Abstract
Introduction
Childhood IgA nephropathy (cIgAN) is a primary glomerulonephritis clinically characterized by microscopic hematuria and proteinuria, the presence of which may potentially overlap with Alport syndrome. Interestingly, earlier studies suggested that familial IgAN could be linked to the chromosome 2q36 region, also the coding region for collagen type 4 alpha 3/4 (COL4A3/A4).
Methods
To investigate a possible relationship or phenocopy between Alport syndrome and cIgAN, COL4A3, COL4A4, and COL4A5 exons were sequenced in 36 cIgAN patients. Clinical data and treatment were collected retrospectively. COL4A3/A4/A5 variants were classified according to American College of Medical Genetics and the Association for Molecular Pathology (ACMG/AMP) guidelines.
Results
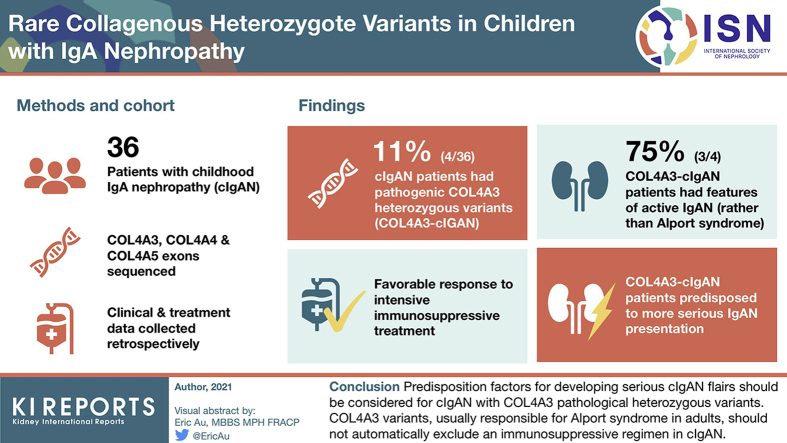
Four of 36 cIgAN patients were affected by ACMG class 4/5 COL4A3 heterozygous variants (COL4A3-cIgAN). We found no COL4A4 or COL4A5 variant. Despite having rare and deleterious COL4A3 variants, 3 of 4 COL4A3-cIgAN children developed clinical and biologic features of active IgAN rather than Alport syndrome. Response to intensive immunosuppressive treatment was favorable, leading to a reduction of endocapillary and extracapillary proliferation lesions. High levels of immune immunoglobulin G and A (IgG/IgA) complexes, reduction of proteinuria, and gradual stabilization of estimated glomerular filtration rate (eGFR) argued against Alport syndrome. Nevertheless, COL4A3-cIgAN patients seemed predisposed to a more serious IgAN presentation compared with the non‒COL4A3-cIgAN group, with more glomerulosclerosis and a lower eGFR over time. One of the 4 patients underwent kidney transplant with subsequent IgAN recurrence.
Conclusions
Predisposition factors for developing serious cIgAN flare-up should be considered for cIgAN with COL4A3 pathologic heterozygous variants. COL4A3 variants, usually responsible for Alport syndrome in adults, should not automatically exclude an immunosuppressive regimen in cIgAN. Moreover, evidence of an ACMG class 4/5 COL4A3 variant in early-stage cIgAN could be a helpful tool for stratifying severity of cIgAN beyond the Oxford classification.
Keywords: Alport syndrome, children, COL4A3 variants, IgA nephropathy, immunosuppressive treatment, Oxford classification
Graphical abstract
Childhood IgA nephropathy (cIgAN) is one of the most common types of glomerulonephritis worldwide.1 Since IgA familial nephropathy has been described,2 predisposing genetic factors have been suggested to play a major role in the development of IgAN. IgAN usually develops in a sporadic form, although familial-related cases have been reported. With the application of genome-wide association studies (GWAS) in large IgAN cohorts compared with controls, the understanding of an IgAN genetic background has improved, despite being related to several loci.
So far, GWAS has identified several susceptibility loci, such as major histocompatability locus, HORMAD2, complement factor H cluster, tumor necrosis factor ligand superfamily member 13 coding for APRIL, and CARD9.3, 4, 5, 6, 7, 8 Three loci could have genetic susceptibility for familial IgAN with IgAN 1 locus on chromosome 6q22-23, IgAN 2 locus on chromosome 4q26-31, and IgAN 3 locus on chromosome 17q12-22.9,10 Interestingly, another locus, 2q36, was identified in a Canadian cohort,3 which is also the coding locus for collagen type 4 alpha 3 or 4 (COLA4 and COL4A3, respectively) implicated in Alport syndrome.
Whole genome or whole exome sequencing, as opposed to GWAS, aims to discover rare or private variants that could be associated with rare phenotypes, such as rare diseases, and this approach could be of interest in familial forms of IgAN.9, 10, 11, 12 IgAN and Alport syndrome could have the same clinical presentation (hematuria, proteinuria, end-stage renal disease [ESRD]), representing potential phenocopies. Moreover, a causal link between these 2 diseases has been proposed recently.13 Intriguingly, glomerular basement membrane injuries (double contours, gaps, thinning with a lamellar and reticular structure, rupture) have been described in both IgAN and Alport syndrome.14,15 Twenty-six years ago, Berthoux et al. studied a cohort of 59 IgAN patients in which 23 patients had a thin glomerular membrane.16
The aim of this work was to study COL4A3, COL4A4, and COL4A5 coding variation in a cIgAN cohort. Genetic variants were classified according to the guidelines of the American College of Medical Genetics and the Association for Molecular Pathology (ACMG/AMP).
Methods
Patients
This longitudinal, multicentric, prospective study was undertaken at 3 university hospitals in Paris (Necker Enfants Malades, Robert-Debré Hôpital, and Trousseau Hôpital) and included 36 consecutive cases of children and adolescents recorded between 1990 and 2019, all <18 years old and newly diagnosed with primary IgAN after renal biopsy. These 36 patients, for whom genetic analysis had been performed, were taken from our cohort of 82 patients, as described elsewhere.17 The study was approved by the ethics committee of St Antoine Hôpital, Paris, France. All parents and children in this study provided informed written consent.
