In working toward a comprehensive strategy to combat the spread of antibiotic resistance, potential farm-to-fork routes of dissemination are gaining attention. The effects of preharvest factors on the microbiota and corresponding antibiotic resistance indicators on the surfaces of produce commonly eaten raw is of special interest.
KEYWORDS: antibiotic resistance, antibiotics, beef, compost, dairy, greenhouse, manure, soil microbiology, vegetables
ABSTRACT
A controlled greenhouse study was performed to determine the effect of manure or compost amendments, derived during or in the absence of antibiotic treatment of beef and dairy cattle, on radish taproot-associated microbiota and indicators of antibiotic resistance when grown in different soil textures. Bacterial beta diversity, determined by 16S rRNA gene amplicon sequencing, bifurcated according to soil texture (P < 0.001, R = 0.501). There was a striking cross-effect in which raw manure from antibiotic-treated and antibiotic-free beef and dairy cattle added to loamy sand (LS) elevated relative (16S rRNA gene-normalized) (by 0.9 to 1.9 log10) and absolute (per-radish) (by 1.1 to 3.0 log10) abundances of intI1 (an integrase gene and indicator of mobile multiantibiotic resistance) on radishes at harvest compared to chemical fertilizer-only control conditions (P < 0.001). Radishes tended to carry fewer copies of intI1 and sul1 when grown in silty clay loam than LS. Composting reduced relative abundance of intI1 on LS-grown radishes (0.6 to 2.4 log10 decrease versus corresponding raw manure; P < 0.001). Effects of antibiotic use were rarely discernible. Heterotrophic plate count bacteria capable of growth on media containing tetracycline, vancomycin, sulfamethazine, or erythromycin tended to increase on radishes grown in turned composted antibiotic-treated dairy or beef control (no antibiotics) manures relative to the corresponding raw manure in LS (0.8- to 2.3-log10 increase; P < 0.001), suggesting that composting sometimes enriches cultivable bacteria with phenotypic resistance. This study demonstrates that combined effects of soil texture and manure-based amendments influence the microbiota of radish surfaces and markers of antibiotic resistance, illuminating future research directions for reducing agricultural sources of antibiotic resistance.
IMPORTANCE In working toward a comprehensive strategy to combat the spread of antibiotic resistance, potential farm-to-fork routes of dissemination are gaining attention. The effects of preharvest factors on the microbiota and corresponding antibiotic resistance indicators on the surfaces of produce commonly eaten raw is of special interest. Here, we conducted a controlled greenhouse study, using radishes as a root vegetable grown in direct contact with soil, and compared the effects of manure-based soil amendments, antibiotic use in the cattle from which the manure was sourced, composting of the manure, and soil texture, with chemical fertilizer only as a control. We noted significant effects of amendment type and soil texture on the composition of the microbiota and genes used as indicators of antibiotic resistance on radish surfaces. The findings take a step toward identifying agricultural practices that aid in reducing carriage of antibiotic resistance and corresponding risks to consumers.
INTRODUCTION
In the United States and many other parts of the world, the majority of antibiotics are administered to livestock (1), raising the concern that this could contribute to the evolution of antibiotic-resistant organisms and the spread of infections with these organisms to human populations (2). Preharvest contamination of vegetables through contact with animal manure-amended soils is a well-recognized source of human pathogens (3) and may also serve as a path for dissemination of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) during cultivation (3, 4). Composting is well known for the benefits it provides for improving the value of manure as a soil amendment (5) and degrading antibiotics (6) and could possibly provide additional benefit by reducing manure-borne ARB and ARG transfer onto produce. Composting alters the abundance and diversity (7–10) of bacteria and is an effective strategy for reducing human pathogens in animal manures. Thus, it has been recommended by the Food Safety Modernization Act (FSMA) that manure should first be composted before being applied as a soil amendment for crops where the soil comes in direct contact with the vegetable (11). However, there is a need to determine the influence of soil amendments on carriage of ARB and ARGs on vegetable surfaces.
Carriage of ARB and ARGs is of special interest for root vegetables, given the direct contact of edible portions with soil and amendments. Soil texture is known to influence the bacterial community structure of the rhizosphere (12); however, the effect of soil texture on carriage of resistant genotypes and phenotypes across bacterial populations is not well described. In addition, secretion of nutrients in the rhizosphere, such as that typical of carrot and radish taproots, provides a unique environment that could also influence abundances of ARB and ARGs (13–15). Nutrients released during growth are known to alter the activity of bacteria within the rhizosphere (16) and also influence the diversity and composition of the rhizosphere bacterial community composition relative to surrounding bulk soil (17, 18).
Among the various means of assessing antibiotic resistance, of particular interest in agroecosystems are holistic measures that broadly capture reservoirs of ARB and ARGs across relevant microbial communities with the potential to be selected or mobilized. In particular, the intI1 integrase gene of class 1 integrons, which are drivers of mobile multidrug resistance in clinical infections, and the sul1 gene are strongly correlated with anthropogenic inputs into the environment (19–22). sul1 is commonly, but not always, found in the 3′ conserved segment of class 1 integrons (19–21). Tetracyclines of various forms are used widely in humans and livestock (23, 24), with numerous genes conferring resistance. tet(W) is especially common in manure (25–27). Thus, these three genes have been targeted as indicators of antibiotic resistance in agroecosystems, wherein their attenuation is assumed to signify reduction in potential for antibiotic resistance to be selected and spread (28–30). Culture-based techniques are also informative in terms of confirming viability and phenotype of ARB, but they cannot access the full microbial community (31).
The overall goal of this study was to determine the effects of amending soil with manure collected during antibiotic use, composting the manure prior to amendment, and soil texture on the associated microbiota and indicators of antibiotic resistance on radish taproot surfaces. Radishes were cultivated in a greenhouse in two different soil textures, loamy sand (LS) or silty clay loam (SCL), amended with raw manure from beef or dairy cattle during typical antibiotic administration (BA and DA, respectively), raw manure from control beef or dairy cattle not given antibiotics during manure collection (BC and DC, respectively), two types of compost (static and turned) derived from the same manures, or chemical fertilizer only as a control. The effects on microbiotas associated with radish surfaces were assessed using 16S rRNA gene amplicon sequencing. sul1 and tet(W) ARGs and the intI1 class I integron were quantified as indicators of antibiotic resistance using quantitative PCR (qPCR). In addition, changes in phenotypic indicators of antibiotic resistance were determined through aerobic heterotrophic plate counts (HPCs) on media containing ceftazidime, clindamycin, erythromycin, vancomycin, tetracycline, or sulfamethoxazole. The findings help inform strategies for mitigating the spread of antibiotic resistance from manure to produce.
RESULTS
Effects of soil texture, amendments, and antibiotic use on the superficial radish microbiota.
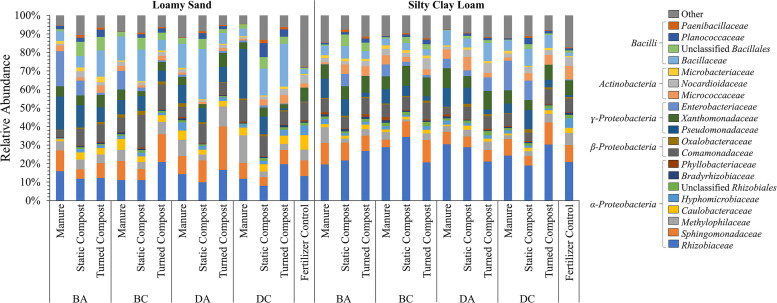
The Earth Microbiome Project protocol using primers 515Fb and 926R for 16S rRNA gene amplicon sequencing (32, 33) was applied to profile the compositions of the superficial radish microbiota (Fig. 1). A total of 1,776,087 nonchloroplast sequences from 77 radish surfaces were generated, which were rarefied to 11,600 bacterial sequences per radish to facilitate statistical comparison (see Fig. S1 in the supplemental material). The taxonomic compositions of the superficial radish microbiota are compared in Fig. 1. Proteobacteria, Firmicutes, and Actinobacteria made up over 96% of all taxa among all radish surfaces, with 32 additional phyla also detected. In descending order of relative abundance, the dominant classes observed across conditions were Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria, Bacilli, and Actinobacteria, which together accounted for over 80% of the total bacterial operational taxonomic units (OTUs) detected, regardless of soil or amendment type. Some of the most abundant families included, in descending order of relative abundance, Rhizobiaceae, Sphingomonadaceae, Bacillaceae, Comamonadaceae, Pseudomonadaceae, Xanthomonadaceae, Methylophilaceae, and Enterobacteriaceae. Together, these families ranged between 39% and 89% of the total relative abundance across individual harvested radishes. There were no measurable differences in relative abundance of among taxonomic groups for radishes grown in different treatments (Kruskal-Wallis, P > 0.050).
FIG 1.
