Abstract
Transposons are genetic elements that change their intracellular genomic position by transposition and are spread horizontally between bacteria when located on plasmids. It was recently discovered that transposition from fully heterologous DNA also occurs in the course of natural transformation. Here, we characterize the molecular details and constraints of this process using the replicative transposon Tn1 and the naturally competent bacterium Acinetobacter baylyi . We find that chromosomal insertion of Tn1 by transposition occurs at low but detectable frequencies and preferably around the A. baylyi terminus of replication. We show that Tn1 transposition is facilitated by transient expression of the transposase and resolvase encoded by the donor DNA. RecA protein is essential for the formation of a circular, double-stranded cytoplasmic intermediate from incoming donor DNA, and RecO is beneficial but not essential in this process. Absence of the recipient RecBCD nuclease stabilizes the double-stranded intermediate. Based on these results, we suggest a mechanistic model for transposition during natural transformation.
Keywords: Acinetobacter baylyi, natural transformation, Tn1, Tn4401, transposition
Introduction
Horizontal gene transfer drives bacterial evolution through the acquisition of novel genetic material and accelerates the spread of adaptive traits such as antimicrobial resistance (AMR) between bacteria [1]. AMR in bacterial pathogens represents a growing public health concern [2] and there is an urgent need to increase our understanding of the basic mechanisms of AMR spread.
Conjugation, transduction and natural transformation are believed to be the main routes of intercellular gene transfer [3]. During conjugation, conjugative plasmids or conjugative transposons move into a recipient bacterium through cell-to-cell contact. Transduction includes the transfer of host DNA, mispackaged into bacteriophage particles during late infection, to a recipient cell. Natural transformation is the active uptake of free DNA from the environment and subsequent genomic integration [4]. Only bacteria that are competent for natural transformation can actively take up free DNA. Competence to undergo natural transformation was experimentally demonstrated in at least 80 bacterial species, both Gram-positive and Gram-negative [5]. The majority of the 12 global priority pathogens are naturally transformable, including those categorized as critically antibiotic-resistant: Acinetobacter baumannii , Pseudomonas aeruginosa and some Enterobacteriaceae [6, 7]. Natural transformation as a pathway for recruiting genetic variation, including AMR genes, was recently demonstrated between different species of the genus Acinetobacter [8] and different genera, for example from carbapenem-resistant Klebsiella pneumoniae to A. baumannii [9].
AMR determinants are frequently captured on mobile genetic platforms such as plasmids and transposons, and in associated mobilizable elements such as integrons, which all represent multidrug resistance-conferring units [10–12]. Transposons are widespread genetic elements in bacteria [13] and in other domains of life. They move intra- and intermolecularly between different genomic positions within a cell in a process called transposition [14]. Through transposition, transposons can be inserted into conjugative plasmids or into bacteriophages, which facilitates the mobility of non-conjugative transposons between cells [12]. Thus, transposon-embedded AMR determinants can be mobilized at multiple hierarchical genetic levels, and intermolecular movement of transposons between different plasmids or between plasmids and chromosomal locations also occurs within clinical pathogens [15–18]. Therefore, the investigation of transposon mobility across different bacterial hosts could aid the understanding of AMR spread.
Stable integration of horizontally acquired mobile genetic elements into the bacterial recipient genome is crucial for long-term inheritance and is facilitated by plasmid establishment or rearrangement events between donor and recipient DNA such as homology-based recombination, site-specific recombination, or transposition [12]. A recent study demonstrated the transposition of a Tn21-like transposon (Tn3-family) of Salmonella enterica serovar Typhimurium 490 into the chromosome of Acinetobacter baylyi ADP1 in the course of natural transformation [19]. Tn21 insertion occurred independently at different loci in several transformants, and target site duplications (TSDs) at the insertion sites strongly suggested DNA recombination by transposition [19]. Since ADP1 harbours no transposons of the Tn3-family [20], these results indicated that the transposase was expressed from the incoming donor DNA. The authors concluded that a cytoplasmic, linear DNA double-strand formed after uptake, allowing gene expression and movement of the structural transposon [19].
In the present study, we used the Tn3-like replicative transposons Tn1 [21] and Tn4401 [22] as donor DNA to quantify and characterize transposition during natural transformation in molecular detail. As a recipient, we employed the naturally competent soil bacterium A. baylyi strain ADP1 also used by Domingues et al. [19]. ADP1 is transformable at high frequency by DNA from any source, including PCR products, which allows the detection and quantification of rare DNA recombination events [23, 24].
Methods
Bacterial strains and growth conditions
A. baylyi ADP1 strain BD413 Rpr (wild-type) [25] and the derivative mutants ΔrecA::tetA (JV37) [26], ΔdprA::aacC1 (NH29) [27], ΔrecO (KOM82) [27] and ΔrecBCD ΔsbcCD (KOM45) [28] were employed as recipient strains in natural transformation assays and have been described previously. The ΔxerC::nptII sacB mutant was constructed as reported previously for A. baylyi deletion strains [29, 30]. Briefly, DNA sequences of about 1000 bp each upstream and downstream of xerC (ACIAD2657) in ADP1 (GenBank CR543861) were PCR-amplified using primers xerC-up-f/xerC-up-r (upstream segment) and xerC-down-f/xerC-down-r (downstream segment; all primer sequences are listed in Table S1 available in the online version of this article). The PCR products were inserted sequentially into the plasmid vector pGT41 upstream and downstream of a nptII sacB selectable marker gene pair. The resulting plasmid contained a ΔxerC::nptII sacB allele embedded into its natural flanking regions and was used to naturally transform ADP1. The resulting XerCD-deficient transformant (kanamycin-resistant) was confirmed phenotypically and by PCR (primers xerC-up-f/xerC-down-r and xerC-ctrl/xerC-down-r; Table S1). PCR assays were carried out with high-fidelity Phusion DNA polymerase (Thermo Scientific) and standard parameters except using an additional 10 % dimethylsulfoxide, or with DreamTaq DNA polymerase (Thermo Scientific).
Escherichia coli strains EC100 (Epicentre), DH5α [31] or SF8 recA [32] were employed as host strains for strain constructions and donor DNA preparations. Strains were grown in Luria–Bertani (LB) medium at either 30 °C ( A. baylyi ) or 37 °C ( E. coli ). For transformant selection, media were supplemented with antibiotics at the following concentrations: ampicillin, 100 mg l−1 (Tn1 in pJK2, pJKH1, pJK7 and pTn4401); streptomycin, 40 mg l−1 (RSF1010); kanamycin, 10 mg l−1 (pKH80-PCR).
