This cohort study performs targeted next-generation sequencing on a cohort of acral nevi to determine their mutational spectrum.
Key Points
Question
What are the mutational profiles of nevi arising on acral skin, and how are these associated with the known genetics of acral melanoma?
Findings
In this cohort study, 50 melanocytic nevi of acral skin were genetically profiled using next-generation sequencing, and results were confirmed with focused immunohistochemistry. Evidence of mutated BRAF was found in the majority (43 of 50) of lesions, with mutually exclusive mutations in NRAS (n = 5) occurring less commonly.
Meaning
Among the nevi specimens profiled, BRAF mutations were overrepresented in acral nevi compared with acral melanoma; findings indicate that acral nevi may not be the precursor lesion for the majority of acral melanomas.
Abstract
Importance
Acral skin may develop nevi, but their mutational status and association with acral melanoma is unclear.
Objective
To perform targeted next-generation sequencing on a cohort of acral nevi to determine their mutational spectrum.
Design, Setting, and Participants
Acral nevi specimens (n = 50) that had been obtained for diagnostic purposes were identified from the pathology archives of a tertiary care academic cancer center and a university dermatology clinic. Next-generation sequencing was performed on DNA extracted from the specimens, and mutations called. A subset of samples was stained immunohistochemically for the BRAF V600E mutation.
Results
A total of 50 nevi from 49 patients (19 males and 30 females; median [range] age, 48 [13-85] years) were examined. Analysis of the sequencing data revealed a high prevalence of BRAF mutations (n = 43), with a lower frequency of NRAS mutations (n = 5). Mutations in BRAF and NRAS were mutually exclusive.
Conclusions and Relevance
In this cohort study, nevi arising on mostly sun-protected acral skin showed a rate of BRAF mutation similar to that of acquired nevi on sun-exposed skin but far higher than that of acral melanoma. These findings are in contrast to the well-characterized mutational landscape of acral melanoma.
Introduction
Acral melanoma is a rare melanocytic tumor that arises on the non–hair-bearing skin of the palms, soles, and nail beds.1 Unlike cutaneous melanoma, acral melanoma is not linked to UV radiation exposure; it exhibits a much lower point mutation burden than cutaneous melanoma but a higher frequency of structural variants and copy number alterations.2,3 The incidence of acral melanoma is uniform across all populations, making it the most common melanoma subtype in individuals of African, Asian, and Hispanic descent.1 Acral melanomas differ in their mutational profiles from cutaneous melanomas, with a lower incidence of BRAF mutations (18%) and NF1 mutations (11%) and the presence of mutations in TYRP1 (8%) and NOTCH2 (4%) and amplification/mutation in the receptor tyrosine kinase c-KIT (approximately 2%-3%).3 Acral melanomas with BRAF mutations harbor fewer genomic amplifications and are more common in patients with European ancestry, possibly constituting a unique subset.4 Melanocytic nevi are benign proliferations of melanocytes; melanomas sometimes arise in a small proportion of melanocytic nevi.5,6,7 Genomic analyses have shown that 100% of nevi on sun-exposed skin harbor mutually exclusive mutations in melanoma driver oncogenes, such as BRAF and NRAS.8,9,10 Although the development of nevi has been linked to sun exposure,10,11 they can also arise on skin with low levels of UV radiation exposure, such as the palms and soles. At this time, little is known about the mutational profiles of acral nevi, which arise on skin with little UV radiation exposure.
Methods
Patient Samples
This study was approved by the University of South Florida institutional review board (protocol No. PRO00036516). Deidentified archival samples were collected under a waiver of informed consent under the Common Rule (45 CFR 46). After institutional review board approval, the pathology databases at 2 institutions were queried for a diagnosis containing the words melanocytic nevus at any acral location (palm, sole, finger, toe, foot, hand). Slides were reviewed and diagnoses verified by the study pathologist (J.L.M.); 49 of 50 cases with greater than 10% nevus cellularity and 1 case with 5% nevus cellularity were submitted for analysis.
DNA Sequencing
Targeted DNA sequencing was performed on 151 cancer-associated genes (eTable 1 in the Supplement) (Agilent SureSelect XT ClearSeq Comprehensive Cancer Panel). For each sample, 75-base paired-end sequence reads were generated using v2.5 chemistry on an Illumina NextSeq 500 sequencer, resulting in average target coverage greater than 150×.
