We quantified rotavirus A (RVA), Rotarix, and RotaTeq strains in oyster and sewage samples during two gastroenteritis seasons and revealed the exact contamination of wild-type RVA by subtracting the quantitative value of rotavirus vaccine strains from that of RVA. The concentration of wild-type RVA was significantly correlated between oysters and sewage, although no significant correlation was seen between wild-type RVA concentration in sewage and the number of rotaviruses detected in patients with gastroenteritis.
KEYWORDS: rotavirus, oyster, bioaccumulation, qPCR, vaccine
ABSTRACT
Rotavirus is one of the major causes of infectious gastroenteritis among infants and children, and live attenuated vaccines for rotavirus A (RVA), namely, Rotarix and RotaTeq, have recently become available in Japan. Rotavirus is known to be excreted from patients and accumulated in oysters similar to norovirus; however, the vaccine strains in aquatic environments or oysters have not yet been analyzed. In this study, we focused on wild-type RVA, which is highly important in considering the risk of infectious diseases. We quantified total RVA, Rotarix, and RotaTeq strains in oyster and sewage samples collected between September 2014 and July 2016 to assess the contamination levels of wild-type RVA by subtracting the quantitative value of rotavirus vaccine strains from that of total RVA. The positive rates of wild-type RVA, Rotarix, and RotaTeq in oysters were 54, 14, and 31%, respectively. These rates were comparable to those of wild-type RVA (57%) and RotaTeq (35%) in sewage; however, Rotarix was not detected in any sewage samples. The comparison of viral concentrations in oysters and sewage suggested more efficient accumulation of the vaccine strains in oysters than the wild-type RVA. The concentration of wild-type RVA in oysters was significantly correlated with that in sewage with a lag time of −6 to 0 weeks which is required for viral transportation from wastewater treatment plants to oysters. On the other hand, no significant correlation was observed between wild-type RVA concentration in sewage and the number of rotavirus-associated gastroenteritis cases, implying the existence of asymptomatic RVA-infected individuals.
IMPORTANCE We quantified rotavirus A (RVA), Rotarix, and RotaTeq strains in oyster and sewage samples during two gastroenteritis seasons and revealed the exact contamination of wild-type RVA by subtracting the quantitative value of rotavirus vaccine strains from that of RVA. The concentration of wild-type RVA was significantly correlated between oysters and sewage, although no significant correlation was seen between wild-type RVA concentration in sewage and the number of rotavirus-associated gastroenteritis cases. This finding suggested the existence of asymptomatic patients and that monitoring of rotavirus vaccine strain could be useful to understand the trend of wild-type RVA and rotavirus outbreak in detail. We believe that our study makes a significant contribution to the literature because it reports the detection of rotavirus vaccine strains in oysters.
INTRODUCTION
Rotavirus is one of the major causes of infectious gastroenteritis worldwide (1). Before the introduction of vaccines, approximately all children under 5 years of age were estimated to have experienced rotavirus infection; 352,000 to 592,000 children under 5 years of age were assumed to die due to rotavirus infections annually, and 82% of these deaths occurred in children from low-income countries (2). Two rotavirus vaccines, Rotarix (GlaxoSmithKline Biologicals, Rixensart, Belgium) and RotaTeq (Merck & Co., Inc., Kenilworth, NJ, USA) were tested for safety and effectiveness and were eventually introduced in 2006 (3, 4). Since 2011 and 2012, these two vaccines have been available in Japan. Kobayashi et al. compared the number of hospitalized pediatric patients with rotavirus infections between 2009 and 2015 and reported a significant decrease in hospitalizations in 2014 and 2015 in the years following the introduction of the vaccines (5).
Both Rotarix and RotaTeq are live attenuated oral vaccines, the strains of which multiply in the intestinal tracts of vaccinated infants. Rotavirus detected in oysters is the cause of oyster-related food poisoning (6, 7). Oysters can accumulate gastroenteritis-causing viruses discharged into seawater through filter-feeding (8). In particular, norovirus accumulation in oysters has been actively studied by many researchers (9–12). Compared to norovirus, rotavirus accumulation in oysters has been relatively less explored (13), despite receiving considerable attention from researchers worldwide.
In our previous study, the concentration of rotavirus A (RVA) in cultured oysters was investigated using quantitative real-time PCR (qPCR) to study the relationship between RVA in oysters and the gastroenteritis epidemic (14). RVA concentrations in oysters were found to increase during winter, which is also known as the gastroenteritis season in Japan. However, RVA in oysters was not related to either RVA concentration in sewage or the number of rotavirus-associated gastroenteritis cases. A possible reason could be the presence of rotavirus vaccine strains together with wild-type RVA. Information regarding the presence of rotavirus vaccine strains in aquatic environments and shellfish is very limited, and further research would be required to reveal the behavior of wild-type RVA, originating from patients with gastroenteritis, in aquatic environments. In this study, we investigated the concentration of wild-type RVA in cultured oysters and sewage water by subtracting the quantitative values of rotavirus vaccine strains, Rotarix and RotaTeq, from that of RVA. Moreover, we performed a correlation analysis for the virus concentration in oysters and sewage, with the number of rotavirus-associated gastroenteritis cases reported from sentinel clinics in the study area.
RESULTS
Quantification of wild-type RVA, Rotarix, and RotaTeq.
Quantitative results of wild-type RVA and vaccine strain concentrations found in oyster and sewage samples are illustrated in Fig. 1. Wild-type RVA and RotaTeq strains were detected in oyster samples during the winter of both years, while the Rotarix strain was detected only in 2016. The highest concentration of each strain was 5.4 log10 copies/g of digestive tissues (DT) for wild-type RVA, 4.2 log10 copies/g of DT for Rotarix, and 4.2 log10 copies/g of DT for RotaTeq. In total, 54 (128 of 237), 14 (34 of 237), and 31% (74 of 237) of oyster samples were positive for wild-type RVA, Rotarix, and RotaTeq, respectively. All strains in oysters appeared to have the same seasonal trends, with the peak in winter known as the epidemic season of gastroenteritis, including rotavirus infection.
