Abstract
Integrity of the intestinal mucosal barrier is closely related to the occurrence of diarrhea. As an important component protein of the intestinal mucosal barrier, Mucin 2 (MUC2) plays a critical role in preventing the invasion of pathogens, toxins, and foreign bodies. In the present study, we preliminary verified the function of the porcine MUC2 gene in resisting porcine epidemic diarrhea virus (PEDV) infection and investigated the effect of DNA methylation in the promoter region on MUC2 gene expression. The results showed that after PEDV infection, the intestinal mucosal barrier was damaged. Moreover, MUC2 expression was significantly higher in PEDV-infected piglets than in healthy piglets (P < 0.01). The mRNA expression of MUC2 was significantly higher in PEDV-infected IPEC-J2 cells than in non-infected IPEC-J2 cells (P < 0.05). Methylation of the mC-5 site in the MUC2 promoter inhibited the binding of Yin Yang 1 (YY1) to the promoter, down regulated the expression of MUC2 and increased the susceptibility of piglets to PEDV. In conclusion, this study suggests that MUC2 plays an essential regulatory role in PEDV infection. High MUC2 expression improves the resistance of pigs to PEDV infection. The binding of YY1 to the MUC2 promoter is hindered by the methylation of the mC-5 site, which downregulates MUC2 expression and ultimately affects the resistance of pigs to PEDV infection.
Keywords: piglet, PEDV, MUC2, DNA methylation, intestinal mucosa
Introduction
Porcine epidemic diarrhea is a highly infectious intestinal disease caused by porcine epidemic diarrhea virus (PEDV). PEDV invades the intestinal epithelial cells and destroys the integrity of the intestinal mucosal barrier, causing severe diarrhea and ultimately resulting in the death of the infected pigs (1). Therefore, the integrity of the intestinal mucosa is essential to resist PEDV infection.
The intestine acts as the body's largest immune organ in addition to its functions of digestion and absorption, and a complete intestinal mucosal barrier can resist 99% of infections (2). Mucin is a type of high-molecular-weight protein present in secreted and membrane-bound forms. Mucin 2 (MUC2) produced by intestinal goblet cells was the major secreted mucin to be identified. Together with antibacterial peptides, antibodies, defensins, and water, MUC2 forms the intestinal mucus layer, which constitutes a chemical barrier against external pathogens (3, 4). The intestinal mucus consists of two mucin layers, both of which are comprised of a large quantity of MUC2. In healthy conditions, the intestinal mucus exhibits antibacterial activity and functions to protect the epithelial cells, which limits the entry of microorganisms into small intestinal epithelial cells (5, 6). It is reported that decreased MUC2 gene expression can induce pathogenic bacteria to invade the biological barrier of the intestinal mucosa (7), indicating that MUC2 plays a decisive part in the biological barrier in the intestine. In addition to form a political protective barrier, MUC2 also has direct antibacterial activity against pathogens. It does not directly kill pathogens or inhaled pollutants but captures them by forming a viscous gel and removes them through mucociliary transport (8). In addition, a previous study has reported an increase in the incidence of colon tumors in MUC2-knockout mice, which is similar to the inflammatory response in mice with adenomatous polyposis coli gene mutation (9, 10), indicating that the loss of MUC2 expression is related to the inflammatory response. Many clinical studies have shown that pathogens and inhaled substances can promote mucin production (8). Overall, overexpression of mucin on the surface of epithelial cells is considered a biomarker of potential disease diagnosis under pathological conditions (11).
The protective roles of mucins against pathogenic infections have been observed in previous reports (12–14). However, there are few studies on the regulatory mechanism of the MUC2 gene expression. DNA methylation is mediated by the DNA methyltransferase family, which does not alter the original DNA sequence. Abnormal DNA methylation can cause a variety of diseases; low methylation leads to abnormal chromatin structure, whereas high methylation may affect gene expression levels (15, 16). DNA methylation modification is one of the main reasons for the decreased expression of many disease resistance genes. In the present study, we analyzed the relationship between MUC2 gene expression level and PEDV infection. Furthermore, we determined the methylation level of the CpG island in the MUC2 promoter region and analyzed the effects of the degree of methylation on MUC2 expression and the key transcription factors in the promoter region. The purpose of this study was to investigate the regulatory effect of MUC2 gene epigenetic modification on PEDV resistance.
Materials and Methods
Ethics Statement
The animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yangzhou University Animal Experiments Ethics Committee (permit number: SYXK (Su) IACUC2012-0029).
Experimental Materials
The experimental pigs were selected from a farm with an outbreak of porcine epidemic diarrhea. We analyzed the pathogens of PEDV, transmissible gastroenteritis virus (TGEV), porcine rotavirus (PoRV), and porcine deltacoronavirus (PDCoV) of 7-day-old piglets with typical clinical symptoms of epidemic diarrhea and selected four 7-day-old piglets infected with PEDV and four healthy piglets feeding in the same conditions. Our previous findings showed that the PEDV detected in pigs is not specific and belongs to the classic strain CV777 (17). The small intestinal tissues were stored in liquid nitrogen for subsequent use.
Total DNA and RNA Extraction and cDNA Synthesis
Total DNA was extracted from the duodenum, jejunum, and ileum using a DNA extraction kit (Beijing Tiangen Biochemical Co., Ltd., Beijing, China). The concentration and purity of DNA were determined using 2% agarose gel electrophoresis and NanoDrop 1000, respectively. Total RNA was extracted from the intestinal tissues and cells using TRIzol (Invitrogen, USA). Total RNA purity and concentration were assessed using NanoDrop 1000, respectively. RNA samples were stored at −70°C. RNA was reversely transcribed into cDNA using a reverse transcription kit (Vazyme Biotech Co., Ltd., Nanjing, China). The cDNA synthesis reaction mixture (10 μl) contained 5 × PrimeScript Buffer (2 μl), PrimeScript RT Enzyme Mix I (0.5 μl), Oligo dT (0.5 μl), Random hexamers (0.5 μl), RNA (500 ng), and RNase-free H2O (up to 10 μl total volume). The thermal conditions used were as follows: 25°C for 10 min, 50°C for 30 min, and 85°C for 5 min; the cDNA samples were stored at 4°C.
