Legionella pneumophila is a pathogen indigenous to natural and large building water systems in the bulk and the biofilm phases. The immediate environment within a system can impact the tolerance of L. pneumophila to environmental stressors, including copper.
KEYWORDS: hot water system, cooling tower, biofilm, copA, premise plumbing, Legionella pneumophila serogroup 5, whole-genome sequencing
ABSTRACT
In large-building water systems, Legionella pneumophila is exposed to common environmental stressors such as copper. The aim of this study was to evaluate the susceptibility to copper of L. pneumophila isolates recovered from various sites: two clinical and seven environmental isolates from hot water system biofilm and water and from cooling tower water. After a 1-week acclimation in simulated drinking water, strains were exposed to various copper concentrations (0.8 to 5 mg/liter) for over 672 h. Complete loss of culturability was observed for three isolates following copper exposure to 5 mg/liter for 672 h. Two sequence type 1427 (ST1427)-like isolates were highly sensitive to copper, while the other two, isolated from biofilm samples, maintained higher culturability. The expression of the copper resistance gene copA evaluated by reverse transcription-quantitative PCR (RT-qPCR) was significantly higher for the biofilm isolates. All four ST1427-like isolates were recovered from the same water system during an outbreak. Whole-genome sequencing results confirmed that the four isolates are very close phylogenetically, differing by only 29 single nucleotide polymorphisms, suggesting in situ adaptation to microenvironmental conditions, possibly due to epigenetic regulation. These results indicate that the immediate environment within a building water distribution system influences the tolerance of L. pneumophila to copper. Increased contact of L. pneumophila biofilm strains with copper piping or copper alloys in the heat exchanger might lead to local adaptation. The phenotypic differences observed between water and biofilm isolates from the hot water system of a health care facility warrants further investigation to assess the relevance of evaluating disinfection performances based on water sampling alone.
IMPORTANCE Legionella pneumophila is a pathogen indigenous to natural and large building water systems in the bulk and the biofilm phases. The immediate environment within a system can impact the tolerance of L. pneumophila to environmental stressors, including copper. In health care facilities, copper levels in water can vary, depending on water quality, plumbing materials, and age. This study evaluated the impact of the isolation site (water versus biofilm, hot water system versus cooling tower) within building water systems. Closely related strains isolated from a health care facility hot water system exhibited variable tolerance to copper stress, shown by differential expression of copA, with biofilm isolates displaying highest expression and tolerance. Relying on the detection of L. pneumophila in water samples following exposure to environmental stressors such as copper may underestimate the prevalence of L. pneumophila, leading to inappropriate risk management strategies and increasing the risk of exposure for vulnerable patients.
INTRODUCTION
Legionella pneumophila is an opportunistic pathogen that is the causative agent of Legionnaires’ disease and Pontiac fever (1). Between 2013 and 2014, 88% of hospitalizations and 100% of deaths resulting from drinking water-associated outbreaks in the United States were attributed to Legionella infections (2). From 2000 to 2014, a 286% increase in legionellosis cases in the United States was reported, further highlighting the importance of this emerging pathogen (3). Similarly Legionnaires’ disease cases have steadily increased in Europe between 2011 and 2016, with 81% of infections attributed to L. pneumophila serogroup 1 (4). While the average mortality rate associated with legionellosis is estimated to be approximately 8% (4, 5), it can reach 25% in health care-associated outbreaks (6). The transmission of L. pneumophila to humans occurs through man-made water systems (1). Indeed, Legionella is known to proliferate in engineered water systems, such as cooling towers and large-building water distribution systems (7). In health care facilities, where the mortality rates are higher, hot water systems feeding taps and showers have a higher prevalence of L. pneumophila than other Legionella species (8–10). Several factors inherent in large-building water systems are favorable to L. pneumophila persistence: lukewarm water temperature, presence of amoebae, use of disinfectants, stagnation, presence of biofilms, and plumbing materials (7, 11).
The impact of plumbing material on microbial growth and development of biofilm is well recognized (12–14). Similar or increased incorporation and persistence of Legionella were measured by non-culture-based methods in copper-grown biofilms compared to those grown on polyvinyl chloride (PVC), unplasticized PVC (uPVC), or cross-linked polyethylene (PEX) (15–17). In contrast, a study reported a mean reduction of 2.5 log of L. pneumophila (measured by gene copies) in water within copper pipes compared to PEX pipes at temperatures of ≤41°C, while total bacterial gene copies remained unchanged (18). Because of its reported bactericidal effect, copper has long been used as a disinfecting material (19, 20). Furthermore, copper-silver ionization treatment has been reported to reduce levels of L. pneumophila in hospital hot water distribution systems (21). Nevertheless, unreliable eradication of L. pneumophila following copper-silver ionization was linked to legionellosis cases (22–24).
In drinking water, copper levels are regulated mostly for esthetic reasons. Regulatory agencies recommend that copper levels be maintained below 1 to 2 mg/liter (25–27). Health effects on healthy adults were only associated with levels exceeding 4 mg/liter (28, 29). However, the state of California revised their public health goal to 0.3 mg/liter in 2008 (30) in response to reported adverse effects on infants and young children at levels below 1 mg/liter (31).
In health care facilities, copper levels in water carried within copper plumbing can vary between <10 μg/liter and 0.8 mg/liter, depending on water corrosiveness and pipe age (9, 32). These levels are even higher in new copper pipes, where dissolved copper can exceed 5 mg/liter (33). Reports of L. pneumophila’s ability to withstand copper stress are contradictory. L. pneumophila has been recovered from water within copper pipe premise plumbing carrying up to 0.8 mg Cu/liter (8, 9), and culturable L. pneumophila has been observed at levels up to 8 mg/liter in a laboratory study (34). Recovery of culturable L. pneumophila in simulated drinking water distribution systems was observed at copper concentrations between 0.8 mg/liter and 3.12 mg/liter (35, 36). Copper has also been reported to inhibit the growth of L. pneumophila (37). These reported differences in copper effect on L. pneumophila could be related to environmental factors such as water chemistry or L. pneumophila growth phase (36), or to intrinsic factors such as the variable ability of each strain to tolerate copper, underlined by the genetic makeup of each strain or adaptive evolution of tolerance due to previous exposure. Indeed, L. pneumophila strains may behave differently depending on the environment from which they are isolated and/or environmental conditions to which they have adapted. Some bacteria can tolerate high copper concentration in water, notably through the mediation of the expression of specific genes. The only gene known to mediate copper resistance in L. pneumophila is the copper-translocating PIB-type ATPase copA (38, 39). copA is encoded on a 100-kb mobile genetic element whose excision is regulated by the small-RNA (sRNA) binding regulator Hfq (39). Its episomal form is linked to higher expression of copA and, therefore, an increased tolerance to copper.
The main objectives of this study were to (i) evaluate the impact of the isolation site (clinical versus environmental, water versus biofilm, and hot water system versus cooling tower) of L. pneumophila strains on their tolerance to various levels of copper in drinking water over a 1-month exposure time and (ii) assess whether isolates recovered from different locations but identified as closely related strains exhibit variable tolerance to copper stress through differential expression of copA.
RESULTS
The culturability of nine L. pneumophila strains (Table 1) was assessed over a 4-week (672-h) period of exposure to various copper concentration (0.8 to 5 mg/liter). Strains were isolated from clinical or environmental samples. Environmental strains were recovered from water and biofilm of the hot water distribution systems in two health care facilities (HCF-A and HCF-B) and from water in two cooling towers (CT-C and CT-D). All strains isolated from the HCF-B hot water system (1427.B.F, 1427.W.HE, and 1427.B.HE) and a patient (1427.C) were isolated following an outbreak and were found to be closely related by sequence-based typing (sequence type 1427 [ST1427]) and pulsed-field gel electrophoresis (24). No infections were associated with the environmental strains isolated from HCF-A (40) and from CT-C. The environmental strain 62.W was isolated from a cooling tower (CT-D) following a large outbreak of L. pneumophila in Québec city (41). Clinical strain 62.C was isolated from a patient during the outbreak associated with CT-D.