Diagnosis of IgAN was based on immunofluorescence microscopy findings, showing mesangial deposition of IgA as the predominant or codominant immunoglobulin. Patients with systemic diseases, such as systemic lupus erythematosus, Henoch-Schoönlein purpura, or chronic liver disease, were excluded.
Clinical Data Set
The following clinical and biologic parameters were collected for each patient at the time of their renal biopsy and at time of immune complex measurement: age, sex, weight, height, clinical presentation at disease onset, time from onset to renal biopsy, presence of a macroscopic hematuria at diagnosis, history of tonsillectomy, familial IgAN history (either proven by renal biopsy or suspected because of a medical history of microscopic and/or macroscopic hematuria in a next-of-kin), systolic and diastolic blood pressure, serum albumin, serum IgA levels, serum creatinine (expressed in micromoles per liter), estimated glomerular filtration rate (eGFR), urine protein-to-creatinine ratio (in grams per millimole, used as an estimation of 24-hour protein excretion adjusted for body surface area).
The eGFR was calculated using the updated bedside Schwartz equation: creatinine clearance (ml/min per 1.73 m2) = K × L / sCr, where L is body length (cm), K = 41.3, and sCr is serum creatinine.18,19
A maximum eGFR set at 120 ml/min per 1.73 m2 was selected because the accuracy of eGFR for higher values is poor and also to avoid a disproportionate impact of small variations in creatinine on the rate of renal function.19
Nephrotic syndrome (NS) was defined as urinary protein excretion ≥ 3 g/g creatinine and serum albumin < 3 g/dL. Acute renal failure was defined as eGFR < 70 ml/min per 1.73 m2 with proteinuria and/or hematuria. ESRD was defined as eGFR < 15 ml/min per 1.73 m2 or the need for renal replacement therapy at diagnosis. Microscopic hematuria was diagnosed when erythrocytes reached >5000/ml and macroscopic hematuria was defined as the presence of gross hematuria. Significant relapse was defined by significant proteinuria > 0.1 g/mmol and/or acute renal failure (increased creatinine by >20%) with hematuria.
There are 3 key factors in the pathogenesis of IgAN: galactose-deficient IgA1 (Gd-IgA1), anti-IgA autoantibodies, and soluble (s) CD89 IgA Fc receptor.20 These molecules form circulating immune complexes that have been identified as potential markers for adult IgAN.21,22 To assess more precisely the IgAN diagnosis, the biomarkers Gd-IgA1, immune complexes (sCD89-IgA1, IgG-IgA), and sCD89 were measured in this cohort.
Gd-IgA1, sCD89, and IgA Complex Measurements
For measurement of Gd-IgA1 levels, an enzyme-linked immunoassay was performed using an anti‒Gd-IgA1 assay (KM55 Assay Kit; IBL, Amsterdam, The Netherlands), according to the manufacturer’s instructions, with a 1:200 plasma dilution. For measurement of free and unbound sCD89, MIP8a anti-human CD89 (1:500; Thermo Fisher, Waltham, MA) was used for coating overnight at 4 °C, followed by blocking for 4 hours at 37 °C and overnight incubation with a 1:5 dilution of plasma at 4 °C. Recognition of free sCD89 was performed using biotinylated monoclonal antibody anti-sCD89 (5 μg/L, clone A77),23 with secondary streptavidin‒horseradish peroxidase (HRP) plus chromogen (TMB) solution. The optic density was read at 450 nm.
For the IgA immune complex measurements, the enzyme-linked immunoassay was performed as follows: coating with goat anti-IgA cross-adsorbed antibody (1:500, in phosphate-buffered saline; Bethyl Laboratories, Montgomery, TX) or with anti-sCD89 (5 μg/ml, clone A3)24 for 4 hours at 37 °C, followed by blocking for 4 hours at 37 °C. Plasma samples (1:5 dilution) were incubated overnight in a 1:10 dilution at 4 °C.
Revelation was performed using an anti-IgG antibody (1:5000; Southern Biotech, Birmingham, AL) coupled with HRP or an anti-IgA antibody (1:2000; BD Biosciences, Le Pont de Claix, France) coupled with alkaline phosphatase, respectively, for IgA-IgG or sCD89-IgA complex detection. The substrates were HRP plus chromogen solution or Sigma fast p-nitrophenylphosphate tablets (Sigma-Aldrich, St Louis, MO). The optical density was read at 405 nm for alkaline phosphatase or 450 nm for HRP.
Histopathology
In this analysis, all biopsy samples were reanalyzed. Some specimens were could not be assessed for all histologic variables. We studied variables of the Oxford classification using different definitions. We studied 5 variables: mesangial hypercellularity (M), scored 1 (M1) if ≥50% of glomeruli had more than 3 cells per mesangial area; segmental glomerulosclerosis or adhesion (S), scored as 0 (S0) if absent or as 1 (S1) if present; endocapillary hypercellularity (E), scored as 0 (E0) if absent or as 1 (E1) if present; extracapillary proliferation (C), including cellular, fibrocellular, or fibrous crescent, scored as 0 (C0) if absent or as 1 (C1) if present; tubular atrophy/interstitial fibrosis (T), modified from the Oxford classification, scored as 1 (T1) if present at between 10% and 25% of the relevant biopsy core and scored as 2 (T2) if present in ≥25% of the relevant biopsy core.
Sequencing and Genetic Analysis of COL4A Genes
We sequenced COL4A3, COL4A5, and COL4A4 coding sequences with targeted capture sequencing (MiSeq; Illumina, San Diego, CA) in cIgAN patients and in both parents when available. For the exome study, genomic DNA was isolated from blood samples, and exons (33 megabases) were captured with the Twist Human Core Exome kit (Twist Bioscience, South San Francisco, CA) and sequenced on a NovaSeq 6000 instrument on Illumina platforms. Data were analyzed using dedicated software (SeqOne, Montpellier, France) to identify diagnostic variants for patients’ renal disease. We used the Geneome Aggregation Database version 2.1 to estimate allele frequency. For the exome data analysis, the assessment was restricted to COL4A3, COL4A5, and COL4A4 genes for consistent comparison with the initial exon capture. Variants were classified according to ACMG/AMP guidelines, whereas collagen variants were classified “pathogenic” or “likely athogenic.”25 Only variant class 4/5 is described in this study.
