Resistance to the critically important antimicrobials vancomycin, teicoplanin, and linezolid is not found in enterococci collected from Australian finisher pigs. However, some antimicrobial resistance was observed.
KEYWORDS: enterococci, pigs, agriculture, genetics, antimicrobial resistance, porcine
ABSTRACT
Enterococci are ubiquitous opportunistic pathogens that have become a major public health issue globally. The increasing prevalence of antimicrobial resistance in hospital-adapted enterococci had been thought to originate from livestock. However, this association between livestock and hospital-adapted enterococci is currently unclear. This study investigates the antimicrobial susceptibilities of enterococci isolated from pig cecal samples and compares the genomic characteristics of Enterococcus faecium from pigs to those of isolates from meat chickens and from human sepsis cases. From 200 cecal samples, antimicrobial susceptibility testing was performed for E. faecium (n = 84), E. hirae (n = 36), and E. faecalis (n = 17). Whole-genome sequencing was performed for all E. faecium isolates, and the sequences were compared to those of previously studied isolates from meat chickens and human sepsis cases through bioinformatics analysis. Resistance (non-wild type) to erythromycin, gentamicin, tetracycline, ampicillin, daptomycin, virginiamycin, and quinupristin-dalfopristin was identified. More importantly, except for a single isolate harboring the vanC operon, no resistance was observed in the three species to vancomycin, teicoplanin, and linezolid, which are critically important antimicrobials used to treat enterococcal infections in humans. The E. faecium isolates from chickens were genetically distinct from human and pig isolates, which were more closely related. Human strains that were closely related to pig strains were not typical “hospital-adapted strains” as previously identified. The results of this study show that enterococci from Australian finisher pigs are not a source of resistance to critically important antimicrobials and that E. faecium from pigs is not part of the current human hospital-adapted population.
IMPORTANCE Resistance to the critically important antimicrobials vancomycin, teicoplanin, and linezolid is not found in enterococci collected from Australian finisher pigs. However, some antimicrobial resistance was observed. In particular, resistance to quinupristin-dalfopristin, a combination of two streptogramin class antimicrobials, was identified despite the absence of streptogramin use Australia-wide since 2005. Other observed resistance among enterococci from pigs include chloramphenicol, erythromycin, and tetracycline resistance. Genomic comparison of E. faecium from Australian pigs to isolates collected from previous studies on chickens and humans indicate that E. faecium from pigs are genetically more similar to those of humans than those from chickens. Despite the increased genetic similarities, E. faecium strains from pigs are phylogenetically distinct and did not belong to the dominant sequence types found in hospital-adapted strains causing sepsis in humans. Therefore, the results indicate that Australian finisher pigs are not a source of hospital-adapted E. faecium in Australia.
INTRODUCTION
Enterococci are ubiquitous bacteria commonly found as commensals of the mammalian and avian gastrointestinal microbiota. However, some enterococci can behave as opportunistic pathogens, causing invasive infections ranging from mild to life-threatening. Within the genus, Enterococcus faecalis and E. faecium are the most frequently reported human pathogens, with E. faecalis appearing more frequently as the agent responsible for infection (1). As opportunistic pathogens, enterococcal infections are generally health care-associated, with few reports of community-acquired infections (2). However, in the past 3 decades, an increase in prevalence of antimicrobial resistance observed in health care-associated enterococci has adversely impacted treatment outcomes (3). In particular, resistance to vancomycin, a critically important glycopeptide antimicrobial used to treat a variety of severe Gram-positive infections, has become a major public health issue globally (1).
Vancomycin resistance in enterococci is conferred by the expression of the van operon, of which the vanA and vanB types are the most clinically relevant (4, 5). The significance of the vanA and vanB operons in enterococci stems from their ability to be horizontally transferred to other strains of enterococci and even to other species of bacteria causing outbreaks to occur (6). Phenotypically, the vanA operon typically confers high levels of resistance against vancomycin and teicoplanin, a glycopeptide antimicrobial with a similar spectrum of activity, whereas the vanB operon typically confers variable levels of resistance to vancomycin only (7). Despite causing a lower number of infections, E. faecium is more often found to harbor resistance to antimicrobials, including vancomycin, than E. faecalis (8).
Globally, the emergence and spread of vancomycin-resistant E. faecium (VREfm) was first reported in the United States and in Europe (9–11). The distribution of VREfm across the two regions has historically been attributed to two different causes. In Europe, the use of avoparcin, a glycopeptide antimicrobial similar to vancomycin, as an in-feed growth promoter was believed to have caused an increase in VREfm colonization in livestock that was thought to have moved into humans in the 1990s (12, 13). Although studies have observed some genetic similarities between enterococci of human and livestock origin and evaluated the possibility of zoonotic transmission (14, 15), robust evidence of this occurring has been sparse. In the United States, where avoparcin has not been used in livestock, it is believed that the large quantity of vancomycin used in the public health system resulted in the high prevalence of VREfm in hospitals (16).