Sequence Data Analysis
Raw sequences were aligned to the human genome reference v37 using the Burrows-Wheeler Aligner (version 0.5.9).12 Genetic variants were identified with GATK UnifiedGenotyper (Broad Institute) simultaneously on all tumor samples (n = 50) and a subset of available normal samples (n = 4). Somatic mutations were enriched according to previously described methods13 by excluding variants seen in 1000 genomes at greater than 1% or seen in more than 1 normal sample from this study. Given the lower pathologic tumor cellularity of most samples (median [range], 25% [5%-80%]), mutations with allelic frequency greater than 40% were filtered out as likely inherited variants. Variants were quality filtered with exclusionary hard filters suggested by GATK Best Practices (MQ0 ≥ 4 && ((MQ0/(1.0 × DP)) > 0.1), DP < 5, QUAL < 30.0, QUAL ≥ 30.0 && QUAL < 50.0, QD < 1.5, or FS > 1000). Variants with genotype quality of 15 or greater were included. Sequence variants were annotated to determine genic context using ANNOVAR software (Center for Applied Genomics, University of Pennsylvania). Mutation summaries and plots were prepared with in-house Perl and R scripts. Mutations in BRAF (V600E) and NRAS (Q61R) were manually examined in all samples and were considered mutated for these 2 genes if the mutation allelic frequency was 1% or greater. One sample with a higher cellularity and a functional EGFR mutation P484L (allelic frequency: 42%) and 1 sample with higher cellularity and BRAF mutation V600E (allelic frequency: 48%) were included in the Figure following manual curation.
Figure. Oncogenic Driver Mutations in Acral Nevi.
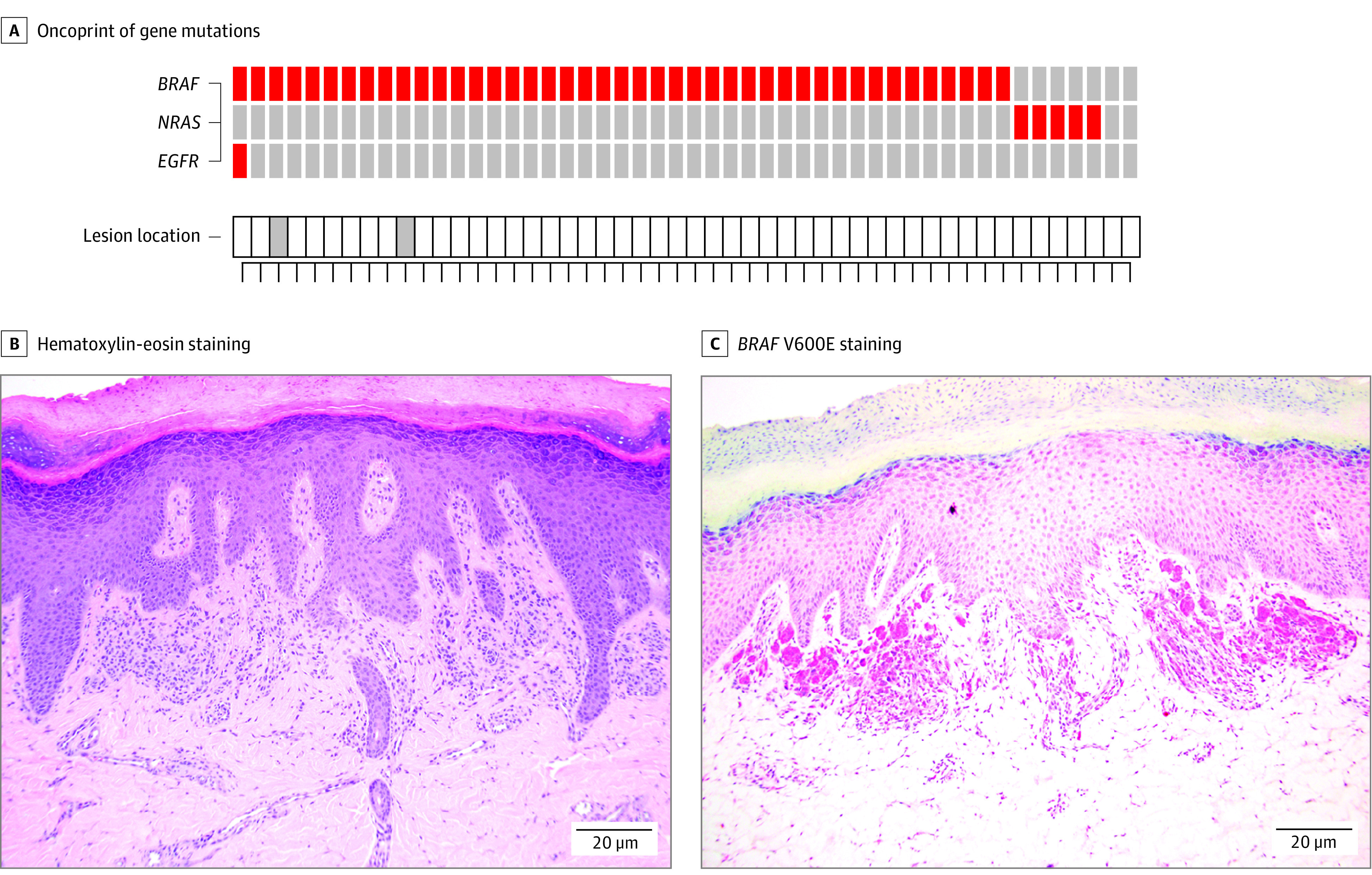
A, Oncoprint of mutations in common acral and cutaneous melanoma genes found in the cohort of acral nevi (n = 50). Data show the identified mutations in each nevus by sequencing. Red boxes indicate protein-altering mutations. Gray boxes indicate lesions from the dorsal foot. Each column represents a different sample. B, Representative immunohistochemical analysis of mutant BRAF V600E staining in acral nevi with a confirmed BRAF V600E mutation by next-generation sequencing. C, BRAF V600E staining.
