This study reveals the subcellular localization of VirB, a key transcriptional regulator of virulence genes found on the large virulence plasmid (pINV) in Shigella. Fluorescent signals generated by an active GFP-VirB fusion form 2, 3, or 4 discrete foci in the bacterial cytoplasm, predominantly at the quarter-cell position. These signals are completely dependent upon VirB interacting with its DNA-binding site found either on the virulence plasmid or an engineered surrogate.
KEYWORDS: VirB, virulence plasmid, Shigella, antisilencing, H-NS, transcriptional silencing, ParB/Spo0J, plasmid partitioning, subcellular localization, GFP fusion
ABSTRACT
VirB is a key regulator of genes located on the large virulence plasmid (pINV) in the bacterial pathogen Shigella flexneri. VirB is unusual, as it is not related to other transcriptional regulators; instead, it belongs to a family of proteins that primarily function in plasmid and chromosome partitioning, which is exemplified by ParB. Despite this, VirB does not function to segregate DNA, but rather counters transcriptional silencing mediated by the histone-like nucleoid structuring protein, H-NS. Since ParB localizes subcellularly as discrete foci in the bacterial cytoplasm, we chose to investigate the subcellular localization of VirB to gain novel insight into how VirB functions as a transcriptional antisilencer. To do this, a green fluorescent protein (GFP)-VirB fusion that retains the regulatory activity of VirB and yet does not undergo significant protein degradation in S. flexneri was used. Surprisingly, discrete fluorescent foci were observed using fluorescence microscopy in live wild-type S. flexneri cells and in an isogenic virB mutant. In contrast, foci were rarely observed (<10%) in pINV-cured cells or in cells expressing a GFP-VirB fusion carrying amino acid substitutions in the VirB DNA-binding domain. Finally, the 25-bp VirB-binding site was demonstrated to be sufficient and necessary for GFP-VirB focus formation using a set of small surrogate plasmids. Combined, these data demonstrate that the VirB-DNA interactions required for the transcriptional antisilencing activity of VirB on pINV are a prerequisite for the subcellular localization of VirB in the bacterial cytoplasm. The significance of these findings, in light of the antisilencing activity of VirB, is discussed.
IMPORTANCE This study reveals the subcellular localization of VirB, a key transcriptional regulator of virulence genes found on the large virulence plasmid (pINV) in Shigella. Fluorescent signals generated by an active GFP-VirB fusion form 2, 3, or 4 discrete foci in the bacterial cytoplasm, predominantly at the quarter-cell position. These signals are completely dependent upon VirB interacting with its DNA-binding site found either on the virulence plasmid or an engineered surrogate. Our findings (i) provide novel insight into VirB-pINV interactions, (ii) suggest that VirB may have utility as a DNA marker, and (iii) raise questions about how and why this antisilencing protein that controls virulence gene expression on pINV of Shigella spp. forms discrete foci/hubs within the bacterial cytoplasm.
INTRODUCTION
Bacterial cells were once viewed as tiny compartments containing mixtures of macromolecules without organization or arrangement. With advances in the creation and use of fluorescent fusion proteins, the discrete subcellular location of many proteins has been revealed (1–6). Such studies have frequently shown that, rather than being confined to a particular subcellular address or target, protein positioning may be dynamic, changing throughout the cell cycle (7–11). As such, subcellular localization studies, when properly controlled, can provide a more holistic view of the molecular biology that occurs within cells and generate new insight into protein function and their interactions with other macromolecules.
In Shigella spp., the causative agents of bacillary dysentery (12), the transcriptional regulator VirB is produced in response to human body temperature (13, 14). At 37°C, VirB upregulates the expression of about 50 genes on the large virulence plasmid pINV (similar effects are seen if virB is artificially induced at lower temperatures [15]). These genes include those encoding the type III secretion system (i.e., the needle and initial effectors), other virulence-associated factors (i.e., IcsP, OspZ, and OspD1 [16–18]), and the transcriptional activator MxiE and its coactivator IpgC (15). Perhaps not surprisingly, VirB is essential for the virulence of this important group of pathogens (13, 19).
As an antisilencing protein, VirB does not function as a canonical transcription factor but rather functions to offset transcriptional silencing mediated by the histone-like nucleoid structuring protein, H-NS, on pINV (16, 20–23). H-NS binds, coats, and condenses AT-rich, horizontally acquired DNA, in a process that has been termed xenogeneic silencing (24–26). Site-specific VirB-DNA interactions likely remodel H-NS–DNA complexes via a poorly understood mechanism, allowing inaccessible DNA or transcriptionally nonpermissive DNA to be accessed/engaged by RNA polymerase so that transcription can occur (27). As such, the bacterial processes of transcriptional silencing and antisilencing are reminiscent of chromatin remodeling in eukaryotic cells. In recent years, it has become clear that transcriptional silencing and antisilencing is widespread in bacteria (28) and that these opposing regulatory processes are necessary for bacterial fitness because they control many aspects of bacterial cell physiology, including virulence in a variety of pathogens (28–34).
Our recent work has focused on gaining mechanistic insight into transcriptional antisilencing by VirB (16, 18, 23, 35, 36). To aid these studies, we have looked to proteins that regulate transcription like VirB but also to proteins that are paralogues of VirB. While classical transcription factors bind to specific DNA motifs to regulate the expression of target genes, how they reach their sites can vary. Some transcription factors are thought to diffuse in three-dimensional space, while others slide along DNA or undergo intersegmental hopping to reach their cognate sites (37). Regardless, the paradigm has been that these proteins are diffuse while searching for their sites and, once they find their sites, which are often located around the genome, are at concentrations too low to be detected by standard fluorescence microscopy. In contrast, some DNA-binding proteins, including members of the ParB/Spo0J protein superfamily, to which VirB belongs, show a discrete subcellular localization pattern in bacteria during plasmid and/or chromosome partitioning (38–41). These proteins bind to their DNA recognition sites, form multimeric complexes, and then, through their association with the ParA ATPase, move to the poles with the DNA prior to cell division, an activity observed in Corynebacterium glutamicum and Bacillus subtilis (42–45).
In this study, we chose to examine the subcellular localization of VirB. Our goal was to broaden our understanding of VirB-DNA interactions on a cellular level so that we might consider how these interactions underpin its antisilencing activity. We were intrigued to know if the signals associated with VirB would resemble those expected for a transcription factor or members of the ParB superfamily. If a subcellular localization signal was detected, we reasoned that this study would become foundational for future work that addresses which attributes of VirB are needed for this localization, whether other macromolecules are required, and ultimately how these factors affect the regulation of virulence genes in this important human pathogen.
RESULTS
Characterization of VirB fused with GFP.