Donor DNA preparations for natural transformation experiments
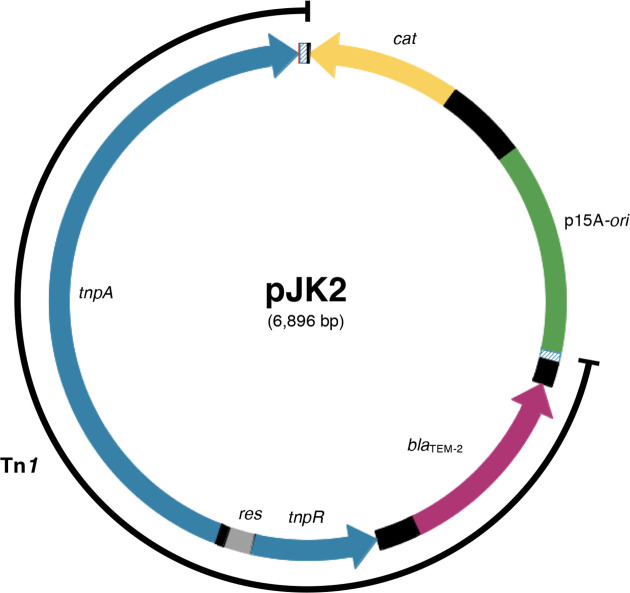
Plasmid pJK2 (6896 bp; Fig. 1) was constructed as follows: transposon Tn1 on plasmid RP4 (4951 bp; GenBank BN000925) was PCR-amplified from the left to the right border without TSDs using DNA from E. coli K12 J53 RP4 [German Collection of Microorganisms and Cell Cultures (DSMZ), DSM 3876] as template and primer Tn1-f+r (Table S1) as forward and reverse primer. The resulting PCR product was 5′-phosphorylated with polynucleotide kinase (Thermo Scientific) and ligated to a 1945 bp PCR product containing the p15A origin of replication and the chloramphenicol resistance gene cat of plasmid pACYC184 [33] (GenBank X06403; primers cat-r and p15A-r1; Table S1) using T4 DNA ligase (Fermentas). pJKH1 (carrying Tn1ΔtnpA; 4289 bp) was derived from pJK2 by digestion with EcoRV and circularization of the large fragment (T4 DNA ligase). The partial deletion of tnpA (869 internal of 1002 codons) was confirmed by restriction analysis. To construct plasmid pJK7 (Tn1ΔtnpR; 6399 bp), we PCR-amplified pJK2 excluding tnpR with primers pJK2-tnpR-del-f and pJK2-tnpR-del-r (Table S1). The PCR-product was 5′-phosphorylated (polynucleotide kinase) and circularized (T4 DNA ligase), and the deletion of tnpR was confirmed by Sanger sequencing. The plasmids RSF1010, pKH80 and pTn4401 have been described previously [28, 34–36]. Plasmid donor DNA was isolated using the Qiagen Plasmid Extraction kit (Qiagen) and genomic donor DNA was purified with the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich).
Fig. 1.
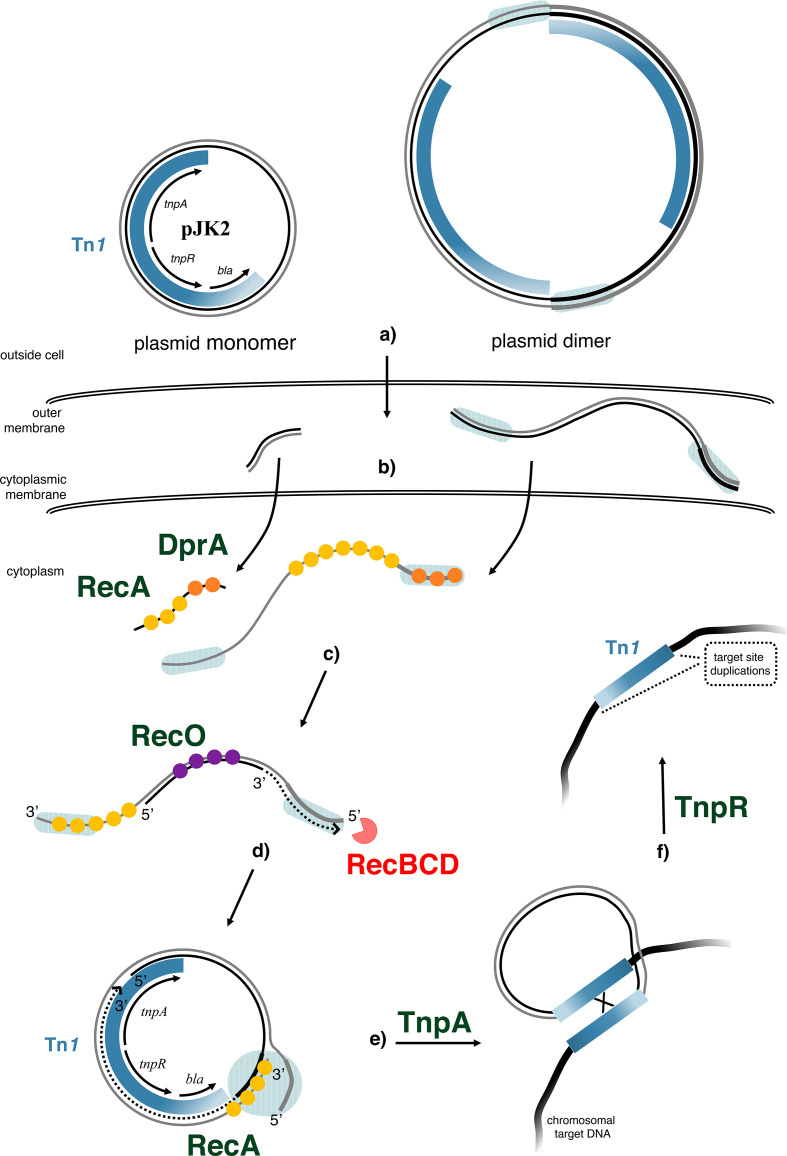
Map of plasmid pJK2. The plasmid backbone contains the p15A origin of replication (green) and the cat gene (yellow; chloramphenicol resistance) of the E. coli cloning vector pACYC184. Transposon Tn1 (4.9 kb) was cloned from plasmid RP4. In Tn1, tnpA and tnpR (blue) encode the transposase and resolvase, respectively. Ampicillin resistance is conferred by bla TEM-2 (magenta). The transposon contains a res site for cointegrate resolution and is flanked by two terminal 38 bp inverted repeats (dashed). Neither Tn1 nor pACYC184 display significant sequence identity with the A. baylyi genome.
When PCR products were employed as donor DNA, PCR reactions were performed using high-fidelity Phusion DNA polymerase. Linear donor DNA substrate pJK2-PCR (6908 bp) was obtained by inverse PCR amplification of pJK2 with primers cat-f and p15A-ori-f-c (Table S1). This PCR product carried a 12 bp overlap at the ends. Donor DNA substrate pKH80-PCR (4760 bp) was PCR-amplified with primers sbcD-up-f and sbcC-down-r (Table S1) from plasmid pKH80. This substrate carried two approximately neighbouring 1.0 kbp DNA stretches from the A. baylyi chromosome covering parts of ACIAD0915 and ACIAD0918, interrupted in the centre by a 2.7 kbp nptII sacB marker gene cassette conferring kanamycin resistance. The PCR products were purified using the Qiaquick PCR purification kit (Qiagen). When indicated, pJK2-PCR was purified by agarose gel electrophoresis (Sea Plaque, LONZA) and subsequent recovery using the Qiaquick Gel Extraction kit (Qiagen).
Natural transformation assays in liquid medium
Preparation of naturally competent cells of A. baylyi as well as natural transformation assays were conducted as described previously [29]. In brief, competent cell stocks were prepared by dilution of an overnight culture 1:100 into liquid LB and growth of that culture under aeration until logarithmic growth (1×109 cells ml−1 determined with a haemocytometer) was reached. The cells were chilled, centrifuged for 10 min at 5000 g and 4 °C, and concentrated 1:10 in LB media containing 20 % glycerol (v/v). Finally, the cells were aliquoted and stored at −80 °C until further use.
