Abstract
Immunocompromised patients, including those with inborn errors of immunity (IEI), may be at increased risk for severe or prolonged infections with SARS-CoV-2 (Zhu et al. N Engl J Med. 382:727–33, 2020; Guan et al. 2020; Minotti et al. J Infect. 81:e61–6, 2020). While antibody and T cell responses to SARS-CoV-2 structural proteins are well described in healthy convalescent donors, adaptive humoral and cellular immunity has not yet been characterized in patients with antibody deficiency (Grifoni et al. Cell. 181:1489–1501 e1415, 2020; Burbelo et al. 2020; Long et al. Nat Med. 26:845–8, 2020; Braun et al. 2020). Herein, we describe the clinical course, antibody, and T cell responses to SARS-CoV-2 structural proteins in a cohort of adult and pediatric patients with antibody deficiencies (n = 5) and controls (related and unrelated) infected with SARS-CoV-2. Five patients within the same family (3 with antibody deficiency, 2 immunocompetent controls) showed antibody responses to nucleocapsid and spike proteins, as well as SARS-CoV-2 specific T cell immunity at days 65–84 from onset of symptoms. No significant difference was identified between immunocompromised patients and controls. Two additional unrelated, adult patients with common variable immune deficiency were assessed. One did not show antibody response, but both demonstrated SARS-CoV-2-specific T cell immunity when evaluated 33 and 76 days, respectively, following SARS-CoV-2 diagnosis. This report is the first to show robust T cell activity and humoral immunity against SARS-CoV-2 structural proteins in some patients with antibody deficiency. Given the reliance on spike protein in most candidate vaccines (Folegatti et al. Lancet. 396:467–78, 2020; Jackson et al. N Engl J Med. 383:1920–31, 2020), the responses are encouraging. Additional studies will be needed to further define the timing of onset of immunity, longevity of the immune response, and variability of response in immunocompromised patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-021-01046-y.
Keywords: Antibody deficiency, adaptive immune response, COVID-19, common variable immunodeficiency, SARS-CoV-2, T cell response
Introduction
Since the start of the COVID-19 pandemic there has been expanding evidence that immunocompromised patients may be at increased risk for severe or prolonged infections with SARS-CoV-2 [1–3]. Clinical descriptions of COVID-19 in patients with T cell and antibody-specific inborn errors of immunity (IEI) are expanding, including reports of worsened disease course in patients with common variable immunodeficiency (CVID) as compared with pure agammaglobulinemia [4–7]. In the largest cohort described to date of patients with IEI and COVID-19, 20% of the cohort required intensive care, with an overall mortality rate of 10%; 6 of 9 deceased patients suffered from an antibody defect [6]. Their findings represent increased morbidity and mortality, especially at younger ages, as compared with the general population. While antibody and T cell responses to SARS-CoV-2 structural proteins are well described in healthy convalescent donors, adaptive humoral and cellular immunity have not yet been characterized in patients with antibody deficiencies [8–11]. Here, we describe five patients affected with antibody deficiencies who developed mild symptoms of COVID-19 and provide comprehensive analysis of their adaptive immune responses.
Methods
All patients provided written informed consent for clinical data and blood sample collection on protocols approved by the National Institutes of Health Institutional Review Board, in concordance with ethical standards as put forth by the Declaration of Helsinki.
Serology Assays
SARS-CoV-2 antibody testing was performed via luciferase immunoprecipitation assay on all subjects as previously described [9]. Briefly, plasma samples were incubated with spike and nucleocapsid proteins fused to Gaussia and Renilla luciferase, respectively. Protein A/G beads were added, and samples were washed prior to addition of coelenterazine substrate (Promega). A Berthold 165 LB 960 Centro Microplate Luminometer was used to measure luciferase activity in light units. Antibody levels were reported as the geometric mean level with 95% confidence interval. As described previously, cutoff limits for determining positive antibodies in the SARS-CoV-2-infected samples were based on the mean plus 3–4 standard deviations of the serum values derived from uninfected blood donor controls for nucleocapsid (125,000 LUs) and spike (45,000 LUs) [9]. Percentages for categorical variables, median, mean, standard deviation and range, and geometric mean were used to describe the data. Unpaired t tests were used for statistical analysis.