FIG 1.
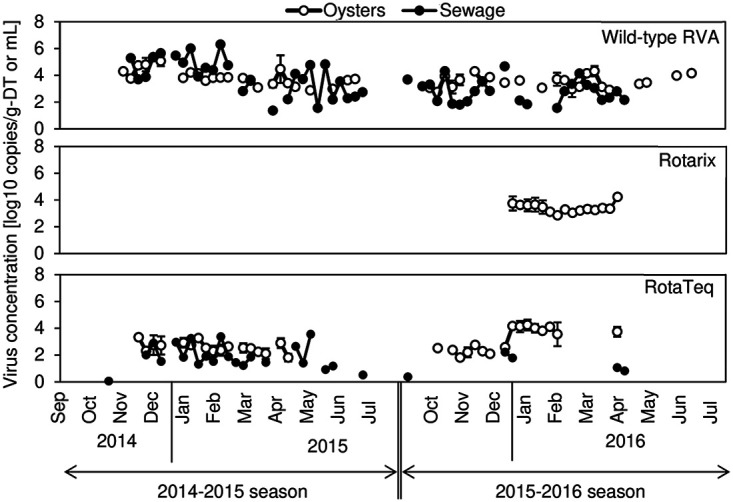
Wild-type RVA, Rotarix, and RotaTeq concentrations in oyster and sewage samples. Each circle represents the geometric mean virus concentration in the composite samples from which the virus was detected.
Wild-type RVA was detected in sewage water as well as in oysters (Fig. 1). Although the RotaTeq strain was detected in sewage, the frequency of detection was lower than that in the oyster samples, especially in the 2015–2016 season (October to July). The Rotarix strain was not detected in any sewage samples. The highest concentration of each strain was 6.3 log10 copies/ml for wild-type RVA and 3.5 log10 copies/ml for RotaTeq. The positive rates of wild-type RVA (57%, 54 of 77) and RotaTeq (35%, 27 of 77) in the sewage samples were comparable to those in the oyster samples.
The proportions of wild-type RVA, Rotarix, and RotaTeq strains were calculated by dividing each concentration by that of the total RVA in oysters (Fig. 2A) or sewage samples (Fig. 2B). The frequency that the RotaTeq strain was detected in oysters was <15%, and most of the RVA detected in the 2014–2015 season were wild-type strains. In the following season, the proportions of Rotarix and RotaTeq strains in oysters increased and exceeded that of the wild-type RVA in 10 weeks from January to April 2016. In contrast, the proportion of the vaccine strains in sewage was quite low (<10%) throughout the study period.
FIG 2.
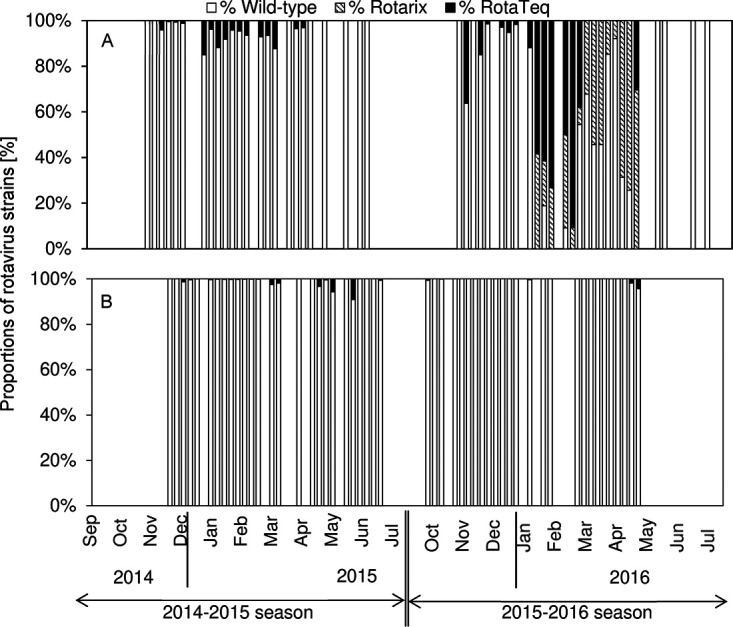
Proportions, or ratios of concentration, of wild-type RVA, Rotarix, and RotaTeq strains to total RVA in oysters (A) and sewage (B).
Correlation among virus concentrations in oysters and sewage and the number of rotavirus-associated gastroenteritis cases.
Table 1 shows the correlation coefficients among rotavirus concentrations in oysters and sewage and the number of rotavirus-associated gastroenteritis cases. As shown in Fig. 1, we obtained a considerable amount of negative data from oyster and sewage samples, which may influence the correlation analysis. The correlation coefficients were calculated in two ways: (i) using only paired positive samples of oyster and sewage (method a) and (ii) using all pairs of oyster and sewage samples by substituting negative data with half of the limit of detection (LOD) (method b) (15, 16). The LOD values for RVA, Rotarix, and RotaTeq in the oyster samples were 8.0 to 16.5, 6.0 to 15.0, and 4.0 to 12.5 copies/g DT, respectively, while those in sewage samples were 4.8 to 13.2, 4.2 to 7.2, and 2.4 to 3.6 copies/ml. The coefficients could not be analyzed for Rotarix due to no positive data from sewage samples (Fig. 1). The analysis using method a found a significant correlation between the concentrations of wild-type RVA in sewage and oysters with a lag time of −6 to 0 weeks (P < 0.05) (Table 1). This indicates the long-term impact of this strain in oyster contamination when discharged from the wastewater treatment plant. On the other hand, the correlation was not significant for the RotaTeq strain. Common to both strains, the viral concentrations in oysters and sewage were not positively related to the number of rotavirus-associated gastroenteritis cases (Table 1). The analysis using method b also revealed a significant correlation between the concentrations of wild-type strains in sewage and oysters with a lag time of −4 to 0 weeks (Table 1). The concentration of RotaTeq was positively correlated (P < 0.05) with the number of rotavirus-associated gastroenteritis cases with a lag time of 1 to 7 weeks. This is likely due to a number of pairs of negative data in sewage and no cases of gastroenteritis reported.