TABLE 1.
Characteristics of L. pneumophila isolates used in this study
| Isolate ID | Isolate type | Serogroup | ST | Sourcea | Sample | Device | Identification codeb | Reference |
|---|---|---|---|---|---|---|---|---|
| JR32 | Laboratory | 1 | JR32 | 72 | ||||
| 67501-2 | Environmental | 1 | 154-like | HCF-A | Water | Faucet | 154.W.F | 40 |
| 87726-5 | Environmental | 2-15 | 378 | HCF-A | Water | Faucet | 378.W.F | 40 |
| 135110 | Clinical | 5 | 1427-like | HCF-B | Clinical | 1427.C | 24 | |
| 76826-3 | Environmental | 5 | 1427-like | HCF-B | Biofilm | Faucet | 1427.B.F | 24 |
| 144141 | Environmental | 5 | 1427-like | HCF-B | Water | Heat exchanger | 1427.W.HE | 24 |
| 144150 | Environmental | 5 | 1427-like | HCF-B | Biofilm | Heat exchanger | 1427.B.HE | 24 |
| 120145 | Environmental | 1 | 284 | CT-C | Water | 284.W | 61 | |
| 120112 | Clinical | 1 | 62 | CT-D | Clinical | 62.C | 61 | |
| 120292 | Environmental | 1 | 62 | CT-D | Water | 62.W | 61 |
HCF-A, health care facility A; HFC-B, health care facility B; CT-C, cooling tower C; CT-D, cooling tower D.
W, water; C, clinical; B, biofilm; F, faucet; HE, heat exchanger.
Prior to copper exposure, strains were adapted in simulated drinking water for 1 week at room temperature, to be representative of temperatures within building plumbing. Following adaptation, the assay was conducted at 36°C, representative of water temperature in temperature-controlled faucets (electronic, foot operated, or thermostatic mixing valve) (42, 43). In order to separate the decline of L. pneumophila culturability due to starvation and due to copper exposure, a control without copper and maintained at 36°C for the duration of the assay was monitored for each strain (Table S1). A general reduction in culturability was observed with increasing copper concentrations (Fig. 1).
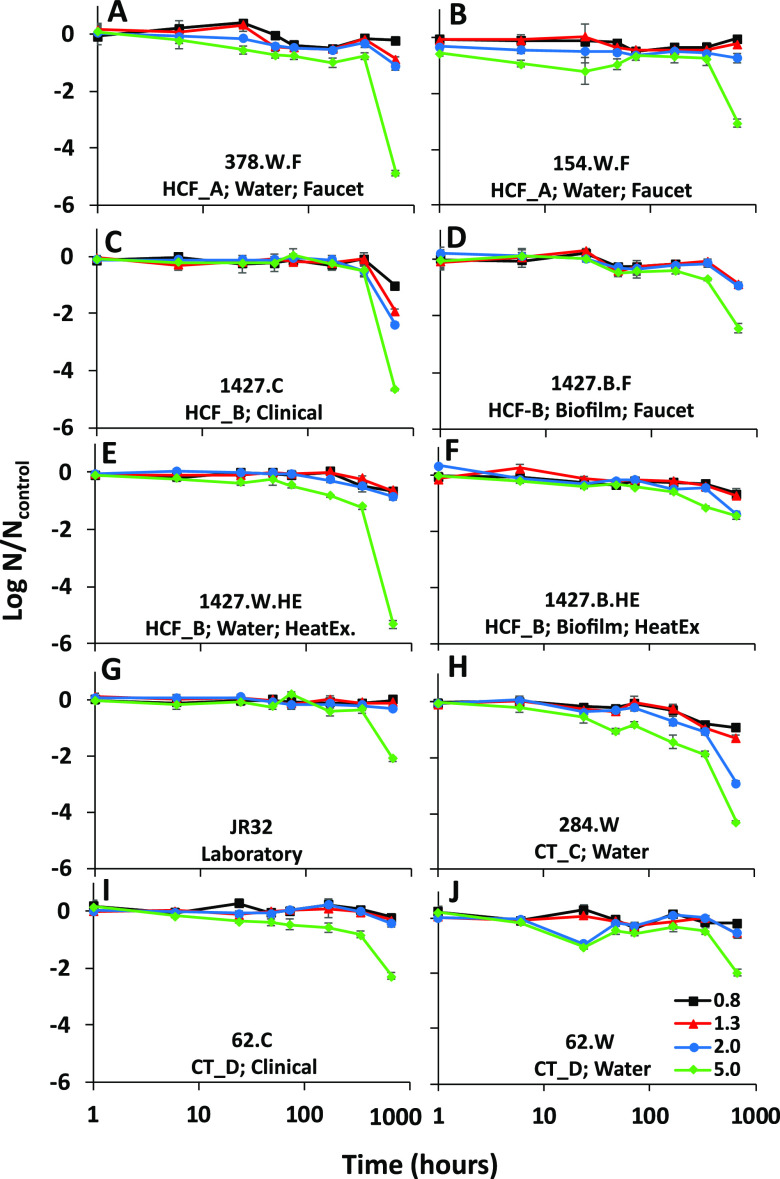
FIG 1.
Survival of L. pneumophila isolates in copper-treated simulated drinking water relative to an untreated control in simulated drinking water. L. pneumophila strains were incubated with different copper concentrations (0.8, 1.3, 2, and 5 mg/liter) for a period of 1 to 672 h. The log reduction in CFU counts was calculated relative to the untreated control for each strain. Results are grouped according to the origin of each isolate. (A and B) Hot water system in health care facility A; (C to F) hot water system in health care facility B; (G) laboratory strain JR32; (H) cooling tower C; (I and J) cooling tower D. Error bars represent standard deviations (n = 3).
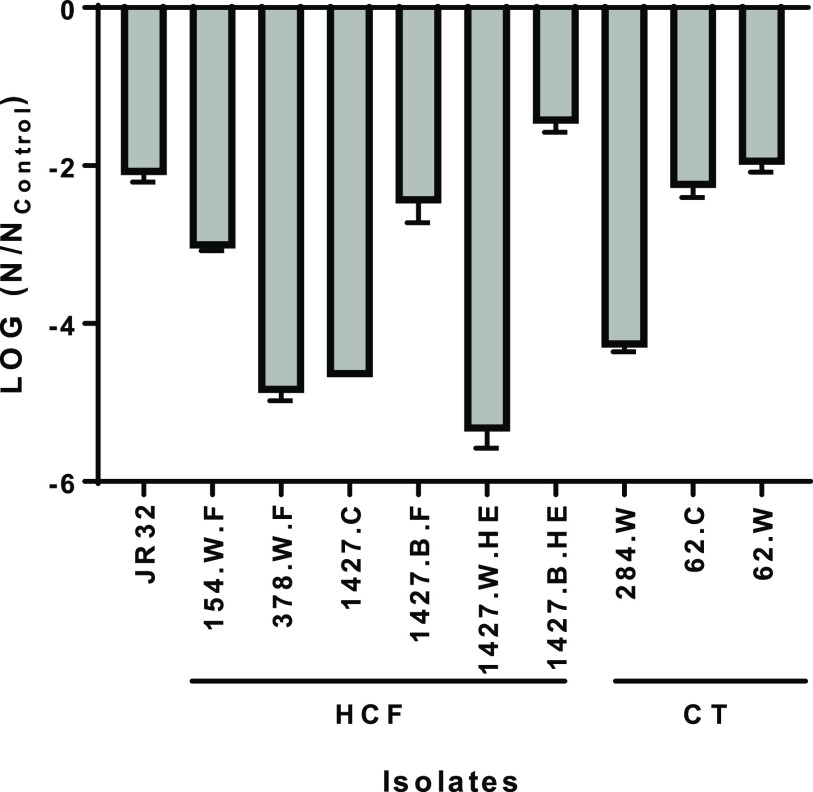
There was no notable change at a concentration of 0.8 mg/liter, with no more than a 1-log reduction compared to the control after 672 h. Similar results were obtained at a concentration of 1.3 mg/liter, except for strain 1427.C, for which a 1.9-log reduction was observed after 672 h. A partial loss of culturability was also observed after 672 h at 2-mg/liter copper concentrations for 1427.C and 284.W strains, with 2.4- and 2.9-log reductions, respectively. The impact on culturability was clearly observed at 5 mg/liter: most isolates showed a slow but steady decline over time, with a sharp drop in culturability between 336 h and 672 h (Fig. 1). Copper tolerance varied greatly between strains after 672 h of exposure (Fig. 2). When isolate pairs were compared by a Kruskal-Wallis analysis, the log reduction was significantly different for 15 of the 45 pairs analyzed (Table S2). The four strains identified as belonging to ST1427 and isolated from the same health care facility clearly displayed different tolerance to copper (Fig. 2). Of note, the strain isolated from the heat exchanger biofilm (1427.B.HE) was significantly more tolerant to copper than the strain isolated from the heat exchanger water (1427.W.HE), (P = 0.0002).
