V. cholerae has a dynamic life cycle, which requires rapid adaptation to changing external conditions. Two-component signal transduction (TCS) systems allow V. cholerae to sense and respond to these environmental changes.
KEYWORDS: Vibrio cholerae, two-component system, VxrAB, VxrA histidine kinase, sensor domain, dimerization, conformational change, signal transduction, X-ray crystallography, mutagenesis
ABSTRACT
VxrA and VxrB are cognate histidine kinase (HK)-response regulator (RR) pairs of a two-component signaling system (TCS) found in Vibrio cholerae, a bacterial pathogen that causes cholera. The VxrAB TCS positively regulates virulence, the type VI secretion system, biofilm formation, and cell wall homeostasis in V. cholerae, providing protection from environmental stresses and contributing to the transmission and virulence of the pathogen. The VxrA HK has a unique periplasmic sensor domain (SD) and, remarkably, lacks a cytoplasmic linker domain between the second transmembrane helix and the dimerization and histidine phosphotransfer (DHp) domain, indicating that this system may utilize a potentially unique signal sensing and transmission TCS mechanism. In this study, we have determined several crystal structures of VxrA-SD and its mutants. These structures reveal a novel structural fold forming an unusual β hairpin-swapped dimer. A conformational change caused by relative rotation of the two monomers in a VxrA-SD dimer could potentially change the association of transmembrane helices and, subsequently, the pairing of cytoplasmic DHp domains. Based on the structural observation, we propose a putative scissor-like closing regulation mechanism for the VxrA HK.
IMPORTANCE V. cholerae has a dynamic life cycle, which requires rapid adaptation to changing external conditions. Two-component signal transduction (TCS) systems allow V. cholerae to sense and respond to these environmental changes. The VxrAB TCS positively regulates a number of important V. cholerae phenotypes, including virulence, the type six secretion system, biofilm formation, and cell wall homeostasis. Here, we provide the crystal structures of the VxrA sensor histidine kinase sensing domain and propose a mechanism for signal transduction. The cognate signal for VxrAB remains unknown; however, in this work we couple our structural analysis with functional assessments of key residues to further our understanding of this important TCS.
INTRODUCTION
Vibrio cholerae, the causative agent of the diarrheal disease cholera, resides in aquatic reservoirs and can cause illness upon the ingestion of contaminated food or water (1). There are approximately 1.3 to 4 million cholera cases annually, resulting in 21,000 to 143,000 deaths worldwide (2). During its life cycle in the aquatic environment and transition to the human host, V. cholerae must sense and respond to many changes in its environment to survive and proliferate. V. cholerae employs two-component systems (TCSs) to sense fluctuating environmental conditions and elicit the appropriate cellular response. One such TCS, the VxrAB system, positively regulates important cellular phenotypes, including virulence, biofilm formation, the type VI secretion system, and cell wall homeostasis. However, its cognate signal sensing and transduction mechanisms are unknown (3–6).
The VxrAB TCS is composed of a sensor histidine kinase (HK) VxrA and a response regulator (RR) VxrB (3). VxrA (VCA0565) is a 499-amino acid (aa) protein encoded on the small chromosome of V. cholerae. At its N terminus, VxrA has a periplasmic sensor domain (VxrA-SD; ∼223 amino acids) flanked by transmembrane helices (Fig. 1A). This sensor domain (SD) harbors a DUF3404 domain (Pfam PF11884), which is uncharacterized and commonly found to be associated with HKs. VxrA-SD is mostly unique to the Vibrio genus and has no significant sequence similarity with any other known proteins. Within the different strains of V. cholerae, Vibrio mimicus, and Vibrio metoecus, the sequences of VxrA-SD are more than 90% identical. The sequence identity with other members of the Vibrio genus is less than 74%, including Vibrio parahaemolyticus (64%) and Vibrio vulnificus (64%) (see Fig. S1 in the supplemental material). The SD is less than 50% identical with other bacteria, including Photobacterium sanctipauli (Fig. S1). After the second transmembrane helix, however, the cytoplasmic portion of VxrA contains a typical histidine kinase A transmitter, including a DHp (dimerization and histidine phosphotransfer) domain and a CA (catalytic ATP-binding) domain. The so-called H box in the DHp domain, as well as the N, G1, F, and G2 boxes in the CA domain, is well defined (7, 8). Most interestingly, unlike most other HKs, the cytoplasmic portion of VxrA does not have a linker domain after the second transmembrane helix. Typical HKs linker domains include a HAMP (histidine kinases, adenylate cyclases, methyl-accepting proteins, and phosphatases) domain (9, 10), a PAS (period circadian protein, aryl hydrocarbon receptor nuclear translocation protein, and single-minded protein) domain (11, 12), or a GAF (cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA) domain (13, 14). This linker domain generally forms a dimer and is critical in transmitting the ligand-binding signal from the periplasmic SD to the cytoplasmic kinase domain (15–17). Without a linker domain in place, it is intriguing to explore the structure of VxrA-SD, its potential conformational change, and the mechanism of signal transduction of the HK.
FIG 1.
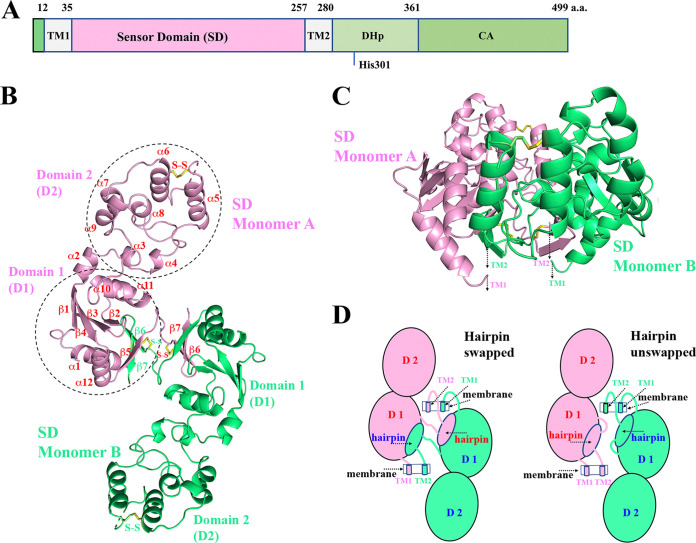
VxrA domain architecture and VxrA-SD dimer structure. (A) Domain arrangement of VxrA. The periplasmic SD is flanked by two transmembrane α-helices (TM1 and TM2). There is not a cytoplasmic linker domain or intracellular signal-transducing domain between TM2 and the DHp domain. The catalytic histidine residue H301 on the DHp domain is labeled. After the DHp domain, the catalytic domain (CA) or the ATP-binding domain is shown. (B) A ribbon diagram of the VxrA-SD dimer in the VxrA-SD_1 structure. Each VxrA-SD monomer can be divided into domain 1 (D1) and domain 2 (D2). D2 (a helical bundle) is inserted within D1 (an α/β fold) through two linkers. Two monomers form a dimer by swapping their two β-strands (a hairpin) from their central β-sheet of D1 (see the text). (C) A side-view of a VxrA-SD_1 dimer, presumably sitting on the membrane with its N and C termini connected to transmembrane helices. (D) A schematic diagram of the dimerization of VxrA-SD. Figure 1B and C, Fig. 2 and 3, a part of Fig. 4, and Fig. S5 are prepared with the program PyMOL (http://PyMOL.org).