Statistical Analysis
Median (interquartile range) values and frequencies are provided for description of continuous and categorical variables, respectively. The Shapiro-Wilk test was used to test the normality of the data distribution for the t test. The mean values and proportions of 2 groups were compared using Mann-Whitney and Fisher exact tests, respectively. All tests were 2-sided, and P < 0.05 was considered statistically significant. All statistical analyses were performed with GraphPad Prism version 7 (GraphPad, La Jolla, CA) and SPSS version 25 (IBM Corp, Armonk, NY).
Results
Analysis of COL4A3/A4/A5 Variants in cIgAN Patients
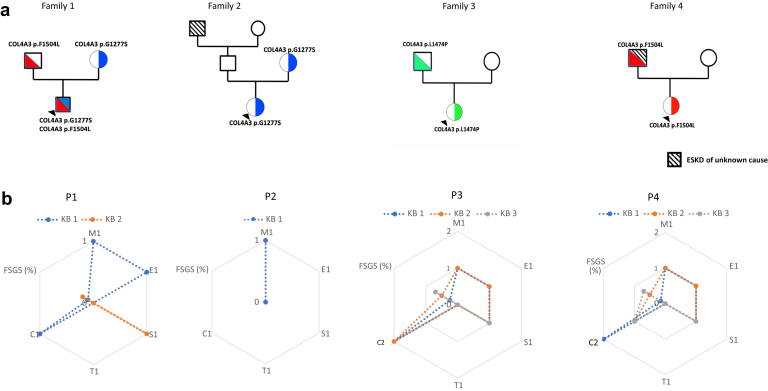
We identified pathogenic or likely pathogenic inherited COL4A3 variants in 4 of 36 patients (11.1%). We found no variants in the COL4A4 and COL4A5 genes. Variation in COL4A3 was defined according to the ACMG classification, as indicated previously. No patients were heterozygous carriers of variants in the COL4A3 gene (families 2, 3, and 4), but 1 patient had an additional COL4A3 variant in trans corresponding to a heterozygous compound equivalent to a recessive disease (family 1) (Table 1 and Figure 1).
Table 1.
Genetic, clinical, biologic, and histologic description of 4 COL4A-cIgAN patients at onset and last follow-upa
| Onset | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Gene p.hgvs | COL4A3, p.G1277S, p.F1504L | COL4A3 p.G1277S | COL4A3 p.L1474P | COL4A3 p.F1504L |
| Sex | Male | Female | Female | Female |
| Familial history | No | Grandfather ESRD | No | Father ESRD |
| Age (years) | 8 | 6 | 14 | 10 |
| Albumin (g/L) | 28 | 36.4 | 15.10 | 29 |
| Proteinuria (g/mmol) | 0.40 | 0.021 | 1.57 | 0.644 |
| eGFR (ml/min per 1.73 m2) | 35.02 | 119.08 | 30.00 | 99.40 |
| Hematuria (cpm/ml) | 106 | 104 | 106 | 106 |
| Oxford classification | M1,E1,S0,T0,C1 | M1,E0,S0,T0,C0 | M1,E1,S1,T0,C2 | M1,E1,S1,T0,C2 |
| Numbers of biopsies | 2 | 1 | 3 | 3 |
| Total treatment | RASB, steroid + i.v. methylprednisolone + i.v. cyclophosphamide | RASB | RASB, steroid + i.v. methylprednisolone + i.v. cyclophosphamide + 10 plasma exchanges | RASB, steroid + i.v. methylprednisolone + i.v. cyclophosphamide |
| Last follow-up before kidney transplantation | ||||
| Follow-up (years) | 8.91 | 5 | 7 | 4 |
| Albumin (g/L) | 41.4 | 49.4 | 43.5 | 36 |
| Proteinuria (g/mmol) | 0.069 | 0.003 | 0.23 | 0.13 |
| eGFR (ml/min per 1.73 m2) | 50 | 100 | 30 | 78 |
| Hematuria (cpm/ml) | <1000 | <1000 | 10,000 | 500,000 |
| Kidney transplantation | No | No | Yes, IgAN recurrence treated | no |
C1, presence of extracapillary proliferation; COL4A3/4, collagen type 4 alpha 3/4; E1, presence of endocapillary hypercellularity; eGFR, estimated glomerular filtration rate; ESRD, end-stage renal disease; IV, intravenous; M1, presence of mesangial hypercellularity; RASB, renin‒angiotensin system blockade; S1, presence of segmental glomerulosclerosis or adhesion; T1, presence of tubular atrophy/interstitial fibrosis.
Gene description of variants according to their p.hgvs for NP_000082.2.
Figure 1.
Description of 4 COL4A3-cIgAN families with the 4 cIgAN (P1‒4). (a) Family trees. (b) Evolution of repeated kidney biopsy for patients 1 to 4 (P1‒P4). C1, presence of extracapillary proliferation; E1, presence of endocapillary hypercellularity; ESKD, end-stage kidney disease; FSGS, focal segmental glomerulosclerosis; M1, presence of mesangial hypercellularity; RASB, renin–angiotensin system blockers; S1, presence of segmental glomerulosclerosis or adhesion; T1, presence of tubular atrophy/interstitial fibrosis.
The patients did not have obvious deafness or known ophthalmologic involvement. No consanguinity had been documented in these families. Two patients were from North Africa and and 2 were Caucasian.
COL4A3 protein was affected either on glycine residue (p.G1277S) affecting a G‒XY‒R triplet motif on the collagenous domain, or on amino acid located in the NC1 domain as p.F1504L and p.L1474P. Patient 4 (family 4) showed inheritance of a COL4A3 p.F1504L variant from his father who had developed ESRD at 40 years of age. This variant was also present in family 1 without a history of ESRD, but, despite this, patient 1 (family 1) developed a severe form of IgAN, as is also seen in patient 4. The former COL4A3 p.G1277S variant was also present in patient 2 (family 2) but with a milder phenotype of renal disease at onset. Taken together, this suggests that p.G1277S and p.F1504L could be associated with a significant part of the phenotype seen in these 4 patients (Table 1), phenocopying Alport syndrome.