Quantitative Real-Time PCR
RT-qPCR was conducted using the AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China). The PCR reaction mixture contained 2 μl cDNA, 0.4 μl of each primer (10 μmol/L), 10 μl 2 × SYBR Premix ExTapTMII, 0.4 μl 50 × ROX Reference Dye II, and RNase-free ddH2O (up to 20 μl total volume). The reaction conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C 10 s and 60°C 34 s. The melting curve analysis was performed at 95°C for 10 s, 60°C for 60 s, and 95°C for 15 s.
Pathogen Identification
We rinsed the intestinal tissue with normal saline, randomly cut the tissue, and extracted the total RNA from the tissue. The samples were tested for the presence of PEDV, TGEV, PoRV, and PDCoV using RT-qPCR. Primer information is provided in Supplementary Table 1.
Hematoxylin-Eosin Staining of the Small Intestinal Tissue
Small intestinal tissues (duodenum, jejunum, and ileum) were fixed with 10% formalin solution, dehydrated with 70, 80, and 90% alcohol, made transparent by treating with xylene solution, embedded with paraffin, sectioned (Leica Rotary Microtome; Leica Biosystems Co., Ltd., Illinois, Buffalo Grove, USA), and stained with hematoxylin-eosin.
MUC2 Expression in Tissues and Cells After Porcine Epidemic Diarrhea Virus Infection
IPEC-J2 cells were cultured at 5% CO2 and 37°C in 12-well plates with Dulbecco's modified Eagle medium (DMEM; Gibco., Grand Island, New York, USA) containing 10% fetal bovine serum. When the cell density reached 80%, the cells were infected with 0.1 MOI PEDV for 12 and 24 h. Non-infected cells were used as the control group. Four replicates were established per group. MUC2 expression in the small intestinal tissue of the diarrhea group and control group and infected cells exposed to different infection periods was detected using RT-qPCR. The fluorescent quantitative primers were designed using Primer 5.0 software; GAPDH and ACTB were used as internal reference genes (Supplementary Table 2).
RNA Interference of MUC2 and Detection of Porcine Epidemic Diarrhea Virus M Gene Expression
Three siRNA sequences (si-MUC2-1, si-MUC2-2, and si-MUC2-3) and negative control sequence (si-MUC2-NC; Supplementary Table 3) were designed and synthesized by Gene Pharma Co., Ltd. (Shanghai, China). The siRNAs and negative control group (NC) were transfected into IPEC-J2 cells using Lipofectamine 2000, with three replicates per treatment group. Cells were incubated overnight and analyzed for expression of fluorescein-labeled siRNAs after 24 h. RNA interference efficiency was determined using RT-qPCR, and the treatment group with the highest interference efficiency was selected for lipofection; NC and Control were used as control groups. Cells were infected with 0.1 MOI PEDV, and the DMEM complete medium was changed after 2 h. The cell morphology was observed under the fluorescence microscope (MoticAE31 inverted fluorescence microscope; Motic China Group Co., Ltd., Xiamen, China). The mRNA expression level of PEDV M gene was determined using RT-qPCR after 24 h.
Methylation of MUC2 Promoter and Its Relationship With mRNA Expression
The promoter region (2,000 bp) of pig MUC2 was determined using NCBI (https://www.ncbi.nlm.nih.gov/) and Ensembl (http://asia.ensembl.org/index.html) databases. Methprimer software (http://www.urogene.org/cgi-bin/me-thprimer/methprimer.cgi) was used to predict the CpG island in the promoter region of the MUC2 gene, and primers used in BSP-PCR were designed [Supplementary Table 4, MUC2 (BSP)]. The Alibaba2 software (http://gene-regulation.com/pub/programs/alibaba2/index.html) was used to predict the transcriptional elements in CpG islands. The genomic DNA was transformed according to the manufacturer instructions of the D5006 bisulfite conversion kit (Beijing Tianmo Technology Development Co., Ltd., Beijing, China). DNA was amplified using the BSP-PCR primers; the PCR reaction mixture (25 μl) included 1 μl of each primer (10 μmol/L), ZYMO Taq Premix (12.5 μl), DNA (2 μl), and ddH2O (up to 25 μl total volume). The reaction conditions were as follows: 95°C for 10 min; 38 cycles of 95°C for 30 s, 52°C annealing for 30 s, and 72°C extension for 35 s; and final extension for 10 min. The PCR product was stored at 4°C. The purified PCR product was ligated into the pMD-19T vector (Bao Bioengineering Dalian Co., Ltd., Dalian, China) overnight at 16°C. The ligation product was transformed into Escherichia coli DH-5α competent cells (Beijing Tiangen Biochemical Co., Ltd., Beijing, China) and plated on ampicillin-containing agar plates at 37°C. After 12-16 h, 20 positive monoclonal colonies were picked out per plate and sent to the company (Songon Biotech Co., Ltd., Shanghai, China) for sequencing. The software QUMA (http://quma.cdb.riken.jp/) was used to determine the methylation ratio of each CpG site, and the positive sequencing results were compared with the reference genome.
