and CCAGG (CCWGG or CCNGG in this study) motifs have been also identified in E. coli [71, 83], showing that plasmids with these RMS and methylation signatures can be shared between environmental and clinical prokaryotes. Therefore, the important role of RMS in horizontal resistomes and virulome transmission and regulation cannot be contradicted. Furthermore, the regulatory roles of RMS in bacterial virulence were recently confirmed in hvKP strains in Taiwan; dam- mutants were found to be less pathogenic in mice and serum than their wild-type dam+. Hence, the presence of these rich RMS in the isolates may facilitate their virulence in human and animal hosts. More concerning is the ability of these RMS to defeat bacteriophage therapy. Indeed, a recent study showed how K. pneumoniae quickly developed resistance to bacteriophages during treatment [85]. Detailed investigations are needed to find ways to protect bacteriophage therapy from destruction or resistance by these RMS systems.
Conclusion
This study has shown the dissemination of bla OXA-48-like and bla NDM carbapenemases in MDR K. pneumoniae isolates in hospitals in Gauteng, South Africa, as well as MDR K. pneumoniae in Africa and globally through self-transmissible IncF, A/C, IncX3 and IncL/M plasmids. The ompk36 porin genes of the six isolates were extensively mutated, suggesting their possible synergistic role in the MDR phenomes of these strains. Notably, the important role of RMS in regulating and facilitating the transcription, transposition and dissemination of resistance and virulence plasmids is revealing and requires further investigation as it also threatens both antimicrobial and bacteriophage chemotherapy. It is essential that rigorous infection prevention and control are instituted to avoid the selection and dissemination of plasmids harbouring RMS, virulence and MDR genes in prokaryotes.
This study showed K. pneumoniae isolates to have resistance profiles to most antibiotics, including colistin. Among all the tested antibiotics, only a few (amikacin, fosfomycin and tigecycline) were still active against these isolates. This raises more concern about treatment options for CRKP, because colistin is one of the last-resort drugs for infections caused by these pathogens. However, it is of note that newer antimicrobial agents such as ceftazidime-avibactam, aztreonam-avibactam and cefiderocol were not tested against these isolates, which could change their resistance outlook. Owing to budget restrictions, we could not sequence all the isolates to provide a broader characterization of their genomes, multilocus sequence types (clones), resistomes, methylomes, virulomes and mobilomes. Hence, the six sequenced isolates are not fully representative of the clones of the 56 isolates although the REP-PCR clustering was used for selection. This is due to the higher resolution of WGS-based phylogenomics over REP-PCR and multilocus sequence typing.
Methods
Bacterial strains and antimicrobial susceptibility testing
Sixty non-repetitive K. pneumoniae isolates with preliminary non-susceptibility to carbapenems were randomly collected from a referral laboratory (National Health Laboratory Service/NHLS) in Pretoria. The VITEK 2 automated system (bioMérieux-Vitek) was used for species identification and antimicrobial susceptibility testing; only those resistant to at least one carbapenem (ertapenem, meropenem, imipenem, doripenem) were included in further analyses (n=56 isolates). The K. pneumoniae isolates were received on blood agar plates (NHLS) and incubated at 37 °C for 24 h. Following incubation, confirmation of the MIC of the isolates was determined using the MicroScan automated system (Beckman Coulter); a colistin-resistant positive control was used to confirm the colistin sensitivity results. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) breakpoints (2019) [86]. Of the 60 isolates, 56 (resistant to at least one carbapenem) were considered for further investigation in this study. MDR K. pneumoniae isolates were defined as showing non-susceptibility to at least one agent in three antibiotic classes; extensively drug-resistant isolates were defined as showing non-susceptibility to at least one agent in all but two antibiotic classes; and pandrug-resistant isolates were defined as showing non-susceptibility to at least one agent in all antibiotic classes.
DNA extraction of CRKP isolates
Total genomic DNA was extracted from all carbapenem-resistant isolates (n=56). The DNA was extracted from an overnight Brain Heart Infusion (BHI) broth using the boiling method. The cells were heated at 95 °C using a digital dry bath (Labnet International) for 15 min and transferred to an ultrasonic bath (Lasec) for another 15 min. The resulting supernatant was stored in a −20 °C freezer until needed for further analysis and it was used as a template for the PCR assays.