TABLE 1.
Correlation coefficients between rotavirus concentrations in sewage and oysters and between the number of rotavirus-associated gastroenteritis cases and virus concentrations in oysters or sewage calculated by two methods
| Methoda and vaccine strain | Correlation | Correlation coefficient at the following lag time (wk)b: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −7 | −6 | −5 | −4 | −3 | −2 | −1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Method a | ||||||||||||||||
| Wild-type | Sewage- oysters |
0.31 | 0.50 | 0.45 | 0.55 | 0.48 | 0.39 | 0.48 | 0.47 | 0.18 | 0.37 | 0.23 | 0.23 | 0.10 | 0.37 | −0.03 |
| Patients- oysters |
0.01 | −0.18 | 0.07 | −0.39 | −0.13 | −0.26 | −0.53 | −0.30 | −0.47 | 0.01 | 0.03 | −0.18 | −0.28 | −0.39 | −0.22 | |
| Patients- sewage |
0.29 | 0.45 | 0.37 | −0.09 | 0.06 | −0.10 | 0.06 | −0.29 | −0.27 | −0.39 | −0.03 | −0.21 | −0.20 | 0.06 | 0.08 | |
| RotaTeq | Sewage- oysters |
−0.01 | −0.33 | 0.45 | 0.13 | −0.13 | −0.01 | −0.35 | −0.25 | 0.22 | 0.13 | 0.20 | −0.12 | −0.06 | 0.13 | 0.32 |
| Patients- oysters |
−0.36 | −0.34 | −0.19 | −0.35 | −0.15 | −0.29 | −0.28 | −0.35 | −0.49 | −0.31 | −0.47 | −0.26 | −0.49 | −0.39 | −0.26 | |
| Patients- sewage |
0.04 | 0.34 | −0.03 | −0.23 | −0.18 | −0.42 | 0.08 | −0.17 | 0.11 | 0.00 | 0.15 | 0.09 | −0.01 | 0.35 | 0.33 | |
| Method b | ||||||||||||||||
| Wild-type | Sewage- oysters |
0.24 | 0.32 | 0.31 | 0.46 | 0.43 | 0.31 | 0.38 | 0.34 | 0.22 | 0.28 | 0.15 | 0.02 | −0.18 | −0.33 | −0.20 |
| Patients- oysters |
0.18 | 0.16 | 0.16 | −0.01 | 0.06 | −0.03 | −0.07 | −0.02 | −0.24 | −0.10 | −0.23 | −0.29 | −0.22 | −0.38 | −0.44 | |
| Patients- sewage |
−0.05 | 0.09 | 0.04 | 0.01 | 0.13 | −0.06 | 0.01 | 0.00 | 0.10 | 0.14 | 0.13 | 0.12 | 0.22 | 0.26 | 0.36 | |
| RotaTeq | Sewage- oysters |
−0.21 | −0.46 | −0.27 | −0.22 | −0.30 | −0.23 | −0.33 | −0.17 | 0.03 | 0.02 | −0.04 | −0.07 | −0.17 | −0.12 | −0.23 |
| Patients- oysters |
−0.23 | −0.31 | −0.31 | −0.31 | −0.31 | −0.45 | −0.33 | −0.34 | −0.42 | −0.29 | −0.32 | −0.24 | −0.26 | −0.23 | −0.28 | |
| Patients- sewage |
0.10 | 0.28 | 0.20 | 0.30 | 0.24 | 0.19 | 0.31 | 0.28 | 0.42 | 0.37 | 0.36 | 0.33 | 0.43 | 0.49 | 0.55 | |
Method a used only paired positive samples of oysters and sewage, and method b used all of the samples with negative data substituted with half of the limit of detection (LOD).
Correlation coefficients between rotavirus concentrations in sewage and oysters and between the number of rotavirus-associated gastroenteritis cases and virus concentrations in oysters or sewage calculated by methods a and b at various time points. The time points are 1 to 7 weeks before (−1 to −7 weeks) or after (+1 to +7 weeks) the day of oyster collection. Statistically significant correlations (P < 0.05) are indicated by boldface values.
DISCUSSION
In this study, we detected and quantified rotavirus vaccine strains (i.e., Rotarix and RotaTeq) in oysters and sewage, and we are not aware of any previous reports of rotavirus vaccine strain detection in oysters. Previous reports focused on the detection of these vaccine strains in feces or environmental water by nucleotide sequencing. Yen et al. reported 21.4% of vaccinated children shed vaccine strains in feces for 3 to 9 days after vaccination (17). Moreover, Bucardo et al. detected the partial VP7 gene of rotavirus in wastewater samples with 98% sequence similarity to the vaccine strain RotaTeq-WI79-4 (18). Based on these studies, rotavirus vaccine strains can be hypothesized to be discharged into the aquatic environment from humans. Not only did the present study support this hypothesis, but it also quantitatively determined temporal changes of the vaccine strains. In conjunction with the quantification results of vaccine strains from oysters, our results will enhance the understanding of the dynamics of vaccine strains in the aquatic environment.