In this study, we report the novel folding of VxrA-SD and its unique dimerization mechanism in five crystal structures. By comparing apo and mutant forms, we test the role of potentially critical residues in both the dimerization region and the putative signaling molecule binding pocket region on the VxrA function. Based on these observations, we propose a signal transduction mechanism associated with the conformational change of VxrA-SD dimer. We also suggest that residues involved in disulfide bonding (C101/C122 and C241/C249), β-linker formation (D238-T240), and the putative ligand-binding pocket (F117, H139, and H180) partially contribute to the function of VxrA but are not entirely essential for VxrA signal sensing and transduction activity.
RESULTS
Summary of experiments.
We first solved the crystal structures of two wild-type VxrA-SD grown under two different conditions (forms 1 and 2, as detailed in “Protein crystallization” in Materials and Methods). The two forms of crystals diffracted to 1.98 Å and 2.25 Å resolution limits, respectively (Table 1). The form 1 structure of VxrA-SD (referred to as VxrA-SD_1 hereafter) was determined using the single-wavelength anomalous dispersion (SAD) method. The structure of form 2 VxrA-SD (referred to as VxrA-SD_2 hereafter) was determined by applying the molecular replacement (MR) method using the VxrA-SD_1 structure as a search template. Subsequently, we obtained crystal structures of three deletion mutants of VxrA-SD. Statistics from data collection and processing and structural determination and refinement, as well as validation, are reported in Table 1. Based on the crystal structures, we also identified residues that could contribute to VxrA function. The contribution of these residues to VxrA function was assayed using known VxrA phenotypes, including the ability of VxrA to positively autoregulate its expression and the contribution of VxrA to enhanced survival in response to β-lactam exposure.
TABLE 1.
Summary of crystallographic dataa
| Data collection statistics | VxrA-SD_1 | VxrA-SD_2 | VxrA-SDΔN239 | VxrA-SDΔN239-T240 | VxrA-SDΔD238-T240 |
|---|---|---|---|---|---|
| Space group | P41212 | P212121 | P41212 | P41212 | P1 |
| Unit cell dimensions | |||||
| a, b, c (Å) | 88.60, 88.60, 107.0 | 112.1, 116.7, 120.7 | 87.65, 87.65, 106.2 | 87.86, 87.86, 107.7 | 45.59, 51.10, 65.63 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 76.11, 73.53, 88.56 |
| Mol wt (Da; residue) | 24,585 (219)b | 24,585 (219)b | 24,472 (218)b | 24,371 (217)b | 24,256 (216)b |
| Mol (AU) | 1 | 4 | 1 | 1 | 2 |
| Se-Met (AU) | 4 | NA | NA | NA | NA |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 0.9792 | 0.9792 |
| Resolution limit (Å) | 1.98 | 2.25 | 2.00 | 2.20 | 1.98 |
| No. of unique reflections | 30,252c | 75,884 | 29,036 | 22,071 | 36,860 |
| Rmerge (%) | 9.7 (64.7)d | 5.1 (80.3)e | 7.1 (78.7)f | 8.8 (86.0)g | 9.1 (78.0)h |
| Completeness (%) | 99.8 (100.0)d | 99.6 (99.9)e | 99.9 (99.8)f | 99.8 (99.9)g | 95.9 (92.8)h |
| CC1/2 | 1.00 (0.94)d | 1.00 (0.72)e | 1.00 (0.75)f | 0.98 (0.74)g | 1.00 (0.55)h |
| Redundancy | 11.5 (11.8)d | 4.3 (4.4)e | 8.0 (7.8)f | 5.7 (6.0)g | 3.6 (3.3)h |
| I/σ (I) | 49.8 (4.0)d | 25.9 (1.3)e | 42.2 (2.4)f | 29.3 (1.7)g | 24.4 (1.7)h |
| Solvent content (%) | 70.6 | 69.4 | 70.0f | 70.7 | 57.4 |
| Phasing | |||||
| Resolution range (Å) | 37.2–1.98 | 46.6–3.00 | 45.4–3.00 | 45.9–3.00 | 44.1–3.00 |
| RCullis (anomalous)i (%) | 0.55 | ||||
| FOM before DM | 0.33 | ||||
| Correlation coefficientj (%) | 51.7 | 56.3 | 59.7 | 44.3 | |
| Refinement | |||||
| Resolution range (Å) | 37.2–1.98 | 46.6–2.25 | 45.4–2.00 | 45.9–2.20 | 44.1–1.98 |
| No. of reflections | 30,175 | 75,798 | 28,978 | 21,978 | 36,789 |
| Completeness (%) | 99.7 | 99.4 | 98.7 | 99.8 | 93.1 |
| Rwork/Rfree (%) | 18.4/20.3 | 20.5/22.7 | 18.1/20.9 | 20.2/22.7 | 19.6/24.4 |
| No. of atoms (protein/HETATM) | 1,636/151 | 6,901/236 | 1,769/149 | 1,779/84 | 3,406/136 |
| RMSD | |||||
| Bond length (Å) | 0.006 | 0.002 | 0.017 | 0.003 | 0.010 |
| Bond angle (°) | 1.000 | 0.464 | 1.524 | 0.601 | 1.049 |
| B-factors (Å2) (main/side chain) | 52.5/56.2 | 70.5/72.0 | 45.0/47.9 | 66.2/68.6 | 48.3/53.8 |
| Wilson B-factor (Å2) | 36.8 | 52.5 | 41.3 | 55.6 | 42.1 |
| MolProbity validation | |||||
| Ramachandran outliers (%) | 0.0 | 0.12 | 0.0 | 0.5 | 0.0 |
| Ramachandran favored (%) | 97.6 | 97.5 | 99.5 | 98.2 | 98.4 |
| Rotamer outliners (%) | 1.2 | 1.7 | 3.7 | 4.2 | 1.6 |
| Clashscore | 1.21 | 2.18 | 3.65 | 1.68 | 2.36 |
| MolProbity score | 0.97 | 1.29 | 1.77 | 1.39 | 1.17 |
| PDB ID | 4R7Q | 7KB3 | 7LA6 | 7KB7 | 7KB9 |
AU, asymmetric units; RMSD, root mean square deviation.
Not including three N-terminal vector-derived residues, SNA.
Including Bijvoet pairs.
Last resolution bin, 1.98 to 2.01 Å.
Last resolution bin, 2.25 to 2.29 Å.
Last resolution bin, 2.00 to 2.03 Å.
Last resolution bin, 2.20 to 2.24 Å.
Last resolution bin, 1.98 to 2.01 Å.
Single-wavelength anomalous diffraction (SAD) method.
Molecular replacement method.
Overall structure of VxrA-SD_1.
There is one VxrA-SD monomer in the asymmetric unit of VxrA-SD_1 crystal (e.g., monomer A in Fig. 1B and C). The VxrA-SD forms a hairpin-swapped 2-fold dimer (Fig. 1B to D) with a symmetry-related VxrA-SD (e.g., monomer B in Fig. 1B to D). Each VxrA-SD monomer can be divided into two domains, domain 1 (D1) and domain 2 (D2) (Fig. 1B to D; see Fig. S2 in the supplemental material). The membrane-proximal domain (D1) contains N and C termini of SD, which are connected to transmembrane helices (Fig. 1C). The membrane-distal domain (D2) (residues P70 to I166) is inserted within the D1 (residues 36 to 62 and 175 to 257) via two linkers (residues 63 to 69 and 167 to 174). The first linker is primarily comprised of the α2 helix. The D2 has a mixed parallel and antiparallel five-helix bundle (α5, α6, α7, α8, and α9) plus two short helices (α3 and α4) in the connecting region. The edge helix, α5, is packed only against the α6 helix. Between the C terminus of α5 and the N terminus of α6, a disulfide bond between C101 and C122 seemingly helps lock the α5 helix to the α6 helix, even though the loop between the two helices is partially disordered.