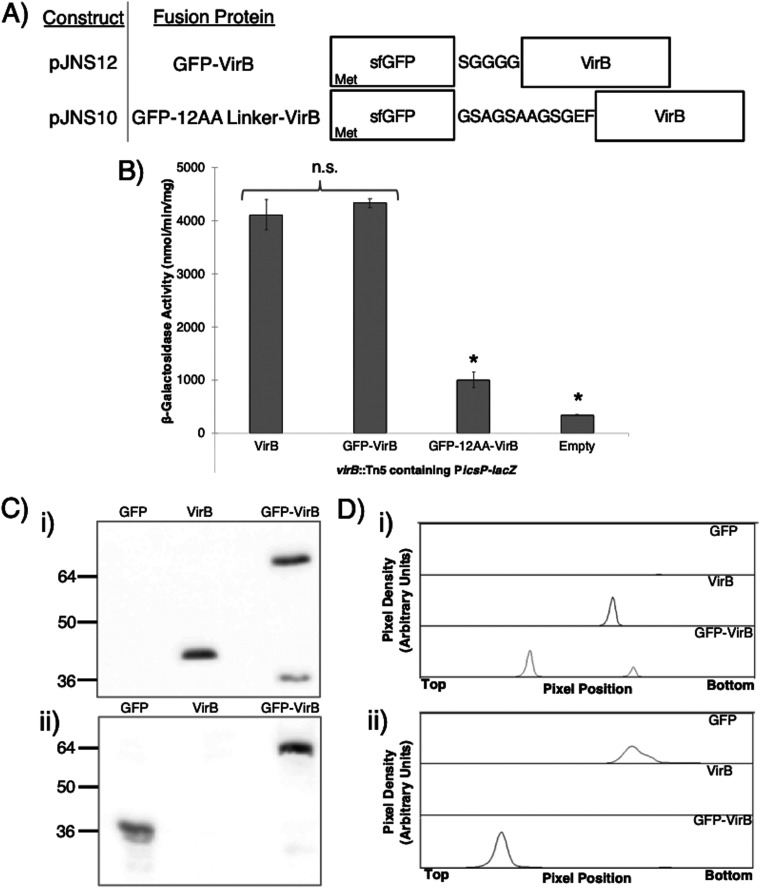
To begin this study, VirB fused to superfolder green fluorescent protein (sfGFP; referred to as GFP for the remainder of this paper) (46, 47) was expressed from the pBAD promoter found on the low-copy-number (∼15 to 20 copies per cell) plasmid, pBAD/His A (Table 1). While N- and C-terminal VirB fusions were engineered and two different linkers were exploited (Fig. 1A) (48, 49), the plasmids generating VirB-GFP were abandoned in the early stages of this work because protein production did not positively correlate with l-arabinose induction (data not shown). Consequently, the two N-terminal fusions (GFP-VirB) were tested and compared to wild-type VirB for their ability to regulate a well-characterized, VirB-dependent promoter (16, 35). To do this, the inducible plasmids pJNS04 (pBAD-VirB), pJNS10 (GFP-12AA-VirB), or pJNS12 (GFP-SGGGG-VirB), were introduced into a virB mutant derivative of Shigella flexneri carrying the PicsP-lacZ transcriptional reporter (17), and β-galactosidase activity was measured in cultures after induction. The data show that the GFP-VirB fusion with the SGGGG linker generated high lacZ expression similar to that observed with inducible VirB, suggesting that VirB remains functional in the context of the GFP fusion protein, whereas the “12AA linker” appeared to interfere with the regulatory activity of VirB (Fig. 1B) (48). Consequently, GFP-SGGGG-VirB (encoded by pJNS12) was used for the remainder of this study.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| S. flexneri strains | ||
| 2457T | Serotype 2a | 82 |
| AWY3 | 2457T virB::Tn5 Knr | 16 |
| BS103 | 2457T cured of the virulence plasmid | 83 |
| Plasmids | ||
| pJNS04 | pBAD-virB; Ampr | This study |
| pJNS10 | pBAD-gfp-12AA linker-virB; Ampr | This study |
| pJNS12 | pBAD-gfp-virB; Ampr | This study |
| pJNS13 | pBAD-gfp-virB K152E; Ampr | This study |
| pJNS18 | pBAD-gfp-virB K152E/R167E; Ampr | This study |
| pJNS22 | pACYC177 carrying 25-bp VirB-binding site derived from pBT-PicsP (36); Cmr | This study |
| pJNS24 | pACYC177 without 25-bp VirB-binding site derived from pBT-empty (36); Cmr | This study |
| pJH66 | pBAD-linker-gfp; Ampr | 79 |
| pBAD/His A | pBR322-derived expression vector allowing regulated and dose-dependent recombinant protein expression; Ampr | Invitrogen |
| pAFW04 | pACYC184-PicsP-lacZ; Cmr | 17 |
| pHJW20 | WT PicsP fused to lacZ; Cmr | 35 |
| pMIC18 | pHJW20 with a mutated VirB-binding site; Cmr | 35 |
| pJAI28 | WT PospD1 fused to lacZ; Cmr | 18 |
| pJAI36 | pJAI28 with mutated VirB-binding sites; Cmr | 18 |
| pATM324 | pBAD18-virB; Ampr | 67 |
Knr, kanamycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
FIG 1.
Constructs and proof of principle experiments to assess the activity and stability of VirB fusion proteins. (A) Constructs producing VirB fused to superfolder GFP (sfGFP) at the N terminus using one of two linkers. (B) β-Galactosidase assay used to assess the regulatory activity of GFP-VirB fusions with either a SGGGG (49) or “12AA” linker (48) at the VirB-dependent icsP promoter (PicsP-lacZ; pAFW04a). Student’s t tests were used to measure statistical significance, *, P < 0.05. Note that β-galactosidase activities generated in the presence of VirB (pJNS04) and GFP-VirB (pJNS12) were equivalent to those achieved with native VirB levels in previous work (35). (C) Western blots to assess GFP-VirB protein stability, probed with an anti-VirB antibody (i) or anti-GFP antibody (ii). For these analyses, phenylmethylsulfonyl fluoride (PMSF) was added during sample preparation. (D) Densitometry of Western blots shown in panel C (subpanels i and ii, respectively). Lanes are labeled accordingly.
To determine whether the observed regulatory activity of GFP-VirB at PicsP-lacZ could be attributed to the full-length fusion protein or to a degradation product, protein production and stability of the GFP-VirB fusion protein was examined. Western blots of whole-cell lysates of cells producing the GFP control (31.5 kDa; pJH66) or GFP-VirB (62.2 kDa; pJNS12), grown using conditions identical to those used in the β-galactosidase assay, were probed using either an anti-VirB antibody (Fig. 1C, subpanel i) or an anti-GFP antibody (Fig. 1C, subpanel ii). The primary bands detected correspond to the GFP-VirB fusion protein and GFP with linker, respectively. In each case, more than 74% and 92% of the total protein detected was found in the respective uppermost band (full-length product), as determined by densitometry (Fig. 1D, subpanels i and ii). While another band in the GFP-VirB sample was detected with the VirB antibody, it migrated faster and was fainter than the VirB protein control, making it unlikely that this minor product was responsible for the β-galactosidase activity associated with the GFP-VirB fusion. In sum, these analyses indicate that the fluorescent protein generated from pJNS12 primarily consists of the full-length GFP-VirB fusion and strongly suggests that the VirB moiety within this fusion is both stable and functional (Fig. 1B), a conclusion further supported by later experiments in this study (see Fig. 5; see also Fig. S4 in the supplemental material).
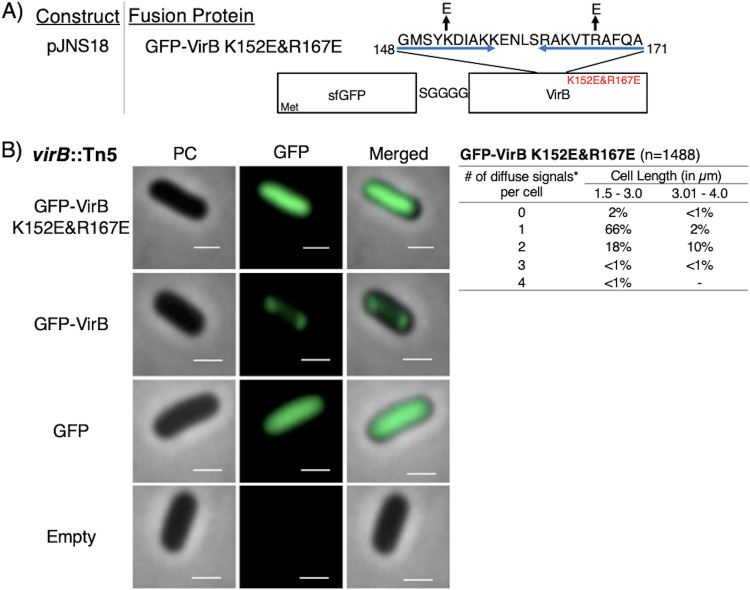
FIG 5.
