The major source of contamination of food with Listeria monocytogenes is thought to be due to biofilm formation and/or persistence in food-processing plants. By establishing as a biofilm, L. monocytogenes cells become harder to eradicate due to their increased resistance to environmental threats.
KEYWORDS: food safety, Listeria monocytogenes, biofilms, mutagenesis
ABSTRACT
Listeria monocytogenes is a ubiquitous foodborne pathogen that results in a high rate of mortality in sensitive and immunocompromised people. Contamination of food with L. monocytogenes is thought to occur during food processing, most often as a result of the pathogen producing a biofilm that persists in the environment and acting as the source for subsequent dispersal of cells onto food. A survey of seafood-processing plants in New Zealand identified the persistent strain 15G01, which has a high capacity to form biofilms. In this study, a transposon library of L. monocytogenes 15G01 was screened for mutants with altered biofilm formation, assessed by a crystal violet assay, to identify genes involved in biofilm formation. This screen identified 36 transposants that showed a significant change in biofilm formation compared to the wild type. The insertion sites were in 27 genes, 20 of which led to decreased biofilm formation and seven to an increase. Two insertions were in intergenic regions. Annotation of the genes suggested that they are involved in diverse cellular processes, including stress response, autolysis, transporter systems, and cell wall/membrane synthesis. Analysis of the biofilms produced by the transposants using scanning electron microscopy and fluorescence microscopy showed notable differences in the structure of the biofilms compared to the wild type. In particular, inactivation of uvrB and mltD produced coccoid-shaped cells and elongated cells in long chains, respectively, and the mgtB mutant produced a unique biofilm with a sandwich structure which was reversed to the wild-type level upon magnesium addition. The mltD transposant was successfully complemented with the wild-type gene, whereas the phenotypes were not or only partially restored for the remaining mutants.
IMPORTANCE The major source of contamination of food with Listeria monocytogenes is thought to be due to biofilm formation and/or persistence in food-processing plants. By establishing as a biofilm, L. monocytogenes cells become harder to eradicate due to their increased resistance to environmental threats. Understanding the genes involved in biofilm formation and their influence on biofilm structure will help identify new ways to eliminate harmful biofilms in food processing environments. To date, multiple genes have been identified as being involved in biofilm formation by L. monocytogenes; however, the exact mechanism remains unclear. This study identified four genes associated with biofilm formation by a persistent strain. Extensive microscopic analysis illustrated the effect of the disruption of mgtB, clsA, uvrB, and mltD and the influence of magnesium on the biofilm structure. The results strongly suggest an involvement in biofilm formation for the four genes and provide a basis for further studies to analyze gene regulation to assess the specific role of these biofilm-associated genes.
INTRODUCTION
The foodborne pathogen Listeria monocytogenes is a serious health threat to immunocompromised people, the elderly, pregnant women, and unborn and newborn babies (1), with a high mortality rate (up to 30%) in those groups (2). L. monocytogenes is ubiquitous in the environment, with contamination of food usually occurring during processing rather than being present in raw food (3, 4). The breadth of foods contaminated with L. monocytogenes is extensive and includes ready-to-eat meat and seafood, as well as vegetables and fruit (5).
L. monocytogenes is motile across a broad range of temperatures and has the capacity to adapt quickly to environmental changes to secure its survival. It can grow over a broad temperature range and survive freezing temperatures (6). L. monocytogenes can tolerate some degree of low pH but is not as acid tolerant as some other foodborne pathogenic bacteria, such as Escherichia coli O157:H7. It is also capable of surviving in high salt concentrations up to 11.5% (6). All of these attributes contribute to the adaptability of this pathogen and impede its control when encountered in food-processing environments.
Surface attachment and biofilm formation are important to the environmental persistence of L. monocytogenes (7). A biofilm is a community of cells that exists in a sessile lifestyle rather than a planktonic one in order to use resources more efficiently and to resist environmental threats. Cells in a biofilm are attached to each other and to a surface. They are held together by an extracellular polymeric matrix that consists of DNA, proteins, lipopolysaccharides, and other substances that contribute to its stability and act as a protective barrier. For L. monocytogenes, the capacity to form biofilms in so-called “harborage” sites enables the pathogen to establish itself and to act as a source for subsequent dispersal of single cells. Management of L. monocytogenes is further impaired because biofilm cells are more resistant to cleaning agents and sanitizers (8), as well as antibiotics (9). As a result, L. monocytogenes is able to contaminate both surfaces and food products.