In Australia, a study of E. faecium isolates collected from 2015 to 2017 in the Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Program (AESOP) reported that approximately 50% of E. faecium isolates from bloodstream infections in hospitals were resistant to vancomycin (17). Based on this report, equal proportions of VREfm in Australia harbor the vanA and vanB operons. This is in contrast to reports from the United States, Europe, and Asia, where either the vanA or the vanB type operon is dominant (18–20).
The livestock industry in Australia is a large and vital sector contributing significantly to Australia’s economy and supplying food for domestic and export consumption. The country’s unique geography, strict quarantine, and importation restrictions protect the industry from many foreign pests and diseases. Moreover, livestock medicinal treatments, including antimicrobials, are strictly regulated by a federal government agency. For example, avoparcin was voluntarily withdrawn from the market in 1999 following recognition that its use gave rise to vancomycin-resistant enterococci in animals in Europe (21). A recent study performed on enterococci from Australian meat chickens found clinical resistance to a number of antimicrobials. The study, however, did not identify any resistance to vancomycin (22), providing evidence of success in antimicrobial stewardship measures. Moreover, E. faecium from meat chickens was found to be genetically distinct from hospital-adapted strains.
Pork is another frequently consumed product, and in Australia pork is second only to chicken in per-capita consumption (23). Swine are commonly colonized by enterococci and, due to the high consumption and potential public health impact, have been scrutinized globally for the presence of antimicrobial-resistant strains. The present study aimed to obtain a collection of enterococci from Australian finisher pigs across all major pig-producing states to investigate the antimicrobial resistance. Due to the clinical importance of E. faecium and VREfm, comprehensive genomic characterization and comparative genomics analyses were analyses performed on E. faecium in order to evaluate production and public health impact.
Based on antimicrobial stewardship practices and Australia’s strict biosecurity procedures both at the international border and at the farm gate, we hypothesize that E. faecium isolates from Australian pigs are genetically distinct from hospital-adapted strains and remain susceptible to important antimicrobials used to treat enterococcal infections in humans.
RESULTS
In total, 146 enterococci consisting of E. faecium (57.5%), E. hirae (24.7%), E. faecalis (11.6%), E. durans (2.7%), E. gallinarum (1.4%), E. hermanniensis (0.7%), E. mundtii (0.7%), and E. avium (0.7%) were identified. All quality control strains examined as part of the antimicrobial susceptibility testing were within stipulated guideline ranges.
E. faecium.
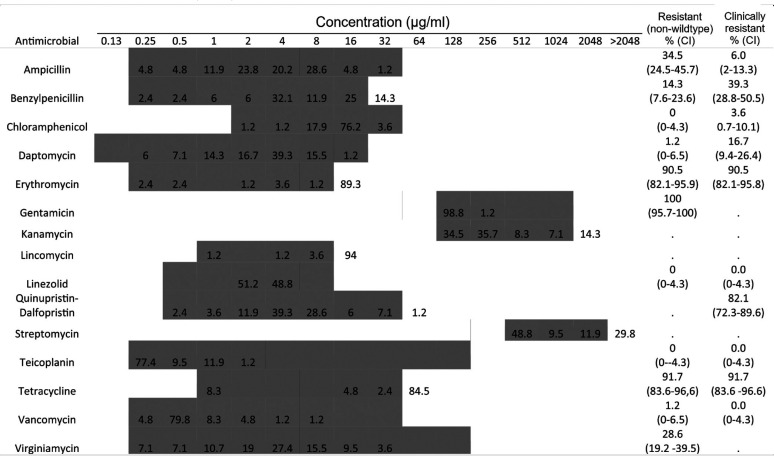
Antimicrobial susceptibility tests were performed on all E. faecium isolates (n = 84) with the MIC distributions shown in Table 1. No resistance (non-wild type) was observed for chloramphenicol, linezolid, or teicoplanin. One isolate (EN060) was resistant to vancomycin. The majority of isolates were resistant to erythromycin (90.5%), high-level gentamicin (100%), and tetracycline (91.7%), with less resistance to ampicillin (34.5%), benzylpenicillin (14.3%), daptomycin (1.2%), and virginiamycin (28.6%). In addition, 82.1% of the isolates were clinically resistant to quinupristin-dalfopristin. All E. faecium isolates were resistant to at least two antimicrobials and 94.1% had (non-wild type) multidrug-resistant (MDR) phenotypes. The largest group of isolates (45.2%) with the same MDR profile were resistant to four antimicrobial classes (aminoglycosides, macrolides, streptogramins, and tetracyclines).