Construction of a GFP-VirB DNA-binding mutant and live-cell imaging of this fusion in a virB mutant strain of S. flexneri. (A) Construct producing GFP-VirB with two amino acid substitutions, K152E and R167E, in the helix-turn-helix DNA-binding domain (denoted with blue arrows; adapted from reference 52). (B) Quantification of fluorescent signals observed during live-cell imaging of GFP-VirBK152E/R167E in virB mutant S. flexneri using MicrobeJ. Note that diffuse signals observed for this fusion were detected as maxima by MicrobeJ. Representative cells are shown. Phase-contrast (left column), fluorescence (middle column) (GFP row, 77-ms exposure; GFP-VirB, GFP-VirB K152E/R167E, and empty rows, 348.8-ms exposure), and merged (right column). Bars, 1 μm. Within the table, a hyphen indicates that no cells fell into this category in any of the images captured; *, maxima detected by MicrobeJ.
GFP-VirB forms discrete foci in Shigella strains.
The next step was to observe the fluorescent signal generated by the GFP-VirB fusion in S. flexneri cells bearing this construct. To do this, live-cell imaging was utilized to avoid artifacts associated with cell fixing. First, we examined the GFP-VirB fusion protein, GFP, or the negative control (empty plasmid control) in a S. flexneri virB mutant strain using identical induction conditions to those used in our previous proof-of-principle assays. Cells were live mounted and imaged using phase-contrast and fluorescence microscopy.
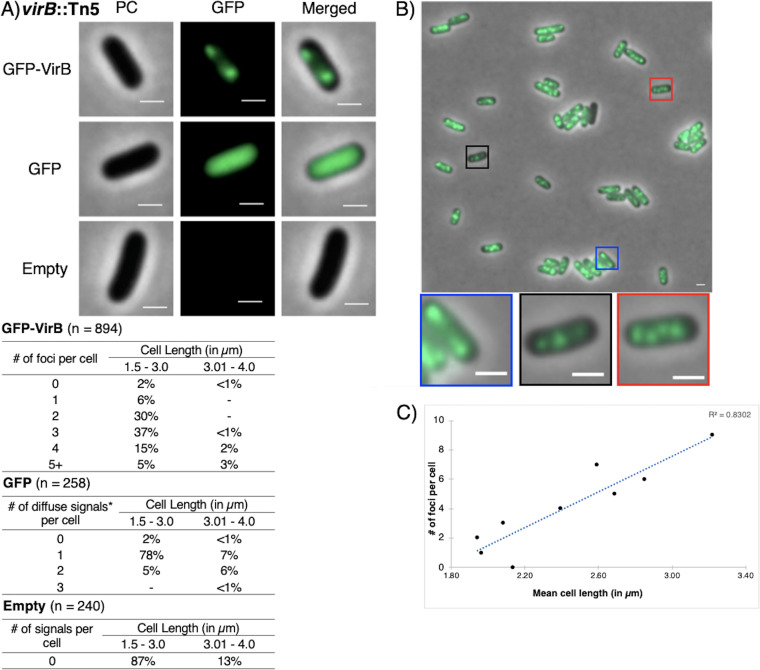
As expected, cells carrying the empty plasmid control did not fluoresce (Fig. 2A, bottom), and most cells producing GFP exhibited a single diffuse fluorescent signal with a single distinct maximum (85% of all cells analyzed) (Fig. 2A, middle; see also Fig. S2 in the supplemental material, left); occasionally, two distinct maxima were seen, likely representing cells undergoing cell division (11% of analyzed cells). Strikingly, when cells producing the GFP-VirB fusion were viewed, discrete fluorescent foci were observed in 98% of analyzed cells (Fig. 2A, top). These foci were routinely seen at the quarter-cell position (Fig. 2B). While the number of foci varied, commonly, two, three, and four foci were detected after image analysis using MicrobeJ (30%, 37%, and 17%, respectively) (Fig. 2A). Notably, a strong positive correlation (R2 = 0.83) between the number of foci and the mean cell length per number of foci was found (Fig. 2C), which suggests that the number of foci increases during cell elongation, prior to cell division.
FIG 2.
Live-cell imaging of GFP-VirB in a virB mutant strain of Shigella flexneri. (A) Quantification of foci observed during live-cell imaging of GFP-VirB in virB mutant S. flexneri using MicrobeJ. Representative cells are shown. PC, phase contrast; GFP, green fluorescent protein fluorescence (for GFP row 67.1-ms exposure; for GFP-VirB and Empty rows, 289.2-ms exposure; Empty, empty plasmid control. Bars, 1 μm in all images. Within tables, a hyphen indicates that no cells fell into this category in any of the images captured; *, maxima detected by MicrobeJ. (B) A representative field of view with magnified cell images below showing the numbers of foci commonly observed. Five fields of view were captured and analyzed across at least three independent replicates. (C) Scatterplot of mean cell length per number of foci observed in cells producing GFP-VirB, showing strong positive correlation between cell length and number of foci.
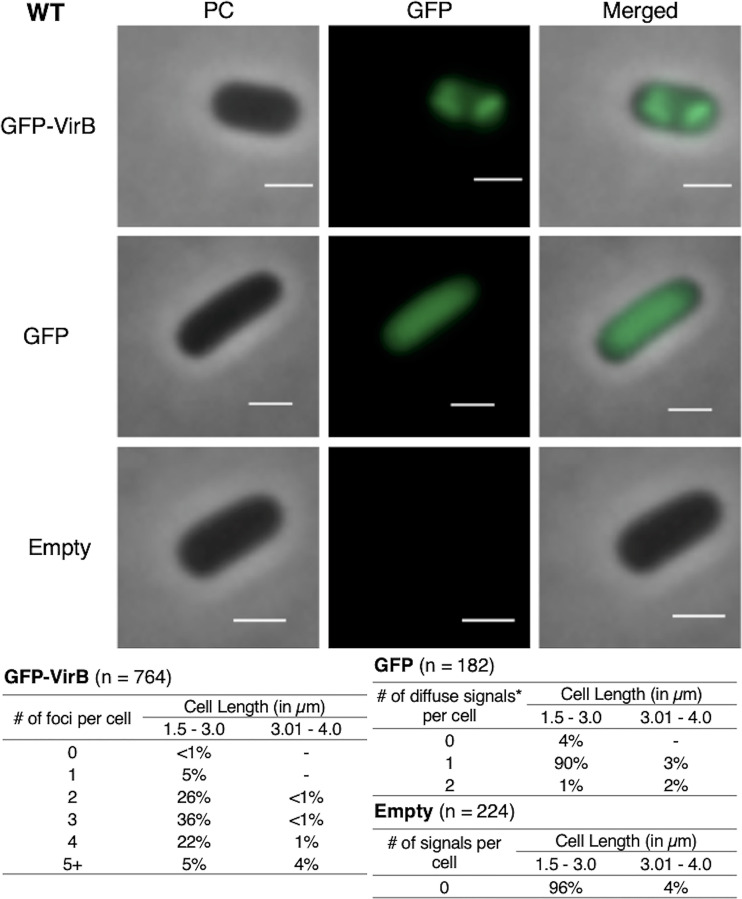
To further investigate the subcellular location of VirB in the Shigella cytoplasm, we next chose to reexamine the GFP-VirB fusion in a wild-type S. flexneri background. Data similar to those gathered in the virB mutant strain were obtained; 99% of cells had 1 or more discrete foci; longer cells had more foci than shorter cells (Fig. 3). This demonstrates that the presence of native VirB does not alter the localization pattern of GFP-VirB, and it also validates the localization pattern observed in the isogenic virB mutant strain of S. flexneri. Consequently, these data (Fig. 2 and 3) together provide novel insight into the VirB protein by finding that GFP-VirB is localized in the bacterial cell cytoplasm into 2 to 4 discrete foci at the quarter cell position. This raises the possibility that VirB forms previously undescribed hubs within the bacterial cytoplasm, which may be critical for its role as a regulator of Shigella virulence.
FIG 3.
Live-cell imaging of GFP-VirB in a wild-type strain of S. flexneri and quantification of foci observed during live-cell imaging of GFP-VirB in wild-type S. flexneri using MicrobeJ. Representative cells are shown. Phase contrast (left column), fluorescence (middle column) (GFP row, 44.5-ms exposure; GFP-VirB and empty, 219-ms exposure), and merged (right column) images of GFP-VirB, GFP, and an empty plasmid control . Bars, 1 μm. Within tables, a hyphen indicates that no cells fell into this category in any of the images captured; *, maxima detected by MicrobeJ.
