Abstract
Siglecs are sialic acid–binding immunoglobulin-like lectins that play vital roles in immune cell signaling. Siglecs help the immune system distinguish between self and nonself through the recognition of glycan ligands. While the primary binding specificities of Siglecs are known to be divergent, their specificities for complex glycans remain unclear. Herein, we determined N-glycan binding profiles of a set of Siglecs by using a complex asymmetric N-glycan microarray. Our results showed that Siglecs had unique terminal epitope-dependent branch preference when recognizing asymmetric N-glycans. Specifically, human Siglec-3, -9, and -10 prefer the α1-3 branch when Siaα2-6Galβ1-4GlcNAc terminal epitope serves as the binding ligand but prefer the opposite α1-6 branch when Siaα2-3Galβ1-4GlcNAc epitope serves as the ligand. Interestingly, Siglec-10 exhibited dramatic binding divergence toward a pair of Neu5Ac-containing asymmetric N-glycan isomers, as well as their Neu5Gc-containing counterparts. This new information on complex glycan recognition by Siglecs provides insights into their biological roles and applications.
Keywords: Siglecs, asymmetric N-glycan, Neu5Gc, Neu5Ac, microarray
Introduction
Sialic acid–binding immunoglobulin-like lectins (Siglecs) are cell-surface transmembrane receptors that are differentially expressed on immune cells (Läubli and Varki, 2020). They play critical roles in immune cell signaling and help the immune system to distinguish self and nonself (Macauley et al., 2014). Most Siglecs, with the only exception being sialoadhesin/Siglec-1, have C-terminal regulatory motifs in their cytoplasmic domains that participate in the regulation of immune systems. On the N-terminal, each Siglec has a V-set immunoglobulin (Ig) domain that recognizes sialic acid–containing glycans (Duan and Paulson, 2020). There are 15 human Siglecs and 9 murine Siglecs. Among those, four are conserved across mammals (Siglec-1, 2, 4, and 15). All remaining Siglecs are named CD33-related Siglecs as they contain less conserved structure between humans and other vertebrates, but all have high homologies to CD33.
Siglecs are immune-modulatory receptors within the mammalian immune system. Most Siglecs have intracellular immunoreceptor tyrosine inhibitory motifs (ITIMs) that can, in principle, participate in inhibitory or activating signals. The binding of anti-Siglec antibodies or multivalent trans-ligand with inhibitory Siglecs can activate/phosphorylate the ITIMs and produce negative signals (Duan and Paulson, 2020). Additionally, some Siglecs are specifically expressed on certain types of immune cells and presented as endocytic receptors. Hence, they were utilized as the desired target for drug development. For example, Siglec-3, also called CD33, is an inhibitory receptor that is relatively specifically expressed on myeloid lineage and endocytosed upon antibody binding, thus serving as a specific target for developing therapeutic antibodies. Gemtuzumab ozogamicin is the first approved CD33-targeting antibody-drug conjugate (ADC) and was used for induction therapy of acute myeloid leukemia (AML) (Laszlo et al., 2014).
Despite the diverse roles that Siglecs play in immune cell regulation and disease processes, their natural ligands, especially the fine binding specificity, toward complex glycans are relatively underinvestigated. Glycan microarray was developed for identifying interactions between glycans and glycan-binding proteins (GBPs) 2 decades ago (Fukui et al., 2002; Palma and Chai, 2019). It enabled simultaneous binding analysis of GBPs to hundreds of glycan structures and had become a major tool to unveil glycan–protein interactions (Gao et al., 2019b). Various versions of glycan microarray were used to investigate interactions between glycans and Siglecs (Blixt et al., 2003; Bochner et al., 2005; Campanero-Rhodes et al., 2006; Rillahan et al., 2012; Rillahan et al., 2013; Gao et al., 2019a). However, the fine specificity details of Siglecs toward natural complex glycans remain largely unknown.
Herein, we investigated the binding specificity of Siglec-3, -9, -10, and -F using a unique glycan microarray containing 98 structurally well-defined complex glycans, revealing a unique terminal epitope-dependent branch preference toward asymmetric N-glycans. Particularly, a dramatic binding divergence of Siglec-10 toward a pair of N-glycan isomers was observed and further confirmed by synthesized Neu5Gc-containing counterparts. Later, quantitative assay by biolayer interferometry analyses suggested a 67-fold avidity difference among the Neu5Gc-containing isomers.
Materials and Methods
Unless otherwise stated, all chemicals were purchased and used without further purification. The 98 N-glycan microarray was prepared as described previously (Supplementary Figure S1) (Li et al., 2019). Sugar nucleotides, including uridine 5′-diphospho-galactose (UDP-Gal) (Muthana et al., 2012), were prepared as described previously. Enzymes including Neisseria meningitides β1-4galactosyltransferase (NmLgtB) (Lau et al., 2010), N. meningitidis CMP-sialic acid synthetase (NmCSS) (Yu et al., 2004), Pasteurella multocida α2-3sialyltransferase mutant M144D (PmST1-M144D) (Sugiarto et al., 2012), and Photobacterium damsela α2-6sialyltransferase (Pd26ST) (Yu et al., 2006) were expressed and purified as previously described.
Chemoenzymatic Synthesis of N-Glycans
N-glycans 38 and 54 were prepared as previously reported (Li et al., 2015). For the α2-6sialylation of 38, 100, 54, and 104, reactions were carried out in reaction systems containing Tris-HCl (100 mM, pH 8.0), an acceptor glycan (10 mM), CTP (15 mM), N-acetylneuraminic acid (Neu5Ac) or N-glycolylneuraminic acid (Neu5Gc) (15 mM), MgCl2 (10 mM), and appropriate amounts of NmCSS and Pd26ST. Reactions were incubated at 37°C for 3 h and monitored by HPLC. After over 95% acceptor was converted, reactions were quenched by the addition of equal volumes of ice-cold ethanol, concentrated, and subject to HPLC separation to afford compounds 99, 101, 103, and 105. Product-containing fractions were pooled and lyophilized for characterization and next step modular assembly. For the β1-4galactosylation of 99 and 103, reactions were performed in mixtures containing Tris-HCl (100 mM, pH 7.5), an acceptor glycan (10 mM), UDP-Gal (15 mM), MgCl2 (10 mM), and an appropriate amount of NmLgtB. Reactions were incubated at 37°C overnight and monitored by HPLC. After over 95% acceptor was converted, reactions were quenched, concentrated, and subject to HPLC separation of compounds 100 and 104. Product-containing fractions were pooled and lyophilized for characterization and subsequent synthesis. The α2-3sialylation of 100 and 104 was carried out in reaction systems containing Tris-HCl (100 mM, pH 8.0), an acceptor glycan (10 mM), CTP (15 mM), Neu5Gc (15 mM), MgCl2 (10 mM), and appropriate amounts of NmCSS and PmST1-M144D. PmST1-M144D-catalyzed reactions were incubated at 37°C for 3 h and monitored by HPLC. After over 90% acceptor was converted, the reaction was quenched, concentrated, and subject to HPLC separation to afford compounds 102 and 106. Product-containing fractions were then pooled and lyophilized for characterization.