T Cell Assays
Testing of T cell responses was performed via stimulation of peripheral blood mononuclear cells with peptide libraries encompassing SARS-CoV-2 structural proteins as previously described [12]. Cells were then cultured for 10 days in 96-well plates with IL-4 (400 IU/mL) and IL-7 (10 ng/mL). On day 10, expanded viral specific T cells (VSTs) were harvested. 2 × 105 VSTs were plated in a 96-well plate and re-stimulated with SARS-CoV-2 structural proteins pooled pepmixes or actin (negative control) with CD28/CD49d (BD Biosciences) and anti-CD107a- Pe-Cy7 antibody. After 1 h of stimulation, brefeldin A (Golgiplug; BD Biosciences, San Jose, CA, USA) and monensin (GolgiStop; BD Biosciences, San Jose, CA, USA) were added. Cells were then incubated for an additional 4 h. Cell viability was assessed using Live-Dead Aqua. Cells were surface stained with fluorophore-conjugated antibodies against CD3-BV785, CD4-BV605, CD8- BV421, TCRαβ-PerCP Cy5.5, TCRγδ- APC-Fire750, CCR7-FITC, CD45-RO-PE Dazzle, HLA-DR-Alexaflour700, and CD56-BV650 (Miltenyi Biotec; BioLegend). Cells were fixed, permeabilized with Cytofix/Cytoperm solution (BD Biosciences), and subsequently stained with IFN-γ-APC and TNF-α-PE (Miltenyi Biotec). All samples were acquired on a CytoFLEX cytometer (Beckman Coulter, Brea, CA, USA). The gating strategy for analysis is presented in Supplemental Fig. 2 .
Results
Clinical History
Immunodeficiency History
The proband of the kindred (P1) is an 11-year-old boy with CVID, atopy, celiac disease, and recurrent infections beginning in the first year of life. He is currently treated with immunoglobulin replacement therapy with improvement in infections (Supplemental Table 1). His twin brother (P2) had a history of periodic fever, apthous stomatitis, pharyngitis, adenitis (PFAPA), recurrent sinusitis, eosinophilic esophagitis, and environmental allergies, as well as hypogammaglobulinemia (Supplemental Fig. 1) [13]. Their younger sister (P3) has history of recurrent sinopulmonary infections, otitis media requiring myringotomy tubes, pharyngitis with subsequent tonsillectomy, as well as atopy with a normal immunologic evaluation [14]. Their mother (P4) also has specific antibody deficiency with recurrent sinopulmonary infection and atopy. She is currently managed with early antibiotic therapy for infections and frequent booster vaccinations. P2, P3, and P4 have never received immunoglobulin therapy. Whole-exome sequencing performed on the proband did not identify any a causative variant (for additional clinical information, see Supplemental data S1).
Patient 6 (P6) is an unrelated 48-year-old male who was diagnosed with CVID at the age of 34 years in the setting of recurrent sinusitis, thrombocytopenia, leukopenia, and splenomegaly (Supplemental Table 2). He is currently treated with immunoglobulin replacement therapy and amantadine prophylaxis, but with ongoing infectious and non-infectious complications. Whole-exome sequencing did not reveal a causal genetic variant.
Patient 7 (P7) is an unrelated 21-year-old female with CVID diagnosed at the age of 14 years in the setting of alopecia areata and idiopathic thrombocytopenic purpura. Her course was complicated by anti-phospholipid syndrome and respiratory infections. She is currently treated with subcutaneous immunoglobulin and hydroxychloroquine. Whole-exome sequencing revealed 2 previously reported compound heterozygous variants in TNFRSF13B (c.310 T > C, p.Cys104Arg and c.260 T > A, p.Ile87Asn) which are believed to be contributory to her CVID [15–17].