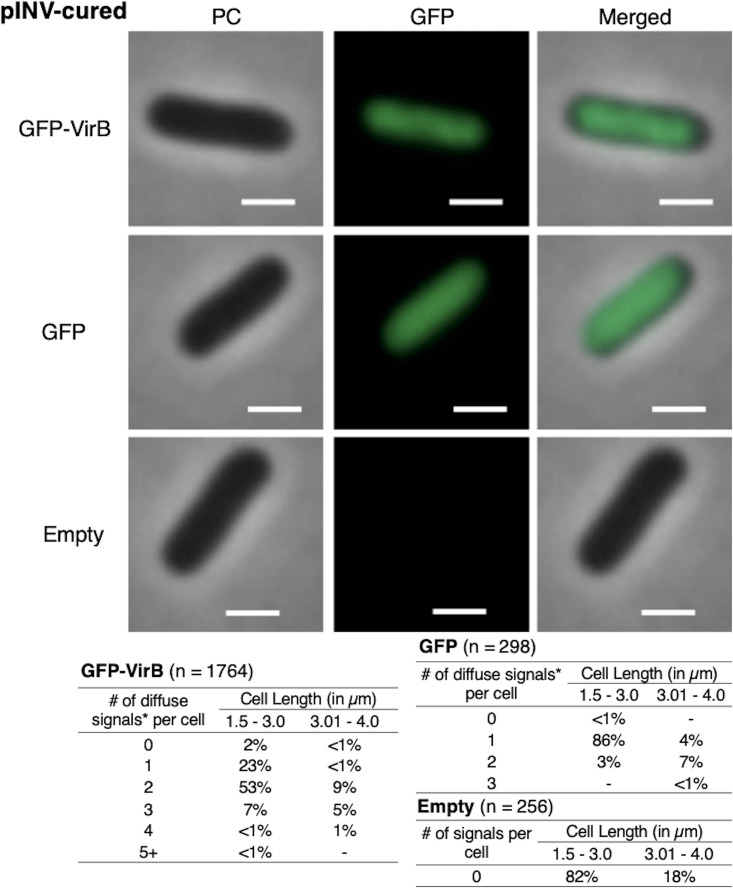
Focus formation of GFP-VirB is lost in the absence of pINV.
Since VirB predominately regulates virulence genes carried by pINV, we next chose to examine GFP-VirB focus formation in a strain lacking this large DNA molecule. Here, we exploited the S. flexneri 2a derivative that has been cured of pINV, BS103. In this strain background, GFP-VirB predominantly formed one or two diffuse fluorescent signals in the cytoplasm (Fig. 4, top; see also Fig. S3 in the supplemental material, left) (24% single maximum and 62% 2 maxima), but critically, these signals did not resemble the tight foci observed in the virB mutant or wild-type strains (Fig. 2A and 3, top, respectively). Instead, the signals were much more diffuse, albeit less uniformly distributed than the GFP control (compare Fig. 4, top to middle; Fig. S3, left to middle panels for field of view). Indeed, in this strain background the GFP-VirB fusion signal appeared to be nucleoid associated, as its broad distribution over much of the cell with the exception of small areas near the cell poles (Fig. 4, top; evident in merged image) is similar to the distribution of DNA in Escherichia coli and Shigella (50, 51). Again, control experiments yielded expected results in these assays (Fig. 4, middle and bottom).
FIG 4.
Live-cell imaging of GFP-VirB in a pINV-cured strain of S. flexneri and quantification of fluorescent signals observed during live-cell imaging of GFP-VirB in pINV-cured S. flexneri using MicrobeJ. Representative cells are shown. Note that only diffuse signals were observed (see also Fig. S3 in the supplemental material). Phase-contrast (left column), fluorescence (middle column) (GFP row, 40.6-ms exposure; GFP-VirB and empty rows, 223.1-ms exposure), and merged (right column) images of GFP-VirB, GFP, and an empty plasmid control. Bars, 1 μm. Within tables, a hyphen indicates that no cells fell into this category in any of the images captured; *, maxima detected by MicrobeJ.
A DNA-binding mutant derivative of GFP-VirB does not form foci in S. flexneri.
The finding that GFP-VirB did not form discrete foci in the absence of pINV raised the possibility that direct VirB interactions with pINV were required for focus formation in wild-type and virB mutant strain backgrounds. To test this hypothesis, the fluorescence of a GFP-VirB fusion carrying amino acid substitutions in the DNA-binding surface of VirB was examined. Two amino acid substitutions, K152E and R167E (52, 53), were introduced into the helix-turn-helix DNA-binding domain of VirB in the fusion (Fig. 5A). Each of these substitutions had previously been shown to prevent VirB from binding to DNA and, hence, prevent regulation by VirB (52, 53). The stability of the resultant fusion protein, GFP-VirB K152E/R167E, and its ability to regulate transcription was then examined (see Fig. S4A to C in the supplemental material). Again, little protein degradation was observed by Western blot analysis (Fig. S4A and B), suggesting that GFP-VirB K152E/R167E was as stable as GFP-VirB (Compare Fig. 1C to Fig. S4A). But, as predicted by earlier studies (52, 53), this protein was unable to regulate PicsP-lacZ. Indeed, levels of β-galactosidase activity in the presence of GFP-VirB K152E/R167E were comparable to those obtained with the empty control (Fig. S4C). The finding that two amino acid substitutions in the DNA-binding domain of the VirB moiety are sufficient to destroy the regulatory activity of this fusion (Fig. S4C) is not only consistent with previous findings (52, 53) but further supports our assertion that the VirB moiety remains properly folded in the context of the fusions used in this study.
When the GFP-VirB KE152/R167E fusion was examined in the virB mutant using fluorescence microscopy, a diffuse fluorescent signal was observed (Fig. 5B, top, and Fig. S2, right) but importantly, once again, no discrete foci were observed. Similar observations were made when this fusion was examined in the wild-type (Fig. S4D) and pINV-cured strains (Fig. S3, right). Consequently, we concluded that an active DNA-binding domain of VirB is required for VirB to form foci in the cell cytoplasm. Moreover, in combination with the findings made with GFP-VirB in a pINV-cured strain, these outcomes strongly suggest that VirB-DNA interactions with pINV are responsible for the focus formation observed in the wild type and virB mutant derivative of S. flexneri and that these foci are not a caused by GFP aggregation. Note that quantification of the mean fluorescence intensity of foci and that for the medial axis of cells for the virB mutant and the pINV-cured strain expressing each fluorescent protein are shown in Fig. S5 in the supplemental material.
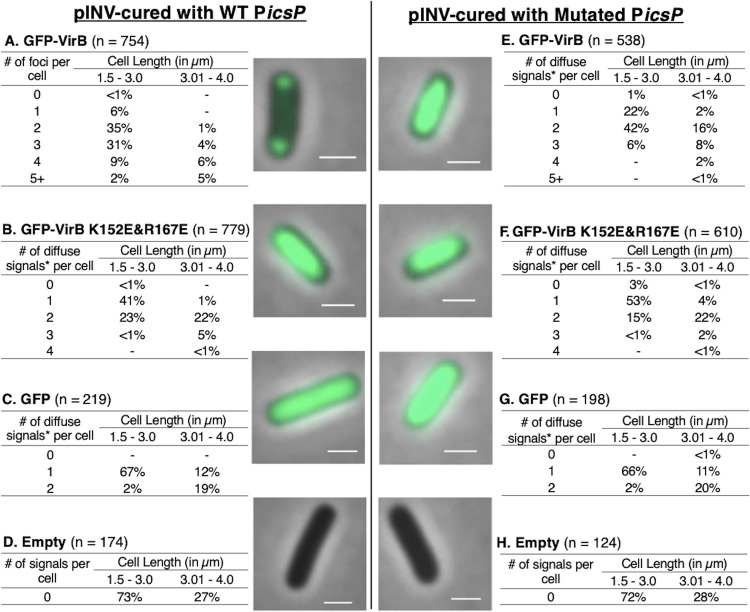
A small plasmid carrying a VirB-binding site restores focus formation in pINV-cured Shigella.