Detection of carbapenemase genes using PCR assays
PCR was used to screen for the presence of six carbapenemase genes: bla IMP, bla KPC, bla VIM, bla OXA-48, bla NDM and bla GES in the 56 isolates. Specifically, multiplex PCR was used to determine the presence of bla VIM, bla OXA-48 and bla NDM while simplex PCR was used for bla IMP, bla KPC and bla GES screening. The oligonucleotide primers were synthesized by Inqaba Biotechnical Industries (Data S1). For the PCR, 1 µl of template DNA was added to 12.5 µl of MyTaq HS mix (Bioline) while 0.4 µM of each primer and nuclease-free water (Qiagen) were added to make up the volume to 25 µl in each PCR tube. The multiplex PCR conditions were as follows: 95 °C for 5 min, followed by 25 cycles of 95 °C for 30 s, 57 °C for 45 s and 72 °C for 30 s, and a final extension step at 72 °C for 7 min. The PCR amplicons were analysed using 1.5 % Seakem agarose gel (Whitehead Scientific) with 5 µl ethidium bromide and visualized under UV light using the Gel Doc EZ Gel (BioRad Laboratories) bioimaging system. A 100 bp ready-to-use DNA ladder (Celtic Molecular Diagnostics) was used to determine the size of the expected genes. All PCR amplicons were run alongside a positive and negative control.
Genotyping using REP-PCR assay
Total genomic DNA from all CRKP isolates (n=56) were used as template in the REP-PCR assay. The primer pair sequences REP 1 (5′-IIIGCGCCGICATCAGGC-3′) and REP 2 (5′-ACGTCTTATCAGGCCTAC-3′) and PCR conditions described previously were used in this assay [87]. For the PCR, 1 µl of template DNA was added to 12.5 µl of MyTaq HS mix (Bioline) while 0.4 µM of each primer and nuclease-free water (Qiagen) was added to make up the volume to 25 µl in each PCR tube. The PCR conditions were as follows: an initial denaturation of 94 °C for 3 min, followed by 30 cycles of 94 °C for 45 s, 45.8 °C for 1 min and 72 °C for 8 min and a final extension step of 72 °C for 16 min. The amplified DNA amplicons (10 µl) were separated by electrophoresis using 1.5 % SeaKem agarose gel (Whitehead Scientific) with 5 µl ethidium bromide. The gels were run for 3 h 20 min at 80 V. The DNA amplicon bands were visualized under UV light using the Gel Doc EZ Gel (BioRad Laboratoriess) bioimaging system and banding patterns were compared to a 1 kb plus ready-to-use DNA ladder (Thermo Fisher Scientific). Analysis of REP-PCR fingerprints was performed using the GelCompare II software (Applied Maths). Relatedness was determined by means of the Dice coefficient and unweighted pair group method with arithmetic mean (UPGMA). In this study a similarity coefficient of 75 % was used to determine different strains of CRKP, i.e. isolates that showed a similarity of 75 % were considered to be part of the same strain.
Plasmid characterization using the PBRT scheme
Plasmid DNA extracted using the plasmid midi kit (Qiagen) was used as template in characterizing plasmids using the PCR-based inc/rep typing scheme in the 56 isolates. These plasmids were typed by targeting 19 replicon groups reported in Enterobacteriaceae and one replicon group targeting a virulence plasmid in K. pneumoniae . This method was carried out as previously described with few modifications [88, 89]. Modifications were made in multiplex 5, where A/C and IncT were detected in a multiplex and IncFII plasmids were detected in a simplex PCR assay instead of multiplex assay. The IncFIIk virulence plasmids in K. pneumoniae were also detected. The PCR assays were performed using a SimpliAmp Thermal cycler mini (Thermo Fisher Scientific) and the PCR conditions used were described previously [88, 89]: briefly, initial denaturation of 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, 60 °C for 30 s and 72 °C for 1 min, and a final extension of 72 °C for 5 min. For IncF, IncFII and IncFIIk plasmids, the same conditions were used except that an annealing temperature of 54 °C for 30 s was used instead. Data S1 shows all the primer sequences that were used for these assays.