Effects of soil amendments and soil texture on the average relative abundances of the 20 most abundant families of bacteria represented by OTUs recovered from the surfaces of radish taproots (n = 3, except DC turned compost in loamy sand, where n = 2) grown in either loamy sand (LS) or silty clay loam (SCL) mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. There were no statistically significant differences in relative abundance of taxonomic groups for radishes grown among all treatments (Kruskal-Wallis, P > 0.05).
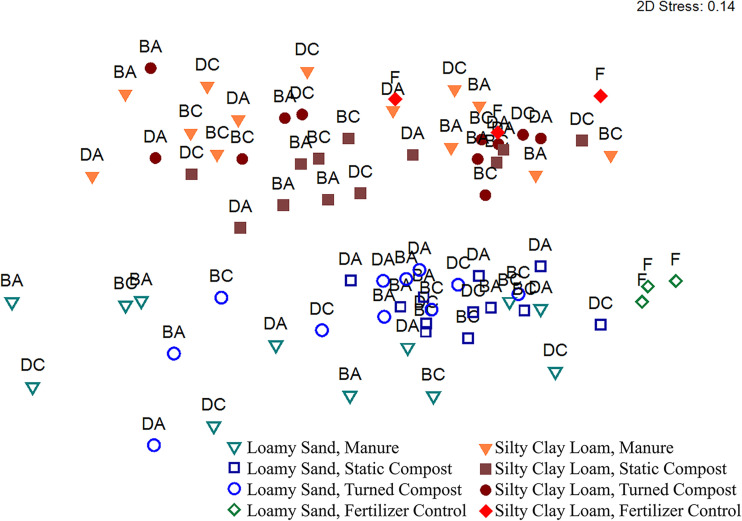
Among the factors investigated in this study, soil texture had the most striking effect on the overall composition of the superficial radish microbiota. Soil texture, crossed with amendment types, resulted in significant differences in bacterial beta diversity between LS and SCL soils (P < 0.001, R = 0.501; analysis of similarity [ANOSIM], unweighted UniFrac) (Fig. 2). Within LS, effects of biological amendment versus chemical fertilizer control were next most apparent (P < 0.013, R > 0.385; ANOSIM, unweighted UniFrac). Notably, radishes grown in LS with chemical fertilizer carried a more diverse microbiota than those grown in the same soil amended with manure or turned compost (P < 0.002; analysis of variance [ANOVA], Shannon index) (Table S1). Effects of composting manure prior to application and type of composting were less apparent. Beta diversity of the radish microbiomes grown in manure versus static compost was distinct and moderately separated in LS soil (P < 0.001, R = 0.449; ANOSIM, unweighted UniFrac). For the SCL soil texture, there was no effect among any amendment conditions on beta diversity (R = 0.124; ANOSIM, unweighted UniFrac) or alpha diversity (P > 0.050; ANOVA, Shannon index). No manure or compost conditions indicated a significant difference in microbiota composition as a function of antibiotic administration (P > 0.050; Wilcoxon). There was also no distinction among any of the beef-versus-dairy conditions (P > 0.050, R = 0.098; ANOSIM, unweighted UniFrac).
FIG 2.
Nonmetric multidimensional scaling plot of unweighted UniFrac distance matrices comparing the effect of soil amendment type, i.e., manure (triangles), static compost (squares), turned compost (circles), or fertilizer control (diamonds), on the microbiota associated with radish taproot surfaces grown in either loamy sand (open symbols) or silty clay loam (closed symbols). Each data point represents an individual radish taproot grown in the corresponding conditions and are labeled as follows: F, chemical fertilizer; BC, BA, DC, and DA, manure-derived amendments originating from control beef cattle, antibiotic-treated beef cattle, control dairy cattle, or antibiotic-treated dairy cattle, respectively. Soil texture, crossed with amendment types, resulted in significant differences in bacterial beta diversity (P < 0.001, R = 0.501; ANOSIM).
Quantification of total bacterial, antibiotic resistance, and mobility markers on radish surfaces.
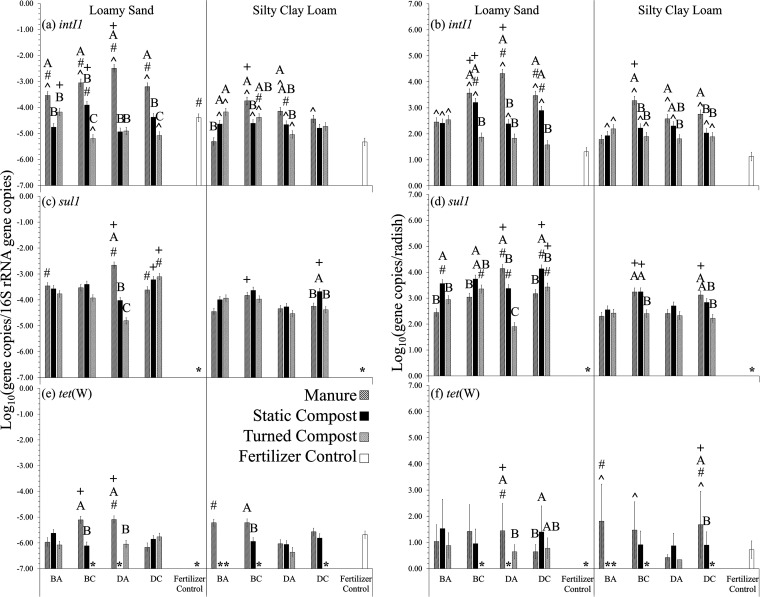
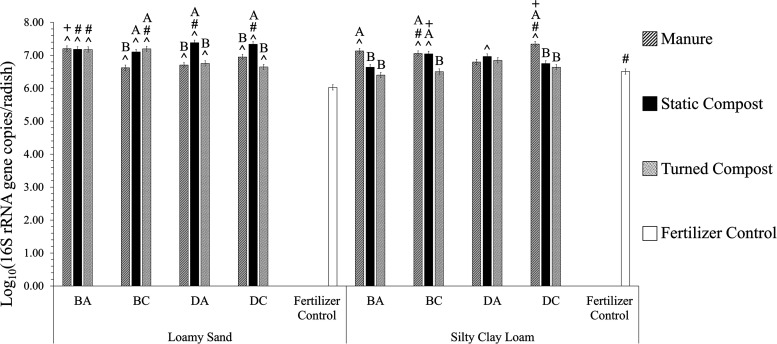
16S rRNA gene copy numbers were quantified as an indicator of total bacterial numbers and also to provide a denominator for measuring relative abundance of tet(W), sul1, and intI1 (Fig. 3a, c, and e; Data Set S1). 16S rRNA gene normalized copy numbers are commonly reported to account for minor variation in DNA recovery during extraction and also as an indicator of selection/enrichment versus attenuation of the genes of interest (33). tet(W), sul1, and intI1 numbers are also reported on a per-radish basis as a measure of net numbers that could potentially carry through the postharvest portion of the food chain (Fig. 3b, d, and f; Data Set S1). 16S rRNA gene copies ranged from 6.03 log10 copies/taproot to 7.38 log10 copies/taproot, with the LS chemical fertilizer control condition yielding the lowest numbers and the LS DA static compost condition the highest (Fig. 4). There were significant differences among many treatments, but the greatest-magnitude difference in 16S rRNA gene copies on radishes was between radishes grown in LS with chemical fertilizer and those grown in LS with DA static compost (1.4 log10; P < 0.001).
FIG 3.
Relative abundances of (a) intI1, (c) sul1, and (e) tet(W) and absolute abundances of (b) intI1, (d) sul1, and (f) tet(W), measured by qPCR, on taproot surfaces of radishes (n = 3) grown in loamy sand (LS) and silty clay loam (SCL) mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. Treatments for which the values for all 3 radishes were below the limit of quantification (asterisks) were excluded from analysis. Survival models were incorporated assuming a Weibull distribution to calculate the means and standard errors (indicated by the error bars). Significant differences (P < 0.050, survival analysis) are indicated as follows: #, for the same amendment, the soil texture that yielded significantly greater gene abundance; ^, for the same soil texture, amendments that yielded gene abundances significantly different from those of the fertilizer control; +, for the same cattle type (beef or dairy), same amendment type, and same soil texture, the antibiotic condition (antibiotic or control) that yielded significantly greater gene abundance. The letters A, B, and C show statistical groupings for the effect of amendment type (manure, static compost, and turned compost) for same cattle and same soil texture.
FIG 4.
Abundances of 16S rRNA genes, enumerated by qPCR, on radish taproot surfaces (n = 3) grown in loamy sand (LS) and silty clay loam (SCL) mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. Standard errors are indicated by the error bars. Significant differences (P < 0.050, survival analysis) are indicated as follows: #, for the same amendment, the soil texture that yielded significantly greater gene abundance; ^, for the same soil texture, amendments that yielded gene abundances significantly different from those of the fertilizer control; +, for the same cattle type (beef or dairy), same amendment type, and same soil texture, the antibiotic condition (antibiotic or control) that yielded significantly greater gene abundance. The letters A and B indicate statistical groupings for the effect of amendment type (manure, static compost, and turned compost) for the same cattle and same soil texture.