FIG 2.
Log reduction of laboratory, environmental, and clinical L. pneumophila isolates after exposure to 5 mg/liter copper for 672 h in simulated drinking water. The log reduction in CFU counts is expressed as a ratio relative to the untreated control for each strain. Error bars represent standard deviations (n = 3).
Viability assays using flow cytometry were conducted in parallel to CFU counts. For the control without copper, the percent viable cells remained stable, with a mean percent viability of 88% ± 11% from the start of the experiment until 672 h (Table S3). For samples with copper, interference between copper and the dye (thiazole orange) was observed. Addition of EDTA to the samples prior to flow cytometry did not solve this issue. Therefore, we were unable to perform viability assays on the copper-treated samples. It is possible that the strains differ in their ability to stay viable after copper treatment.
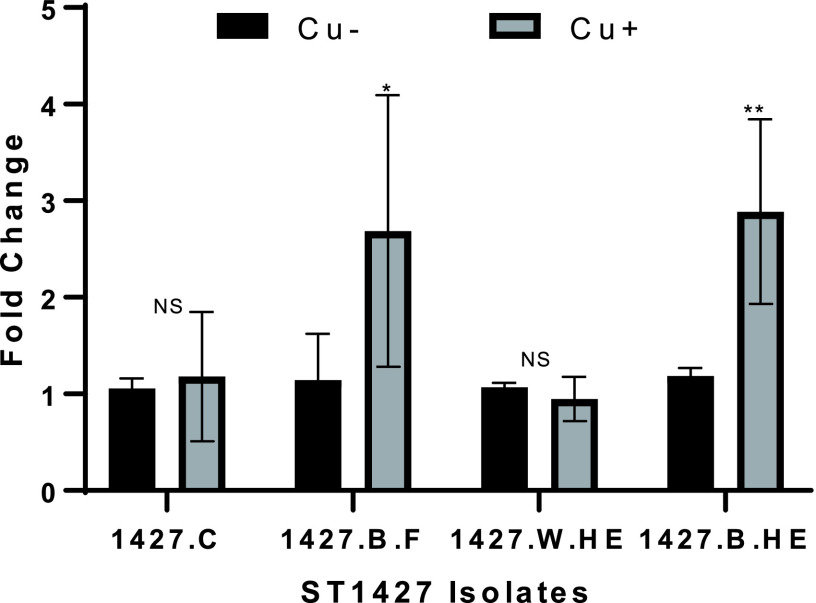
Since the four strains belonging to the same ST displayed variable tolerance to copper, a more detailed investigation was conducted to test if the expression of copA gene was comparable between the isolates. RT-qPCR was used to test expression of copA after 30 min exposure to copper. Strains 1427.B.F and 1427.B.HE induced strong expression of copA after copper exposure, while 1427.HE and 1427.C did not (Fig. 3). The results for expression of copA are in agreement with the resistance of the strains to copper exposure: higher log reduction of culturability was associated with no induction of copA after exposure to copper.
FIG 3.
Expression of copA after a 30-min exposure to copper concentration of 5 mg/liter in simulated drinking water at 36°C. RT-qPCR was used to test expression of copA in untreated (Cu−) and treated (Cu+) L. pneumophila. The fold change is the change in expression between 30 min and before treatment. Three replicates were used. We used an unpaired Student's t test to assess statistical significance for each time point. *, P ≤ 0.05; **, P ≤ 0.001 (treated versus untreated).
Whole-genome sequencing was conducted to investigate the phylogenetic relationship between the strains and identify genetic determinants of the phenotypic difference between the strains. The Microbial Genome Atlas was used to find the closest strains in the database (44). Based on amino acid identity, 1427.B.F was more similar to L. pneumophila strain Thunder Bay (98.5% for 92.32% of protein shared) and to L. pneumophila strain Philadelphia 1 (98.4% for 94.11% proteins shared). This was supported by Mash (45), with k-mers from contigs of the four isolates assembled by SPAdes (46) most closely matching L. pneumophila Thunder Bay out of 96 full L. pneumophila genomes (Table S6).
A high-level phylogenetic tree was constructed using RAxML-ng (47) with single nucleotide polymorphisms (SNPs) situated outside predicted recombinant regions called by Snippy and Gubbins (48, 49) to place the four isolates in a genomic context among 96 fully completed L. pneumophila genomes (Fig. S1). The four strains sequenced in this study formed a clade embedded in a cluster of Philadelphia-1 like strains. A higher-resolution tree was constructed using the same process to place the 1427-like isolates among closely related strains (Fig. 4), showing them to form a sister clade to a clade of strains rooted by L. pneumophila Thunder Bay (50).
FIG 4.
