Abstract
Eukaryotic transcription requires the assembly of a multi-subunit pre-initiation complex (PIC) comprised of RNA polymerase II (Pol II) and the general transcription factors. The co-activator Mediator is recruited by transcription factors, facilitates the assembly of the PIC, and stimulates phosphorylation of the Pol II C-terminal domain (CTD) by the TFIIH subunit CDK7. Here, we present the cryo-electron microscopy structure of the human Mediator-bound PIC at sub-4 Å. Transcription factor binding sites within Mediator are primarily flexibly tethered to the tail module. CDK7 is stabilized by multiple contacts with Mediator. Two binding sites exist for the Pol II CTD, one between the head and middle modules of Mediator and the other in the active site of CDK7, providing structural evidence for Pol II CTD phosphorylation within the Mediator-bound PIC.
One Sentence Summary:
Cryo-electron microscopy reveals the high-resolution structure of the human Mediator-bound transcription pre-initiation complex.
Transcription of all messenger RNA (mRNA) in eukaryotes is carried out by RNA polymerase II (Pol II) (1). Pol II cannot by itself locate the transcription start site (TSS), open a transcription bubble to expose the template strand, and transition to an elongation state. Instead, DNA-bound transcription factors position the co-activator complex Mediator to facilitate the assembly of the pre-initiation complex (PIC) consisting of Pol II and the general transcription factors (GTF) TFIIA, TFIIB, TFIID (TBP), TFIIE, TFIIF, and TFIIH (2). These GTFs help position Pol II to initiate transcription at the correct genomic locus, and TFIIH feeds DNA into the active site of Pol II, generating force against the TBP/TFIIB/TFIIA lobe to unwind the DNA and expose the template strand (3). The entire Mediator-bound PIC (Med-PIC) is 2.7 megadaltons (MDa) in size, contains 56 polypeptides, and represents a unique challenge for structural characterization due to difficulties in obtaining and assembling these complexes, as well as to their inherent flexibility (4–9).
The largest subunit of Pol II, RPB1, contains a long, repetitive C-terminal domain (CTD) connected by a flexible linker region to the rest of the subunit (10). The CTD consists of 26 repeats in yeast and 52 repeats in humans of the consensus YSPTSPS heptamer sequence. Phosphorylation of the serine at position 5 (Ser5) by cyclin-dependent kinase 7 (CDK7) during transcription initiation leads to the recruitment of the 5’ capping enzymes that are indispensable for growth (11). Neither the CTD nor the 80-residue linker is visible in structures of the PIC due to their mobility (12). CDK7 is part of the cyclin-activated kinase (CAK) module of TFIIH, together with cyclin-H and Mat1. Mounting evidence indicates that Mediator recruits TFIIH to the PIC and stimulates the CAK module’s ability to phosphorylate Ser5 (13, 14).
Atomic models of Med-PICs are currently limited to yeast (yMed-PIC) (6–9). Mediator is divided into four modules: the head (MedHead) interacts with Pol II, the middle (MedMiddle) primarily serves a structural role, the tail (MedTail) serves as a hub for the binding of transcription factors, and the dissociable kinase module (MedKinase) is found at enhancers and prevents interaction with the PIC (15, 16). The existing high-resolution models of yMed-PIC highlight differences in how MedHead interacts with Pol II, suggesting that the interface between Mediator and Pol II is not rigid (6, 7). MedHead is capable of binding the CTD, shown in a co-crystal structure (17). Based on the location of the CTD modeled into the full yeast Mediator complex, the CTD also likely serves to stabilize the interface between MedHead and MedMiddle (8, 9). Functional and structural studies have identified a minimal core Mediator (cMed), devoid of both MedKinase and MedTail (18, 19). Structures of Mediator have poorly defined density for MedTail, leaving open the question of where most transcription factors bind (5, 6, 8). A second highly flexible part of Med-PIC is the CAK module of TFIIH. It has been localized to the end of the hook domain of Mediator, directly above core TFIIH (cTFIIH), to which it remains flexibly tethered (7, 8). However, the orientation of the individual subunits within this density has not yet been determined.
In this study, we present the structure of the human Med-PIC assembled on a closed promoter DNA construct, with the TBP subunit of TFIID replacing the full TFIID complex. Human Mediator is held together by a central scaffold subunit, Med14, which forms two contact sites with MedTail. The precise orientation of the CAK module within Med-PIC is revealed, with clear density for the Pol II CTD in the active site. A second CTD binding site between MedHead and MedMiddle shows how Mediator positions the rest of the CTD for phosphorylation by CDK7. Many regions of Mediator that interact with transcription factors are flexibly tethered, facilitating its assembly. The structure also provides key insights into the conformational landscape of Mediator relative to the PIC.
Structural characterization of the human Med-PIC
The Med-PIC complex was assembled by extending our previous protocol for assembling the closed complex PIC from purified factors to accommodate the addition of Mediator (Fig. S1A) (12). In contrast to previous protocols where factors were added in a stepwise manner, three subcomplexes, DNA-TBP-TFIIB-TFIIA, Pol II-TFIIF, and TFIIE-TFIIH-Mediator, were first assembled and were next incubated together. Negative stain electron microscopy (EM) of assembled complexes indicated that a subset of particles contained all components of Med-PIC and that significant conformational heterogeneity exists (Fig. S1B).
A cryo-EM data set was collected, and 2D classification in Relion-3 showed many classes representing the full complex (Fig. S1C, Table S1) (20). A subset of 156,383 particles refined to a resolution of 4.8 Å, but due to the high intrinsic flexibility of Med-PIC, distal regions including MedMiddle, MedTail, and TFIIH, are significantly averaged out in the post-processed map. Focused refinements on subcomplexes were used to improve the resolution of all portions of the density compared to the full complex (Fig. S2, S3). These regions were chosen because either the subcomplex behaves like a rigid body within the full complex, as is the case for the core PIC (cPIC), cTFIIH, MedHead, MedTail, and MedMiddle-CAK, or to center a region within the box to improve its resolution, as in the case of Med1 and Med14C. These refinements improved the resolution of the vast majority of MedTail, Med14, MedHead, and Pol II to 3.5 Å or better (Fig. S3) and that of flexible regions, including Med1, MedMiddle-CAK, and cTFIIH, to 5.8, 6.5, and 7.1 Å, respectively. These improvements allowed the building, refining, or flexible fitting of atomic models for nearly the entire complex (Fig. 1A–B, S4–7, Video S1, Table S2). Overall, the structure of Med-PIC is highly similar to previous human PIC complexes and yeast Med-PIC complexes (Fig. S8) (7, 8, 12). The presence of Mediator does not cause significant changes in the structures of Pol II or the GTFs, including TFIIB, TBP, TFIIA, TFIIE, and TFIIF. Med-PIC is compatible with the incorporation of TFIID as no clashes are observed upon superimposing the structure of TFIID-TFIIA-DNA (Fig. S8) (21).