For transformation assays, aliquoted competent cells were thawed on ice, diluted 1:40 in liquid LB to 2.5×108 cells ml−1, and DNA was added at 100 ng ml−1 unless indicated otherwise. The assays were incubated under aeration for 90 min before the cells were chilled on ice, washed and resuspended in phosphate-buffered saline. Appropriate dilutions were plated on LB (recipient titre) and media containing selective antibiotics (transformant titre). LB plates were incubated for 24 h while transformant plates were incubated 48 to 72 h. Colonies were counted and transformation frequencies were calculated as transformants per recipient. If no transformant colonies were obtained, the limit of detection was calculated instead. In control experiments, recipient cells were incubated without donor DNA to identify resistant mutants arising and unselective and selective plating were performed according to the respective assay with donor DNA.
Verification of transposition events
To identify transposition events, ampicillin-resistant transformant colonies were picked and restreaked three times on selective medium. In some cases, an intermittent cultivation step on non-selective medium was carried out. Typically, isolates were investigated after the final restreak by PCR for the presence of Tn1 (primer Tn1-f+r) and pJK2 vector backbone DNA (primers cat-r/p15A-r1), or for the presence of Tn4401 (primers KPC-A/KPC-B) and pTn4401 vector backbone DNA (primers cat-r/p15A-r1), respectively (Table S1). Occasionally, ampicillin-resistant mutants arose during these cultivation steps, presumably through mutations. These isolates typically displayed an aberrant colony phenotype and were distinguished by the absence of transposon and pJK2 vector DNA in PCR analyses. These procedures clearly separated so-termed transposants from transient (unstable) transformants and mutants. Divergent transformant types were found in experiments using wild-type ADP1 with pJK7 donor DNA, or strain ΔrecBCD ΔsbcCD with pJK2 donor DNA (see the Results and Discussion section for details).
To verify transposants, genomic DNA was isolated from stable ampicillin-resistant isolates that were PCR-positive for Tn1 and PCR-negative for pJK2 vector backbone, using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich). Purified DNA was used as template for Sanger DNA sequencing (BigDye 3.1 technology; Applied Biosystems) as follows: 1 µg genomic DNA was mixed with 4 µl BigDye, 2 µl BigDye buffer and 10 µM of primer (either bla-ins-f or Tn1-tnpA-ins-f; Table S1) in a volume of 20 µl. Assays were denatured for 5 min at 95 °C, followed by 99 cycles of 30 s at 95 °C, 10 s at 55 °C and 4 min at 60 °C, and subsequently analysed on an Applied Biosystems 3130xl Genetic Analyzer (in-house sequencing facility). blast was used to identify both recombinant joints of Tn1 with the chromosome of ADP1 (GenBank CR543861.1) and to verify the TSDs (Tables S2 and S3). The transposition frequency was calculated as transposants per transformant isolates multiplied by the transformation frequency.
Characterization of target site sequences
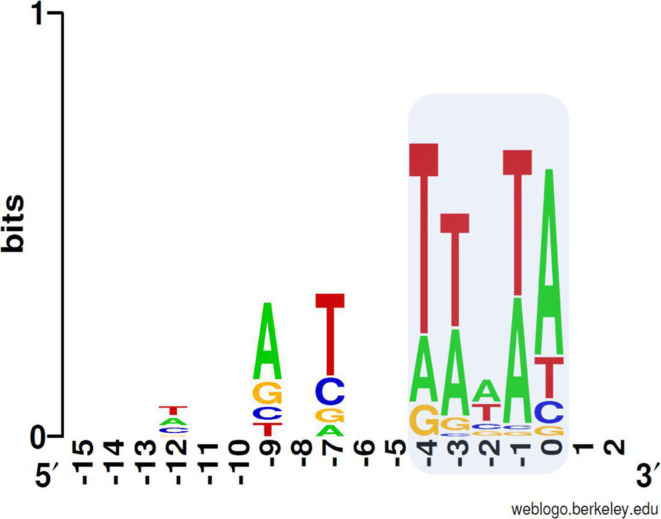
A consensus DNA sequence logo was generated by multiple sequence alignment and visualized using WebLogo software [37] (Figs 2 and Fig. S1) The aligned regions covered the 5 bp TSD plus 45 nucleotides upstream and downstream of the duplication (Table S4). The total number and positions of this TSD consensus sequence (TTWTA) were determined for the chromosome of ADP1 using Gene Construction kit v 4.0.3 (Textco BioSoftware, Inc.). Linear regression analysis of the TSD consensus sequence distribution over the chromosome of ADP1 was performed using GraphPad v 8.4.2 (GraphPad Software; P *** <0.001, ** <0.01, * <0.05).
Fig. 2.
Consensus DNA sequence logo for Tn1-insertions into the chromosome of ADP1. Blue background colour indicates the target site duplication (TSD) consensus sequence at positions −4 to 0. Additional conserved positions around the TSD are shown. The TSD sequence motif (TTWTA; W: T or A) and preference for T at position −7 are typical for this transposon family. The logo was generated using WebLogo [37].
Susceptibility testing
The minimal inhibitory concentrations (MICs) were determined using gradient diffusion strips (Liofilchem) following the manufacturer’s instructions. Briefly, 0.5 McFarland solutions in 0.9 % saline (w/v) were prepared from freshly grown colonies on LB and spread evenly onto Müller–Hinton agar plates with a sterile cotton swab. A gradient strip for ampicillin was applied and plates were incubated at 30 °C for 18 h. Results were read at 100 % growth inhibition (n=1 per strain).
Electroporation assays
Electrocompetent cells of A. baylyi were prepared as published for E. coli [38] with modifications. Briefly, a logarithmic culture of A. baylyi was grown at 30 °C in LB to 2.5×108 cells ml−1. The cells were chilled, washed twice with ice-cold distilled water, and concentrated 500 times in 10 % ice-cold glycerol solution (v/v). A 40 µl aliquot was mixed with DNA (400 ng pJK2 or 200 ng RSF1010 DNA), transferred into a 2 mm gap electroporation cuvette and pulsed with 12.5 kV cm−1 (25 µF, 200 Ω) using a BioRad electroporator. Next, the cells were suspended in 1 ml prewarmed SOC (2 % tryptone, 0.5 % yeast, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) and aerated for 1 h at 30 °C. Appropriate dilutions were plated on LB (recipient titre) and LB containing antibiotics (transformants) and the plates were incubated at 30 °C for 16 to 20 h or 36 to 48 h, respectively.