Newly synthesized N-glycans were purified by HPLC using a Waters XBridge BEH amide column (130 Å, 5 μm, 10 mm × 250 mm) under a gradient running condition (solvent A: water or 100 mM ammonium formate; solvent B: acetonitrile; flow rate: 4.5 ml/min, B%: 65–50% in 30 min) and monitored by UV absorbance at 210 nm. MALDI-TOF MS analyses were performed on UltrafleXtreme MALDI TOF/TOF Mass Spectrometer (Bruker). Scan range of MS was set according to molecular weight, and reflector mode was used for analysis. Mass spectra were obtained in negative extraction mode with the following voltage settings: ion source 1 (19.0 kV), ion source 2 (15.9 kV), and lens (9.3 kV). The reflector voltage was set to 20 kV. The laser was pulsed at 7 Hz and the pulsed ion extraction time was set at 400 ns. The laser power was kept in the range of 40–90%. 1H NMR spectra were recorded on a Bruker AVANCE 600 (600 MHz) spectrometer at 25°C. All 1H Chemical shifts (in ppm) were assigned according to D2O (δ = 4.79 ppm).
Compound 99, white power (0.92 mg). 1H NMR (600 MHz, D2O) δ 5.05 (s, 1H), 4.83 (d, J = 1.7 Hz, 1H), 4.52 (dd, J = 8.0, 2.8 Hz, 2H), 4.47 (d, J = 8.4 Hz, 1H), 4.36 (d, J = 7.9 Hz, 1H), 4.17 (d, J = 1.9 Hz, 1H), 4.11 (dd, J = 3.5, 1.6 Hz, 1H), 4.02 (d, J = 7.3 Hz, 2H), 3.95–3.29 (m, 42H), 2.60 (dd, J = 12.4, 4.7 Hz, 1H), 2.02–1.92 (m, 9H), 1.65 (t, J = 12.2 Hz, 1H). MALDI-MS: C67H111N5O50, calc. for 1785.6297, found [M-H]− 1784.765.
Compound 100, white power (0.74 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.7 Hz, 1H), 5.05 (s, 1H), 4.85 (d, J = 1.7 Hz, 1H), 4.54–4.46 (m, 3H), 4.38 (dd, J = 13.8, 7.9 Hz, 2H), 4.17 (s, 1H), 4.11 (dd, J = 3.6, 1.6 Hz, 1H), 4.03 (d, J = 4.4 Hz, 3H), 3.97–3.35 (m, 56H), 2.60 (dd, J = 12.4, 4.7 Hz, 1H), 2.03–1.91 (m, 11H), 1.65 (t, J = 12.2 Hz, 1H). MALDI-MS: C73H121N5O55, calc. for 1947.6825, found [M-H]− 1946.883.
Compound 101, white power (0.61 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.8 Hz, 1H), 5.05 (s, 1H), 4.86 (s, 1H), 4.55–4.48 (m, 3H), 4.36 (d, J = 7.9 Hz, 2H), 4.17 (s, 1H), 4.11 (dd, J = 3.6, 1.5 Hz, 1H), 4.03 (s, 3H), 3.95–3.35 (m, 65H), 2.59 (ddd, J = 12.2, 7.4, 4.7 Hz, 2H), 2.03–1.91 (m, 15H), 1.64 (q, J = 11.8 Hz, 2H). MALDI-MS: C84H138N6O63, calc. for 2238.7779, found [M-2H + Na]− 2259.918.
Compound 102, white power (0.36 mg). 1H NMR (600 MHz, D2O) δ 5.05 (s, 1H), 4.84 (s, 1H), 4.55–4.43 (m, 4H), 4.36 (d, J = 7.9 Hz, 1H), 4.17 (d, J = 2.8 Hz, 1H), 4.11 (d, J = 3.4 Hz, 1H), 4.03 (s, 6H), 3.95–3.36 (m, 72H), 2.69 (dd, J = 12.4, 4.6 Hz, 1H), 2.60 (dd, J = 12.4, 4.7 Hz, 1H), 2.01–1.91 (m, 12H), 1.73 (t, J = 12.2 Hz, 1H), 1.65 (t, J = 12.2 Hz, 1H). MALDI-MS: C84H138N6O64, calc. for 2254.7728, found [M-2H + Na]− 2276.097.
Compound 103, white power (1.21 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.6 Hz, 1H), 5.03 (s, 1H), 4.86 (s, 1H), 4.52 (t, J = 6.4 Hz, 2H), 4.47 (d, J = 8.4 Hz, 1H), 4.37 (d, J = 7.9 Hz, 1H), 4.17 (s, 1H), 4.10 (dd, J = 3.4, 1.6 Hz, 1H), 4.03 (s, 3H), 3.95–3.32 (m, 56H), 2.60 (dd, J = 12.3, 4.7 Hz, 1H), 2.02–1.91 (m, 12H), 1.65 (t, J = 12.2 Hz, 1H). MALDI-MS: C67H111N5O50, calc. for 1785.6297, found [M-H]− 1784.867.
Compound 104, white power (0.87 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.7 Hz, 1H), 5.03 (s, 1H), 4.86 (d, J = 1.8 Hz, 1H), 4.55–4.48 (m, 3H), 4.37 (t, J = 7.8 Hz, 2H), 4.16 (d, J = 2.5 Hz, 1H), 4.11 (dd, J = 3.4, 1.6 Hz, 1H), 4.03 (d, J = 3.6 Hz, 3H), 3.95–3.37 (m, 55H), 2.60 (dd, J = 12.4, 4.7 Hz, 1H), 2.03–1.93 (m, 12H), 1.65 (t, J = 12.2 Hz, 1H), 1.24 (d, J = 6.9 Hz, 1H). MALDI-MS: C73H121N5O55, calc. for 1947.6825, found [M-H]− 1947.017.