SARS-CoV-2 History
With regard to the kindred, the healthy father (P5) had onset of fever progressing to fatigue, anosmia, and cough in August of 2020. Symptoms persisted for 14 days, with a normal chest X-ray during his disease course. SARS-CoV-2 PCR was positive on the second day of illness. A day later, two of the children (P2 with hypogammaglobulinemia and P3) developed 2 days of fever without respiratory symptoms. P3 had persistent anosmia lasting weeks. Twenty days after the first family member became ill, the mother with SAD (P4) developed fever, fatigue, severe headache, and anosmia, with persistent symptoms over several weeks. SARS-CoV-2 testing was not performed on the other family members at the time of illness. Patient 1 with CVID remained asymptomatic when his family was ill. All family members recovered without need for hospitalization or treatment.
In November 2020, P6 with CVID (unrelated) was incidentally found to be SARS-CoV-2 positive on admission for post-operative bleeding after surgery for benign prostatic hypertrophy. His only symptoms of SARS-CoV-2 infection were dry mouth and cough 3 days after diagnosis. His course was otherwise uncomplicated.
P7 with CVID (unrelated) developed nasal and sinus congestion, mild anosmia, and fatigue in November of 2020. SARS-CoV-2 PCR was positive on day 4 of illness, with return to baseline 10 days after testing.
Five additional pediatric and adult immunocompetent controls with mild (n = 4) to severe (n = 1) symptoms of SARS-CoV-2 infection were included in the analysis for comparison.
Serologic Responses
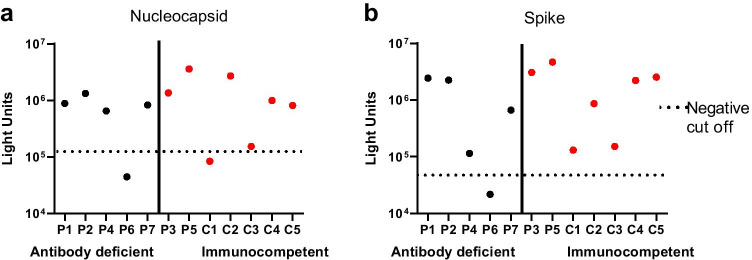
SARS-CoV-2 antibody testing was performed via luciferase immunoprecipitation assay on the kindred 84 days after the first family member developed symptoms (P5). At that time, P2, P3 and P4 were 79 days, 80 days, and 65 days, respectively, from the onset of their own symptoms. Patient 6 was evaluated 33 days following detection of SARS-CoV-2 infection by PCR testing. Patient 7 was evaluated 80 days after first developing symptoms. All five subjects in our kindred (P1–P5) had detectable antibodies targeting both spike (median LU = 2.43 × 106) and nucleocapsid (median LU = 1.33 × 106, Fig. 1) proteins of SARS-CoV-2. In contrast, P6 is seronegative for spike (LU = 2.16 × 104) and nucleocapsid (LU = 4.45 × 104), while P7 is seropositive for spike (LU = 6.61 × 105) and nucleocapsid (LU = 8.29 × 105) (Fig. 1). Antibody levels of additional pediatric and adult immunocompetent controls evaluated after SARS-CoV-2 infection are included for comparison (Fig. 1).
Fig. 1.