Resistance plasmid transferability/mobility
Transferability of resistance plasmids was determined using conjugation experiments. The experiments were performed on 26 isolates showing reduced susceptibility to meropenem using a broth mating method. The meropenem-resistant isolates were used as plasmid donors and the E. coli J53-Ar (sodium-azide-resistant) strain served as a recipient strain. For broth mating, 3 h growth cultures of donor and recipient strains grown in Luria Bertani (LB) broth (VWR international) were mixed with each other at a ratio of 1 : 4 (donor to recipient) and incubated at 37 °C for 3 h. Grown cells (200 µl) of the mixtures were spread onto Mueller-Hinton agar (Sigma-Aldrich) containing 0.5 µg ml−1 meropenem (Sigma-Aldrich) and 100 µg ml−1 sodium azide (VWR international) to select only for plasmid-encoded carbapenem resistance and then incubated at 37 °C for 24 or 48 h. A PCR assay was used to confirm carbapenemase gene (bla NDM-1 and/or bla OXA-48) carriage by transconjugants.
Whole-genome sequencing of K. pneumoniae isolates
Six representative isolates, based on their carbapenemase gene, REP pattern, plasmid number and type, were selected for WGS. Genomic DNA was extracted from the K. pneumoniae isolates Kp8, Kp10, Kp15, Kp29, Kp32 and Kp33 using a Zymo Research Fungal/Bacterial kit (Inqaba Biotec) according to the manufacturer’s instructions. Genomic DNA was sent for sequencing at Inqaba Biotec on the PacBio RSII sequencer (Pacific Biosciences) at an average coverage of 90×.
Genomic analyses and annotation
PacBio’s SMRT Link v8.0 software suite was used for trimming the raw reads, assembling with HGAP, and determining methylation modifications and motifs (using MotifMaker: https://github.com/PacificBiosciences/MotifMaker) in the genomic sequences. DNA methylases (MTases), restriction endonucleases (REases) and their motifs were searched from the Restriction Enzyme Database (REBASE) [90]. Complete genomic annotations were done with NCBIs PGAP [91]. Clonal STs, resistance and virulence genes, plasmid typing, integrons, transposons, ISs and prophages were determined using online databases including MLST2.0 [92], ResFinder [93], BacWGSTdb [94], Plasmidfinder [95], INTEGRALL (http://integrall.bio.ua.pt/), ISFinder [96] and PHASTER [97], respectively. Whole-genome-based K. pneumoniae capsule polysaccharide-based typing (K-type) was performed using the Kaptive Web database [98].
Chromosomal colistin and fluoroquinolone resistance mutations
Mutations conferring resistance to colistin and fluoroquinolones as well as in ompK36 porin genes were determined from the assembled genomes using blastn. Briefly, mgrB, crrB, kpnEF, phoPQ, pmrAB, gyrA, gyrB parC, parE and ompK36 genes in reference strain K. pneumoniae ATCC 13883 (PRJNA244567) were aligned with this study’s genomes using blastn. The mutations in the genomes of this study’s isolate were manually curated and tabulated in Data S2.2.
Phylogenomic, phylogeography and resistome analyses
Genome sequences of CRKP strains from South Africa (n=88), Africa (n=380) and globally (n=343) were downloaded from the PATRIC website (https://www.patricbrc.org/); carbapenemase-producing K. pneumoniae genomes (n=190) were further culled from the global CRKP genomes for the global carbapenemase-producing K. pneumoniae phylogenomics. These genomes, alongside those from this study, were used for the whole-genome phylogenomics. Four phylogenomic trees, one for this study’s genomes, one for South African genomes, one for African genomes and one for global genomes, were drawn using RAxML’s maximum-likelihood-based phylogenetic inference. A bootstrap reassessment of 1000× was used and the trees were annotated using Figtree (http://tree.bio.ed.ac.uk/software/figtree/). Isolates with strong bootstrap support values (>50) were clustered into a clade and highlighted with the same colour. The resistomes of these genomes were downloaded from NCBI’s Pathogen/Isolate Browser database (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/search/) and the clades’ phylogeographies were manually mapped.