Part of the center of the D1 is an antiparallel four-stranded β-sheet (β1, β4, β3, and β2), flanked by two helices (α10 and α11) on one side of the β-sheet (Fig. 1A). The helices α10 and α11 are packed against the helical D2. There is an additional antiparallel three-stranded β-sheet (β5, β7, and β6). The two strands β6 and β7 come from the second VxrA-SD (e.g., monomer B in Fig. 1B) of a VxrA-SD dimer. The two VxrA-SDs swapped their β6 and β7 strands, or rather β6-β7 hairpin, to form a dimer. The loop that connects β5 and β6 or instead connects β5 and β6-β7 hairpin is 9 residues long. For the convenience of description, we will refer to this loop as “β-hairpin linker.” The two β-sheets in the D1 are nearly perpendicular to each other in an arrangement like the two lateral faces of a right triangular prism. In contrast, the third lateral face is formed by two antiparallel helices (α1 and α12). The core of the prism-like arrangement is highly hydrophobic, including residues L40, I44, F47, M198, L205, W221, A225, Y232, and L234, which interact with the β6-β7 hairpin (Fig. 2B and C).
FIG 2.
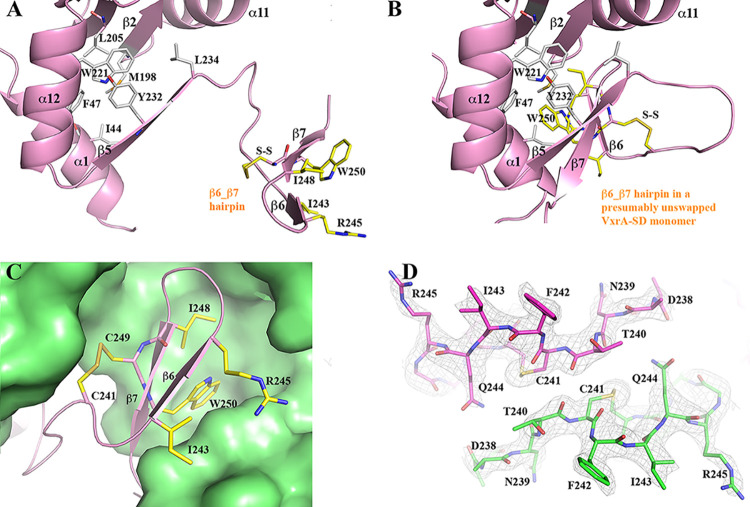
Formation of β-hairpin-swapped VxrA-SD dimer. (A) The core of D1 and the β6-β7 hairpin of one of the VxrA-SD monomers in the VxrA-SD_1 structure. The residues contributing to the core and the key residues of the β hairpin are drawn in stick format and colored in gray and yellow, respectively. (B) A presumably unswapped VxrA-SD. (C) A zoom-in view of the docking of a swapping β hairpin from one VxrA-SD onto the core of D1 of its pairing VxrA-SD. The disulfide bond (C241/C249) apparently helps the stability of the β hairpin. (D) The two antiparallel β-linkers between two monomers in a VxrA-SD dimer. Also shown is a σA-weighted 2mFo-DFc electron density map associated with the β-linkers. The map is drawn in mesh at a contour level of 1σ, and it validates the swapping of β-hairpin at the β-linker region.
A Dali structural homology search (18) using either a VxrA-SD monomer or a presumably unswapped VxrA-SD monomer results in no significant hits. The closest match is an unrelated type III secretion system needle protein with a helical bundle (PDB entry 2P58, Z-score 4.4, root-mean-square deviation [r.m.s.d.] value of 3.2 Å, and length of alignment of 77 aa with a sequence identity of 4%) that aligns part of helices of VxrA-SD. The low structural homology of VxrA-SD to currently known protein structures supports the novelty of VxrA-SD fold.
β6-β7 hairpin and VxrA-SD dimerization.
In a presumably unswapped VxrA-SD (Fig. 2B), the strands β6 and β7 are two regular strands of the β-sheet (β5, β7, and β6), which forms a lateral face of the prism-like arrangement mentioned above. The two strands contribute I243, I248, W250, and the aliphatic portion of the sidechain of R245 to the hydrophobic core of the prism (Fig. 2C). Among them, the indole side chain of the highly conserved W250 is more prominent. The indole ring does not form any specific parallel or T-shaped π-π stacking with its three neighboring aromatic rings from F47, W221, and Y232, respectively. There is a disulfide bond between C241 and C249, which tightens the loop part before the β6 strand to the middle of the β7 strand and seems to be unnecessary in the stabilization of the prism-like arrangement. However, in a swapped VxrA-SD (Fig. 2A), the disulfide bond may play a role in the stability of the swapped β6-β7 hairpin, in addition to the three main chain hydrogen bonds between the antiparallel β6 and β7 strands. The two cysteine residues are highly conserved in VxrA throughout different species (Fig. S1). When the β6-β7 hairpin from one VxrA-SD docks on the D1 of its pairing VxrA-SD (Fig. 2C), the hairpin contributes not only to the formation of a three-stranded β -sheet through main-chain–main-chain hydrogen bonds but also to the hydrophobic core of the D1. This finding indicates that each individual swapped VxrA-SD is structurally stable. It is noticeable that the two swapped VxrA-SDs have no direct contact with each other except in their crossover region, which is formed by two β-hairpin linkers, running in opposite directions (Fig. 1B and 2D). Moreover, there is no specific interaction between the two linkers (Fig. 2D). Therefore, it is possible that the two swapped VxrA-SDs in a dimer can move relative to each other, changing the conformation of the sensor domain dimer.
A second VxrA-SD structure captures a different conformation.
During the cocrystallization trials of VxrA-SD with potential signal molecules, we determined a second crystal structure of VxrA-SD referred to as VxrA-SD_2. There are four VxrA-SD monomers in one asymmetric unit, forming a dimer of dimers, or a tetramer (not shown in the figure). Each dimer is similarly a β6-β7 hairpin-swapped dimer (Fig. 3A and B). However, the VxrA-SD shows a dimer form on size-exclusion chromatography (Fig. S3); therefore, we believe that the VxrA-SD tetramer observed in the crystal is simply from the crystal packing effect.
FIG 3.