Results and Discussion
Natural transformation by transposon-containing DNA
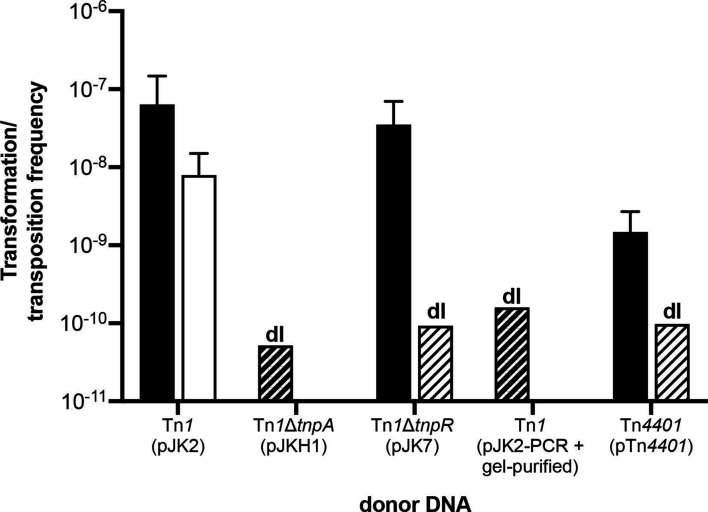
We inserted the replicative transposon Tn1 into the narrow-host-range plasmid vector pACYC184 that replicates in Enterobacteriaceae but not in A. baylyi . The resulting circular plasmid pJK2 (Fig. 1) was used as donor DNA to naturally transform A. baylyi ADP1 wild-type cells. Ampicillin-resistant transformant colonies were obtained at a frequency of (6.4±8.4)×10−8 per recipient cell (Fig. 3).
Fig. 3.
Frequencies of transformation (black bars) and transposition (white bars) in A. baylyi wild-type using different transposon-containing donor DNA substrates: pJK2 (Tn1; n=3), pJKH1 (Tn1ΔtnpA; n=4), pJK7 (Tn1ΔtnpR; n=3), pJK2-PCR (agarose gel-purified; n=3) and pTn4401 (Tn4401; n=9). Bars represent the mean with standard deviation. Striped bars indicate the detection limit (dl) when no transformation or transposition events were observed.
Analyses of resistant colonies revealed two distinguishable groups of transformants. In the first group, ampicillin resistance was stably maintained after repeated restreaking on selective medium and also after intermittent non-selective cultivation. PCR analyses showed the presence of Tn1 but the absence of pJK2 vector backbone DNA. This result indicates that Tn1 was chromosomally acquired by DNA uptake and transposition. In contrast, the ampicillin resistance was unstable in transformants of the second group, resulting in heterogeneous colony size and shape when restreaked. The resistance was generally lost after repeated purification on selective medium or after intermittent non-selective cultivation, and both Tn1 and pJK2 backbone DNA could be PCR-amplified from cell material of the first, and with decreasing frequency of subsequent recultivations. This result suggests that transformants of group 2 received the pJK2 plasmid and became resistant through transient expression of bla TEM-2, but lost the plasmid and resistance with further cultivation due to the inability of pJK2 to propagate stably in A. baylyi .
To confirm that transformants of the first group occurred by transposition of Tn1 into the recipient chromosome, we determined the DNA sequences upstream and downstream of Tn1 by Sanger sequencing of genomic DNA. We analysed 20 group 1 isolates with circular pJK2 DNA and an additional 8 isolates obtained with linear pJK2-derivatives (Supplemental Information: Supplemental Results). Altogether, we identified 29 transposition events of Tn1 into the chromosome of ADP1, including one isolate with two transposons (Table S2). In all transposition transformants (‘transposants’), Tn1 was found as a chromosomal insert with a 5 bp TSD at the recombinant joints, which is a hallmark of transposase activity. The consensus sequence of the TSD is in agreement with previous reports on Tn3-family transposons (Fig. 2) [39, 40]. We did not identify fragments of pACYC184 vector DNA or indications of non-transposition recombination events. The MIC of ADP1 for ampicillin (2 μg ml−1) was increased to >256 μg ml−1 in eight randomly chosen transposants (Table S2). Together, these results confirm that the transposants have acquired Tn1 during natural transformation through transposition.
The frequency of transposants using pJK2 donor DNA was (7.9±7.1)×10−9 (Fig. 3). To compare this transposition frequency to extra-chromosomal plasmid establishment or to homologous recombination during natural transformation, we used circular plasmid DNA (RSF1010) or linear homologous DNA (pKH80-PCR) in transformation assays with A. baylyi and obtained frequencies of (5.4±1.1)×10−6 and (1.3±0.5)×10−4, respectively. These data indicate that Tn1 transposition in the course of natural transformation is about 680 times lower than plasmid aquisition and about 16 000 times lower than homologous recombination during natural transformation. Nonetheless, the transposant frequency was higher than illegitimate recombination frequencies with fully heterologous donor DNA in A. baylyi [24, 41]. We conclude that transposon spread through natural transformation can occur at biologically relevant frequencies, although reports showing this process in situ are currently lacking. The importance of transposant formation in the environment or for AMR spread in the hospital is unknown but probably low compared with conjugation.
Chromosomal distribution of transposition events
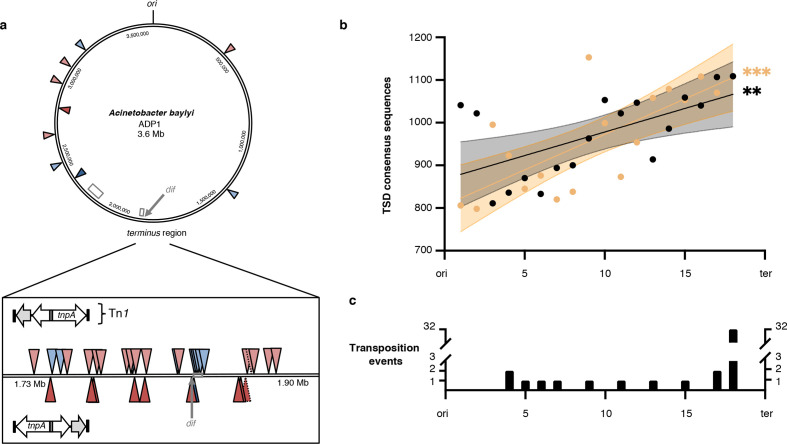
All but one of the Tn1 insertions occurred in nonessential genes [42] or in intergenic regions of the ADP1 chromosome (Tables S2 and S3). However, the distribution of insertions was nonrandom, and 24 of 29 transpositions clustered within a 170 kbp sector around the assumed terminus of replication (Fig. 4a) [20]. This distribution was supported by 14 additional unique Tn1 insertions recovered from an A. baylyi ΔrecBCD ΔsbcCD strain (Supplemental Information: Supplemental Results and Fig. 4a). To further characterize the distribution bias, we determined the total number of TSD consensus sequence sites and their positions across the A. baylyi chromosome. The TSD consensus sequence positions (n=34 863) followed a normal distribution, indicated by a small difference between the median and mean (0.16 %) relative to genome size. Although the incidence of TSD consensus sequence hits increases significantly along the origin–terminus axis of replication (Fig. 4b), even the highest increase in hit counts (30 %) does not explain why we observed more than 80 % of the verified Tn1 insertions in a region representing only 5 % of the ADP1 chromosome (Fig. 4c).
Fig. 4.