Fig. 1.
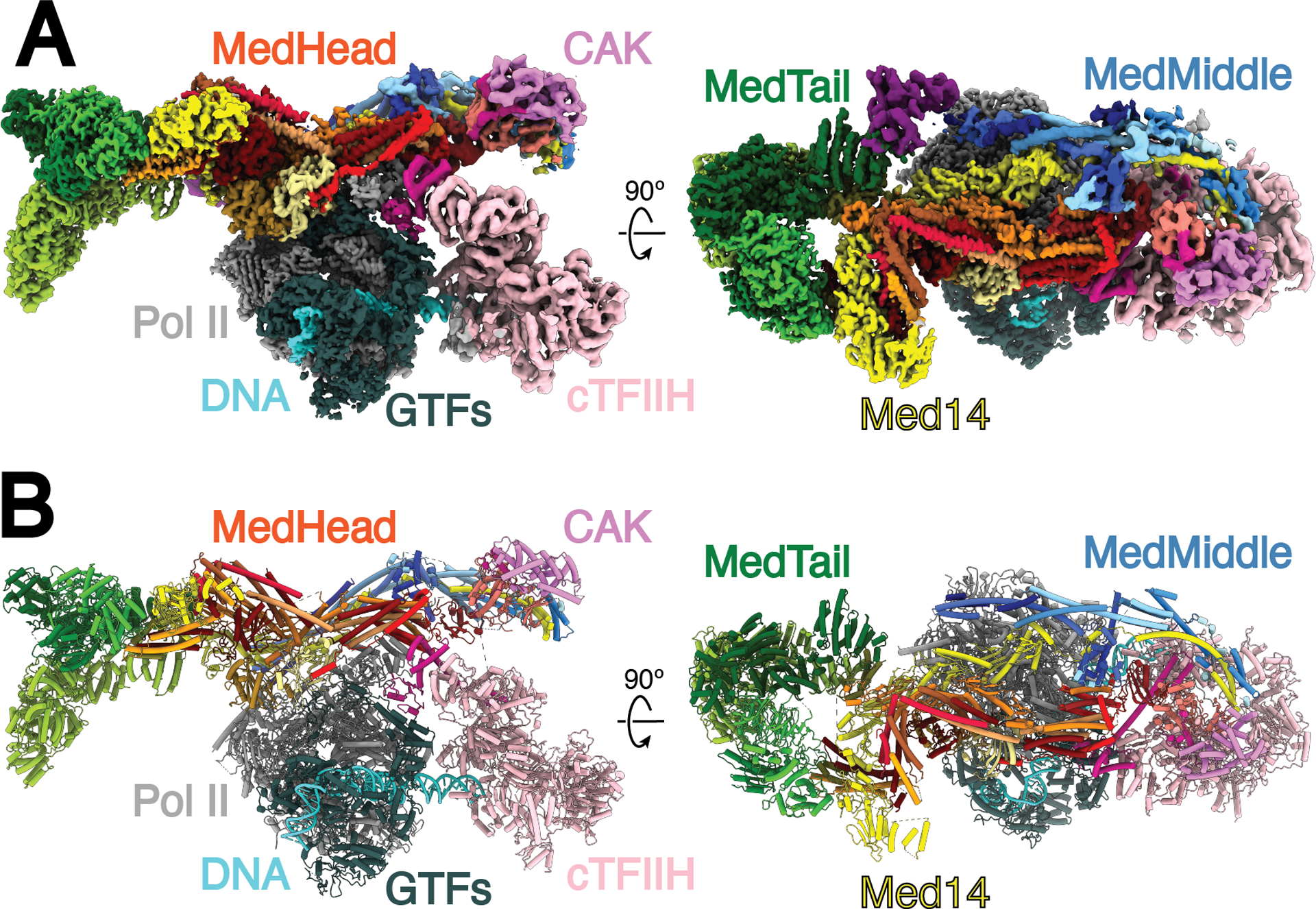
Structure of the human Mediator-bound pre-initiation complex. A) Composite density map for Med-PIC built from the focused refinement maps for cPIC, cTFIIH, MedHead, MedMiddle-CAK, Med14C, MedTail, and Med1. The colors of the subunits will be repeated throughout the manuscript. B) Model of the human Mediator-bound pre-initiation complex. Gray, Pol II; Dark Gray, general transcription factors; Pink, TFIIH core; Salmon, CDK7; Violet, cyclin H; Medium Violet Red, Mat1; Cyan, DNA, Reds, MedHead; Blues, MedMiddle; Yellow, Med14; Greens, MedTail.
Structure of human Mediator
The human Mediator complex within Med-PIC is divided into three modules, held together by the central Med14 scaffold subunit (Fig. 2). MedMiddle closely resembles the structure of its yeast counterpart (7, 8). Homology models for the human MedMiddle subunits Med4, 7, 9, 10, 19, 21, and 31, based on the S. cerevisiae ortholog structures, were built using the MedMiddle-CAK map (Fig. 2A, S4). The N-terminal 200 residues of Med14 were modeled similarly. Additional density near the connector domain of MedMiddle could be assigned to Med26, a metazoan-specific subunit, that has been shown to localize in this part of Mediator and interact with Med4, 7, and 19 (Fig. 2A) (15). The C-terminus of Med26 is sufficient to interact with Mediator, strongly suggesting the C-terminus of Med26 is what can be seen, leaving the N-terminus flexibly attached. The N-terminus has been shown to interact with the super elongation complex (SEC), which is responsible for the release of paused Pol II through phosphorylation of the Pol II CTD and SPT5 by CDK9 (22). Additional unmodeled density attributed to the N-terminus of Med1 is located between the plank domain (Med4 and Med9) and MedTail subunit Med24 (Fig. 2A, S9A). This is consistent with the location of Med1 shown in both yeast and humans previously (15). Density for the plank, Med1, and the N-terminus of Med24 is significantly worse than surrounding areas, indicating that this portion of Mediator moves independently of MedMiddle and MedTail. Previous structures of yeast Med-PICs show interactions between Med9 and the foot domain of Pol II (Fig. S10A–B) (6, 7). In S. pombe, Med4 and Med9 also interact with Med1, but there is no change in the overall structure compared to S. cerevisiae, where Med1 was not included during complex assembly. The contact between Med9 and the foot domain of Pol II is broken in the human Med-PIC. Instead, Med9 is very close to RPB8, and the interactions between Med4 and Med9 with Med1 are retained (Fig. S10C). These differences are likely driven by the presence of the larger MedTail in the human Med-PIC, which positions Med1 further away from the plank through interactions with Med24.