TABLE 1.
MICs of E. faecium isolates (n = 84)a
The shaded areas indicate the dilution ranges tested. Vertical bars indicate the resistance breakpoints. Values for MIC distributions and frequencies of resistance are represented as percentages of isolates. Confidence intervals (CI) for non-wild-type and clinically resistant isolates are indicated in parentheses.
E. hirae.
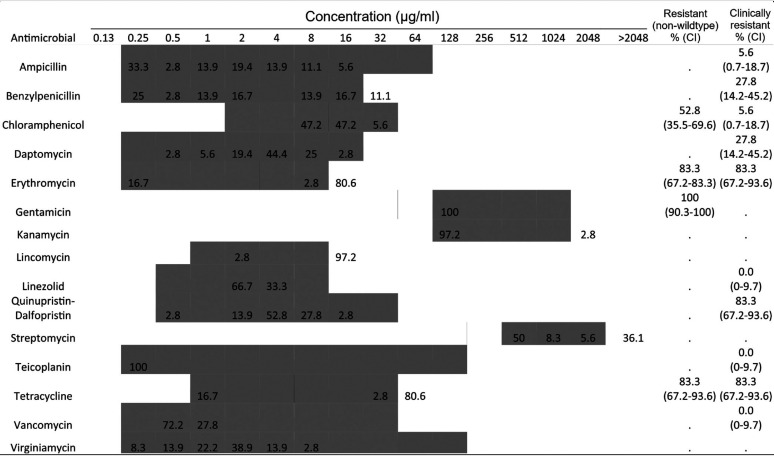
Antimicrobial susceptibility tests were performed on all E. hirae isolates (n = 36) with MIC distributions shown in Table 2. No clinical resistance was observed for linezolid, teicoplanin, or vancomycin. The majority of isolates were resistant to chloramphenicol (52.8%), erythromycin (83.3%), gentamicin (100%), and tetracycline (83.3%). All isolates were resistant to at least two antimicrobials tested, with 97.2% showing MDR phenotypes. The largest group of isolates (19.4%) with the same MDR profile were resistant to six antimicrobial classes (aminoglycosides, lipopeptides, macrolides, phenicols, streptogramins, and tetracyclines).
TABLE 2.
MICs of E. hirae isolates (n = 36)a
The shaded areas indicate the dilution ranges tested. Vertical bars indicate the resistance breakpoints. Values for MIC distributions and frequencies of resistance are represented as percentages of isolates. Confidence intervals for non-wild-type and clinically resistant isolates are indicated in parentheses.
E. faecalis.
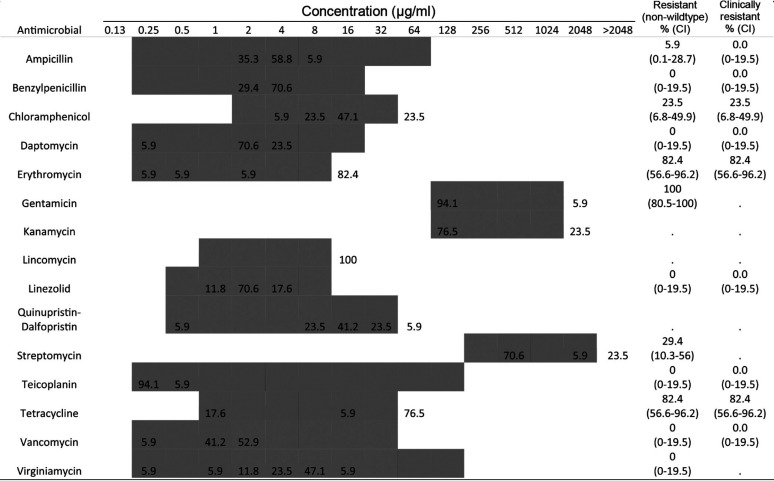
Antimicrobial susceptibility of 17 E. faecalis isolates was determined with MIC distributions shown in Table 3. No resistance was observed for benzylpenicillin, daptomycin, linezolid, teicoplanin, vancomycin, or virginiamycin. The majority of isolates were resistant to erythromycin (82.4%), gentamicin (100%), and tetracycline (82.4%). All isolates were resistant to at least one antimicrobial tested, with 76.5% showing MDR phenotypes. The largest group of isolates (47.1%) with the same MDR profile were resistant to three antimicrobial classes (aminoglycosides, macrolides, and tetracyclines).
TABLE 3.
MICs of E. faecalis isolates (n = 17)a
The shaded areas indicate the dilution ranges tested. Vertical bars indicate the resistance breakpoints. Values for MIC distributions and frequencies of resistance are represented as percentages of isolates. Confidence intervals for non-wild-type and clinically resistant isolates are indicated in parentheses.