Characterization of Tn1 insertion sites. (a) Chromosome map of ADP1 with a magnified section of ∼170 kbp (box) around the approximate terminus of replication (ter; G-C/G+C skew inversion site). This region contains 174 open reading frames, of which 12 are essential [42]. The two major prophage regions of ADP1 and the dif site for chromosome partitioning by XerCD are indicated by grey open boxes and grey arrows, respectively [20, 47]. Tn1 insertions are marked as triangles (red, wild-type; blue, ΔrecBCD ΔsbcCD). The orientation of Tn1 is indicated by the colour intensity (light, transcriptional orientation of tnpA in sense; dark, tnpA in antisense). A dashed triangle edge specifies insertion sites in a double transposant. Thirty-four out of 43 insertions occurred around the terminus (24 in wild-type and 10 in the ΔrecBCD ΔsbcCD strain). (b) Number and distribution of TSD consensus sequences across the chromosome of ADP1, which was bi-directionally grouped into 2×18 100 kb segments from origin (ori) to ter. The number of TSD consensus sequence hits per group is plotted along the ori–ter axis (black and orange, first and second replichore, respectively). For both directions, linear regression showed a significant increase in the number of hits (first replichore: P=0.0076, R 2=0.5544; second replichore: P=0.0004, R 2=0.3683; 95 % confidence intervals are indicated); lowest hit number: 798 (group 2, second replichore), highest hit number: 1153 (group 9, second replichore). (c) Number of experimentally observed transposition events per region.
A comparable transposon insertion bias has been observed previously, although not in the context of natural transformation. Tn917 is a Tn3-like transposon in Gram-positive bacteria and preferentially inserts at the terminus in B. subtilis [43] and Entercoccus faecalis [44], but not in staphylococci [45, 46]. The bias is less strong in B. subtilis cells lacking functions involved in postreplicational chromosome segregation (RipX, SpoIIIE) or lacking the replication termination protein RTP [43]. In B. subtilis, codV and ripX encode the dif-directed chromosome partitioning recombinase often termed XerCD in Gram-negative bacteria [47]. We hypothesized that the absence of XerCD would alter the observed insertion bias of Tn1 in A. baylyi . We naturally transformed an ADP1 ΔxerC mutant by pJK2 donor DNA but did not obtain any transformants (detection limit: 2.2×10−10; n=3). The reason for this result is unclear. Further experimental investigation was impeded by the genetic limitations of the employed recipient strain: A. baylyi carries no gene homologous to rtp (tus in E. coli ) [24], and the orthologue of spoIIIE (ftsK) is essential in A. baylyi [42]. Using RSF1010 as donor DNA for the A. baylyi ΔxerC strain, the transformation frequency was (1.4±0.7)×10−6 (n=3), which was only four times decreased compared to the wild-type.
Finally, we hypothesized that the distribution of Tn1 insertions would be different using artificial transformation (electroporation). During electroporation, circular double-stranded DNA is directly delivered to the cytoplasm. However, no ampicillin-resistant isolates were obtained after electroporation of A. baylyi by circular pJK2 DNA (detection limit: 3.6×10−10; n=3). In contrast, electroporation by RSF1010 (n=1) resulted in streptomycin-resistant transformants at a frequency of 5.6×10−5, which was 10 times higher than the frequency observed with natural transformation of A. baylyi . The reason for the distribution bias of transposition events remains unknown.
Requirement of transposon genes for transposition
Tn1 carries two genes involved in transposition: tnpA is the transposase gene, and tnpR encodes the resolvase for cointegrate intermediates created by TnpA. TnpR also acts as a repressor for both tnpA and tnpR [48] (Fig. 1). We hypothesized that transposition and cointegrate resolution during natural transformation were conferred by the tnpA and tnpR gene products encoded by the donor DNA and that deletion of these genes would decrease transposant frequency. We removed the tnpA gene from Tn1 of pJK2, and natural transformation of A. baylyi by the resulting plasmid pJKH1 yielded no ampicillin-resistant transformants (detection limit: 5.2×10−11; Fig. 3), confirming that tnpA of Tn1 but no recipient functions act as transposase for Tn1.
We also used plasmid pJK7 (carrying Tn1ΔtnpR) as donor DNA for the transformation of A. baylyi and the transformation frequency was similar to that obtained with pJK2 [(3.5±3.5)×10−8; Fig. 3]. We PCR-screened 150 transformant isolates that were stably resistant after three consecutive streakouts and found that Tn1 and pJK2 vector backbone was detectable in all isolates. This transformant type was not detected with pJK2 but is consistent with unresolved cointegrate intermediates. We conclude that in the absence of TnpR, the derepressed TnpA efficiently recombined Tn1 with the recipient and formed cointegrates that remained unresolved. Recipient functions such as RecA may contribute to their eventual resolution, but, taken together, these findings indicate that the main resolvase is the TnpR encoded by the donor DNA.
Formation of double-stranded cytoplasmic intermediates
Taken together, the presented results demonstrate that tnpA and tnpR genes as well as bla TEM-2- in the transient transformants of group 2 described above are expressed after uptake of DNA into the cytoplasm. Moreover, our data suggest that a circular intermediate is involved in the first (cointegration) step of transposition. However, the donor DNA is transported as linear single-strands into the cytoplasm during natural transformation [49, 50]. We hypothesized that the uptake of two complementary DNA single-strands is required for double-strand formation by hybridization. The resulting linear DNA double-strands, however, would be susceptible to degradation by exonucleases such as RecBCD [51, 52-]. Subsequent circularization by annealing at overlapping ends, followed by fill-in DNA synthesis or gap repair, would protect these double-strands from exonucleolytic degradation [53].
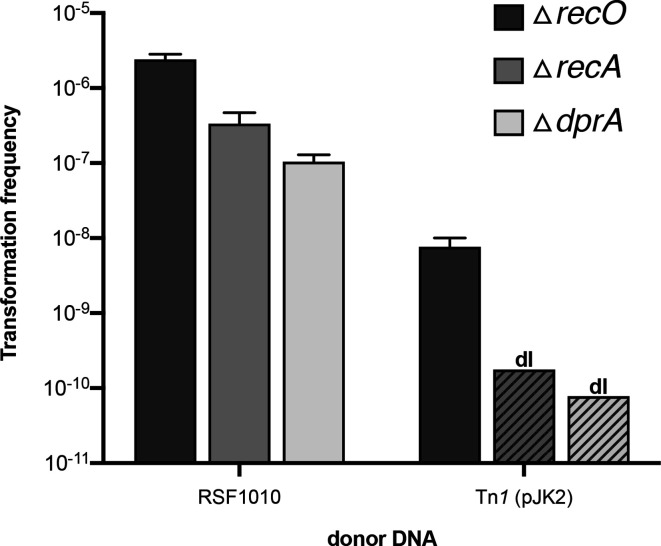
In B. subtilis, recO is required for efficient plasmid transformation [54], which can be explained by the activity of RecO to hybridize complementary DNA single-strands [55]. To investigate the role of RecO in double-strand formation in A. baylyi, we transformed a ΔrecO strain by circular pJK2 DNA. The resulting frequency of ampicillin-resistant transformants was eight times lower than in wild-type A. baylyi [(7.7±2.4)×10−9; Fig. 5]. With RSF1010 as donor DNA, the frequency was (2.4±0.4)×10−6, which was two times lower than in wild-type A. baylyi (Fig. 5).
Fig. 5.
Transformation frequencies of A. baylyi mutant strains (ΔrecO, ΔrecA or ΔdprA) by circular donor DNA substrates (RSF1010 or pJK2). Bars represent the mean with standard deviation from three to five experiments. Striped bars indicate the detection limit (dl) when no transformation or transposition events were observed.