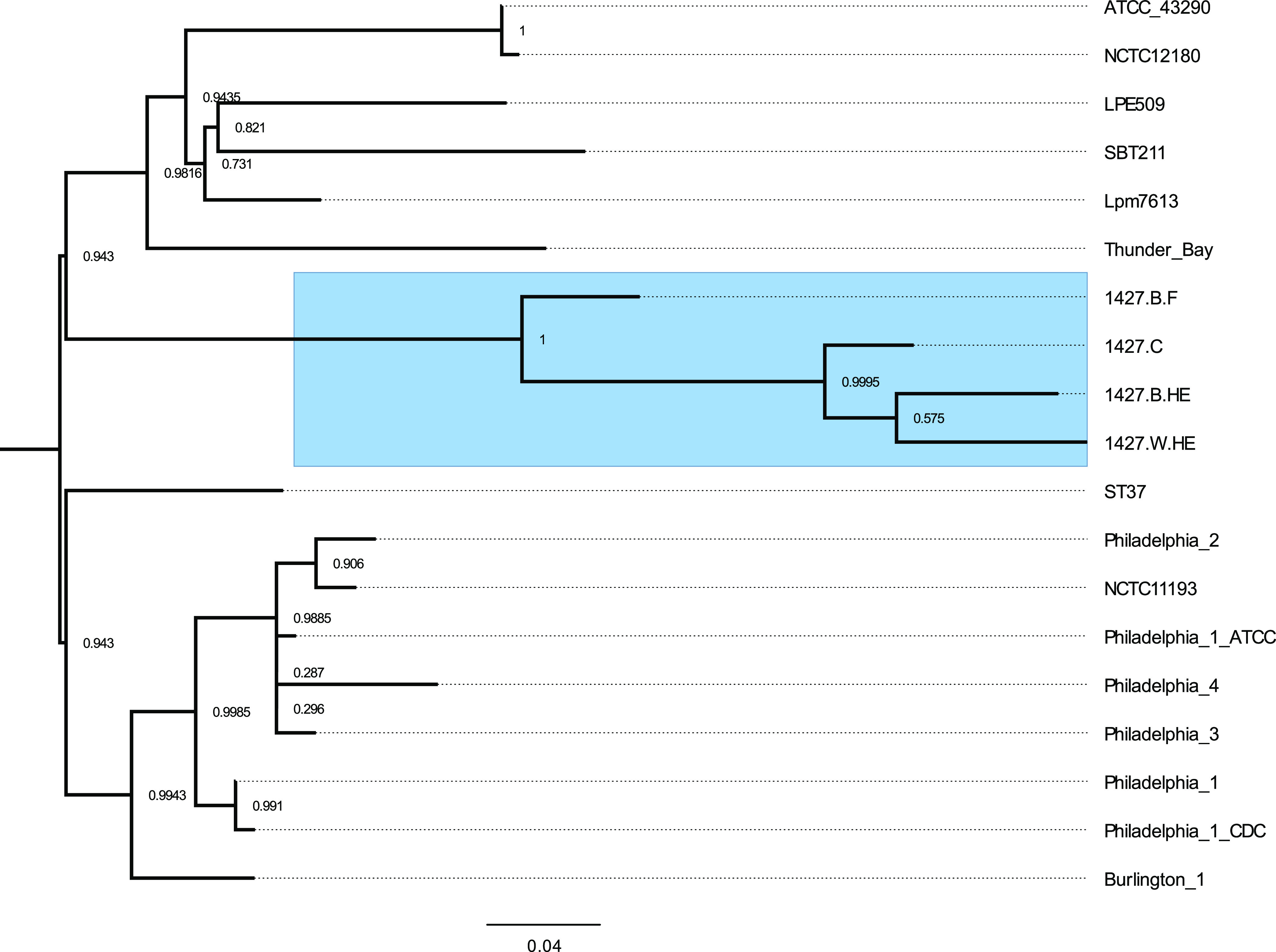
Whole-genome-based phylogeny of the 1427-like isolates. Two hundred SNPs falling outside recombinant regions were used to generate a phylogenetic tree of the 4 new isolates in the context of closely related Philadelphia-like L. pneumophila strains. The four 1427-like isolates are highlighted in blue. Branch support was evaluated using 1,000 bootstrap replicates in RAxML-ng transfer bootstrap expectation mode. The scale bar shows mean nucleotide substitutions per site.
The pangenome of the four 1427-like isolates was then analyzed using ROARY (51). Eight genes were identified as being present in only a subset of the isolates, but manual inspection showed these to be likely false negatives introduced during gene annotation. Only 29 SNPs were identified between the strains, confirming that the isolates are closely related (Table S4). Interestingly, the isolates from the heat exchanger (1427.B.HE and 1427.W.HE) were the most genetically similar to the clinical isolate (1427.C).
DISCUSSION
The tolerance to copper of nine L. pneumophila isolates recovered from environmental or clinical sources, in biofilm or water, belonging to various serogroup and sequence types was evaluated in simulated drinking water.
Strains were cultured and acclimatized in simulated drinking water to mimic building water system conditions encountered by L. pneumophila in the planktonic state. First, cells were grown to the post-exponential phase, representative of the transmissive phase of L. pneumophila. In an engineered water system context, L. pneumophila would likely enter water after being expelled from a host cell, in the transmissive state. Post-exponential-phase cells are also markedly more resistant to stress than exponential-phase bacteria (36, 52–55). Second, cells were acclimatized in simulated drinking water at room temperature for 1 week prior to copper exposure. Starvation has been shown to increase the tolerance of L. pneumophila to numerous stresses (52, 56–59). Following acclimation for 1 week in conditions representative of drinking water distribution systems, the tolerance to copper was tested in conditions simulating faucet temperatures, where a mix of cold and hot water is used and results in lukewarm temperatures. For this reason, a temperature of 36°C was selected for the assay.
In this study, different L. pneumophila isolates displayed variable copper susceptibility over time when exposed to high concentrations of copper. At copper concentrations typically found in drinking water (≤2 mg/liter), all tested isolates were tolerant to copper and remained culturable. In contrast, distinct groups were observed at higher concentrations of 5 mg/liter for prolonged contact time. The first group, including isolates from water and clinical sources (378.W.F, 1427.W.HE, 1427.C, and 284.W), had lower tolerance to high copper doses and showed >4-log reductions in culturability after 672 h of exposure. The second group included isolates from water, clinical, and biofilm sources (JR32, 154.W.F, 1427.B.F, 62.C, and 62.W) and displayed 2- to 3-log reductions in culturability after 672 h at 5 mg/liter. Finally, the isolate 1427.B.HE was considered resistant to copper stress, as it showed a <2-log reduction in culturability after 672 h. No trend was observed suggesting that copper susceptibility is linked to sequence type, although the number of strains from each ST was not sufficient to draw any conclusion. While two isolates (patient-derived 62.C and cooling water 62.W) presented similar copper tolerance profiles at all tested concentrations (Fig. 2), the four ST1427 strains had different tolerance profiles. In both cases, isolates were recovered during the investigation of L. pneumophila outbreaks (24, 60). Compared to a strain isolated from cooling tower water unrelated to an outbreak (284.W and ST284), the outbreak isolate displayed more resistance to copper (Fig. 2). As previously determined, the outbreak-associated strain (62.W) was significantly more infectious for human macrophages, whereas the infectivity for Acanthamoeba castellanii was comparable (61).
The nature of the isolation site (environmental versus clinical, hot water system versus cooling towers, and biofilm versus water) is also an important factor to consider when the copper resistance of environmental strains is being evaluated. The compositions of drinking water and plumbing material have been shown to impact downstream microbial communities and the integration of L. pneumophila into biofilm (62). In addition, Legionella may also resist environmental stressors through internalization within free-living amoebae, which have the capability to become established within biofilms grown on copper surfaces of drinking water systems (63). In this study, four isolates were recovered during an L. pneumophila outbreak investigation in a health care facility (HCF-B) (24). The source of the outbreak was identified as the hot water system from which isolates 1427.B.F, 1427.W.HE, and 1427.B.HE were recovered. These environmental isolates belong to the same sequence type (ST1427) (24). In this study, whole-genome sequencing analysis was performed on these four strains and confirmed that they are closely related. Despite genomic similarities, we report that tolerance to copper stress was highly variable between the four isolates (Fig. 2). In this study, tolerance to copper stress was evaluated based on the impact on culturability. However, cells could still be in a viable-but-nonculturable (VBNC) state. The absence of culturable L. pneumophila in water collected from building copper piping water systems might suggest a low risk level, but the presence of VBNC cells and their ability to recover culturability in the presence of host cells or favorable environmental conditions may still present a risk.
Microevolution within the population of L. pneumophila within hospitals water distribution systems was previously studied by whole-genome sequencing (64). Strains isolated from a single hospital generally clustered according to the location. For examples, isolates from the same wards clustered together. However, all isolates from one hospital were related and most likely originated from a single ancestor. Therefore, it seems that water systems are seeded by one or a few strains that colonize the system and evolve within it, generating some limited diversity over time (64). Our results are in agreement with this hypothesis. Presumably, the water system was colonized initially by one 1427-like strain, which evolved within it over time, generating isolates with different phenotypes. The number of SNPs identified between these strains are in the range of SNPs found between isolates from the same water systems (64, 65).