Fig. 2.
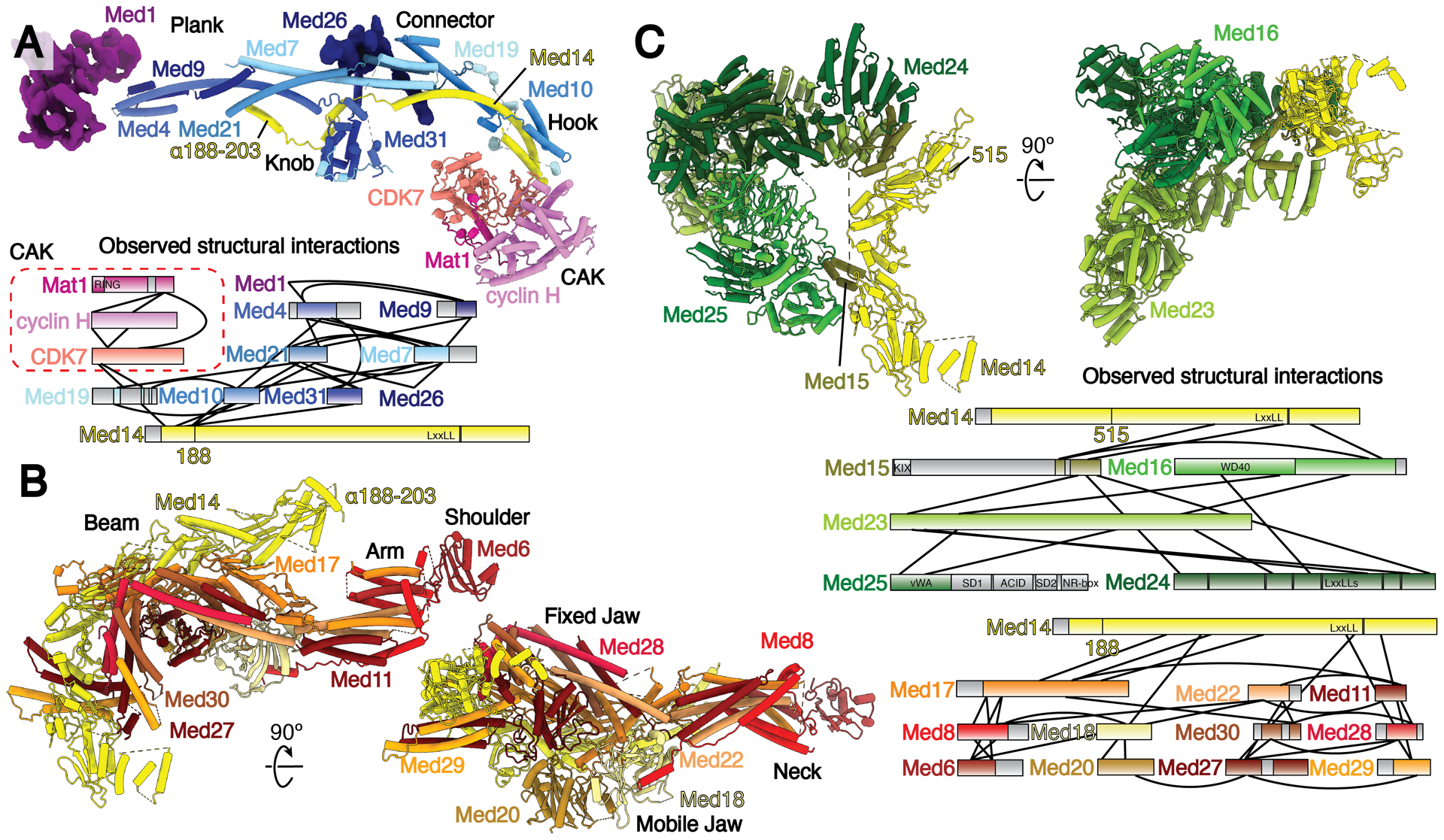
Models and observed structural interactions for human Mediator. (A-C) Model and observed structural interaction diagram for MedMiddle and the CAK module of TFIIH (A), MedHead (B), and MedTail (C). The N-terminus of the scaffold subunit Med14 extends the length of MedMiddle. Putative density for Med1 and Med26 are shown and colored purple and dark blue, respectively. The C-terminus of Med14 forms extensive interactions with MedHead. MedTail also interacts with the C-terminus of Med14, but on the opposite face. Portions for which models were built are shown in color; unmodeled sections are shown in gray. Known domains are shown with a light-to-dark (top-to-bottom) gradient. Everything else is shown with a dark-to-light gradient. Models colored as in Fig. 1.
MedHead adopts a very similar structure to the yeast model except for the presence of the additional subunits Med27, 28, 29, and 30 (Fig. 2B, S5). These subunits, which were assigned previously to either MedHead or MedTail (15, 23), exhibit extensive interactions with the fixed jaw of MedHead and were therefore assigned to MedHead. The C-terminus of the scaffold subunit Med14 extends the RM1 and RM2 repeats visible in yeast structures and wraps around MedHead, serving as a clear divider between MedHead and MedTail. Med17, a scaffold subunit within MedHead, stabilizes the fixed jaw on one face and interacts with the RM1 and RM2 repeats of Med14 on the other (Fig. 2B, S9B).
MedTail connects to the rest of Mediator through two relatively small interfaces with MedHead and Med14. Two C-terminal domains of Med15 are crucial for forming both interfaces. The first contact site is located near the C-terminus of Med14. Two helices each from Med27 and Med29 project underneath Med14, with two helices of Med15 (residues 617–649) wedged between them (Fig. S9C). A concave surface on Med16 makes contact with both this site and Med14. The second site is formed by a C-terminal extension between β18 and β19 (residues 596–620) of Med17 that interacts with the Ring-WD40-DEAD domain (RWD) of Med15 (residues 674–692) (Fig. S5, S9D). The RWD domain of Med15 is wedged in a large cavity between Med23 and Med24.