Genomic plasmid typing, evolution and resistome analyses
Plasmid genomes (n=26) from this study were aligned with muscle. The aligned files were used to draw a phylogenetic tree with PhyML (http://www.phylogeny.fr/index.cgi), using a bootstrap sampling of 100×. The newick tree file was annotated with Figtree. The genomes of the 26 plasmids were aligned with those of closely related plasmids using progressive Mauve [98]. The carbapenemase-bearing plasmids of this study were parsed through the BacWGSTdb database/server to determine other closely related plasmids, their source, incompatibility and geographical locations; 130 similar plasmids were obtained and used for the plasmid resistome analyses [94]. Carbapenemase-bearing plasmid genomes (n=592) and metadata were downloaded from NCBI and PATRIC (https://www.patricbrc.org/). Their plasmid replicons and resistomes were determined using PlasmidFinder [95] and ResFinder [93]. These were arranged according to their replicons/incompatibility groups and mapped to show their geographical distribution worldwide on maps.
Supplementary Data
Funding information
Funding for this study was provided by the NHLS, NRF (National Research Foundation) and the University of Pretoria.
Acknowledgements
We are grateful to Sebastien Santini – CNRS/AMU IGS UMR7256 for curating the phylogeny (http://www.phylogeny.fr/index.cgi) database.
Author contributions
K.K. undertook the laboratory work, data curation, initial descriptive statistics and initial draft of the manuscript for her MSc work; N.M.M. (https://orcid.org/0000-0001-8890-2663) supervised the study and provided funding; J.O.S. (https://orcid.org/0000-0002-9508-984X) conceived, designed and supervised the study, undertook bio-informatics analyses and descriptive statistics, data curation, image designs, and the complete write-up, review and formatting of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethical approval was provided by the Faculty of Health Sciences Research Ethics Committee (209/2018). All protocols and consent forms were executed according to the agreed ethical approval terms and conditions. All clinical samples were obtained from a reference laboratory and not directly from patients, who agreed to our using their specimens for this research. The guidelines stated by the Declaration of Helsinki for involving human participants were followed in the study.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ARG, antimicrobial resistance gene; CRKP, carbapenem-resistant Klebsiella pneumoniae; ESBL, extended-spectrum β-lactamase; HGT, horizontal gene transfer; IS, insertion sequence; MDR, multi-drug resistance; MGE, mobile genetic element; MIC, minimum inhibitory concentration; RE, restriction endonuclease; REP-PCR, repetitive element palindromic PCR; RMS, restriction modification systems; RND, resistance-nodulation-division; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Fourteen supplementary figures, six supplementary tables and three supplementary data files are available with the online version of this article.
References
- 1.Friedlaender C. Ueber die Schizomyceten bei Der acuten fibrösen Pneumonie. Arch für Pathol Anat und Physiol und für Klin Med. 1882;87:319–324. [Google Scholar]
- 2.Ashurst J, Dawson A. Klebsiella pneumonia. Treasure Island: StatPearls Publishing; 2018. [PubMed] [Google Scholar]
- 3.Jondle CN, Gupta K, Mishra BB, Sharma J. Klebsiella pneumoniae infection of murine neutrophils impairs their efferocytic clearance by modulating cell death machinery. PLOS Pathog. 2018;14:e1007338. doi: 10.1371/journal.ppat.1007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osei Sekyere J, Mensah E. Molecular epidemiology and mechanisms of antibiotic resistance in Enterococcus spp., Staphylococcus spp., and Streptococcus spp. in Africa: a systematic review from a one health perspective. Ann N Y Acad Sci. 2020;1465:29–58. doi: 10.1111/nyas.14254. [DOI] [PubMed] [Google Scholar]
- 5.Kidd TJ, Mills G, Sá-Pessoa J, Dumigan A, Frank CG, et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh N. How often are antibiotic-resistant bacteria Said to "Evolve" in the News? PLoS One. 2016;11:e0150396. doi: 10.1371/journal.pone.0150396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodd MC. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J Environ Monit. 2012;14:1754. doi: 10.1039/c2em00006g. [DOI] [PubMed] [Google Scholar]