Effect of DNA Methylation on the MUC2 Gene Promoter Activity
We constructed a recombinant plasmid containing the MUC2 promoter, and primers to amplify the promoter were designed (Supplementary Table 4, MUC2-1). The pGL3-basic vector and PCR products were digested using SpeI and NcoI, and the purified products were ligated and transformed into Escherichia coli DH-5α competent cells. Monoclonal colonies were picked out for sequencing, and it was verified whether the recombinant plasmid was successfully constructed. The recombinant plasmid pGL3-MUC2 was methylated with methyltransferase M.SssI, and the product was purified and recovered using the TIANquick Midi purification kit (Beijing Tiangen Biochemical Co., Ltd., Beijing, China). Next, 100 ng M.SssI-methylated plasmid pGL3-MUC2, untreated pGL3-MUC2 plasmid, empty plasmid pGL3-basic, and 2 ng Renilla luciferase reporter plasmid (pRL-TK) were transfected into IPEC-J2 cells cultured in 12-well plates at 37°C for 48 h, and the fluorescence activity was detected using the dual-luciferase reporter gene system.
Chromatin Immunoprecipitation-PCR
The Pierce Agarose ChIP Kit (Thermo Fisher Scientific, USA) was used for the chromatin immunoprecipitation (ChIP)-PCR assay. Approximately 80 mg of jejunum tissue samples were cut and placed in pre-chilled phosphate-buffered saline; the tissue samples were then fixed with formaldehyde to lyse the tissue and cross-link protein to DNA. The chromatin was enzymatically digested and incubated with specific anti-YY1 antibodies (Proteintech Group, Inc., USA). The protein-DNA complexes were precipitated, eluted, and de-crosslinked; proteins were digested, and enriched DNA was purified and finally precipitated and eluted for PCR amplification. ChIP-PCR was used to detect the enriched precipitated DNA. The positive control using amplified the DNA precipitated by anti-RNA Polymerase II antibody precipitation obtained DNA as a template, the negative control using rabbit Anti-IgG antibody as a template, whereas the input test group amplified MUC2 gene using unprecipitated DNA as template. The antibodies used in the positive and negative controls were from the Pierce Agarose ChIP Kit. The primers designed for ChIP-PCR are presented in Supplementary Table 4 [MUC2 (CHIP-PCR)]. The reaction mixture (20 μl) contained 1 μl of each primer (10 μmol/L), 1 μl DNA, 10 μl 2 × Taq Master Mix, and 7 μl ddH2O. The reaction procedure was as follows: 95°C for 5 min; 38 cycles of 95°C for 30 s, 60°C annealing for 30 s, and 72°C for 30 s; and 72°C for 10 min. PCR products were detected by 2% agarose gel electrophoresis and compared with the target sequence on the basis of the sequencing results.
RNA Interference Transcription Factor YY1 and Detection of MUC2 Gene Expression
Three siRNA sequences and one negative control sequence were designed for the transcription factor Yin Yang 1 (Supplementary Table 3). Primers were designed and synthesized by Gene Pharma Co. Ltd. (Shanghai, China). si-YY1-1, si-YY1-2, si-YY1-3, and si-YY1-NC were transfected into IPEC-J2 cells, with three replicates in each treatment group, and the expression of fluorescein-labeled siRNAs in the cells was assessed after 24 h. The relative expression of the YY1 gene was detected by RT-qPCR, and the RNA interference efficiency was determined. The treatment group with the highest interference efficiency was selected for further analysis.
Statistical Analysis
The RT-qPCR results were analyzed using the 2−ΔΔCt method; SPSS 18.0 software was used to analyze the data. The relative quantitative results were analyzed by using independent sample t-test for the three replicates of each set; the results are presented as mean ± standard deviation (Mean ± SD). The correlation between promoter methylation and mRNA expression levels was analyzed using Pearson's correlation test. The relative mRNA expression was analyzed by t-test using SPSS 18.0 software.
Results
Identification of PEDV
In this study, PCR was used to ensure that the piglets in the normal group were not infected with PEDV and the piglets in the diarrhea group were only infected with PEDV. The results showed that the bands in the diarrhea group were consistent with the expected product size (216 bp), while the normal piglet samples showed no obvious bands (Supplementary Figure 1A). In order to eliminate the interference of other pathogens, we employed agarose gel electrophoresis to detect the other common viral diarrhea pathogens (TGEV, PoRV, PDCoV), and the result showed no obvious bands (Supplementary Figure 1B). These results suggested that the piglets in the diarrhea group were only infected with PEDV, whereas the piglets in the healthy group were not infected with PEDV. Therefore, the samples were deemed suitable to use in the subsequent experiments.