While there were quantifiable measurements of tet(W), sul1, and intI1 among all treatments, there were some exceptions where measurements were below the limit of quantification or detection (Fig. 3; Data Set S1). Survival models were therefore applied to statistically comparing the quantities of these three ARGs (survival analysis; n = 3, P < 0.050). The strongest trends were observed for sul1 and intI1 (Fig. 3a to d), with only a few instances of statistically significant differences in comparisons of treatments with respect to tet(W) (Fig. 3e and f).
Comparison of surface-associated markers of antibiotic resistance and mobility on radishes grown in different soil textures.
Soil texture had a marked effect on levels of sul1 and intI1 relative to 16S rRNA gene copies measured on radish surfaces, with the majority of statistical comparisons indicating greater numbers in the LS than the SCL conditions (Fig. 3a and c, number symbols). The greatest-magnitude differences occurred when BA manure and DA manure were added to LS. This resulted in 1.8-log10-greater intI1 and 1.0-log10-greater sul1 levels when LS was amended with BA manure than when BA manure was added to SCL (P < 0.001) and 1.7-log10-greater intI1 and 1.7-log10-greater sul1 levels when LS was amended with BA manure on radishes than when DA manure was added to SCL (P < 0.001).
Similar trends were observed on a per-radish basis when DA manure was used to amend LS, resulting in 1.7-log10- and 1.7-log10-greater intI1 and sul1 levels, respectively, than when it was used to amend SCL (Fig. 3b and d, number symbols; P < 0.001). There was no effect of LS versus SCL soil on a per-radish basis for the beef manure conditions.
Effects of biological soil amendments on radish surface-associated markers of antibiotic resistance and mobility compared to fertilizer control soils.
Relative abundances of intI1 were greater on radishes grown in all four LS conditions where manure was added than on those grown with the chemical fertilizer (BA manure by 0.9 log10, BC manure by 1.3 log10, DA manure by 1.9 log10, and DC manure by 1.2 log10) (Fig. 3a, carets). In SCL conditions, BC manure (1.6 log10), DA manure (1.2 log10), and DC manure (0.9 log10) (but not BA manure) also increased intI1 on radishes compared to chemical fertilizer.
On a per-radish basis, compared to chemical fertilizer-only controls, addition of biological amendments had a marked effect in terms of elevating intI1 in all cases, except when BC, DA, or DC turned compost was added to LS (Fig. 3b, carets). All four manure conditions resulted in greater quantities of intI1 on radishes grown in LS than the chemical fertilizer: BA manure by 1.1 log10, BC manure by 2.3 log10, DA manure by 3.0 log10, and DC manure by 2.2 log10 (P < 0.001). Biological amendments also increased intI1 levels per radish in the SCL conditions BC manure (2.2 log10), DA manure (1.5 log10), and DC manure (1.6 log10), but not BA manure (P < 0.001).
sul1 was consistently undetectable or below the limit of detection on radishes grown in the fertilizer control soils but was readily detected at levels of >1.9 log10 copies/radish in conditions receiving biological amendment (Fig. 3d, carets). However, the inability to detect sul1 in fertilizer control soils prevented statistical comparison.
Like sul1, tet(W) was below the limit of detection on radishes grown in LS with chemical fertilizer only but was quantifiable in SCL (Fig. 3f, carets). BA, BC, and DC manure amendments increased tet(W) levels per radish by less than 1.1 log10 in SCL compared to chemical fertilizer only (P < 0.001).
Comparison of compost versus raw manure on radish surface-associated markers of antibiotic resistance and mobility.
On radishes grown in LS soil, turned composting of the DA manure and BC manure decreased intI1 relative abundance by over 2.1 log10 compared to the corresponding raw manures (Fig. 3a, bars with the same letters; P < 0.001). Static composting of the DA manure also decreased intI1 abundance, by over 2.4 log10 (P < 0.001). Static composting also decreased intI1 abundance in LS amended with BA, BC, and DC manures by 0.9 to 1.2 log10. Similarly, on a per-radish basis in LS soil, turned composting of the DA, DC, and BC manures decreased intI1 levels on radishes by over 1.7 log10 (Fig. 3b, bars with the same letters; P < 0.001). Static composting of the DA manure decreased intI1 by 1.9 log10 (P < 0.001). There were some effects of composting manure prior to amendment on radishes grown in SCL, but the magnitude of reduction of intI1 was less. Most notably, both composting methods for BC manure decreased intI1 per radish by 1.0 to 1.4 log10 (P < 0.001).
Static and turned composting of the DA manure also decreased relative abundance of sul1 by 1.4 and 2.2 log10, respectively, on the LS-grown radishes (Fig. 3c, bars with the same letters; P < 0.001). In contrast, there were few instances where static composting of the manure increased sul1 per radish grown in LS (BA, BC, and DC manures), by 1.1, 0.7, and 1.0 log10, respectively (Fig. 3d, bars with the same letters; P < 0.001).
For all three genes, most differences that were significant in comparisons of static versus turned compost were <0.9 log10, with a few exceptions. For growth in LS, turned composting of the BC manure resulted in greater reduction of relative and per-radish abundance of intI1 compared to static composting, by 1.3 log10 (Fig. 3a and b, bars with the same letters; P < 0.001). Turned composting of the DC manure also resulted in greater reduction of intI1 levels per radish than static composting, by 1.3 log10 (P < 0.001). Turned composting of the DA manure resulted in 1.5 log10 less sul1 per radish grown in LS than static composting (Fig. 3c, bars with the same letters; P < 0.001). In most cases, if there were differences, turned composting decreased carriage of the gene targets on radishes more than static composting.
Comparison of antibiotic administration versus control conditions on radish surface-associated markers of antibiotic resistance and mobility.
One key question was whether collecting the manure during antibiotic administration ultimately influenced the carriage of markers of antibiotic resistance and mobility on surfaces of radishes grown in soils receiving corresponding amendments.
According to expectation, the DA manure conditions tended to yield higher relative and absolute abundances of both intI1 and sul1 on radishes grown in LS than amendments with DC manure (Fig. 3a to d, plus signs; e.g., 0.7 to 0.8 log10 difference in intI1 and 1.0 difference in sul1 [P < 0.001]). However, for growth in SCL, there was no effect when DA and DC conditions were compared, except that DC manure and DC static compost amendments resulted in slightly higher per-radish (0.7 log10) and relative (0.6 log10) abundances of sul1, respectively. Turned composting of the DC manure tended to increase both relative abundance and per-radish abundance of sul1 on LS-grown radishes (e.g., DC versus DA turned compost, by 1.7 and 1.5 log10, respectively; P < 0.001).
For beef manure, effects of antibiotic use were not particularly striking. The greatest effect was when BC manure was applied to SCL, which resulted in 1.6-log10-greater relative abundance of intI1 on radishes than the BA manure (Fig. 3a, plus signs; P < 0.001). On a per-radish basis, amended SCL with BC manure resulted in 1.5-log10-greater intI1 on radishes than BA manure (Fig. 3b, plus signs; P < 0.001). Both of these examples were contrary to the expectation of antibiotic use elevating antibiotic resistance markers on radishes.
Comparison of surface-associated HPCs on radishes grown in different soil textures.
To assess the response of a culture-based indicator of antibiotic resistance to manure-based amendments, HPCs associated with radish taproot surfaces were enumerated on antibiotic-free R2A “control” plates (Fig. 5) and R2A plates supplemented with clindamycin, ceftazidime, erythromycin, sulfamethoxazole, tetracycline, or vancomycin at clinically relevant concentrations (Fig. 6). Counts on plates with antibiotics normalized to those on antibiotic-free plates were also considered an indicator of the proportion of CFU that were potentially resistant to the antibiotics (Fig. 7), both intrinsically and through acquisition of ARGs. HPCs were selected as a target because R2A is a broadly applied medium to enumerate oligotrophic bacteria across multiple relevant environments, including water, soil, and produce (34–36). Note that further characterization (e.g., taxonomic identification to verify that it is an intended target of the antibiotic and MIC determination) would be needed to verify that the colonies are actually resistant; therefore, we refer here to HPC organisms “capable of growth in the presence of antibiotics.”
FIG 5.