The evolution of the 1427-like population was driven by the persistence of the most adapted variants in the different microenvironments present in the water distribution systems, such as the tap or heat exchanger. Isolates 1427.B.HE and 1427.B.F displayed a response to the presence of copper by significantly inducing higher expression of copA (Fig. 3). Those two strains were recovered from environmental biofilm growing on metallic surfaces: stainless steel heat exchanger plates containing chromium and nickel and an internal faucet surface. The ability of 1427.B.HE and 1427.B.F to maintain culturability in the presence of copper could be linked to their ability to form biofilm and their previous exposure to heavy metals from the substrata. Indeed, biofilm-derived cells have an increased resistance to heavy metals compared to suspended cells (66). In the present case, the exposure of L. pneumophila to chrome and nickel during stagnation within the heat exchanger may have triggered integration into the biofilm, thereby activating metal resistance mechanisms, including those against copper (67). For example, the L. pneumophila gold-induced-gene (GIG) operon was previously reported to be induced following exposure to copper (34), but the functional importance of this locus for copper resistance is unknown. The four strains that were sequenced did not possess the GIG operon. The reported increased tolerance to copper, lead, and zinc of bacteria in drinking water versus bacteria in raw water further highlights the critical role of the environment in acquiring resistance (68).
Differences in the ability of studied strains to maintain culturability was observed only at high copper concentration (5 mg/liter), after prolonged exposure (672 h) (Fig. 1). This variation may be caused by differential copA expression as shown in Fig. 2. However, tolerance could also be associated with the presence of other, yet-unidentified copper resistance genes. Unfortunately, no SNPs were found exclusively in the two most resistant strains. In addition, no nonsynonymous mutation was found in genes associated with copper resistance. Therefore, SNPs and gene content cannot explain the difference in copper resistance and the difference in the expression of copA seen between the isolates, suggesting that these phenotypic differences are mediated by an epigenetic mechanism.
Epigenetic regulation in bacteria involves DNA methylation, which affects the binding of transcriptional regulator to promoters (69). Epigenetic regulation can change gene expression without the need for DNA mutation. This process is mediated by DNA methyltransferases such as Dam (69). The role of DNA methylation in gene regulation in bacteria is still poorly understood and has not been described yet in L. pneumophila. Nevertheless, evidence of Dam-mediated methylation of L. pneumophila genomic DNA was reported in 1996 (70). To our knowledge, this is the only study of DNA methylation in L. pneumophila. In addition, DNA methyltransferases are encoded in L. pneumophila genomes. copA is transcribed together with the upstream gene (lpg1034) from a transcription start site located at position −97 (71). However, the promoter region does not contain GATC Dam methylation motifs, suggesting that a transcriptional regulator of copA might be under epigenetic regulation. Nevertheless, epigenetic regulation of copA is consistent with the transcriptional analysis showing that the resistant strains are responsive to the presence of copper but the susceptible strains are not. Further study of epigenetic regulation in L. pneumophila will be required to investigate this possibility, which is outside the scope of this report.
These results suggest that the immediate environment within the building plumbing system can have a significant impact on the copper tolerance of closely related isolates. Exposure to copper, like many selective pressures bacteria are exposed to, can lead to alterations of the genome of L. pneumophila (Table S3), which can translate into behavioral modifications that may have clinical and environmental consequences. This is critical information to consider when one is selecting a sampling strategy for the monitoring and control of L. pneumophila. The selection of sampling sites within a given building’s water system and the choice to sample water versus biofilm may affect the evaluation of prevalence, and therefore, impact the subsequent risk assessment.
The loss of culturability suffered by some strain during exposure to high levels of copper does not necessarily equate to a loss of viability. However, an alternative detection method is needed to measure VBNC cells in high-copper-concentration environments. In the context of an environmental investigation, there is a need to carefully account for information such as site of isolation of the strain and to consider the possible VBNC state. This is especially the case for strains isolated from water with a lesser ability to remain culturable when exposed to copper stress over time. Also, the higher resistance observed for the biofilm-derived strains supports the role of biofilm grown on metallic surfaces in promoting metal resistance. Importantly, an increase in resistance to metals is known to simultaneously lead to increased antibiotic resistance (66, 67). The difference observed between water and biofilm isolates from a hot water system in a health care facility questions the ability to monitor a water system based on water samples alone. An underestimation of L. pneumophila prevalence combined with the exposure of vulnerable patients may lead to inappropriate risk management strategies.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Experiments were performed using nine L. pneumophila strains isolated from clinical or environmental samples and L. pneumophila JR32, a Philadelphia-1 laboratory-adapted strain resistant to streptomycin (72). A summary of isolates used in this study and their characteristics is presented in Table 1. The clinical strains (1427.C and 62.C) and environmental strains (1427.W.HE, 1427.B.HE, 284.W, and 62.W) were provided by the Laboratoire de Santé Publique du Québec. The average copper concentration in water from HCF-A and HCF-B were 0.57 mg/liter and 0.40 mg/liter, respectively (32, 73). Isolates 154.W.F, 1427.B.F, 378.W.F, 1427.W.HE, 1427.C, and 1427.B.HE were typed by sequence-based typing, according to the European Working Group for Legionella Infections (EWGLI) protocol (74, 75). Sanger sequencing results were analyzed, and an ST was assigned according to the EWGLI database.
Strains stored in 60% glycerol at −80°C were grown on buffered charcoal yeast extract (BCYE; Oxoid) agar for 3 days at 36°C. Resulting colonies were inoculated overnight at 36°C into sterile yeast extract broth (pH 6.88) with the growth supplement SR0110 (Oxoid). Cells were harvested by centrifugation (3,000 × g for 30 min), washed twice with sterile synthetic water, and suspended in the same medium to an initial concentration of 108 cells/ml. Prior to copper exposure, cells were starved in sterile synthetic water for 1 week at room temperature (22°C), to simulate environmental conditions in the drinking water pipes of municipal distribution systems and building main distribution systems.
Synthetic-water preparation.
Synthetic water was prepared by adding salts (final salt concentrations found in Table S5) to 0.22-μm-filtered and autoclaved Milli-Q water. The water was buffered with 3-(N-morpholino)propanesulfonic acid (MOPS; Sigma-Aldrich) to a final concentration of 1 mM. The salts and their respective concentrations were selected to mimic Montreal tap water as per the Annual Drinking Water Quality Report (76). The synthetic water was adjusted to an alkalinity of 60 mg CaCO3/liter and a pH of 7.8. Individual stock salt solutions were prepared in sterile ultrapure water. The MOPS buffer was selected due to its target pH range and its low rate of complexation with metal ions and since it does not interfere with cell growth (77). The final pH was adjusted to 7.8 using 0.1 M NaOH.
Experimental conditions.
Each strain was starved in synthetic water for 1 week and subsequently diluted in 20 ml of sterile synthetic water (final concentration of approximately 108 cells/ml), in a 50-ml sterile propylene tube, providing a large volume of air for aeration. Different copper concentrations were added to the cell suspension: 0, 0.8, 1.3, 2, and 5 mg/liter. Concentrations of total copper were measured at the beginning of the experiment and after 28 days, using inductively coupled plasma mass spectrometry (ICPMS) after acidification with 0.5% HNO3. Copper chloride was selected as the copper source, since it has the highest dissolved-to-total copper ratio in water (71 to 98% for copper chloride, compared to 51 to 67% for copper sulfate and 63% for copper nitrate). A high dissolved-to-total ion ratio was used to represent the behavior of copper in drinking water, which is found mostly in the dissolved form. As an example, in HCF-A, the mean dissolved-to-total ratio was 98% (n = 75; 87% to 100%).
