CONSPECTUS:
Inhibitor discovery for protein–protein interactions has proven difficult due to the large protein surface areas and dynamic interfaces involved. This is particularly the case when targeting transcription-factor–protein interactions. To address this challenge, structural biology approaches for ligand discovery using X-ray crystallography, mass spectrometry, and nuclear magnetic resonance (NMR) spectroscopy have had a significant impact on advancing small molecule inhibitors into the clinic, including the U.S. Food and Drug Administration approved drug, Venetoclax. Inspired by the protein-observed NMR approach using 1H─15N-HSQC NMR which detects chemical shift perturbations of 15N-labeled amides, we have applied a complementary protein-observed 19F NMR approach using 19F-labeled side-chains that are enriched at protein–protein-interaction interfaces. This protein-observed 19F NMR assay is abbreviated PrOF NMR to distinguish the experiment from the more commonly employed ligand-observed 19F NMR methods.
In this Account, we describe our efforts using PrOF NMR as a ligand discovery tool, particularly for fragment-based ligand discovery (FBLD). We metabolically label the aromatic amino acids on proteins due to the enrichment of aromatic residues at protein interfaces. We choose the 19F nucleus due to its high signal sensitivity and the hyperresponsiveness of 19F to changes in chemical environment. Simultaneous labeling with two different types of fluorinated aromatic amino acids for PrOF NMR has also been achieved. We first describe the technical aspects of considering the application of PrOF NMR for characterizing native protein–protein interactions and for ligand screening. Several test cases are further described with a focus on a transcription factor coactivator interaction with the KIX domain of CBP/p300 and two epigenetic regulatory domains, the bromodomains of BRD4 and BPTF. Through these case studies, we highlight medicinal chemistry applications in FBLD, selectivity screens, structure–activity relationship (SAR) studies, and ligand deconstruction approaches. These studies have led to the discovery of some of the first inhibitors for BPTF and a novel inhibitor class for the N-terminal bromodomain of BRD4. The speed, ease of interpretation, and relatively low concentration of protein needed for NMR-based binding experiments affords a rapid, structural biology-based method to discover and characterize both native and new ligands for bromodomains, and it may find utility in the study of additional epigenetic proteins and transcription-factor–protein interactions.
Graphical Abstract
1. INTRODUCTION
Organofluorine research continues to open up new avenues of inquiry at the interface of chemistry and biology despite the dearth of naturally occurring organofluorine compounds.1 As an example of biomedical impact, 98% of positron emission tomography scans use 18F, primarily from fluorodeoxyglucose.2 Fluorochemicals are highly prevalent as anesthetics,3 and the percentages of fluorine-containing pharmaceuticals continues to increase; 47% of the 2018 U.S. Food and Drug Administration approved drugs contained fluorine.4 This increase is in part from improved metabolic stability from strong C─F bonds, beneficial tuning of physicochemical properties, and noncovalent interactions.4,5 In some cases, fluorination has led to an unusual mechanism of action, such as in the estrogen receptor-degrading drug, Fulvestrant.6
Nuclear magnetic resonance (NMR) spectroscopy has also been impacted by organofluorine compounds. Some of the earliest chemical shifts analyzed used the 19F nucleus, providing insights on shielding and coupling.7,8 The first fluorinated ligands, N-acetyl-fluorophenylalanines, binding to chymotrypsin were detected by 19F NMR in 1967,9 and the first fluorinated protein spectrum was obtained in 1974.10 19F magnetic resonance has now been used for molecular interaction studies,11 magnetic resonance imaging,12 and biomolecular structural analysis.13 In this Account, we describe our ligand-discovery applications using 19F NMR for the study of protein–protein interactions (PPIs). We refer to this approach as protein-observed 19F NMR (PrOF NMR) to distinguish the experiment from more heavily implemented ligand-observed 19F NMR methods.14
The estimated 650 000 PPIs in human cells are essential nodes in all biological processes.15 Detailed understanding of the molecular mechanisms of PPIs and their dynamic regulation is thus necessary for determining the fundamental rules governing their function. Transcription factor PPIs are particularly challenging to both study and perturb their function given their binding promiscuity, transient interactions, and in some cases high regions of disorder; they are often termed “undruggable proteins”.16,17 However, this protein descriptor can evolve over time.
Although many methods have been developed for PPI ligand discovery, our lab was inspired by how structural biology enabled inhibitor development for challenging proteins. One method for screening low complexity molecules, termed fragments, uses protein-observed 1H─15N-HSQC NMR through analysis of protein amide resonances. Fesik and co-workers developed this approach in their seminal “SAR by NMR” (SAR: structure–activity relationship) report.18 We sought to develop a complementary NMR approach using side-chain labeling with fluorine. Although it was clear from prior studies that 19F would be a sensitive reporter of ligand binding,19,20 it was uncertain if such a sparse labeling strategy would be useful for ligand discovery, particularly in the case of identifying low molecular weight fragments. Here, we first describe our initial insights for using PrOF NMR and the technical aspects of running a PrOF NMR experiment. Second, we describe our PrOF NMR ligand discovery efforts for the transcription factor interaction domain, KIX, of the coactivator CBP and its paralogue p300 (CBP/p300), and two epigenetic proteins, BRD4 and BPTF. We conclude the Account regarding future areas for developing PrOF NMR.
2. WHY CHOOSE A PrOF NMR EXPERIMENT?
2.1. Magnetic Resonance Properties of the 19F Nucleus
The 19F nucleus has several favorable properties for biomolecular NMR. The first is a high gyromagnetic ratio. 19F is 83% as sensitive as 1H and thus the second most sensitive stable NMR-active nucleus.11 Unlike protons, due to the absence of fluorine in common buffers, detergents, and naturally occurring biopolymers, experimental setup for PrOF NMR can be performed rapidly without the need for background signal suppression, allowing for a broad array of solution conditions.21
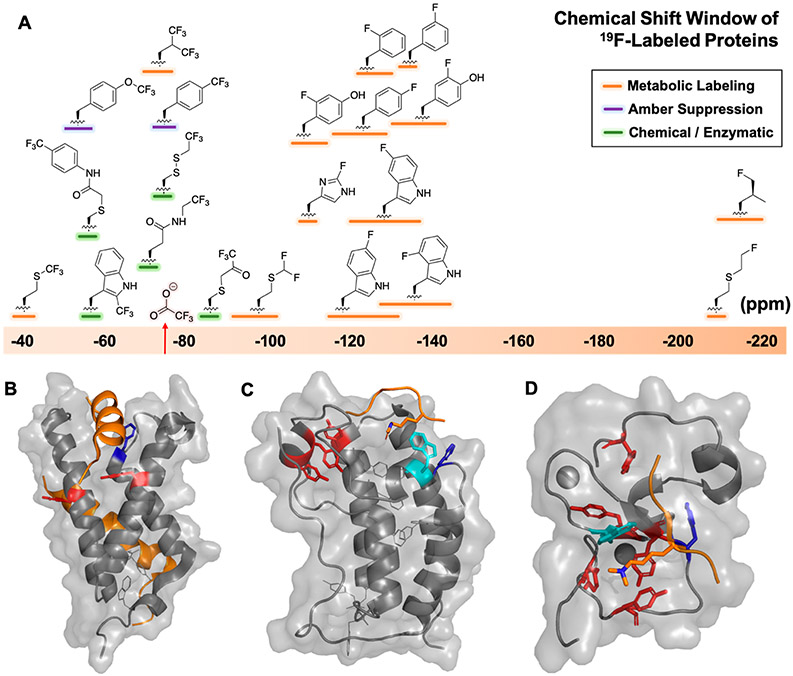
An additional attribute of the 19F nucleus is a broad observable chemical shift range of fluorinated functional groups (Figure 1A). This is an indication of the responsiveness of the fluorine nucleus to subtle changes in chemical environment. Using a range of solvent conditions on model fluorinated compounds, we demonstrated that, under nonpolar conditions, such as those found in a protein interior, the change in chemical shift of the resonance for a 3-fluorotyrosine (3FY) analogue was 2.7-fold greater than the other tyrosine and tryptophan analogues studied.22 We attributed this to dipolar interactions of fluorine with the ortho-substituted hydroxyl group. The environmental responsiveness is also seen in the chemical shift dispersion in folded fluorinated proteins. For example, Lian et al. reported a 14.6 ppm shift range in 4-fluorophenylalanine (4FF)-labeled lysozyme.19 Seven 3FY-residues incorporated into the first bromodomain of protein BRD4, spanned 12 ppm.23 Regarding chemical shift perturbations from ligand interactions, we showed the chemical shift response of the fluorine resonance of 5-fluorotryptophan (5FW) was 6–20-fold more responsive than the indole NH.24 These results showcase how installing fluorine atoms into proteins leads to sensitive and highly responsive NMR probes for ligand-interaction studies.