At the onset of the disease, 3 of 4 patients with COL4A3 presented with NS, including 2 patients with low eGFR. All patients had macroscopic hematuria. Patients with NS have the full spectrum of proliferative lesions according to the Oxford classification (M1, E1, and C1‒C2 lesions). No patient had tubular atrophy/interstitial fibrosis (T1 lesion) and 2 patients had segmental glomerulosclerosis or adhesion (S1 lesion). No biopsy was assessed using electron microscopy. Figure 1 and Table 1 show patient pedigree and phenotype data.
The patients with synpharyngitic hematuria and NS were treated with an immunosuppressive regimen comprised of pulse steroid therapy (i.v. methylprednisolone 500 mg/m2 or 1000 mg/1.73 m2 for 3 consecutive days) followed by oral steroid therapy for between 12 and 18 months. If there was significant extracapillary proliferation on kidney biopsy (C1‒C2), cyclophosphamide (1, 3, or 6 i.v. pulses of 600 or 700 mg/m2) could be added to the immunosuppressive regimen. One patient underwent plasma exchanges. All patients received renin‒angiotensin system blockade (RASB) (i.e., with angiotensin-converting enzyme inhibitors and/or angiotensin receptor blocker) (Table 1).
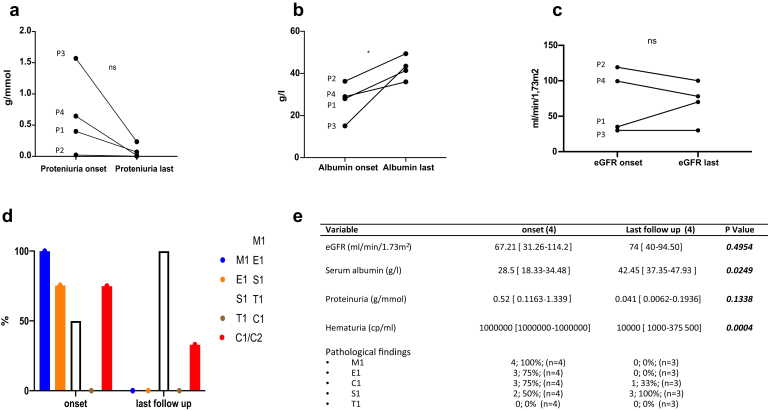
During follow-up, 3 patients had repeat renal biopsies. Descriptions of the evolution of kidney biopsies are given in Figure 1 and Table 1. The decision to perform repeat biopsy was based on clinical judgment and not on a pre-established protocol. All kidney biopsies showed IgA deposits. The evolution of biologic and histologic characteristics is shown in Figure 2 and Table 1. We observed a diminution of proteinuria (Figure 2a) and an increase in serum albumin (Figure 2b) in the 4 patients. For patient (P)1 and P3, eGFR remained stable or increased, whereas it decreased in P2 and P4 (Figure 2c). Repeat kidney biopsy showed a final reduction of active lesions and a progression of glomerulosclerosis (Figure 2d,e).
Figure 2.
Evolution of biologic and histologic characteristics COL4A3-cIgAN (P1‒P4) between onset and last follow-up (before kidney transplantation). (a) Serum albumin at onset and last follow-up. (b) Proteinuria at onset and last follow-up. (c) Estimated glomerular filtration rate (eGFR) at onset and last follow-up. (d) Evolution of histologic lesions. (e) Table of clinical, biologic, and histologic characteristic of COL4A3-cIgAN. For quantitative variables, values are expressed as median [interquartile range]. For qualitative variables, values are expressed as number (%). C1/C2, presence of extracapillary proliferation; E1, presence of endocapillary hypercellularity; M1, presence of mesangial hypercellularity; S1, presence of segmental glomerulosclerosis or adhesion; T1, presence of tubular atrophy/interstitial fibrosis.
Despite the fact that patient 1 (family 1) presented with NS and acute renal failure at onset and with a COL4A3 heterozygous compound mutation, equivalent to recessive disease, eGFR improved significantly after immunosuppressive therapy (oral steroids, 3 perfusions of methylprednisolone, and 1 injection of cyclophosphamide). The first renal biopsy showed M1,E1,S0,T0,C1. A second kidney biopsy was performed for persistent mild proteinuria (0.1 g/mmol) without hematuria (0 cpm/ml). Optical microscopy showed an S1 lesion according to the Oxford classification (M0,E0,S1,T0,C0). He was eventually treated with RASB. During follow-up he had no further relapses and proteinuria remained low. At the last follow-up, there was no microscopic hematuria, with minimal proteinuria (0.07 g/mmol) and an eGFR 50 ml/min per 1.73 m2.
Patient 2 had the least severe presentation. She had at least 1 COL4A3 variant, similar to family 1 (p.G1277S), with a clearly different evolution of renal disease. The first kidney biopsy was performed due to the recurrence of macroscopic hematuria with normal eGFR and low proteinuria (0.021 g/mmol). Kidney biopsy showed only M1. She received only RASB treatment. At last follow-up, eGFR was normal with no proteinuria and no hematuria.
Patient 3 presented a more severe clinical (including NS with acute renal failure) and histologic (M1,E1,S1[15%],T0,C2) profile compared with the other families reported. She was initially treated with pulse IV and oral steroids with cyclophosphamide and plasma exchange. Evolution of the renal disease was marked by eGFR deterioration with a full spectrum of proliferative lesion persistence as per the Oxford classification, with occurrence of S1 lesions during a second kidney biopsy (M1,E1,S1[60%],T0,C2). This led to continuation of treatment with 5 additional cyclophosphamide injections and 10 plasma exchanges. A third kidney biopsy 3 months after the end of the cyclophosphamide cycle for therapeutic evaluation showed persistence of the M1 lesion and an extensive S1 lesion, whereas E1 and C1 were no longer apparent (M1,E0,S1[80%],T0,C0). Due to extensive S1, immunosuppressive treatment was then stopped and eGFR eventually declined. Patient 3 reached ESRD and eventually received a pre-emptive kidney transplant from a deceased donor 6 years after the initial diagnosis and flare-up of IgAN. She presented with histologic and clinical IgAN recurrence 8 months after her kidney transplant with acute renal failure and proteinuria (0.2 g/mmol) without hematuria. The kidney graft biopsy showed glomerulitis (g1 according to the Banff lesion score) and endocapillary and extracapillary proliferation. Immunofluorescence microscopy showed mesangial IgA deposits, and C4d staining was negative. The patient received 3 perfusions of methylprednisolone with 10 plasma exchanges and 1 injection of rituximab at 375 mg/m2 (Figure 1 and Tables 2 and 3). At last follow-up after IgAN recurrence, we observed amelioration of eGFR (77 ml/min per 1.73 m2) and proteinuria (0.1 g/mmol).