Compound 105, white power (0.59 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.8 Hz, 1H), 5.05 (s, 1H), 4.86 (s, 1H), 4.56–4.48 (m, 3H), 4.39–4.32 (m, 2H), 4.17 (s, 1H), 4.11 (dd, J = 3.5, 1.6 Hz, 1H), 4.03 (s, 3H), 3.95–3.38 (m, 58H), 2.59 (td, J = 12.9, 4.6 Hz, 2H), 2.04–1.90 (m, 12H), 1.64 (td, J = 12.2, 9.2 Hz, 2H). MALDI-MS: C84H138N6O63, calc. for 2238.7779, found [M-2H + Na]− 2260.103.
Compound 106, white power (0.43 mg). 1H NMR (600 MHz, D2O) δ 5.10 (d, J = 2.7 Hz, 1H), 5.03 (s, 1H), 4.86 (s, 1H), 4.54–4.44 (m, 4H), 4.37 (d, J = 7.9 Hz, 1H), 4.17 (d, J = 2.5 Hz, 1H), 4.14–4.09 (m, 1H), 4.03 (s, 6H), 3.94–3.38 (m, 40H), 2.69 (dd, J = 12.4, 4.6 Hz, 1H), 2.60 (dd, J = 12.4, 4.7 Hz, 1H), 2.01–1.94 (m, 12H), 1.73 (t, J = 12.2 Hz, 1H), 1.65 (t, J = 12.2 Hz, 1H). MALDI-MS: C84H138N6O64, calc. for 2254.7728, found [M-2H + Na]− 2276.203.
Glycan Derivatization and Quantification
All synthesized glycans with free reducing-end were derivatized by reductive amination using 2-amino-N-(2-aminoethyl)-benzamide (AEAB) as previously described (Song et al., 2009). Labeled glycans were further purified by HPLC to homogeneity using a porous graphitic carbon column (5 μm, 4.6 mm × 150 mm) under a gradient running condition (solvent A: 0.1% TFA in water; solvent B: 0.1% TFA in acetonitrile; flow rate: 1 ml/min, B%: 15–45% in 30 min) and monitored by UV absorbance at 330 nm. Product-containing fractions were pooled and lyophilized. The quantifications of AEAB-labeled glycans were conducted as previously described (Li et al., 2019).
Neu5Gc-N-glycan Microarray Fabrication
The AEAB labeled–glycans were prepared at a concentration of 100 μM in the printing buffer (150 mM phosphate, pH 8.5), and printed on multivalent NHS-derivatized microscope-glass slides (Z Biotech, LLC), each for 400 pL in replicates of six, as described previously (Heimburg-Molinaro et al., 2011). Noncontact printing was performed at room temperature with a humidity of 60% by a sciFLEXARRAYER S3 spotter (Scienion) with two PDC 80 Piezo Dispense Capillary. After overnight dehumidification under room temperature, the slides were washed with MilliQ water and subsequently blocked with 50 mM ethanolamine in 100 mM Tris-HCl (pH 9.0) for 2 h. The blocked slides were then washed with MilliQ water twice, dried, and stored desiccated at −20°C until use.
Microarray Assay of Siglecs With N-Glycan Microarrays
The 98 N-glycan microarray slide (Li et al., 2019) was fitted with a ProPlate 8-well microarray module (Sigma-Aldrich), and the subarrays were then rehydrated for 10 min with 200 μL of Buffer TSMTB (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% (v/v) Tween-20, and 1% (w/v) BSA) at room temperature. Then, the buffer was drained and 200 μL of Siglec-3, -8, -9, -10, and -F (R&D Systems) (20 µg/ml) in TSMTB were added into each subarray, sealed, and incubated at room temperature for 1 h with gentle shaking. Slides were then washed with Buffer TSMT (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, and 0.05% (v/v) Tween-20) for four times. Next, 200 μL of 5 µg/ml goat anti-human IgG Fc antibody cross-adsorbed, DyLight® 650 (Thermo Fisher) was added into each subarray, sealed, and incubated at room temperature for 1 h with gentle shaking. Finally, slides were washed with TSMT, TSM (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, and 2 mM MgCl2) and MilliQ water, four times for each buffer, respectively, and dried by brief centrifugation. Slides were scanned at a resolution of 10 μm using a Genepix 4100 A microarray scanner (Molecular Devices Corp) with 500 or 600 PMT gains and 80% power. Image analyses were carried out using Genepix Pro 6.0 as previously reported (Li et al., 2019). Spots were defined as circular features with a variable radius as determined by the Genepix scanning software, and local background subtraction was performed. Similarly, Siglec-10 was analyzed using the newly fabricated Neu5Gc-N-glycan array at concentrations of 1 and 5 μg/ml.
Biolayer Interferometry Receptor Binding Assay and Data Analysis
The AEAB-labeled glycan 102 and 106 were labeled with Biotin by using the reagent EZ-Link™ NHS-Biotin (Thermo Fisher). In detail, 1 mM AEAB-labeled glycan was incubated with 10 mM NHS-Biotin at room temperature for 10 min. Then, labeled glycans were purified by HPLC to homogeneity using an ODS4 column (5 μm, 4.6 mm × 150 mm) under a gradient running condition (solvent A: 0.1% TFA in water; solvent B: 0.1% TFA in acetonitrile; flow rate: 1 ml/min, B%: 5–50% in 30 min), monitored by UV absorbance at 330 nm. Product-containing fractions were pooled and lyophilized for storage. The purified Biotin-labeled glycans were quantified by HPLC as described above.
Avidities were measured by biolayer interferometry using an Octet RED instrument (Pall FortéBio, Fremont, CA, United States). The prepared biotinylated glycans were preloaded onto streptavidin-coated biosensors at up to 100 nM for 3 min in 1× kinetic buffer (Pall FortéBio, Menlo Park, CA, United States). Siglec-10 was diluted to concentrations of 1 μM, 500 nM, and 250 nM with 1× kinetic buffer, respectively. The glycan-loaded biosensors were submerged in wells containing different concentrations of Siglec-10 for 5 min followed by 15 min of dissociation in 1× kinetic buffer at 25°C with the orbital shake speed of 1000 rpm. As a reference control for subtraction, glycan-loaded biosensors were also dipped in wells containing 1× kinetic buffer. The binding kinetics data were processed by the ForteBio data analysis software (version 11.1). The association and dissociation curves were fitted, and the avidity values were calculated by using a heterogeneous ligand (2:1) model.