Figure 1.
(A) Chemical shift ranges of fluorine-labels incorporated in proteins, referenced to trifluoroacetate (red). Representative proteins (gray) and ligands (orange) for PrOF NMR in (B) KIX with MLL and c-Myb (PDB: 2AGH), (C) BRD4-D1 with acetylated histone H4 (PDB: 3QZT), and (D) PHD finger of BPTF with trimethylated histone H3 (PDB: 2F6J). Tyrosines (red), tryptophans (cyan), and phenylalanines (blue) within 10 Å of binding sites.
One notable challenge for PrOF NMR is the high chemical shift anisotropy (CSA) of the 19F nucleus. CSA can be the dominant source of resonance broadening for medium to high molecular weight proteins.13 The CSA scales with the square of the field strength, limiting sensitivity gains at high magnetic fields. Despite these CSA concerns, we were attracted to this approach, as the side-chains of interest for mediating PPIs are primarily at the protein surface.25 The surface accessibility leads to side-chain flexibility, helping to reduce resonance broadening. This property was apparent from the original NMR spectrum of 86 kDa alkaline phosphatase, where all 11 tyrosine resonances are observable, albeit as broad resonances.10
2.2. Metabolic Fluorine-Labeling of Proteins and Spectral Acquisition
We favor sequence-selective 19F incorporation via metabolic labeling for PrOF NMR over site-selective methods such as cysteine bioconjugation or amber suppression, as typically only one or a few sites are labeled in these cases. With an unbiased labeling strategy, a more global coverage can lead to detecting orthosteric binding and the discovery of novel interaction sites.
We metabolically label proteins with fluorine-labeled aromatic amino acids. The enrichment of aromatic amino acids at PPI interfaces (e.g., Figure 1B-D),11,25 and their relatively low abundance, leads to a “goldilocks” phenomenon where a minimal set of fluorinated amino acids can be incorporated at important sites for protein function while reducing potential perturbation from fluorine incorporation. In some cases, however, even a small number of fluoroaromatics can stabilize protein structure, which we found for KIX,26 or destabilize such as for 3FY-labeled caldmodulin.27 In the latter study, fractional labeling was recommended.
The metabolic labeling of proteins using fluorinated amino acids is well-established.21,28 For phenylalanine and tyrosine, auxotrophic bacterial strains, such as DL39(DE3), can be used. Alternatively, glyphosate inhibits the shikimate pathway prohibiting tyrosine, phenylalanine, and tryptophan biosynthesis.29 For fluorinated tryptophan incorporation, we favor the approach of Crowley et al., via the addition of fluoroindoles to the growth media. This produces fluorinated-tryptophan from serine via tryptophan synthase.30 This method is compatible with standard laboratory strains (e.g., BL21(DE3)). In many cases, highly fluorinated proteins (>90% label incorporation) are obtained in yields within 2–4-fold of the unlabeled protein expression.21 We have used these approaches to incorporate 2-fluorotyrosine (2FY), 3FY, 5FW, and 4FF. Additional fluorinated probes incorporated into proteins by us and others can be seen in Figure 1A.
Acquiring a PrOF NMR spectrum is generally straightforward. We find 500–600 MHz field strengths to be the most suitable for PrOF NMR for globular proteins, although 700 MHz magnets have been used for studying G protein-coupled receptors with CF3-lableled side chains which have more narrow resonances resulting from fast bond rotation of the CF3 group.31 Having a dedicated channel that tunes to 19F or 1H/19F is ideal. While the latter prohibits H─F heteronuclear experiments and 1H decoupling, the quality of spectra tends not to benefit from signal enhancement from decoupling based on the already broad resonances. The PrOF NMR experiments described below make use of chemical shift perturbations to analyze ligand binding sites, selectivity, and ligand affinities. However, additional structural and dynamics analyses have been conducted by others including 19F─19F NOE experiments,32 dynamics studies of low populated protein states through saturation transfer and CPMG-relaxation dispersion,34 and solvent accessibility using solvent isotope effects.33
3. CBP/p300 INTERACTIONS AND FRAGMENT SCREENING
3.1. KIX PPI Analysis by PrOF NMR
For developing PrOF NMR as a ligand discovery tool, we were drawn to the KIX domain of CBP/p300 (Figure 1B). These proteins are master coactivators of the cell,35 serving as central hubs for transcription factors and epigenetic regulators via their histone acetyl transferase (HAT) activity.36 The CBP/p300 KIX domain is an 80 amino acid helical protein, bound by over a dozen transcription factors including the mixed lineage leukemia protein (MLL),37 c-Myb,38 and the phosphorylation-dependent kinase inducible domain of CREB (pKID).39 The biological outcomes through CBP/p300-PPIs range from hematopoiesis,40 long-term memory formation,41 and cell-cycle regulation,42 many of which are mediated by the KIX domain. Despite varied biological functions of KIX–transcription-factor interactions and disease relevance, prior ligand discovery approaches have led to few small molecule inhibitors, consistent with the known difficulties of targeting dynamic coactivator–transcription-factor interfaces.36 As prior efforts using 1H─15N-HSQC NMR yielded few ligands,43 we reasoned that a biophysical approach using 19F-labeled protein side chains might provide a new handle for ligand discovery and allow us to test the practical limits of PrOF NMR.
Given the responsiveness of fluorine resonances to differing chemical environments, we anticipated a PrOF NMR approach would provide a simplified experiment to characterize native PPIs in this dynamic protein, and to sensitively report on small molecule interactions where weak affinities were anticipated. In KIX, seven aromatic amino acids are present. The majority of the aromatic amino acids are important for regulating KIX PPIs or protein stability and would cover both known binding sites for MLL, c-Myb, and pKID. Using a DL39(DE3) cell line, we labeled KIX with 3FY. Y631 is in the MLL binding site, and Y649, Y650, and Y658 are located around the second binding site for c-Myb and pKID. Prior to PrOF NMR analysis, we noted only a minimal perturbation from fluorine incorporation to both KIX structure and function based on native ligand binding (~2-fold changes in Kd) and a similar α-helical content by circular dichroism (CD).26
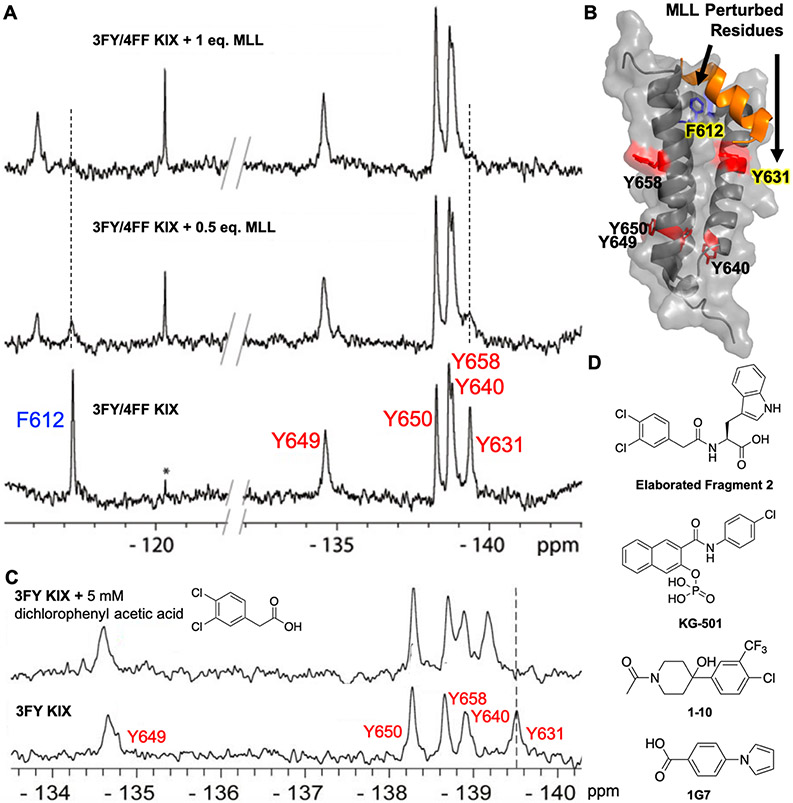
After assigning the NMR spectrum, we determined the binding footprints of MLL, pKID, and c-Myb by monitoring chemical shift perturbations. Despite a sparse labeling strategy, we identified differences in interactions with pKID and c-Myb which have distinct but overlapping binding sites.26 Labeling at F612 with 4FF was introduced to confirm binding of MLL. We have since shown that two fluorinated amino acids can be incorporated simultaneously for better coverage of the MLL-binding site (Figure 2A,B).44
Figure 2.