OncoScan Copy Number Variation Analysis
OncoScan copy number variation arrays (Applied Biosystems) were analyzed on 18 cases using chromosome analysis software (ChAS Suite, version 4.0 [Thermo Fisher Scientific]). Median of absolute values of all pairwise differences, normal diploid waviness SD, and single nucleotide polymorphism quality control of normal diploid markers were analyzed for quality control. Karyotypes with labeled copy number gain and loss segments were generated by ChAS. Two samples were excluded owing to quality control concerns.
Immunohistochemistry
Twelve samples with sufficient remaining tissue were subjected to immunohistochemical (IHC) staining for BRAF. The mouse primary against BRAF V600E (ab228461; Abcam) was used. Positive staining was defined as 2 to 3+ positivity in greater than 50% of lesional nuclei.
Results
We performed targeted next-generation sequencing on 50 acral nevi from 19 males and 30 females. The racial breakdown was 44 White patients, 1 Black patient, and 4 patients for whom race/ethnicity was unknown. Of the 50 lesions, 23 were located on the plantar surface; 24 were located on sun-protected acral areas of the instep, arch of foot, or interdigital space; 2 were on the dorsal foot; and 1 was on the palm. The most common diagnosis was compound or junctional melanocytic nevus of the acral type (n = 38), followed by compound or intradermal nevus with congenital features (n = 3), nevus with atypical features/dysplastic nevus (n = 3), compound melanocytic nevus (n = 2), combined nevus (n = 2), and intradermal melanocytic nevus (n = 2) (Table).
Table. Overview of Acral Nevi Samples Analyzed.
| Sample | Location | Diagnosis | Mutationa |
|---|---|---|---|
| 1 | Medial toe | CMN, acral type with congenital features | BRAF |
| 2 | Medial foot | CMN, acral type | BRAF |
| 3 | Medial foot | CMN, acral type | BRAF |
| 4 | Sole of foot | CMN, acral type with atypical features | BRAF |
| 5 | Plantar foot | CMN, acral type | BRAF |
| 6 | Heel | CMN, acral type | BRAF |
| 7 | Toe | CMN, acral type | BRAF |
| 8 | Dorsal foot | CMN, acral type | BRAF |
| 9 | Plantar foot | Combined nevus (CMN, acral type and dendritic blue nevus) | NRAS |
| 10 | Interdigital toe web | CMN, acral type | BRAF |
| 11 | Interdigital toe skin | CMN, acral type | BRAF |
| 12 | Instep of foot | CMN, acral type | BRAF |
| 13 | Right plantar foot | CMN, acral type | BRAF |
| 14 | Inner foot | CMN, acral type | BRAF |
| 15 | Plantar surface | CMN, acral type | BRAF |
| 16 | Interdigital space | CMN, acral type | BRAF |
| 17 | Planta toe base | Combined nevus (CMN, acral type and dendritic blue nevus) | NRAS |
| 18 | Medial sole | CMN, acral type, with atypical features | BRAF |
| 19 | Third toe | JMN, acral type | ND |
| 20 | Plantar surface | CMN, acral type | BRAF |
| 21 | Foot | CMN, acral type | BRAF |
| 22 | Palm | IDN | BRAF |
| 23 | Arch of foot | IDN | NRAS |
| 24 | Lateral foot | CMN, acral type | BRAF |
| 25 | Interdigital space | CMN, acral type | BRAF |
| 26 | Lateral foot | CMN, acral type | BRAF |
| 27 | Foot instep | CMN, acral type | BRAF |
| 28 | Medial foot | CMN, acral type | BRAF |
| 29 | Medial foot | CMN, acral type | ND |
| 30 | Inner toe | CMN, acral type | BRAF |
| 31 | Lateral heel | CMN, acral type | BRAF |
| 32 | Foot | CMN, acral type | BRAF |
| 33 | Plantar surface foot | CMN | BRAF |
| 34 | Plantar heel | CMN, acral type | BRAF |
| 35 | Plantar foot | CMN, acral type | BRAF |
| 36 | Plantar foot | CMN, acral type | BRAF |
| 37 | Hand | CMN, acral type | BRAF |
| 38 | Dorsal foot | CMN, acral type | BRAF |