The rest of MedTail is formed by subunits Med16, Med23, Med24, and Med25. Med16 is divided into N-terminal and C-terminal domains, with the N-terminus forming a 7-blade WD-40 domain and the C-terminus forming a mostly helical domain that constitutes much of the first interface with MedHead described above (Fig. S7, S9B). The N-terminus of Med24 interacts with Med1 and is much more flexible than the rest of MedTail. We could only identify a single domain of Med25, the von Willebrand factor type A (vWA) domain, wedged in a pocket formed by Med16 and Med23 (Fig. S9E).
Almost all domains that are bound by transcription factors in Mediator, including the N-terminus of Med15, the N-terminus of Med25, and the C-terminus of Med1, are flexibly attached to the main body and not visible in the density map (Fig. 3). The first visible portion of Med15 is located underneath MedTail, near the upstream DNA, allowing its N-terminus to easily engage with DNA-bound transcription factors. The C-terminus of Med1 contains the NR-boxes important for nuclear receptor (NR) binding (24). Many NRs also bind to a C-terminal fragment (1147–1454) of Med14 (25–27). These two binding interfaces for NRs are quite far from each other (Fig. 3). The NR AF-1 and AF-2 domains that mediate these interactions are at opposite ends of NR sequences, suggesting that NRs might have to stably associate with the full complex to bridge these two interfaces.
Fig. 3.
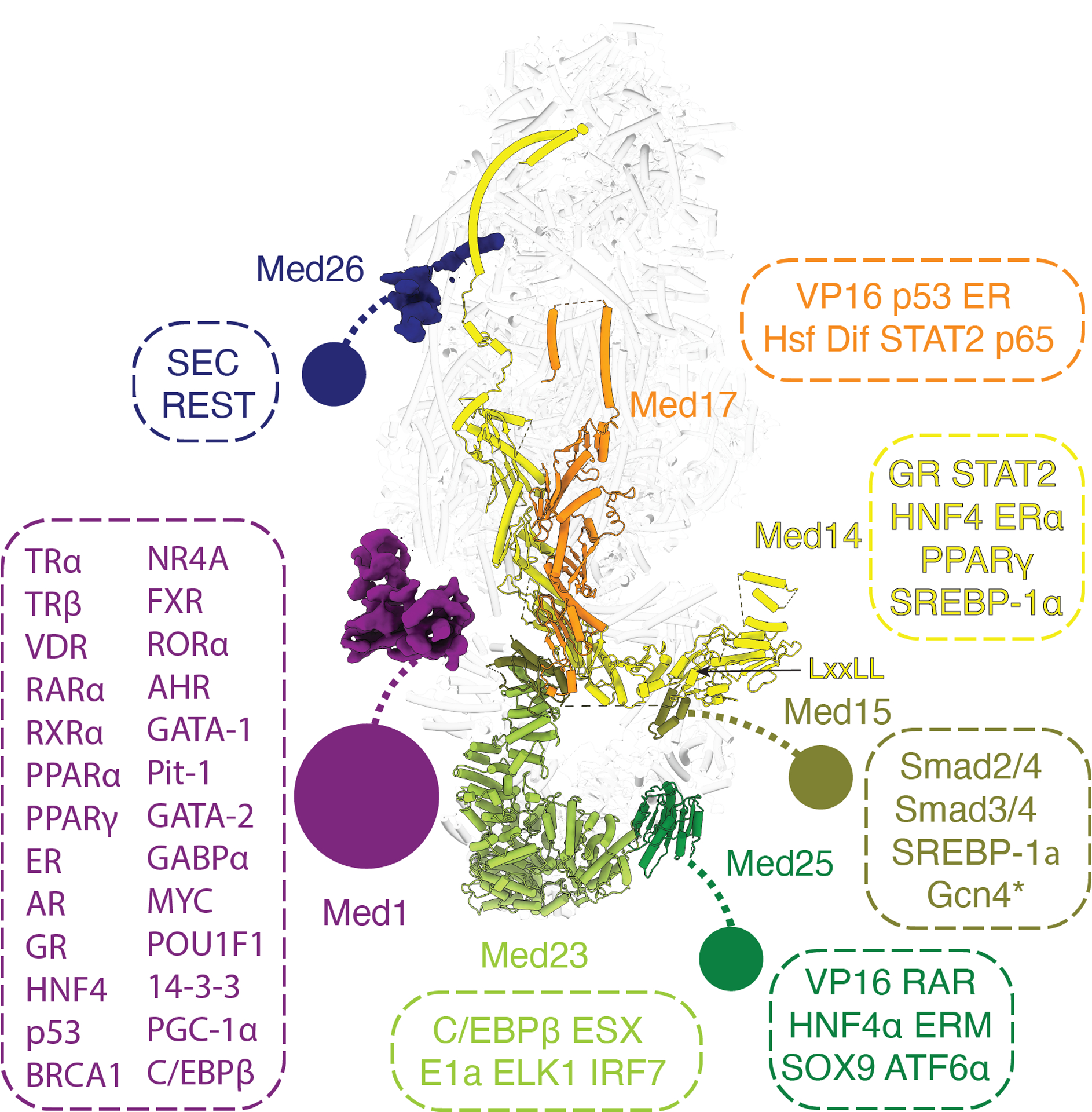
Location of Mediator domains and subunits that interact with transcriptional activators or elongation factors. Flexible tethered domains are indicated by solid circles connected by dashed lines. All interactions shown are between human factors except Gcn4 which is from yeast and indicated by an asterisk.
The VP16 activation domain (AD) used to purify Mediator for this study binds to the ACID domain located at the N-terminus of Med25 (28, 29). The VP16 AD appears to stay bound to Mediator during complex assembly. Due to the absence of density of the ACID domain bound to the VP16 AD in this structure, we can conclude that the ACID domain remains flexibly tethered upon activator binding. It has been hypothesized that conformational changes following activator binding to Mediator could lead to the activation of Med-PIC (30–32). Given that so many of the activator-binding domains within Mediator are flexibly tethered to the main body, it is unlikely that this is a universal mechanism for activating Med-PIC for transcription.
The overall architecture of Mediator appears highly conserved based on recent structures of mouse and Chaetomium thermophilum Mediator (33, 34). The putative locations of Med1 and Med26 described earlier are in agreement with the mouse structure. The loss of subunits present in other species, including Med27, Med28, Med29, Med30, Med23, and Med24, appears to increase the flexibility of MedTail in the Chaetomium thermophilum structure.