Transposon mutagenesis has proven to be a successful tool for identification of genes involved in biofilm formation by L. monocytogenes (10, 11). As a result, to date, four research groups have successfully identified genes involved in biofilm formation by L. monocytogenes using the Himar1-based transposition system (12–15). The majority of the identified genes were associated with biosynthesis or motility. This transposition system has also been used to identify the genetic factors underlying other phenotypic changes, such as desiccation survival or nisin sensitivity (16, 17).
A survey of seafood-processing plants in New Zealand identified four persistent strains (persisted in factories for at least 6 months), classified by their unique pulsotypes (18). One of these pulsotypes (type 5132), represented by L. monocytogenes 15G01, was shown to have a high capacity to form biofilms in in vitro assays (18–20). Further genomic studies revealed that L. monocytogenes 15G01 (lineage II genome) belongs to the sequence type ST-321, determined using MLST, and is lacking the φtRNA-Ser prophage (20). This isolate exhibited low invasion in mammalian cell cultures (20), which is linked to truncation of the primary virulence factor internalin A (InlA) (21). Since the capacity to form biofilms is believed to be a major contributing factor in persistence of L. monocytogenes and subsequent contamination of food in food-processing premises, a library of mutants of L. monocytogenes 15G01, previously generated using the Himar1 mariner-based transposition system (22), was screened for mutants with altered biofilm formation using the crystal violet assay. This study aimed to reveal additional genes in L. monocytogenes 15G01 that are associated with biofilm formation and to further characterize the role of these genes through visualization of the structure by using scanning electron microscopy (SEM) and fluorescence microscopy to visualize the structure of the altered biofilms.
RESULTS
An in vitro biofilm assay identified multiple mutants that have either more or less biofilm formation than does the wild type.
A set of 4,500 mutants of a transposant library of approximately 6,500 mutants of L. monocytogenes 15G01, created with the mariner transposition system, was screened at 30°C in modified Welshimer’s broth (MWB) for mutants with altered biofilm formation using the 1% aqueous crystal violet (CV) assay (a set of 2,000 mutants has been analyzed previously (20). These conditions were used since they had been previously shown to induce biofilm formation by L. monocytogenes 15G01 (19). In total, 36 mutants were found with greater (10 mutants) or lower (26 mutants) biofilm formation ability under these conditions compared to the wild type (Table 1), with the average optical density at 595 nm (OD595) of the mutants being at least two standard deviations (SD) below or above the average OD595 value (1.197) of the wild-type strain, L. monocytogenes 15G01. The growth of the mutants was compared to the growth of the wild-type strain in MWB during the screen to confirm that the altered biofilm formation observed was not due to differences in the ability of the mutants to grow in this media, and only mutants showing equal or higher OD595 than the wild type after 48 h were included.
TABLE 1.
Biofilm-related genes identified in L. monocytogenes 15G01 through transposon insertions based on DNA homologies with the L. monocytogenes EGD genome database (accession number HG421741)a
| Function group and putative gene function | Transposon insertion site | Mean biofilm mass (%) relative to WT ± SD | No. of hits | Coordinate(s) for insertion in L. monocytogenes EGD | Orientation of transposon insertion | Mutant strainb |