Results
Fine Specificity of Human Siglecs Toward the 98 N-Glycan Microarray
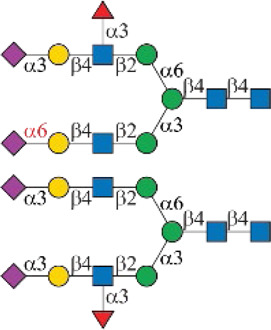
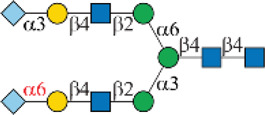
The primary glycan ligands of Siglecs were reported and well summarized (Macauley et al., 2014; Duan and Paulson, 2020). Human Siglec-3, -8, -9, and -10 and mouse Siglec-F recognize Neu5Acα2-6Galβ1-4GlcNAc (Ac6LN) and/or Neu5Acα2-3Galβ1-4GlcNAc (Ac3LN), which are often identified as terminal epitopes on complex glycans found on mammalian cells. To explore fine binding specificities of Siglecs, these Siglecs were analyzed against a previously fabricated microarray containing 98 structurally well-defined complex glycans (Supplementary Figure S1) (Li et al., 2019).
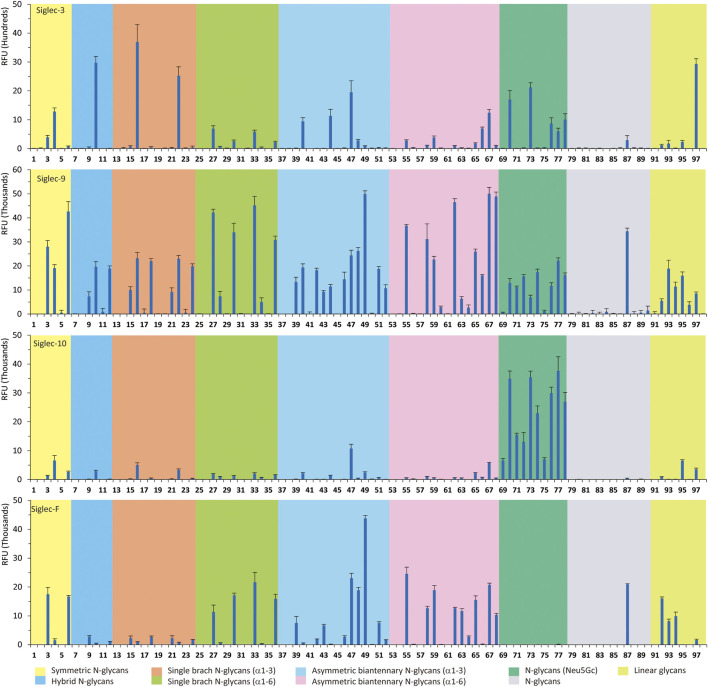
As shown in Figure 1, human Siglec-3 gave lower binding signals toward N-glycans compared with Siglec-9, -10, and -F. Siglec-3, which is found on myeloid cells, is associated with acute myeloid leukemia (AML) and Alzheimer’s disease (Freeman et al., 1995; Zhao, 2019) and was reported to prefer the Ac6LN trisaccharide, plus relatively weak affinity to Gc6LN (Neu5Gcα2-3Galβ1-4GlcNAc) and Ac3LN (Blixt et al., 2003). Recently, Rodrigues et al. (2020) reported that Siglec-3 could recognize both α2-3 and α2-6sialosides in solution and on cells. This is consistent with our results that it bound to N-glycans with terminal epitopes Ac3LN (compound 3), Ac6LN (compound 4), Gc3LN (compound 77), and Gc6LN (compound 78). In addition, the RFUs of Siglec-3 to α2-6sialosides (4 and 78) are higher than those of α2-3sialosides (3 and 77), again consistent with a previous report toward O-mannosyl glycans (Meng et al., 2018). In addition, a slight preference toward Neu5Ac over Neu5Gc was observed, as binding signals of glycans with Neu5Ac residues (4, 10, and 16) were greater than those of their Neu5Gc-containing counterparts (70, 73, and 78). Furthermore, high to moderate bindings were observed toward N-glycans carrying Ac3LN on the α1-3 branch (16, 33, 40, 44, 47, 66, and 67), whereas no meaningful binding signals were observed to their positional isomers (28, 34, 50, 51, 56, 60, and 63). These data suggested that Siglec-3 had an apparent preference toward the α1-3 branch when terminal epitope Ac6LN serves as the binding ligand. On the other hand, Siglec-3 exhibited an opposite branch preference toward the α1-6 branch when terminal epitope Ac3LN served as the binding ligand; for example, it bound to 27, 33, 55, and 59, but failed to bind their positional isomers (15, 21, 39, and 43). Such a unique terminal epitope-dependent branch preference was double evidenced by strong binding to 47, which presents terminal epitopes on preferred branches (Ac6LN on the α1-3 branch and Ac3LN on the α1-6 branch), but no binding to 63 that presents terminal epitopes on nonpreferred branches (Ac3LN on the α1-3 branch and Ac6LN on the α1-6 branch).
FIGURE 1.
Selective recognition of Sia-containing N-glycans by human and mouse Siglecs.
Siglec-9 was reported to bind to both α2-3 and α2-6sialosides, with a high affinity to epitope Neu5Acα2-3Galβ1–4(6-sulfo)GlcNAc (Zhang et al., 2000; Rillahan et al., 2012; Duan and Paulson, 2020). Our microarray results are consistent with previous reports as all related N-glycans showed binding signals. N-glycans with sialyl Lewis X (sLeX) epitopes showed the highest binding signals, including compounds 6, 27, 33, 49, 67, and 68. Additionally, glycans with the Ac3LN epitope (3, 27, and 33) exhibited higher bindings than those with the Ac6LN epitope (4, 28, and 34). Interestingly, the same terminal epitope-dependent branch preference for Siglec-3 was also observed for Siglec-9. When bound to glycans with the terminal epitope Ac6LN, Siglec-9 showed an apparent preference toward the α1-3 branch (16, 22, 40, and 44) over the α1-6 branch (28, 34, 66, and 60) of N-glycans. In contrast, an opposite branch preference was found when bound to glycans with the terminal epitope Ac3LN and sLeX. Lastly, it is observed that Siglec-9 has a slight preference to Neu5Ac-containing N-glycans (9, 10, 12, 15, 16, 18, 39, and 40) over their Neu5Gc-containing counterparts (69–76).