- 8.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Wang G, Sebra R, Zhuge J, Yin C, et al. Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both bla KPC and bla CTX-M Integrated in the Chromosome. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00076-17. 27 06 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osei Sekyere J, Amoako DG. Carbonyl cyanide m-Chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae . Front Microbiol. 2017;8:228. doi: 10.3389/fmicb.2017.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong H, Zhang S, Pan H, Cai T. Influence of induced ciprofloxacin resistance on efflux pump activity of Klebsiella pneumoniae . J Zhejiang Univ Sci B. 2013;14:837. doi: 10.1631/jzus.B1200221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu S, Klein EY, Chun BC. Temporal association between antibiotic use and resistance in Klebsiella pneumoniae at a tertiary care hospital. Antimicrob Resist Infect Control. 2018;7:83. doi: 10.1186/s13756-018-0373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mbelle NM, Feldman C, Sekyere JO, Maningi NE, Modipane L, et al. Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci Rep. 2020;10:1–17. doi: 10.1038/s41598-020-58012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y-T, Su C-F, Chuang C, Lin J-C, Lu P-L, et al. Appropriate treatment for bloodstream infections due to carbapenem-resistant Klebsiella pneumoniae and Escherichia coli: a nationwide multicenter study in Taiwan. Open Forum Infect Dis. 2019;6 doi: 10.1093/ofid/ofy336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, et al. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae . Antimicrob Agents Chemother. 2010;54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osei Sekyere J, Maningi NE, Modipane L, Mbelle NM. Emergence of mcr-9.1 in ESBL-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics. Resistome and Mobilome mSystems. 2020;5:e00148–20. doi: 10.1128/mSystems.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4:2. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, Landolsi S, Ferjani S, et al. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae . Int J Antimicrob Agents. 2018;52:898–905. doi: 10.1016/j.ijantimicag.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg Infect Dis. 2019;25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen T, Sekyere JO, Govinden U, Moodley K, Sivertsen A, et al. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and Pandrug-Resistant clinical Enterobacteriaceae in Durban, South Africa. Antimicrob Agents Chemother. 2018;62:e02178–17. doi: 10.1128/AAC.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan L, Wang S, Guo Y, Jin Y, Duan J, et al. Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front Microbiol. 2018;9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Nava RG, Oliveira-Silva M, Nakamura-Silva R, Pitondo-Silva A, Vespero EC. New sequence type in multidrug-resistant Klebsiella pneumoniae harboring the blaNDM-1-encoding gene in Brazil. Int J Infect Dis. 2019;79:101–103. doi: 10.1016/j.ijid.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Sidjabat H, Nimmo GR, Walsh TR, Binotto E, Htin A, et al. Carbapenem resistance in Klebsiella pneumoniae due to the new Delhi metallo-β-lactamase. Clin Infect Dis. 2011;52:481–484. doi: 10.1093/cid/ciq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osei Sekyere J, Govinden U, Essack S. The molecular epidemiology and genetic environment of carbapenemases detected in Africa. Microb Drug Resist. 2015;22:59–68. doi: 10.1089/mdr.2015.0053. [DOI] [PubMed] [Google Scholar]
- 27.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17 doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somboro AM, Osei Sekyere J, Amoako DG, Essack SY, Bester LA. Diversity and proliferation of metallo-β-lactamases: a clarion call for clinically effective metallo-β-lactamase inhibitors. Appl Environ Microbiol AEM. 2018;27:00698–18. doi: 10.1128/AEM.00698-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somboro AM, Amoako DG, Osei Sekyere J, Kumalo HM, Khan R, et al. 1,4,7-Triazacyclononane Restores the Activity of β-Lactam Antibiotics against Metallo-β-Lactamase-Producing Enterobacteriaceae: Exploration of Potential Metallo-β-Lactamase Inhibitors. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.02077-18. 