|---|---|---|---|---|---|---|
| Biosynthesis | ||||||
| Adenylosuccinate synthase | LMON_0057 (purA) | 135.7 ± 5.12 | 1 | 59174 | 3′–5′ | 24A10 |
| Dihydroxyacetone kinase family protein | LMON_1882 | 173.03 ± 7.82 | 1 | 1888930 | 3′–5′ | 35H9 |
| Acetyltransferase, GNAT family | LMON_2362 | 27.32 ± 6.92 | 1 | 2377834 | 5′–3′ | 31E7 |
| Glycosyl hydrolase, family 31 | LMON_2457 | 19.90 ± 4.93 | 1 | –d | 3′–5′ | 32E5 |
| Cell wall/membrane | ||||||
| Sortase A, LPXTG-specific SrtA | LMON_0935 | 7.49 ± 0.59 | 1 | 948604 | 3′–5′ | 31C7 |
| Glycosyltransferase | LMON_0939 | 18.74 ± 3.36 | 1 | – | 5′–3′ | 35C1 |
| Putative peptidoglycan-bound protein (LPXTG motif) Lmo1666 homologue | LMON_1733 | 29.70 ± 5.66 | 3 | 1723653, 1721718, 1721318 | 5′–3′, 3′–5′, 3′–5′ | 24H3, 25D2, 64B3 |
| Cardiolipin synthetase ClsA | LMON_2515 | 163.02 ± 15.27 | 2 | 2538121, 2538525 | 3′–5′, 5′–3′ | 30A9, 34F11 |
| Glycosyltransferase LafA | LMON_2570 | 16.10 ± 2.43 | 2 | 2591134, 2590370 | 3′–5′, 3′–5′ | 28G11, 32A9 |
| Translation and transcription | ||||||
| GTP-binding protein HflX | LMON_1358 | 16.06 ± 6.92 | 1 | – | 3′–5′ | 43C9 |
| SSU ribosomal protein S1p | LMON_2007 (rpsA) | 140.68 ± 10.74 | 1 | 2012734 | 3′–5′ | 28A2 |
| Transporter systems | ||||||
| Manganese ABC transporter, ATP-binding protein SitB | LMON_1917 | 133.83 ± 7.38 | 1 | 1925244 | 3′–5′ | 47H11 |
| Bacitracin export ATP-binding protein BceA | LMON_2188 | 138.01 ± 1.19 | 1 | 2194403 | 3′–5′ | 31G1 |
| ABC transporter, permease protein EscB | LMON_2290 | 142.51 ± 7.66 | 2 | 2305608, 2305511 | 3′–5′, 3′–5′ | 32E2, 47H10 |
| Phosphate ABC transporter, periplasmic phosphate-binding protein PstS | LMON_2511 | 30.81 ± 0.50 | 1 | 2532962 | 3′–5′ | 36A2 |
| PTS system, IIA component | LMON_2675 | 32.69 ± 3.94 | 1 | 2692293 | 3′–5′ | 26C10 |
| Mg2+ transport ATPase, P-type | LMON_2712 | 7.16 ± 1.47 | 2 | 2731046, 2731895 | 3′–5′, 3′–5′ | 30H2, 44D3 |
| ABC transporter, ATP-binding protein | LMON_2792 | 6.93 ± 0.27 | 1 | 2816670 | 3′–5′ | 28D10 |
| Motility | ||||||
| Flagellin protein FlaA | LMON_0695 (flaA) | 23.74 ± 5.93 | 1 | – | 3′–5′ | 41H7 |
| Autolysis | ||||||
| Membrane-bound lytic murein transglycosylase D precursor MltD | LMON_2714 | 2.11 ± 0.35 | 1 | 2734305 | 5′–3′ | 39G5 |
| DNA repair and stress response | ||||||
| Glutamate decarboxylase | LMON_2376 | 6.81 ± 0.93 | 1 | 2393310 | 3′–5′ | 35F3 |
| Excinuclease ABC subunit B UvrB | LMON_2501 (uvrB) | 18.33 ± 2.58 | 2 | 2525427, 2525564 | 3′–5′, 3′–5′ | 33E11, 42G3 |
| Unknown | ||||||
| FIG00774663: hypothetical protein | LMON_1212 | 21.32 ± 0.69 | 1 | – | 5′–3′ | 36B9 |
| FIG00774466: hypothetical protein | LMON_2144 | 28.89 ± 3.37 | 1 | 2153693 | 5′–3′ | 26H1 |
| COG1801: uncharacterized conserved protein | LMON_2417 | 15.37 ± 0.51 | 1 | – | 3′–5′ | 40C12 |
| Hypothetical protein (mutant strain 44F5) | NCc | 10.70 ± 5.02 | 1 | – | 3′–5′ | 44F5 |
| Hypothetical protein (mutant strain 41A8) | NC | 21.88 ± 7.19 | 1 | – | 3′–5′ | 41A8 |
| Intergenice | ||||||
| Methionine ABC transporter ATP-binding protein and hypothetical protein | LMON_2430 and LMON_2431 | 18.27 ± 1.96 | 1 | 2450096 | 3′–5′ | 67C5 |
| Dihydroxyacetone kinase family protein and putative alkaline-shock protein | LMON_1882 and LMON_1883 | 173.44 ± 15.52 | 1 | 1889436 | 3′–5′ | 33F8 |
Homologies were identified by Megablast and Geneious 7 software using default settings. The biofilm mass produced by the mutants (CV assay) was calculated relative to the biofilm formation by the wild type in two independent experiments with eight replicates. Putative gene functions are based on information from http://www.genome.jp/.