The specificity of Siglec-10 was previously profiled as having a high affinity to Gc6LN, with moderate and weak affinity to Ac6LN and Ac3LN, respectively (Crocker et al., 2007). In our array, as expected, Siglec-10 exhibited strong bindings to the majority of Neu5Gc-terminated N-glycans (69–78). In addition, bindings of Siglec-10 toward various glycan ligands showed gradient diminished signals (Gc6LN >> Gc3LN > Ac6LN >> Ac3LN). For example, the binding signals of Siglec-10 to complex type N-glycans 76 (Gc6LN) >> 75 (Gc3LC) > 40 (Ac6LC) >> 39 (Ac3LN) and hybrid type N-glycans 70 (Gc6LN) >> 69 (Gc3LC) > 10 (Ac6LC) >> 9 (Ac3LN) clearly showed this trend. Siglec-10 also exhibited a terminal epitope-dependent branch preference toward asymmetric N-glycans. For example, high to moderate bindings were observed when epitope Ac6LN was presented on the terminal of the α1-3 branch (16, 22, 66, 40, and 44), while no meaningful signals could be observed to their positional isomers where Ac6LN was presented on the α1-6 branch (28, 34, 50, 56, and 60). This result suggested that Siglec-10 had an apparent preference toward the α1-3 branch when α2-6sialylated glycans served as ligands. In contrast, Siglec-10 exhibited an opposite branch preference toward the α1-6 branch when terminal epitope Ac3LN or sLeX served as the binding ligand. This is evidenced by relatively weak bindings toward compounds 27, 30, 33, and 36, which present Ac3LN or sLeX on the α1-6 branch but no bindings to their positional isomers. This terminal epitope-dependent branch preference is identical to that of Siglec-3 and Siglec-9.
Human Siglec-8 did not bind to any glycans on the array (data not shown), which is consistent with previous observations that Siglec-8 specifically recognizes Neu5Acα2-3 (6-sulfo)Galβ1–4GlcNAc (6-sulfo-sLeX) (Bochner et al., 2005). Mouse Siglec-F is a functional paralogue of human Siglec-8, and it was reported to bind to Ac3LN and 6'-sulfo-sLeX (Tateno et al., 2005). As depicted in Figure 1, Siglec-F could recognize N-glycans with Ac3LN and sLeX epitopes, such as 3, 6, and 49, whereas no binding was observed toward any Neu5Gc-containing glycans, suggesting a strict preference toward Neu5Ac. In addition, Siglec-F showed an apparent α1-6 branch preference. For example, high binding signals were observed for glycans 27, 30, 33, and 36, but very low bindings were observed for their positional isomers 15, 18, 21, and 24. Interestingly, compound 49, which contains sLeX on the α1-3 branch and Ac3LN on the α1-3 branch, showed the strongest binding signals.
Chemoenzymatic Synthesis of Neu5Gc-Containing N-Glycans
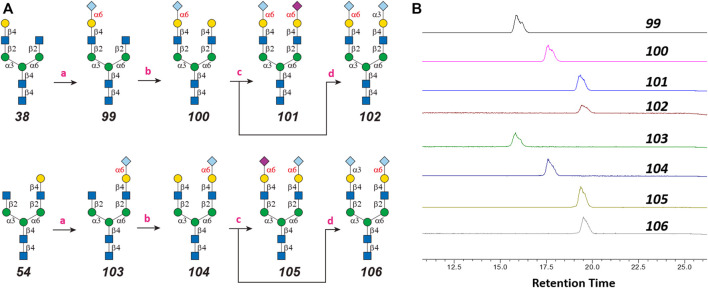
One interesting observation is that Siglec-10 showed high binding to an asymmetric N-glycan 47 but no binding to its positional isomer 63 (Figure 1). Such a dramatic binding divergence can be explained by its terminal epitope-dependent branch preference, as both ligands (Ac3LN and Ac6LN) on 47 are located on the terminal of favored branches, whereas both ligands are located on the unfavored branches of 63. Because Siglec-10 strongly prefers Neu5Gc-containing N-glycans, we speculate that a Neu5Gc-modified counterpart of 47 (Figure 2A, compound 102) may be of higher affinity and a most favorable N-glycan ligand of Siglec-10. To test this hypothesis and to further validate the terminal epitope-dependent branch preference of Siglec-10, we enzymatically synthesized eight Neu5Gc-containing N-glycans (Figure 2A). In detail, compounds 99 to 102 were assembled starting from previously prepared glycan 38 (Li et al., 2015). First, α2-6Neu5Gc was installed onto the α1-3 branch to achieve 99 by Pd26ST-catalyzed α2-6sialylation in the presence of cytidine-5′-triphosphate (CTP), Neu5Gc, and NmCSS for the in situ generation of the sugar donor CMP-Neu5Gc. Then, β1-4Gal was installed onto the α1-6 branch by NmLgtB-catalyzed reaction in the presence of UDP-Gal to provide 100. The addition of α2-6Neu5Ac to the α1-6 branch of 100 by Pd26ST then provided 101. On the other hand, the addition of α2-3Neu5Gc to this branch by PmST1-M144D-catalyzed α2-3sialylation gave the desired asymmetric N-glycan 102. In the same synthetic manner, another four asymmetric N-glycans 103, 104, 105, and 106 were assembled starting from N-glycan 54. All compounds were purified and characterized by HPLC (Figure 2B), mass spectrometry, and NMR (supporting information).
FIGURE 2.
Enzymatic synthesis of Neu5Gc-containing asymmetric N-glycans: (A) a, α2-6sialylation with Pd26ST, NmCSS, CTP, and Neu5Gc; b, β1-4galactosylation with NmLgtB and UDP-Gal; c, α2-6sialylation with Pd26ST, NmCSS, CTP, and Neu5Ac; d, α2-3sialylation with PmST1-M144D, NmCSS, CTP, and Neu5Gc; (B) HPLC analysis of purified N-glycans.