Because focus formation was dependent on the presence of pINV, we next chose to examine if small surrogate plasmids carrying a single well-characterized VirB-regulated promoter sourced from pINV would allow fluorescent foci to form in a pINV-cured background of S. flexneri. Initially, to test this, the low-copy-number plasmid pHJW20 (35), carrying a well-characterized VirB-binding site in the context of PicsP (22, 35, 36), or a derivative pMIC18 (35) with a mutated VirB-binding site was introduced into BS103 that was carrying either pJNS12, pJNS18, or one of the control plasmids (pJH66 or pBAD/His A). Remarkably, foci formed in the presence of the small surrogate plasmid carrying the wild-type VirB-binding site (Fig. 6A) but not with the derivative carrying the mutated site (Fig. 6E). Furthermore, the DNA-binding mutant derivative gave a diffuse signal regardless of whether a wild-type or mutated site was present (Fig. 6B and F), thus strengthening support for the previous interpretation that the ability of VirB to interact with DNA is absolutely necessary in order for foci to form; controls behaved as expected (Fig. 6C, D, G, H). Similar results were observed when this experiment was repeated using another small plasmid carrying the well-characterized VirB-dependent promoter ospD1 (18) (see Fig. S6 in the supplemental material).
FIG 6.
Live-cell imaging of GFP-VirB and controls in a pINV-cured strain of S. flexneri carrying a plasmid bearing a wild-type or mutated VirB-binding site and quantification of fluorescent foci of GFP-VirB (A) or maxima (*) (B, C, and E to G) associated with diffuse signals observed during live-cell imaging using MicrobeJ. Representative merged cell images of pINV-cured S. flexneri cells containing pHJW20 (WT PicsP) (A to D) or pMIC18 (Mutated PicsP) (E to H) are shown. No fluorescent signal was detected for the empty controls (D and H) (GFP, 157.6-ms exposure; GFP-VirB and empty, 485.3-ms exposure). Bars, 1 μm. Within tables, a hyphen indicates that no cells fell into this category in any of the images captured.
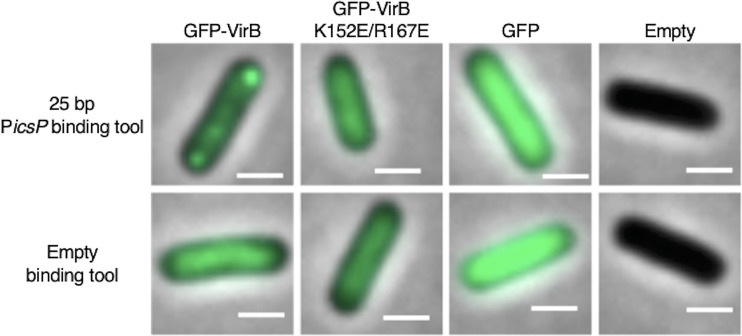
Finally, to investigate if the VirB-binding site alone was sufficient for focus formation, small plasmids carrying just the 25-bp sequence that contains the VirB-binding site (pJNS22) or a “no site” control (pJNS24) (36), were introduced into BS103, and the fusion proteins or controls were induced as described previously. Strikingly, fluorescent foci were observed in cells producing GFP-VirB in the presence of the 25-bp VirB-binding site-containing plasmid, but were not observed in cells carrying the “no site” control (Fig. 7). In contrast, cells producing the GFP-VirB DNA-binding mutant derivative did not form foci in the presence of the 25-bp VirB-binding site-containing plasmid or the “no site” control. Again, all other controls behaved as seen previously.
FIG 7.
Live-cell imaging of GFP-VirB and controls in a pINV-cured strain of S. flexneri carrying a plasmid bearing a 25-bp VirB binding site or empty (“no-site”) binding tool control. Representative merged cell images of pINV-cured S. flexneri carrying pJNS22 (25-bp VirB-binding site) or pJNS24 (“no site” control) with either GFP-VirB, GFP-VirB K152E/R167E, GFP, or an empty plasmid control are shown (GFP, 45.1-ms exposure; GFP-VirB and empty, 297.6-ms exposure). Bars, 1 μm.
Taken together, our findings demonstrate that interactions between VirB and its cognate DNA-binding sequence are necessary and sufficient for the formation of discrete cytoplasmic GFP-VirB foci in S. flexneri. In addition to providing new information on VirB activity, these observations raise the possibility that this fluorescent fusion protein could be used in conjunction with its cognate binding sequence as a plasmid or genome marker for fluorescence microscopy experiments in Enterobacteriaceae and other species.
DISCUSSION
In this study, we have investigated the subcellular location of VirB, a key transcriptional regulator of Shigella virulence plasmid genes (13, 19), using a GFP-VirB fusion protein (Fig. 1). Our data show that GFP-VirB forms discrete foci in the Shigella cytoplasm (Fig. 2 and 3). Three lines of evidence demonstrate that these foci are completely dependent upon VirB engaging its DNA-binding site, namely, (i) the loss of GFP-VirB foci in pINV-cured cells (Fig. 4), (ii) the loss of foci when a DNA-binding mutant derivative of GFP-VirB is produced in a virB mutant derivative (Fig. 5), and (iii) the restoration of GFP-VirB foci when small surrogate plasmids bearing a VirB recognition site are introduced into pINV-cured cells (Fig. 6 and 7). Throughout this study, GFP-VirB levels were matched to native levels of VirB (see Materials and Methods; see also Fig. S1 in the supplemental material) and the GFP-VirB fusions were found to remain stable and active under the conditions of our assays (Fig. 1C and B, respectively). Thus, our work strongly suggests that VirB forms functional hubs on DNA after engaging its recognition site, and this occurs in the absence of any other pINV-encoded factor. The reason for hub formation, the macromolecular nature of these hubs, and the role that this plays in transcriptional antisilencing of virulence genes on pINV are discussed below.
Even though the regulatory actions of VirB are thought to be limited to the Shigella virulence plasmid, pINV (13, 15, 54), we were surprised to find that VirB formed foci for several reasons. First, pINV is large (230 kb) (55, 56), approximately 1/20 of the Shigella chromosome, and is thought to be present at only 1 or 2 copies per chromosome (57–59). Second, VirB upregulates at least 50 virulence-associated genes that are scattered around pINV (16, 60, 61) and are found within the large ipa-mxi-spa operons (15, 22, 62, 63). Third, another transcription factor that binds to pINV, VirF (13, 15, 16), does not form foci in Shigella when fused to mCherry (see Fig. S7 in the supplemental material). Although transcriptional regulators have been severely understudied in terms of their localization, especially in prokaryotic systems (37), the paradigm has always been that these proteins are found throughout the bacterial nucleoid at concentrations too low to be detected by standard fluorescence microscopy. Consequently, our finding that GFP-VirB forms DNA-dependent fluorescent foci in the bacterial cytoplasm is intriguing because it suggests that VirB forms multimeric complexes on DNA.
So, why then would a transcriptional regulator of virulence genes in Shigella form multimeric complexes when it engages DNA? A clue came from the evolutionary lineage of VirB. VirB belongs to the ParB superfamily (19, 52), a family of proteins where the vast majority play central roles in plasmid or chromosome segregation. Examples of this family, including Spo0J, KorB, SopB, and RepB, have been studied in depth and are widespread among well-studied bacterial phyla (38, 42, 45). These proteins display a discrete subcellular localization in the bacterial cytoplasm and routinely form 2, 3, or 4 foci in the bacterial cytoplasm. Like VirB, the number of foci appears to correlate with cell length (41). Initially, ParB was thought to engage its parS site and then spread along DNA to form a DNA-ParB filament (43, 64, 65). More recently, however, single-molecule approaches (45) and super-resolution microscopy (66) have revealed that ParB-ParB interactions, described as ParB bridges, are commonplace on DNA (45). In vivo, almost all ParB molecules are actively confined around parS sites by a network of synergistic protein-protein and protein-DNA interactions (66). Thus, ParB seems to form stochastic DNA cages when ParB proteins interact from distal parS sites (66). Given the relatedness of VirB to ParB, the dynamic multimeric ParB complexes and ParB-DNA lattices reported by Sanchez et al. are intriguing because they raise the possibility that similar VirB complexes are responsible for the GFP-VirB foci that we observed in this study. At this stage, it remains unclear if VirB spreads along DNA once bound to its recognition sites (36) or if it forms multimeric complexes with itself once bound to DNA akin to the ParB-mediated bridging or caging described previously (45, 66). However, the number of fluorescent foci per cell observed in our assays is far fewer than the estimated number of VirB-regulated loci on pINV, and so we currently favor the VirB bridging/caging hypothesis.