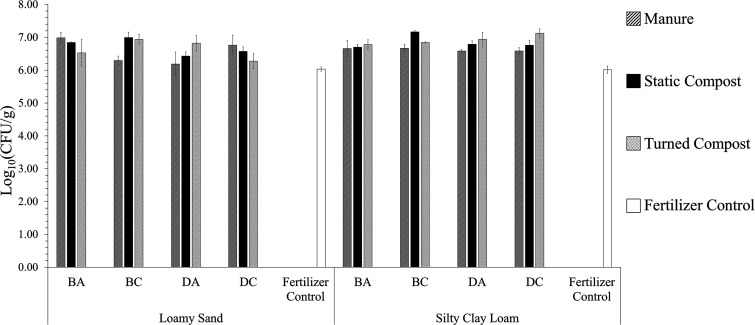
Aerobic heterotrophic bacterial counts (HPCs) from radish taproot surfaces (n = 3) grown in loamy sand (LS) or silty clay loam (SCL) mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with a chemical fertilizer only as a control on R2A control. Standard errors are indicated by the error bars.
FIG 6.
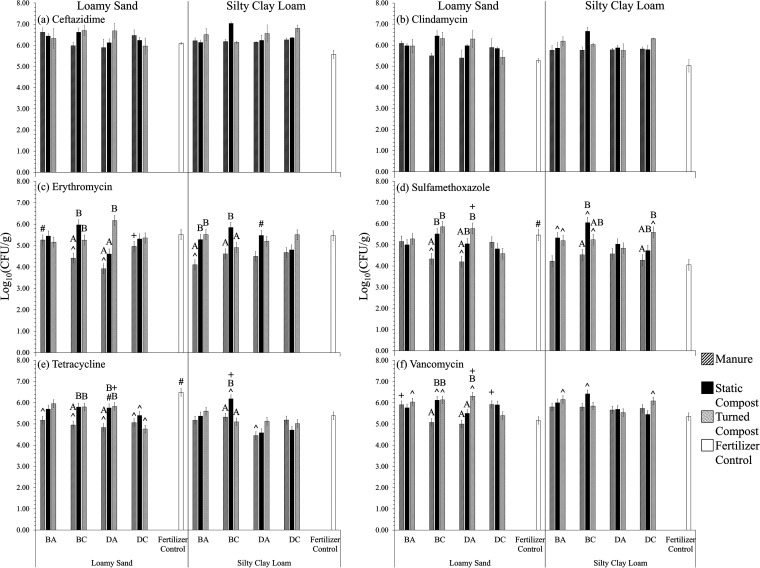
Aerobic heterotrophic bacterial counts (HPCs) from radish taproot surfaces (n = 3) on R2A supplemented with (a) ceftazidime (25 μg/ml), (b) clindamycin (10 μg/ml), (c) erythromycin (25 μg/ml), (d) sulfamethoxazole (100 μg/ml), (e) tetracycline (3 μg/ml), and (f) vancomycin (11 μg/ml). Loamy sand (LS) and silty clay loam (SCL) were mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. Survival models were incorporated assuming a Weibull distribution to calculate the means and standard errors for Em, Smz, Tc, and Vm (indicated by the error bars). Standard errors for Cm and Cz are indicated by the error bars. Significant differences (P < 0.050, survival analysis) are indicated as follows: #, for the same amendment, the soil texture that yielded significantly greater gene abundance; ^, for the same soil texture, amendments that yielded gene abundances significantly different from that of the fertilizer control; +, for the same cattle type (beef or dairy), same amendment type, and same soil texture, the antibiotic condition (antibiotic or control) that yielded significantly greater gene abundance. The letters A and B indicate statistical groupings for effect of amendment type (manure, static compost, and turned compost) for the same cattle and same soil texture.
FIG 7.
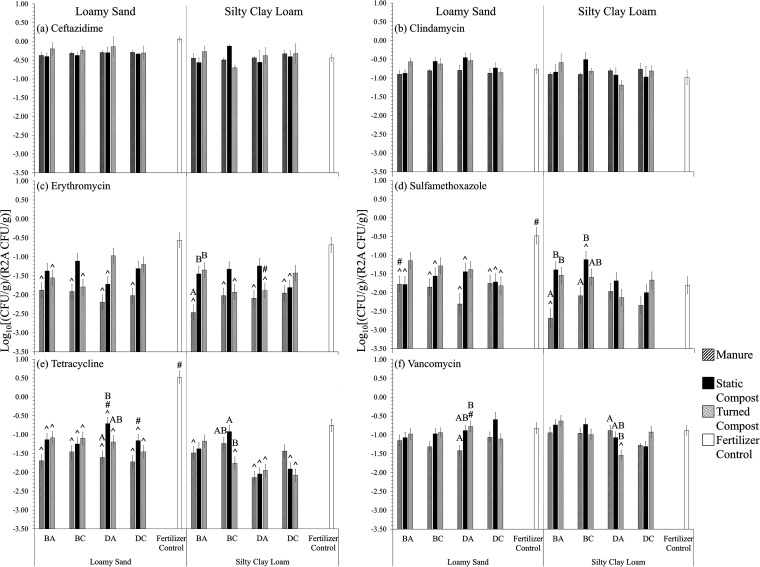
Aerobic heterotrophic bacterial counts (HPCs) from radish taproot surfaces (n = 3) on R2A supplemented with (a) ceftazidime (25 μg/ml), (b) clindamycin (10 μg/ml), (c) erythromycin (25 μg/ml), (d) sulfamethoxazole (100 μg/ml), (e) tetracycline (3 μg/ml), and (f) vancomycin (11 μg/ml), relative to HPCs on R2A medium without antibiotics. Loamy sand (LS) and silty clay loam (SCL) were mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. Survival models were incorporated assuming a Weibull distribution to calculate the means and standard errors for Em, Smz, Tc, and Vm (indicated by the error bars). Standard errors for Cm and Cz are indicated by the error bars. Significant differences (P < 0.050, survival analysis) are indicated as follows: #, for the same amendment, the soil texture that yielded significantly greater gene abundance; ^, for the same soil texture, amendments that yielded gene abundances significantly different from that of the fertilizer control. The letters A and B indicate statistical groupings for effect of amendment type (manure, static compost, and turned compost) for the same cattle and same soil texture.
There were only few instances where HPCs from radish surfaces on media containing antibiotics were significantly different between soil textures. When there were differences, HPCs were greater on LS-grown radishes than on radishes grown in corresponding SCL conditions (Fig. 6 and 7, number signs). Most notably, radishes grown in the LS fertilizer control condition itself produced about 1.4 and 1.1 log10 more CFU/g on plates containing sulfamethoxazole and tetracycline, respectively (P < 0.001) (Fig. 6d and e, number signs). Normalizing to antibiotic-free R2A control plates tended to result in similar differences in LS versus SCL conditions on plates with sulfamethoxazole and tetracycline, by 3.0 log10 more (Fig. 7d and e, number signs). Soil texture had no effect on the numbers of HPC organisms recovered from radish surfaces and grown on R2A control plates, R2A with ceftazidime, or R2A with clindamycin (Fig. 5; Fig. 6a and b, number signs; P > 0.050, ANOVA).
Effects of biological amendments on radish surface-associated HPCs compared to fertilizer-only control soils.
Contrary to expectation, it was often the case that the absolute and normalized HPCs recovered from radishes grown in the chemical fertilizer-only control condition on antibiotic-containing plates were significantly greater than when radishes were grown in soils with biological amendments (Fig. 6 and 7). The addition of BC and DA manure decreased HPCs on plates containing erythromycin, sulfamethoxazole, and tetracycline by 1.1 to 1.7 log10 compared to fertilizer control radishes grown in LS (P < 0.001) (Fig. 6c to e, carets). Further, amendment of BA manure to SCL decreased HPCs recovered from radishes grown on plates containing erythromycin by about 1.4 log10, compared to the fertilizer control condition (Fig. 6c; P < 0.001). All manure types decreased HPCs on plates containing tetracycline, by 1.3 to 1.7 log10, compared to fertilizer control LS-grown radishes (P < 0.001) (Fig. 6e, carets). Both DC static and turned compost types also decreased HPCs on plates containing tetracycline, by 1.1 and 1.7 log10 respectively, compared to fertilizer control LS-grown radishes (P < 0.001) (Fig. 6e, carets). Exceptions were BA and BC static and turned composts and DC turned compost in SCL, which were associated with an increase in HPCs on plates containing sulfamethoxazole, by 1.1 to 2.0 log10, compared to fertilizer control SCL-grown radishes (P < 0.001) (Fig. 6d, carets). Many biological amendments in both soils decreased HPCs on plates with erythromycin and tetracycline when normalized to the HPCs on media without antibiotics (Fig. 7c and e, carets).
Comparison of compost versus raw manure on radish surface-associated HPCs.