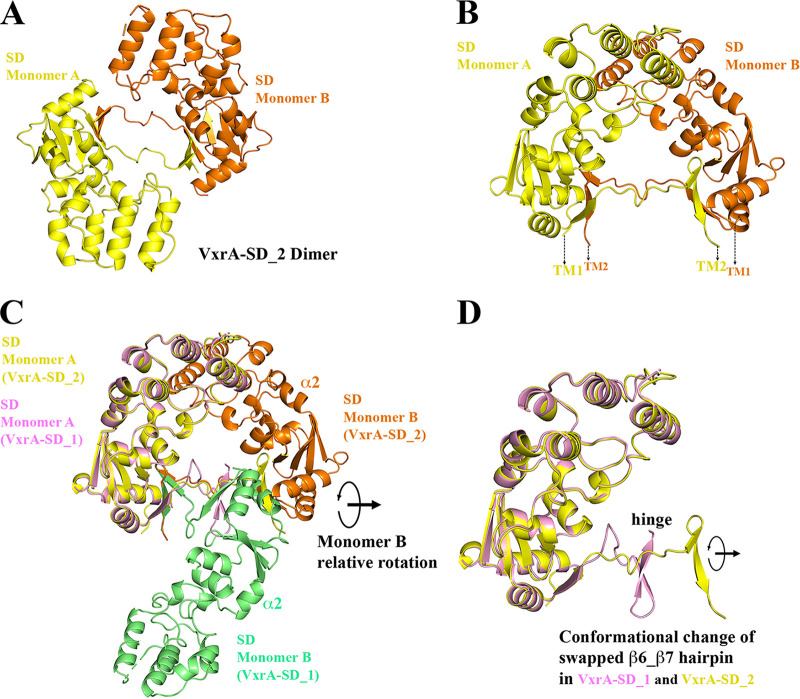
VxrA-SD in VxrA-SD_2 structure and its comparison to VxrA-SD_1 structure. (A) A VxrA-SD dimer in VxrA-SD_2 structure, in which two VxrA-SD dimers form a dimer of dimer, a tetramer (not shown in the figure). (B) A side view of a VxrA-SD_2 dimer, presumably sitting on the membrane with its N and C termini connected to transmembrane helices. (C) A structural alignment of one of the VxrA-SD monomers (monomer A) from VxrA-SD dimers from VxrA-SD_1 and _2 structures, respectively. This figure shows that the main body of VxrA-SD is quite rigid without significant conformational change. However, when one monomer (monomer A) is aligned, the other monomer (monomer B) shows a large conformational change with a relative rotation of about 150 degrees. (D) The relative rotation is apparently associated with the rotation of β hairpin while the β-linker serves as the rotating henge. The flexibility of the β-linker makes a scissoring-like motion of two pairing VxrA-SDs (blades) possible.
In the VxrA-SD_2 structure, a VxrA-SD monomer can be superimposed onto one VxrA-SD_1 monomer, with an r.m.s.d. value of 0.79 Å. This suggests that the main body of VxrA-SD is relatively rigid, given that it has no significant variation within the two crystal structures (Fig. 3C). The apparent difference is that when one monomer from each dimer is aligned, the second monomers from two dimers are oriented to each other with a rotation of about 150 degrees. The two β-linkers between two monomers are like the rotating hinge between two blades (monomers). The scissoring-like rotation can also be visualized from the relative rotation of the β6-β7 hairpin in the two dimer structures (Fig. 3D). Because of the rotation, the β6-β7 hairpin in VxrA-SD_2 is stretched further away from the main body of VxrA-SD. This relative conformational change of the β hairpin with respect to the main body of VxrA-SD implies that the two β hairpin-swapped VxrA-SDs can have variable conformations, like a scissor.
The implication of a signal transduction mechanism.
Conformational dynamics of the sensor domain of HKs and their role in signal transduction across the membrane have been extensively explored (16, 19, 20). The uncommon VxrA-SD folding and the unusually large conformational change of its β6-β7 hairpin-swapped VxrA-SD dimer makes VxrA an exciting system to study further, particularly considering its unusual cytoplasmic domain organization. The cytoplasmic domain of VxrA does not include a HAMP domain, which has been shown to be very important to the signal transduction of HK.
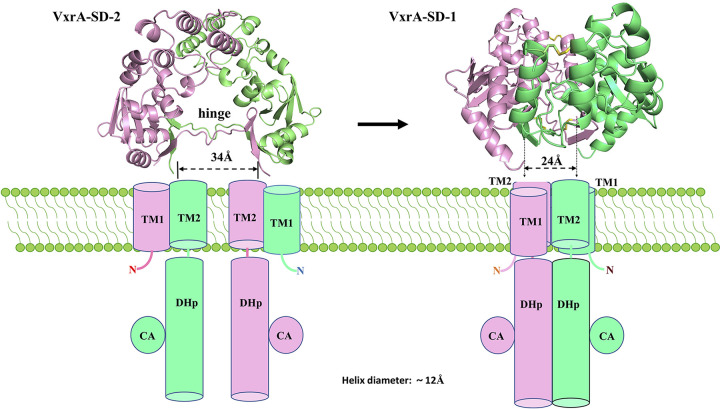
To further understand the consequences of the conformational change of VxrA-SD, we modeled VxrA-SDs from two structures sitting on a membrane in such a way that the N and C termini of each VxrA-SD are presumably connected to their transmembrane helices (Fig. 4). Therefore, based on the positions of the N and C termini of VxrA-SD, the positions of two transmembrane helices (TM1 and TM2) can be estimated. Since the position of TM2 mainly decides the status of its downstream helical DHp, the relative positions of two TM2 helices will then determine the relative positions of two N-terminal helices of DHp and their packing. To approximate the relative distance between two TM2, or rather the relative distance between DHp domains, the distance between the last residue of the β7-strand (V252 Cα) of two VxrA-SDs of a dimer was measured. The distance is about 24 Å in the dimer in VxrA-SD_1 and about 34 Å in the dimer in VxrA-SD_2 (Fig. 4). In the VxrA-SD_1 structure, considering that the diameter of a classic alpha-helix is about 12 Å, the two cytoplasmic DHp domains may form a helical bundle without stretching any connecting regions (loops) between either SD and TM2 or TM2 and DHp (Fig. 4, right panel). However, in the VxrA-SD_2 structure, the distance between two cytoplasmic DHp domains will be about 34 Å (Fig. 4, left panel). Therefore, the two DHp domains, probably including their connecting loops, will likely be separated. We propose that the conformation and the conformational change of the periplasmic VxrA-SD dictate the conformational properties of cytoplasmic DHp, leading to signal transduction through the membrane. Though we have no data to suggest either how DHp responds to the conformational change of VxrA-SD or what the activation status of VxrA is in each conformation shown in Fig. 4, we propose a scissor-like closing mechanism for signaling transduction from periplasmic sensor domain to cytoplasmic DHp domain, as elaborated on in the Discussion.
FIG 4.
A cartoon diagram of proposed signal transduction mechanism for VxrA. Side views of VxrA-SD_1 and _2 dimers being placed on the cell membrane. The two views are adapted from Fig. 1C and 3B, respectively. The cylinders represent the transmembrane helices and the helical DHp domains. CA domains that are attached to DHp domains are represented by circles. We propose that the conformational change of VxrA-SD dimer from VxrA-SD_2 to VxrA-SD_1 closes the transmembrane helices TM1 and TM2 and subsequently the HDp domains in a scissor-like mechanism, which changes the activation status of kinase domains.
Mutagenesis of predicted signaling molecule binding site.