(A) Titration of dual 3FY/4FF-labeled KIX with MLL; perturbed residues are highlighted in yellow in (B) (PDB: 2AGH). (C) PrOF NMR screening identifies a fragment interaction near Y631. Adapted with permission from ref 44. Copyright 2018 John Wiley & Sons. (D) Ligands characterized using 3FY-KIX.
Prior research showed transcription factors bind to KIX through two distinct binding sites and potentiate binding affinities for ternary complex formation ~2-fold in most cases,39 but as high as 20-fold in others.45 NMR analysis has focused on the backbone amides and aliphatic side chains.46-48 Using PrOF NMR, we showed that the aromatic amino acid side chains also report on allosteric interactions. For c-Myb- and pKID-binding, a flexible loop in the MLL binding site has differential responses suggesting subtle differences in binding mechanisms.26 Allosteric effects in the reverse direction were more difficult to observe, in which case 3FY-labeled KIX showed no side chain movements in the c-Myb/pKID site.26 Using 2FY-labeled KIX, we reasoned this protein construct would behave more similarly to unlabeled KIX given the higher pKa of 2FY (9.2 vs 8.4 for 3FY and 10.1 for tyrosine).22 Indeed, this protein can now report on MLL-induced allosteric effects at Y650.22 We anticipate this labeling approach will be useful for studying positive and negative allosteric regulators of KIX.
3.2. Putting PrOF NMR into Practice for Ligand Discovery of the CBP/p300 KIX Domain
Our initial report used fluorinated KIX as a protein-based NMR tool to also discover small molecule interactions.26 The area of fragment-based ligand discovery (FBLD) has emerged as one promising approach for PPIs, and such a study was an excellent test case for a sparse labeling strategy to detect both small and weak interacting ligands. Although a pilot study of 50 fragments supported the suitability of this approach,26 Gee et al. were the first to put fragment screening into practice by screening 508 fragments against the 3FY-KIX domain and monitoring chemical shift perturbation at a single concentration.49 Mixtures resulting in chemical shift perturbations were deconvoluted by testing each compound to reproduce the binding behavior (e.g., Figure 2C). An advantage of PrOF NMR is the opportunity for rapid ligand titrations by monitoring a change in chemical shift for Kd determination. For low affinity ligands, fast chemical exchange kinetics are typically observed under the conditions of eq 1, where the chemical shift at a given ligand concentration is a weighted average of the bound (νA) and unbound states (νB). The Kd is determined with eq 2 by nonlinear regression using the total protein and ligand concentrations (P and L) and the chemical shift perturbation (Δδexp). A 1H─15N-HSQC experiment and SAR-by-commerce was conducted to verify and follow-up hits. These results demonstrated the use of PrOF NMR for FBLD and enabled ligandability analysis of the KIX domain. In this instance, ligands were only found to affect resonances near the MLL site and not the c-Myb/pKID site, suggesting a peptidomimetic approach might be better for the latter due to the low ligandability.
| (1) |
| (2) |
An unexpected finding using 4FF/3FY dual-labeled-KIX revealed one of the fragments binding to a third site near Y631 on the protein.44 Using PrOF NMR, 1H─15N-HSQC NMR, in-silico solvent mapping, and small molecule docking, we identified a previously uncharacterized small molecule binding site with an elaborated fragment 2 (Kd = 740 μM, Figure 2D) and KG-501,43 a known CREB-KIX inhibitor of moderate affinity (Kd = 115 μM). This new binding site offers the potential to negatively regulate transcription factor binding without competing with native PPIs.
Given the speed of the experiment (~4–5 min at 50 μM KIX), our KIX ligand discovery efforts have translated into the classroom for use of fragment screening in a curriculum-based undergraduate research experience (CURE).50 We use Ni(II) chelates as T1 relaxation agents to reduce experiment time and increase the experiment accessibility in a CURE setting.51 We can further reduce experiment times to 1–2 min with cryoprobes, or a 30 min experiment using a room temperature broadband probe at 400 MHz with T1 relaxation agents.
4. PrOF NMR APPLICATIONS IN EPIGENETICS
Epigenetic mechanisms of controlling gene expression are defined as heritable modifications to DNA, RNA, and proteins.52 Our lab has developed chemical tools to study the recognition of histone lysine acetylation, which facilitates the accessibility of chromatin and can be dysregulated in disease. Using PrOF NMR, we focused on identifying the substrate specificity of effector proteins involved in recognition of histone acetylation, and chemical probe development for bromodomain-containing proteins.
Bromodomains are left-handed helical bundles that recognize lysine acylation states.53 Histone hyperacetylation correlates with activated transcription, with bromodomain-containing proteins playing a role in recruiting transcription factors to hyperacetylated loci.54 Substrate recognition by most bromodomains is via conserved asparagine and tyrosine residues in the binding pocket, which form direct and water-mediated hydrogen bonds to acetyl-lysine.55 When we became interested in studying bromodomains, few selective chemical probes were available to distinguish the diverse functions of the 46 human bromodomain-containing proteins.55 Given the transient interactions between bromodomains and their native targets, assay techniques to study bromodomain–ligand interactions were limited. Our initial structural analysis of the 61 human bromodomains found that the binding sites of bromodomains were enriched with aromatic amino acids and nearly half of the human bromodomains contained greater than four aromatic residues.23 This suggested PrOF NMR may be generally suitable for bromodomain analyses.
4.1. Fluorinated Amino Acids are Sensitive Probes for BRD4 Ligand Discovery
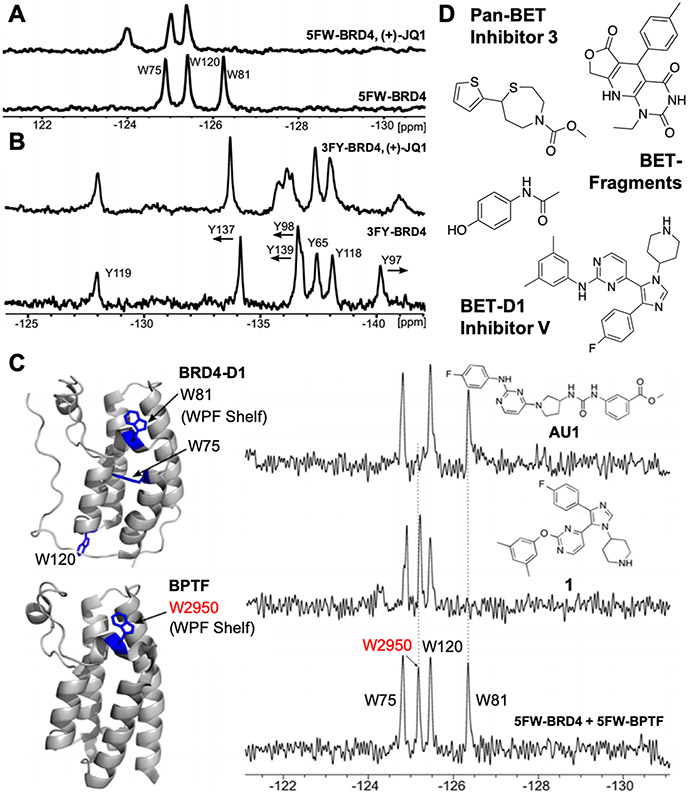
We initially applied our approach to the first bromodomain of BRD4 (BRD4-D1), a member of the bromodomain and extraterminal (BET) family of proteins. BRD4 is an emerging drug target, in part due to its regulation of Myc expression.56,57 BRD4-D1 has three tryptophan and seven tyrosine residues; five of these aromatic residues are within 10 Å of the binding site (Figure 1C) and serve as reporters of small-molecule binding. Three tryptophan residues are distributed on the protein with W81 in the “WPF-shelf” region of the binding site, W75, an intermediate distance away, and W120 is on the opposite side of the protein. In the BRD4-D1 PrOF NMR spectra, the three resonances for 5FW are dispersed over 2 ppm and 3FY residues span over 12 ppm (Figure 3A, B).23 We determined fluorination of these proteins caused minimal perturbation using isothermal titration calorimetry (ITC), CD, and protein crystallography, supporting comparable ligand binding in unlabeled bromodomains to that of 19F-labeled proteins.