Internal numbering of the transposon library.
NC, no counterpart in L. monocytogenes EGD but present in the parental strain.
–, the exact insertion site could not be determined.
Defined by the two genes at each boundary of the intergenic space. 67C5 is 217 bp downstream of LMON_2430 and 100 bp upstream of LMON_2431, while 33F8 insertion is 7 bp downstream of LMON_1882 and 373 bp upstream of LMON _1883.
Characterization of the transposon insertion sites in mutants with altered biofilm formation identified 27 loci potentially involved in this process.
Nested semiarbitrary PCR enabled the amplification of the genome sequences flanking the transposons in the 36 mutants. DNA sequencing of the flanking regions and subsequent BLASTN and Megablast comparisons of the sequences with the genome of reference strain L. monocytogenes EGD permitted the locations and directions of the insertions in 15G01 to be estimated for all mutants, with identification of the exact insertion sites for 28 mutants (Table 1). Transposon insertions seemed to predominantly occur in the second half of the genome raising doubts as to the random nature of transposition (see Fig. S3a in the supplemental material). In the 36 mutants examined, 27 genes were disrupted by a transposon insertion, with six loci disrupted in two independent mutants and one disrupted in three independent mutants (Table 1). Two mutants had an insertion of the transposon in an intergenic region (Table 1), and a further two mutants had insertions in genes that were not present in the reference genome of L. monocytogenes EGD but in the parental strain L. monocytogenes 15G01.
Functional analysis of the disrupted genes identified multiple functional groups involved in biofilm formation by L. monocytogenes 15G01.
Comparison of the disrupted genes in each of the mutants affected in biofilm formation with homologous genes in L. monocytogenes EGD and other L. monocytogenes strains in the GenBank database divided them into seven diverse functional groups (Table 1). Of particular note, five genes annotated as being involved in cell wall/membrane synthesis or integrity were identified, including genes encoding a putative peptidoglycan-bound protein disrupted in three mutants that had low biofilm formation, cardiolipin synthetase (designated clsA), disrupted in two mutants with greater biofilm formation and the glycosyltransferase LafA in two mutants with low biofilm formation.
Genes involved in transport systems, stress response, autolysis, and motility also influenced biofilm formation, including a gene predicted to encode a P-type Mg2+ transport ATPase (designated mgtB) and a gene annotated as encoding the ABC transporter permease protein EscB, which were both disrupted in two mutants. Disruption of the P-type Mg2+ transport ATPase resulted in decreased biofilm formation, while the insertion of the transposon into escB resulted in increased biofilm formation.
Complementation of selected genes confirms a role for mltD in biofilm formation by L. monocytogenes 15G01.
Four mutants (33E11, 34F11, 39G5, and 44D3) were examined further to confirm the role of the disrupted genes in biofilm formation by L. monocytogenes 15G01 (Table 2). The first, the clsA mutant (34F11), was selected because this gene had been disrupted in multiple mutants with increased biofilm formation and because the clsA gene was known to be involved in biofilm formation by other bacterial species (23–25). The uvrB mutant (33E11) was selected since multiple mutants with a transposon insertion in the gene coding for the excision nuclease ABC subunit B were identified in the screen and uvrB is part of an operon with uvrA, which, when disrupted in L. monocytogenes in a previous study, resulted in increased biofilm formation at 15°C (15). The mltD mutant (39G5) was studied because it was predicted to affect autolysis, which has been implicated in changes in biofilm formation by L. monocytogenes (26, 27) and other bacteria (28), while the mgtB mutant (44D3) was included because it has also been disrupted in multiple mutants and has not been associated with biofilm formation by L. monocytogenes before.
TABLE 2.