Observation of Small Intestine Tissue and IPEC-J2 Cells After PEDV Infection
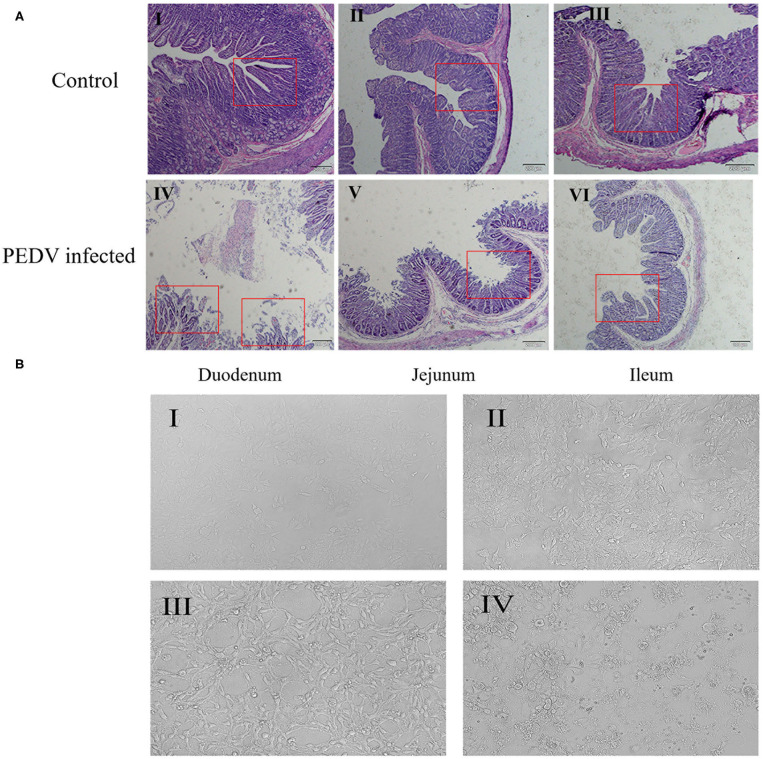
PEDV can cause damage to the intestinal barrier, in the present study, we want to verify whether PEDV can also damage the intestinal barrier in pigs. So, we observed the intestinal damage caused by PEDV infection under microscope. As shown in Figure 1A, the intestinal villi of the diarrhea group piglets were structurally damaged: the villous density and height were decreased, the villi become sparse, the lamina propria was exposed, and the intestinal glands were atrophic. In contrast, the intestinal mucosal barrier of healthy piglet was intact, and the intestinal mucosal epithelial cells were well-organized, clear in outline, and regular in arrangement.
Figure 1.
Effects of PEDV infection on intestinal epithelium and IPEC-J2 cells. (A) Hematoxylin-eosin staining and microscopic examination of small intestinal tissues (10×). The control group represents the Duodenum (I), Jejunum (II), Ileum (III) of normal piglets, and the PEDV infected group represents the Duodenum (IV), Jejunum (V), Ileum (VI) of diarrheic piglets. The red square marks the changes of intestinal villi. (B) Cell morphology of PEDV-infected IPEC-J2 cells at different time periods observed under a light microscope (100×). (I) 0 h, (II) 12 h, (III) 24 h, (IV) 48 h.
At the same time, we observed the damage caused by PEDV to IPEC-J2 cells using optical microscopy. The result showed that, after 24 h of PEDV infection, cell morphology began to change, and numbers of atrophied cell increased (Figure 1B-III), whereas after 48 h of PEDV infection, cell lesions occurred, normal cell morphology was completely lost, and extensive cell death occurred (Figure 1B-IV). These results showed that PEDV could cause damage to IPEC-J2 cells and small intestine tissues in piglets.
MUC2 Is Up-Regulated in PEDV-Infected Tissues and Cells
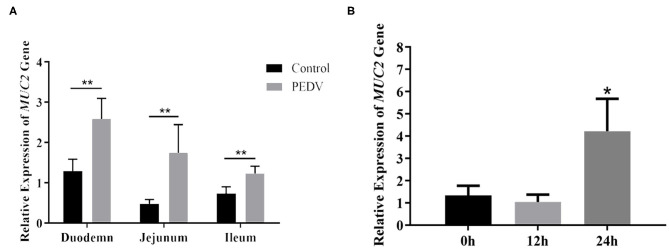
MUC2 protects the gut from pathogenic microorganisms, so does MUC2 play a role in PEDV infestation? To investigate that, the mRNA level of MUC2 was detected at the cellular and organizational levels after PEDV infection. The results showed that the MUC2 expression level in the small intestine (duodenum, jejunum, and ileum) of piglets from the diarrhea group was significantly higher than that from the healthy group (P < 0.01; Figure 2A). In addition, the results of PEDV infection of IPEC-J2 cells at different time periods showed that MUC2 expression was significantly up-regulated at 24 h (P < 0.05; Figure 2B). Overall, these results suggested that MUC2 may play a significant role in PEDV infection.
Figure 2.
Changes of MUC2 expression after PEDV infection. (A) MUC2 expression in the small intestine of the diarrhea group piglets and healthy piglets. (B) MUC2 expression in PEDV-infected IPEC-J2 cells at different time periods. **P < 0.01,*P < 0.05.
MUC2 Knockdown Contributes to PEDV Replication
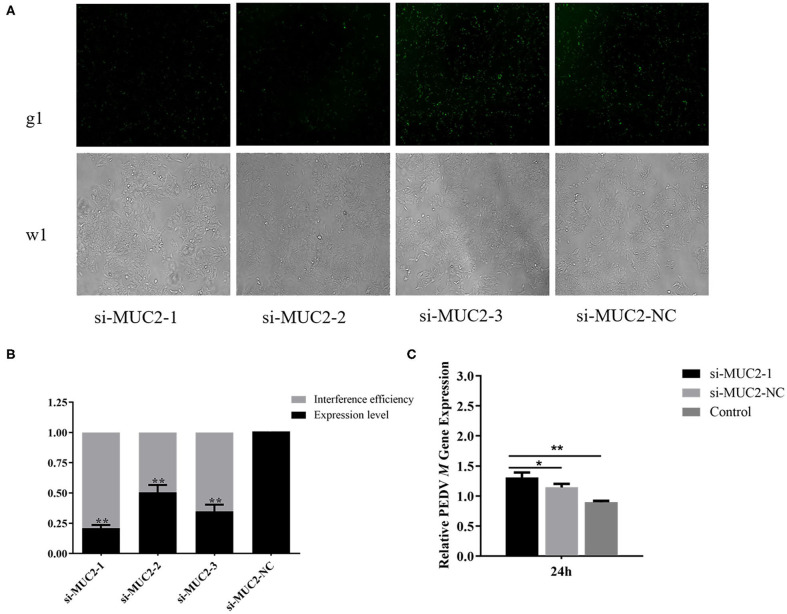
To further investigate the role of MUC2 in PEDV infection, we used RNA interference to down regulate MUC2 expression. The fluorescein markers, used to monitor siRNA transfection, were highly expressed after 24 h, indicating that siRNAs had been successfully transfected (Figure 3A). The treatment group with the highest interference efficiency (si-MUC2-1) was selected for the subsequent experiments (Figure 3B). Then we detected the PEDV M gene to investigated the effect of MUC2 down regulation on PEDV replication. The result showed that after 24 h of PEDV infection, M gene expression in the MUC2-RNAi group was significantly higher than that in the MUC2-NC group (P < 0.05), and significantly higher than that in the Control group (P < 0.01; Figure 3C). In general, these results showed that down regulation of MUC2 expression may contribute to PEDV replication.