Although wild-type RVA and RotaTeq were detected in oysters in both seasons (2014–2015 and 2015–2016), Rotarix was not detected in the 2014–2015 season. In addition, RotaTeq was not detected in sewage as often as in oysters, and Rotarix was not detected in sewage water throughout the study period. According to the data of rotavirus vaccine usage in the area studied, provided by GlaxoSmithKline K.K. and MSD K.K. (a subsidiary of Merck & Co., Inc.), the number of infants vaccinated with Rotarix and RotaTeq in the 2014–2015 season was not significantly different from that in the 2015–2016 season. Thus, the difference in trends between Rotarix and RotaTeq detection could be due to the difference in the usage of Rotarix and RotaTeq in Japan. Rotarix is inoculated in infants twice until the age of 24 weeks, while RotaTeq is inoculated thrice until the age of 32 weeks. Weaning of infants is generally initiated approximately at 20 to 24 weeks of age in Japan. Feces of infants receiving weaning foods are flushed through the toilet and flow into the sewage, as with those of the adults. Therefore, there may be a greater inflow of the RotaTeq strain into sewage compared with the Rotarix strain. Both vaccine strains were detected from oysters even when they were not detected in sewage, suggesting that the strains existing in sewage at concentrations lower than the detection limit of qPCR were accumulated in oyster DT as the result of bioconcentration.
The positive rates of wild-type RVA, Rotarix, and RotaTeq in oysters were 54, 14, and 31%, respectively. It is difficult to compare this result with previous studies because, as per our knowledge, there has been no research on the detection of vaccine strains in oysters; however, our positive rate of wild-type RVA was found to be higher than that of the total RVA from previous studies conducted in Brazil (19%) (13) and Thailand (13%) (19). This variability may be explained by differences in the sampling area, study season, and detection methods. Particularly, our method of virus extraction from oysters was developed to improve the sensitivity of virus detection (20) by the decomposition of the organic matter in oysters using amylase, along with efficient precipitation of residues using citrate buffer for virus elution. This method achieved a 6.7 times higher extraction efficiency of norovirus from oyster samples than the previous method, which used sterilized water to elute viruses (20). The positive rates of wild-type RVA and RotaTeq in sewage were 57 and 35%, respectively. These values are similar to those reported by other studies targeting total RVA (21, 22).
Although no data were available, we believe children continually shed vaccine strains throughout the year. The seasonal trends commonly observed for vaccine and wild-type strains are probably due to other factors in oysters and environment. For example, Maalouf et al. detected a seasonal effect on the expression of ligands implicated in norovirus attachment to oyster tissues, which peaks in late winter and spring (23). The proportion of vaccine strains in oysters in the 2015–2016 season was much higher than that during the previous season. One of the reasons for this difference could be the number of patients with gastroenteritis in the monitored seasons as the source of wild-type RVA detected in the oysters. Indeed, only 12 patients with rotavirus were reported in the 2015–2016 season, which was much less than that in the 2014–2015 season (80 cases). In contrast, the proportion of vaccine strains in sewage was lower than 10% throughout the study period. This result may suggest a more efficient accumulation of vaccine strains into oysters than wild-type RVA due to their decreased removal in wastewater treatment, longer survival in natural water, higher affinity for and/or more stability in oysters. Rotarix strain belongs to a serotype G1P[8], while RotaTeq strain is a reassortant targeting five serotypes (G1, G2, G3, G4, and P[8]). The efficient accumulation of vaccine strains in oysters, suggested by the results and considerations above, could be explained by the difference in serotypes from the wild type, although the underlying factors should be analyzed in further studies.
Furthermore, we tried to identify the relationship between rotaviruses in oysters and sewage and the number of rotavirus-associated gastroenteritis cases. In both methods of data analysis in this study, the correlation between wild-type RVA in sewage and the number of rotavirus-associated gastroenteritis cases was not significant, hence suggesting that there were many asymptomatic cases that were undiagnosed (24, 25). This observation is also consistent with those of another study by Ruggeri et al. reporting that the seasonal trend of RVA detection in hospitalized patients with rotavirus was different from that in sewage, implying the circulation of RVA in asymptomatic populations (26). Wild-type RVA in oysters and sewage were significantly correlated at a lag time of −6 to 0 weeks using method a (P < 0.05) and that of −4 to 0 weeks using method b, similar to our findings with norovirus (9). It would take the obtained lag time for wild-type RVA discharged from both symptomatic and asymptomatic cases to reach oysters through the sewage system and natural aquatic environment in the study area. In contrast, the RotaTeq strain in oysters was not correlated with that in the sewage by both methods of data analysis, although its more efficient accumulation in oysters compared with that of wild-type strains is indicated above. This contradiction can be resolved by further studies.
In conclusion, this study demonstrated that the detection and quantification of rotavirus vaccine strains can provide us with more detailed information about the dynamics of wild-type strains which cannot usually be obtained by detecting only total RVA. The concentration of wild-type strains (originally discharged from RVA-infected individuals) in oysters correlated with those in sewage. Monitoring of wild-type strains in oysters enables us to not only supply safer oysters to consumers but to understand the epidemiology of rotavirus infections, including the prevalence of asymptomatic cases. The advent of vaccines has decreased the occurrence of rotavirus infection-related illnesses. However, we believe rotavirus is still a safety issue in Japan, where rotavirus vaccines have recently been included in the routine vaccination supported by municipal/prefectural governments in October 2020. Moreover, the vaccines are effective only for a part of serotypes of rotavirus, which frequently have caused illness. If new virulent serotypes that are not covered by the vaccines emerge, the number of illnesses will increase again and reach the level seen prior to vaccination. Some studies (27–29) reported the emergence of different serotypes of rotavirus from those belonging to the genogroup Wa, which had been previously epidemic, just after the introduction of vaccines. Furthermore, in the present study, wild-type RVA and vaccine strains were detected simultaneously from 87 oyster samples (37%) and 23 sewage samples (30%), implying a risk of coinfection in those exposed to oysters and sewage, followed by reassortment between wild-type and vaccine strains as previously reported (30–32). While monitoring of rotavirus in oysters, particular attention should be paid to the change in epidemic strains and the emergence of virulent strains generated by such a genetic reassortment.