Our studies of VirB-dependent transcriptional regulation in Shigella have already revealed that VirB is not a traditional transcription factor (23). VirB can regulate transcription from remotely located binding sites >1 kb upstream of a promoter (18, 23, 35), and instead of promoting the recruitment or activity of RNA polymerase, like other transcription factors, VirB functions to offset or counteract transcriptional silencing imparted by the pervasive nucleoid structuring protein H-NS (15, 16, 22, 23). The finding that VirB forms foci in the bacterial cytoplasm and the parallels between VirB and ParB may provide new insight into the transcriptional antisilencing mechanism imparted by VirB. For instance, large-scale molecular interactions between VirB molecules docked on pINV may lead to the remodeling of the AT-rich DNA molecule pINV, leading to the eviction or remodeling of H-NS–DNA interactions such that gene expression occurs. Ongoing research in our laboratory is exploring these hypotheses.
In addition, the relatedness of VirB to ParB also raises questions about the potential for VirB to function in plasmid partitioning. Although VirB is a close homologue of the ParB encoded by S. flexneri pINV, ParBSf (37% identical), there is currently no evidence that these proteins functionally substitute for one another: in cells lacking virB, virulence gene expression does not occur (13, 15) (even though ParBSf is encoded by pINV) and the plasmid, pINV, is stably maintained (67). Additionally, the VirB protein does not contain the N-terminal domain found in ParB that is required for interaction with the ATPase partner protein, ParA (22, 68), and no parA gene is found in the vicinity of the virB gene or elsewhere on pINV (with the exception of the native parA encoding the ParASf that works with the designated ParBSf). Instead, all evidence indicates that the role of VirB is to modulate gene expression by antagonizing H-NS-mediated silencing (22, 69). Nevertheless, based on findings presented in this study, the interplay between VirB and the ParABSf system does seem worthy of further exploration because work by others suggests that overexpression of VirB leads to destabilization of pINV (66). For instance, it would be interesting to determine if VirB and ParB colocalize in the S. flexneri cytoplasm and if there is any evidence of molecular cross talk between VirB and the native ParAB system that causes VirB-mediated interference in pINV partitioning.
Our finding that GFP-VirB foci still form in Shigella when a small surrogate plasmid bearing a single VirB recognition site was introduced into pINV-cured cells, demonstrated that focus formation was not reliant on anything found on pINV, except the VirB-binding site(s). At this stage, it remains unclear why foci were seen to form on these small pACYC plasmids. These foci could be caused by VirB-VirB bridging interactions across different copies of pACYC184 (a medium-copy-number plasmid), the clustering of medium-copy-number plasmids that has been previously reported (70, 71), or some combination thereof. Regardless, it is clear from our work that the DNA-binding activity of VirB is required for these foci to appear and that specific DNA-VirB interactions are sufficient and necessary for these foci to form. These observations raise the possibility that GFP-VirB and its recognition site could be used in combination as markers of DNA molecules, thus expanding the repertoire of ParB/parS-like markers that can be used in this manner (72). Indeed, unique features of the VirB-binding site system may avoid lethal segregation defects that appear when parS sites are built into bacterial chromosomes at a distance (73). Since the established VirB recognition site is only 25-bp long, much less foreign DNA would need to be introduced into a chromosome or plasmid of interest (tandem arrays of >250 lacO sites have previously been built on bacterial genomes to generate a focus of fluorescently tagged LacI [71, 74, 75]). Future work will test if this potential marker of DNA works reliably in closely related enteric organisms and other more distantly related bacteria.
In sum, this study provides a new view of a major regulator of virulence gene expression in Shigella spp. The formation of GFP-VirB foci strongly suggests that VirB forms large VirB-VirB and VirB-DNA complexes once it engages its DNA recognition sites. This is tantalizing given that the established role of VirB is to counteract transcriptional silencing imparted by another DNA structuring protein, H-NS. In addition, our work introduces the idea that fluorescently tagged VirB and its binding site may have utility as a DNA marker, allowing plasmid or perhaps even chromosomal loci to be tracked in cells. Finally, this work also fosters questions about the evolution of VirB to its role as a transcriptional antisilencer from a DNA partitioning protein. We anticipate that the work presented here will be foundational for future studies that explore the relationship between virulence gene regulation, plasmid partitioning, and topological remodeling of plasmids in bacterial cells and think this study, once again, highlights the merits of taking a holistic view of molecular biology by examining the subcellular localization of proteins like VirB in whole, living cells.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
Shigella flexneri strains were routinely grown on trypticase soy agar (TSA; trypticase soy broth [TSB] containing 1.5% [wt/vol] agar). When necessary, to ensure maintenance of the virulence plasmid, Congo red binding was examined on TSA plates containing 0.01% [wt/vol] Congo red. Depending on the assay, liquid cultures of S. flexneri strains were routinely grown overnight at 30°C in either Luria-Bertani (LB) broth or minimal medium, to limit growth medium autofluorescence during imaging (M9 minimal medium supplemented with 0.4% d-glucose, 0.4% Casamino Acids, 0.01 mg/ml nicotinic acid, 0.01 mg/ml tryptophan, 0.01 mg/ml thiamine, 0.1 mM CaCl2, and 0.5 mM MgSO4) (76, 77). Overnight cultures were then diluted 1:100 and subcultured at 37°C with aeration in the specified medium (note that 40 mM glycerol replaced d-glucose in minimal medium to allow induction of pBAD to occur [78]). Where appropriate and throughout this study, diluted cultures were induced with 0.02% l-arabinose for the last 3 h of a 5-h growth period. These conditions were chosen because they allowed fusion proteins levels to mirror levels of native VirB (see Fig. S1A in the supplemental material) at their peak in early-stationary-phase cultures (5-h growth) (Fig. S1B) based on Western blot analysis. To ensure plasmid maintenance, antibiotics were added to growth medium at the following final concentrations: ampicillin, 100 μg/ml, and chloramphenicol, 25 μg/ml. The bacterial strains and plasmids used in this study are listed in Table 1.
Plasmid constructs.
The gfp used throughout this work encodes a superfolder GFP which contains key substitutions (see Table S1 in the supplemental material) to minimize protein aggregation and enhance folding of both the fluorescent protein and the tagged protein of interest (46, 47). All pBAD-inducible plasmids generated in this work were derived from pBAD/His A (Invitrogen). pJNS12 is pBAD-gfp-linker-virB flanked by NcoI and HindIII restriction sites. The virB gene in this plasmid, and others created in this work, was sourced from pATM324, while the linker and the gfp genes were identical to those found in pJH66 (79). The native translation start site of the virB gene has been removed in pJNS12 to allow the entire fusion protein to be made from the gfp translation start site. pJNS18 is pBAD-gfp-linker-virB K152E/R167E and is derived from pJNS12. Each of the mutations encoding the substitutions are contained in a region flanked by BglII and PsiI sites in this construct. pJNS04 is pBAD-virB, where the virB gene is flanked by NheI and HindIII sites. Finally, pJNS10 is pBAD-gfp-12AA-virB, which is similar to pJNS12; however, the 12AA linker (48) replaces the linker that is found in pJNS12. All plasmid constructs are listed in Table 1 and all were verified by Sanger dideoxy sequencing. The sequences of primers used in this study are available upon request.
Analysis of fusion protein stability.