Figure 3.
Effects of MUC2 gene knockdown on PEDV replication. (A) The expression of fluorescein markers in IPEC-J2 cells under fluorescence microscope (g1) (100×); the lower panels (w1) are the corresponding cells in the absence of fluorescence exposure, which shows the overall density of cells. (B) The mRNA expression of MUC2 and analysis of interference efficiency. (C) Detection of PEDV M gene expression after interference with MUC2 expression. **P < 0.01, *P < 0.05.
Analysis of DNA Methylation in the Promoter Region of the MUC2 Gene
To investigate whether DNA methylation is regulating the expression of MUC2, we predicted the CpG island in the MUC2 promoter and the results showed that there was a CpG island ~208 bp in size (−324 to −116 bp; Supplementary Figure 2A). The CpG island was amplified by PCR, and the PCR product was detected by 1% agarose gel electrophoresis. The result showed a single bright band, and the size was as expected (Supplementary Figure 2B).
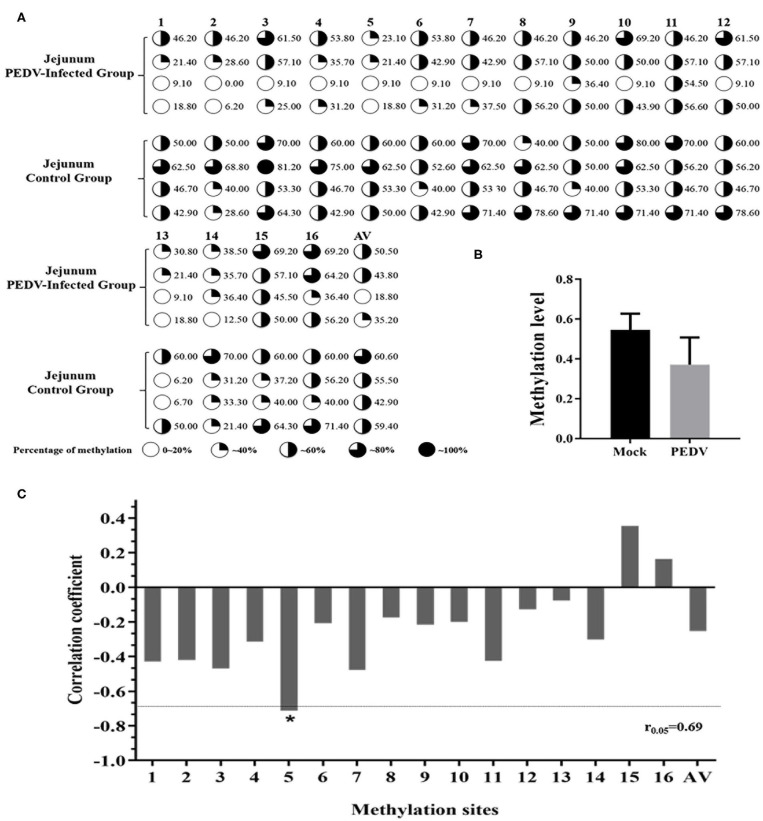
In order to further explore the effect of DNA methylation on MUC2 expression. We analyzed the correlation between CpG island methylation in MUC2 promoter and mRNA expression. The results of the sequencing showed that there were 16 CG sites in the CpG island of the MUC2 promoter region, all of which showed different degrees of methylation (Figure 4A); the methylation level in the diarrhea group was lower than that in the healthy group (P > 0.05; Figure 4B). The methylation levels of most sites were negatively correlated with mRNA expression, except mC-15 and mC-16; among these sites, the methylation level of mC-5 was significantly correlated with mRNA expression (P < 0.05; Figure 4C).
Figure 4.
Correlation between the degree of methylation of the CpG island in MUC2 promoter region and mRNA expression. (A) The methylation level of each site of the CpG island of the MUC2 gene promoter. (B) Degree of methylation between the two groups. (C) Correlation between the degree of methylation of the CpG island in MUC2 gene promoter region and mRNA expression. *P < 0.05.
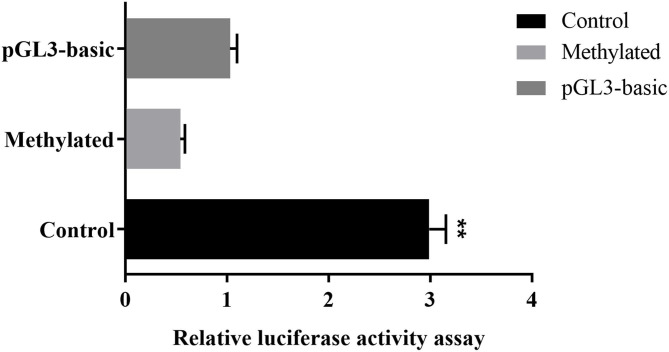
To further prove that DNA methylation can affect MUC2 expression. We detected the effect of promoter methylation of MUC2 gene on promoter activity using Dual-luciferase reporter assay. The results showed that the methylation of the CpG island of the MUC2 promoter significantly reduced the luciferase activity (P < 0.01; Figure 5), suggesting that DNA methylation inhibited the transcriptional activity of MUC2.