MATERIALS AND METHODS
Sample collection.
Municipal sewage (1 liter) flowing into a sewage treatment plant was collected every week from 24 September 2014 to 28 April 2016, while nine cultured oysters (Crassostrea gigas) were collected every week until 28 July 2016 in a bay used for harvesting market oysters in Miyagi Prefecture, Japan. The treatment plant, which locates 1.8 km upstream from the mouth of a river flowing into the bay, receives approximately 4,800 m3 of sewage per day from 67.4% of the population (14,367 inhabitants) of Matsushima town and treats it with an oxidation ditch process followed by chlorine disinfection. Domestic wastewater from the remaining population is treated with septic tanks and discharged into the above river. The oyster samples were collected in the bay (3.2 km, far from the river mouth), delivered to the laboratory on ice, and shucked within 6 h of arrival. Digestive tissues (DT) were excised from oysters and introduced into 1.5-ml microtubes individually. Sewage and DT samples were stored at −80°C until analyzed.
Virus concentration.
Virus particles were concentrated from sewage samples using the polyethylene glycol (PEG) precipitation method (33). Briefly, 3.2 g of PEG 6000 (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) and 0.92 g of NaCl (Kanto Chemical Co., Tokyo, Japan) were added to 40 ml of sewage sample. Ten million (1.0 × 107) copies of murine norovirus (MNV) strain S7-PP3, kindly provided by Yukinobu Tohya (Nihon University, Japan), were added to the samples as process control and mixed for 12 h at 4°C. The samples were then centrifuged at 9,000 × g for 30 min at 4°C, and the supernatant was discarded. Then, 1 ml of sterilized ultrapure water was added to each pellet and vortexed for 1 min. Suspensions of each pellet were centrifuged at 10,000 × g for 10 min at 4°C, and 1 ml of each supernatant was recovered as virus concentrate.
Viruses in oyster samples were extracted by a previously described method (34). Briefly, 1 ml of enzyme solution containing 6.3 mg/ml of amylase (Sigma-Aldrich, St. Louis, MO, USA), 6.3 mg/ml of lipase (Sigma-Aldrich, St. Louis, MO, USA), and 0.25 mg/ml of proteinase K (Roche, Penzberg, Germany) was added to each DT sample, which was then mashed using two stainless beads (3.2 mm in diameter) on Micro Smash-100 (TOMY, Tokyo, Japan) at 4,200 rpm for 60 s. Ten million (1.0 × 107) copies of MNV were added to the samples as process control. The DT samples were incubated at 37°C for 1 h and 60°C for 15 min and centrifuged thereafter at 9,100 × g for 12 min. Supernatants were recovered as virus concentrates. Three concentrates from DT were pooled in a 5.0-ml tube to make a composite sample; three composites were obtained each week. To remove organic matter from oyster DT, 500 μl of citric acid buffer (400 mM, pH 2.5) was added to the same amount of DT composites and centrifuged at 9,100 × g for 12 min (34). Supernatants of each composite were used for RNA extraction. Viral RNA from sewage concentrates was extracted using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany), and viral RNA from DT composites was extracted using the NucliSENS miniMAG RNA extraction kit (bioMérieux, Marcy l'Etoile, France), following the manufacturer’s instructions. A reverse transcription (RT) reaction of RNA was performed to prepare cDNA using 20 μl of the iScript advanced cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) and 20 μl of RNA samples as described previously (34).
Virus quantification.
RVA, Rotarix, and RotaTeq genomes in collected samples were quantitatively detected using qPCR following the methods described by Miura et al. (35) for RVA and Gautam et al. (36) for vaccine strains. Gautam et al. (36) designed specific primers and TaqMan probes for the NSP2 and VP4 genes of Rotarix and the VP6 and VP3 genes of RotaTeq. They evaluated the developed RT-qPCR assays using RVA-positive stool samples containing Rotarix, RotaTeq, or wild-type RVA and RVA-negative stool samples to check the sensitivity and specificity of each primer set. The sensitivity was calculated by dividing the number of samples detected with each primer set by the number of positive stool samples, while the specificity was calculated by dividing the number of samples not detected with each primer set by the number of negative stool samples. On the basis of their results, we selected primer sets (Table 2) of VP4 for Rotarix with 92% sensitivity and 100% specificity and VP6 for RotaTeq with 100% sensitivity and 98.6% specificity (36).
TABLE 2.
Sequences of primers and probes used in this study
| Target | Primer or probea | Sequenceb |
|---|---|---|
| RVA | Rota NVP3-F1 | ACCATCTACACATGACCCTC |
| Rota NVP3-F2 | ACCATCTTCACGTAACCCTC | |
| Rota NVP3-R | GGTCACATAACGCCCC | |
| Rota NVP3-TP | FAM-TGAGCACAATAGTTAAAAGC-MGB-NFQ | |
| Rotarix | Rotarix VP4-F | TGTGAGTAA“C”GATTCAAATAAATGGAAGTT |
| Rotarix VP4-R | TCACCATGAAATGTCCATACTCTTCCACCA | |
| Rotarix VP4-P | FAM-ATA{C}CAGA{C}TTGTAGGAATAYTTAAATA-BHQ1 | |
| RotaTeq | RotaTeq-VP6-F | GCGGCGTTATTTCCAAATGCACAG |
| RotaTeq-VP6-R | CGTCGGCAAGCACTGATTCACAAA | |
| RotaTeq-VP6-P | FAM-ATCACGCAA“C”AGTAGGACT“C”ACGCTT-BHQ1 | |
Forward and reverse primers are indicated by F and R at the end of the designation. Probes are indicated by P at the end of the designation.