Table 2.
Comparison of clinical, biologic, and immunologic characteristics between COL4A3-cIgAN and non‒COL4A3-cIgAN patients at onset and last follow-up (before kidney transplantation)
| COL4A3-cIgAN (4) | Non‒COL4A3-cIgAN (32) | P value | |
|---|---|---|---|
| Variables at onset | |||
| Age (years) | 9 (6.5–13) | 11.10 (8.66–15.12) | |
| Familial IgAN history | 2 (50%) | 4 (10%) | |
| Female | 3 (75%) | 11 (34%) | |
| eGFR (ml/min per 1.73 m2) | 67.21 (31.26–114.2) | 88.44 (78.88–104.1) | 0.1418 |
| Serum albumin (g/L) | 28.5 (18.33–34.48) | 36.60 (30.55–40) | 0.0424 |
| Proteinuria (g/mmol) | 0.52 (0.1163–1.339) | 0.125(0.04–0.3375) | 0.003 |
| IgA1 | 2.62 (2.038–3.3) | 2.6 (2.038–3.300) | 0.2734 |
| Gd-IgA1 (μg/ml) | 6.139 (1.884–6.282) | 4.866 (2.856–7.378) | 0.7061 |
| sCD89-IgA (OD) | 0.441 (0.0259–0.9377) | 0.08 (0–0.2893) | 0.2758 |
| IgG-IgA (OD) | 0.1831 (0.0636–0.2606) | 0.1145 (0.04195–0.2379) | 0.6854 |
| Free sCD89 (μg/ml) | 0.5373 (0.08–1.236) | 0.4282 (0.1508–0.6710) | 0.6789 |
| Pathologic findings | |||
| M1 | 4; 100% (n = 4) | 25 80% (n = 31) | |
| E1 | 3; 75% (n = 4) | 27 87% (n = 31) | |
| C1 | 3; 75% (n = 4) | 15; 48% (n = 31) | |
| S1 | 2; 50% (n = 4) | 26; 80% (n = 31) | |
| T1 | 0; 0% (n = 4) | 2; 6% (n = 40) | |
| Variables at last follow-up | |||
| eGFR (ml/min per 1.73 m2) | 90 (57–117.5) | 105.4 (99.49–120.0) | 0.027 |
| Serum albumin (g/L) | 42.45 (37.35–47.93) | 36.5 (27.03–37.95) | 0.0696 |
| Proteinuria (g/mmol) | 0.041 (0.0062–0.936) | 0.03 (0.0100–0.060) | 0.3415 |
C1, presence of extracapillary proliferation; cIgAN, childhood IgA nephropathy; COL4A3/4, collagen type 4 alpha 3/4; Gd-IgA1, galactose-deficient IgA1; sCD89-IgA and IgG-IgA, anti-IgA autoantibodies; sCD89, soluble CD89 IgA Fc receptor; E1, presence of endocapillary hypercellularity; eGFR, estimated glomerular filtration rate; M1, presence of mesangial hypercellularity; S1, presence of segmental glomerulosclerosis or adhesion; T1, presence of tubular atrophy/interstitial fibrosis.
Data expressed as median (interquartile range) for quantitative variables and as number (%) for qualitative variables.
Table 3.
Comparison of clinical and biologic characteristics between COL4A3-cIgAN and non‒COL4A-cIgAN RKB patients at onset and last follow-up (before kidney transplantation)
| COL4A3-cIgAN (4) | Non‒COL4A3-cIgAN RKB (5) | P value | |
|---|---|---|---|
| Variables at onset | |||
| Age (years) | 9 (6.5‒13) | 11.10 (8.669‒15.12) | |
| Familial IgAN history | 2 (50%) | 0 (0%) | |
| Female | 3 (75%) | 3 (60%) | |
| eGFR (ml/min per 1.73 m2) | 67.21 (31.26–114.2) | 1131 (63.88–113.8) | 0.3772 |
| Serum albumin (g/L) | 28.5 (18.33–34.48) | 30 (26–44.1) | 0.5556 |
| Proteinuria (g/mmol) | 0.52 (0.1163–1.339) | 0.1074 (0.049–0.1935) | 0.1905 |
| Pathologic findings | |||
| M1 | 4; 100% (n = 4) | 3; 60% (n = 5) | |
| E1 | 3; 75% (n = 4) | 5; 100% (n = 5) | |
| C1 | 3; 75% (n = 4) | 4; 80% (n = 5) | |
| S1 | 2; 50% (n = 4) | 4; 80% (n = 5) | |
| T1 | 0; 0% (n = 4) | 2; 40% (n = 5) | |
| Variables at last follow-up | |||
| eGFR (ml/min per 1.73 m2) | 90 (57–117.5) | 77 (30–85) | 0.3091 |
| Serum albumin (g/L) | 42.45 (37.35–47.93) | 32.3 (30.50–39.10) | 0.0635 |
| Proteinuria (g/mmol) | 0.041 (0.0062–0.936) | 0.2 (0.1333–0.25) | 0.3810 |
| Pathologic findings | |||
| M1 | 0; 0% (n = 3) | 4; 80% (n = 5) | |
| E1 | 0; 0% (n = 3) | 5; 100% (n = 5) | |
| C1 | 1; 33% (n = 3) | 1; 20% (n = 5) | |
| S1 | 3; 100% (n = 3) | 3; 60% (n = 5) | |
| T1 | 0; 0% (n = 4) | 2; 40% (n = 5) |
C1, extracapillary proliferation; cIgAN, childhood IgA nephropathy; COL4A3/4, collagen type 4 alpha 3/4; E1, endocapillary hypercellularity; eGFR, estimated glomerular filtration rate; M1, mesangial hypercellularity; RKB, repeat kidney biopsy; S1, segmental glomerulosclerosis or adhesion; T1, tubular atrophy/interstitial fibrosis.