Figure 5.
Effects of DNA methylation on the MUC2 gene promoter activity. pGL3-basic: empty plasmid; Methylated: pGL3-MUC2 recombinant plasmid were treated with M.SssI; Control: pGL3-MUC2 recombinant plasmid was not methylated. Both Methylated group and Control group contained the promoter sequence analyzed in this study. **P < 0.01.
Transcription Factor-Binding Sites Prediction and Confirmation
We next predicted transcription factors in CpG island of MUC2 promoter region to investigate how the mC-5 site inhibits MUC2 expression. The result showed that the CpG island of MUC2 promoter has 14 transcription factors. A Pearson correlation analysis suggested that the methylation level of the mC-5 site in the binding domain of the transcription factor YY1 was significantly negatively correlated with gene expression (P < 0.05; Supplementary Figure 3).
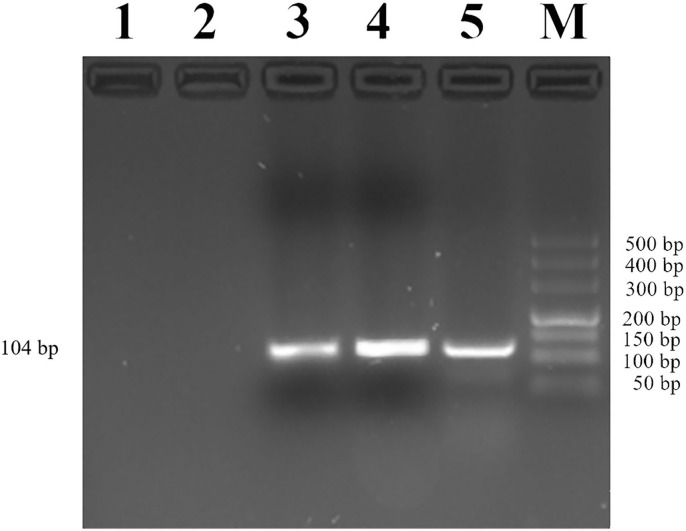
Then, we used CHIP-PCR to investigate whether transcription factor YY1 binds to the promoter region of MUC2. The result showed a single bright band (104 bp), which was consistent with the target fragment in size and with the sequencing results (Figure 6). The results suggested that transcription factor YY1 specifically binds to the mC-5 site of MUC2 gene promoter region.
Figure 6.
Identification of YY1 binding to the MUC2 gene promoter. Lane 1: PCR amplification products with RNase-free water (Blank control without DNA template). Lane 2: Rabbit IgG antibody was added before PCR amplification (negative control). Lane 3: Anti-YY1 antibody was added before PCR amplification. Lane 4: PCR amplification product with input. Lane 5: Anti-RNA Polymerase II antibody was added before PCR amplification (positive control). Lane M: DL500 DNA marker.
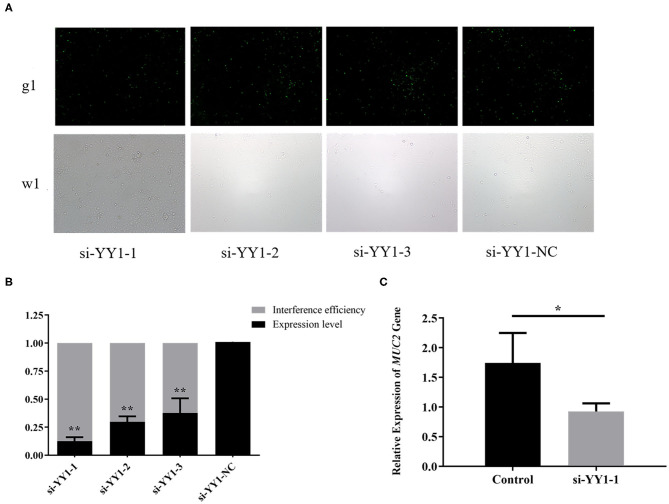
YY1 Knockdown Down-Regulates MUC2 Expression
To explore the effect of transcription factor YY1 on MUC2 gene expression, we constructed the RNA interference vector of YY1 and transfected it into IPEC-J2 cells. The fluorescein markers were detected to be highly expressed after 24 h, indicating that siRNAs were successfully transfected (Figure 7A). The treatment group with the highest interference efficiency (si-YY1-1) was used to interfere MUC2 expression. (Figure 7B) RT-qPCR results showed that the expression level of MUC2 gene decreased significantly (P < 0.05; Figure 7C) suggesting that interfering transcription factor YY1 can down regulate MUC2 gene expression.
Figure 7.
Effects of YY1 gene knockdown on MUC2 expression. (A) The expression of fluorescein markers in IPEC-J2 cells under fluorescence microscope (g1) (100×); the lower panels (w1) are the corresponding cells in the absence of fluorescence exposure, which shows the overall density of cells. (B) The mRNA expression of YY1 and analysis of interference efficiency. (C) The mRNA expression of MUC2 gene after RNA interference. **P < 0.01, *P < 0.05.