Antibody responses as measured by LIPS assay for patients with antibody deficiencies (P1, P2, P4, P6, P7) in black compared to immunocompetent controls (P3, P5, C1-C5) in red for nucleocapsid (a) and spike (b). Negative cutoff values (denoted by the dotted line) are based on uninfected negative controls as previously described [9]
T Cell Responses
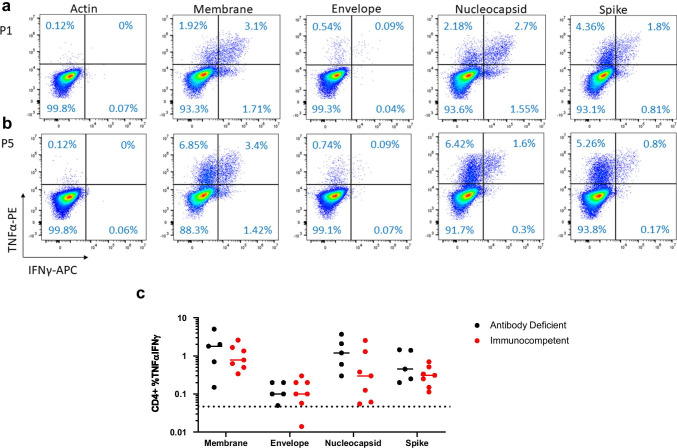
Intracellular cytokine staining demonstrated specific CD4+ T cell responses in all affected patients (n = 5) targeting spike (mean IFN-γ/TNF-α+ 0.75%; standard deviation [SD] 0.62), membrane (mean IFN-γ/TNF-α+ 1.94%; SD 1.9), and nucleocapsid (mean IFN-γ/TNF-α+ 1.58%; SD 1.37) (Fig. 2a, Supplemental Fig. 3). All immunocompetent control patients (n = 7) demonstrated specific CD4+ T cell responses to spike (mean IFN-γ/TNF-α+ 0.33%; SD 0.20), membrane (mean IFN-γ/TNF-α+ 1.12%; SD 0.80), and nucleocapsid (mean IFN-γ/TNF-α+ 0.75%; SD 0.89) (Fig. 2b, Supplemental Fig. 3). Specificity was determined as a response > 2 × the mean of the negative control, actin (mean IFN-γ/TNF-α+ 0.025%; SD 0.05). There is no statistically significant difference between the affected patients and control groups with respect to CD4+ T cell responses to actin (p = 0.67) or any of the SARS-CoV-2 proteins: membrane (p = 0.32), envelope (p = 0.86), nucleocapsid (p = 0.23), or spike (p = 0.13) (Fig. 2c). Single IFN-γ+ and TNF-α+ CD4+ populations are reported in Supplemental Table 3. Affected and control patients did not show appreciable CD8+ T cell responses (Supplemental Fig. 4). CD107a expression was minimal and did not differ between patients with antibody deficiency compared to immunocompetent controls (data not shown).
Fig. 2.
Flow cytometry of CD4+ cells positive for IFN-γ and TNF- for actin (negative control), and for SARS-CoV-2 membrane, envelope, nucleocapsid, and spike for (a) affected patient (P1) and (c) immunocompetent control patient (P5). Mean percent positive IFN-γ and TNF-α responses by intracellular flow cytometry for CD4+ cells for membrane, envelope, nucleocapsid, and spike are presented graphically (c) for antibody-deficient (in black) and immunocompetent (in red) patients. Specificity was determined as a response > 2 × the mean of the negative control (actin) as denoted by the dotted line. No significant difference between affected and control CD4 T cell response for actin (p = 0.67), membrane (p = 0.32), envelope (p = 0.86), nucleocapsid (p = 0.23), and spike (p = 0.13) was found by unpaired t test. Flow plots for other patients and controls are shown in supplemental Fig. 3
Memory T Cell Phenotype
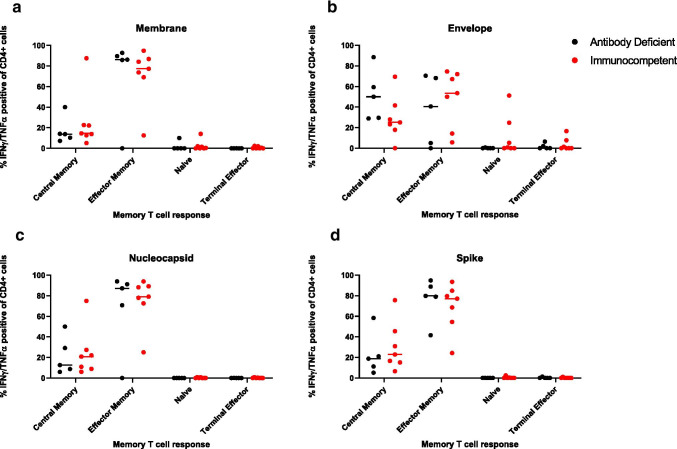
Memory T cell phenotype of SARS-CoV-2-specific cells was evaluated after 10 days of VST microexpansion. In P1-P5, SARS-CoV-2-specific CD3+ cells were primarily effector (mean 78.59%; SD 10.7) and central memory (mean 20.83%; SD 10.5) T cells. Patient 6 (unrelated) also had detectable SARS-CoV-2-specific CD3+ cells comprising both effector (14.74%) and central memory T cell (55.21%) populations despite an undetectable humoral response. Patient 7 had detectable SARS-CoV-2-specific CD3+ cells comprising effector (51.92%), central memory (26.71%), naïve (16.92%), and terminal effector (4.45%) memory T cell populations. The specific CD4+ T cell memory response in the affected patients was predominantly effector memory for membrane (mean 70.89%; SD 39.72), nucleocapsid (mean 68.63%; SD 