Genomic characteristics of E. faecium from pigs.
Of the 84 E. faecium isolates sequenced, 71 passed quality control and were included for analysis. Multilocus sequence typing (MLST) of E. faecium from Australian pigs returned 24 known sequence types (STs) from 44 isolates and 23 unknown STs from 27 isolates. The most frequent STs identified were ST5 (12.7%), ST185 (7.0%), and ST133 (5.6%), with the majority of STs consisting of only one isolate.
With regard to the putative antimicrobial resistance genes identified, one isolate, EN060, demonstrating resistance to vancomycin harbored the vanC operon. The aac(6′)-li gene, conferring aminoglycoside resistance, and the msrC gene, conferring resistance to macrolides and streptogramin B, were identified in all isolates. The ermB gene, conferring resistance to macrolides, lincosamides, and streptogramin B (MLSB), was identified in 85.9% of isolates. No cfrC gene, conferring resistance to linezolid and phenicol antimicrobials, was identified.
Comparative genomics of E. faecium from multiple hosts (pigs, poultry, and human sepsis cases).
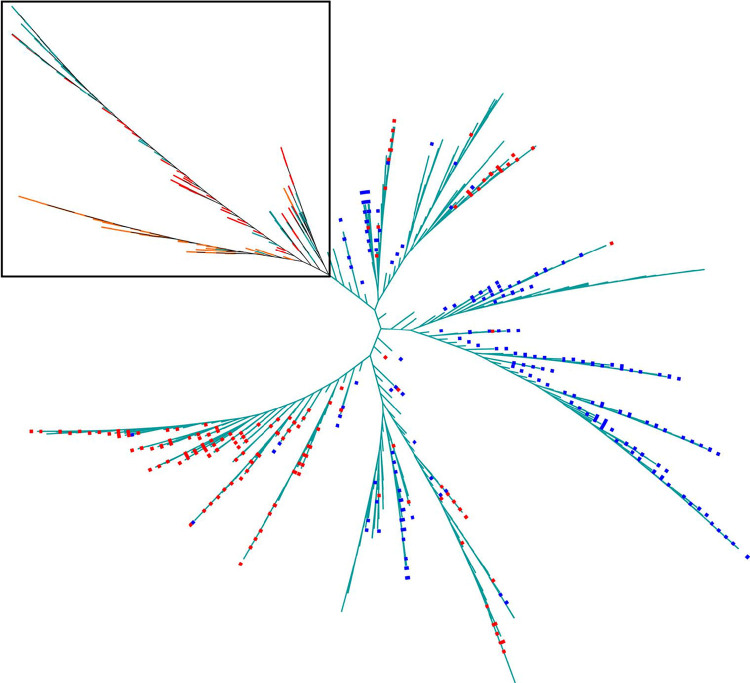
Genomic analysis of 1,161 E. faecium isolates from pigs (n = 71), poultry (n = 65), and human sepsis cases (n = 1,025) collected in 2016, 2016, and 2015 to 2017, respectively, in Australia identified approximately 26,000 single nucleotide polymorphisms (SNPs) among isolates after recombination removal. An SNP-based maximum-likelihood phylogenetic tree showed several clusters of isolates (Fig. 1). Most poultry isolates were genetically distinct, whereas pig and human isolates were mostly intermixed. There was no human sepsis isolate on the same phylogenetic branch since pigs harbored the van operon. Four STs (ST22, ST32, ST94, and ST116) were shared among E. faecium isolates from pigs and humans.
FIG 1.
Maximum-likelihood phylogenetic tree based on single nucleotide polymorphisms (branch lengths are not to scale). The tree includes 1,162 E. faecium isolates from pigs (red branches, n = 71) and poultry (orange branches, n = 65), as well as human sepsis isolates (teal branches, n = 1,025). Red squares indicate isolates with the vanA operon, and blue squares indicate isolates with the vanB operon. Isolates from pigs and chickens are contained within the black square. No van operon was identified in isolates within the black square.