Mediator stabilizes the CAK module of TFIIH
While previous structural studies of Med-PICs established that the CAK module of TFIIH occupies a position between the shoulder and hook domains of Mediator, the position and orientation of each CAK module subunit could not be determined (6–9). Rigid body docking of the human CAK module structure into our density led to an unambiguous orientation of the CAK module with the active site of CDK7 facing the hook domain of MedMiddle (Fig. 4A) (35). Mediator stabilizes the CAK module through interactions involving Med6, the N-terminus of Med14, and a small fragment of Med19 (~133–148) with CDK7 (Fig. 4A). This orientation of the CAK module positions the C-terminus of Mat1 ~50 Å from the N-terminus bound to cTFIIH, a distance easily spanned by the small fragment of Mat1 (211–243) missing in the structure (Fig. 4B).
Fig. 4.
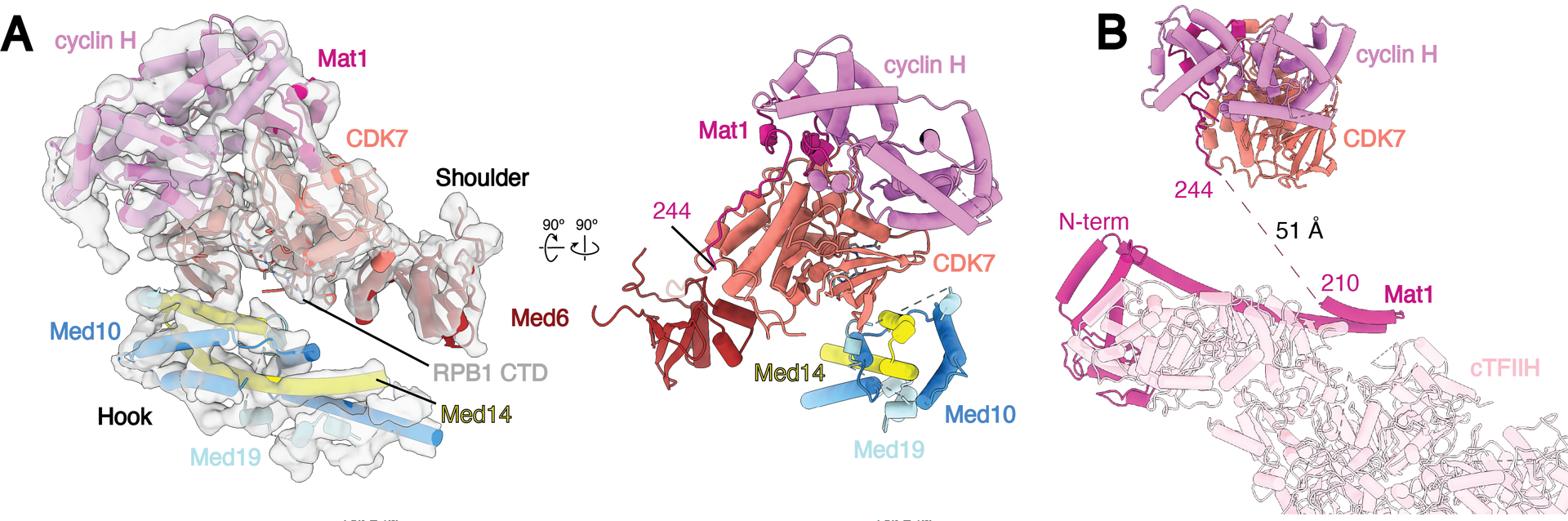
Structure of TFIIH within Med-PIC A) Docking of the CAK module (CDK7, cyclin-H, and Mat1) within the MedMiddle-CAK density. The CAK module of TFIIH is stabilized in the Med-PIC by interactions between CDK7 and Med6, the N-terminus of Med14, and a small fragment of Med19. B) The model of the complete human TFIIH complex places the two modeled segments of Mat1 (1–210, 244–308) close to each other. The missing 34 residues can easily span the 51 Å distance between the termini. Models are colored as in Figure 1.
CDK7 adopts the active conformation seen in the human CAK module structure, with the T-loop projecting towards Mat1 and away from the active site (Fig. 5A–B) (35). Clear electron density in the CDK7 active site closely matches the location of the substrate peptide in the CDK2-cyclin-A-substrate peptide complex (36) (Fig. 5A–C). This peptide shares the identical serine-proline sequence that is found in the RPB1 CTD targeted by the kinase. Therefore, we built a model for the RPB1 CTD in the active site that we designate as CDKCTD.
Fig. 5.