39.40), and spike (mean 76.86%; SD 20.75) (Fig. 3). The specific CD4+ T cell memory response in the control patients is predominantly effector for membrane (mean 90.87%; SD 5.62), envelope (mean 73.32%; SD 1.80), nucleocapsid (mean 91.1%; SD 4.03), and spike (mean 85.32%; SD 11.56) (Fig. 3). There was no significant difference in CD4+ T cell memory response for spike between affected and control patients with respect to naïve (p = 1.0), central memory (p = 0.63), effector memory (p = 0.62), and terminal effector (p = 0.57) T cells. Overall, the T cell responses in all the CVID patients were not significantly different from healthy adult and pediatric convalescent subjects (additional data not shown).
Fig. 3.
Flow cytometry memory phenotype of CD4+ cells positive for IFN-γ and TNF- in antibody-deficient (in black) and immunocompetent control (in red) patients. Percent distribution and mean (horizontal lines) are shown for each T cell phenotype (naïve, central memory, effector memory and terminal effector) based on stimulation with each peptide library evaluated (membrane (a), envelope (b), nucleocapsid (c), spike (d))
Discussion
To date, there are very little data on adaptive immune responses to SARS-CoV-2 in patients with IEI. Though it may be expected that antibody responses could be impaired in patients with various forms of antibody deficiency, it has been demonstrated that some patients with CVID do have detectable primary antibody responses to viral antigens (e.g., influenza) as well as memory B cell responses [18, 19]. Furthermore, patients with many forms of antibody deficiency can demonstrate cellular responses to antigens which impact clinical decision-making regarding inactivated vaccine administration to patients on immunoglobulin therapy [18–21]. Here, we demonstrate that 3 members of a family with varying degrees of antibody deficiency and 2 unrelated patients with CVID all had a robust adaptive immune response to SARS-CoV-2 following asymptomatic or mild disease. While supplemental immunoglobulin therapy has been shown to potentially contain some anti-SAR-CoV-2 antibodies, the high antibody titers in the proband (P1) and unrelated P7 suggest that this was in fact a primary immune response. Furthermore, the type and magnitude of B and T cell response was similar between this small group of antibody-deficient patients and healthy controls. Of note, the LIPS assay used for this study has been compared to the commercially available Roche assay for nucleocapsid [22], with result concordance in 383 of 400 tested samples. Similar to the administration of influenza vaccination in patients with IEI, these findings provide some preliminary support for vaccination in the management of patients with antibody deficiencies.
In contrast, an unrelated adult (P6) with CVID receiving immunoglobulin supplementation who was positive for SARS-CoV-2 by PCR testing did not have a demonstrable antibody response at 33 days after diagnosis but did have a detectable SARS-CoV-2-specific T cell response. Lack of a humoral response following relatively asymptomatic infection (similar to this patient’s course) has been described, which may have contributed to these findings [23]. However, given the spectrum of severity of patients with CVID and related antibody disorders, it also stands to reason that not all patients will have robust B cell and/or T cell responses to SARS-CoV-2 following infection or vaccination.