01 02 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann New. 2019;1457:61–91. doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 31.Mbelle NM, Feldman C, Osei Sekyere J, Maningi NE, Modipane L, et al. The resistome, mobilome, Virulome and phylogenomics of multidrug-resistant Escherichia coli clinical isolates from Pretoria, South Africa. Sci Rep. 2019;9:1–43. doi: 10.1038/s41598-019-52859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skálová A. Molecular characterization of OXA-48-like-producing Enterobacteriaceae in the Czech Republic: evidence for horizontal transfer of pOXA-48-like plasmids. Antimicrob Agents Chemother. 2016:01889–16. doi: 10.1128/AAC.01889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Power K, Wang J, Karczmarczyk M, Crowley B, Cotter M, et al. Molecular analysis of OXA-48-carrying conjugative IncL/M-like plasmids in clinical isolates of Klebsiella pneumoniae in Ireland. Microb Drug Resist. 2014;20:270–274. doi: 10.1089/mdr.2013.0022. [DOI] [PubMed] [Google Scholar]
- 34.Bonnin RA, Nordmann P, Carattoli A, Poirel L. Comparative genomics of IncL/M-type plasmids: evolution by acquisition of resistance genes and insertion sequences. Antimicrob Agents Chemother. 2013;57:674–676. doi: 10.1128/AAC.01086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Chavda KD, Melano RG, Hong T, Rojtman AD, et al. Molecular survey of the blaKPC-harboring IncFIA plasmids in New Jersey and new York hospitals. Antimicrob Agents Chemother. 2014;58:2289–2294. doi: 10.1128/AAC.02749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papagiannitsis CC, Di Pilato V, Giani T, Giakkoupi P, Riccobono E, et al. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother. 2016;71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 37.Huang T-W, Chen T-L, Chen Y-T, Lauderdale T-L, Liao T-L, et al. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One. 2013;8:e62774. doi: 10.1371/journal.pone.0062774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada S, Doia Y. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.00959-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen D, et al. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare blaCTX-M-24-harboring virulence plasmid. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, et al. Colistin resistance gene mcr-1 mediates cell permeability and resistance to hydrophobic antibiotics. Front Microbiol. 2020;10 doi: 10.3389/fmicb.2019.03015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang CT, Yi WC, Shun CT, Tsai SF. DNA adenine methylation modulates pathogenicity of Klebsiella pneumoniae genotype K1. J Microbiol Immunol Infect. 2017;50:471–477. doi: 10.1016/j.jmii.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 42.Hiraoka S, et al. Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community. Nat. Commun. 2019;10 doi: 10.1038/s41467-018-08103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blow MJ, Clark TA, Daum CG, Deutschbauer AM, Fomenkov A, et al. The epigenomic landscape of prokaryotes. PLoS Genet. 2016;12:e1005854. doi: 10.1371/journal.pgen.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asante J, Osei Sekyere J. Understanding antimicrobial discovery and resistance from a metagenomic and metatranscriptomic perspective: advances and applications. Environ Microbiol Rep. 2019;11:62–86. doi: 10.1111/1758-2229.12735. [DOI] [PubMed] [Google Scholar]
- 45.Beaulaurier J, Zhu S, Deikus G, Mogno I, Zhang X-S, et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol. 2018;36:61–69. doi: 10.1038/nbt.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anonymous Sixth baby dies in Klebsiella outbreak. news24. 2020 [Google Scholar]
- 47.Anonymous Six babies have died from hospital superbug. Timeslive. Available at https://www.timeslive.co.za/news/south-africa/2018-09-16-six-babies-have-died-from-hospital-superbug (Accessed: 19th June 2020) 2018
- 48.Jacobson RK, et al. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase-producing isolates of Klebsiella pneumoniae in South Africa. S Afr Med J. 2015;105:1030–1035. doi: 10.7196/SAMJ.2015.v105i12.9926. [DOI] [PubMed] [Google Scholar]
- 49.Osei Sekyere J. Current state of resistance to antibiotics of Last-Resort in South Africa: a review from a public health perspective. Front Public Heal. 2016;4:209. doi: 10.3389/fpubh.2016.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perovic O, et al. Antimicrobial resistance surveillance in the South African private sector report for 2016. South African J Infect Dis. 2018;33:114–117. [Google Scholar]