Figure 3.
PrOF NMR titrations of (+)-JQ1 in BRD4-D1 labeled with (A) 5FW and (B) 3FY. (C) Screening of kinase inhibitors against mixtures of 5FW-BRD4-D1 and 5FW-BPTF (adapted with permission from refs 23 and 60. Copyright 2014 and 2015 American Chemical Society, respectively). (D) Bromodomain inhibitors and fragments characterized by PrOF NMR.23,61-63
In ligand-binding experiments, W81 functions as a reliable reporter for ligand binding due to its proximity to the binding site. Our initial binding experiments used a high affinity chemical probe, (+)-JQ1.58 19F resonances for W81 exhibited the largest chemical shift and slow chemical exchange binding effects (Figure 3A).23 In this case, both the unbound and bound resonances corresponding to the protein binding site remained well-resolved, consistent with a long residence time on the protein. 3FY-labeled BRD4-D1 gave similar results affecting resonances Y97, Y98, and Y139 consistent with the (+)-JQ1-binding footprint and Y137 near, but outside the binding site (Figure 3B). We characterized the small molecule acetaminophen, which has the molecular weight of a fragment molecule (151 g/mol, Figure 3D),23 and whose affinity was unable to be characterized by fluorescence anisotropy (FA) and TR-FRET due to its weak affinity, despite several cocrystal structures being obtained.59 By PrOF NMR, a highly responsive binding isotherm could be obtained for 3FY- and 5FW-labeled BRD4-D1, yielding a Kd of 290 and 230 μM, respectively, based on the resonances for Y139 and W81.23 Quantification of such a small molecule interaction motivated our FBLD-bromodomain studies.
In collaboration with Eli Lilly and Co. we compared PrOF NMR with a ligand-observed NMR experiment, 1H─CPMG, to evaluate the strengths and weaknesses of both methods. In the 930-fragment screen, there was >80% agreement between the methods.24 However, in the case of hits that were competitively displaced with a known inhibitor, 9% of the hits were not detected by PrOF NMR, indicating potential false negatives from fluorine perturbations to binding, or a binding event unable to induce a fluorine chemical shift. Two of the top binding hits were evaluated by ITC using fluorinated and unlabeled proteins and shown to have comparable Kd’s. However, we observed fluorine-labeling to prevent fragment binding in other studies.62
We further combined PrOF NMR and 1H─CPMG to test the suitability of screening more 3D-enriched fragments for discovering selective bromodomain inhibitors.62 After conducting a round of SAR by PrOF NMR, we identified an underrepresented thiazapane scaffold as a new BRD4-D1 ligand (Figure 3D). PrOF NMR characterized a thiazapane with a 10-fold affinity enhancement and good ligand efficiency (Kd = 20 μM, 0.40 respectively)
To place PrOF NMR in context with existing approaches, 1H-CPMG had the advantage of avoiding the deconvolution step to identify hits, and PrOF NMR provided a more rapid experiment for the 15 kDa bromodomain and led to reliable Kd determinations for rank-ordering compounds. Using these methods in parallel is more robust for screening and is rapid for ligand deconvolution.
SOFAST-HMQC experiments with bromodomains can be acquired in ~7 min under similar conditions.64 PrOF NMR can be obtained ~3–4 times faster on fluorine-tuned cryoprobes, although amide-detected experiments have better surface coverage. 1D 1H NMR using indole NH experiments was also compared and showed the 1H chemical shift response was 6–20-fold lower.24 In addition, we find fluorine chemical shift measurements to be highly reproducible and find changes larger than 0.03 ppm to be reliable for reporting on binding.24 We thus find PrOF NMR to be a valuable and complementary experiment for fragment screening.
4.2. Screening Protein Mixtures by PrOF NMR
An important finding from our PrOF NMR study with BRD4-D1 was that fluorinated proteins could be screened in mixtures.60 A relatively simple spectrum of sufficiently resolved resonances was observed from a mixture of two 5FW-labeled bromodomains from BRD4-D1 and BPTF (Figure 3C, bottom). Although bromodomains are similar in sequence around the binding site, the sensitivity of the 19F nucleus to differing chemical environments still led to good resonance dispersion. Few selective inhibitors were known at the time for non-BET family bromodomains, such as BPTF, while a surprising number of kinase inhibitors were shown to inhibit BET-family bromodomains.65,66 Screening two bromodomains at once allowed for assessing selectivity at the outset for both proteins. As a proof of principle, we characterized several known kinase-bromodomain inhibitors which bound to either BRD4 or BPTF, demonstrating the utility of analyzing two proteins in the same NMR experiment.23
In a follow-up report, we screened a library of kinase inhibitors against a mixture of the BPTF and BRD4-D1 bromodomains to identify molecules that were selective for either BRD4-D1 or BPTF.60 From this 229-compound screen, we identified three selective BPTF hits and 15 selective BRD4-D1 hits. These binders were characterized using ITC, FA and differential scanning fluorimetry to yield the first selective inhibitor of BPTF, an arylurea AU1, and a series of trisubstituted imidazoles with selectivity for BRD4-D1 over BPTF (Figure 3C).
4.3. PrOF NMR Enables D1-Selective BET-Inhibitor Discovery
We identified the BET-bromodomain inhibitor, 1,4,5-trisubstituted imidazole SB-284851-BT, as a dual p38α/BRD4 ligand. Similar p38α inhibitors were identified from two prior reports.65,67 While these studies and our dual screen showed these molecules bound BRD4-D1, the selectivity profiles within the BET family were not evaluated. We followed-up testing for intra-BET selectivity using new analogues including molecule V (Figure 3D) and found a >55-fold selectivity for BRD4-D1 over BRD4-D2.63 This trend was consistent between the two bromodomains of BRDT. Using a cocrystal structure of V, we identified a conserved structured water from the binding site being displaced by the fluorophenyl group of V, a surprising finding given the stability of this water network and its role in substrate recognition.68 This result in part supported a smaller energetic penalty of displacing this structured water in BRD4-D1 than in BRD4-D2.63 To demonstrate the efficacy of a BET-D1 inhibitor, we showed compound V reduces proliferation of MM.1S, a BRD4-sensitive cell line, through inhibiting c-Myc expression.63 Encouraged by the lack of available BET-D1 selective molecules, V, is a promising lead for future chemical probe development.
To date, PrOF NMR has been well-established for FBLD for BET bromodomains and is a complementary biophysical experiment to ligand-observed NMR.21,23,24 The rapid Kd determination has been used for SAR generation and rank-ordering compounds.24,51 Finally, a novel multiprotein screening approach has led to a new inhibitor class with an unexplored binding interaction for tailoring BET bromodomain selectivity.60,63
4.4. BPTF Bromodomain Inhibitors Developed by PrOF NMR
Our PrOF NMR ligand discovery efforts have also been applied toward the bromodomain and plant homeodomain (PHD) containing transcription factor (BPTF). BPTF is the largest component of the nucleosome remodeling factor (NURF). NURF regulates gene expression by altering chromatin accessibility and recruitment to promoter regions via BPTF.69 BPTF dysfunction has been linked to cancer.70-72 The first indications for a role in cancer were found by Rotter and co-workers demonstrating that BPTF overexpression was linked to a malignant phenotype and showing that knockdown of BPTF prevented cell proliferation in lung cancer.72 More recently, an additional oncogenic role of BPTF is through regulating Myc function via a Myc-BPTF PPI.73
Given the biological significance of BPTF, we were drawn to this protein based on a lack of chemical inhibitors for its bromodomain. A WPF shelf motif, found in ~30% of bromodomains such as BRD4, is present in BPTF.55 This motif has a key role in the analysis of binding to bromodomains, and proteins can be fluorinated at the tryptophan for PrOF NMR.