To our knowledge, this is the first report showing robust T cell activity and humoral responses against SARS-CoV-2 structural proteins in patients with antibody deficiency. Given the reliance on spike protein in most candidate vaccines [24, 25], the responses demonstrated are encouraging, though additional studies will be needed to further define the quality of the antibody response and the longevity of immune responses against SARS-CoV-2 in immunocompromised patients compared with healthy donors.
Supplementary Information
Below is the link to the electronic supplementary material.
(PDF 645 kb)
Acknowledgements
The authors would like to thank the staff of the Center for Cancer and Immunology Research and the NIAID Laboratory of Infectious Disease for enabling this work. We also are very grateful to our patients for participating in this work. We would like to acknowledge the National Institute of Allergy and Infectious Diseases (NIAID) Centralized Sequencing Program (CSP), including Morgan Similuk, Jia Yan, Rajarshi Ghosh, Bryce Seifert, Michael Setzer, Michael Kamen, Colleen Jodarski, Kathleen Jevtich, Yunting Yu, and Rylee Duncan; the Department of Laboratory Medicine (DLM) staff members, including Adrienne Borges, Josephine Geh, and Julie Niemela; NIAID Collaborative Bioinformatics Resource (NCBR) and Frederick National Laboratory for Cancer Research (FNLCR), including Justin B. Lack and Vasudev Kuram; and the Genomic Research Integration System (GRIS) team, including Sandhya Xirasagar, Jason Barnett, Xi Cheng, Yongjie Fan, Ke Huang, Krishnaveni Kaladi, Eric Karlins , Zhiwen Li, Joseph Mackey, Andrew Oler, Daniel Veltri, Lingwen Zhang, Satishkumar Ranganathan, Nikita Vlassenko, Smilee Samuel, and Robert Gilmore, Office of Cyber Infrastructure and Computational Biology, NIAID, NIH, Rockville, Maryland.
Abbreviations
- CVID
Common variable immunodeficiency
- IEI
Inborn errors of immunity
- IgA
Immunoglobulin A
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- LU
Light units
- PFAPA
Periodic fever, aphthous stomatitis, pharyngitis, adenitis
- SAD
Specific antibody deficiency
- VST
Viral specific T cell
Authors’ Contribution
HK, JDS, CAL, JC, and CMB conceived and designed the experiments; HK, JDS, MJW, VK, KW, CAL, and PB conducted the research; HK, JDS, CAL, JIC, CMB, and MDK analyzed data; HK, JDS, CAL, JIC, CMB, and MDK wrote the manuscript; IF, AK, and KMR provided patient samples and history; LN and MW provided genetic sequencing support. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the Jeffrey Modell Foundation, the Board of Visitors of the Children’s National Health System, the Katzen Foundation, the Connor Family Foundation, and the intramural research programs of the National Institute of Allergy and Infectious Diseases and the National Institute of Dental and Craniofacial Research.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024, Task Order No. 75N91020F00005.
The GRIS system was developed in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under BCBB Support Services Contract HHSN316201300006W/HHSN27200002 to MSC, Inc.
Data Availability
This data or associated data is not in a data repository.
Code Availability
Not applicable.
Declarations
Ethics Approval
All patients provided written informed consent on protocols approved by the National Institutes of Health Institutional Review Board.
Consent to Participate
See above.
Consent for Publication
All authors concur with the submission of this manuscript, and the material submitted for publication is original research and has not been previously reported and is not under consideration for publication elsewhere.
Conflict of Interest
CMB is co-founder and on the scientific advisory boards for Catamaran Bio and Mana Therapeutics with stock and/or ownership, is on the Board of Directors for Caballeta Bio with stock options and has stock in Neximmune and Repertoire Immune Medicine.MDK is on a scientific advisory panel for Enzyvant Therapeutics. The other authors declare that there are no conflicts of interests.