Somewhat surprisingly, compost amendment tended to increase the number of HPCs on radishes capable of growth on R2A plates in certain cases for erythromycin, vancomycin, tetracycline, and sulfamethoxazole, relative to amendment with the raw manure (Fig. 6 and 7c to f, bars with the same letters). Specifically, DA turned compost and BC static compost increased HPCs on plates containing these four antibiotics compared to manure-amended LS-grown radishes, by 0.9 to 2.3 log10 (P < 0.001) (Fig. 6c to f, bars with the same letters). In SCL, BA and BC static compost also increased HPCs on plates containing erythromycin and sulfamethoxazole, by 1.1 to 1.5 log10, compared to manure-amended SCL-grown radishes (P < 0.001) (Fig. 6c and d, bars with the same letters). The trend toward higher HPCs of organisms capable of growth in the presence of antibiotics for radishes grown with compost amendments was not likely due to higher overall HPCs for compost-amended radishes, as these were not significantly higher in the compost treatments than the others (Fig. 5; P > 0.050, ANOVA). Composting had no effect on the numbers of CFU observed on R2A with ceftazidime or clindamycin (Fig. 6a and b; P > 0.050, ANOVA). In terms of normalizing data to antibiotic-free plates, BA composts increased normalized HPCs on plates containing erythromycin and sulfamethoxazole by over 2.3 log10 compared to manure-amended SCL-grown radishes (Fig. 7c and d, bars with the same letters; P < 0.001).
Composting method itself (static versus turned) had little effect, with only five cases in which there was a significant difference in HPCs capable of growth in the presence of an antibiotic (Fig. 6c, e, and f, bars with the same letters; P < 0.001).
Comparison of antibiotic administration versus control conditions on radish surface-associated HPCs.
For both dairy cattle- and beef cattle-derived amendments, administration of antibiotics did not appear to be a major factor in the carriage of HPCs capable of growth on antibiotic media on radish surfaces (Fig. 6 and 7). There were only three exceptions where there were differences that were >1.0 log10 in magnitude. There were 1.2-log10 and 1.1-log10 increases HPCs on sulfamethoxazole and tetracycline plates, respectively, when DA turned compost was applied to LS compared to DC turned compost (Fig. 6e and f, plus signs; P < 0.001). HPCs on erythromycin plates increased by 1.1 log10 for radishes grown in LS amended with DC manure compared to the DA manure (Fig. 6c, plus signs; P < 0.001). No differences were observed among the R2A control (Fig. 5), R2A with ceftazidime, and R2A with clindamycin (Fig. 6a and b; P > 0.050, ANOVA).
DISCUSSION
This study provides a comprehensive and integrated comparison of the effects of amending soil with beef or dairy manure, collected during antibiotic administration or from control cattle not undergoing antibiotic treatment, on carriage of indicators of antibiotic resistance on radish surfaces and whether composting the manure alters the effects. In particular, the greenhouse study enabled replicated assessment of potential interactive effects between biological amendments and soil properties while controlling for potential background effects. In the present study, the patterns revealed via 16S rRNA gene amplicon sequencing were largely consistent with those observed from qPCR and culture: soil texture was a major overarching factor, with effects of manure and compost amendment types most observable in the LS conditions.
A parallel study reported metagenomic profiling of the “resistomes” (i.e., total ARGs) of the radishes grown in the dairy manure/compost conditions that were the subject of this study (37). Specific findings that were consistent between the two studies are that soil texture and static composting of dairy manure can cause shifts in the compositions of both the microbiota and markers of antibiotic resistance. However, both studies also found that there were no measurable shifts in the microbiota or antibiotic resistance metrics associated with collecting manure during administration of antibiotics. The present study expands on the first, providing analysis of beef manure/compost-amended conditions and applying qPCR as a sensitive and quantitative tool for comparing responses of sul1, tet(W), and intI1 as markers of antibiotic resistance. While metagenomic sequencing is an emerging and promising technology, it is costly, has high detection limits, and does not provide strict quantitative information in the manner that qPCR does, and meaningful ways of interpreting metagenomic data are still evolving (38). Further, the culture-based approach applied in the present study provided insight into phenotypic responses of viable and culturable bacteria on the radish surfaces. Taken together, the findings of the two studies are largely consistent, with both highlighting soil texture and biological amendments as overarching drivers shaping beta diversity of the radish microbiota and resistomes (37).
Here, precise quantitation of tet(W), sul1, and intI1 as indicators of antibiotic resistance was applied, providing an accessible and quantitative approach to assessing effects of multiple agricultural management conditions on the potential for antibiotic resistance to spread (20, 22). Notably, both sul1 and intI1 were quite sensitive to the experimental conditions applied in this study and behaved in a manner largely consistent with expectation, i.e., generally higher in manure, equal or lower in composted conditions, and lowest or undetectable in chemical fertilizer controls (Fig. 3). sul1 was a particularly sensitive indicator of biological amendment effects and was consistently below the limit of quantification on radishes grown in the fertilizer-only control soils. On the other hand, tet(W) was sparsely detected and had low sensitivity, making it a poor indicator for the effects of the conditions (Fig. 3). This illustrates the value of selecting target indicator genes that are highly abundant, which would not likely be the case if ARGs of the most urgent clinical relevance, such as vanA, mcr1, or blaNDM-1, encoding resistance to vancomycin, colistin, and extended-spectrum beta-lactam antibiotics, respectively, were directly targeted.
The addition of biological amendment almost always significantly increased intI1 and sul1 on the radishes compared to the chemical fertilizer control for both soil textures, especially with raw manure. Interestingly, though, where there were statistically significant effects between corresponding conditions, it was not always the condition with antibiotic treatment that yielded higher markers of antibiotic resistance relative to the no-antibiotic control (Fig. 3). This indicates that the effects of residual antibiotics in the manure are more complex than a simple selection pressure paradigm and that there are more nuanced microbial ecological impacts of the antibiotics as well, particularly when manure is composted. Similarly to the present study, it was reported in a prior field study in Canada that radishes grown in raw dairy manure carried more copies of sul1 than those grown in control plots with no biological soil amendment, but there was no significant difference between these two conditions in terms of copies of intI1 (30). Thus, although sul1 is often present in the class 1 integron, the present study and the prior study both indicate that sul1 and intI1 can behave independently. Therefore, it may be of value to monitor both genes if possible.
Interestingly, there was a strong, overarching effect of soil texture throughout this study, where effects of manure-derived amendments were much more pronounced in LS than in SCL soil (Fig. 3). While farmers generally cannot change the composition of their soils, they can still factor in knowledge of the soil texture to select appropriate manure management practices to mitigate the spread of antimicrobial resistance. For example, this study suggests that composting may be more beneficial for soils higher in sand content. Future studies to better understand the precise mechanisms by which soil textures may amplify or attenuate the effects of biological amendments on antibiotic resistance markers on vegetables would be of interest. Various differences between LS and SCL could have contributed to the differences observed in this study, including more oligotrophic conditions in LS being more strongly influenced by the influx of nutrients in the manure as well as a lower sorptive capacity making any residual antibiotics more bioavailable. Multiple soil studies have reported higher levels of ARGs associated with more abundant nutrient content (29, 39–41), which is consistent with higher levels in this study when there was a substantial influx of manure-derived amendment. Relative content of silt and sand is also known to be associated with differences in bacterial community structure (12), which in turn can shift in distinct ways in response to various stimuli and ultimately influence the levels of ARGs (40). Consistent with this, we observed here that the beta diversity of the radish taproot microbiota clearly bifurcated based on soil texture, and effects of amendment types were more distinctive in the LS than in the SCL soil (Fig. 2).
Another important factor is that selective agents, including antibiotics and their degradation intermediates, are generally more bioavailable and capable of inducing resistance in sandy soils (42). In a companion study of the soils in which the radishes grew, there were no significant differences in levels of antibiotics detected among the two soil textures (15); however, bioavailability of the antibiotics can vary by soil texture, other environmental conditions, and the antibiotic itself (43). Additionally, the locally available soils were selected for contrasting physical properties, resulting in their origination from two distinct regions, which would harbor correspondingly distinct microbiota (39, 41) available for colonizing the radish taproots. These soils also had a history of agricultural use; thus, it is not surprising that fecal bacteria such as Enterobacteriaceae were found even on radishes grown in the chemical fertilizer-only control soils (44). The pH values of the biological amendments were slightly higher than those of the soils (15), which could have also contributed to observed shifts in microbiota.
Because qPCR cannot directly distinguish live/dead organisms or phenotype, HPC organisms associated with radish taproot surfaces were also cultured on antibiotic-containing R2A plates. Given that the vast majority of environmental bacteria are not amenable to growth on standard lab media (45, 46), R2A was selected because it is versatile and it can be applied to both vegetables and soil (34, 35). Relative to other media, the oligotrophic nature of R2A allows it to cultivate a broader range of bacteria, e.g., capturing over 8 log CFU/g from the tomato plant rhizosphere (35). Still, because R2A captures phylogenetically diverse bacteria, it is important to recognize that it cannot distinguish intrinsically resistant strains from strains that are resistant because of carriage of ARGs. However, the intention of the present study was to apply the method consistently in order to facilitate comparisons. Notably, overarching effects of soil texture were also noted in terms of effects of biological amendments on proportions of HPCs capable of growth on antibiotic-containing media (Fig. 7). Interestingly, some amendments added to LS actually resulted in fewer HPCs on sulfamethoxazole-containing plates, further reinforcing the idea that soil texture is a key mediating factor in terms of potential for manure-borne antibiotic resistance to spread. Overall, it is advised that HPCs on antibiotic media should be interpreted with caution, given the confounding factor of not being able to distinguish intrinsic resistance. As observed in this study, relative comparisons were consistent with qPCR-based measures for stronger effects (e.g., soil texture), but generally, it is important to consider that the two methods measure distinct dimensions of the microbiome and resistome.