Unlike recO, the recA gene is not required for plasmid transformation of B. subtilis [54], suggesting that RecO is sufficient to restore circular plasmids from two complementary single-strands in this organism. We tested whether this was also the case in A. baylyi . In a ΔrecA mutant transformed by circular RSF1010 DNA, plasmid transformants occurred at a frequency of (3.4±1.3)×10−7 (Fig. 5), which was ~16 times lower than in wild-type A. baylyi . With pJK2 as donor DNA, no transformants were obtained (detection limit: 1.8×10−10; Fig. 5). This result suggests an unexpected role of RecA in plasmid circularization in A. baylyi .
The DprA (DNA processing A) protein is thought to load incoming DNA single-strands with RecA protein [56]. In many naturally competent bacteria, deletion of dprA abolishes or severely reduces natural transformation [27, 57–60]. We investigated whether natural transformation by circular extrachromosomal DNA was affected in an A. baylyi ΔdprA mutant. Using pJK2 as donor DNA, the transformation frequencies of the ΔdprA strain dropped below detection limit (7.8×10−11) (Fig. 5). With RSF1010, the transformation frequency was (1.4±0.2)×10−7 and thus approximately 50 times lower than that of the wild-type (Fig. 5). These frequencies are comparable with those using the ΔrecA strain as recipient. The results support the assumption that DprA acts upstream of the RecA recombination pathway. They also demonstrate that plasmid transformation (RSF1010) is not abolished in the absence of DprA.
Our findings show that deficiencies of RecA, DprA and, to a lesser degree, RecO reduce transformation of circular extrachromosomal DNA in A. baylyi , suggesting a role of these functions in plasmid circularization. In our experiments, the non-replicative plasmid pJK2 generally yielded poorer transformation frequencies than the replicative plasmid RSF1010. We cannot exclude the possibility that some or all of the investigated genes also modulate the transposition efficiency in addition to double-strand conversion and circularization.
Next we tested whether circularization is a requirement for transposons to jump during natural transformation. We used an inverse PCR product of pJK2 as donor DNA with Tn1 approximately in the centre (pJK2-PCR). The substrate was agarose gel-purified to eliminate all traces of contaminating template DNA (confirmed by PCR; Supplemental Information: Supplemental Results). Using this linear DNA substrate for transformation of wild-type A. baylyi , no ampicillin-resistant transformants were obtained (detection limit: 1.6×10−10, Fig. 3), suggesting that pJK2-PCR is insufficient to produce both transient transformants and transposants, presumably due to the cytoplasmic instability of linear double-stranded intermediates.
Antagonistic activity of RecBCD
Cytoplasmic linear double-stranded DNA is target for degradation by the RecBCD exonuclease in many bacteria [52] and the absence of the nuclease may protect linear intermediates and enhance plasmid circularization. In A. baylyi , RecBCD is thought to destroy donor DNA following uptake into the cytoplasm after the creation of double-stranded ends during homologous recombination [30]. However, in the context of plasmid transformation, in B. subtilis strains lacking AddAB, the functional equivalent of RecBCD, transformation frequencies are somewhat reduced [61]. To investigate the effect of RecBCD deficiency on plasmid transformation in A. baylyi , we transformed a ΔrecBCD ΔsbcCD strain by RSF1010 and obtained a 70 times increased transformation frequency [(3.9±0.4)×10−5] compared with the wild-type (Fig. 6). We also transformed the ΔrecBCD ΔsbcCD strain by Tn1-containing donor substrates. Using a gel-purified linear pJK2-PCR substrate, no transformants were obtained (detection limit: 4.9×10−10; n=3), strongly suggesting that a circular intermediate is required for transposition and transformation. With circular pJK2 donor DNA the frequency of ampicillin-resistant transformants was (6.8×±5.6)×10−4 (Fig. 6), which was four orders of magnitude higher than that of the wild-type. Analysis of 40 ampicillin-resistant isolates revealed stable resistance even after repeated recultivation. PCR analyses verified the presence of Tn1 and pJK2 vector backbone DNA in all isolates and circular pJK2 plasmid DNA could be isolated from cell material of restreaks, indicating extrachromosomal DNA rather than cointegrates. No transposants were identified during our standard screening, but a transformational screening approach revealed 14 unique chromosomal insertions (Supplemental Information: Supplemental Results and Table S3). These results were unexpected, and we concluded that established pJK2 replicated relatively stably in A. baylyi in the absence of RecBCD. The parental plasmid of pJK2, pACYC184 [33], propagates as a theta replicon in E. coli and in other Enterobacteriaceae [62]. It is possible that absence of RecBCD allows rolling circle replication of pACYC184 in A. baylyi , as in E. coli RecBCD-deficient mutants [62]. In wild-type E. coli , the exonucleolytic activity of RecBCD is thought to degrade the nascent multimers of the rolling circle, forcing the plasmid into theta replication. In contrast, loss of RecBCD allows rolling circle replication [62].
Fig. 6.
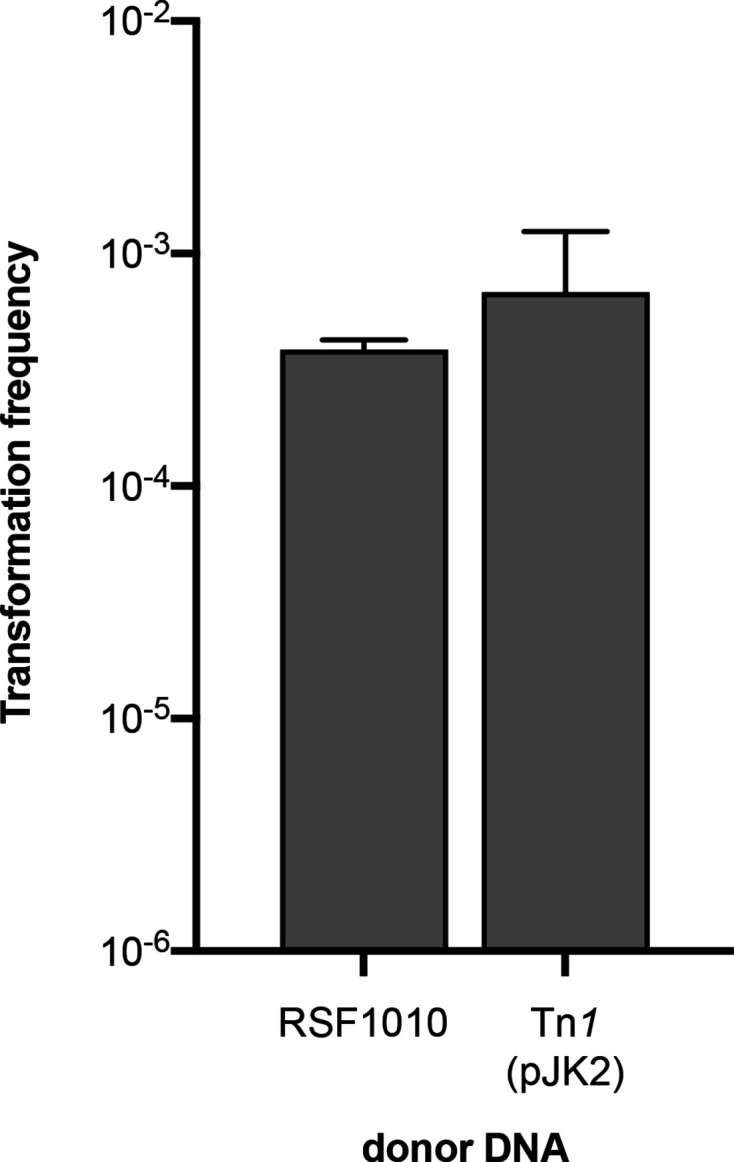
Transformation frequencies of the ΔrecBCD ΔsbcCD strain using plasmid DNA substrates (RSF1010 or pJK2; n=5). Bars represent the mean with standard deviation.