Footnotes
Hannah Kinoshita and Jessica Durkee-Shock are co-first authors
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, I. China Novel Coronavirus, T Research, A novel coronavirus from patients with pneumonia in china, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, Liu XQ, Chen RC, Tang CL, Wang T, Ou CQ, Li L, Chen PY, Sang L, Wang W, Li JF, Li CC, Ou LM, Cheng B, Xiong S, Ni ZY, Xiang J, Hu Y, Liu L, Shan H, Lei CL, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Cheng LL, Ye F, Li SY, Zheng JP, Zhang NF, Zhong NS, He JX, C. China Medical Treatment Expert Group for, Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):2000547. 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed]
- 3.Minotti C, Tirelli F, Barbieri E, Giaquinto C, Dona D. How is immunosuppressive status affecting children and adults in SARS-CoV-2 infection? A systematic review. J Infect. 2020;81:e61–e66. doi: 10.1016/j.jinf.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho HE, Mathew S, Peluso MJ, Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy ClinImmunolPract. 2021;9:490–493 e492. doi: 10.1016/j.jaip.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields AM, Burns SO, Savic S, Richter AG, UPC-Consortium. COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147(3):870–875.e1. [DOI] [PMC free article] [PubMed]
- 6.Meyts I, Bucciol G, Quinti I, Neven B, Fischer A, Seoane E, Lopez-Granados E, Gianelli C, Robles-Marhuenda A, Jeandel PY, Paillard C, Sankaran VG, Demirdag YY, Lougaris V, Aiuti A, Plebani A, Milito C, Dalm VA, Guevara-Hoyer K, Sanchez-Ramon S, Bezrodnik L, Barzaghi F, Gonzalez-Granado LI, Hayman GR, Uzel G, Mendonca LO, Agostini C, Spadaro G, Badolato R, Soresina A, Vermeulen F, Bosteels C, Lambrecht BN, Keller M, Mustillo PJ, Abraham RS, Gupta S, Ozen A, Karakoc-Aydiner E, Baris S, Freeman A, Yamazaki-Nakashimada M, Scheffler-Mendoza S, Espinosa-Padilla S, Gennery AR, Jolles S, Espinoza Y, Poli MC, Fieschi C, Hauck F, Cunningham-Rundles C, Mahlaoui N, ICoIEo Immunity, Warnatz K, Sullivan KE, Tangye SG. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147(2):520–31. 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed]
- 7.Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, Mastroianni CM, Turriziani O, Bondioni MP, Filippini M, Soresina A, Spadaro G, Agostini C, Carsetti R, Plebani A. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy ClinImmunol. 2020;146:211–213 e214. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501 e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT Jr, Cohen JI. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. J Infect Dis. 2020;222:206–13. 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed]
- 10.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 11.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall MA, Beyer K, Rohmel J, Voigt S, Schmitz J, Miltenyi S, Demuth I, Muller MA, Hocke A, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander LE, Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–4. [DOI] [PubMed]
- 12.Keller MD, Harris KM, Jensen-Wachspress MA, Kankate VV, Lang H, Lazarski CA, Durkee-Shock J, Lee PH, Chaudhry K, Webber K, Datar A, Terpilowski M, Reynolds EK, Stevenson EM, Val S, Shancer Z, Zhang N, Ulrey R, Ekanem U, Stanojevic M, Geiger A, Liang H, Hoq F, Abraham AA, Hanley PJ, Cruz CR, Ferrer K, Dropulic L, Gangler K, Burbelo PD, Jones RB, Cohen JI, Bollard CM. SARS-CoV-2-specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein. Blood. 2020;136:2905–2917. doi: 10.1182/blood.2020008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez E, Bonilla FA, Orange JS, Ballow M. Specific antibody deficiency: controversies in diagnosis and management. Front Immunol. 