Composting manure before adding it to either soil resulted in reduction in carriage of intI1 on radishes in the majority of cases, while this was also true for sul1 to a lesser extent (Fig. 3). This is interesting given that composting generally elevated sul1 levels relative to the raw manure used in this study (7), suggesting that the addition of compost to soil and cultivation subsequently attenuated these genes. The low numbers of tet(W) measured in this study were consistent with the low numbers measured following composting (7). Contrary to expectation, composting increased proportions of HPC organisms capable of growth on certain antibiotic-containing media. This may be a result of the oligotrophic R2A medium being more suited to cultivation of microbes originating from compost, which is much more depleted of nutrients than manure. The HPC results are an indication that, although there were many positive signs in this study that composting reduces certain indicators of antibiotic resistance, implementing additional barriers to the spread of antibiotic resistance beyond composting would be wise (47).
Overall, the results of this study indicate that soil texture and manure management influence levels of intI1 and sul1 on radish surfaces. In particular, the intI1 integrase gene of the class 1 integron appears especially promising as an antimicrobial resistance indicator gene because of its ability to capture hundreds of ARGs, association with transposases that facilitate their HGT, prominent clinical relevance, and strong relationship with anthropogenic inputs into the environment (20, 21, 48, 49). The overall findings highlight the need for future research to further clarify which combinations of on-farm management approaches most effectively attenuate the potential for antibiotic resistance to spread to vegetables and under which circumstances they are most effective. This study suggests that the role of different soil textures, in particular, is worthy of further examination. Given that radishes represent vegetables that are often eaten raw and have a high degree of soil contact, further research, such as human health risk assessment, is also warranted to better understand the potential role of produce as a source of antibiotic-resistant infections in humans.
MATERIALS AND METHODS
Antibiotic administration.
Administration of antibiotics to beef and dairy cattle was described by Ray et al. (6). In brief, healthy lactating cows were administered standard therapeutic doses of pirlimycin (two doses of 50 mg each, 24 h apart; n = 3), and end-of-lactation cows received preventative doses of cephapirin (single dose of 300 mg into each of four quarters; n = 3). Three additional control cows did not receive antibiotics. Healthy steers were administered in-feed chlortetracycline and sulfamethazine (350 mg each antibiotic each day for 7 days) together (n = 3), and other healthy steers were administered tylosin alone (11 mg/kg in feed) (n = 3). The manure from these six steers was collected as manure from beef cattle administered antibiotics. Three additional control steers were fed similar diets but without medication, and manure from this group of steers was applied as treatments from control beef cattle. Prior to this study, beef steers had not received any antibiotics, while dairy cows had not received antibiotics within the previous 12 months.
Manure collection and composting.
Manure, consisting of feces and urine excreted within the pen, was collected days 1 to 3 and 5 to 7 from the nine dairy cows and nine beef steers, respectively, after the start of antibiotic administration, as described by Ray et al. (6). This was to collect from the peak antibiotic excretion periods for the respective cattle groups. Manure from the six pirlimycin- and cephapirin-treated dairy cows were mixed to represent dairy manure with antibiotics (DA), while manure from the six chlortetracycline-, sulfamethazine-, and tylosin-treated beef steers were mixed to represent beef manure with antibiotics (BA). Manure from the dairy (n = 3) and beef (n = 3) cattle that had not received antibiotics was also collected on the same days and designated control dairy (DC) manure and control beef (BC) manure, respectively. Manure was stored at 4°C for about 9 months prior to composting.
Manure samples were mixed with appropriate amounts of grass hay and mulch to achieve target C/N ratios and subjected to one of two methods of composting: static or turned (6). To accommodate multiple conditions, composting was conducted on a small scale, in tumblers (71-cm length, 64-cm diameter; n = 3). After mixing, static composters remained unturned, whereas turned composters were turned daily. The static composting achieved the target temperature of 55°C for 3 days as described in the FSMA (11). However, while the turned compost also achieved a temperature of 55°C for 3 days, this was below the FSMA-required allotted time for that specific composting method (6), which is a common limitation of small-scale composting. All eight compost treatments were performed in triplicate and cured for 42 days prior to storage at 4°C for about 8 months and mixed before being applied to soils.
Radish production and harvest.
The greenhouse study was described previously, in part, in a parallel study of the rhizosphere soil (15) and metagenomes of radishes grown in dairy amendments (37). Silty clay loam was collected in Blacksburg, VA (37°12′54.5″N, 80°26′32.4″W), and loamy sand was collected in Suffolk, VA (36°41′2.749″N, 76°46′5.023″W) (Table S2). In each round pot (6-in. diameter, 5-in. height; Poppelmann GmbH & Co., Lohne, Germany), 947 g of loamy sand soil (15% water content) or 1,023 g of silty clay loam soil (20% water content) was hand mixed with 26.7 g compost (65% water content) or 40.9 g manure (80% water content) (Table S3) and then sowed with radish seeds (Raphanus sativus, Crunchy Royal; Johnny’s Seeds, Inc., Albion, ME). Four replicate pots of each treatment for each of the two soil textures were mixed and cultivated in a greenhouse at Virginia Tech (Blacksburg, VA). Municipal water was filtered using granulated activated carbon to reduce free chlorine and chloramines, and pots were hand watered, weighed, and measured with a moisture meter (Etekcity, Anaheim, CA) to maintain 50 to 70% field capacity moisture. Water-soluble 20-20-20 fertilizer (JR Peters Inc., Allentown, PA) was applied as necessary. Hairnets were used to cover pots during growth to reduce the potential for cross-contamination. Radishes were harvested after 60 days at maturity (Table S4), and taproots were subjected to bacterial enumeration and DNA extraction. Three radishes representative of market size were selected for harvest across three of the four replicate pots to reduce variance based on radish size.
During harvesting of the radishes, gloves were worn to prevent cross contamination between plants and soil. Taproots were separated from the top leaves and the root hairs and brushed with a small, soft paintbrush that was rinsed with 70% ethanol and air dried between uses. The brushed taproots were then placed in individual sterile plastic bags for subsequent culturing and bacterial-DNA analysis.
Enumeration of aerobic bacteria on radish taproots by HPC.
Taproots were weighed individually in 710-ml sterile filter bags with adequate sterile peptone buffer (Becton Dickinson and Company, Franklin Lakes, NJ) with Tween 80 (Fisher Scientific, Waltham, MA) (0.1% each) to create a 1/10 (wt/wt) dilution. The mass (in grams) of the taproots served as a proxy for the approximate surface area and was used for HPC data (Table S4). Bacteria were removed from the radish surface by shaking at 220 rpm for 5 min using a benchtop rotator, alternating with hand massaging for 2 min, and shaking again for an additional 5 min. The bacterial suspensions were then serially diluted and spread plated onto seven medium types: unamended R2A (Becton Dickinson and Company, Franklin Lakes, NJ) and R2A supplemented with 10 μg/ml ceftazidime (Cz; Dot Scientific, Inc., Burton, MI), 25 μg/ml clindamycin (Cm; Sigma-Aldrich, St. Louis, MO), 25 μg/ml erythromycin (Em; Sigma-Aldrich, St. Louis, MO), 100 μg/ml sulfamethoxazole (Smz; Dot Scientific, Inc., Burton, MI), 3 μg/ml tetracycline (Tc; Dot Scientific, Inc., Burton, MI), or 11 μg/ml vancomycin (Vm; Sigma-Aldrich, St. Louis, MO). Antibiotic concentrations were selected based on trial enumerations of bacterial colonies from compost from dairy cattle treated with antibiotics on R2A at different concentrations. Concentrations were chosen by an observed decrease in CFU from the lowest antibiotic concentration tested. Plates were incubated at 37°C for 24 h.
DNA extraction.
Following enumeration, the remaining radish bacterial suspension was filtered through 0.22-μm-pore-size, 47-mm mixed-cellulose-ester membranes (EMD Millipore, Burlington, MA) to collect all microbial cells dissociated from the radish taproot surfaces. Filters were then folded four times, torn, and stored in sterile, DNase-free, O-ring screw-cap tubes at −80°C until DNA extraction, within 5 months.