- 51.Habeeb MA, Haque A, Nematzadeh S, Iversen A, Giske CG. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int J Antimicrob Agents. 2013;41:524–526. doi: 10.1016/j.ijantimicag.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 52.Ocampo AM, Chen L, Cienfuegos AV, Roncancio G, Chavda KD, et al. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of Non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother. 2016;60:332–342. doi: 10.1128/AAC.01775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novović K, Trudić A, Brkić S, Vasiljević Z, Kojić M, et al. Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02550-16. 24 04 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bocanegra-Ibarias P, Garza-González E, Morfín-Otero R, Barrios H, Villarreal-Treviño L, et al. Molecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in Mexico. PLoS One. 2017;12:e0179651. doi: 10.1371/journal.pone.0179651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saavedra SY, Diaz L, Wiesner M, Correa A, Arévalo SA, et al. Genomic and Molecular Characterization of Clinical Isolates of Enterobacteriaceae Harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00841-17. 22 11 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peltier F, Choquet M, Decroix V, Adjidé CC, Castelain S, et al. Characterization of a multidrug-resistant Klebsiella pneumoniae ST607-K25 clone responsible for a nosocomial outbreak in a neonatal intensive care unit. J Med Microbiol. 2019;68:67–76. doi: 10.1099/jmm.0.000884. [DOI] [PubMed] [Google Scholar]
- 57.Ekwanzala MD, Budeli P, Dewar JB, Kamika I, Momba MNB. Draft Genome Sequences of Novel Sequence Type 3559 Carbapenem-Resistant Klebsiella pneumoniae Isolates Recovered from the Environment. Microbiol Resour Announc. 2019;8 doi: 10.1128/MRA.00518-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynch T, Chen L, Peirano G, Gregson DB, Church DL, et al. Molecular evolution of a Klebsiella pneumoniae ST278 isolate harboring blaNDM-7 and involved in nosocomial transmission. J Infect Dis. 2016;214:798–806. doi: 10.1093/infdis/jiw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shankar C, Kumar S, Venkatesan M, Veeraraghavan B. Emergence of ST147 Klebsiella pneumoniae carrying blaNDM-7 on IncA/C2 with ompK35 and OmpK36 mutations in India. J Infect Public Health. 2019;12:741–743. doi: 10.1016/j.jiph.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 60.Chou A, Roa M, Evangelista MA, Sulit AK, Lagamayo E, et al. Emergence of Klebsiella pneumoniae ST273 Carrying bla NDM-7 and ST656 Carrying bla NDM-1 in Manila, Philippines. Microb Drug Resist. 2016;22:585–588. doi: 10.1089/mdr.2015.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee C-S, Vasoo S, Hu F, Patel R, Doi Y. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol. 2014;52:4109–4110. doi: 10.1128/JCM.01404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moussounda M, Diene SM, Dos Santos S, Goudeau A, François P, et al. Emergence of blaNDM-7-Producing Enterobacteriaceae in Gabon, 2016. Emerg Infect Dis. 2017;23:356–358. doi: 10.3201/eid2302.161182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuon FF, Graf ME, Merlini A, Rocha JL, Stallbaum S, et al. Risk factors for mortality in patients with ventilator-associated pneumonia caused by carbapenem-resistant Enterobacteriaceae . Braz J Infect Dis. 2017;21:1–6. doi: 10.1016/j.bjid.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan YJ, Lin T-L, Chen C-T, Chen Y-Y, Hsieh P-F, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 2015;5:15573. doi: 10.1038/srep15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordon D. The diversity of lipopolysaccharide (o) and capsular polysaccharide (K) antigens of invasive Klebsiella pneumoniae in a Multi-Country collection. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae . Future Microbiol. 2014;9:1071–1081. doi: 10.2217/fmb.14.48. [DOI] [PubMed] [Google Scholar]
- 67.Liu B-T, et al. Characteristics of carbapenem-resistant Enterobacteriaceae in ready-to-eat vegetables in China. Front Microbiol. 2018;9:1147. doi: 10.3389/fmicb.2018.01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mei Y, et al. Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM–5 in China. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong N, Yang X, Zhang R, Chan EW-C, Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect. 2018;7:146. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Korotetskiy IS. Differential gene expression and alternation of patterns of DNA methylation in the multidrug resistant strain Escherichia coli ATCC BAA-196 caused by iodine-containing nano-micelle drug FS-1 that induces antibiotic resistance reversion. bioRxiv. 2020 [Google Scholar]