| 39 | Second toe | CMN | BRAF |
| 40 | Plantar foot | CMN, acral type | BRAF |
| 41 | Plantar foot | CMN, acral type | NRAS |
| 42 | Foot | CMN, acral type with congenital features | BRAF |
| 43 | Plantar surface | CMN, acral type | BRAF |
| 44 | Plantar foot | CMN, acral type | BRAF |
| 45 | Plantar foot | CMN, acral type | BRAF |
| 46 | Plantar surface | CMN, dysplastic type | BRAF |
| 47 | Plantar foot | CMN, acral type | BRAF |
| 48 | Foot medial plantar surface | CMN, acral type | BRAF |
| 49 | Foot arch | CMN, acral type | NRAS |
| 50 | Plantar foot | CMN with congenital features | BRAF |
Abbreviations: CMN, compound melanocytic nevus; IDN, intradermal melanocytic nevus; JMN, junctional melanocytic nevus; ND, no driver mutations detected.
Mutations in BRAF or NRAS are noted if mutant allelic frequency was 1% or greater.
The range of all mutations identified was 15 to 76 (16-83 mutations/megabase), median: 35 (38 mutations/megabase) (eTable 2 in the Supplement). Among the pathogenic protein-altering mutations identified, activating mutations (V600E) in BRAF were the most frequent, with the automated calling algorithm detecting these in 50% (25 of 50) of all acral nevi. Because nevi tend to be small lesions with low cellularity, we next manually inspected the sequencing data and found evidence of the BRAF V600E mutation in 1 sample with 48% allelic frequency and a further 17 samples with lower allelic frequency (Figure, A). Allelic frequency varied across samples (median [range], 10% [2%-48%]). The allelic frequency of BRAF mutations was associated with pathologic cellularity of the samples (eFigure 1 in the Supplement). In total, mutant BRAF was observed in 43 of 50 specimens (86%; 95% CI, 73%-94%). The presence of the BRAF V600E mutation was confirmed by IHC staining in 12 cases, including some with low allelic frequency (Figure, B and C). An additional 5 of 50 (10%; CI, 3%-22%) acral nevi harbored activating NRAS mutations (all Q61R; 3 identified automatically, 2 after manual review) (Figure, A). The NRAS-mutant nevi stained negatively for BRAF V600E by IHC (eFigure 2 in the Supplement). One of the lesions with an NRAS mutation had congenital features histologically. In all cases, BRAF and NRAS driver mutations were mutually exclusive. Other key findings included a noncanonical mutation in EGFR (p.P848L; allelic frequency: 42%), which is known to be pathogenic, that overlapped with a BRAF V600E mutation.14
Copy number alterations are more common in acral melanoma than cutaneous melanoma.3 Surprisingly, very few copy number alterations were detected in the present study. One of 17 samples showed a single gain, and 2 of 17 samples showed losses (eTable 3 in the Supplement). Genes commonly observed to have copy number variations in acral melanoma (CCND1, CDKN2A, TP53) were not altered.
Discussion
Our study represents, to our knowledge, the largest series of acral nevi that have been sequenced to date. Most of the patient cohort was White, while previous studies were predominantly performed in Asian populations, where there is a much higher frequency of acral nevi. The high prevalence of BRAF mutations in the present study’s population was surprising, given the overall lower frequency of BRAF mutations in acral melanoma. As there is little evidence that BRAF mutations are UV radiation signature mutations, the presence of BRAF-mutant nevi on non–sun-exposed skin is entirely plausible. This finding calls into question whether acral nevi are the precursor lesion for most acral melanomas.