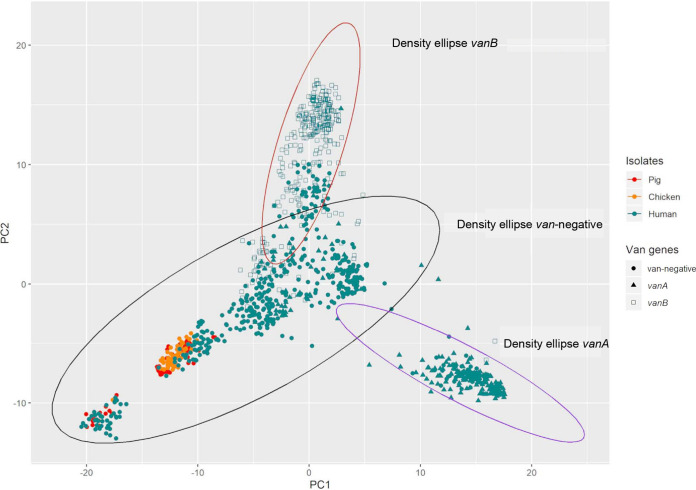
Based on the presence or absence of 19,218 genes identified, principal components 1 and 2 showed E. faecium isolates from humans had the most genetic diversity, forming three overlapping clusters (Fig. 2). The three clusters were observed to be made up of isolates based on the absence or presence and the type of van operon identified (van negative, vanA positive, and vanB positive). The 95% density ellipses of vanA-positive and vanB-positive isolates were mutually exclusive (Fig. 2), but each overlapped with the van-negative ellipse located between them. Poultry and pig E. faecium isolates were genetically similar, forming two clusters that shared similarities with van-negative isolates from humans. The two poultry and pig clusters of isolates were located at the distal end of the van-negative ellipse, farthest from the vanA-positive and vanB-positive ellipses. No chicken or pig isolates were located within the vanA-positive and vanB-positive 95% density ellipse.
FIG 2.
PCA of the total gene content of E. faecium isolates from pigs (red, n = 71), poultry (orange, n = 65), and teal (human, n = 1,025). Isolates harboring the vanA operon (solid triangles) or the vanB operon (hollow squares), as well as van-negative isolates (solid circles), are drawn with a 95% density ellipse.
DISCUSSION
In this study on enterococci from Australian pigs, resistance (non-wild type) was observed against a number of antimicrobial classes, including aminoglycosides, macrolides, streptogramins, and tetracycline. However, more importantly, aside from a single vancomycin-resistant isolate, resistance to critical antimicrobials such as teicoplanin and linezolid was not identified. The results from this study provide strong evidence that in Australia, enterococci from pigs are not a reservoir for critically important antimicrobial resistance.
MLST analysis of the pig E. faecium isolates, which showed large variations in STs, consisted of four STs associated with the human sepsis isolates. However, shared STs only harbored vancomycin-susceptible isolates and were not typical of hospital-adapted strains. Nevertheless, the sharing of similar STs between isolates from pigs and those from human sepsis cases suggests the possibility of coevolution or bidirectional transmission of E. faecium between the two hosts or the presence of another source of infection to which humans and pigs were both exposed. Further molecular investigation showed that this is likely the case. Principal-component analysis (PCA) based on the total gene content showed that isolates from pigs and poultry had genetic compositions highly similar to that of human vancomycin-susceptible E. faecium. However, the SNP-derived maximum-likelihood phylogenetic tree based on the core genes indicated isolates from pigs were similar to isolates from humans while isolates from poultry were genetically more distinct from the other two host species. It is possible that this observation is caused by the colonization of E. faecium strains that have adapted to different host physiologies, with pig and human physiologies being much more similar to each other than to that of avian species. However, this finding requires further investigation.
In this study, antimicrobial resistance was determined using ECOFF breakpoints and clinical resistance defined by Clinical and Laboratory Standards Institute (CLSI) breakpoints. Antimicrobial resistance to tetracycline, erythromycin, and quinupristin-dalfopristin was observed in a large proportion of enterococcus isolates. Tetracycline and tylosin are occasionally used as in-feed or in-water medication to treat specific respiratory or enteric diseases in pigs in Australia. Tylosin, a macrolide antimicrobial agent, has been shown to coselect for erythromycin resistance through the ermB gene (24). In the United States, the prevalence of tetracycline resistance in E. faecium from pigs ranged from 58.97% in 2017 and >70% for E. faecalis from 2013 to 2017 (25). The prevalences of erythromycin resistance were 17.95% in E. faecium and 59.15% in E. faecalis in 2017 (25). Here, we observed a relatively high and similar prevalence (approximately 80 to 90%) of tetracycline and erythromycin resistance for all three of the most common enterococcus species isolated. In addition to E. faecalis, which is intrinsically resistant to quinupristin-dalfopristin, a large proportion of E. faecium and E. hirae isolates were also resistant to quinupristin-dalfopristin. In Australia, quinupristin-dalfopristin is not registered for use in pigs, and the use of virginiamycin, another streptogramin class antimicrobial, was withdrawn from the market for pigs in 2005 (21). Molecular investigation demonstrated that the majority of isolates harbored the msrC and ermB genes, which confer resistance to macrolides and streptogramin B. Although the combination of quinupristin-dalfopristin consists of a streptogramin A and a streptogramin B compound, a study has previously shown that resistance to the combination drug in E. faecium can also be conferred by the msrC and ermB genes. This result suggests the coselection of quinupristin-dalfopristin-resistant enterococci in pigs through use of tylosin (26).