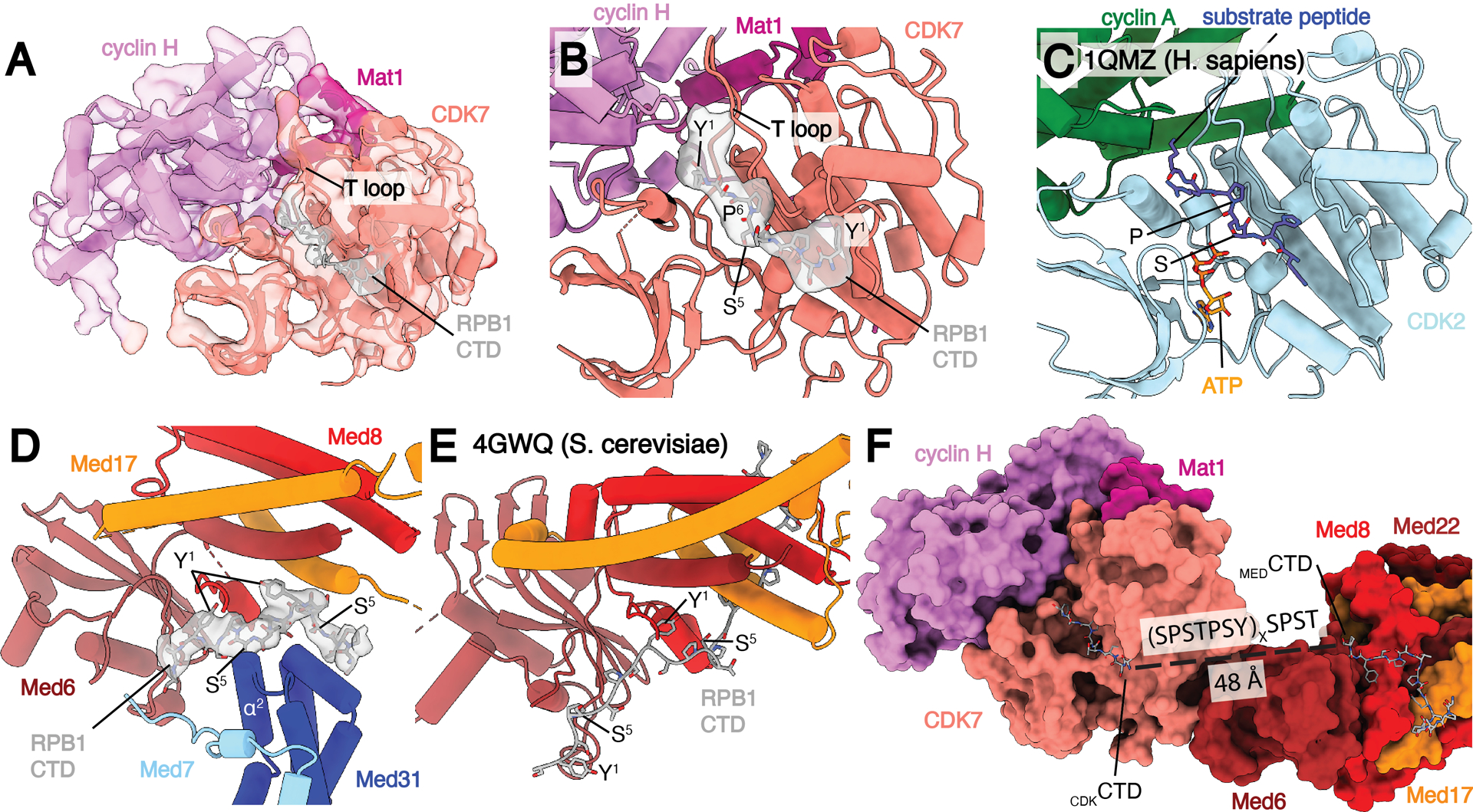
Location of RPB1 CTD binding in Med-PIC. A) Structure of the TFIIH CAK module. Segmented map of MedMiddle-CAK shows clear density representing an active conformation of the T-loop of CDK7 and density for Pol II CTD in the active site of CDK7. B) Model of the CAK module with density observed for the CDKCTD in the active site. A consensus sequence of the Pol II CTD is modeled due to limited resolution. The T-loop is in the extended, active conformation. C) Model of the CDK2-cyclin A-substrate peptide structure shows high similarity to the CAK module structure with the conserved SP motif that is common to substrates of both enzymes. D) Model and density of MEDCTD with interacting subunits of MedHead and MedMiddle. S5 makes close contacts with α2 of Med31, preventing binding of phosphorylated repeats in this location. E) Model of MEDCTD in the yeast MedHead crystal structure shows a more extensive interface between MEDCTD and MedHead than in the Med-PIC, likely due to the presence of MedMiddle in the Med-PIC. F) View of CDKCTD and MEDCTD within the human Med-PIC structure. Based on the directionality of the CTD, CDKCTD is C-terminal to MEDCTD, and the gap between them would require at least two repeats of the CTD. MedMiddle is hidden for easier visibility. Models are colored as in Figure 1. Annotated domains of Mediator are labeled in black.
S. cerevisiae MedHead (scMedHead) was co-crystallized with a short peptide of the RPB1 CTD, which shows slightly more than three full repeats engaged with scMedHead at the shoulder and neck domains (17). We observe additional electron density in this same location and used the S. cerevisiae structure to build a model for this portion of the CTD that we will refer to as MEDCTD (Fig. 5D–E). MEDCTD is 16 residues long, slightly more than two full repeats, and adopts a somewhat different path than the yeast structure, likely due to the presence of Med31, which interacts with the other side of MEDCTD. In scMedHead, the elongated structure of the N-terminal portion of MEDCTD forms extensive interactions with Med17. In contrast, we see clear density for MEDCTD starting to wrap around Med31. The C-terminal end of MEDCTD also does not form as extensive of an interface with Mediator as in scMedHead, due to a clash with the Med7 N-terminus. Experiments in S. pombe show that the CTD is necessary for interaction between MedHead and Pol II in vitro, suggesting that MEDCTD is critical for this interaction (8). MEDCTD binding to Mediator would likely be disrupted following phosphorylation of Ser5 due to close interactions between Ser5 and the end of Med31 helix α2 (Fig. 5D).
The directionality of MEDCTD and CDKCTD is the same, with the N-terminal end of MEDCTD pointing towards Pol II and the C-terminal end of CDKCTD leading away from Med-PIC (Fig. 5F). This observation strongly suggests that MEDCTD is N-terminal to CDKCTD within the full CTD sequence. The distance between the termini of those two CTD fragments is 48 Å. In an elongated state, one repeat of the CTD can span approximately 25 Å (37), so while two repeats of the CTD may be sufficient to bridge that gap, we would likely see better-defined density for the CTD in that case. Therefore, we suspect that three or more repeats are likely looped out between MEDCTD and CDKCTD.
The role of MEDCTD binding is likely to capture the CTD and position it in the correct direction and close to the active site of CDK7 to facilitate pS5 formation. Mass spectrometry experiments with both yeast and human complexes show that pSer5 can be found within any repeat of the CTD except the final repeat (38, 39). However, the phosphorylation patterns of individual CTD peptides and the direction that sequential phosphorylation can occur remain unknown. Two possibilities exist for the direction of sequential phosphorylation that generates different outcomes (Fig. 6). If the CTD is phosphorylated in a C- to N-terminal direction, binding at MEDCTD precedes phosphorylation, and it is not clear how Pol II would dissociate from Mediator given that the CTD is threaded through a hole in Mediator formed by the hook, knob, and shoulder domains and the CAK module of TFIIH. Phosphorylated repeats would also be located far from the nascent RNA that needs to be capped.
Fig. 6.