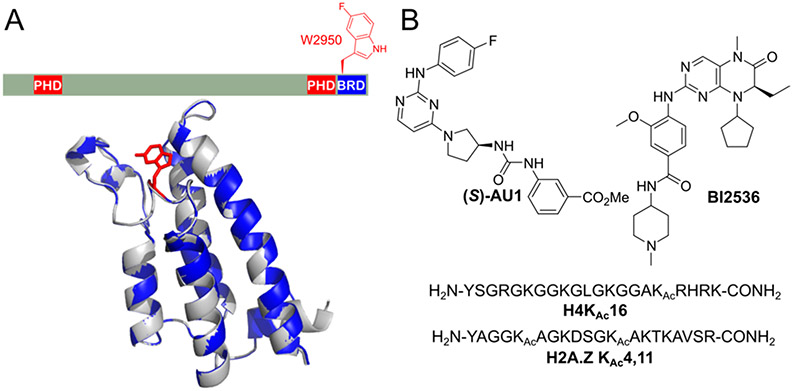
Our first attempt at fluorinating the BPTF bromodomain focused on 5FW-labeling the WPF shelf (W2950 or W2824 based on PDB nomenclature).23 Fluorination does not significantly affect helicity or thermal stability as assessed by CD, nor overall fold based on X-ray crystallography (Figure 4A).60,74 Given the weak affinity of histones for bromodomains, we quantified the affinity of native histone H4 acetylated at lysine 16 (H4Kac16) by PrOF NMR, showing binding consistent with prior reports.75 PrOF NMR was also used to quantify new bromodomain interactions with acetylated peptides based on isoform I of the histone variant H2AZ.1 (Figure 4B), and to characterize bromodomain selectivity for H2AZ.1 via PrOF NMR using a panel of fluorinated bromodomains.75 This result was notable as the first characterization of a direct H2AZ-bromodomain interaction.
Figure 4.
(A) Structural representation of BPTF domains and overlay of 5FW-labeled and unlabeled BPTF bromodomains (Reproduced with permission from ref 74. Copyright 2019 The Royal Society of Chemistry). (B) BPTF ligands characterized by PrOF NMR.
For small molecule discovery, early results from our dual screening assay with BRD4 and BPTF as described above were encouraging.60 The identification of a dual kinase-bromodomain ligand, BI2536 for BPTF (Kd = 37 μM, Figure 4B)60 whose binding interaction had gone undetected in thermal shift and Bromoscan assays,65,66 demonstrated the sensitivity of PrOF NMR. In addition an amino oxadiazole and arylurea, AU1, were found to bind the bromodomain of BPTF over BRD4 (Figure 4B).60 AU1 was the first reported small molecule ligand for the bromodomain of BPTF with selectivity over the BET bromodomain BRD4-BD1. PrOF NMR was unable to quantify affinity based on an intermediate chemical exchange process, so ITC was used to determine a Kd of 2.8 μM. A BPTF-dependent-luciferase reporter assay with AU1 demonstrated the importance of the bromodomain in regulating transcription in live cells.
After the discovery of AU1, efforts turned to using PrOF NMR for medicinal chemistry optimization to improve the performance of AU1 as a BPTF bromodomain ligand, including both SAR and ligand deconstruction (Figure 5).74 During the SAR phase, the (S)-enantiomer of AU1 was determined to be responsible for bromodomain binding, and numerous analogues were developed to probe whether improved protein contacts could be made. As a second application of PrOF NMR, a quantitative ligand-deconstruction approach was used to optimize important sites for binding to the bromodomain.74 Most of the deconstructed fragments exhibited binding with similar ligand efficiencies to the bromodomain of BPTF, suggesting that the efficacy of AU1 toward binding was the result of several weak contacts dispersed throughout the molecule. Although significant improvements in affinity were not achieved, this study demonstrated the suitability of PrOF NMR for ligand discovery and hit-to-lead optimization studies.
Figure 5.
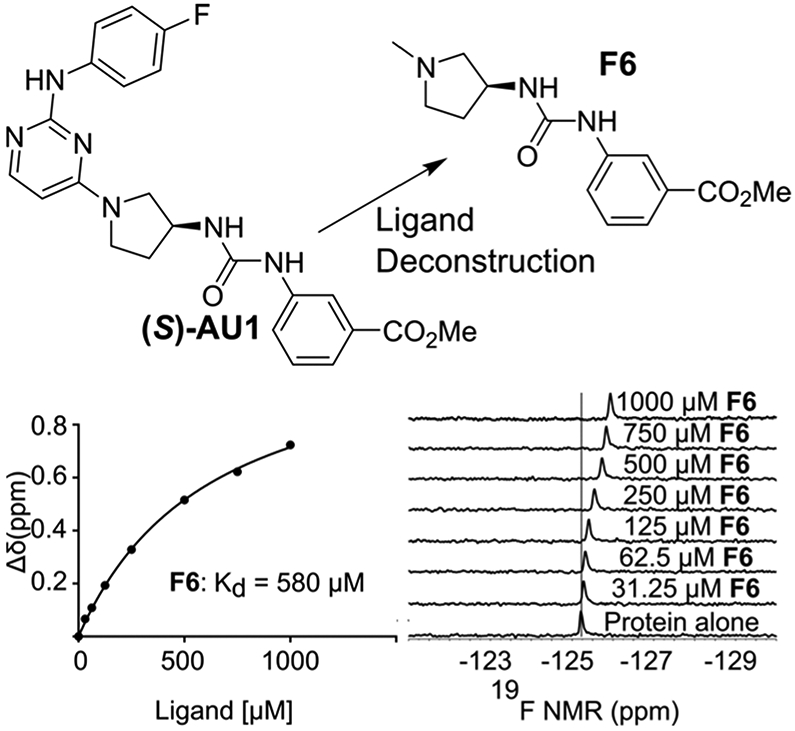
Ligand deconstruction of (S)-AU1. A representative PrOF NMR titration for Kd determination of F6 (Reproduced with permission from ref 74. Copyright 2019 The Royal Society of Chemistry).
As the first BPTF ligand, AU1 was studied in a cellular context. In Eph4 mammary epithelial cells, Frey et al. demonstrated AU1-dependent BPTF inhibitory effects on chromatin accessibility and cell survival.76 Treatment of Eph4 cells with AU1 decreased c-Myc DNA occupancy consistent with disrupting a BPTF-Myc PPI. AU1 treatment also led to cell cycle arrest in the G1 phase, reducing proliferation of Eph4 cells. ATAC-Seq experiments showed a change in chromatin accessibility, consistent with BPTF knockdown experiments. Although AU1 remains imperfect with additional kinase off-targets, Kirberger et al. identified a chronic mylogenous leukemia cell line, K562, that was BPTF-sensitive using a combination of CRISPR-Cas9 gene deletion studies and activity with only the active (S)-enantiomer of AU1, opening up a new area of inquiry for BPTF inhibitors.74 Given the success of PrOF NMR-mediated inhibitor discovery for the BPTF bromodomain, future experiments will study the PHD domain, which contains an aromatic box for recognizing histones (Figure 1D).69
5. CONCLUSION AND 19FUTURE OUTLOOK
PrOF NMR has provided valuable structural information on challenging proteins since 1974, and, encouragingly, research laboratories have started to evaluate its promise against a diverse array of targets.11-13,77-80 However, as the concept of employing PrOF NMR for medicinal chemistry is relatively new, we anticipate the two significant challenges of protein size and resonance assignment will be addressed soon. Although CSA effects are limiting for screening large proteins, exciting progress has been made in developing TROSY-based experiments leading to the study of 180 kDa proteins.81 To address sequence assignments of fluorinated proteins, pulse sequences can be applied if the backbone amides are assigned.82 We and others have attempted computational predictions, but the success has been limited;83-85 we anticipate new approaches to address this challenge will be enabling. Given the simplicity of PrOF NMR experiments, future impactful findings may lie in the analysis of complicated biomolecular systems, whether in cellular environments77 or of large multidomain proteins. For those that are attracted to this biorthogonal atom, its potential continues to excite.
ACKNOWLEDGMENTS
We gratefully acknowledge past and present Pomerantz lab members for their contributions and the NIH (R01GM121414, 5T32GM132029-01) for financial support.
Biography
Anand Divakaran is a Ph.D. student with Professors Will Pomerantz and Dan Harki at the University of Minnesota. After earning a B.Sc. in Chemistry at Calvin College, he pursued industrial research at Perrigo pharmaceuticals and 3M prior to graduate school. He is interested in studying epigenetic post-translational modifications and currently works on selectively inhibiting bromodomains.