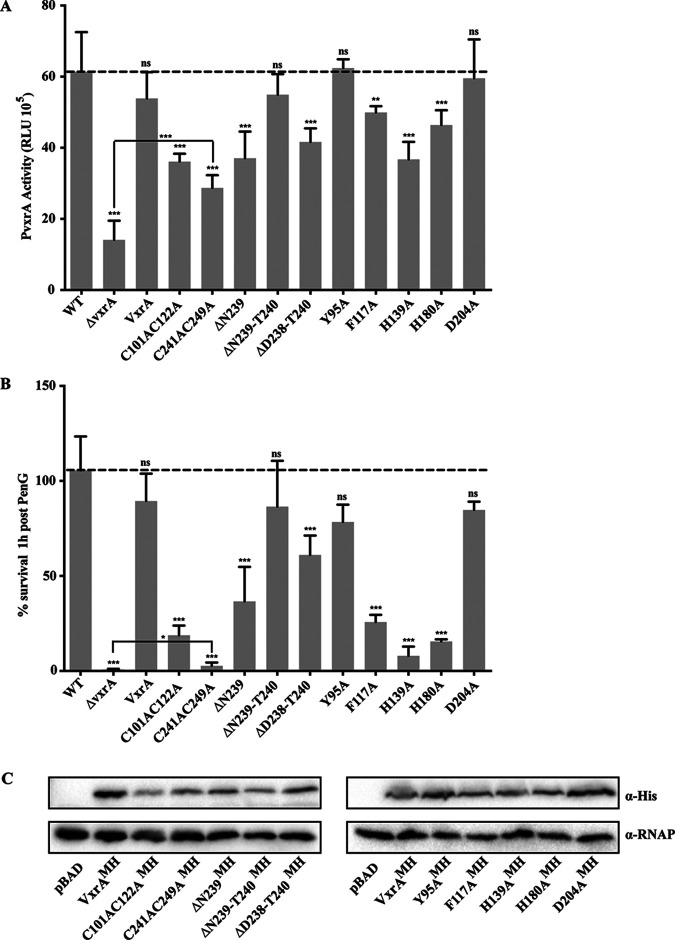
There is a pocket in the D2 domain with a surface area of about 75.2 Å2 and a volume of about 21.5 Å3 (21), shown in pink in Fig. S4. Several prominent residues, including Y95, F117, and H139, contribute to the formation of the pocket, which is predicted to be a part of the binding site for signal antibiotics of VbrK from V. parahaemolyticus (22), a homolog of VxrA, with a sequence identity of 64.1% in their SDs (Fig. S1). In the D1 domain on the interface between D1 and D2, there are three interconnected small pockets with a total surface area of about 136.1 Å2 and a total volume of about 21.5 Å3 (21), shown in red in Fig. S4. The H180 and D204 are two critical pocket-forming residues of the small pocket accessible from D1 (Fig. S4). To assess the impact of the putative binding pockets of VxrA, we generated the following mutated versions of VxrA, under the control of the native VxrA promoter: vxrAMyc-His (MH), Y95A, vxrAMH, F117A, vxrAMH, H139A, vxrAMH, H180A, and vxrAMH, D204A. Since the Vxr system positively regulates its expression, we assessed VxrA function in these mutants by measuring vxrA expression using a transcriptional reporter (4). VxrA contributes to the resistance to β-lactam treatment; thus, we additionally tested the effect of these mutations on penicillin G resistance (5). For the mutations in D2, in comparison to vxrA expression in the wild type, an F117A mutation resulted in a 1.2-fold reduction in vxrA expression and an H139A mutation resulted in a 1.7-fold decrease in vxrA expression. In contrast, a Y95A mutation appeared to have no effect on VxrA function (Fig. 5A). The residue Y95 is located at the entrance of the pocket in D2 (Fig. S4). Similarly, when the effect of these mutations on tolerance to penicillin G treatment was analyzed, an F117A mutation resulted in a 3.6-fold and an H139A mutation resulted in a 11.9-fold decrease in survival compared to that of the wild type. In contrast, a strain harboring a Y95A mutation was able to survive as well as or better than a strain expressing a wild-type VxrA (Fig. 5B).
FIG 5.
Functional analysis of proposed key VxrA-SD residues. (A) Autoregulation of the vxr operon was used as a readout of VxrA functionality. A transcriptional reporter harboring the regulatory region of vxrA upstream of a promoterless luciferase (lux) reporter (PvxrA-lux) was used to analyze the expression of vxr genes in wild-type and ΔvxrA strains or strains harboring mutated versions of VxrA in the Tn-7 site, under the control of vxrA promoter. Cultures were grown to exponential phase, and luminescence was measured. The graph represents the averages and standard deviations of relative light units (RLU) obtained from at least three biological replicates, normalized to wild-type levels. RLU are reported in luminescence counts · min−1 · ml−1 · OD600−1. Data were analyzed using a one-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test. ***, P < 0.0001; **, P < 0.005; ns, not significant. (B) Penicillin G resistance was used as a readout of VxrA functionality. Cells were grown to exponential phase and then treated with 100 μg/ml penicillin G for 1 h. CFU were determined pre- and posttreatment and were used to calculate the percentage of cells surviving treatment. Wild-type and ΔvxrA strains were compared to strains harboring wild-type or mutated versions of VxrA in the Tn-7 site, under the control of its own promoter. The graph represents the averages and standard deviations obtained from two technical replicates and three biological replicates. Data were analyzed using a one-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test. ***, P < 0.0001; *, P < 0.01; ns, not significant. (C) Protein stability was analyzed from whole-cell lysates. Overnight grown cultures of V. cholerae ΔvxrA cells harboring pBAD empty vector or vxrA constructs under the control of an arabinose inducible promoter were grown to an OD600 of 0.1, and then 0.2% arabinose was added to induce production of wild-type or mutated VxrA. The anti-myc antibody was used to probe for tagged VxrA and anti-RNA polymerase to probe for RNA polymerase as a loading control.
For the interconnected pocket that is partially formed in the D1 domain, an H180A mutation resulted in a 1.3-fold reduction in vxrA expression and a 6.3-fold reduction in survival following penicillin G treatment, while the D204A mutation showed little impact (Fig. 5). None of these mutations resulted in the complete abolition of VxrA activity, as a ΔvxrA mutant resulted in a 4.4-fold reduction in vxrA expression and a 134.8-fold reduction in survival following penicillin G treatment compared to those of the wild type (Fig. 5). Overall, this suggests that the signal molecule binding site of VxrA may be associated with the pocket in D2 domain, similar to what was reported for VbrK (22).
Mutagenesis of disulfide bonds and β-hairpin linker.
There are two conserved disulfide bonds in VxrA-SD structure. The C101/C122 disulfide bond is thought to stabilize the D2 domain of VxrA-SD, which is predicted to be the binding site of antibiotics (22). The C241/C249 disulfide bond may stabilize the β6-β7 hairpin and may help in the stabilization of the β6-β7 hairpin in swapped VxrA-SD dimer. To evaluate the functionality of the two disulfide bonds, we generated mutated versions of VxrA, vxrAMH, C101AC122A, and vxrAMH, C241AC249A and expressed them under the control of the native vxrA promoter. We assessed VxrA function in these mutants by measuring vxrA expression and penicillin G resistance.
As shown in Fig. 5A, vxrA expression was reduced approximately 1.7-fold for vxrAMH, C101AC122A and 2.1-fold for vxrAMH, C241AC249A. As shown in Fig. 5B, survival following penicillin G treatment was reduced approximately 4.9-fold for vxrAMH, C101AC122A and 11.7-fold for vxrAMH, C241AC249A. The dramatically affected activity and survival of vxrAMH, C241AC249A were near to those of the ΔvxrA mutant; however, some activity and growth were detectable at levels significantly above those of the ΔvxrA mutant. These findings indicate that both disulfide bonds, which are predicted to help stabilize a part of D2 and the swapped β hairpin, do affect VxrA activity. However, the disulfide bonds are not absolutely essential, as some activity and growth were detected in the absence of each disulfide bond.