Genes encoding aminoglycoside resistance were identified in all E. faecium sequences from pigs, supporting the high prevalence of non-wild-type gentamicin susceptibility observed. Although gentamicin is not registered in Australia for use in food animals, another aminoglycoside, neomycin, is available in a number of products for oral and parenteral use in pigs. High-level aminoglycoside resistance is common in human E. faecium isolates which have been categorized as intrinsically resistant by the CLSI (27).
The widespread use of beta-lactam antimicrobials such as ampicillin to treat enterococcus infections in humans has resulted in a high prevalence of ampicillin resistance observed in E. faecium. In Australia, the 2018 AGAR AESOP reported that 89.4% of E. faecium isolates responsible for causing sepsis in humans were not susceptible to ampicillin (28). In the present study, we observed that 34.5% of E. faecium isolates from pigs were not susceptible to ampicillin (ECOFF for ampicillin is similar to nonsusceptible for CLSI), indicating a reduced proportion of isolates compared to those from the AESOP collection.
A single vancomycin-resistant isolate harbored the vanC operon. Genetically, the vanC operon is located on the chromosome and encodes a different ligase (d-Ala–d-Ser) affecting the binding of vancomycin compared to the dominant vanA and vanB operons (d-Ala–d-Ala) observed clinically (7). The vanC operon is typically transmitted vertically from parent to daughter cells and confers lower levels of vancomycin resistance than that conferred by the vanA or vanB operon. Although the vanC operon is almost exclusively found in E. gallinarum, E. casseliflavus, and E. flavescens (8), as observed in this study, the vanC operon can also occasionally be found in E. faecium.
Although low susceptibility for critical antimicrobials in enterococci was found in pigs, a limitation of this study is the isolation of only a single representative enterococcus isolate from each sample. This sampling strategy does not capture all the strains within each sample and, as such, there is a possibility that strains with low prevalence are missed. In addition, we are unable to comment on the distribution of species and strains within each of the animals sampled.
In conclusion, this study has provided significant insights into the characteristics of three commonly identified enterococci from Australian finisher pigs. Except for a single isolate harboring the vanC operon, no resistance was observed in all three species against vancomycin, teicoplanin, and linezolid, which are critically important antimicrobials used to treat enterococcal infections in humans. E. faecium in pigs did not harbor the vanA or vanB operons, which are commonly found in the major hospital-adapted strains. As hypothesized in a recent study by O’Dea et al., it appears that the genetic makeup of this subset of enterococci found in chickens, humans, and now pigs does not favor the uptake of vanA or vanB operon-containing genetic elements (22). Moreover, MLST, PCA, and SNP-based phylogenetic analysis indicate some similarities within the populations of E. faecium in pigs and humans. Overall, this study provides phenotypic and genotypic evidence that E. faecium isolates from pigs are not a source and reservoir of hospital-adapted VREfm or vancomycin resistance-conferring genetic elements in Australia.
MATERIALS AND METHODS
Sampling.
Cecal samples were collected as part of a nationwide surveillance study on antimicrobial resistance in enteric commensals and pathogens in Australian pigs. From January to June 2016, 200 samples from finisher pigs were collected from 31 farms across Australia. The sampling strategy was designed to capture as many representatives as possible of the various production systems utilized in the Australian pig industry. Sampling at abattoirs was performed according to the U.S. Department of Agriculture Food Safety and Inspection Service directive by trained personnel (29). Sampling at eligible abattoirs gave access to approximately 85% of pigs slaughtered in Australia. The interval between collections of individual samples was established at each facility as a function of chain speed, daily throughput and shift length to prevent sampling bias. Samples were collected from the cecum and placed in a sterile pot. Once collected, specimens were stored at 2 to 4°C and transported to the primary laboratory within 24 h of collection. All animal fecal samples were acquired postmortem at processing facilities; therefore, no ethics approval was required.
Bacterial isolation.
Isolation of enterococci was performed with 10 g of cecal contents mixed in 7 ml of 0.1% buffered peptone water (Thermo Fisher Scientific, Australia) followed by inoculation of Slanetz & Bartley agar plates (Thermo Fisher Scientific) that were then incubated at 42°C for 48 h. Three well-isolated colonies were subcultured onto sheep blood agar (Thermo Fisher Scientific) and incubated at 37°C for 24 h. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Microflex) was performed for species identification. One enterococcus isolate per sample was selected for the study. Antimicrobial susceptibility testing was performed by broth microdilution using Veterinary Reference Card panels (Sensititre; Trek Diagnostics, East Grinstead, UK) in the CMV3AGPF format for Enterococcus spp. according to the manufacturer’s protocols. Quality control was performed using E. faecalis ATCC 29212 and Staphylococcus aureus ATCC 29213. The MIC results were accepted only if ATCC strains passed quality control (QC) ranges recommended by CLSI guidelines. No QC values were available for lincomycin, streptomycin, and virginiamycin (27). MICs were interpreted using CLSI breakpoints into susceptible (S), intermediate (I), or resistant (R) categories and, when available, the EUCAST ECOFF breakpoints into wild-type and non-wild-type categories (27, 30). In this study, isolates classified as “non-wild type” based on ECOFF breakpoints are referred to as “resistant,” and isolates classified as “resistant” based on CLSI breakpoints are explicitly referred to as “clinically resistant.” Isolates that were resistant to three or more antimicrobial classes were classified as MDR.
