Abstract
The family Roniviridae includes the genus Okavirus for three species of viruses with enveloped, rod-shaped virions. The monopartite, positive-sense ssRNA genome (26–27 kb) contains five canonical long open reading frames (ORFs). ORF1a encodes polyprotein pp1a containing proteinase domains. ORF1b is expressed as a large polyprotein pp1ab by ribosomal frameshifting from ORF1a and encodes replication enzymes. ORF2 encodes the nucleoprotein. ORF3 encodes two envelope glycoproteins. ORFX encodes a putative double membrane-spanning protein. Roniviruses infect shrimp but only yellow head virus is highly pathogenic. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Roniviridae, which is available at ictv.global/report/roniviridae.
Keywords: Roniviridae, Okavirus, shrimp, ICTV Report, taxonomy
Virion
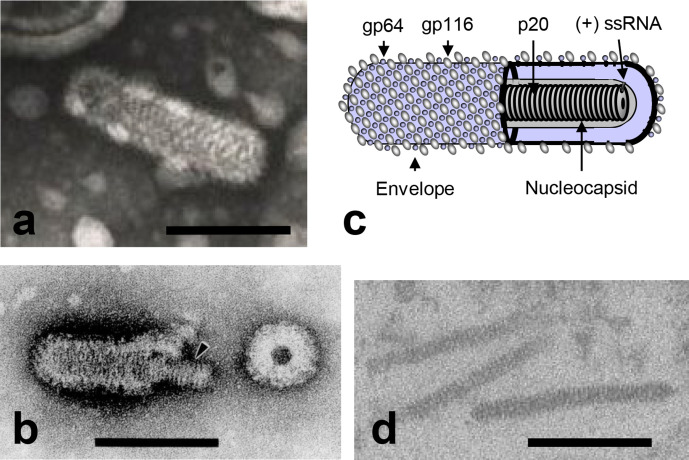
Virions are enveloped, rod-shaped particles (40–60 nm in diameter and 150–200 nm in length) containing three structural proteins [1] (Table 1, Fig. 1). The nucleoprotein (p20) complexes with the RNA genome to form the helical nucleocapsid. Two transmembrane glycoproteins (gp116 and gp64) form prominent peplomers on the virion surface.
Table 1.
Characteristics of members of the family Roniviridae
|
Typical member: |
gill-associated virus (AF227196), species Gill-associated virus, genus Okavirus |
|---|---|
|
Virion |
Enveloped, rod-shaped particles 150–200 nm in length and 40–60 nm in diameter with a helical nucleocapsid composed of the nucleocapsid protein (p20); the lipid envelope contains two transmembrane glycoproteins (gp64 and gp116) |
|
Genome |
Positive-sense, single-stranded RNA (26–27 kb) containing 5 or 6 long open reading frames |
|
Replication |
Cytoplasmic; nucleocapsids bud at membranes of the endoplasmic reticulum/Golgi complex to form mature virions |
|
Translation |
From a nested set of 5′-capped and 3′-co-terminal polyadenylated mRNAs transcribed from genomic RNA |
|
Host Range |
Penaeid shrimp are natural hosts; experimental infection reported in penaeid and palaemonid shrimp of various species |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Nidovirales; the subfamily Okanivirinae includes the genus Okavirus, the subgenus Tipravirus and three species |
Fig. 1.
(a) Negative-contrast electron micrograph of gill-associated virus. (b) Negative-contrast electron micrograph of a partially disrupted yellow head virus virion displaying the internal nucleocapsid. (c) Schematic illustration of a ronivirus virion. (d) Thin-section electron micrograph of unenveloped cytoplasmic nucleocapsids in gill-associated virus-infected shrimp cells. The bars represent 100 nm. Courtesy of K. M. Spann, P. Loh, J. A. Cowley and R. J. McCulloch; panels (a), (b) and (c) reproduced with permission from [2].
Genome
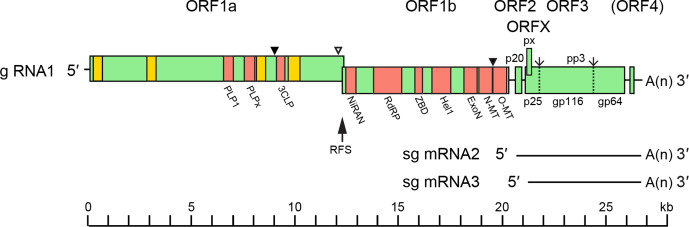
The ronivirus genome is a linear, positive-sense ssRNA (26–27 kb) with a 5′-methylated cap and 3′-polyadenylated tail (Fig. 2) [2]. The genome contains five canonical long open reading frames (ORFs), which, in order from the 5′-terminus, include: ORF1a, and the overlapping ORF1b, encoding replicase enzymes; ORF2 encoding the nucleoprotein (p20); ORF3 encoding the precursor polyprotein (pp3) from which the envelope glycoproteins gp116 and gp64 are derived; and alternative ORFX, which commences three nucleotides downstream of the pp3 initiation codon and encodes a putative small double membrane-spanning protein (px). In gill-associated virus, ORF3 is followed by ORF4 but it is severely truncated in other roniviruses and evidence for its expression is poor.