We additionally assessed the importance of the β-linker, shortening the linker by generating vxrAMH, ΔN239, vxrAMH, ΔN239-T240, and vxrAMH, ΔD238-T240 mutants. Of these, only vxrAMH, ΔN239 and vxrAMH, ΔD238-T240 showed a decrease in vxrA expression (1.7-fold and 1.5-fold, respectively) or penicillin G resistance (2.5-fold and 9.2-fold reduction in survival), indicating that partial shortening of the loop affects function but that an intermediary shortening does not appear to affect function (Fig. 5). Western blotting was used to determine that the modified variants of VxrA are, in fact, produced (Fig. 5C). While slight variations in the protein levels of the VxrA mutants were observed, each protein was produced at detectable levels. Given that the transcription of vxrA is decreased in some vxrA mutant backgrounds, the lower levels of VxrA protein observed in Fig. 5C are not unexpected. In some mutants, the slight variation observed in protein production did not appear to affect function, as measured by vxrA transcription and penicillin G resistance, suggesting that these residues are not essential for VxrA function.
We were able to crystallize versions of VxrA-SD harboring deletion mutations and determined their structures, as shown in Fig. S5. All three VxrA-SD mutants form the β hairpin-swapped dimers, similar to the wild-type versions, VxrA-SD_1 and _2. Additionally, the deletion mutants VxrA-SDΔN239 and VxrA-SDΔN239-T240 have similar conformations to that of the VxrA-SD_1, while the three-residue deletion mutant VxrA-SDΔD238-T240 has a conformation between those of VxrA-SD_1 and _2 in terms of relative distance and relative orientation between two VxrA-SDs (Fig. S5). These structures suggest that the shortening of the β-linker (up to three residues) does not disrupt the formation of the β hairpin-swapped dimer, the important primary structural feature of VxrA-SD. The shortening may, however, limit the range of conformational change to some extent but results in no significant effect on function. The structural implication is seemingly consistent with functional measurements, given that shortening the linker slightly affected function but did not abolish the function of VxrA.
DISCUSSION
We have determined crystal structures of VxrA-SD, which harbors a domain of unknown function (DUF3404) in two distinct conformations. In both conformations, the periplasmic VxrA-SD forms a dimer, as is the case for most TCS SDs. For VxrA-SD, the dimerization arises through the swapping of a β hairpin between two SDs, which may represent a class of HK-SD dimerization mechanism. Another example of SD dimerization through swapping secondary structural elements has been observed in KinB-SD, in which a helix is swapped to form a helix-bundle dimer (23). In VxrA-SD, the long and flexible linker between the β hairpin and SD main body allows for the possibility of extensive conformational changes of the VxrA-SD dimer. Shortening this linker up to three residues does appear to affect VxrA function, though it does not entirely ablate VxrA function. The conformational change between two VxrA-SD structures, as reported here, can be characterized by a relative rotation of about 150 degrees of two VxrA-SDs (blades) approximately through their β-hairpin linkers (hinge) (Fig. 3C) in a scissor-like fashion. This rotation leads to a change in the distance between the two C-terminal helices of the VxrA-SD dimer (Fig. 4). The change of the interhelix space can lead to a change in the distance between the two downstream transmembrane helices and, subsequently, to a change in the association of two cytoplasmic DHp bundles (Fig. 4). The association of the two DHp bundles is believed to correlate to HK’s catalytic event, starting with the autophosphorylation of the conserved histidine of the DHp domain (20, 24, 25).
The signal transduction mechanism from periplasmic SD to cytoplasmic DHp has been extensively studied (16, 19, 26, 27). A conformational change of SD induced by signal molecule binding initiates the signal transduction cascades. Several models, including piston-like shift (28–30), rotational displacement (31, 32), scissor-like closing (33), and their combinations (19), have been proposed to describe how SD conformational change transmits a signal across the membrane. These mechanisms generally suggest a conformational change of SD induced by ligand binding that is propagated through the membrane. The conformational change of VxrA-SD dimer observed in this study can be described using a scissor-like closing model with the two β-hairpin linkers functioning as a hinge of two SD blades. Relative rotation of two blades can cause the β-hairpin linkers to twist in a different extension, thus regulating the distance between two SD blades. Subsequently, the linkages downstream of the SD and eventually DHp activity are controlled, as illustrated in Fig. 4. To some extent, the scissor-like mechanism is similar to the closing mechanism proposed for a hybrid TCS BT4663 from Bacteroides thetaiotaomicron (33). BT4663 SD is much larger than VxrA-SD, containing two 7-blade β-propellers followed by one Ig-like Y_Y_Y domain. Binding of unsaturated disaccharides (the signal sensed by the system) at BT4663 SD dimer interface closes the two C termini of BT4663 SD dimer in a scissoring-like motion. Subsequently, it activates the cytoplasmic HK domains (33). It is important to note that there is also no HAMP domain or another cytoplasmic linker domain between the transmembrane helix and the DHp domain in BT4663.
The signal recognized by VxrA remains unknown, as does what triggers the large conformational change between two VxrA-SD dimers. Considering the flexibility of the β-hairpin linker, we do not exclude potential impacts from molecular packing in crystals, especially in the 2nd form of VxrA-SD crystal, in which the sensor domain forms a dimer of dimers. Though we do not yet know the extent of conformational changes, an in vivo scissor-like motion of a VxrA-SD dimer may be taking place. It is possible that the two conformations can exist in the absence of the signaling molecule and that the binding ligand changes the equilibrium between these two states, stabilizing the signal transduction state.
Considering the sequence identity (64.1%) between VxrA-SD and VbrK-SD (Fig. S1), we initially thought that VxrA-SD could bind to certain antibiotics, as suggested by the studies on VbrK-SD (22). Cocrystallization of VxrA-SD with antibiotics, including penicillin G, ampicillin, and carbenicillin, produced crystals but without any antibiotics bound. Soaking VxrA-SD apo crystals with these antibiotics up to 50 mM concentration did not result in complex crystals formation. A fluorescence thermal shift small molecule binding screen, which included penicillin and related antibiotics, also revealed no significant hits. These findings suggest that the VxrA cognate signal molecule could be different from the VbrK cognate signal molecule.
While this paper was in final preparation and review, two groups reported the structure of V. parahaemolyticus VbrK-SD, which is 63% identical to VxrA (34, 35). They found VbrK-SD in monomeric form in both structures and solution. It is not clear how monomeric VbrK-SD can form a dimer for signal transduction through the membrane, as is typical of the HK of TCS, or if it maintains its monomeric state and utilizes a completely unknown signing mechanism. One of the studies also demonstrated that that VbrK-SD does not directly bind antibiotics (35). This could suggest that the VxrA-SD also utilizes an indirect mechanism to sense antibiotic stress, but further study is needed.
There is a predicted binding pocket on the D2 domain of VxrA-SD and an interconnected pocket partially on the D1 domain; however, site-directed mutagenesis on these pockets demonstrated that no single residue appeared to be fully responsible for VxrA function. It is not clear if either pocket is a validated binding site for the signal molecule of VxrA-SD. Additionally, the mechanism by which signal molecule binding at this membrane-distal site could induce a necessary conformational change of VxrA-SD dimer remains to be understood.
In addition to VxrA and BT4663 mentioned above, only a few HKs without intermediate linker domains between the transmembrane helices and DHp domain have been characterized. These include DesK from Bacillus subtilis (36), which senses membrane fluidity, BinK, a hybrid HK from Vibrio fischeri (37), and ammonium sensor HK Ks-Amt5 from the anammox bacterium “Candidatus Kuenenia stuttgartiensis” (38). HAMP domains, for example, as intermediate linker domains, have been more extensively studied (15, 27, 39, 40). Without a signal-amplifier linker domain, like a HAMP domain, the conformational change of SD needs to be sufficient to induce the necessary rearrangement of transmembrane helices and active cytoplasmic HK domains directly (38). VxrA, lacking a linker domain and showing a large conformational change of its SD, seemingly falls into this type of category of HK in TCS.