- 71.Decano AG. Plasmids shape the diverse accessory resistomes of Escherichia coli ST131. bioRxiv. 2020 doi: 10.1099/acmi.0.000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugawara Y, Akeda Y, Sakamoto N, Takeuchi D, Motooka D, et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One. 2017;12:e0184720. doi: 10.1371/journal.pone.0184720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali Hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother. 2014;58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du H, Chen L, Chavda KD, Pandey R, Zhang H, et al. Genomic characterization of Enterobacter cloacae isolates from China that Coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother. 2016;60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poirel L, Aires-de-Sousa M, Kudyba P, Kieffer N, Nordmann P. Screening and characterization of multidrug-resistant gram-negative bacteria from a remote African area, São Tomé and Príncipe. Antimicrob Agents Chemother. 2018;62:e01021–18. doi: 10.1128/AAC.01021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, et al. Escherichia coli sequence type 410 is causing new International high-risk clones. mSphere. 2018;3:e00337-18–18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dortet L, Flonta M, Boudehen Y-M, Creton E, Bernabeu S, et al. Dissemination of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in Romania. Antimicrob Agents Chemother. 2015;59:7100–7103. doi: 10.1128/AAC.01512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lomonaco S, Crawford MA, Lascols C, Timme RE, Anderson K, et al. Resistome of carbapenem- and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One. 2018;13:e0198526. doi: 10.1371/journal.pone.0198526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muggeo A, Guillard T, Klein F, Reffuveille F, François C, et al. Spread of Klebsiella pneumoniae ST395 non-susceptible to carbapenems and resistant to fluoroquinolones in north-eastern France. J Glob Antimicrob Resist. 2018;13:98–103. doi: 10.1016/j.jgar.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 80.Bedenić B, Slade M, Starčević Lidija Žele, Sardelić S, Vranić-Ladavac M, et al. Epidemic spread of OXA-48 beta-lactamase in Croatia. J Med Microbiol. 2018;67:1031–1041. doi: 10.1099/jmm.0.000777. [DOI] [PubMed] [Google Scholar]
- 81.Conlan S, et al. Plasmid dynamics in KPC-Positive <span class="named-content genus-species" id="named-content-1">Klebsiella pneumoniae</span> during long-term patient colonization. mBio. 2016;7:e00742–16. doi: 10.1128/mBio.00742-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ashcroft MM, et al. Strain and lineage-level methylome heterogeneity in the multi-drug resistant pathogenic Escherichia coli ST101 clone. bioRxiv. 2020 [Google Scholar]
- 83.Huang Y, Li G, Li M, Wang Y, Yang Z. The high-risk KPC-producing Klebsiella pneumoniae lack type I R-M systems. Int J Antimicrob Agents. 2020;106050 doi: 10.1016/j.ijantimicag.2020.106050. [DOI] [PubMed] [Google Scholar]
- 84.Bao J, Wu N, Zeng Y, Chen L, Li L, et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae . Emerg Microbes Infect. 2020;9:771–774. doi: 10.1080/22221751.2020.1747950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.CLSI & Clinical and Laboratory Standards Institute (CLSI) Twenty-Seventh Informational Supplement M100-S27. USA: CLSI, Wayne, PA; 2019. Performance standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 86.Iraz M, Özad Düzgün A, Sandallı C, Doymaz MZ, Akkoyunlu Y, et al. Distribution of β-lactamase genes among carbapenem-resistant Klebsiella pneumoniae strains isolated from patients in turkey. Ann Lab Med. 2015;35:595–601. doi: 10.3343/alm.2015.35.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 89.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Larsen M, V, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruan Z, Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44:682–687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends in Microbiology. 2016;24 doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 98.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.