Neu5Gc N-Glycan Microarray Fabrication and Assay With Human Siglec-10
The Neu5Gc-containing N-glycans were labeled with AEAB to provide an amino group for microarray fabrication as previously reported (Li et al., 2019). The Neu5Gc N-glycan microarray was then constructed on NHS glass slides with four additional sialylated N-glycans (3, 4, 77, and 78) and four sialylated linear glycans (93, 95, 96, and 97). The recognition by Siglec-10 was then assayed at the concentrations of 1 µg/ml and 5 µg/ml (Figure 3). As shown in Figure 3, di-sialylated glycans (78, 101, 102, and 105) showed high RFU compared with mono-sialylated glycans (100and 104) and linear glycan (93, 95, 96, and 97). The preferences of Siglec-10 toward Neu5Gc (77, 78) over Neu5Ac (3 and 4) and α2-6sialosides (4, 78) over α2-3sialosides (3, 77) were further confirmed by this focused array. In addition, when terminal epitope Gc6LN served as the binding ligand, Siglec-10 preferred the α1-3 branch (100) over the α1-6 branch (104). And as expected, Siglec-10 showed the highest binding signal to glycan 102 (the Neu5Gc modified counterpart of 47), whereas its positional isomer 106 only showed comparable bindings as that of mono-sialylated 100. These results further confirmed the terminal epitope-dependent branch preference; that is, Siglec-10 prefers α2-6sialosides on the α1-3 branch and α2-3sialosides on the α1-6 branch of N-glycans. Interesting, the binding signals of Siglec-10 toward the four α2-6sialyated N-glycans with Neu5Ac/Neu5Gc chimeras (4, Neu5Ac on both branches; 78, Neu5Gc on both branches; 101, Neu5Ac on α1-6 branch, Neu5Gc on α1-3 branch; 105, Neu5Gc on α1-6 branch, and Neu5Ac on α1-3 branch) are distinct, indicating that minor structural divergence in complex glycan may cause substantial changes in glycan-protein interactions.
FIGURE 3.
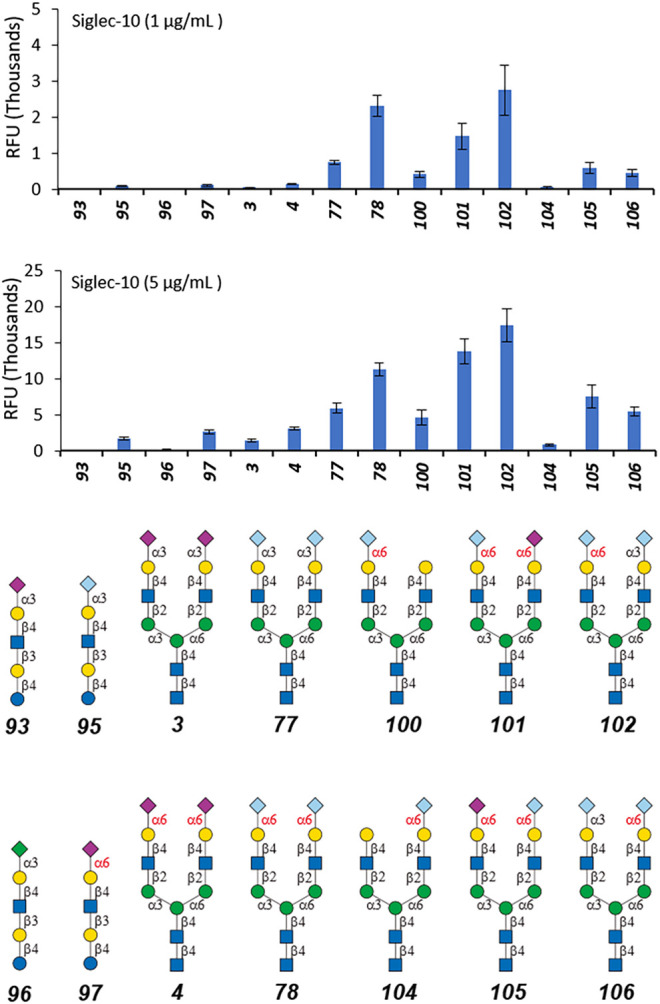
Selective recognition of Neu5Ac-containing N-glycans by human Siglec-10.
Avidity of Siglec-10 to N-Glycans 102 and 106
As shown in Figure 3, the binding signals of Siglec-10 to 102 is around 5-fold stronger than to its positional isomer 106 and 20-fold stronger than to linear glycans 97, suggesting compound 102 as a potential high-affinity ligand of Siglec-10. The avidity of Siglec-10 toward 102 and 106 was thus measured by biolayer interferometry (BLI). AEAB labeled 102 and 106 were further conjugated with NHS-Biotin and purified with HPLC, and then immobilized onto streptavidin-coated biosensors for BLI assay (Figure 4). The association and dissociation curves were fitted, and the avidity values were calculated with the consideration of the bivalency of the Siglec-10-Fc chimera protein. The avidity values of Siglec-10 toward 102 and 106 were 0.11 μM and 7.34 μM, respectively, indicating a 67-fold higher avidity of 102 than 106. The result further confirmed the terminal epitope-dependent branch preference and revealed a high avidity glycan-binding partner (102) of human Siglec-10.
FIGURE 4.
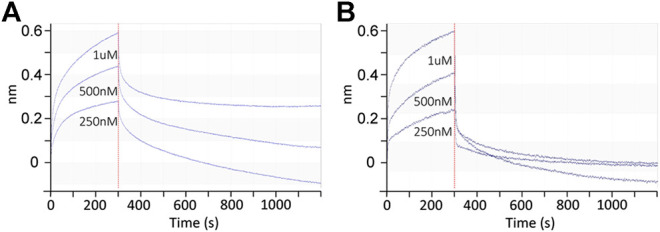
Binding kinetics between the Siglec-10-Fc chimera protein homodimer and Neu5Gc-containing N-glycans 102 (A) and 106 (B) determined by BLI. Association and dissociation phases are shown and separated by the red dashed line at 300 s.
Discussion and Conclusion
Siglecs are attractive therapeutic targets and several related antibody-based therapies had been developed for the treatment of immune-related diseases. In certain applications, glycan ligands have an advantage over antibodies, such as their ability to dissociate from their target once endocytosed. However, glycan-based therapeutic strategies for cargo delivery and immunomodulation are underinvestigated due to the lack of suitable ligands (Angata et al., 2015). A comprehensive understanding of glycan recognition details by Siglecs is essential toward the discovery and designing of efficient ligands. In fact, recent advances in glycobiology have prompted such applications. For example, high specific efficient N-glycan ligands with chemical modifications toward Siglec-2 were reported (Peng and Paulson, 2017). Conjugates of toxins with this novel ligand could be efficiently internalized via Siglec-2, resulting in the killing of B-cell lymphoma cells.