Discussion
The mortality of PEDV infection in piglets within 7 days or even a few hours after birth is 100% (18, 19), which leads to severe economic losses to the pig industry. The small intestine plays a significant role in absorbing nutrients and blocking various antigens entering through the villi and crypts of the intestinal epithelium. PEDV infection can lead to acute and massive necrosis of epithelial cells and severe atrophy of intestinal villi (20–22). Previous studies have revealed that the small intestine of PEDV-infected piglets appears thin and transparent (23), intestinal cells are shed, and enteritis occurs (24). These results are consistent with the observations of intestinal sections in the present study, indicating that PEDV-infected piglets have lesions in the small intestine. Additionally, PEDV causes structural damage to the small intestinal villi and decreases villous height, damaging the intestinal mucosal barrier. Furthermore, PEDV can affect the immune, digestive and absorptive functions of the intestine and eventually cause diarrhea. The results further indicate that the pathogenicity of PEDV depends on the integrity of intestinal mucosal barrier in piglets to a certain extent.
MUC2 is an essential structural component of the intestinal mucosal barrier. An intact mucosal barrier can prevent pathogenic microorganisms in the intestinal cavity from contacting the epithelium. It plays an anti-infective role and regulates the balance between intestinal immunity and external stimuli (25, 26). In the present study, the MUC2 gene expression in the small intestinal tissue of PEDV-infected piglets was noted to be significantly higher than that in the intestine of healthy piglets, indicating that PEDV infection was associated with high MUC2 expression. In addition, 24 h after the PEDV infection of IPEC-J2 cells, the MUC2 expression was significantly up-regulated. Moreover, at this time point, the cell morphology began to change. These results indicated that the up-regulated expression of MUC2 is advantageous in repairing the damaged intestinal mucosal barrier. And the MUC2 protein in the intestinal lumen can remove the pathogens to protect the intestinal epithelium from pathogen invasion. Furthermore, it was observed that the expression level of PEDV M gene in IPEC-J2 cells significantly increased after MUC2 knockdown. Previous studies have shown that after MUC2 knockdown, intestinal MUC2 secretion is reduced, and the mucus layer becomes thin and discontinuous (27). Our results suggested that the lack of MUC2 protein leads to impaired function of the intestinal mucosal barrier and induces PEDV to invade the mucosal lamina propria, resulting in a surge of PEDV replication. Overall, our data indicated that high MUC2 expression is beneficial to resist PEDV infection.
In the present study, we found that there were 16 CG sites in the promoter region of MUC2 and all of them had different degrees of methylation. Correlation analysis of MUC2 mRNA expression showed that the methylation of most sites was negatively correlated with mRNA expression. Among these sites, the methylation of mC-5 site was significantly negatively correlated with MUC2 mRNA expression. The result of dual-luciferase reporter assay indicated that DNA methylation inhibits the transcription of MUC2. These results suggested that the mC-5 site may be an important methylation site that regulates MUC2 gene transcription.
DNA methylation is associated with gene transcription silencing (28–30). Blocking the binding of transcription factors and promoters is one of the mechanisms that inhibit gene expression. By inhibiting the binding of methylation-sensitive transcription factors to DNA or hindering the binding of methylation-insensitive transcription factors through proteins, the transcription and expression of the gene can be regulated by DNA methylation (31). In the present study, we identified 14 potential transcription factor-binding domains in the CpG island region of the MUC2 promoter, of which the mC-5 site was located in the transcription factor YY1-binding domain. YY1 is a zinc finger transcription factor widely expressed in various tissues and participates in various biological processes such as embryonic development and differentiation, tumor metastasis, and cell proliferation (32). As a transcription factor, YY1 can act as an activation factor to activate gene expression or as an inhibitor to suppress gene transcription (33, 34). In the present study, the result of ChIP-PCR confirmed that the transcription factor YY1 specifically binds to the MUC2 gene promoter. Moreover, the expression level of MUC2 decreased with the down-regulation of YY1 expression. This indicated that methylation of the mC-5 site hinders the binding of the transcription factor YY1 to the promoter target sequence, thereby inhibiting the expression of MUC2.
In this study, we verified that high expression of MUC2 contributes to the resistance of piglets to PEDV infection. Meanwhile, DNA methylation was found to be associated with MUC2 expression. It is worth noting that methylation of mC-5 site which is located in the binding area of YY1 was significantly negatively correlated with MUC2 expression. We speculated that DNA methylation reduces the transcriptional activity in the promoter region of MUC2, and the mC-5 methylation of the MUC2 promoter inhibits its binding with YY1, which further inhibits MUC2 expression and ultimately regulates the ability of piglets to resist PEDV infection. This study verified the role of MUC2 in PEDV infection, and revealed the regulatory mechanism of MUC2, which lay the foundation for deeply exploring the regulatory mechanism of MUC2, and further functional and mechanism studies on this gene in pigs will be our further endeavor.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yangzhou University Animal Experiments Ethics Committee (permit number: SYXK (Su) IACUC2012-0029). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
WB and SW conceived and supervised the study. YX and WB designed the experiments. YX and YZ performed the experiments. YX and SS analyzed the data. YX, HW, and WB contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was funded by the National Natural Science Foundation of China (31972535 and 31702082), Key Research and Development Project (Modern Agriculture) of Jiangsu Province (BE2019344 and BE2019341), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.646408/full#supplementary-material
References