{C} and “C” denote AP-dC (G-clamp) and C-5 propynyl-dC, respectively. FAM, MGB, NFQ, and BHQ in the probes stand for 5-carboxyfluorescein, minor groove binder, nonfluorescent quencher, and black hole quencher, respectively.
To determine concentrations of viral genomes, the amplification reaction was performed on a CFX96 touch real-time PCR system (Bio-Rad, Hercules, CA, USA), with 20 μl of reaction mixture containing 5 μl of cDNA, 10 μl of SsoAdvanced universal probe supermix (Bio-Rad, Hercules, CA, USA), and primers (0.4 μl) and probes (0.3 μl for Rota NVP3-TP and 0.2 μl for Rotarix VP4-P and RotaTeq-VP6-P) which are listed in Table 2. The PCR cycling conditions were 30 s at 95°C, followed by 50 cycles, with 1 cycle consisting of 15 s at 95°C and 60 s at 60°C. A 10-fold dilution of each cDNA sample was also analyzed by qPCR in the same manner to detect possible inhibition during the PCR (9). Samples with quantification cycles (Cq values) below 40 were considered positive in accordance with the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines (37). The standard curve was generated from 10-fold serial dilutions (101 to 105 copies per well) of oligonucleotide DNA containing target sequences (38–40) synthesized, as shown in Table 3. Concentration of wild-type RVA was calculated by subtracting the number of vaccine strains from the total RVA.
TABLE 3.
Sequences of oligonucleotide DNA used as the standards for RVA, Rotarix, and RotaTeq
| Strain | Oligonucleotide sequence |
|---|---|
| RVA | ACCATCTACACATGACCCTCTATGAGCACAATAGTTAAAAGCTAACACTGTCAAAAACCTAAATGGCTATAGGGGCGTTATGTGACC |
| Rotarix | GTGAGTAACGATTCAAATAAATGGAAGTTTTTAGAAATGTTTAGAAGCAGTAGTCAAAATGAATTTTATAATAGACGTACATTAACTTCTGATACCAGACTTGTAGGAATATTTAAATATGGTGGAAGAGTATGGACATTTCATGGTGA |
| RotaTeq | GCGGCGTTATTTCCAAATGCACAGCCATTTGAACATCACGCAACAGTAGGACTCACGCTTAGAATTGAATCTGCAGTTTGTGAATCAGTGCTTGCCGACG |
MNV added to all oyster and sewage samples as the process control was also quantified by qPCR, and the recovery rate was obtained by dividing the MNV amount measured by qPCR by the amount added to each sample (9). If their recovery rates were less than 1%, suggesting significant loss of viruses in the PEG precipitation or inhibition in RT-qPCR, the oyster and sewage samples were excluded from further analyses.
Correlation analysis.
Correlations among rotavirus concentrations in oysters and sewage and the number of rotavirus-associated gastroenteritis cases, which were reported weekly from the sentinel clinics, were analyzed. Since cases reported in Matsushima town covered by the studied wastewater treatment plant were rare, we used the number of cases reported in a big city close to the town. In Japan, it is not required that all children born before 1 August 2020 are vaccinated against rotavirus; however, vaccination effectiveness was revealed by a survey covering 2.3% of the country’s population, demonstrating a significant decrease in hospitalized children in 2014 and 2015 (5). This trend should be common to this study area, although the data were not available there before October 2013. Considering the lag time required for the transmission of the rotavirus from infected individuals to oysters, Pearson’s correlation coefficient was calculated using the virus concentration in sewage, and the number of cases reported 1 to 7 weeks before (−1 to −7 weeks) or after (+1 to +7 weeks) the day of oyster collection. Finally, the null hypothesis of no correlation was tested with a significance level of 0.05.
ACKNOWLEDGMENTS
This study was supported by the Japan Science and Technology Agency (JST) through a Core Research for Evolutionary Science and Technology (CREST) program, “Innovation of water monitoring system with rapid, highly precise and exhaustive pathogen detection technologies” and JSPS KAKENHI grant 18H03792.