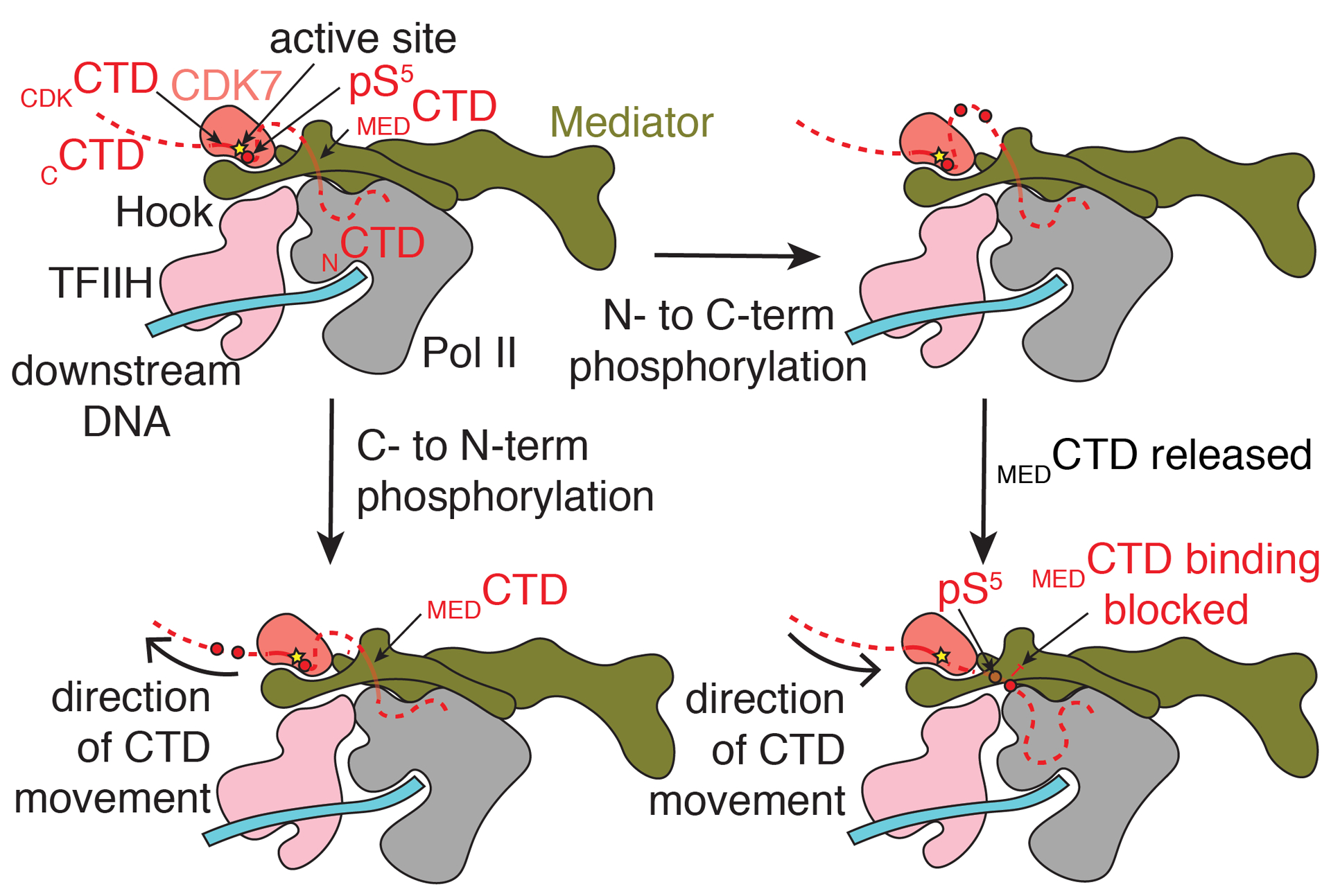
Model for phosphorylation of the Pol II CTD by CDK7. MEDCTD binding positions the rest of the CTD in the CDK7 active site. Following phosphorylation, indicated by a red circle, translocation of the CTD towards the N-terminus (bottom) would place phosphorylated repeats further from the nascent RNA emerging from Pol II. Separation of Mediator and Pol II would be difficult without separation of the CAK module and Mediator. Translocation of the CTD towards the C-terminus would position phosphorylated repeats to block binding of the CTD at MEDCTD, a possible way to favor disassembly of Med-PIC. Phosphorylated repeats would also be significantly closer to the RNA exit tunnel of Pol II to recruit the capping complex properly. CAK = cyclin-activated kinase module; CTD = C-terminal domain of RPB1; pS5 = phosphorylated serine 5 residue (red circle).
If the CTD is phosphorylated in an N- to C-terminal direction, C-terminal phosphorylated repeats would not be able to bind at MEDCTD due to steric clashes that would arise with the added phosphates. Given that the CTD is important for Pol II-Mediator interaction and phosphorylation of the CTD leads to dissociation of Pol II and Mediator, we find this mechanism more likely (8, 40). Separation of MedHead and Pol II would place the phosphorylated CTD close to the nascent RNA for capping to occur.
Given the large movements of MedMiddle and the CAK module of TFIIH relative to the PIC, we speculate that these conformational changes play an important role in the sequential phosphorylation of the CTD. The intrinsic flexibility of Mediator has been linked to the opening and closing of the MEDCTD binding site on Mediator (5, 6), and if this movement is tied to binding and release of the CTD at MEDCTD, it could also facilitate the progression of CDK7 along the CTD.
Supplementary Material
Acknowledgments:
We thank past and present lab members for advice, assistance, and comments on the manuscript. We thank Jason Pattie for computer support. We thank Janette Meyers, Rose Marie Haynes, and Harry Scott at the PNCC for data collection support. We are grateful to Amy Rosenzweig, Ishwar Radhakrishnan, and Jason Brickner for helpful discussion and comments on the manuscript. We thank the staff at the Structural Biology Facility (SBF) of Northwestern University for technical support.
Funding:
This work was supported by a Cornew Innovation Award from the Chemistry of Life Processes Institute at Northwestern University (to Y He), a Catalyst Award by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to Y He), an Institutional Research Grant from the American Cancer Society (IRG-15-173-21 to Y He), an H Foundation Core Facility Pilot Project Award (to Y He), and a Pilot Project Award under U54-CA193419 (to Y He). Y He is supported by R01-GM135651 and P01-CA092584 from the NIH. R Abdella and A Talyzina are supported by the Molecular Biophysics Training Program from NIGMS/NIH (T32-GM008382). A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. R Tjian is a Howard Hughes Medical Institute Investigator. This work used the Sapphire imager from the Northwestern University Keck Biophysics Facility funded by NIH grant 1S10OD026963-01, as well as the resources of the Northwestern University Structural Biology Facility, which is generously supported by the NCI CCSG P30 CA060553 grant awarded to the Robert H. Lurie Comprehensive Cancer Center.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: Electron density maps and coordinates for the Med-PIC have been deposited in the Electron Microscopy Data Bank (EMDB) with ID code EMDB-23255 and the Protein Data Bank (PDB) with ID code 7LBM, respectively. Electron density maps for the focused refinements on cPIC, cTFIIH, MedHead, MedMiddle-CAK, Med14C, MedTail, and Med1 have been deposited in the EMDB with ID codes EMDB-23256, EMDB-23257, EMDB-23258, EMDB-23259, EMDB-23260, EMDB-23261, and EMDB-23262, respectively.