2017;8:586. doi: 10.3389/fimmu.2017.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morbach H, Eichhorn EM, Liese JG, Girschick HJ. Reference values for B cell subpopulations from infancy to adulthood. Clin Exp Immunol. 2010;162:271–279. doi: 10.1111/j.1365-2249.2010.04206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freiberger T, Ravcukova B, Grodecka L, Pikulova Z, Stikarovska D, Pesak S, Kuklinek P, Jarkovsky J, Salzer U, Litzman J. Sequence variants of the TNFRSF13B gene in Czech CVID and IgAD patients in the context of other populations. Hum Immunol. 2012;73:1147–1154. doi: 10.1016/j.humimm.2012.07.342. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Gallo M, Radigan L, Almejun MB, Martinez-Pomar N, Matamoros N, Cunningham-Rundles C. TACI mutations and impaired B-cell function in subjects with CVID and healthy heterozygotes. J Allergy Clin Immunol. 2013;131:468–476. doi: 10.1016/j.jaci.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzer U, Bacchelli C, Buckridge S, Pan-Hammarstrom Q, Jennings S, Lougaris V, Bergbreiter A, Hagena T, Birmelin J, Plebani A, Webster AD, Peter HH, Suez D, Chapel H, McLean-Tooke A, Spickett GP, Anover-Sombke S, Ochs HD, Urschel S, Belohradsky BH, Ugrinovic S, Kumararatne DS, Lawrence TC, Holm AM, Franco JL, Schulze I, Schneider P, Gertz EM, Schaffer AA, Hammarstrom L, Thrasher AJ, Gaspar HB, Grimbacher B. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113:1967–1976. doi: 10.1182/blood-2008-02-141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartley GE, Edwards ESJ, Bosco JJ, Ojaimi S, Stirling RG, Cameron PU, Flanagan K, Plebanski M, Hogarth PM, O'Hehir RE, van Zelm MC. Influenza-specific IgG1(+) memory B-cell numbers increase upon booster vaccination in healthy adults but not in patients with predominantly antibody deficiency. Clin Transl Immunology. 2020;9:e1199. doi: 10.1002/cti2.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedmann D, Goldacker S, Peter HH, Warnatz K. Preserved cellular immunity upon influenza vaccination in most patients with common variable immunodeficiency. J Allergy ClinImmunolPract. 2020;8:2332–2340 e2335. doi: 10.1016/j.jaip.2020.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Paroli M, Accapezzato D, Francavilla V, Insalaco A, Plebani A, Balsano F, Barnaba V. Long-lasting memory-resting and memory-effector CD4+ T cells in human X-linked agammaglobulinemia. Blood. 2002;99:2131–2137. doi: 10.1182/blood.V99.6.2131. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Wu Y, Lam KT, Lee PP, Tu W, Lau YL. Dendritic and T cell response to influenza is normal in the patients with X-linked agammaglobulinemia. J Clin Immunol. 2012;32:421–429. doi: 10.1007/s10875-011-9639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elecsys® Anti-SARS-CoV-2. Package Insert 2020–07, V4.0; Material Numbers 09203095190 and 09203079190.
- 23.Jiang C, Wang Y, Hu M, Wen L, Wen C, Wang Y, Zhu W, Tai S, Jiang Z, Xiao K, Faria NR, De Clercq E, Xu J, Li G. Antibody seroconversion in asymptomatic and symptomatic patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Transl Immunology. 2020;9:e1182. doi: 10.1002/cti2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, Dold C, Faust SN, Finn A, Flaxman AL, Hallis B, Heath P, Jenkin D, Lazarus R, Makinson R, Minassian AM, Pollock KM, Ramasamy M, Robinson H, Snape M, Tarrant R, Voysey M, Green C, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, Oxford CVTG. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O'Dell S, Schmidt SD, Swanson PA, II, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, R.N.A.S.G An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 645 kb)
Data Availability Statement
This data or associated data is not in a data repository.
Not applicable.