REFERENCES
- 1.Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Reiner RC, Fullman N, Thompson RL, Abajobir A, Ahmed M, Alemayohu MA, Alvis-Guzman N, Amare AT, Antonio CA, Asayesh H, Avokpaho E, Awasthi A, Bacha U, Barac A, Betsue BD, Beyene AS, Boneya DJ, Malta DC, Dandona L, Dandona R, Dubey M, Eshrati B, Fitchett JRA, Gebrehiwot TT, Hailu GB, Horino M, Hotez PJ, Jibat T, Jonas JB, Kasaeian A, Kissoon N, Kotloff K, Koyanagi A, Kumar GA, Rai RK, Lal A, El Razek HMA, Mengistie MA, Moe C, Patton G, Platts-Mills JA, Qorbani M, Ram U, Roba HS, Sanabria J, Sartorius B, et al. 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis 9:565–572. doi: 10.3201/eid0905.020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, Cervantes Y, Linhares AC, López P, Macías-Parra M, Ortega-Barría E, Richardson V, Rivera-Medina DM, Rivera L, Salinas B, Pavía-Ruz N, Salmerón J, Rüttimann R, Tinoco JC, Rubio P, Nuñez E, Guerrero ML, Yarzábal JP, Damaso S, Tornieporth N, Sáez-Llorens X, Vergara RF, Vesikari T, Bouckenooghe A, Clemens R, De Vos B, O’Ryan M. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- 4.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, Shinefield HR, Christie CDC, Ylitalo S, Itzler RF, Coia ML, Onorato MT, Adeyi BA, Marshall GS, Gothefors L, Campens D, Karvonen A, Watt JP, O’Brien KL, DiNubile MJ, Clark HF, Boslego JW, Offit PA, Heaton PM, Rotavirus Efficacy and Safety Trial (REST) Study Team. 2006. Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi M, Adachi N, Miyazaki M, Tatsumi M. 2018. Decline of rotavirus-coded hospitalizations in children under 5 years: a report from Japan where rotavirus vaccines are self-financed. Vaccine 36:2727–2732. doi: 10.1016/j.vaccine.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Le Guyader FS, Le Saux JC, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P, Atmar RL. 2008. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J Clin Microbiol 46:4011–4017. doi: 10.1128/JCM.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iritani N, Kaida A, Abe N, Kubo H, Sekiguchi J-I, Yamamoto SP, Goto K, Tanaka T, Noda M. 2014. Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J Med Virol 86:2019–2025. doi: 10.1002/jmv.23883. [DOI] [PubMed] [Google Scholar]
- 8.Ueki Y, Sano D, Watanabe T, Akiyama K, Omura T. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res 39:4271–4280. doi: 10.1016/j.watres.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Pu J, Miura T, Kazama S, Konta Y, Azraini ND, Ito E, Ito H, Omura T, Watanabe T. 2018. Weekly variations in norovirus genogroup II genotypes in Japanese oysters. Int J Food Microbiol 284:48–55. doi: 10.1016/j.ijfoodmicro.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Campos CJA, Kershaw S, Morgan OC, Lees DN. 2017. Risk factors for norovirus contamination of shellfish water catchments in England and Wales. Int J Food Microbiol 241:318–324. doi: 10.1016/j.ijfoodmicro.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Le Guyader FS, Parnaudeau S, Schaeffer J, Bosch A, Loisy F, Pommepuy M, Atmar RL. 2009. Detection and quantification of noroviruses in shellfish. Appl Environ Microbiol 75:618–624. doi: 10.1128/AEM.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajko-Nenow P, Keaveney S, Flannery J, O’Flaherty V, Doré W. 2012. Characterisation of norovirus contamination in an Irish shellfishery using real-time RT-qPCR and sequencing analysis. Int J Food Microbiol 160:105–112. doi: 10.1016/j.ijfoodmicro.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Rigotto C, Victoria M, Moresco V, Kolesnikovas CKK, Corrêa AA, Souza D, Miagostovich MPP, Simões CMO, Barardi CRM. 2010. Assessment of adenovirus, hepatitis A virus and rotavirus presence in environmental samples in Florianopolis, South Brazil. J Appl Microbiol 109:1979–1987. doi: 10.1111/j.1365-2672.2010.04827.x. [DOI] [PubMed] [Google Scholar]
- 14.Ito E, Pu J, Miura T, Kazama S, Nishiyama M, Ito H, Konta Y, Nguyen GT, Omura T, Watanabe T. 2019. Weekly variation of rotavirus A concentrations in sewage and oysters in Japan, 2014–2016. Pathogens 8:89. doi: 10.3390/pathogens8030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafleur B, Lee W, Billhiemer D, Lockhart C, Liu J, Merchant N. 2011. Statistical methods for assays with limits of detection: serum bile acid as a differentiator between patients with normal colons, adenomas, and colorectal cancer. J Carcinog 10:12. doi: 10.4103/1477-3163.79681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitcomb BW, Schisterman EF. 2008. Assays with lower detection limits: implications for epidemiological investigations. Paediatr Perinat Epidemiol 22:597–602. doi: 10.1111/j.1365-3016.2008.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen C, Jakob K, Esona MD, Peckham X, Rausch J, Hull JJ, Whittier S, Gentsch JR, LaRussa P. 2011. Detection of fecal shedding of rotavirus vaccine in infants following their first dose of pentavalent rotavirus vaccine. Vaccine 29:4151–4155. doi: 10.1016/j.vaccine.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucardo F, Lindgren P-E, Svensson L, Nordgren J. 2011. Low prevalence of rotavirus and high prevalence of norovirus in hospital and community wastewater after introduction of rotavirus vaccine in Nicaragua. PLoS One 6:e25962. doi: 10.1371/journal.pone.0025962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittigul L, Singhaboot Y, Chavalitshewinkoon-Petmitr P, Pombubpa K, Hirunpetcharat C. 2015. A comparison of virus concentration methods for molecular detection and characterization of rotavirus in bivalve shellfish species. Food Microbiol 46:161–167. doi: 10.1016/j.fm.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Masago Y, Ueki Y, Watanabe T. 2013. Detection and quantification of norovirus in oysters using low pH elution and enzymatic virus elution methods. J Jpn Soc Civ Eng Ser G Environ Res 69:III_657–III_665. doi: 10.2208/jscejer.69.III_657. [DOI] [Google Scholar]