Steve Kirberger completed his B.S. in chemistry at the University of Minnesota followed by a a Ph.D. in Chemistry in the lab of Prof. Will Pomerantz. His studies involved development of peptide-based 19F MRI contrast agents and bromodomain inhibitors.
William C. K. Pomerantz received his B.S. in chemistry from Ithaca College in 2002, followed by a Fulbright Fellowship at ETH, Zurich with Professors François Diederich and Jack Dunitz. He obtained a Ph.D. in chemistry under Professors Sam Gellman and Nick Abbott at the University of Wisconsin—Madison and was a postdoctoral fellow under Prof. Anna Mapp at the University of Michigan. He joined the chemistry faculty at the University of Minnesota in 2012. His research focuses on the development of chemical biology and medicinal chemistry approaches for modulating transcription factor function.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Murphy CD; Schaffrath C; O’Hagan D Fluorinated Natural Products: The Biosynthesis of Fluoroacetate and 4-Fluorothreonine in Streptomyces Cattleya. Chemosphere 2003, 52, 455–461. [DOI] [PubMed] [Google Scholar]
- (2).Nuclear Medicine/ Radiopharmaceuticals Market by Type (Diagnostic (SPECT (Technetium), PET (F-18)), Therapeutic (Beta Emitters (I-131), Alpha Emitters, Brachytherapy(Y-90), Application (Oncology, Thyroid, Cardiology) - Global Forecasts to 2023. Markets and Markets; https://www.marketsandmarkets.com/Market-Reports/radiopharmaceuticals-market-417.html (accessed Jul 8, 2019).
- (3).Halpern DF Fluorinated Inhalation Anesthetics. In Organofluorine Chemistry; Banks RE, Smart BE, Tatlow JC, Eds.; Springer US: Boston, MA, 1994; pp 543–554. [Google Scholar]
- (4).Pan Y The Dark Side of Fluorine. ACS Med. Chem. Lett 2019, 10, 1016–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gillis EP; Eastman KJ; Hill MD; Donnelly DJ; Meanwell NA Applications of Fluorine in Medicinal Chemistry. J. Med. Chem 2015, 58, 8315–8359. [DOI] [PubMed] [Google Scholar]
- (6).Nathan MR; Schmid P A Review of Fulvestrant in Breast Cancer. Oncol. Ther 2017, 5, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gutowsky HS; Hoffman CJ Nuclear Magnetic Shielding in Fluorine and Hydrogen Compounds. J. Chem. Phys 1951, 19, 1259–1267. [Google Scholar]
- (8).Gutowsky HS; Hoffman CJ Chemical Shifts in the Magnetic Resonance of F19. Phys. Rev 1950, 80, 110–111. [Google Scholar]
- (9).Spotswood TML; Evans JM; Richards JH Enzyme-Substrate Interaction by Nuclear Magnetic Resonance. J. Am. Chem. Soc 1967, 89, 5052–5054. [DOI] [PubMed] [Google Scholar]
- (10).Hull WE; Sykes BD Fluorotyrosine Alkaline Phosphatase. 19F Nuclear Magnetic Resonance Relaxation Times and Molecular Motion of the Individual Fluorotyrosines. Biochemistry 1974, 13, 3431–3437. [DOI] [PubMed] [Google Scholar]
- (11).Arntson KE; Pomerantz WCK Protein-Observed Fluorine NMR: A Bioorthogonal Approach for Small Molecule Discovery. J. Med. Chem 2016, 59, 5158–5171. [DOI] [PubMed] [Google Scholar]
- (12).Tirotta I; Dichiarante V; Pigliacelli C; Cavallo G; Terraneo G; Baldelli Bombelli F; Metrangolo P; Resnati G 19F Magnetic Resonance Imaging (MRI): From Design of Materials to Clinical Applications. Chem. Rev 2015, 115, 1106–1129. [DOI] [PubMed] [Google Scholar]
- (13).Kitevski-LeBlanc JL; Prosser RS Current Applications of 19F NMR to Studies of Protein Structure and Dynamics. Prog. Nucl. Magn. Reson. Spectrosc 2012, 62, 1–33. [DOI] [PubMed] [Google Scholar]
- (14).Dalvit C; Vulpetti A Ligand-Based Fluorine NMR Screening: Principles and Applications in Drug Discovery Projects. J. Med. Chem 2019, 62, 2218–2244. [DOI] [PubMed] [Google Scholar]
- (15).Stumpf MPH; Thorne T; de Silva E; Stewart R; An HJ; Lappe M; Wiuf C Estimating the Size of the Human Interactome. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 6959–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Darnell JE Transcription Factors as Targets for Cancer Therapy. Nat. Rev. Cancer 2002, 2, 740–749. [DOI] [PubMed] [Google Scholar]
- (17).Koehler AN A Complex Task? Direct Modulation of Transcription Factors with Small Molecules. Curr. Opin. Chem. Biol 2010, 14, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Shuker SB; Hajduk PJ; Meadows RP; Fesik SW Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science 1996, 274, 1531–1534. [DOI] [PubMed] [Google Scholar]
- (19).Lian C; Le H; Montez B; Patterson J; Harrell S; Laws D; Pearson J; Oldfield E; Matsumura I Fluorine-19 Nuclear Magnetic Resonance Spectroscopic Study of Fluorophenylalanine- and Fluorotryptophan-Labeled Avian Egg White Lysozymes. Biochemistry 1994, 33, 5238–5245. [DOI] [PubMed] [Google Scholar]
- (20).Hoeltzli SD; Frieden C 19F NMR Spectroscopy of [6–19F]Tryptophan-Labeled Escherichia Coli Dihydrofolate Reductase: Equilibrium Folding and Ligand Binding Studies. Biochemistry 1994, 33, 5502–5509. [DOI] [PubMed] [Google Scholar]
- (21).Gee CT; Arntson KE; Urick AK; Mishra NK; Hawk LML; Wisniewski AJ; Pomerantz WCK Protein-Observed 19F-NMR for Fragment Screening, Affinity Quantification and Druggability Assessment. Nat. Protoc 2016, 11, 1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ycas PD; Wagner N; Fu R; Pomerantz WCK Comparative Assessment of Monofluorinated Tyrosine Isomers for Protein Observed 19F-NMR. J. Biomol. NMR, in press. [Google Scholar]
- (23).Mishra NK; Urick AK; Ember SWJ; Schonbrunn E; Pomerantz WC Fluorinated Aromatic Amino Acids Are Sensitive 19f NMR Probes for Bromodomain-Ligand Interactions. ACS Chem. Biol 2014, 9, 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Urick AK; Calle LP; Espinosa JF; Hu H; Pomerantz WCK Protein-Observed Fluorine NMR Is a Complementary Ligand Discovery Method to 1H CPMG Ligand-Observed NMR. ACS Chem. Biol 2016, 11, 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bogan AA; Thorn KS Anatomy of Hot Spots in Protein Interfaces. J. Mol. Biol 1998, 280, 1–9. [DOI] [PubMed] [Google Scholar]
- (26).Pomerantz WC; Wang N; Lipinski AK; Wang R; Cierpicki T; Mapp AK Profiling the Dynamic Interfaces of Fluorinated Transcription Complexes for Ligand Discovery and Characterization. ACS Chem. Biol 2012, 7, 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kitevski-Leblanc JL; Evanics F; Scott Prosser R Optimizing 19F NMR Protein Spectroscopy by Fractional Biosynthetic Labeling. J. Biomol. NMR 2010, 48, 113–121. [DOI] [PubMed] [Google Scholar]
- (28).Frieden C; Hoeltzli SD; Bann JG The Preparation of 19F-Labeled Proteins for NMR Studies. Methods Enzymol 2004, 380, 400–415. [DOI] [PubMed] [Google Scholar]
- (29).Kim H-W; Perez JA; Ferguson SJ; Campbell ID The Specific Incorporation of Labelled Aromatic Amino Acids into Proteins through Growth of Bacteria in the Presence of Glyphosate. FEBS Lett. 1990, 272, 34–36. [DOI] [PubMed] [Google Scholar]
- (30).Crowley PB; Kyne C; Monteith WB Simple and Inexpensive Incorporation of 19F-Tryptophan for Protein NMR Spectroscopy. Chem. Commun 2012, 48, 10681–10683. [DOI] [PubMed] [Google Scholar]