Overnight cultures were diluted in LB and then induced, as described previously. Cells were normalized to cell density (optical density at 600 nm [OD600]), harvested, and washed with 0.2 M Tris buffer (pH 8.0), prior to resuspension in 400 μl 10 mM Tris (pH 7.4), and 100 μl 4× SDS-PAGE buffer (containing β-mercaptoethanol). When necessary and as noted, 4 μl of 100 mM phenylmethylsulfonyl fluoride (PMSF) was added to resuspended samples prior to SDS-PAGE buffer addition to limit proteolysis. Equal volumes of each normalized, boiled protein preparation were electrophoresed on 12.5% SDS-PAGE gels. For Western blot analyses, VirB was detected using an affinity purified anti-VirB antibody obtained from Pacific Immunology, and GFP was detected using a polyclonal anti-GFP antibody obtained from Molecular Probes. In both cases, a GE anti-rabit IgG–horseradish peroxidase (HRP; NA9340) secondary antibody was used. All blots were imaged using the auto exposure setting with chemiluminescence detection (Azure 500; Azure Biosystems). Densitometry was done using the lane and band detection features of the AzureSpot analysis software.
Quantification of VirB fusion protein activity.
For these assays, overnight cultures were diluted in LB and induced (as described above) prior to being assayed. Cultures were lysed and β-galactosidase activity was measured using a modified Miller protocol (16, 80).
Visualization of fusion proteins and quantification of foci.
Overnight cultures were diluted 1:100 in 2.5 ml minimal medium and induced as described previously. Cells were immobilized in 1% agarose containing minimal medium on glass slides before imaging. Data acquisition was performed using transmitted light and fluorescence microscopy (Zeiss Axio Imager M2) equipped with oil immersion (EC Plan-Neofluar 63×/Ph3). Fluorescence excitation was performed (X-cite 120LED) at a range of 470 to 525 nm using 24% exposure for all captured images; cells bearing GFP-VirB, GFP-VirB K152E/R167E, and the empty control routinely required longer exposures than the GFP control (this is common for fusion proteins; exposure times are provided in the figure legends). Images were captured by an Orca Flash 4.0 LT monochromatic digital CMOS camera. Routinely, 5 fields of view were captured at random (solely using phase contrast during image focusing) for each experimental strain, with roughly 60 cells per field of view. Image analysis was completed using MicrobeJ (81). All cells in each field of view were counted and analyzed unless they did not fit the predetermined parameters (i.e., cell length, >4 μm [less than 3% of all cells analyzed], or cells on the edges of fields of view). Cell outlines were reviewed manually to ensure automatic segmentation was detected accurately. Also, focus detection was reviewed manually to determine whether signals were true foci or diffuse signals with maxima. Data were ordered by the number of foci/maxima detected followed by cell length, which was measured by the medial axis function in MicrobeJ. The average fluorescence intensity (“mean.channel 2”) of diffuse signals along the medial axis of cells and foci were quantified for representative field of view images using the bacteria and maximum intensity functions of MicrobeJ, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Monika M. Karney for technical assistance with Western blots, Boo Shan Tseng for microscope support, and Amei Amei for guidance with our statistical analyses. We are grateful to members of the Wing lab past and present for many helpful discussions on this research topic. The University of Nevada Las Vegas (UNLV) Genomics Core Facility (sponsored by the National Institutes of General Medical Sciences; P20GM103440) provided sequencing services.
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (NIH), grant R15 AI090573 (to H.J.W.) and by the National Institute of General Medical Sciences at the National Institutes of Health (NIH) grant R01GM118792-01 (to G.R.B.). J.N.S. was the recipient of a GPSA travel grant that supported travel to the University of Wyoming for image capture training appropriate for this study.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nevo-Dinur K, Govindarajan S, Amster-Choder O. 2012. Subcellular localization of RNA and proteins in prokaryotes. Trends Genet 28:314–322. 10.1016/j.tig.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228. 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snapp E. 2005. Design and use of fluorescent fusion proteins in cell biology. Curr Protoc Cell Biol Chapter 21:21 4 1–21 4 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crivat G, Taraska JW. 2012. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol 30:8–16. 10.1016/j.tibtech.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laloux G, Jacobs-Wagner C. 2014. How do bacteria localize proteins to the cell pole? J Cell Sci 127:11–19. 10.1242/jcs.138628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudner DZ, Losick R. 2010. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol 2:a000307–a000307. 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman GR, Perez AM, Ptacin JL, Ighodaro E, Folta-Stogniew E, Comolli LR, Shapiro L. 2013. Oligomerization and higher-order assembly contribute to sub-cellular localization of a bacterial scaffold. Mol Microbiol 90:776–795. 10.1111/mmi.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelastro PS, Sliusarenko O, Jacobs-Wagner C. 2010. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J Bacteriol 192:539–552. 10.1128/JB.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raskin DM, de Boer PA. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181:6419–6424. 10.1128/JB.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Lutkenhaus J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34:82–90. 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 12.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. 2018. Shigellosis. Lancet 391:801–812. 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 13.Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol 3:627–635. 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 14.Tobe T, Nagai S, Okada N, Adler B, Yoshikawa M, Sasakawa C. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol Microbiol 5:887–893. 10.1111/j.1365-2958.1991.tb00762.x. [DOI] [PubMed] [Google Scholar]
- 15.Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology (Reading) 151:951–962. 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 16.Wing HJ, Yan AW, Goldman SR, Goldberg MB. 2004. Regulation of IcsP, the outer membrane protease of the Shigella actin tail assembly protein IcsA, by virulence plasmid regulators VirF and VirB. J Bacteriol 186:699–705. 10.1128/jb.186.3.699-705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basta DW, Pew KL, Immak JA, Park HS, Picker MA, Wigley AF, Hensley CT, Pearson JS, Hartland EL, Wing HJ. 2013. Characterization of the ospZ promoter in Shigella flexneri and its regulation by VirB and H-NS. J Bacteriol 195:2562–2572. 10.1128/JB.00212-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna JA, Wing HJ. 2020. The antiactivator of type III Secretion, OspD1, is transcriptionally regulated by VirB and H-NS from remote sequences in Shigella flexneri. J Bacteriol 202. 10.1128/JB.00072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A. 1990. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J Bacteriol 172:619–629. 10.1128/jb.172.2.619-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol 175:6142–6149. 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter ME, Dorman CJ. 1997. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol Gen Genet 256:93–103. 10.1007/s004380050550. [DOI] [PubMed] [Google Scholar]
- 22.Turner EC, Dorman CJ. 2007. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J Bacteriol 189:3403–3413. 10.1128/JB.01813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weatherspoon-Griffin N, Picker MA, Pew KL, Park HS, Ginete DR, Karney MM, Usufzy P, Castellanos MI, Duhart JC, Harrison DJ, Socea JN, Karabachev AD, Hensley CT, Howerton AJ, Ojeda-Daulo R, Immak JA, Wing HJ. 2018. Insights into transcriptional silencing and anti-silencing in Shigella flexneri: a detailed molecular analysis of the icsP virulence locus. Mol Microbiol 108:505–518. 10.1111/mmi.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 25.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 26.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13:773–780. 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Grainger DC, Hurd D, Goldberg MD, Busby SJ. 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res 34:4642–4652. 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoebel DM, Free A, Dorman CJ. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology (Reading) 154:2533–2545. 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 29.Ayala JC, Silva AJ, Benitez JA. 2017. H-NS: an overarching regulator of the Vibrio cholerae life cycle. Res Microbiol 168:16–25. 10.1016/j.resmic.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picker MA, Wing HJ. 2016. H-NS, its family members and their regulation of virulence genes in Shigella species. Genes (Basel) 7:112. 10.3390/genes7120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustamante VH, Santana FJ, Calva E, Puente JL. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol 39:664–678. 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- 32.De la Cruz MA, Fernandez-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vazquez A, Calva E. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol 66:727–743. 