For quantitative variables, values are expressed as median (interquartile range). For qualitative variables, values are expressed as number (%).
Patient 4 also had 1 COL4A3 variant, similar to that seen in family 1 (p.F1504L), but different from that in family 2. Patient 4 also had a serious evolution of renal disease, as in family 1. Like patient 1 at onset, patient 4 presented with significant proliferative lesions (M1,E1,S1,T0,C2). She was treated initially by oral steroids and 3 perfusions of methylprednisolone. In family 4, a second biopsy was performed for NS relapse with macroscopic hematuria showing M1,E1,C1 lesion persistence with an increased proportion of S1 lesions (M1,E1,S1[50%],T0,C1) (Table 2). Treatment was done with oral steroids and IV methylprednisolone for 3 consecutive days. Remission of NS was eventually achieved. Six months later, another NS relapse with gross hematuria led to a third biopsy, which showed continued C1 lesions with extensive S1 lesions (>50%) (M0,E0,S1,T0,C1). She was treated with 6 injections of cyclophosphamide. The most recent data showed no relapse, with a persistence of microscopic hematuria and proteinuria (0.13 g/mmol). As a secondary outcome, eGFR decreased but remained stable from the outset (78 ml/min per 1.73 m2).
Comparison of COL4A3-cIgAN and Non‒COL4A3-cIgAN at Onset
Thirty-two children were included in the non‒COL4A3-cIgAN group. Age at diagnosis was 9 (range, 6.5‒13) years in the COL4A3-cIgAN group and 11.10 (range, 8.66‒15.12) years in the non‒COL4A3-cIgAN group. Two patients (50%) had a suspected or proven family history of IgAN COL4A3-cIgAN compared with 4 patients (10%) in the non‒COL4A3-cIgAN group. Proteinuria was higher and serum albumin level lower in the COL4A3-cIgAN cohort compared with the non‒COL4A3-cIgAN cohort (proteinuria: 0.52 vs. 0.125 g/mmol of creatinuria, P = 0.003; serum albumin: 28.5 vs. 36.60 g/dL, P = 0.0043). No difference was observed for eGFR, but eGFR was even lower in the COL4A3-cIgAN group (67.2 vs. 88.44 ml/min per 1.73 m2, P = 0.1418) (Table 2).
We analyzed total IgA1 and immune complexes Gd‒IgA1, sCD89, sCD89‒IgA, and IgG‒IgA,21,26,27 and no difference was observed between the COL4A3-cIgAN and non‒COL4A3-cIgAN groups (Table 2).
Analysis of the kidney biopsy findings revealed the same relative proportion of M1,E1,C1,S1,T1 in the COL4A3-cIgAN group when compared with the non‒COL4A3-cIgAN group (Table 2).
Comparison of COL4A3-cIgAN and Non‒COL4A3-cIgAN at Last Follow-up
We compared the evolution at last follow-up for the COL4A3-cIgAN (before P3 kidney transplantation) patients and the 32 non‒COL4A3-cIgAN patients. With a median follow-up of 5.5 years and 6.88 years, respectively, we found that eGFR was lower in the COL4A3-cIgAN group compared with the non‒COL4A3-cIgAN group (90 vs. 105.4 ml/min per 1.73 m2, P = 0.027).
The degrees of proteinuria (0.041 vs. 0.03 g/mmol, P = 0.3415) and serum albumin (42.45 vs. 36.5 g/L, P = 0.0696) were not different between the 2 groups (Table 2).
Comparison of COL4A3-cIgAN and Non‒COL4A3-cIgAN With Repeat Kidney Biopsy Group
We decided to compare COL4A3-cIgAN vs. severe non‒COL4A3-cIgAN patients who received repeat kidney biopsy (RKB). The RKB group was composed of 5 cIgAN patients who received repeat biopsies for serious relapses (defined earlier). There were no biologic differences between the 2 groups (Table 3). Two children from the RKB cohort initially presented with nephrotic syndrome and 2 with acute renal failure, whereas there were 3 patients from the COL4A3-cIgAN group with nephrotic syndrome and 2 with acute renal failure.
On first biopsy, there were no histologic differences between the 2 groups. The proportions of M1, E1, C1, and S1 was the same, except for T1 (absent in COL4A3-cIgAN) (Table 3).
There was a difference, however, for the last kidney biopsy. For COL4A3-cIgAN, M1 and E1 were absent and C1 had diminished, whereas, in the RKB group, the M1 proportion had increased, E1 remained the same, and C1 was reduced.
COL4A3-cIgAN demonstrated a more significant progression to glomerulosclerosis compared with the RKB group. One patient in the COL4A3-cIgAN group and 2 in the RKB group progressed to ESRD (Table 3). One patient each from the COL4A3-cIgAN and RKB-cIgAN groups underwent kidney transplantation.
Discussion
To our knowledge, this is the first report of collagen variant sequencing in cIgAN in pediatric patients. We found 11% of collagen variants in this cohort of 36 patients. Because Alport syndrome can phenocopy IgAN,29 it seems important to distinguish these 2 disease entities to allow better treatments and avoid potential aggressive immunosuppressive treatments in Alport syndrome patients. Recently, a type IV collagen mutation was described in adult familial IgAN, suggesting a possible Alport family with IgA deposits.30 Unfortunately, a detailed histopathology was not available for the families reported in the study. Interestingly, as far back as 1995, Berthoux et al. also described an IgAN cohort, which showed a notable proportion (40%) of patients with a thin glomerular membrane, indicating either a specific subgroup (with an unfavorable prognosis) or an association with thin membrane nephropathy.16
Herein we have described 4 cIgAN with ACMG class 4/5 COL4A3 heterozygous variants with clinical and biologic biomarkers and histologic characteristics that, in retrospect and given their genetic status, had a heavy immunosuppressive regimen.
In COL4A3-cIgAN, the clinical, biologic, and---most importantly---the histologic characteristics, corresponded to the clinical presentation of IgAN for 3 patients with highly active disease. The cIgAN patient 2/family 2 with p.G1277S had a presentation featuring macroscopic hematuria and slight proteinuria that is potentially compatible with Alport syndrome. Low grade of proliferation, low Gd‒IgA1 level, and absence of plasma immune complexes were observed. At the last follow-up, patient 2 had only microscopic hematuria, with a normal eGFR.