References and Notes:
- 1.Thomas MC, Chiang C-M, The general transcription machinery and general cofactors. Critical reviews in biochemistry and molecular biology 41, 105--178 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Soutourina J, Transcription regulation by the Mediator complex. Nature Publishing Group 34, 1--13 (2017). [Google Scholar]
- 3.Nogales E, Louder RK, He Y, Structural Insights into the Eukaryotic Transcription Initiation Machinery. Annual review of biophysics 46, 59--83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ, Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol 9, e1000603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Khattabi L et al. , A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 178, 1145--1158.e1120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai K-L et al. , Mediator structure and rearrangements required for holoenzyme formation. Nature 142, 749--201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilbach S et al. , Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature Publishing Group 551, 204--209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson PJ et al. , Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 166, 1411--1422.e1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plaschka C et al. , Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P, A structural perspective of CTD function. Genes Dev 19, 1401--1415 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Ghosh A, Lima CD, Enzymology of RNA cap synthesis. Wiley Interdisciplinary Reviews: RNA 1, 152--172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y et al. , Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359--365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair D, Kim Y, Myers LC, Mediator and TFIIH govern carboxyl-terminal domain-dependent transcription in yeast extracts. Journal of Biological Chemistry 280, 33739--33748 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Esnault C et al. , Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Molecular Cell 31, 337--346 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Tsai K-L et al. , Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430--1444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrenko N, Jin Y, Wong KH, Struhl K, Mediator Undergoes a Compositional Change during Transcriptional Activation. Molecular Cell 64, 443--454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD, Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America 109, 17931--17935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaschka C, Nozawa K, Cramer P, Mediator architecture and RNA polymerase II interaction. Journal of molecular biology 428, 2569--2574 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Cevher MA et al. , Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol 21, 1028–1034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakane T, Kimanius D, Lindahl E, Scheres SHW, Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louder RK et al. , Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604--609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi H et al. , Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92--104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeronimo C, Robert F, The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends in Cell Biology, 1--19 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Belorusova AY et al. , Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer. Nucleic acids research 48, 11199–11213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ, Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J 18, 5380–5388 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik S, Wallberg AE, Kang YK, Roeder RG, TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol 22, 5626–5637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S, MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol 30, 2155–2169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojnic E et al. , Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nature structural \& molecular biology 18, 404--409 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Milbradt AG et al. , Structure of the VP16 transactivator target in the Mediator. Nature structural \& molecular biology 18, 410--415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer KD, Lin S.-c., Bernecky C, Gao Y, Taatjes DJ, p53 activates transcription by directing structural shifts in Mediator. Nature structural \& molecular biology 17, 753--760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R, Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058--1062 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Bernecky C, Taatjes DJ, Activator-mediator binding stabilizes RNA polymerase II orientation within the human mediator-RNA polymerase II-TFIIF assembly. Journal of molecular biology 417, 387–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H et al. , Structure of mammalian Mediator complex reveals Tail module architecture and interaction with a conserved core. Nature Communications 12, 1355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H et al. , Mediator structure and conformation change. Mol Cell, (2021). [DOI] [PubMed] [Google Scholar]
- 35.Greber BJ et al. , The cryoelectron microscopy structure of the human CDK-activating kinase. Proceedings of the National Academy of Sciences of the United States of America 117, 22849–22857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown NR, Noble ME, Endicott JA, Johnson LN, The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nature cell biology 1, 438–443 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Jasnovidova O, Stefl R, The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA 4, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Suh H et al. , Direct Analysis of Phosphorylation Sites on the Rpb1 C-Terminal Domain of RNA Polymerase II. Molecular Cell 61, 297--304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schüller R, Forn, Heptad-Specific Phosphorylation of RNA Polymerase~II CTD. Molecular Cell 61, 305--314 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Sogaard TM, Svejstrup JQ, Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem 282, 14113–14120 (2007). [DOI] [PubMed] [Google Scholar]
- 41.He Y, Fang J, Taatjes DJ, Nogales E, Structural visualization of key steps in human transcription initiation. Nature 495, 481--486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taatjes DJ, Tjian R, Structure and function of CRSP/Med2: a promoter-selective transcriptional coactivator complex. Molecular Cell 14, 675--683 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41--60 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Patel A, Toso D, Litvak A, Nogales E, Efficient graphene oxide coating improves cryo-EM sample preparation and data collection from tilted grids. bioRxiv, 2021.2003.2008.434344 (2021). [Google Scholar]
- 45.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Ogura T, Iwasaki K, Sato C, Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. Journal of Structural Biology 143, 185--200 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Lander GC et al. , Appion: An integrated, database-driven pipeline to facilitate EM image processing. Journal of Structural Biology 166, 95--102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B, DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. Journal of Structural Biology 166, 205--213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N, “CTFFIND4: Fast and accurate defocus estimation from electron micrographs,” (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G et al. , EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38--46 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1--12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henderson R et al. , Outcome of the first electron microscopy validation task force meeting., 205--214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Garcia R et al. , DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. bioRxiv, 2020.2006.2012.148296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Goddard TD et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Miwa K et al. , Crystal Structure of Human General Transcription Factor TFIIE at Atomic Resolution. Journal of molecular biology 428, 4258–4266 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Liebschner D et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann L et al. , A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. Journal of molecular biology 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Drozdetskiy A, Cole C, Procter J, Barton GJ, JPred4: a protein secondary structure prediction server. Nucleic acids research 43, W389–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb B, Sali A, Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 54, 5 6 1–5 6 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lariviere L et al. , Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol 13, 895–901 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Monte D et al. , Crystal structure of human Mediator subunit MED23. Nat Commun 9, 3389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taschner M, Mourao A, Awasthi M, Basquin J, Lorentzen E, Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J Biol Chem 292, 7462–7473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kidmose RT et al. , Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greber BJ, Toso DB, Fang J, Nogales E, The complete structure of the human TFIIH core complex. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