- 21.Fumian TM, Leite JPG, Castello AA, Gaggero A, de Caillou MS, Miagostovich MP. 2010. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J Virol Methods 170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Hassine-Zaafrane M, Kaplon J, Ben Salem I, Sdiri-Loulizi K, Sakly N, Pothier P, Aouni M, Ambert-Balay K. 2015. Detection and genotyping of group A rotaviruses isolated from sewage samples in Monastir, Tunisia between April 2007 and April 2010. J Appl Microbiol 119:1443–1453. doi: 10.1111/jam.12920. [DOI] [PubMed] [Google Scholar]
- 23.Maalouf H, Zakhour M, Le Pendu J, Le Saux JC, Atmar RL, Le Guyader FS. 2010. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl Environ Microbiol 76:5621–5630. doi: 10.1128/AEM.00148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips G, Lopman B, Rodrigues LC, Tam CC. 2010. Asymptomatic rotavirus infections in England: prevalence, characteristics, and risk factors. Am J Epidemiol 171:1023–1030. doi: 10.1093/aje/kwq050. [DOI] [PubMed] [Google Scholar]
- 25.Paul A, Gladstone BP, Mukhopadhya I, Kang G. 2014. Rotavirus infections in a community based cohort in Vellore, India. Vaccine 32:A49–A54. doi: 10.1016/j.vaccine.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 26.Ruggeri FM, Bonomo P, Ianiro G, Battistone A, Delogu R, Germinario C, Chironna M, Triassi M, Campagnuolo R, Cicala A, Giammanco GM, Castiglia P, Serra C, Gaggioli A, Fiore L. 2015. Rotavirus genotypes in sewage treatment plants and in children hospitalized with acute diarrhea in Italy in 2010 and 2011. Appl Environ Microbiol 81:241–249. doi: 10.1128/AEM.02695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 28.Komoto S, Tacharoenmuang R, Guntapong R, Ide T, Haga K, Katayama K, Kato T, Ouchi Y, Kurahashi H, Tsuji T, Sangkitporn S, Taniguchi K. 2015. Emergence and characterization of unusual DS-1-like G1P[8] rotavirus strains in children with diarrhea in Thailand. PLoS One 10:e0141739. doi: 10.1371/journal.pone.0141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luchs A, da Costa AC, Cilli A, Komninakis SCV, Carmona RCC, Morillo SG, Sabino EC, Timenetsky MCST. 2019. First detection of DS-1-like G1P[8] double-gene reassortant rotavirus strains on the American continent, Brazil, 2013. Sci Rep 9:2210. doi: 10.1038/s41598-019-38703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose TL, Marques da Silva MF, Goméz MM, Resque HR, Ichihara MYT, de Mello Volotão E, Leite JPG. 2013. Evidence of vaccine-related reassortment of rotavirus, Brazil, 2008–2010. Emerg Infect Dis 19:1843–1846. doi: 10.3201/eid1911.121407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jere KC, Chaguza C, Bar-Zeev N, Lowe J, Peno C, Kumwenda B, Nakagomi O, Tate JE, Parashar UD, Heyderman RS, French N, Cunliffe NA, Iturriza-Gomara M. 2017. Emergence of double- and triple-gene reassortant G1P[8] rotaviruses possessing a DS-1-like backbone after rotavirus vaccine introduction in Malawi. J Virol 92:1246–1263. doi: 10.1128/JVI.01246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donato CM, Ch’ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. 2012. Identification of strains of rotateq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J Infect Dis 206:377–383. doi: 10.1093/infdis/jis361. [DOI] [PubMed] [Google Scholar]
- 33.Kazama S, Masago Y, Tohma K, Souma N, Imagawa T, Suzuki A, Liu X, Saito M, Oshitani H, Omura T. 2016. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res 92:244–253. doi: 10.1016/j.watres.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Pu J, Kazama S, Miura T, Azraini ND, Konta Y, Ito H, Ueki Y, Cahyaningrum EE, Omura T, Watanabe T. 2016. Pyrosequencing analysis of norovirus genogroup II distribution in sewage and oysters: first detection of GII.17 Kawasaki 2014 in oysters. Food Environ Virol 8:310–312. doi: 10.1007/s12560-016-9261-5. [DOI] [PubMed] [Google Scholar]
- 35.Miura T, Schaeffer J, Le Saux JC, Le Mehaute P, Le Guyader FS. 2018. Virus type-specific removal in a full-scale membrane bioreactor treatment process. Food Environ Virol 10:176–186. doi: 10.1007/s12560-017-9330-4. [DOI] [PubMed] [Google Scholar]
- 36.Gautam R, Esona MD, Mijatovic-Rustempasic S, Tam KI, Gentsch JR, Bowen MD. 2014. Real-time RT-PCR assays to differentiate wild-type group A rotavirus strains from Rotarix® and RotaTeq® vaccine strains in stool samples. Hum Vaccin Immunother 10:767–777. doi: 10.4161/hv.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee A, Mullick S, Deb AK, Panda S, Chawla-Sarkar M. 2013. First report of human rotavirus G8P[4] gastroenteritis in India: evidence of ruminants-to-human zoonotic transmission. J Med Virol 85:537–545. doi: 10.1002/jmv.23483. [DOI] [PubMed] [Google Scholar]
- 39.Ferguson AS, Layton AC, Mailloux BJ, Culligan PJ, Williams DE, Smartt AE, Sayler GS, Feighery J, McKay LD, Knappett PSK, Alexandrova E, Arbit T, Emch M, Escamilla V, Ahmed KM, Alam MJ, Streatfield PK, Yunus M, van Geen A. 2012. Comparison of fecal indicators with pathogenic bacteria and rotavirus in groundwater. Sci Total Environ 431:314–322. doi: 10.1016/j.scitotenv.2012.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W, Gu AZ, Zeng SY, Li D, He M, Shi HC. 2011. Development of a combined immunomagnetic separation and quantitative reverse transcription-PCR assay for sensitive detection of infectious rotavirus in water samples. J Microbiol Methods 84:447–453. doi: 10.1016/j.mimet.2011.01.011. [DOI] [PubMed] [Google Scholar]