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- 33.Biswas M, Maqani N, Rai R, Kumaran SP, Iyer KR, Sendinc E, Smith JS, Laloraya S. 2009. Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae. Mol Cell Biol 29:2889–2898. 10.1128/MCB.00728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams McMackin EA, Marsden AE, Yahr TL. 2019. H-NS family members MvaT and MvaU regulate the Pseudomonas aeruginosa type III secretion system. J Bacteriol 201 10.1128/JB.00054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castellanos MI, Harrison DJ, Smith JM, Labahn SK, Levy KM, Wing HJ. 2009. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. J Bacteriol 191:4047–4050. 10.1128/JB.00313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karney MM, McKenna JA, Weatherspoon-Griffin N, Karabachev AD, Millar ME, Potocek EA, Wing HJ. 2019. Investigating the DNA-binding site for VirB, a key transcriptional regulator of Shigella virulence genes, using an in vivo binding tool. Genes (Basel) 10:149. 10.3390/genes10020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suter DM. 2020. Transcription tactors and DNA play hide and seek. Trends Cell Biol 30:491–500. 10.1016/j.tcb.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Kusiak M, Gapczynska A, Plochocka D, Thomas CM, Jagura-Burdzy G. 2011. Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J Bacteriol 193:3342–3355. 10.1128/JB.00328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breier AM, Grossman AD. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol 64:703–718. 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 40.Erdmann N, Petroff T, Funnell BE. 1999. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc Natl Acad Sci U S A 96:14905–14910. 10.1073/pnas.96.26.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta M, Nielsen HJ, Youngren B, Austin S. 2010. P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J Bacteriol 192:1175–1183. 10.1128/JB.01245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donovan C, Schwaiger A, Kramer R, Bramkamp M. 2010. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J Bacteriol 192:3441–3451. 10.1128/JB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray H, Ferreira H, Errington J. 2006. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol 61:1352–1361. 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 44.Broedersz CP, Wang X, Meir Y, Loparo JJ, Rudner DZ, Wingreen NS. 2014. Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc Natl Acad Sci U S A 111:8809–8814. 10.1073/pnas.1402529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham TG, Wang X, Song D, Etson CM, van Oijen AM, Rudner DZ, Loparo JJ. 2014. ParB spreading requires DNA bridging. Genes Dev 28:1228–1238. 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 47.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat Methods 9:480–482. 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. 1999. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 17:691–695. 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 49.Argos P. 1990. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J Mol Biol 211:943–958. 10.1016/0022-2836(90)90085-Z. [DOI] [PubMed] [Google Scholar]
- 50.Collet C, Thomassin J-L, Francetic O, Genevaux P, Tran Van Nhieu G. 2018. Protein polarization driven by nucleoid exclusion of DnaK(HSP70)–substrate complexes. Nat Commun 9:2027. 10.1038/s41467-018-04414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huls PG, Vischer NOE, Woldringh CL. 2018. Different amounts of DNA in newborn cells of Escherichia coli preclude a role for the chromosome in size control according to the “adder” model. Front Microbiol 9:664. 10.3389/fmicb.2018.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beloin C, McKenna S, Dorman CJ. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J Biol Chem 277:15333–15344. 10.1074/jbc.M111429200. [DOI] [PubMed] [Google Scholar]
- 53.Gao X, Zou T, Mu Z, Qin B, Yang J, Waltersperger S, Wang M, Cui S, Jin Q. 2013. Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes. Nucleic Acids Res 41:10529–10541. 10.1093/nar/gkt748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weatherspoon-Griffin N, Picker MA, Wing HJ. 2016. The genetic organization and transcriptional regulation of Shigella virulence genes, p 65–107. In Picking WD, Picking WL (ed), Shigella: molecular and cellular biology. Caister Academic Press, Poole, UK. [Google Scholar]
- 55.Venkatesan MM, Goldberg MB, Rose DJ, Grotbeck EJ, Burland V, Blattner FR. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun 69:3271–3285. 10.1128/IAI.69.5.3271-3285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, Zhang X, Zhang J, Yang G, Wu H, Qu D, Dong J, Sun L, Xue Y, Zhao A, Gao Y, Zhu J, Kan B, Ding K, Chen S, Cheng H, Yao Z, He B, Chen R, Ma D, Qiang B, Wen Y, Hou Y, Yu J. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res 30:4432–4441. 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D'Hauteville H, Kunst F, Sansonetti P, Parsot C. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38:760–771. 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 58.McVicker G, Hollingshead S, Pilla G, Tang CM. 2019. Maintenance of the virulence plasmid in Shigella flexneri is influenced by Lon and two functional partitioning systems. Mol Microbiol 111:1355–1366. 10.1111/mmi.14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radnedge L, Davis MA, Youngren B, Austin SJ. 1997. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol 179:3670–3675. 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchiya K, Tobe T, Komatsu K, Suzuki T, Watarai M, Fukuda I, Yoshikawa M, Sasakawa C. 1995. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol 17:241–250. 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- 61.Santapaola D, Del Chierico F, Petrucca A, Uzzau S, Casalino M, Colonna B, Sessa R, Berlutti F, Nicoletti M. 2006. Apyrase, the product of the virulence plasmid-encoded phoN2 (apy) gene of Shigella flexneri, is necessary for proper unipolar IcsA localization and for efficient intercellular spread. J Bacteriol 188:1620–1627. 10.1128/JB.188.4.1620-1627.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorman CJ, Porter ME. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol 29:677–684. 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 63.Dorman CJ, McKenna S, Beloin C. 2001. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int J Med Microbiol 291:89–96. 10.1078/1438-4221-00105. [DOI] [PubMed] [Google Scholar]
- 64.Rodionov O, Yarmolinsky M. 2004. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol Microbiol 52:1215–1223. 10.1111/j.1365-2958.2004.04055.x. [DOI] [PubMed] [Google Scholar]
- 65.Bingle LE, Macartney DP, Fantozzi A, Manzoor SE, Thomas CM. 2005. Flexibility in repression and cooperativity by KorB of broad host range IncP-1 plasmid RK2. J Mol Biol 349:302–316. 10.1016/j.jmb.2005.03.062. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez A, Cattoni DI, Walter JC, Rech J, Parmeggiani A, Nollmann M, Bouet JY. 2015. Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst 1:163–173. 10.1016/j.cels.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Schuch R, Maurelli AT. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect Immun 65:3686–3692. 10.1128/IAI.65.9.3686-3692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schumacher MA, Mansoor A, Funnell BE. 2007. Structure of a four-way bridged ParB-DNA complex provides insight into P1 segrosome assembly. J Biol Chem 282:10456–10464. 10.1074/jbc.M610603200. [DOI] [PubMed] [Google Scholar]
- 69.Beloin C, Dorman CJ. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol Microbiol 47:825–838. 10.1046/j.1365-2958.2003.03347.x. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y. 2017. Spatial distribution of high copy number plasmids in bacteria. Plasmid 91:2–8. 10.1016/j.plasmid.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Pogliano J, Ho TQ, Zhong Z, Helinski DR. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci U S A 98:4486–4491. 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62:331–338. 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 73.Toro E, Hong SH, McAdams HH, Shapiro L. 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci U S A 105:15435–15440. 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135:1685–1700. 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sasmor HM, Betz JL. 1990. Specific binding of lac repressor to linear versus circular polyoperator molecules. Biochemistry 29:9023–9028. 10.1021/bi00490a020. [DOI] [PubMed] [Google Scholar]
- 76.Brandon LD, Goldberg MB. 2001. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J Bacteriol 183:951–958. 10.1128/JB.183.3.951-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 78.Weatherspoon-Griffin N, Wing HJ. 2016. Characterization of SlyA in Shigella flexneri identifies a novel role in virulence. Infect Immun 84:1073–1082. 10.1128/IAI.00806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmes JA, Follett SE, Wang H, Meadows CP, Varga K, Bowman GR. 2016. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci U S A 113:12490–12495. 10.1073/pnas.1602380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 81.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Formal SB, Dammin GJ, Labrec EH, Schneider H. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol 75:604–610. 10.1128/JB.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maurelli AT, Blackmon B, Curtiss R, 3rd, 1984. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun 43:397–401. 10.1128/IAI.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.