Exposure to copper was carried out at 36°C to simulate temperature at the tap when hot and cold water are mixed. The survival of L. pneumophila isolates exposed to copper was monitored over time (1 h, 6 h, 24 h, 48 h, 72 h, 168 h, 336 h, and 672 h) by culturing. Before each sampling, the samples were vigorously agitated. Water without a bacterial inoculum was used as a negative control. For each condition and time point, cell culturability was evaluated in triplicate on BCYE agar. Cell viability was evaluated by flow cytometry assay (FCA), using an Accury-C6 BD flow cytometer. FCA profiles were obtained with a combination of two nucleic acid fluorescent dyes: thiazole orange (TO) for cells in any physiological state and propidium iodide (PI) for cells with a damaged membrane (BD cell viability kit). In order to avoid copper-PI interferences, copper was quenched immediately prior to cell staining by adding excess EDTA at a final concentration between 0.25 M and 1.66 M, for copper concentrations between 0.8 mg/liter and 5 mg/liter. TO and PI were added at final concentrations of 420 nM and 43 μM, respectively. The samples were incubated for 5 min at room temperature, as recommended by the manufacturer. The green fluorescence from TO was acquired by the FL1 channel (533/30 nm), and the red fluorescence from PI was acquired by the FL3 channel (670 nm). A threshold was applied to the FL1 channel to eliminate background noise, as defined by the negative controls: sterile synthetic water stained with both dyes and unstained cells in sterile synthetic water. A dead-cell control (heat killed at 90°C for 2 min) was used to define the dead-cell region for each strain. Analyses were performed on the high-rate setting (66 μl/min) and analyzed with BD Accuri C6 software.
copA expression.
Analysis of copA gene expression was performed by reverse transcription-quantitative PCR (RT-qPCR). The isolates 1427.C, 1427.B.F, 1427.W.HE, and 1427.B.HE were starved in synthetic drinking water (Table S5). Cells were collected prior and after exposure to 5 mg/liter of copper for 30 min. RNA was extracted with TRIzol reagent according to the manufacturer’s protocol (Invitrogen). One microgram of RNA was then converted to cDNA by using random nonamers and ProtoScript II reverse transcriptase (New England Biolabs) according to the manufacturer’s instructions. For each sample, a negative control without reverse transcriptase was used. qPCRs were then performed with 1 μl of cDNA using Universal SYBR green Supermix (Bio-Rad). Primers used for real-time PCR analysis are qcopA forward (CAATACCCTGGTGGTCGATAAAAC) and qcopA reverse (TGCCGCAGCTAATGCTAAAG) (39). A relative quantification strategy was used to perform analysis of the qPCR data. Transcript levels were normalized to 16S rRNA in each sample, using primers 16S_QF and 16S_QR (78), and changes in the gene expression data from prior copper exposure to after exposure were calculated as fold change (79). Statistical analysis between the strains exposed and the wild-type control was performed using an unpaired one-tailed Student's t test.
Whole-genome sequencing of Legionella pneumophila strains.
DNA libraries for sequencing on the MiSeq platform were made using the Nextera XT DNA library prep kit (Illumina). Briefly, the Wizard genomic DNA purification kit (Promega) was used to extract and purify the DNA from four Legionella pneumophila isolates, 1427_B_F, 1427_W_HE, 1427_C, and 1427_B_HE (Table 1). DNA quality and concentration were evaluated using a 0.8% agarose gel and the Quant-iT PicoGreen double-stranded-DNA (dsDNA) assay kit (Thermo Fisher). After the quality check, the genomic samples were prepared following the Nextera XT manufacturer’s instructions. To ensure proper fragmentation of the genomic DNA, the DNA samples were evaluated on an Agilent Technology 2100 Bioanalyzer (Agilent). Once the DNA passed this quality control measure, the DNA samples were diluted to 2 nM and pooled to create the DNA library for sequencing. The library was frozen until further use (no more than 3 days). On the day of the sequencing run, 0.2 N NaOH was used to denature the DNA library. This denatured library was diluted to a 12 pM loading concentration with HT1 buffer (Illumina). A PhiX control (20 pM) was spiked at 1% into the library. The library was sequenced on an Illumina MiSeq platform using a V3 MiSeq reagent kit (600 cycles).
WGS read analysis and phylogeny construction.
Reads were preprocessed for quality control and adapter trimming using fastp 0.20.0 (80). SPAdes 3.13.1 (46) was used to assemble each of the four isolates into draft collections of contigs. These were compared using Mash 2.2 (45) to all (96, as of 22 January 2020 [Table S6]) complete L. pneumophila genomes available through RefSeq. The four isolates were shown to form a clade which was most like strain Thunder Bay.
Analysis was conducted using Thunder Bay (NC_021350) as a reference genome for mapping and annotation. The Snippy 4.4.5 (48) pipeline (81, 82) was used to call variant sites in the four isolates. Gubbins 2.4.1 (49) was used to filter out probable recombinant sites from the SNP alignments. Phylogenies were constructed from the remaining SNP sites using RaxML-NG 0.9.0 (47) with the model GTR+Gamma. To increase resolution, the same method was used to produce a phylogenetic tree using a subset of closely related genomes (Table S6). Branch support was estimated using 1,000 bootstrap replicates in transfer bootstrap expectation mode. Finally, Prokka 1.14.3 (48) was used to annotate the genomes of the four isolates, and pangenome analysis was conducted using Roary 3.12.0 (51).
Data availability.
The raw reads of this sequencing project have been uploaded to the NCBI SRA database under the BioProject number PRJNA610262.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a NSERC Discovery Grant (RGPIN/04499-2018) to S.P.F. J.L. is the recipient of an Alexander Graham Bell Canada Graduate Scholarships–Master’s. This study was supported by the partners of the NSERC Industrial Chair on Drinking Water.
We thank Chair staff, especially Jacinthe Mailly, Mélanie Rivard, Marie-Ève Benoit, and Wendy Andriantsarafara.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15:506–526. 10.1128/cmr.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedict K, Reses H, Vigar M, Roth DM, Roberts VA, Mattioli M, Cooley LA, Hilborn ED, Wade TJ, Fullerton K, Yoder J, Hill VR. 2017. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2013–2014. MMWR Morb Mortal Wkly Rep 66:1216–1220. 10.15585/mmwr.mm6644a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrison LE, Kunz JM, Cooley LA, Moore MR, Lucas C, Schrag S, Sarisky J, Whitney CG. 2016. Vital signs: deficiencies in environmental control identified in outbreaks of Legionnaires’ disease—North America, 2000–2014. MMWR Morb Mortal Wkly Rep 65:576–584. 10.15585/mmwr.mm6522e1. [DOI] [PubMed] [Google Scholar]
- 4.European Centre for Disease Prevention and Control. 2017. Legionnaires' disease. In ECDC annual epidemiological report for 2015. European Centre for Disease Prevention and Control, Stockholm, Sweden. https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2015-legionnaires-disease_0.pdf.
- 5.Centers for Disease Control and Prevention. 2017. Active bacterial core surveillance (ABCs) report emerging infections program network: legionellosis, 2015. CDC, Atlanta, GA.