- (31).Liu JJ; Horst R; Katritch V; Stevens RC; Wüthrich K Biased Signaling Pathways in B2-Adrenergic Receptor Characterized by 19F-NMR. Science 2012, 335, 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yu L; Hajduk PJ; Mack J; Olejniczak ET Structural Studies of Bcl-XL/Ligand Complexes Using 19F NMR. J. Biomol. NMR 2006, 34, 221–227. [DOI] [PubMed] [Google Scholar]
- (33).Chrisman IM; Nemetchek MD; De Vera IMS; Shang J; Heidari Z; Long Y; Reyes-Caballero H; Galindo-Murillo R; Cheatham TE; Blayo AL; Shin Y; Fuhrmann J; Griffin PR; Kamenecka TM; Kojetin DJ; Hughes TS Defining a Conformational Ensemble That Directs Activation of PPARγ. Nat. Commun 2018, 9, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Manglik A; Kim TH; Masureel M; Altenbach C; Yang Z; Hilger D; Lerch MT; Kobilka TS; Thian FS; Hubbell WL; Prosser RS; Kobilka BK Structural Insights into the Dynamic Process of B2-Adrenergic Receptor Signaling. Cell 2015, 161, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Goodman RH; Smolik S CBP/P300 in Cell Growth, Transformation, and Development. Genes Dev. 2000, 1553–1577. [PubMed] [Google Scholar]
- (36).Breen ME; Mapp AK Modulating the Masters: Chemical Tools to Dissect CBP and P300 Function. Curr. Opin. Chem. Biol 2018, 45, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ernst P; Wang J; Huang M; Goodman RH; Korsmeyer SJ MLL and CREB Bind Cooperatively to the Nuclear Coactivator CREB-Binding Protein. Mol. Cell. Biol 2001, 21, 2249–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Parker D; Rivera M; Zor T; Henrion-Caude A; Radhakrishnan I; Kumar A; Shapiro LH; Wright PE; Montminy M; Brindle PK Role of Secondary Structure in Discrimination between Constitutive and Inducible Activators. Mol. Cell. Biol 1999, 19, 5601–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Goto NK; Zor T; Martinez-Yamout M; Dyson HJ; Wright PE Cooperativity in Transcription Factor Binding to the Coactivator CREB-Binding Protein (CBP): The Mixed Lineage Leukemia Protein (MLL) Activation Domain Binds to an Allosteric Site on the KIX Domain. J. Biol. Chem 2002, 277, 43168–43174. [DOI] [PubMed] [Google Scholar]
- (40).Kasper LH; Boussouar F; Ney PA; Jackson CW; Rehg J; Van Deursen JM; Brindle PK A Transcription-Factor-Binding Surface of Coactivator P300 Is Required for Haematopoiesis. Nature 2002, 419, 738–743. [DOI] [PubMed] [Google Scholar]
- (41).Oliveira AMM; Abel T; Brindle PK; Wood MA Differential Role for CBP and P300 CREB-Binding Domain in Motor Skill Learning. Behav. Neurosci 2006, 120, 724–729. [DOI] [PubMed] [Google Scholar]
- (42).Wang R; He Y; Robinson V; Yang Z; Hessler P; Lasko LM; Lu X; Bhathena A; Lai A; Uziel T; Lam LT Targeting Lineage-Specific MITF Pathway in Human Melanoma Cell Lines by A-485, the Selective Small-Molecule Inhibitor of P300/CBP. Mol. Cancer Ther 2018, 17, 2543–2550. [DOI] [PubMed] [Google Scholar]
- (43).Best JL; Amezcua CA; Mayr B; Flechner L; Murawsky CM; Emerson B; Zor T; Gardner KH; Montminy M Identification of Small-Molecule Antagonists That Inhibit an Activator:Coactivator Interaction. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 17622–17627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gee CT; Arntson KE; Koleski EJ; Staebell RL; Pomerantz WCK Dual Labeling of the CBP/P300 KIX Domain for 19F NMR Leads to Identification of a New Small-Molecule Binding Site. ChemBioChem 2018, 19, 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yang K; Stanfield RL; Martinez-Yamout MA; Dyson HJ; Wilson IA; Wright PE Structural Basis for Cooperative Regulation of KIX-Mediated Transcription Pathways by the HTLV-1 HBZ. Activation Domain. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 10040–10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Brüschweiler S; Schanda P; Kloiber K; Brutscher B; Kontaxis G; Konrat R; Tollinger M Direct Observation of the Dynamic Process Underlying Allosteric Signed Transmission. J. Am. Chem. Soc 2009, 131, 3063–3068. [DOI] [PubMed] [Google Scholar]
- (47).Palazzesi F; Barducci A; Tollinger M; Parrinello M The Allosteric Communication Pathways in KIX Domain of CBP. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 14237–14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sugase K; Dyson HJ; Wright PE Mechanism of Coupled Folding and Binding of an Intrinsically Disordered Protein. Nature 2007, 447, 1021–1025. [DOI] [PubMed] [Google Scholar]
- (49).Gee CT; Koleski EJ; Pomerantz WCK Fragment Screening and Druggability Assessment for the CBP/P300 KIX Domain through Protein-Observed 19F NMR Spectroscopy. Angew. Chem., Int. Ed 2015, 54, 3735–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Heemstra JM; Waterman R; Antos JM; Beuning PJ; Bur SK; Columbus L; Feig AL; Fuller AA; Gillmore JG; Leconte AM; Londergan CH; Pomerantz WCK; Prescher JA; Stanley LM Throwing Away the Cookbook: Implementing Course-Based Undergraduate Research Experiences (CUREs) in Chemistry. ACS Symp. Ser 2017, 1248, 33. [Google Scholar]
- (51).Hawk LML; Gee CT; Urick AK; Hu H; Pomerantz WCK Paramagnetic Relaxation Enhancement for Protein-Observed 19F NMR as an Enabling Approach for Efficient Fragment Screening. RSC Adv. 2016, 6, 95715–95721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Berger SL; Kouzarides T; Shiekhattar R; Shilatifard A An Operational Definition of Epigenetics. Genes Dev. 2009, 23, 781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Filippakopoulos P; Knapp S The Bromodomain Interaction Module. FEBS Lett. 2012, 586, 2692–2704. [DOI] [PubMed] [Google Scholar]
- (54).Kurdistani SK; Tavazoie S; Grunstein M Mapping Global Histone Acetylation Patterns to Gene Expression. Cell 2004, 117, 721–733. [DOI] [PubMed] [Google Scholar]
- (55).Filippakopoulos P; Picaud S; Mangos M; Keates T; Lambert JP; Barsyte-Lovejoy D; Felletar I; Volkmer R; Müller S; Pawson T; Gingras AC; Arrowsmith CH; Knapp S Histone Recognition and Large-Scale Structural Analysis of the Human Bromodomain Family. Cell 2012, 149, 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Delmore JE; Issa GC; Lemieux ME; Rahl PB; Shi J; Jacobs HM; Kastritis E; Gilpatrick T; Paranal RM; Qi J; Chesi M; Schinzel AC; McKeown MR; Heffernan TP; Vakoc CR; Bergsagel PL; Ghobrial IM; Richardson PG; Young RA; Hahn WC; Anderson KC; Kung AL; Bradner JE; Mitsiades CS BET Bromodomain Inhibition as a Therapeutic Strategy to Target C-Myc. Cell 2011, 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Mertz JA; Conery AR; Bryant BM; Sandy P; Balasubramanian S; Mele DA; Bergeron L; Sims RJ Targeting MYC Dependence in Cancer by Inhibiting BET Bromodomains. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Filippakopoulos P; Qi J; Picaud S; Shen Y; Smith WB; Fedorov O; Morse EM; Keates T; Hickman TT; Felletar I; Philpott M; Munro S; McKeown MR; Wang Y; Christie AL; West N; Cameron MJ; Schwartz B; Heightman TD; La Thangue N; French CA; Wiest O; Kung AL; Knapp S; Bradner JE Selective Inhibition of BET Bromodomains. Nature 2010, 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chung CW; Dean AW; Woolven JM; Bamborough P Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery. J. Med. Chem 2012, 55, 576–586. [DOI] [PubMed] [Google Scholar]