Families 1, 3, and 4 showed a high proportion of endocapillary and extracapillary proliferation, which is not usually described in Alport syndrome. Patients with the COL4A3 ACMG class 4/5 variant obviously responded to an aggressive immunosuppressive regimen with reverse endocapillary and extracapillary proliferation on kidney biopsy and a decrease of proteinuria, increase of albumin serum, and eGFR stabilization. This does not, however, rule-out worse renal function in adulthood.
The evolution and variation of histologic proliferation of lesions on RKBs and the response to immunosuppression do not support Alport syndrome when the form is indolent.
Moreover, detection of high levels of Gd-IgA1, IgG-IgA, sCD89-IgA, and free sCD89 was observed and supported the IgAN diagnosis. These immune complexes are specific to IgAN.21,28 The clinical and histologic evolution together with the immunosuppression response and detection of a high level of specific immune complexes (Gd-IgA1, IgG-IgA, sCD89-IgA, and free sCD89) may support a superimposed Alport syndrome in patients with IgAN.
There should be cautious interpretation of the COL4A3 variant in such clinical circumstances. In other circumstances with the described phenotype, these children would have been classified in the Alport syndrome category and would have received straightforward RASB therapy, because they carry the pathogen classified as a COL4A3 variant. However, given our results showing the evolution of proliferative lesions in RKB and the response after immunosuppressive therapy, it appears that these patients belong more to the cIgAN category and therefore require immunosuppressive treatment. Acute renal failure with many relapses and the relatively favorable evolution after immunosuppressive therapy (except in patient 3) suggest that the immune response plays a significant role in the disease and it should thus be treated; this does not support the evolution of a balanced Alport syndrome.
Moreover, cIgAN COL4A3 heterozygous variants seem to become predisposed to serious IgAN presentation. The clinical presentation of COL4A3-cIgAN patients was more severe at onset compared with the non‒COL4A3-cIgAN patients, with a more profound nephrotic syndrome. COL4A3-cIgAN patients had many relapses and received repeat kidney biopsies, demonstrating development of a high proportion of glomerulosclerosis at the last follow-up associated with a lower eGFR compared with non‒COL4A3-cIgAN patients. Compared with with cIgAN in the RKB group, the COL4A3-cIgAN group had more glomerulosclerosis at last kidney biopsy follow-up. COL4A3 heterozygous variants in cIgAN seem to develop more chronic lesions after the various relapses of cIgAN. We have already shown that cIgAN demonstrated more histologic inflammation than for adult IgAN, with a greater number of chronic lesions.23 Monitoring of RKB patients is essential, especially in cIgAN. Kidney biopsy monitoring in cIgAN has allowed us to follow the response to immunosuppressive treatment and the development of chronic lesions such as glomerulosclerosis. Knowing the status of the collagen variant would help us to monitor these severely affected patients. Integrative analysis of the Oxford classification with the collagenous variant may help us to understand the wide spectrum of IgAN presentation and prognosis. Following these results, it would be of interest to screen patients diagnosed with severe IgAN at the onset for collagenous variants.
Risk stratification could be based on the existence of a rare genetic variant of the COL4A3 gene. In this cohort, the majority of cases showed an autosomal inheritance with heterozygous COL4A3 variants, as has been described in familial focal segmental glomerulosclerosis.31 Histopathologic images provide valuable information for diagnosis, staging, and prognosis for IgAN and are being used extensively by clinicians in clinical practice. Molecular characteristics, such as collagenous mutation, may be of interest and there is some question as to whether the integration of collagenous features could lead to more accurate prediction of patient outcome. Despite being classified as potentially pathogenic, the role of these collagenous variants affecting COL4A3 is still unknown but confirms the heterogeneous nature of IgAN. Although a proportion of families who present with IgAN may carry a disease-causing variant in COL4A3, the majority do not. Interestingly, locus 2q36, which encompasses the COL4A3/A4 locus, has been identified as a disease-susceptibility locus for IgAN.3
We believe that COL4A3 heterozygous variants in cIgAN should be considered more as a predisposition factor to develop a serious flare-up of cIgAN rather than considered per se as authentic dominant Alport syndrome. Sequencing the COL4A3 variant at the beginning of cIgAN can stratify the risk for severe flare-up, especially when nephrotic syndrome is seen at presentation. The collagenous variant may help us to better understand the wide spectrum of IgAN presentation and prognosis.
For these reasons, intense genetic screening for related living kidney donors seems logical in kidney transplantation in order to select an appropriate donor and thus minimize the risk of relapse due to the COL4A3 trigger and any consequent deleterious effects to the donor.32
The cIgAN with COL4A3 pathologic heterozygous variants should be considered as having a predisposition factor to develop serious flare-ups of cIgAN, rather than as dominant Alport syndrome that affects adults later in life. The observation of COL4A3 variants usually responsible for Alport syndrome in adults should not be a reason for avoiding an immunosuppressive regimen in cIgAN. Moreover, evidence of the ACMG class 4/5 COL4A3 variant at an early stage of cIgAN may be a helpful tool for categorizing the severity of cIgAN beyond the Oxford classification.
Similarly to newly discovered immune biomarkers based on IgA complexes, such as sCD89-IgA, free sCD89, Gd-IgA1, and IgG-IgA, and based on our findings and to be confirmed in a larger study, predictive and diagnostic factors could be uncovered using DNA sequencing during initial management of cIgAN. Indeed, ACMG evaluation of variants affecting COL4A3 potentially represents an indicative prognostic biomarker of cIgAN suitable for risk stratification.
Disclosure
All the authors declared no competing interests.
Funding Statement
This work has been funded in large part by a research grant from the Association pour l’Information et la recherche sur les maladies renales genetiques (https://www.airg-france.fr/resultat-de-lappel-a-projet-2017).
Footnotes
Figure S1. Description of COL4A3-cIgAN families.
Contributor Information
Alexandra Cambier, Email: alexandra.cambier@aphp.fr.
Laurent Mesnard, Email: laurent.mesnard@aphp.fr.
Supplementary Material
Figure S1. Description of COL4A3-cIgAN families.