MATERIALS AND METHODS
Protein cloning, expression, and purification.
The vxrA (VCA0565) gene corresponding to residues D38 to S256 was PCR amplified from V. cholerae N16961 chromosomal DNA and cloned into expression vector pMCSG7 using ligation independent cloning (41). The expression vector was then transformed into Escherichia coli BL21(DE3)-Magic (42). The deletion mutants VxrA-SDΔN239, VxrA-SDΔN239-T240, and VxrA-SDΔD238-T240 were created according to protocol described in reference 43, which is a modification of polymerase incomplete primer extension (PIPE) cloning (44). The details of producing selenomethionine (Se-Met)-labeled VxrA-SD have been described earlier (45). The production of unlabeled VxrA-SD proteins including three deletion mutants and the purification of either Se-Met labeled or unlabeled proteins are described in an earlier publication (46).
Strain and plasmid construction.
Plasmids for expression of VxrA in V. cholerae were constructed using the Gibson Assembly recombinant DNA technique (New England BioLabs, Ipswich, MA). Mutations in vxrA were introduced using the Q5 site-directed mutagenesis kit (New England BioLabs, Ipswich, MA). Plasmid sequences were confirmed via Sanger sequencing (UC Berkeley DNA Sequencing Facility, Berkeley, CA). Complementation of ΔvxrA with either wild-type vxrA harboring a Myc-His tag or mutated versions of vxrA harboring a Myc-His tag under the control of the native vxrA promoter was carried out using a Tn7-based system, as previously described (4). Briefly, triparental matings with donor E. coli S17λpir carrying pGP704-Tn7 with the gene of interest, helper E. coli S17λpir harboring pUX-BF13, and V. cholerae deletion strains were carried out by mixing all three strains and incubating mating mixtures on LB agar plates for 18 h at 30°C. Transconjugants were selected on thiosulfate-citrate-bile salts-sucrose (TCBS; BD Difco, Franklin Lakes, NJ) agar medium containing gentamicin (15 μg/μl) at 30°C. Insertion of the complementation construct to the Tn7 site was verified by PCR.
Size-exclusion chromatography of VxrA-SD dimer.
To determine the oligomeric state of VxrA-SD in solution, size-exclusion chromatography was performed on an SRT SEC-150 column (Sepax Technologies) as we described previously (47). Premixed protein standards, including RNase (13.7 kDa), carbonic anhydrase (29 kDa), albumin (66 kDa), and ferritin (440 kDa), were used for the calibration of the SRT SEC-150 column.
Protein crystallization.
The screening of crystallization condition for either Se-Met-labeled or native VxrA-SD (including mutants) was performed as described previously (47). Diffraction quality crystals of Se-Met-labeled VxrA-SD appeared under the following condition: 0.8 M lithium sulfate monohydrate, 0.1 M TRIS (pH 8.5) for VxrA-SD_1. Diffraction quality crystals of wild-type VxrA-SD appeared under multiple conditions, including 0.1 M sodium acetate-HCl (pH 4.6) and 1.5 M ammonium chloride for VxrA-SD_2. Crystallization conditions for mutants are 0.2 M lithium sulfate, 0.1 M sodium cacodylate-HCl (pH 6.5), and 30% (wt/vol) polyethylene glycol (PEG) 400 for VxrA-SDΔN239, 0.1 M Tris-HCl (pH 8.5) and 1 M magnesium sulfate for VxrA-SDΔN239-T240, and 0.2 M potassium thiocyanate and 20% (wt/vol) PEG 3350 for VxrA-SDΔD238-T240. Prior to X-ray diffraction data collection, all crystals were treated with cryoprotectants (25% glycerol in crystal mother liquor) and cryocooled directly in liquid nitrogen.
X-ray diffraction and structure determination.
One set of single-wavelength anomalous dispersion (SAD) data were collected near the selenium absorption peak (12.66 keV) at 100 K from one Se-Met-labeled VxrA-SD_1 crystal. The data were obtained at the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory using the program SBCcollect (48). The intensities of the data set were integrated and scaled with the HKL3000 program suite (49) (Table 1). In the crystal, four Se sites were located using the program SHELXD (50), and they were used for phasing with the program MLPHARE (51). After density modification (DM) (51), a partial model (including 90% of total 222 residues correctly docked) was built in cycles of a model building using HKL Builder. All of the above programs are integrated within the program suite HKL3000 (49). The model of the VxrA-SD_1 crystal structure was completed manually using the program COOT (52). Its final refinements were performed using the program Phenix.refine (53) (Table 1). The diffraction data of wild-type VxrA-SD_2 crystal and three mutant crystals were collected at the same beamline as that mentioned above. Their structures were determined using the MR method (51) with a monomer structure of VxrA-SD_1 as the searching template. Their model rebuildings and completions as well as structural refinements were similar to those described above for VxrA-SD_1 crystal. The structural validations of all structures were performed using the program MolProbity (54).
Luminescence assays.
Luminescence assays were performed as previously described (3). Briefly, overnight cultures of V. cholerae cells were diluted 1:500 in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl [pH 7.5]) containing chloramphenicol (5 μg/ml). Cells were then grown aerobically at 30°C to an optical density at 600 nm (OD600) of 0.4 to 0.5. Then, the luminescence of cells was measured using a Victor3 multilabel counter (PerkinElmer, Waltham, MA). Luciferase activity is reported as counts per minute per milliliter per OD600 unit, shown as relative light units (RLU). Assays were repeated with three biological replicates. Three technical replicates were measured for all assays. Statistical analysis was performed using one-way analysis of variance (ANOVA) and Bonferroni’s multiple-comparison test.
Penicillin G resistance assays.
Cells were grown with shaking at 30°C to exponential phase (OD600 ∼ 0.3) in 10 ml LB broth in 50-ml flasks, followed by the addition of 100 μg/ml of penicillin G. CFU per milliliter were enumerated by serial dilution and spot plating in technical triplicate at exponential phase, followed by the addition of penicillin G. CFU per milliliter were enumerated by serial dilution and spot plating in technical triplicate after 1 h of penicillin G treatment. Percent survival was calculated by dividing CFU per milliliter after 1 h of treatment by CFU per milliliter before treatment. Three biological replicates were performed. Statistical analyses were performed using Prism 8 software (GraphPad Software, Inc., San Diego, CA) using ANOVA assuming equal distribution and Dunnett’s multiple-comparison test. P values of <0.05 were determined to be statistically significant.
Analysis of VxrA production.
Overnight cultures of V. cholerae cells were diluted 1:500 in LB broth, and then cells were grown aerobically at 30°C to OD600 of 0.4 to 0.5. Western blotting was performed as previously described (4). Briefly, whole-cell extracts were suspended in 2% SDS. A Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL) was used to estimate protein concentrations. Equal amounts of total protein (determined by BCA assay) were loaded onto an SDS 12% polyacrylamide gel. After the blot was probed for His, the blot was stripped of all antibodies using Western Blot Stripping Buffer (GM Biosciences, MD), and the same blot was used to probe for RNA polymerase.
Supplementary Material
ACKNOWLEDGMENTS
We thank Albin Cordona-Correa for technical assistance. We thank members of the Structural Biology Center at Argonne National Laboratory for their help in conducting X-ray diffraction data collection.