- (60).Urick AK; Hawk LML; Cassel MK; Mishra NK; Liu S; Adhikari N; Zhang W; Dos Santos CO; Hall JL; Pomerantz WCK Dual Screening of BPTF and Brd4 Using Protein-Observed Fluorine NMR Uncovers New Bromodomain Probe Molecules. ACS Chem. Biol 2015, 10, 2246–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ayoub AM; Hawk LML; Herzig RJ; Jiang J; Wisniewski AJ; Gee CT; Zhao P; Zhu JY; Berndt N; Offei-Addo NK; Scott TG; Qi J; Bradner JE; Ward TR; Schönbrunn E; Georg GI; Pomerantz WCK BET Bromodomain Inhibitors with One-Step Synthesis Discovered from Virtual Screen. J. Med. Chem 2017, 60, 4805–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Johnson JA; Nicolaou CA; Hu H; Pomerantz WCK The Advantages of Using 3D-Enriched Fragments for Targeting Bromodomains. ACS Med. Chem. Lett, submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Divakaran A; Talluri SK; Ayoub AM; Mishra NK; Cui H; Widen JC; Berndt N; Zhu J-Y; Carlson AS; Topczewski JJ; Schonbrunn EK; Harki DA; Pomerantz WCK Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase–Bromodomain Inhibitor. J. Med. Chem 2018, 61, 9316–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Harner MJ; Chauder BA; Phan J; Fesik SW Fragment-Based Screening of the Bromodomain of ATAD2. J. Med. Chem 2014, 57, 9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Ciceri P; Müller S; O’Mahony A; Fedorov O; Filippakopoulos P; Hunt JP; Lasater EA; Pallares G; Picaud S; Wells C; Martin S; Wodicka LM; Shah NP; Treiber DK; Knapp S Dual Kinase-Bromodomain Inhibitors for Rationally Designed Polypharmacology. Nat. Chem. Biol 2014, 10, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ember SWJ; Zhu JY; Olesen SH; Martin MP; Becker A; Berndt N; Georg GI; Schonbrunn E Acetyl-Lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors. ACS Chem. Biol 2014, 9, 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Elkins JM; Fedele V; Szklarz M; Abdul Azeez KR; Salah E; Mikolajczyk J; Romanov S; Sepetov N; Huang X-P; Roth BL,; Al Haj Zen A; Fourches D; Muratov E; Tropsha A; Morris J; Teicher BA; Kunkel M; Polley E; Lackey KE; Atkinson FL; Overington JP; Bamborough P; Müller S; Price DJ; Willson TM; Drewry DH; Knapp S; Zuercher WJ Comprehensive Characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol 2016, 34, 95–103. [DOI] [PubMed] [Google Scholar]
- (68).Aldeghi M; Ross GA; Bodkin MJ; Essex JW; Knapp S; Biggin PC Large-Scale Analysis of Water Stability in Bromodomain Binding Pockets with Grand Canonical Monte Carlo. Commun. Chem 2018, 1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wysocka J; Swigut T; Xiao H; Milne TA; Kwon SY; Landry J; Kauer M; Tackett AJ; Chait BT; Badenhorst P; Wu C; Allis CD A PHD Finger of NURF Couples Histone H3 Lysine 4 Trimethylation with Chromatin Remodelling. Nature 2006, 442, 86–90. [DOI] [PubMed] [Google Scholar]
- (70).Xiao S; Liu L; Fang M; Zhou X; Peng X; Long J; Lu X BPTF Associated with EMT Indicates Negative Prognosis in Patients with Hepatocellular Carcinoma. Dig. Dis. Sci 2015, 60, 910–918. [DOI] [PubMed] [Google Scholar]
- (71).Dai M; Lu J-J; Guo W; Yu W; Wang Q; Tang R; Tang Z; Xiao Y; Li Z; Sun W; Sun X; Qin Y; Huang W; Deng W; Wu T BPTF Promotes Tumor Growth and Predicts Poor Prognosis in Lung Adenocarcinomas. Oncotarget 2015, 6, 33878–33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Buganim Y; Goldstein I; Lipson D; Milyavsky M; Polak-Charcon S; Mardoukh C; Solomon H; Kalo E; Madar S; Brosh R; Perelman M; Navon R; Goldfinger N; Barshack I; Yakhini Z; Rotter V A Novel Translocation Breakpoint within the BPTF Gene Is Associated with a Pre-Malignant Phenotype. PLoS One 2010, 5, e9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Richart L; Carrillo-De Santa Pau E; Río-Machín A; De Andrés MP; Cigudosa JC; Lobo VJSA; Real FX BPTF Is Required for C-MYC Transcriptional Activity and in Vivo Tumori-genesis. Nat. Commun 2016, 7, 10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Kirberger SE; Ycas PD; Johnson JA; Chen C; Ciccone MF; Woo RWL; Urick AK; Zahid H; Shi K; Aihara H; McAllister SD; Kashani-Sabet M; Shi J; Dickson A; dos Santos CO; Pomerantz WCK Selectivity, Ligand Deconstruction, and Cellular Activity Analysis of a BPTF Bromodomain Inhibitor. Org. Biomol Chem 2019, 17, 2020–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Perell GT; Mishra NK; Sudhamalla B; Ycas PD; Islam K; Pomerantz WCK Specific Acetylation Patterns of H2A.Z Form Transient Interactions with the BPTF Bromodomain. Biochemistry 2017, 56, 4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Frey WD; Chaudhry A; Slepicka PF; Ouellette AM; Kirberger SE; Pomerantz WCK; Hannon GJ; dos Santos CO BPTF Maintains Chromatin Accessibility and the Self-Renewal Capacity of Mammary Gland Stem Cells. Stem Cell Rep. 2017, 9, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Li C; Wang GF; Wang Y; Creager-Allen R; Lutz EA; Scronce H; Slade KM; Ruf RAS; Mehl RA; Pielak GJ Protein 19F NMR in Escherichia Coli. J. Am. Chem. Soc 2010, 132, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Norton RS; Leung EWW; Chandrashekaran IR; MacRaild CA Applications of 19F-NMR in Fragment-Based Drug Discovery. Molecules 2016, 21, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kim TH; Mehrabi P; Ren Z; Sljoka A; Ing C; Bezginov A; Ye L; Pomès R; Prosser RS; Pai EF The Role of Dimer Asymmetry and Protomer Dynamics in Enzyme Catalysis. Science 2017, 355, eaag2355. [DOI] [PubMed] [Google Scholar]
- (80).Sharaf NG; Gronenborn AM 19F-Modified Proteins and 19F-Containing Ligands as Tools in Solution NMR Studies of Protein Interactions. Methods Enzymol 2015, 565, 67–95. [DOI] [PubMed] [Google Scholar]
- (81).Boeszoermenyi A; Chhabra S; Dubey A; Radeva DL; Burdzhiev NT; Chanev CD; Petrov OI; Gelev VM; Zhang M; Anklin C; Kovacs H; Wagner G; Kuprov I; Takeuchi K; Arthanari H Aromatic 19 F- 13 C TROSY: A Background-Free Approach to Probe Biomolecular Structure, Function, and Dynamics. Nat. Methods 2019, 16, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kitevski-LeBlanc JL; Al-Abdul-Wahid MS; Prosser RS A Mutagenesis-Free Approach to Assignment of19F NMR Resonances in Biosynthetically Labeled Proteins. J. Am. Chem. Soc 2009, 131, 2054–2055. [DOI] [PubMed] [Google Scholar]
- (83).Gregory DH; Gerig JT Prediction of Fluorine Chemical Shifts in Proteins. Biopolymers 1991, 31, 845–858. [DOI] [PubMed] [Google Scholar]
- (84).Pearson JG; Oldfield E; Lee FS; Warshel A Chemical Shifts in Proteins: A Shielding Trajectory Analysis of the Fluorine Nuclear Magnetic Resonance Spectrum of the Escherichia Coli Galactose Binding Protein Using a Multipole Shielding Polarizability-Local Reaction Field-Molecular Dynamics Approach. J. Am. Chem. Soc 1993, 115, 6851–6862. [Google Scholar]
- (85).Isley WC; Urick AK; Pomerantz WCK; Cramer CJ Prediction of 19F NMR Chemical Shifts in Labeled Proteins: Computational Protocol and Case Study. Mol. Pharmaceutics 2016, 13, 2376–2386. [DOI] [PubMed] [Google Scholar]