Limitations
One caveat to the present study is the lack of matching normal DNA for many of the specimens. Our use of the 1000 Genomes Project database and allelic frequency filtering excluded many inherited variants, but some likely remained. We note that our filters likely did not remove all rare inherited variants, making it difficult to reliably distinguish a somatic mutation from a germline polymorphism. In addition, the samples in this study were formalin fixed, which is also thought to increase mutational frequency. Together, these may have resulted in artificial mutations leading to the elevated mutational burden that we report.
Conclusions
Acral nevi demonstrated a mutational spectrum similar to that of nevi on sun-exposed skin, suggesting that they are unlikely to be the precursor lesion for the majority of acral melanomas. The study’s patient population was mostly White, in contrast with other data from predominantly Asian populations.15 The present study’s findings in acral nevi contrast with the well-known mutational spectrum of acral melanoma.
eFigure 1. Correlation between nevus cellularity and allelic frequency for BRAF and NRAS mutations
eFigure 2. Staining of NRAS mutant acral nevi with BRAF-V600E specific antibody
eTable 1. List of genes in the NGS panel
eTable 2. List of identified mutations
eTable 3. Copy number alterations
References
- 1.Chen YA, Teer JK, Eroglu Z, et al. Translational pathology, genomics and the development of systemic therapies for acral melanoma. Semin Cancer Biol. 2020;61:149-157. doi: 10.1016/j.semcancer.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135-2147. doi: 10.1056/NEJMoa050092 [DOI] [PubMed] [Google Scholar]
- 3.Newell F, Wilmott JS, Johansson PA, et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat Commun. 2020;11(1):5259. doi: 10.1038/s41467-020-18988-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh I, Jorgenson E, Shen L, et al. Targeted genomic profiling of acral melanoma. J Natl Cancer Inst. 2019;111(10):1068-1077. doi: 10.1093/jnci/djz005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127(1):179-182. doi: 10.1038/sj.jid.5700490 [DOI] [PubMed] [Google Scholar]
- 6.Tsao H, Bevona C, Goggins W, Quinn T. The transformation rate of moles (melanocytic nevi) into cutaneous melanoma: a population-based estimate. Arch Dermatol. 2003;139(3):282-288. doi: 10.1001/archderm.139.3.282 [DOI] [PubMed] [Google Scholar]
- 7.Shain AH, Joseph NM, Yu R, et al. Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell. 2018;34(1):45-55.e4. doi: 10.1016/j.ccell.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19-20. doi: 10.1038/ng1054 [DOI] [PubMed] [Google Scholar]
- 9.Colebatch AJ, Ferguson P, Newell F, et al. Molecular genomic profiling of melanocytic nevi. J Invest Dermatol. 2019;139(8):1762-1768. doi: 10.1016/j.jid.2018.12.033 [DOI] [PubMed] [Google Scholar]
- 10.Stark MS, Tan JM, Tom L, et al. Whole-exome sequencing of acquired nevi identifies mechanisms for development and maintenance of benign neoplasms. J Invest Dermatol. 2018;138(7):1636-1644. doi: 10.1016/j.jid.2018.02.012 [DOI] [PubMed] [Google Scholar]
- 11.Dodd AT, Morelli J, Mokrohisky ST, Asdigian N, Byers TE, Crane LA. Melanocytic nevi and sun exposure in a cohort of Colorado children: anatomic distribution and site-specific sunburn. Cancer Epidemiol Biomarkers Prev. 2007;16(10):2136-2143. doi: 10.1158/1055-9965.EPI-07-0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25(14):1754-1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teer JK, Zhang Y, Chen L, et al. Evaluating somatic tumor mutation detection without matched normal samples. Hum Genomics. 2017;11(1):22. doi: 10.1186/s40246-017-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarcar B, Gimbrone NT, Wright G, et al. Characterization of epidermal growth factor receptor (EGFR) P848L, an unusual EGFR variant present in lung cancer patients, in a murine Ba/F3 model. FEBS Open Bio. 2019;9(10):1689-1704. doi: 10.1002/2211-5463.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon KR, Choi YD, Kim JM, et al. Genetic alterations in primary acral melanoma and acral melanocytic nevus in Korea: common mutated genes show distinct cytomorphological features. J Invest Dermatol. 2018;138(4):933-945. doi: 10.1016/j.jid.2017.11.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Correlation between nevus cellularity and allelic frequency for BRAF and NRAS mutations
eFigure 2. Staining of NRAS mutant acral nevi with BRAF-V600E specific antibody
eTable 1. List of genes in the NGS panel
eTable 2. List of identified mutations
eTable 3. Copy number alterations


