Abstract
Members of the family Pospiviroidae have single-stranded circular RNA genomes that adopt a rod-like or a quasi-rod-like conformation. These genomes contain a central conserved region that is involved in replication in the nucleus through an asymmetric RNA–RNA rolling-circle mechanism. Members of the family Pospiviroidae lack the hammerhead ribozymes that are typical of viroids classified in the family Avsunviroidae. The family Pospiviroidae includes the genera Apscaviroid, Cocadviroid, Coleviroid, Hostuviroid and Pospiviroid, with >25 species. This is a summary of the ICTV Report on the family Pospiviroidae, which is available at ictv.global/report/pospiviroidae.
Keywords: Pospiviroidae, ICTV Report, taxonomy
Genome
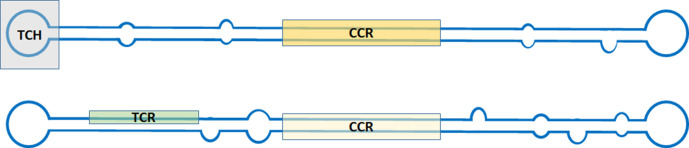
Members of the family Pospiviroidae have circular single-stranded RNA genomes of a few hundred nucleotides. They may assume rod-like or quasi-rod-like conformations containing a central conserved region (CCR) and a terminal conserved hairpin (TCH) or a terminal conserved region (TCR) (Table 1, Fig. 1) [1–3]. The G+C content is >50 %. The genome of viroids does not encode any proteins.
Table 1.
Characteristics of members of the family Pospiviroidae
|
Example: |
potato spindle tuber viroid (V01465), species Potato spindle tuber viroid, genus Pospiviroid |
|---|---|
|
Genome |
Single-stranded circular RNA of 246–375 nt that adopts a rod-like or quasi-rod-like conformation of minimum free energy and contains typical conserved motifs |
|
Replication |
Mediated by nuclear DNA-dependent RNA polymerase II, with oligomeric RNAs of (+) polarity cleaved by a type III RNase and circularized by DNA ligase 1 |
|
Translation |
Absent |
|
Host range |
Plants (dicotyledons and some monocotyledons) |
|
Taxonomy |
Several genera including >25 species |
Fig. 1.
Rod-like structure models for viroids, The positions of the central conserved region (CCR), the terminal conserved region (TCR) and the terminal conserved hairpin (TCH) are indicated by shading. The sequence-specific TCH and TCR elements have never been found together in the same viroid.
Replication
Replication is nuclear and mediated by DNA-dependent RNA polymerase II, which is redirected to use RNA templates through an asymmetric RNA–RNA rolling-circle mechanism. Circular RNA molecules of (+) polarity (by convention the most abundant strand in vivo) are repeatedly transcribed into oligomeric complementary (−) RNAs. Such intermediates serve as templates for generating oligomeric (+) RNAs that are cleaved by a host enzyme of the RNase III class. The termini of the resulting linear monomers are ligated by the host DNA ligase 1 to generate the mature circular viroid RNA [4]. In contrast to members of the family Avsunviroidae, the (−) oligomeric RNAs of members of the family Pospiviroidae are not cleaved and do not generate the corresponding circular forms.
Taxonomy
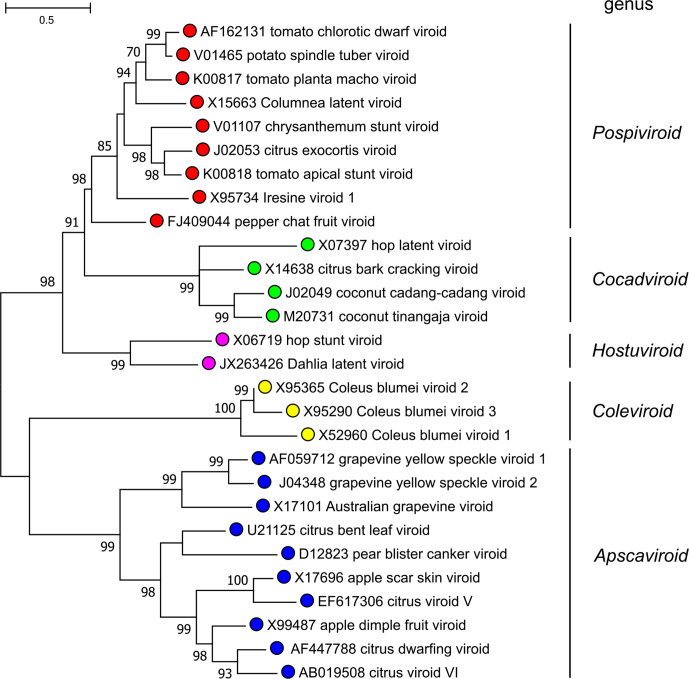
Current taxonomy: ictv.global/taxonomy. Demarcation of genera is based upon the type of CCR and the presence of a TCH or TCR (Fig. 1), as well as phylogenetic clustering in trees based upon whole-genome sequences (Fig. 2). Species demarcation criteria include there being <90 % sequence identity and distinct biological properties with respect to other members of the genus [5]. Members of the genus Pospiviroid, such as potato spindle tuber viroid, share the same CCR and have a TCR. Most infect herbaceous hosts, mainly solanaceous species. Hostuviroids, such as hop stunt viroid, share the same CCR and have a TCH, except for members of the species Dahlia latent viroid, which have a TCR instead of the TCH. Hop stunt viroid has a wide natural host range, while dahlia latent viroid is restricted to Dahlia spp. Cocadviroids, such as coconut cadang-cadang viroid, share the same CCR and have a TCH. Some members infect monocotyledons, while others can only infect dicotyledons. Apscaviroids, such as apple scar skin viroid, share the same CCR and have a TCR. Apscaviroids mainly infect woody plants. Coleviroids, such as Coleus blumei viroid 1, share the same CCR and have a TCR or a TCH. The natural host range of coleviroids is restricted to species in the genus Coleus.
Fig. 2.
Phylogenetic tree of viroid sequences. Maximum-likelihood analysis was conducted with megax [6]. Nodes are labelled with bootstrap support (1000 replicates) where this was >70%.
Resources
Full ICTV Report on the family Pospiviroidae: ictv.global/report/pospiviroidae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
We thank Beatriz Navarro and Michela Chiumenti for help with the phylogenetic analyses and preparation of the corresponding figures and files. Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Donald B. Smith, Richard J. Orton and F. Murilo Zerbini.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CCR, central conserved region; TCH, terminal conserved hairpin; TCR, terminal conserved region.
References
- 1.Steger G, Perreault JP. Structure and associated biological functions of viroids. Adv Virus Res. 2016;94:141–172. doi: 10.1016/bs.aivir.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Giguère T, Raj Adkar-Purushothama C, Perreault J-P. Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae . PLoS One. 2014;9:e98655. doi: 10.1371/journal.pone.0098655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Carrasco A, Flores R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo : A “naked” rod-like conformation similar but not identical to that observed in vitro . RNA Biol. 2017;14:1046–1054. doi: 10.1080/15476286.2016.1223005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores R, Gago-Zachert S, Serra P, Sanjuán R, Elena SF. Viroids: survivors from the RNA world? Annu Rev Microbiol. 2014;68:395–414. doi: 10.1146/annurev-micro-091313-103416. [DOI] [PubMed] [Google Scholar]
- 5.Di Serio F, Flores R, Verhoeven JTJ, Li S-F, Pallás V, et al. Current status of viroid taxonomy. Arch Virol. 2014;159:3467–3478. doi: 10.1007/s00705-014-2200-6. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]