The frozen filters were added directly to lysing matrix E tubes from the FastDNA spin kit for soil (MP Biomedicals, Solon, OH) and subjected to DNA extraction following the manufacturer’s instructions, with the exception of an additional bead-beating step and a 2-h incubation (chemical lysis). After resuspension of the DNA with 100 μl DNase-/pyrogen-free water, the tubes were incubated at 55°C for 5 min before elution of the DNA. The newly eluted DNA was then applied to the OneStep PCR inhibitor removal kit (Zymo Research Corporation, Irvine, CA) before being stored at −80°C.
16S rRNA gene amplicon sequencing.
After 16S rRNA gene copy numbers per microliter of each DNA extract were determined, all template DNA from each radish surface sample was diluted to 3 × 104 16S rRNA gene copies/μl. The Earth Microbiome Project protocol for 16S rRNA gene amplicon sequencing for bacterial communities was followed using primers 515fB and 926r and (32, 33). Briefly, triplicate 25-μl reaction mixtures contained 10 μl 2.5× 5PRIME HotMaster mix (QuantaBio, Beverly, MA), 0.5 μl primer 515fB (10 μM), 0.5 μl primer 926r (10-μM), and 14 μl water and template DNA. Thermocycler conditions on the CFX Connect (Bio-Rad Laboratories, Hercules, CA) were 1 cycle of 94°C for 3 min, 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s, and a final extension cycle at 72°C for 10 min. All PCR products were measured using Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA) and pooled equally at 240 ng each. Products were then purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Libraries were prepped by the Biocomplexity Institute of Virginia Tech using the MiSeq reagent kit v3 (Illumina, Inc., San Diego, CA), and 2 × 300-bp paired-end reads were produced using the MiSeq system (Illumina, Inc., San Diego, CA). Paired ends were then stitched at 372 to 375 bp and a score of ≥0.90 using PANDASeq (50), and OTUs were picked de novo using QIIME (51) from the GreenGenes database at a 97% cutoff. Chimera sequences were discarded using ChimeraSlayer with QIIME (52). Chloroplast and mitochondrial sequences were also discarded. All samples were rarefied to 11,600 sequences for analysis.
Quantitative PCR.
Quantitative real-time PCR (qPCR) was used to quantify the 16S rRNA gene (53), sul1 (54), tet(W) (55), and intI1 (56) copy numbers of DNA extracts from each radish sample (Table S5). DNA extracts for 16S rRNA gene were diluted 1/10, or 1/2 for intI1, to reduce PCR inhibition. For each gene target, 10-μl reaction mixtures consisted of 5 μl 2× SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA), 0.8 μl 5 μM forward primer (400 nM), 0.8 μl 5 μM reverse primer (400 nM), 1 μl of DNA template, and 2.4 μl molecular-grade water (Sigma-Aldrich, St. Louis, MO) and were run on the CFX Connect system (Bio-Rad Laboratories, Hercules, CA). The protocol consisted of 1 cycle of 98°C for 2 min, 40 cycles of 98°C for 5 s, and annealing temperature and time (Table 1), followed by a melting curve. Standard curves consisted of 7 10-fold dilutions (108 to 102 copies/μl for the 16S rRNA gene and 107 to 101 copies/μl for sul1, tet(W), and intI1, indicating quantifiable limits.
TABLE 1.
Primers used for qPCR targeting the 16S rRNA gene and ARGs
| Gene target | Size of PCR product (bp) | Annealing temp (°C) | Extension time (s) | Position | Forward or reverse primer (5′→3′) | Reference |
|---|---|---|---|---|---|---|
| 16S rRNA gene | 124 | 55 | 5 | 1369–1492 | CGGTGAATACGTTCYCGG | 53 |
| GGWTACCTTGTTACGACTT | ||||||
| sul1 | 162 | 71 | 7 | 133–295 | CGCACCGGAAACATCGCTGCAC | 54 |
| TGAAGTTCCGCCGCAAGGCTCG | ||||||
| tet(W) | 167 | 61 | 7 | 161–328 | GAGAGCCTGCTATATGCCAGC | 55 |
| GGGCGTATCCACAATGTTAAC | ||||||
| intI1 | 67 | 66 | 5 | 737–803 | TCGTGCGTCGCCATCACA | 56 |
| GCTTGTTCTACGGCACGTTTGA |
Statistical analysis.
For qPCR data, several treatments had values below the limits of quantification (the lowest measurement of the standard curve) and detection (values that could not be measured due to lack of fluorescence), which were defined as left-censored data (57). For the 16S rRNA gene, the quantification limit was 1,000 copies/taproot; for tet(W) and sul1, the quantification limit was 10 copies/taproot, and for intI1, the quantification limit was 20 copies/taproot. Treatments where all values were below quantification (i.e., all values were defined as left censored) were excluded from analysis. All other left-censored data were arbitrarily defined as 0.6 copy/taproot, with 1 added to all values in order to fit the model, and a Weibull distribution was assumed for all gene targets. The R (v 3.4.3) packages NADA, pscl, and survival were used for the statistical analysis. Left-censored events from qPCR and CFU data were input as “2,” while all other quantifiable events were input as “1.” Means and standard errors were calculated from the survival model.
Because different treatments were excluded depending on the target gene, different factors of differing levels were used. For tet(W), a one-way analysis was used where 1 factor with 19 levels was used. For sul1 and intI1, two-way analysis was used where soil (LS and SCL) was crossed with amendment: DA manure, DA static compost, DA turned compost, DC manure, DC static compost, DC turned compost, BA manure, BA static compost, BA turned compost, BC manure, BC static compost, BC turned compost, and chemical fertilizer control (intI1 only since sul1 was not detected on chemical fertilizer control radishes).
JMP Pro 12/13 (SAS Institute Inc., Cary, NC) was used for ANOVA, Wilcoxon, and Kruskal-Wallis analyses and to generate figures. The significance cutoff was selected at a P value of <0.050 with the Bonferroni correction applied. R2A HPCs between 25 and 250 CFU/plate were used, and counts from R2A control, R2A with clindamycin, and R2A with ceftazidime were log-transformed to achieve normal distribution. A stepwise two-way ANOVA was used to compare the CFU on R2A supplemented with antibiotics between two factors at a time for each antibiotic plate type and whether there were interacting effects. HPCs from each antibiotic plate were independent of each other. CFU enumerated on R2A plates containing clindamycin and ceftazidime were also analyzed as normalized to HPCs from R2A without antibiotics for corresponding samples and analyzed using one-way Wilcoxon tests. Because of values below the quantifiable limit of 25 CFU/plate, HPCs for R2A agar supplemented with erythromycin, sulfamethoxazole, tetracycline, and vancomycin were also analyzed using the 2-way survival analysis assuming a Weibull distribution.
Analyses of similarities (ANOSIM) of unweighted UniFrac distances (58) were performed to improve discrimination between transferred bacteria and horizontally acquired bacteria. A nonmetric multidimensional scaling plot was generated using PRIMER-E 6.1.13 (Quest Research Limited, Auckland, New Zealand).
Ethics approval.
All animal studies were approved by IACUC protocol DASC 13–145 (PI, K. Knowlton).
Data availability.
Raw 16S rRNA gene amplicon reads were deposited in the NCBI Sequence Read Archive under SRA accession no. SRP126729.
Supplementary Material
ACKNOWLEDGMENTS
Funding was supported by USDA NIFA-AFRI no. 2014-05280 and 2017-68003-26498. In addition, M.A.P. was supported in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. The Virginia Tech Advanced Research and Computing network and National Science Foundation NNCI award 1542100 provided internal support for this work.