Whole-genome sequencing.
Whole-genome sequencing was performed on all E. faecium isolates as previously described (31). Genomic data used for comparison was obtained from the AESOP from 2015 to 2017 (17) and Australian meat chickens sequenced previously in 2016 (22). All sequence data obtained from this study were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA639902. Sequencing of DNA libraries was performed on the Illumina NextSeq platform using a 300-cycle mid-output v2 reagent kit. Output sequences were analyzed using SPAdes v3.14.1 for assembly (32), mlst (33) for MLST identification in association with the pubMLST database (https://pubmlst.org/efaecium, accessed 23 November 2020), kraken2 v2.1.1 for species identification (34), and prokka v1.14.6 for gene annotation (35). Assembled sequences with <40× average depth were excluded from further analysis. Resistance and virulence genes were identified using ABRicate v1.0.1 (36) with the ResFinder (accessed 23 November 2020) (37) and VFDB (23 November 2020) (38) databases, respectively. The identified resistance and virulence genes are considered present at >95% coverage and identity.
Comparative study.
Genomic data were compared to those from E. faecium sequenced as part of the AESOP from 2015 to 2017 (17) and Australian meat chickens sequenced previously in 2016 (22) (see Table S1 in the supplemental material). Phylogenetic trees were constructed using SNPs identified within the core genome with Snippy v4.1.0 (https://github.com/tseemann/snippy), followed by recombination removal with ClonalframeML v1.11 (39). Maximum-likelihood phylogenetic trees were constructed in RAxML v0.9.0 (40) using the generalized time-reversible model set at 1,000 bootstraps and visualized in iTOL (https://itol.embl.de/) (41).
Statistical analysis.
Exact confidence intervals for proportions were derived by the Clopper-Pearson method in Stata v14.2 (StataCorp LLC, College Station, TX). PCA of binomial variables was performed in R for determination of associations by total gene content (Fig. 2). The 95% density ellipses were calculated (within R) from the specified correlation matrix (i.e., the first two components) and plotted using GGPlot2 (42).
Data availability.
Sequence data were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA639902.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Australian Pork Limited for facilitating sample collection.
This project was funded by the Australian Government’s Department of Agriculture and Water Resources’ Animal Biosecurity and Response Reform Program.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lee T, Pang S, Abraham S, Coombs GW. 2019. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present, and the future. J Glob Antimicrob Resist 16:36–47. 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Murray BE. 1990. The life and times of the enterococcus. Clin Microbiol Rev 3:46–65. 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmeli Y, Eliopoulos G, Mozaffari E, Samore M. 2002. Health and economic outcomes of vancomycin-resistant enterococci. Arch Intern Med 162:2223–2228. 10.1001/archinte.162.19.2223. [DOI] [PubMed] [Google Scholar]
- 4.Werner G, Coque T, Hammerum A, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson K, Leclercq R, Lester C. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance 13:19046. [PubMed] [Google Scholar]
- 5.Freitas AR, Tedim AP, Francia MV, Jensen LB, Novais C, Peixe L, Sánchez-Valenzuela A, Sundsfjord A, Hegstad K, Werner G, Sadowy E, Hammerum AM, Garcia-Migura L, Willems RJ, Baquero F, Coque TM. 2016. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986–2012). J Antimicrob Chemother 71:3351–3366. 10.1093/jac/dkw312. [DOI] [PubMed] [Google Scholar]
- 6.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial-resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 7.Faron ML, Ledeboer NA, Buchan BW. 2016. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant enterococcus in the health care setting. J Clin Microbiol 54:2436–2447. 10.1128/JCM.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med 319:157–161. 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 10.Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet i:57–58. 10.1016/S0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 11.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 33:1588–1591. 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aarestrup FM. 1995. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist 1:255–257. 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 13.Bager F, Madsen M, Christensen J, Aarestrup FM. 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med 31:95–112. 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 14.Lester CH, Frimodt-Møller N, Sørensen TL, Monnet DL, Hammerum AM. 2006. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob Agents Chemother 50:596–599. 10.1128/AAC.50.2.596-599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerum AM. 2012. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect 18:619–625. 10.1111/j.1469-0691.2012.03829.x. [DOI] [PubMed] [Google Scholar]