- 6.Soda EA, Barskey AE, Shah PP, Schrag S, Whitney CG, Arduino MJ, Reddy SC, Kunz JM, Hunter CM, Raphael BH, Cooley LA. 2017. Vital signs: health care–associated Legionnaires’ disease surveillance data from 20 states and a large metropolitan area—United States, 2015. MMWR Morb Mortal Wkly Rep 66:584–589. 10.15585/mmwr.mm6622e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buse HY, Schoen ME, Ashbolt NJ. 2012. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res 46:921–933. 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Boppe I, Bédard E, Taillandier C, Lecellier D, Nantel-Gauvin M-A, Villion M, Laferrière C, Prévost M. 2016. Investigative approach to improve hot water system hydraulics through temperature monitoring to reduce building environmental quality hazard associated to Legionella. Building Environ 108:230–239. 10.1016/j.buildenv.2016.08.038. [DOI] [Google Scholar]
- 9.Bargellini A, Marchesi I, Righi E, Ferrari A, Cencetti S, Borella P, Rovesti S. 2011. Parameters predictive of Legionella contamination in hot water systems: association with trace elements and heterotrophic plate counts. Water Res 45:2315–2321. 10.1016/j.watres.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Marchesi I, Marchegiano P, Bargellini A, Cencetti S, Frezza G, Miselli M, Borella P. 2011. Effectiveness of different methods to control legionella in the water supply: ten-year experience in an Italian university hospital. J Hosp Infect 77:47–51. 10.1016/j.jhin.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 11.National Academies of Sciences, Engineering, and Medicine. 2019. Management of Legionella in water systems. The National Academies Press, Washington, DC, USA. 10.17226/25474. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Masters S, Edwards MA, Falkinham JO, III, Pruden A. 2014. Effect of disinfectant, water age, and pipe materials on bacterial and eukaryotic community structure in drinking water biofilm. Environ Sci Technol 48:1426–1435. 10.1021/es402636u. [DOI] [PubMed] [Google Scholar]
- 13.Proctor CR, Gächter M, Kötzsch S, Rölli F, Sigrist R, Walser J-C, Hammes F. 2016. Biofilms in shower hoses—choice of pipe material influences bacterial growth and communities. Environ Sci Water Res Technol 2:670–682. 10.1039/C6EW00016A. [DOI] [Google Scholar]
- 14.Yu J, Kim D, Lee T. 2010. Microbial diversity in biofilms on water distribution pipes of different materials. Water Sci Technol 61:163–171. 10.2166/wst.2010.813. [DOI] [PubMed] [Google Scholar]
- 15.Buse HY, Lu J, Struewing IT, Ashbolt NJ. 2014. Preferential colonization and release of Legionella pneumophila from mature drinking water biofilms grown on copper versus unplasticized polyvinylchloride coupons. Int J Hyg Environ Health 217:219–225. 10.1016/j.ijheh.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Giao MS, Wilks SA, Keevil CW. 2015. Influence of copper surfaces on biofilm formation by Legionella pneumophila in potable water. Biometals 28:329–339. 10.1007/s10534-015-9835-y. [DOI] [PubMed] [Google Scholar]
- 17.Moritz MM, Flemming HC, Wingender J. 2010. Integration of Pseudomonas aeruginosa and Legionella pneumophila in drinking water biofilms grown on domestic plumbing materials. Int J Hyg Environ Health 213:190–197. 10.1016/j.ijheh.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Proctor CR, Dai D, Edwards MA, Pruden A. 2017. Interactive effects of temperature, organic carbon, and pipe material on microbiota composition and Legionella pneumophila in hot water plumbing systems. Microbiome 5:130. 10.1186/s40168-017-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casey AL, Adams D, Karpanen TJ, Lambert PA, Cookson BD, Nightingale P, Miruszenko L, Shillam R, Christian P, Elliott TS. 2010. Role of copper in reducing hospital environment contamination. J Hosp Infect 74:72–77. 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt MG, Attaway HH, Sharpe PA, John J, Jr, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, Freeman KD, Michels HT, Salgado CD. 2012. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J Clin Microbiol 50:2217–2223. 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YE, Stout JE, Yu VL. 2011. Controlling Legionella in hospital drinking water: an evidence-based review of disinfection methods. Infect Control Hosp Epidemiol 32:166–173. 10.1086/657934. [DOI] [PubMed] [Google Scholar]
- 22.Demirjian A, Lucas CE, Garrison LE, Kozak-Muiznieks NA, States S, Brown EW, Wortham JM, Beaudoin A, Casey ML, Marriott C, Ludwig AM, Sonel AF, Muder RR, Hicks LA. 2015. The importance of clinical surveillance in detecting Legionnaires' disease outbreaks: a large outbreak in a hospital with a Legionella disinfection system—Pennsylvania, 2011–2012. Clin Infect Dis 60:1596–1602. 10.1093/cid/civ153. [DOI] [PubMed] [Google Scholar]
- 23.Blanc DS, Carrara P, Zanetti G, Francioli P. 2005. Water disinfection with ozone, copper and silver ions, and temperature increase to control Legionella: seven years of experience in a university teaching hospital. J Hosp Infect 60:69–72. 10.1016/j.jhin.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Bédard E, Lévesque S, Martin P, Pinsonneault L, Paranjape K, Lalancette C, Dolcé C-É, Villion M, Valiquette L, Faucher SP, Prévost M. 2016. Energy conservation and the promotion of Legionella pneumophila growth: the probable role of heat exchangers in a nosocomial outbreak. Infect Control Hosp Epidemiol 37:1475–1480. 10.1017/ice.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. 2011. Guidelines for drinking-water quality, 4th ed. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 26.Health Canada. 2014. Summary table, p 25. In Guidelines for Canadian drinking water quality. Federal-Provincial-Territorial Committee on Drinking Water of the Federal-Provincial-Territorial Committee on Health and the Environment, Ottawa, Ontario, Canada. [Google Scholar]
- 27.United States Environmental Protection Agency. 2009. National recommended water quality criteria. Office of Water, Office of Science and Technology, Washington, DC, USA. [Google Scholar]
- 28.Olivares M, Araya M, Pizarro F, Uauy R. 2001. Nausea threshold in apparently healthy individuals who drink fluids containing graded concentrations of copper. Regul Toxicol Pharmacol 33:271–275. 10.1006/rtph.2000.1440. [DOI] [PubMed] [Google Scholar]
- 29.Araya M, Chen B, Klevay LM, Strain JJ, Johnson L, Robson P, Shi W, Nielsen F, Zhu H, Olivares M, Pizarro F, Haber LT. 2003. Confirmation of an acute no-observed-adverse-effect and low-observed-adverse-effect level for copper in bottled drinking water in a multi-site international study. Regul Toxicol Pharmacol 38:389–399. 10.1016/j.yrtph.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 30.California Environmental Protection Agency, Pesticide and Environmental Toxicology Branch, Office of Environmental Health Hazard Assessment. 2008. Public health goals for copper in drinking water. California Environmental Protection Agency, Sacramento, CA. https://oehha.ca.gov/media/downloads/water/chemicals/phg/copperphg020808.pdf.