In this study, we screened binding profiles of Siglec-3, -9, -10, and -F against a comprehensive N-glycan microarray to reveal glycan recognition details of Siglecs (Table 1). The results showed a surprising terminal epitope-dependent branch preference toward N-glycans by Siglec-3, -9, and -10. These Siglecs prefer the α1-3 branch of N-glycans when α2-6sialylated epitopes serve as binding ligands, while they have an opposite preference to the α1-6 branch when α2-3sialylated epitopes serve as ligands. Such a feature could assist in designing high-affinity binding partners of Siglecs. For example, we designed and synthesized an asymmetric N-glycan (102) with much higher avidity than its positional isomer toward Siglec-10. Note that recombinant Siglec-Fc chimera proteins in the form of disulfide-linked homodimer were used in this study instead of native Siglecs. Even though such chimera proteins were widely used to reveal the glycan recognition of Siglecs and other human GBPs (Blixt et al., 2003; Bochner et al., 2005; Campanero-Rhodes et al., 2006; Rillahan et al., 2012; Rillahan et al., 2013; Gao et al., 2019b; Rodrigues et al., 2020), the nonnatural bivalent form could possibly influence their fine specificity toward glycan-binding partners.
TABLE 1.
Binding specificity of Siglecs toward N-glycans observed in this study. Recombinant Siglec-Fc chimera proteins were used in this study.
| Siglec | Ligand (preference) | Branch Preference | Strongest N-glycan Binder |
|---|---|---|---|
| Siglec-3 |
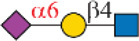 (+++);
(+++);
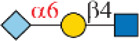 (+++)
(+++)
|
α1-3 branch |
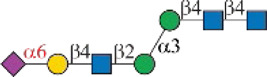
|
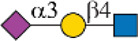 (++)
(++)
|
α1-6 branch | ||
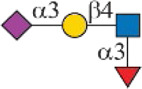
| Siglec-9 |
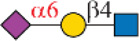 (+++);
(+++);
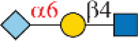 (++)
(++)
|
α1-3 branch |
|
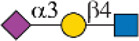 (+++);
(+++);
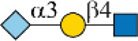 (++)
(++)
|
α1-6 branch | ||
 (+++);
(+++);
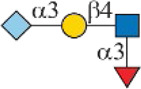 (++)
(++)
|
α1-6 branch | ||
| Siglec-10 |
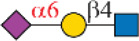 (+);
(+);
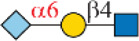 (+++)
(+++)
|
α1-3 branch |
|
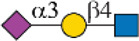 (weak);
(weak);
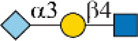 (++)
(++)
|
α1-6 branch | ||
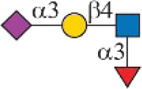 (weak);
(weak);
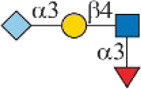 (++)
(++)
|
α1-6 branch | ||
| Siglec-F |
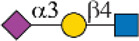 (++);
(++);
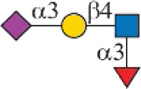 (++)
(++)
|
α1-3 branch |
|
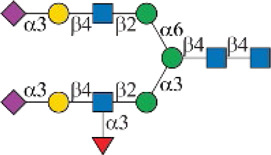
Symbols:  GlcNAc GlcNAc  Man Man  Gal Gal  L-Fuc L-Fuc  Neu5Ac Neu5Ac  Neu5Gc Neu5Gc | |||
High-avidity binding partners of Siglecs could lead to extensive academic and clinical implementations. For example, tumor cells can escape the surveillance of the immune system via inhibition of immune cells through immune checkpoints and their ligands. A promising therapeutic approach for cancer is to block these immune checkpoints, for example, the programmed cell death ligand 1 (PD-L1) and cytotoxic T lymphocyte-associated protein 4 (CTLA-4) (Leach et al., 1996; Topalian et al., 2012). A recent report showed that CD24–Siglec-10 interaction is an innate immune checkpoint that is essential for mediating antitumor immunity and can promote tumor immune escape. The modulation of this interaction is expected to become a new target for tumor therapy (Barkal et al., 2019). The high sialylated CD24 that overexpressed on tumor cells functions as the main ligand of Siglec-10. It induces the inhibition of the immune system and promotes tumor immune escape. Additionally, CD24–Siglec-10 interaction could suppress the immune response to the danger-associated molecular pattern (DAMP) (Cai et al., 2009; Rillahan et al., 2012). It is thus tempting to speculate that the strong Siglec-10 binding partner 102, with or without further modification, may serve as an invaluable reagent to block this immune checkpoint.
Acknowledgments
We thank Z biotech LLC (Aurora, CO) for microarray fabrication (supported by NIH R43GM123820).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LL conceived and designed the project. SW,CC and LL performed glycan synthesis and glycan microarray assays, MG and DL performed kinetic assay, SW wrote the manuscript, and LL and XW revised the manuscript, which was edited and approved by all authors.
Funding
This work was supported by the National Institutes of Health (Grant No. U54HL142019 and U01GM125288 to LL). XW and MG were partially supported by the National Institutes of Health (Grant No. R21AI144433 to XW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.645999/full#supplementary-material.
References
- Angata T., Nycholat C. M., Macauley M. S. (2015). Therapeutic targeting of siglecs using antibody- and glycan-based approaches. Trends Pharmacol. Sci. 36 (10), 645–660. 10.1016/j.tips.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkal A. A., Brewer R. E., Markovic M., Kowarsky M., Barkal S. A., Zaro B. W., et al. (2019). CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572 (7769), 392–396. 10.1038/s41586-019-1456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O., Collins B. E., van den Nieuwenhof I. M., Crocker P. R., Paulson J. C. (2003). Sialoside specificity of the siglec family assessed using novel multivalent probes: identification of potent inhibitors of myelin-associated glycoprotein. J. Biol. Chem. 278 (33), 31007–31019. 10.1074/jbc.M304331200 [DOI] [PubMed] [Google Scholar]
- Bochner B. S., Alvarez R. A., Mehta P., Bovin N. V., Blixt O., White J. R., et al. (2005). Glycan array screening reveals a candidate ligand for siglec-8. J. Biol. Chem. 280 (6), 4307–4312. 10.1074/jbc.M412378200 [DOI] [PubMed] [Google Scholar]
- Cai L., Guan W., Kitaoka M., Shen J., Xia C., Chen W., et al. (2009). A chemoenzymatic route to N-acetylglucosamine-1-phosphate analogues: substrate specificity investigations of N-acetylhexosamine 1-kinase. Chem. Commun. (Camb) 20, 2944–2946. 10.1039/b904853g [DOI] [PubMed] [Google Scholar]
- Campanero-Rhodes M. A., Childs R. A., Kiso M., Komba S., Le Narvor C., Warren J., et al. (2006). Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem. Biophys. Res. Commun. 344 (4), 1141–1146. 10.1016/j.bbrc.2006.03.223 [DOI] [PubMed] [Google Scholar]
- Crocker P. R., Paulson J. C., Varki A. (2007). Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7 (4), 255–266. 10.1038/nri2056 [DOI] [PubMed] [Google Scholar]