A model for natural transformation by transposons
In this study we experimentally verify that bacteria can acquire transposons horizontally through transposition in the course of natural transformation, as observed previously for a Tn21 transposon [19]. In control experiments, we investigated the circularization of a replicative plasmid in A. baylyi during transformation, which has been studied before in B. subtilis [54]. Our results indicate differences in plasmid transformation between the two species. In contrast to our results, absence of RecA in B. subtilis did not affect plasmid establishment; instead RecO protein was required for that process [54].
Based on our findings, we propose a model for plasmid transformation and transposition in A. baylyi (Fig. 7). We suggest that sporadic dimeric forms of plasmid DNA with redundant ends are the main substrate for cytoplasmic donor DNA circularization. To initiate circularization, a small single-strand fragment is annealed with a large dimeric complementary strand with 5′- and 3′-overhangs. DNA fill-in synthesis converts the 5′-overhang into a double-strand (using the 3′-recessed end as primer), while the 3′-overhang is charged with RecA protein. It is consistent with our findings that DprA has a RecA-loading function [56]. The resulting nucleoprotein filament can undergo homology search. When it finds the homology of the redundant double-stranded end, the result is a circular intermediate with a displaced strand, and textbook DNA repair and gap synthesis generate a circular double-strand. This model is supported by lack of transformants with linear pJK2-PCR as donor DNA, since this substrate did not contain dimeric DNA molecules. The resulting circular double-stranded intermediate is temporally protected from DNA degradation (Fig. 7). Finally, the tnpA and tnpR genes are expressed and can lead to transposition of Tn1.
Fig. 7.
Model of Tn1 transposition during natural transformation of A. baylyi . Proteins with beneficial or necessary functions are represented in green, and proteins with antagonistic functions are red; exemplary sequence segments providing homology are indicated by a shaded background. (a) Circular monomeric as well as sporadic dimeric forms of plasmid DNA are available for uptake into the periplasm. (b) DNA single-strands are taken up into the cytoplasm and protected from degradation by DprA (orange). DprA also loads RecA (yellow) onto single-stranded DNA. (c) RecO (purple) is involved in the annealing of DNA single-strands [55], in initiating or improving gap repair as part of RecFOR or RecOR [63, 64], or both. The linear double-stranded molecule is susceptible to degradation by RecBCD exonuclease (red). (d) The RecA-loaded DNA 3’-single-strand end invades the double-stranded fragment at its terminal homology, and circularization of the donor plasmid is completed by DNA repair and gap synthesis. (e) From the established circularized plasmid, the genes necessary for transposition (tnpA, tnpR) are expressed, and TnpA together with a recipient DNA polymerase form a transposon–target DNA cointegrate and generate the TSD. (f) TnpR resolves the cointegrate, and the recipient chromosome contains now a copy of Tn1.
Is circular double-stranded DNA in general a necessary requirement for transposition in the course of natural transformation? Probably not. Domingues et al. obtained transposants using chromosomal DNA for natural transformation of A. baylyi [19]. Hypothetically, such DNA can form circular intermediates at direct repeats, and transposons surrounded by direct repeats may be stabilized through DNA circularization. It is also conceivable that Chi sequences surrounding the transposon protect the double-stranded intermediate from exonucleolytic degradation [51].
To put the results of this study into broader perspective, we employed a published transposon-containing plasmid as donor DNA for transformation of ADP1. The pACYC184 derivative pTn4401 (15.6 kbp) [35] carries the replicative transposon Tn4401 (~10 kbp) of the Tn3-family that contains the carbapenemase gene bla KPC-2 [22]. Ampicillin-resistant transformants formed at a frequency of (1.5±1.2)×10−9 (Fig. 3), which was 40 times lower than the frequency observed with pJK2 DNA. Among 60 investigated transformants, no transposants were found (detection limit: 9.8×10−11). Several explanations for the lack of transposition of Tn4401 are conceivable: the MIC conferred by KPC-2 may be too low to detect transposants in this organism. The plasmid size possibly reduces the dimer/monomer ratio. Alternatively, the repression of tnpA may be tighter in Tn4401 than in Tn1. Taken together, this result shows that additional constraints exist for transposition during natural transformation, and further investigations are needed.
In conclusion, we showed that transposition of Tn1 in the course of natural transformation occurs at biologically relevant frequencies. Our results open up the possibility that transposable elements can even spread from dead cells, where fully heterologous, transposon-containing free DNA can transform naturally competent bacteria, albeit at low frequencies. Transposons play an important role in the dissemination of multi-drug resistance, and the identification of both the transferred resistance genes and the genetic context that was transferred is crucial to understand AMR spread.
Supplementary Data
Funding information
J. K. received a project stipend from The Norwegian Pharmaceutical Society. K. H. was funded by the Research Council of Norway (grant number 275672).
Acknowledgements
Plasmid pTn4401 was a kind gift from Gaëlle Cuzon and Thierry Naas. We are grateful to João A. Gama for help with target site distribution and Kaare M. Nielsen and Sören Abel for helpful discussions.
Author contributions
K. H. conceived the project; J. K. performed the experiments; J. K. and K. H. analysed the data; J. K. and K. H. wrote the paper with input from P. J. J. and K. H.; and P. J. J. supervised the project. All authors approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; dl, detection limit; LB, Luria-Bertani; MIC, minimal inhibitory concentration; TSD, target site duplication.
One supplementary figure and four supplementary tables are available with the online version of this article.