This work was supported by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under contract numbers HHSN272201200026C (to W.F.A.) and HHSN272201700060C (to K.J.F.S.) and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) grant R01AI055987 to F.H.Y. The Structural Biology Center beamlines are supported by U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. The submitted paper was created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (Argonne). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under contract number DE-AC02-06CH11357.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Charles RC, Ryan ET. 2011. Cholera in the 21st century. Curr Opin Infect Dis 24:472–477. 10.1097/QCO.0b013e32834a88af. [DOI] [PubMed] [Google Scholar]
- 2.Ali M, Nelson AR, Lopez AL, Sack DA. 2015. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis 9:e0003832. 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teschler JK, Cheng AT, Yildiz FH. 2017. The two-component signal transduction system VxrAB positively regulates Vibrio cholerae biofilm formation. J Bacteriol 199. 10.1128/JB.00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorr T, Alvarez L, Delgado F, Davis BM, Cava F, Waldor MK. 2016. A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta-lactam tolerance. Proc Natl Acad Sci U S A 113:404–409. 10.1073/pnas.1520333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin JH, Choe D, Ransegnola B, Hong HR, Onyekwere I, Cho BK, Dorr T. 2019. A multifaceted cellular damage repair and prevention pathway promotes high level tolerance to β-lactam antibiotics. bioRxiv [DOI] [PMC free article] [PubMed]
- 7.Parkinson JS, Kofoid EC. 1992. Communication modules in bacterial signaling proteins. Annu Rev Genet 26:71–112. 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 8.Grebe TW, Stock JB. 1999. The histidine protein kinase superfamily. Adv Microb Physiol 41:139–227. 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Ponting CP. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett 176:111–116. 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 10.Williams SB, Stewart V. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol 33:1093–1102. 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 11.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravind L, Ponting CP. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22:458–459. 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 14.Ho YS, Burden LM, Hurley JH. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J 19:5288–5299. 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson JS. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol 64:101–122. 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 16.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob-Dubuisson F, Mechaly A, Betton JM, Antoine R. 2018. Structural insights into the signalling mechanisms of two-component systems. Nat Rev Microbiol 16:585–593. 10.1038/s41579-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gushchin I, Gordeliy V. 2018. Transmembrane signal transduction in two-component systems: piston, scissoring, or helical rotation? Bioessays 40:1700197. 10.1002/bies.201700197. [DOI] [PubMed] [Google Scholar]
- 20.Bhate MP, Molnar KS, Goulian M, DeGrado WF. 2015. Signal transduction in histidine kinases: insights from new structures. Structure 23:981–994. 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian W, Chen C, Lei X, Zhao J, Liang J. 2018. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367. 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Wang Q, Zhang H, Yang M, Khan MI, Zhou X. 2016. Sensor histidine kinase is a beta-lactam receptor and induces resistance to beta-lactam antibiotics. Proc Natl Acad Sci U S A 113:1648–1653. 10.1073/pnas.1520300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan K, Chhor G, Binkowski TA, Jedrzejczak RP, Makowska-Grzyska M, Joachimiak A. 2014. Sensor domain of histidine kinase KinB of Pseudomonas: a helix-swapped dimer. J Biol Chem 289:12232–12244. 10.1074/jbc.M113.514836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mechaly AE, Sassoon N, Betton JM, Alzari PM. 2014. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol 12:e1001776. 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, Senadheera DB, Cvitkovitch DG, Chen L, Goodman SD, Han A. 2013. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol 11:e1001493. 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews EE, Zoonens M, Engelman DM. 2006. Dynamic helix interactions in transmembrane signaling. Cell 127:447–450. 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Gushchin I, Melnikov I, Polovinkin V, Ishchenko A, Yuzhakova A, Buslaev P, Bourenkov G, Grudinin S, Round E, Balandin T, Borshchevskiy V, Willbold D, Leonard G, Buldt G, Popov A, Gordeliy V. 2017. Mechanism of transmembrane signaling by sensor histidine kinases. Science 356:eaah6345. 10.1126/science.aah6345. [DOI] [PubMed] [Google Scholar]
- 28.Scott WG, Stoddard BL. 1994. Transmembrane signalling and the aspartate receptor. Structure 2:877–887. 10.1016/s0969-2126(94)00088-3. [DOI] [PubMed] [Google Scholar]
- 29.Ottemann KM, Xiao W, Shin YK, Koshland DE. Jr.. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751–1754. 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 30.Falke JJ, Hazelbauer GL. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265. 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126:1095–1108. 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940. 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 33.Lowe EC, Basle A, Czjzek M, Firbank SJ, Bolam DN. 2012. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci U S A 109:7298–7303. 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho SY, Yoon SI. 2020. Structural analysis of the sensor domain of the beta-lactam antibiotic receptor VbrK from Vibrio parahaemolyticus. Biochem Biophys Res Commun 533:155–161. 10.1016/j.bbrc.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Goh BC, Chua YK, Qian X, Lin J, Savko M, Dedon PC, Lescar J. 2020. Crystal structure of the periplasmic sensor domain of histidine kinase VbrK suggests indirect sensing of beta-lactam antibiotics. J Struct Biol 212:107610. 10.1016/j.jsb.2020.107610. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez P, Porrini L, Albanesi D, Abriata LA, Dal Peraro M, de Mendoza D, Mansilla MC. 2019. Transmembrane prolines mediate signal sensing and decoding in Bacillus subtilis DesK histidine kinase. mBio 10. 10.1128/mBio.02564-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks JF, Mandel MJ. 2016. The histidine kinase BinK is a negative regulator of biofilm formation and squid colonization. J Bacteriol 198:2596–2607. 10.1128/JB.00037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfluger T, Hernandez CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA. 2018. Signaling ammonium across membranes through an ammonium sensor histidine kinase. Nat Commun 9:164. 10.1038/s41467-017-02637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris HU, Dunin-Horkawicz S, Mondejar LG, Hulko M, Hantke K, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. 2011. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure 19:378–385. 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Dunin-Horkawicz S, Lupas AN. 2010. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J Mol Biol 397:1156–1174. 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 41.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. 2009. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 498:105–115. 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu N, Christendat D, Dharamsi A, Pai EF. 2000. Purification, crystallization and preliminary X-ray study of orotidine 5'-monophosphate decarboxylase. Acta Crystallogr D Biol Crystallogr 56:912–914. 10.1107/s090744490000576x. [DOI] [PubMed] [Google Scholar]
- 43.Nocek B, Stein AJ, Jedrzejczak R, Cuff ME, Li H, Volkart L, Joachimiak A. 2011. Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity. J Mol Biol 406:325–342. 10.1016/j.jmb.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klock HE, Lesley SA. 2009. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol 498:91–103. 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 45.Tan K, Deatherage Kaiser BL, Wu R, Cuff M, Fan Y, Bigelow L, Jedrzejczak RP, Adkins JN, Cort JR, Babnigg G, Joachimiak A. 2017. Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi. Protein Sci 26:1738–1748. 10.1002/pro.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. 2004. Automation of protein purification for structural genomics. J Struct Funct Genomics 5:111–118. 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan Y, Tan K, Chhor G, Butler EK, Jedrzejczak RP, Missiakas D, Joachimiak A. 2015. EsxB, a secreted protein from Bacillus anthracis forms two distinct helical bundles. Protein Sci 24:1389–1400. 10.1002/pro.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. 2006. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat 13:30–45. 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 50.Schneider TR, Sheldrick GM. 2002. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58:1772–1779. 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 51.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:4. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.