Fig. 2.
Yellow head virus genome (g RNA1, 26 662 nt) and the two 3′-coterminal sub-genomic RNAs, sg mRNA2 and sg mRNA3. ORF1a hydrophobic regions HD1–HD4 (yellow). Functional domains (pink) - ORF1a: 3C-like protease (3CLP) and papain-like protease domains PLP1 and PLPx; ORF1b: nidovirus RdRP-associated nucleotidyltransferase (NiRAN); RNA-directed RNA polymerase (RdRP); cysteine- and histidine-rich zinc-binding domain (ZBD); superfamily 1 helicase (Hel1); exoribonuclease (ExoN); guanosine N7-methyltransferase (N-MT); and ribose 2′-O-methyl transferase (O-MT). ORF2 encodes the nucleoprotein (p20). ORF3 encodes a precursor polyprotein (pp3), which undergoes post-translation processing to generate envelope glycoproteins (gp116 and gp64) and an N-terminal triple membrane-spanning fragment (p25). ORFX is an alternative reading frame in ORF3 encoding a small double membrane-spanning protein (px). RFS - ribosomal frameshift site upstream of the ORF1a stop codon that allows translation of pp1ab. Known (▼) and likely (∇) sites of proteolytic cleavage of pp1a and pp1ab; signal peptidase type 1 cleavage sites in pp3 (↓).
Replication
Ronivirus replication is cytoplasmic. Elongated nucleocapsids are visible in infected cells and bud at membranes of the endoplasmic reticulum/Golgi complex to form mature virions. During infection, genome length RNA (g RNA1) and two 3′-coterminal subgenomic mRNAs (sg mRNA2 and sg mRNA3) are transcribed, each with a 5′-methylated cap and a 3′-poly(A) tail [3]. Double-stranded RNAs of equivalent size appear to be replicative intermediates. Ronivirus g RNA1, sg mRNA2 and sg mRNA3 each initiate with a 5′-AC dinucleotide and lack a common leader sequence. Roniviruses appear to differ from coronaviruses and arteriviruses by not using a discontinuous transcription, but rather a continuous transcription strategy similar to that utilized by toroviruses.
Taxonomy
Current taxonomy: ictv.global/taxonomy. The family Roniviridae includes the genus Okavirus with the species Yellow head virus, Gill-associated virus and Okavirus 1. Viruses in these species represent only three of eight okavirus genotypes that have been identified in penaeid shrimp [4, 5]. Yellow head virus (species Yellow head virus) is assigned to genotype 1 with two subtypes (1a and 1b), gill-associated virus (species Gill-associated virus) is assigned to genotype 2 and yellow head virus-8 (species Okavirus 1) is assigned to genotype 8. Roniviruses are most closely related to other nidoviruses infecting arthropods, including members of the families Mesoniviridae (from mosquitoes) and Euroniviridae (from crustaceans).
Resources
Current ICTV Report on the family Roniviridae: ictv.global/report/roniviridae
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Donald B. Smith, Richard J. Orton and Nick J. Knowles.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: g RNA, genome length RNA; ORF, open reading frame; sg RNA, subgenomic RNA; ssRNA, single-stranded RNA.
References
- 1.Chantanachookin C, Boonyaratpalin S, Kasornchandra J, Direkbusarakom S, Ekpanithanpong U, et al. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis Aquat Org. 1993;17:145–157. doi: 10.3354/dao017145. [DOI] [Google Scholar]
- 2.Dhar AK, Cowley JA, Hasson KW, Walker PJ. Genomic organization, biology, and diagnosis of Taura syndrome virus and yellowhead virus of penaeid shrimp. Adv Virus Res. 2004;63:353–421. doi: 10.1016/S0065-3527(04)63006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowley JA, Walker PJ. Molecular biology and pathogenesis of roniviruses. In: Perlman S, Gallagher T, Snijder EJ, editors. Nidoviruses. American Society of Microbiology; 2008. pp. 361–377. [Google Scholar]
- 4.Wijegoonawardane PKM, Cowley JA, Phan T, Hodgson RAJ, Nielsen L, et al. Genetic diversity in the yellow head nidovirus complex. Virology. 2008;380:213–225. doi: 10.1016/j.virol.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong X, Liu S, Zhu L, Wan X, Liu Q, et al. Complete genome sequence of an isolate of a novel genotype of yellow head virus from Fenneropenaeus chinensis indigenous in China. Arch Virol. 2017;162:1149–1152. doi: 10.1007/s00705-016-3203-2. [DOI] [PubMed] [Google Scholar]