- 16.Leclercq R, Courvalin P. 1997. Resistance to glycopeptides in enterococci. Clin Infect Dis 24:545–556. 10.1093/clind/24.4.545. [DOI] [PubMed] [Google Scholar]
- 17.Lee T, Pang S, Stegger M, Sahibzada S, Abraham S, Daley D, Coombs G, on behalf of the Australian Group on Antimicrobial Resistance. 2020. A three-year whole-genome sequencing perspective of Enterococcus faecium sepsis in Australia. PLoS One 15:e0228781. 10.1371/journal.pone.0228781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goossens H, Jabes D, Rossi R, Lammens C, Privitera G, Courvalin P. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J Antimicrob Chemother 51(Suppl 3):iii5–iii12. 10.1093/jac/dkg271. [DOI] [PubMed] [Google Scholar]
- 19.Song JY, Hwang IS, Eom JS, Cheong HJ, Bae WK, Park YH, Kim WJ. 2005. Prevalence and molecular epidemiology of vancomycin-resistant enterococci (VRE) strains isolated from animals and humans in Korea. Korean J Intern Med 20:55–62. 10.3904/kjim.2005.20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerum AM, Justesen US, Pinholt M, Roer L, Kaya H, Worning P, Nygaard S, Kemp M, Clausen ME, Nielsen KL, Samulioniené J, Kjærsgaard M, Østergaard C, Coia J, Søndergaard TS, Gaini S, Schønning K, Westh H, Hasman H, Holzknecht BJ. 2019. Surveillance of vancomycin-resistant enterococci reveals shift in dominating clones and national spread of a vancomycin-variable vanA Enterococcus faecium ST1421-CT1134 clone, Denmark, 2015 to March 2019. Eurosurveillance 24:1900503. 10.2807/1560-7917.ES.2019.24.34.1900503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Commonwealth of Australia. 2004. Findings of the reconsideration of the registration of products containing virginiamycin and their labels. Commonwealth of Australia, Canberra, Australia. https://apvma.gov.au/sites/default/files/publication/14376-avoparcin-status-document.pdf. [Google Scholar]
- 22.O’Dea M, Sahibzada S, Jordan D, Laird T, Lee T, Hewson K, Pang S, Abraham R, Coombs GW, Harris T, Pavic A, Abraham S. 2019. Genomic, antimicrobial resistance, and public health insights into spp. from Australian chickens. J Clin Microbiol 57:e00319-19. 10.1128/JCM.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meat & Livestock Australia. 2019. State of the industry report. Meat & Livestock Australia, North Sydney, Australia. [Google Scholar]
- 24.Jackson CR, Fedorka-Cray PJ, Barrett JB, Ladely SR. 2004. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl Environ Microbiol 70:4205–4210. 10.1128/AEM.70.7.4205-4210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Food and Drug Administration. 2021. NARMS now: integrated data. U.S. Department of Health and Human Services, Rockville, MD. https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data. [Google Scholar]
- 26.Wang S, Guo Y, Lv J, Qi X, Li D, Chen Z, Zhang X, Wang L, Yu F. 2016. Characteristic of Enterococcus faecium clinical isolates with quinupristin/dalfopristin resistance in China. BMC Microbiol 16:246. 10.1186/s12866-016-0863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Coombs GW, Daley DA, Mowlaboccus S, Lee YT, Pang S, Australian Group on Antimicrobial Resistance. 2020. Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2018. Commun Dis Intell (2018) 44. 10.33321/cdi.2020.44.19. [DOI] [PubMed] [Google Scholar]
- 29.Food Safety and Inspection Service. Methods: The National Antimicrobial Resistance Monitoring System—enteric bacteria. U.S. Department of Health and Human Services, Rockville, MD. https://www.fda.gov/media/101741/download. [Google Scholar]
- 30.European Committee on Antimicrobial Susceptibility Testing. 2020. Antimicrobial wild type distributions of microorganisms. EUCAST, Basel, Switzerland. https://mic.eucast.org/. Accessed 6 December 2020. [Google Scholar]
- 31.Abraham S, Kirkwood RN, Laird T, Saputra S, Mitchell T, Singh M, Linn B, Abraham RJ, Pang S, Gordon DM, Trott DJ, O’Dea M. 2018. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J 12:2352–2362. 10.1038/s41396-018-0200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. 2020. mlst. https://github.com/tseemann/mlst.
- 34.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Seemann T. 2020. Abricate. https://github.com/tseemann/abricate.
- 37.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA639902.