References
- 1.von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 4.Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/MMBR.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P, et al. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 6.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 7.Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019;65:34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- 8.Domingues S, Rosário N, Cândido Â, Neto D, Nielsen KM, et al. Competence for Natural Transformation Is Common among Clinical Strains of Resistant Acinetobacter spp. Microorganisms. 2019;7:E30. doi: 10.3390/microorganisms7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traglia GM, Place K, Dotto C, Fernandez JS, Montaña S, et al. Interspecies DNA acquisition by a naturally competent Acinetobacter baumannii strain. Int J Antimicrob Agents. 2019;53:483–490. doi: 10.1016/j.ijantimicag.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/MMBR.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A. Resistance plasmid families in Enterobacteriaceae . Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31:e00088–17.:UNSP. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tansirichaiya S, Rahman MA, Roberts AP. The transposon registry. Mob DNA. 2019;10:40. doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman AB, Dyda F. Mechanisms of DNA transposition. Microbiol Spectr. 2015;3:MDNA3-0034–3-2014. doi: 10.1128/microbiolspec.MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez T, Vázquez GJ, Aquino EE, Martínez I, Robledo IE, et al. ISEcp1-mediated transposition of bla KPC into the chromosome of a clinical isolate of Acinetobacter baumannii from Puerto Rico. J Med Microbiol. 2014;63:1644–1648. doi: 10.1099/jmm.0.080721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, et al. Nested russian Doll-Like genetic mobility drives rapid dissemination of the carbapenem resistance gene bla KPC . Antimicrob Agents Chemother. 2016;60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brolund A, Rajer F, Giske CG, Melefors Ö, Titelman E, et al. Dynamics of resistance plasmids in extended-spectrum-β-lactamase-producing Enterobacteriaceae during postinfection colonization. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.02201-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans DR, Griffith MP, Sundermann AJ, Shutt KA, Saul MI, et al. Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single Hospital. eLife. 2020;9 doi: 10.7554/eLife.53886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, et al. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012;8:e1002837. doi: 10.1371/journal.ppat.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, et al. The Tn3-family of replicative transposons. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- 22.Naas T, Cuzon G, Villegas M-V, Lartigue M-F, Quinn JP, et al. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott KT, Neidle EL. Acinetobacter baylyi ADP1: transforming the choice of model organism. IUBMB Life. 2011;63:1075–1080. doi: 10.1002/iub.530. [DOI] [PubMed] [Google Scholar]
- 24.Harms K, Lunnan A, Hülter N, Mourier T, Vinner L, et al. Substitutions of short heterologous DNA segments of intragenomic or extragenomic origins produce clustered genomic polymorphisms. Proc Natl Acad Sci U S A. 2016;113:15066–15071. doi: 10.1073/pnas.1615819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen KM, van Weerelt MD, Berg TN, Bones AM, Hagler AN, et al. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:1945–1952. doi: 10.1128/AEM.63.5.1945-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overballe-Petersen S, Harms K, Orlando LAA, Mayar JVM, Rasmussen S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci U S A. 2013;110:19860–19865. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hülter N, Sørum V, Borch-Pedersen K, Liljegren MM, Utnes ALG, et al. Costs and benefits of natural transformation in Acinetobacter baylyi . BMC Microbiol. 2017;17:34. doi: 10.1186/s12866-017-0953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harms K, Wackernagel W. The RecBCD and SbcCD DNases suppress homology-facilitated illegitimate recombination during natural transformation of Acinetobacter baylyi . Microbiology. 2008;154:2437–2445. doi: 10.1099/mic.0.2008/018382-0. [DOI] [PubMed] [Google Scholar]
- 29.Harms K, Schön V, Kickstein E, Wackernagel W.The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi .Mol Microbiol 200764691–702. 10.1111/j.1365-2958.2007.05692.x [DOI] [PubMed] [Google Scholar]
- 30.Kickstein E, Harms K, Wackernagel W. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi . Microbiology. 2007;153:2259–2270. doi: 10.1099/mic.0.2007/005256-0. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Romanowski G, Lorenz MG, Wackernagel W. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl Environ Microbiol. 1993;59:3438–3446. doi: 10.1128/AEM.59.10.3438-3446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/JB.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, et al. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 35.Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in bla KPC gene mobilization. Antimicrob Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer JT, van Tuijl JJ, Finnerty WR. Transformation and mobilization of cloning vectors in Acinetobacter spp. J Bacteriol. 1986;165:301–303. doi: 10.1128/JB.165.1.301-303.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia J-M, Brenner SE. Weblogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seringhaus M, Kumar A, Hartigan J, Snyder M, Gerstein M, et al. Genomic analysis of insertion behavior and target specificity of mini-Tn7 and Tn3 transposons in Saccharomyces cerevisiae . Nucleic Acids Res. 2006;34:e57. doi: 10.1093/nar/gkl184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies CJ, Hutchison CA.Insertion site specificity of the transposon Tn3 .Nucleic Acids Res 199523507–514. 10.1093/nar/23.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hülter N, Wackernagel W. Double illegitimate recombination events integrate DNA segments through two different mechanisms during natural transformation of Acinetobacter baylyi . Mol Microbiol. 2008;67:984–995. doi: 10.1111/j.1365-2958.2007.06096.x. [DOI] [PubMed] [Google Scholar]
- 42.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Q, Huguet-Tapia JC, Peters JE. Tn917 targets the region where DNA replication terminates in Bacillus subtilis, highlighting a difference in chromosome processing in the firmicutes. J Bacteriol. 2009;191:7623–7627. doi: 10.1128/JB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garsin DA, Urbach J, Huguet-Tapia JC, Peters JE, Ausubel FM, et al. Construction of an Enterococcus faecalis Tn917-mediated-gene-disruption library offers insight into Tn917 insertion patterns .J Bacteriol 20041867280–7289. 10.1128/JB.186.21.7280-7289.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grueter L, Koenig O, Laufs R. Transposon mutagenesis in Staphylococcus epidermidis using the Enterococcus faecalis transposon Tn917 . FEMS Microbiol Lett. 1991;66:215–218. doi: 10.1016/0378-1097(91)90335-8. [DOI] [PubMed] [Google Scholar]
- 46.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing .Proc Natl Acad Sci U S A 200410112312–12317. 10.1073/pnas.0404728101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castillo F, Benmohamed A, Szatmari G. Xer site specific recombination: double and single recombinase systems. Front Microbiol. 2017;8:453. doi: 10.3389/fmicb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casadaban MJ, Chou J, Cohen SN. Overproduction of the Tn3 transposition protein and its role in DNA transposition. Cell. 1982;28:345–354. doi: 10.1016/0092-8674(82)90352-X. [DOI] [PubMed] [Google Scholar]
- 49.Palmen R, Vosman B, Buijsman P, Breek CK, Hellingwerf KJ, et al. Physiological characterization of natural transformation in Acinetobacter calcoaceticus . J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 50.de Vries J, Wackernagel W. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc Natl Acad Sci U S A. 2002;99:2094–2099. doi: 10.1073/pnas.042263399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amundsen SK, Neiman AM, Thibodeaux SM, Smith GR. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics. 1990;126:25–40. doi: 10.1093/genetics/126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saunders CW, Guild WR. Monomer plasmid DNA transforms Streptococcus pneumoniae . Mol Gen Genet. 1981;181:57–62. doi: 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- 54.Kidane D, Carrasco B, Manfredi C, Rothmaier K, Ayora S, et al. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 2009;5:e1000630. doi: 10.1371/journal.pgen.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryzhikov M, Gupta R, Glickman M, Korolev S. RecO protein initiates DNA recombination and strand annealing through two alternative DNA binding mechanisms. J Biol Chem. 2014;289:28846–28855. doi: 10.1074/jbc.M114.585117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mortier-Barrière I, Velten M, Dupaigne P, Mirouze N, Piétrement O, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 57.Karudapuram S, Zhao X, Barcak GJ. DNA sequence and characterization of Haemophilus influenzae dprA +, a gene required for chromosomal but not plasmid DNA transformation. J Bacteriol. 1995;177:3235–3240. doi: 10.1128/JB.177.11.3235-3240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ando T, Israel DA, Kusugami K, Blaser MJ. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori . J Bacteriol. 1999;181:5572–5580. doi: 10.1128/JB.181.18.5572-5580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergé M, Mortier-Barrière I, Martin B, Claverys J-P. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol Microbiol. 2003;50:527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 60.Friedrich A, Prust C, Hartsch T, Henne A, Averhoff B, et al. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl Environ Microbiol. 2002;68:745–755. doi: 10.1128/AEM.68.2.745-755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol Gen Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 62.Cohen A, Clark AJ. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986;167:327–335. doi: 10.1128/JB.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 64.Sakai A, Cox MM. RecFOR and RecOR as distinct RecA loading pathways. J Biol Chem. 2009;284:3264–3272. doi: 10.1074/jbc.M807220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.