- 1.Wood EN. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec. (1977) 12:243. 10.1136/vr.100.12.243 [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Liao SF. Physiological effects of dietary amino acids on gut health and functions of swine. Front Vet Sci. (2019) 6:169. 10.3389/fvets.2019.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N, Tang XC. Research progress of mucin-2 and intestinal mucosal barrier damage. Basic Clin Med. (2015) 35:985–988. 10.16352/j.issn.1001-6325.2015.07.031 [DOI] [Google Scholar]
- 4.Liu Y, Yu XJ, Zhao JX, Zhang H, Zhai QX, Chen W. The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression. Int J Biol Macromol. (2020) 164:884–91. 10.1016/j.ijbiomac.2020.07.191 [DOI] [PubMed] [Google Scholar]
- 5.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. (2014) 260:8–20. 10.1111/imr.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ermund A, Schütte A, Johansson MEV, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastroint Liver Physiol. (2013) 305:G341–7. 10.1152/ajpgi.00046.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. (2014) 63:281–91. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basbaum C, Lemjabbar H, Longphre M, Li D, Gensh E, McNamara N. Control of mucin transcription by diverse injury-induced signaling pathways. Am J Respir Crit Care Med. (1999) 160:S44–8. 10.1164/ajrccm.160.supplement_1.12 [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. (2006) 127:469–80. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 10.Yang K, Popova NV, Yang WC, Lozonschi L, Tadesse S, Kent S, et al. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res. (2008) 68:7313–22. 10.1158/0008-5472.CAN-08-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauhan SC, Singh AP, Ruiz F, Johansson SL, Jain M, Smith LM, et al. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125). Mod Pathol. (2006) 19:1386–94. 10.1038/modpathol.3800646 [DOI] [PubMed] [Google Scholar]
- 12.Knight P, Brown J, Pemberton A. Innate immune response mechanisms in the intestinal epithelium: potential roles for mast cells and goblet cells in the expulsion of adult Trichinella spiralis. Parasitology. (2008) 135:655–70. 10.1017/S0031182008004319 [DOI] [PubMed] [Google Scholar]
- 13.Hasnain SZ, Wang H, Ghia J, Haq N, Deng Y, Velcich A, et al. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. (2010) 138:1763–71. 10.1053/j.gastro.2010.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim CH, Kim D, Ha Y, Cho KD, Lee BH, Seo IW, et al. Expression of mucins and trefoil factor family protein-1 in the colon of pigs naturally infected with Salmonella typhimurium. J Comp Pathol. (2009) 140:38–42. 10.1016/j.jcpa.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Soto D, Song C, Mc Laughlin-Drubin ME. Epigenetic alterations in human papillomavirus- associated cancers. Viruses. (2017) 9:E248. 10.3390/v9090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JY, Wang F, Wu ZC, Wu SL, Bao WB. Regulatory effect of methylation of the porcine AQP3 gene promoter region on its expression level and porcine epidemic diarrhea virus resistance. Genes. (2020) 11:1167. 10.3390/genes11101167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong QF, Huang YJ, Wu LS, Wu ZC, Wu SL, Bao WB. Effects of porcine epidemic diarrhea virus infection on tight junction protein gene expression and morphology of the intestinal mucosa in pigs. Pol J Vet Sci. (2019) 22:345–353. 10.24425/pjvs.2019.129226 [DOI] [PubMed] [Google Scholar]
- 18.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. (2012) 18:161. 10.3201/eid1801.111259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Cao Y, Yang Q. Transferrin receptor 1 levels at the cell surface influence the susceptibility of newborn piglets to PEDV infection. PLoS Pathog. (2020) 16:e1008682. 10.1371/journal.ppat.1008682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonpaew S, Teeravechyan S, Frantz PN, Chailangkarn T, Jongkaewwattana A. PEDV and PDCoV pathogenesis: the interplay between host innate immune responses and porcine enteric coronaviruses. Front Vet Sci. (2019) 6:34. 10.3389/fvets.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. (2014) 460:180–93. 10.1016/j.virol.2014.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducatelle R, Coussement W, Debouck P, Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. II. Electron microscopic study. Vet Pathol. (1982) 19:57–66. 10.1177/030098588201900109 [DOI] [PubMed] [Google Scholar]
- 23.Jung K, Eyerly B, Annamalai T, Lu Z, Saif L J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet Microbiol. (2015) 177:373–8. 10.1016/j.vetmic.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Gauger P, Stafne M, Joseph T, Arruda P, Eric B, et al. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. (2015) 482:51–9. 10.1016/j.virol.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperandio B, Fischer N, Sansonetti PJ. Mucosal physical and chemical innate barriers: lessons from microbial evasion strategies. Semin Immunol. (2015) 27:111–8. 10.1016/j.smim.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 26.Hu GQ, Yang YJ, Qin XX, Qi S, Zhang J, Yu SX. Salmonella outer protein B suppresses colitis development via protecting cell from necroptosis. Front Cell Infect Microbiol. (2019) 9:87. 10.3389/fcimb.2019.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. (2014) 20:1165. 10.3748/wjg.v20.i5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compere SJ, Palmiter RD. DNA methylation controls the inducibility of the mouse metallothionein-I gene in lymphoid cells. Cell. (1981) 25:233–40. 10.1016/0092-8674(81)90248-8 [DOI] [PubMed] [Google Scholar]
- 29.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. (1997) 7:157–65. 10.1016/S0960-9822(97)70086-1 [DOI] [PubMed] [Google Scholar]
- 30.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nature Genet. (1999) 22:203–6. 10.1038/9727 [DOI] [PubMed] [Google Scholar]
- 31.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genet. (2005) 37:853–62. 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Liao W, Liu XD. Transcription factor YY1: structure, regulation mechanism and the advances in hematologic malignancies. Prog Mod Biomed. (2015) 15:2596–600. 10.13241/j.cnki.pmb.2015.13.052 [DOI] [Google Scholar]
- 33.Wang JW. The relevant studies on how transcription factor YY1 inhibits FEN1 expression and causes drug resistance in breast cancer cells[D]. Zhejiang Univ. (2015). 10.1186/s12885-015-1043-1 [DOI] [Google Scholar]
- 34.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. (2006) 25:1125–42. 10.1038/sj.onc.1209080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.