References
- 1.Wyatt R.J., Kritchevsky S.B., Woodford S.Y. IgA nephropathy: long-term prognosis for pediatric patients. J Pediatr. 1995;127:913–919. doi: 10.1016/s0022-3476(95)70027-7. [DOI] [PubMed] [Google Scholar]
- 2.Kiryluk K., Julian B.A., Wyatt R.J. Genetic studies of IgA nephropathy: past, present, and future. Pediatr Nephrol. 2010;25:2257–2268. doi: 10.1007/s00467-010-1500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson A.D., Li X.Q., Wang K. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol. 2007;18:2408–2415. doi: 10.1681/ASN.2007020241. [DOI] [PubMed] [Google Scholar]
- 4.Gharavi A.G., Kiryluk K., Choi M. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X.Q., Li M., Zhang H. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2011;44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 6.Sallustio F., Cox S.N., Serino G. Genome-wide scan identifies a copy number variable region at 3p21.1 that influences the TLR9 expression levels in IgA nephropathy patients. Eur J Hum Genet. 2015;23:940–948. doi: 10.1038/ejhg.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiryluk K., Li Y., Sanna-Cherchi S. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feehally J., Farrall M., Boland A. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21:1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gharavi A.G., Yan Y., Scolari F. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22-23. Nat Genet. 2000;26:354–357. doi: 10.1038/81677. [DOI] [PubMed] [Google Scholar]
- 10.Bisceglia L., Cerullo G., Forabosco P. Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet. 2006;79:1130–1134. doi: 10.1086/510135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julian B.A., Quiggins P.A., Thompson J.S. Familial IgA nephropathy. Evidence of an inherited mechanism of disease. N Engl J Med. 1985;312:202–208. doi: 10.1056/NEJM198501243120403. [DOI] [PubMed] [Google Scholar]
- 12.Scolari F., Amoroso A., Savoldi S. Familial clustering of IgA nephropathy: further evidence in an Italian population. Am J Kidney Dis. 1999;33:857–865. doi: 10.1016/s0272-6386(99)70417-8. [DOI] [PubMed] [Google Scholar]
- 13.Packham D.K. Thin basement membrane nephropathy and IgA glomerulonephritis: can they be distinguished without renal biopsy? Nephrology (Carlton) 2007;12:481–486. doi: 10.1111/j.1440-1797.2007.00813.x. [DOI] [PubMed] [Google Scholar]
- 14.Savige J., Rana K., Tonna S. Thin basement membrane nephropathy. Kidney Int. 2003;64:1169–1178. doi: 10.1046/j.1523-1755.2003.00234.x. [DOI] [PubMed] [Google Scholar]
- 15.Masuda Y., Yamanaka N., Ishikawa A. Glomerular basement membrane injuries in IgA nephropathy evaluated by double immunostaining for alpha5(IV) and alpha2(IV) chains of type IV collagen and low-vacuum scanning electron microscopy. Clin Exp Nephrol. 2015;19:427–435. doi: 10.1007/s10157-014-1008-8. [DOI] [PubMed] [Google Scholar]
- 16.Berthoux F.C., Laurent B., Koller J.M. Primary IgA glomerulonephritis with thin glomerular basement membrane: a peculiar pathological marker versus thin membrane nephropathy association. Contrib Nephrol. 1995;111:1–7. doi: 10.1159/000423868. [DOI] [PubMed] [Google Scholar]
- 17.Cambier A., Rabant M., Peuchmaur M. Immunosuppressive treatment in children with IgA nephropathy and the clinical value of podocytopathic features. Kidney Int Rep. 2018;3:916–925. doi: 10.1016/j.ekir.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz G.J., Muñoz A., Schneider M.F. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20:629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pottel H. Measuring and estimating glomerular filtration rate in children. Pediatr Nephrol. 2017;34:249–263. doi: 10.1007/s00467-016-3373-x. [DOI] [PubMed] [Google Scholar]
- 20.Robert T., Berthelot L., Cambier A. Molecular insights into the pathogenesis of IgA nephropathy. Trends Mol Med. 2015;21:762–775. doi: 10.1016/j.molmed.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Vuong M.T., Hahn-Zoric M., Lundberg S. Association of soluble CD89 levels with disease progression but not susceptibility in IgA nephropathy. Kidney Int. 2010;8:1281–1287. doi: 10.1038/ki.2010.314. [DOI] [PubMed] [Google Scholar]
- 22.Berthoux F., Suzuki H., Thibaudin L. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol. 2012;23:1579–1587. doi: 10.1681/ASN.2012010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cambier A., Rabant M., El Karoui K. Clinical and histological differences between adults and children in new onset IgA nephropathy. Pediatr Nephrol. 2020;35:1897–1905. doi: 10.1007/s00467-020-04614-3. [DOI] [PubMed] [Google Scholar]
- 24.Monteiro R.C., Cooper M.D., Kubagawa H. Molecular heterogeneity of Fc alpha receptors detected by receptor-specific monoclonal antibodies. J Immunol. 1992;148:1764–1770. [PubMed] [Google Scholar]
- 25.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berthelot L., Papista C., Maciel T.T. Transglutaminase is essential for IgA nephropathy development acting through IgA receptors. J Exp Med. 2012;209:793–806. doi: 10.1084/jem.20112005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro R.C., Van De Winkel J.G.J. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 28.Pillebout E., Jamin A., Ayari H. Biomarkers of IgA vasculitis nephritis in children. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savige J., Ariani F., Mari F. Expert consensus guidelines for the genetic diagnosis of Alport syndrome. Pediatr Nephrol. 2019;34:1175–1189. doi: 10.1007/s00467-018-3985-4. [DOI] [PubMed] [Google Scholar]
- 30.Li Y., Groopman E.E., D’Agati V. Type IV collagen mutations in familial IgA nephropathy. Kidney Int Rep. 2020;5:1075–1078. doi: 10.1016/j.ekir.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone A.F., Phelan P.J., Hall G. Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int. 2014;86:1253–1259. doi: 10.1038/ki.2014.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashtan C.E. Renal transplantation in patients with Alport syndrome: patient selection, outcomes, and donor evaluation. Int J Nephrol Renovasc Dis. 2018;11:267–270. doi: 10.2147/IJNRD.S150539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.