We thank Katharine Knowlton, Partha Ray, and Chrissy Teets for providing the cattle-based soil amendments from their work with the animal studies and composting. We also acknowledge Kang Xia and Mike Brosius for their assistance in the greenhouse, as well as Leigh Anne Krometis, W. Cully Hession, Allen Straw, Hunter Frame, Hanh Le, Lauren Wind, Kyle Jacobs, Matthew Blair, and Oladayo Omosa for additional assistance and consultation during vegetable growth and harvesting. We also thank Kim Waterman, Vaishali Dharmarha, Kendall Fogler, Robert Williams, Christine Pankow, Natalie Pulido, Trevor Zamora, Nicholas Poe, Michelle Stark, and Meghan Ruppel for their assistance in the laboratory. Last, we thank Pan Ji and Emily Garner for assistance with data analysis.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall BM, Levy SB. 2011. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 24:718–733. 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliveira M, Viñas I, Usall J, Anguera M, Abadias M. 2012. Presence and survival of Escherichia coli O157:H7 on lettuce leaves and in soil treated with contaminated compost and irrigation water. Int J Food Microbiol 156:133–140. 10.1016/j.ijfoodmicro.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Marti R, Scott A, Tien YC, Murray R, Sabourin L, Zhang Y, Topp E. 2013. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl Environ Microbiol 79:5701–5709. 10.1128/AEM.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youngquist CP, Mitchell SM, Cogger CG. 2016. Fate of antibiotics and antibiotic resistance during digestion and composting: a review. J Environ Qual 45:537–545. 10.2134/jeq2015.05.0256. [DOI] [PubMed] [Google Scholar]
- 6.Ray P, Chen C, Knowlton KF, Pruden A, Xia K. 2017. Fate and effect of antibiotics in beef and dairy manure during static and turned composting. J Environ Qual 46:45–54. 10.2134/jeq2016.07.0269. [DOI] [PubMed] [Google Scholar]
- 7.Keenum IM, Williams R, Ray P, Garner ED, Knowlton KF, Pruden A. Combined effects of composting and antibiotic administration on cattle manure-borne antibiotic resistance genes. Microbiome, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Gutek A, Grewal S, Michel FC, Yu Z. 2015. Changes in diversity of cultured bacteria resistant to erythromycin and tetracycline in swine manure during simulated composting and lagoon storage. Lett Appl Microbiol 61:245–251. 10.1111/lam.12450. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Sangwan N, Li HY, Su JQ, Oyang WY, Zhang ZJ, Gilbert JA, Zhu YG, Ping F, Zhang HL. 2017. The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J 11:100–111. 10.1038/ismej.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storteboom HN, Kim S-C, Doesken KC, Carlson KH, Davis JG, Pruden A. 2007. Response of antibiotics and resistance genes to high-intensity and low-intensity manure management. J Environ Qual 36:1695–1703. 10.2134/jeq2007.0006. [DOI] [PubMed] [Google Scholar]
- 11.Food and Drug Administration. 2015. Standards for growing, harvesting, packing, and holding of produce for human consumption, p 74353–74568. In Food Safety Modernization Act. Food and Drug Administration, College Park, MD. [Google Scholar]
- 12.Allard SM, Walsh CS, Wallis AE, Ottesen AR, Brown EW, Micallef SA. 2016. Solanum lycopersicum (tomato) hosts robust phyllosphere and rhizosphere bacterial communities when grown in soil amended with various organic and synthetic fertilizers. Sci Total Environ 573:555–563. 10.1016/j.scitotenv.2016.08.157. [DOI] [PubMed] [Google Scholar]
- 13.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 14.Prashar P, Kapoor N, Sachdeva S. 2014. Rhizosphere: its structure, bacterial, diversity and significance. Rev Environ Sci Biotechnol 13:63–77. 10.1007/s11157-013-9317-z. [DOI] [Google Scholar]
- 15.Chen C, Guron GK, Pruden A, Ponder M, Du P, Xia K. 2018. Antibiotics and antibiotic resistance genes in bulk and rhizosphere soils subject to manure amendment and vegetable cultivation. J Environ Qual 47:1318–1326. 10.2134/jeq2018.02.0078. [DOI] [PubMed] [Google Scholar]
- 16.Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13. 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 17.Jechalke S, Kopmann C, Rosendahl I, Groeneweg J, Weichelt V, Krögerrecklenfort E, Brandes N, Nordwig M, Ding GC, Siemens J, Heuer H, Smalla K. 2013. Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl Environ Microbiol 79:1704–1711. 10.1128/AEM.03172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen QL, An XL, Zhu YG, Su JQ, Gillings MR, Ye ZL, Cui L. 2017. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ Sci Technol 51:8149–8157. 10.1021/acs.est.7b01420. [DOI] [PubMed] [Google Scholar]
- 19.Hall RM, Stokes HW. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115–132. 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 20.Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu YG. 2015. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J 9:1269–1279. 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. 2017. Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS One 12:e0179169. 10.1371/journal.pone.0179169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruden A, Arabi M, Storteboom HN. 2012. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol 46:11541–11549. 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 23.Mathew AG, Cissell R, Liamthong S. 2007. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 4:115–133. 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- 24.Sarmah AK, Meyer MT, Boxall AB. 2006. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759. 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Fahrenfeld N, Knowlton K, Krometis LA, Hession WC, Xia K, Lipscomb E, Libuit K, Green BL, Pruden A. 2014. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: field-scale mass balance approach. Environ Sci Technol 48:2643–2650. 10.1021/es404988k. [DOI] [PubMed] [Google Scholar]
- 26.Wallace JS, Garner E, Pruden A, Aga DS. 2018. Occurrence and transformation of veterinary antibiotics and antibiotic resistance genes in dairy manure treated by advanced anaerobic digestion and conventional treatment methods. Environ Pollut 236:764–772. 10.1016/j.envpol.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luby E, Ibekwe AM, Zilles J, Pruden A. 2016. Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: prospects and challenges. J Environ Qual 45:441–453. 10.2134/jeq2015.07.0367. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, An X, Li H, Su J, Ma Y, Zhu YG. 2016. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int 92-93:1–10. 10.1016/j.envint.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Tien YC, Li B, Zhang T, Scott A, Murray R, Sabourin L, Marti R, Topp E. 2017. Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci Total Environ 581-582:32–39. 10.1016/j.scitotenv.2016.12.138. [DOI] [PubMed] [Google Scholar]
- 31.Kampfer P, Erhart R, Beimfohr C, Bohringer J, Wagner M, Amann R. 1996. Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb Ecol 32:101–121. 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- 32.Caporaso GJ, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters W, R Hyde E, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson CR, Randolph KC, Osborn SL, Tyler HL. 2013. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol 13:274. 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SA, Park J, Chu B, Kim JM, Joa JH, Sang MK, Song J, Weon HY. 2016. Comparative analysis of bacterial diversity in the rhizosphere of tomato by culture-dependent and -independent approaches. J Microbiol 54:823–831. 10.1007/s12275-016-6410-3. [DOI] [PubMed] [Google Scholar]
- 36.van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. 2012. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci U S A 109:1159–1164. 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guron GKP, Arango-Argoty G, Zhang L, Pruden A, Ponder MA. 2019. Effects of dairy manure-based amendments and soil texture on lettuce- and radish-associated microbiota and resistomes. mSphere 4:e00239-19. 10.1128/mSphere.00239-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruden A, Alcalde RE, Alvarez PJJ, Ashbolt N, Bischel H, Capiro NL, Crossette E, Frigon D, Kassandra G, Haas CN, Ikuma K, Kappell A, LaPara T, Kimbell L, Li M, Li X, McNamara P, Seo Y, Sobsey MD, Sozzi E, Navab-Daneshmand T, Raskin L, Riquelme MV, Vikesland P, Wigginton K, Zhou Z. 2018. An environmental science and engineering framework for combating antimicrobial resistance. Environ Eng Sci 35:1001–1011. [Google Scholar]
- 39.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udikovic-Kolic N, Wichmann F, Broderick NA, Handelsman J. 2014. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc Natl Acad Sci U S A 111:15202–15207. 10.1073/pnas.1409836111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G. 2014. Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616. 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Zhang W, Wang G, Zhang Y, Gao Y, Boyd SA, Teppen BJ, Tiedje JM, Zhu D, Li H. 2017. Bioavailability of soil-sorbed tetracycline to Escherichia coli under unsaturated conditions. Environ Sci Technol 51:6165–6173. 10.1021/acs.est.7b00590. [DOI] [PubMed] [Google Scholar]
- 43.Aga DS, Lenczewski M, Snow D, Muurinen J, Sallach JB, Wallace JS. 2016. Challenges in the measurement of antibiotics and in evaluating their impacts in agroecosystems: a critical review. J Environ Qual 45:407–419. 10.2134/jeq2015.07.0393. [DOI] [PubMed] [Google Scholar]
- 44.Brandl MT. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol 44:367–392. 10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- 45.Pham VH, Kim J. 2012. Cultivation of unculturable soil bacteria. Trends Biotechnol 30:475–484. 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 47.Dharmarha V, Guron G, Boyer RR, Niemira BA, Pruden A, Strawn LK, Ponder MA. 2019. Gamma irradiation influences the survival and regrowth of antibiotic-resistant bacteria and antibiotic-resistance genes on romaine lettuce. Front Microbiol 10:710. 10.3389/fmicb.2019.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes HW. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190:5095–5100. 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC. 2012. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol 3:119. 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW, Human Microbiome Consortium . 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki MT, Taylor LT, DeLong EF. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays. Appl Environ Microbiol 66:4605–4614. 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei R, Kim SC, Carlson KH, Pruden A. 2006. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435. 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 55.Aminov RI, Garrigues-Jeanjean N, Mackie RI. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 67:22–32. 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall AB, Wellington EM. 2011. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5:1253–1261. 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chambers DL, Wojdak JM, Du P, Belden LK. 2011. Corticosterone level changes throughout larval development in the amphibians Rana sylvatica and Ambystoma jeffersonianum reared under laboratory, mesocosm, or free-living conditions. Copeia 2011:530–538. 10.1643/CP-09-180. [DOI] [Google Scholar]
- 58.Lozupone C, Hamady M, Knight R. 2006. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA gene amplicon reads were deposited in the NCBI Sequence Read Archive under SRA accession no. SRP126729.