- Duan S., Paulson J. C. (2020). Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 38 (1), 365–395. 10.1146/annurev-immunol-102419-035900 [DOI] [PubMed] [Google Scholar]
- Freeman S. D., Kelm S., Barber E. K., Crocker P. R. (1995). Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood 85 (8), 2005–2012. 10.1182/blood.v85.8.2005.bloodjournal8582005 [DOI] [PubMed] [Google Scholar]
- Fukui S., Feizi T., Galustian C., Lawson A. M., Chai W. (2002). Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat. Biotechnol. 20 (10), 1011–1017. 10.1038/nbt735 [DOI] [PubMed] [Google Scholar]
- Gao C., Hanes M. S., Byrd-Leotis L. A., Wei M., Jia N., Kardish R. J., et al. (2019a). Unique binding specificities of proteins toward isomeric asparagine-linked glycans. Cell Chem. Biol 26 (4), 535–547. 10.1016/j.chembiol.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Wei M., McKitrick T. R., McQuillan A. M., Heimburg-Molinaro J., Cummings R. D. (2019b). Glycan microarrays as chemical tools for identifying glycan recognition by immune proteins. Front. Chem. 7, 833. 10.3389/fchem.2019.00833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg-Molinaro J., Song X., Smith D. F., Cummings R. D. (2011). Preparation and analysis of glycan microarrays. Curr. Protoc. Protein Sci. Chapter 12, Unit12.10. 10.1002/0471140864.ps1210s64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo G. S., Estey E. H., Walter R. B. (2014). The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Rev. 28 (4), 143–153. 10.1016/j.blre.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Lau K., Thon V., Yu H., Ding L., Chen Y., Muthana M. M., et al. (2010). Highly efficient chemoenzymatic synthesis of β1-4-linked galactosides with promiscuous bacterial β1-4-galactosyltransferases. Chem. Commun. 46 (33), 6066–6068. 10.1039/C0CC01381A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läubli H., Varki A. (2020). Sialic acid-binding immunoglobulin-like lectins (siglecs) detect self-associated molecular patterns to regulate immune responses. Cell. Mol. Life Sci. 77 (4), 593–605. 10.1007/s00018-019-03288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D. R., Krummel M. F., Allison J. P. (1996). Enhancement of antitumor immunity by CTLA-4 blockade. Science 271 (5256), 1734–1736. 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- Li L., Liu Y., Ma C., Qu J., Calderon A. D., Wu B., et al. (2015). Efficient chemoenzymatic synthesis of an N-glycan isomer library. Chem. Sci. 6 (10), 5652–5661. 10.1039/C5SC02025E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Guan W., Zhang G., Wu Z., Yu H., Chen X., et al. (2019). Microarray analyses of closely related glycoforms reveal different accessibilities of glycan determinants on N-glycan branches. Glycobiology 30 (5), 334–345. 10.1093/glycob/cwz100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S., Crocker P. R., Paulson J. C. (2014). Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14 (10), 653–666. 10.1038/nri3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C., Sasmal A., Zhang Y., Gao T., Liu C. C., Khan N., et al. (2018). Chemoenzymatic assembly of mammalian O-mannose glycans. Angew. Chem. Int. Ed. Engl. 57 (29), 9003–9007. 10.1002/anie.201804373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthana M. M., Qu J., Li Y., Zhang L., Yu H., Ding L., et al. (2012). Efficient one-pot multienzyme synthesis of UDP-sugars using a promiscuous UDP-sugar pyrophosphorylase from Bifidobacterium longum (BLUSP). Chem. Commun. (Camb) 48 (21), 2728–2730. 10.1039/c2cc17577k [DOI] [PubMed] [Google Scholar]
- Palma A. S., Chai W. (2019). “Chapter 18 glycan microarrays with semi-synthetic neoglycoconjugate probes in understanding glycobiology,” in Synthetic glycomes, Editors Li L., Wang P. G., Guan W. (Croydon, London, United Kingdom: The Royal Society of Chemistry; ), 421–446. 10.1039/9781788016575-00421 [DOI] [Google Scholar]
- Peng W., Paulson J. C. (2017). CD22 ligands on a natural N-glycan scaffold efficiently deliver toxins to B-lymphoma cells. J. Am. Chem. Soc. 139 (36), 12450–12458. 10.1021/jacs.7b03208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan C. D., Schwartz E., McBride R., Fokin V. V., Paulson J. C. (2012). Click and pick: identification of sialoside analogues for siglec-based cell targeting. Angew. Chem. Int. Ed. Engl. 51 (44), 11014–11018. 10.1002/anie.201205831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan C. D., Schwartz E., Rademacher C., McBride R., Rangarajan J., Fokin V. V., et al. (2013). On-chip synthesis and screening of a sialoside library yields a high affinity ligand for siglec-7. ACS Chem. Biol. 8 (7), 1417–1422. 10.1021/cb400125w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues E., Jung J., Park H., Loo C., Soukhtehzari S., Kitova E. N., et al. (2020). A versatile soluble siglec scaffold for sensitive and quantitative detection of glycan ligands. Nat. Commun. 11 (1), 5091. 10.1038/s41467-020-18907-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., Cummings R. D. (2009). Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16 (1), 36–47. 10.1016/j.chembiol.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto G., Lau K., Qu J., Li Y., Lim S., Mu S., et al. (2012). A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating lewisx. ACS Chem. Biol. 7 (7), 1232–1240. 10.1021/cb300125k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H., Crocker P. R., Paulson J. C. (2005). Mouse siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6'-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology 15 (11), 1125–1135. 10.1093/glycob/cwi097 [DOI] [PubMed] [Google Scholar]
- Topalian S. L., Drake C. G., Pardoll D. M. (2012). Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr. Opin. Immunol. 24 (2), 207–212. 10.1016/j.coi.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Huang S., Chokhawala H., Sun M., Zheng H., Chen X. (2006). Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem. Int. Ed. Engl. 45 (24), 3938–3944. 10.1002/anie.200600572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Yu H., Karpel R., Chen X. (2004). Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg. Med. Chem. 12 (24), 6427–6435. 10.1016/j.bmc.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Zhang J. Q., Nicoll G., Jones C., Crocker P. R. (2000). Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J. Biol. Chem. 275 (29), 22121–22126. 10.1074/jbc.M002788200 [DOI] [PubMed] [Google Scholar]
- Zhao L. (2019). CD33 in alzheimer’s disease—biology, pathogenesis, and therapeutics: a mini-review. Gerontology 65 (4), 323–331. 10.1159/000492596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.