- 31.Stenhammar L. 1999. Diarrhoea following contamination of drinking water with copper. Eur J Med Res 4:217–218. [PubMed] [Google Scholar]
- 32.Bedard E, Laferriere C, Charron D, Lalancette C, Renaud C, Desmarais N, Deziel E, Prevost M. 2015. Post-outbreak investigation of Pseudomonas aeruginosa faucet contamination by quantitative polymerase chain reaction and environmental factors affecting positivity. Infect Control Hosp Epidemiol 36:1337–1343. 10.1017/ice.2015.168. [DOI] [PubMed] [Google Scholar]
- 33.Edwards M, Parks J, Griffin A, Raetz M, Martin A, Scardina P, Elfland C. 2011. Lead and copper corrosion control in new construction. Water Research Foundation, Denver, CO, USA. [Google Scholar]
- 34.Jwanoswki K, Wells C, Bruce T, Rutt J, Banks T, McNealy TL. 2017. The Legionella pneumophila GIG operon responds to gold and copper in planktonic and biofilm cultures. PLoS One 12:e0174245. 10.1371/journal.pone.0174245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin RL, Strom OR, Pruden A, Edwards MA. 2020. Interactive effects of copper pipe, stagnation, corrosion control, and disinfectant residual influenced reduction of legionella pneumophila during simulations of the Flint water crisis. Pathogens 9:730. 10.3390/pathogens9090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Pruden A, Edwards MA, Rhoads WJ. 2021. Natural organic matter, orthophosphate, pH, and growth phase can limit copper antimicrobial efficacy for Legionella in drinking water. Environ Sci Technol 55:1759–1768. 10.1021/acs.est.0c06804. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y-SE, Vidic RD, Stout JE, Yu VL. 1996. Individual and combined effects of copper and silver ions on inactivation of Legionella pneumophila. Water Res 30:1905–1913. 10.1016/0043-1354(96)00077-2. [DOI] [Google Scholar]
- 38.Kim EH, Charpentier X, Torres-Urquidy O, McEvoy MM, Rensing C. 2009. The metal efflux island of Legionella pneumophila is not required for survival in macrophages and amoebas. FEMS Microbiol Lett 301:164–170. 10.1111/j.1574-6968.2009.01813.x. [DOI] [PubMed] [Google Scholar]
- 39.Trigui H, Dudyk P, Sum J, Shuman HA, Faucher SP. 2013. Analysis of the transcriptome of Legionella pneumophila hfq mutant reveals a new mobile genetic element. Microbiology (Reading) 159:1649–1660. 10.1099/mic.0.067983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bédard E, Paranjape K, Lalancette C, Villion M, Quach C, Laferrière C, Faucher SP, Prévost M. 2019. Legionella pneumophila levels and sequence-type distribution in hospital hot water samples from faucets to connecting pipes. Water Res 156:277–286. 10.1016/j.watres.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Lévesque B, Gervais MC, Chevalier P, Gauvin D, Anassour-Laouan-Sidi E, Gingras S, Fortin N, Brisson G, Greer C, Bird D. 2014. Prospective study of acute health effects in relation to exposure to cyanobacteria. Sci Total Environ 466–467:397–403. 10.1016/j.scitotenv.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 42.Sydnor ER, Bova G, Gimburg A, Cosgrove SE, Perl TM, Maragakis LL. 2012. Electronic-eye faucets: Legionella species contamination in healthcare settings. Infect Control Hosp Epidemiol 33:235–240. 10.1086/664047. [DOI] [PubMed] [Google Scholar]
- 43.Bédard E, Prévost M, Déziel E. 2016. Pseudomonas aeruginosa in premise plumbing of large buildings. MicrobiologyOpen 5:937–956. 10.1002/mbo3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-R LM, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, Cole JR, Konstantinidis KT. 2018. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res 46:W282–W288. 10.1093/nar/gky467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan MA, Knox N, Prashar A, Alexander D, Abdel-Nour M, Duncan C, Tang P, Amatullah H, Dos Santos CC, Tijet N, Low DE, Pourcel C, Van Domselaar G, Terebiznik M, Ensminger AW, Guyard C. 2013. Comparative genomics reveal that host-innate immune responses influence the clinical prevalence of Legionella pneumophila serogroups. PLoS One 8:e67298. 10.1371/journal.pone.0067298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66:3029–3034. 10.1128/IAI.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Bana BH, Haddad MT, Garduno RA. 2014. Stationary phase and mature infectious forms of Legionella pneumophila produce distinct viable but non-culturable cells. Environ Microbiol 16:382–395. 10.1111/1462-2920.12219. [DOI] [PubMed] [Google Scholar]
- 54.Hales LM, Shuman HA. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol 181:4879–4889. 10.1128/JB.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol 44:107–118. 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 56.Chang CW, Hwang YH, Cheng WY, Chang CP. 2007. Effects of chlorination and heat disinfection on long-term starved Legionella pneumophila in warm water. J Appl Microbiol 102:1636–1644. 10.1111/j.1365-2672.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. 2014. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol 5:20. 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandyopadhyay P, Xiao H, Coleman HA, Price-Whelan A, Steinman HM. 2004. Icm/dot-independent entry of Legionella pneumophila into amoeba and macrophage hosts. Infect Immun 72:4541–4551. 10.1128/IAI.72.8.4541-4551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch D, Fieser N, Glöggler K, Forsbach-Birk V, Marre R. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol Lett 219:241–248. 10.1016/S0378-1097(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 60.Levesque S, Lalancette C, Bernard K, Pacheco AL, Dion R, Longtin J, Tremblay C. 2016. Molecular typing of Legionella pneumophila isolates in the province of Quebec from 2005 to 2015. PLoS One 11:e0163818. 10.1371/journal.pone.0163818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levesque S, Plante PL, Mendis N, Cantin P, Marchand G, Charest H, Raymond F, Huot C, Goupil-Sormany I, Desbiens F, Faucher SP, Corbeil J, Tremblay C. 2014. Genomic characterization of a large outbreak of Legionella pneumophila serogroup 1 strains in Quebec city, 2012. PLoS One 9:e103852. 10.1371/journal.pone.0103852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu J, Buse HY, Gomez-Alvarez V, Struewing I, Santo Domingo J, Ashbolt NJ. 2014. Impact of drinking water conditions and copper materials on downstream biofilm microbial communities and Legionella pneumophila colonization. J Appl Microbiol 117:905–918. 10.1111/jam.12578. [DOI] [PubMed] [Google Scholar]
- 63.Thomas V, Bouchez T, Nicolas V, Robert S, Loret JF, Levi Y. 2004. Amoebae in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence. J Appl Microbiol 97:950–963. 10.1111/j.1365-2672.2004.02391.x. [DOI] [PubMed] [Google Scholar]
- 64.David S, Afshar B, Mentasti M, Ginevra C, Podglajen I, Harris SR, Chalker VJ, Jarraud S, Harrison TG, Parkhill J. 2017. Seeding and Establishment of Legionella pneumophila in hospitals: implications for genomic investigations of nosocomial Legionnaires' disease. Clin Infect Dis 64:1251–1259. 10.1093/cid/cix153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartley PB, Ben Zakour NL, Stanton-Cook M, Muguli R, Prado L, Garnys V, Taylor K, Barnett TC, Pinna G, Robson J, Paterson DL, Walker MJ, Schembri MA, Beatson SA. 2016. Hospital-wide eradication of a nosocomial Legionella pneumophila serogroup 1 outbreak. Clin Infect Dis 62:273–279. 10.1093/cid/civ870. [DOI] [PubMed] [Google Scholar]
- 66.Harrison JJ, Ceri H, Turner RJ. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol 5:928–938. 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 67.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 68.Calomiris JJ, Armstrong JL, Seidler RJ. 1984. Association of metal tolerance with multiple antibiotic resistance of bacteriaisolated from drinking water. Appl Environ Microbiol 47:1238–1242. 10.1128/AEM.47.6.1238-1242.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sánchez-Romero MA, Casadesús J. 2020. The bacterial epigenome. Nat Rev Microbiol 18:7–20. 10.1038/s41579-019-0286-2. [DOI] [PubMed] [Google Scholar]
- 70.Lema MW, Brown A. 1996. Dam-like methylation in Legionellae. Curr Microbiol 32:64–66. 10.1007/s002849900011. [DOI] [PubMed] [Google Scholar]
- 71.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee J-Y, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol 9:503–519. 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 72.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. 10.1128/IAI.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bédard E, Boppe I, Kouamé S, Martin P, Pinsonneault L, Valiquette L, Racine J, Prévost M. 2016. Combination of heat shock and enhanced thermal regime to control the growth of a persistent Legionella pneumophila strain. Pathogens 5:35. 10.3390/pathogens5020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ratzow S, Gaia V, Helbig JH, Fry NK, Luck PC. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol 45:1965–1968. 10.1128/JCM.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaia V, Fry NK, Afshar B, Luck PC, Meugnier H, Etienne J, Peduzzi R, Harrison TG. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol 43:2047–2052. 10.1128/JCM.43.5.2047-2052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ville de Montreal. 2015. Qualité de l’eau potable produite par les usines Atwater et Charles-J.-Des-Baillets et distribuée en réseau. Ville de Montreal Division de l’expertise technique, Montreal, Quebec, Canada.
- 77.Ferreira CMH, Pinto ISS, Soares EV, Soares HMVM. 2015. (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions—a review. RSC Adv 5:30989–31003. 10.1039/C4RA15453C. [DOI] [Google Scholar]
- 78.Trigui H, Dudyk P, Oh J, Hong J-I, Faucher SP. 2015. A regulatory feedback loop between RpoS and SpoT supports the survival of Legionella pneumophila in water. Appl Environ Microbiol 81:918. 10.1128/AEM.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907. https://arxiv.org/pdf/1207.3907.pdf.
- 82.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. https://arxiv.org/pdf/1303.3997.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads of this sequencing project have been uploaded to the NCBI SRA database under the BioProject number PRJNA610262.