L. monocytogenes strains and plasmids used in this study
| Strain or plasmid | Description | Mutation | Source or reference |
|---|---|---|---|
| Strains | |||
| 15G01 | Wild-type serotype 1/2a; Ems Kans | 18 | |
| 34F11 | 15G01 with transposon inserted in the clsA gene (LMON_2515); Emr Kans | 15G01 clsA::himar1 | This study |
| 33E11 | 15G01 with transposon inserted in the uvrB gene (LMON_2501); Emr Kans | 15G01 uvrB::himar1 | This study |
| 33E11-C | 33E11 containing the pIMK-uvrB plasmid; Emr Kanr | 15G01 uvrB::himar1/pIMKuvrB | This study |
| 33E11-EV | 33E11 containing the pIMK plasmid; Emr Kanr | 15G01 uvrB::himar1/pIMK | This study |
| 39G5 | 15G01 with transposon inserted in the mltD gene (LMON_2714); Emr Kans | 15G01 mltD::himar1 | This study |
| 39G5-C | 39G5 containing the pIMK-mltD plasmid; Emr Kanr | 15G01 mltD::himar1/pIMKmltD | This study |
| 39G5-EV | 39G5 containing the pIMK plasmid; Emr Kanr | 15G01 mltD::himar1/pIMK | This study |
| 44D3 | 15G01 with transposon inserted in the mgtB gene (LMON_2712); Emr Kans | 15G01 mgtB::himar1 | This study |
| 41H7 | 15G01 with transposon inserted in the flaA gene (LMON_0695); Emr Kans | 15G01 flaA::himar1 | This study |
| Plasmids | |||
| pIMK | Site-specific listerial integrative vector, 5.1 kb; Kanr | 80 | |
| pIMK-uvrB | Site-specific plasmid carrying the LMON_2501 gene; Kanr | This study | |
| pIMK-mltD | Site-specific plasmid carrying the LMON_2714 gene; Kanr | This study |
Growth curves of the selected 36 mutants in MWB at 30°C were then produced by manual measurements to further examine growth behavior (see Table S1 and Fig. S1 in the supplemental material) in the presence of erythromycin (the transposon contains an erythromycin resistance gene). In addition, growth studies with an automated plate reader were carried out for the four selected mutants without selective antibiotics to further rule out the possibility that the changes in biofilm formation were due to impaired growth (see Fig. S2). The two mutants 39G5 and 44D3 formed cell aggregates during growth which is reflected by the high OD600 values; however, the growth pattern was not affected. 39G5 had an extended exponential phase compared to the other strains, but all five examined strains were in stationary phase at 48 h, the time point at which biofilm formation was measured.
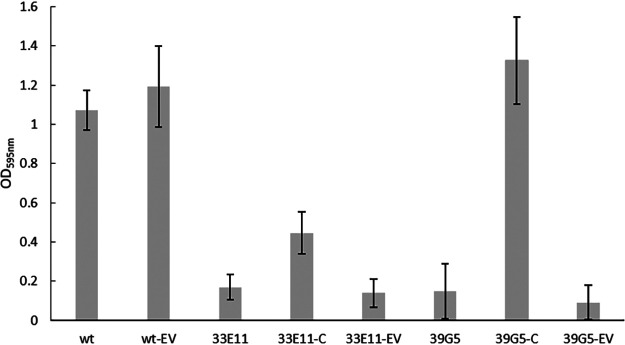
Complementation studies were subsequently carried out on the four mutants. In these studies, introduction of the pIMK vector containing the wild-type mltD gene into the mltD mutant resulted in the restoration of biofilm formation to levels produced by the wild type (Fig. 1). Conjugation of the empty vector into this mutant had no obvious effect on biofilm formation (Fig. 1). These data confirmed that the change in biofilm formation by the mltD (a homologue of the peptidoglycan hydrolase murA) mutant was a result of inactivation of the gene. Thus, mltD is required for biofilm formation by L. monocytogenes 15G01.
FIG 1.
Comparison of biofilm formation by L. monocytogenes 15G01 (wt), the uvrB and mltD mutants (33E11 and 39G5), and mutants containing a wild-type copy of the corresponding gene (complemented strains [-C]) or the empty vector pIMK (-EV). Error bars represent the SD for three independent experiments (n = 6). Biofilm formation was determined by measuring the OD595 as part of the CV assay.
In contrast to the successful complementation of the mltD mutant (39G5), introduction of the pIMK vector containing the wild-type uvrB gene into the uvrB mutant only partially restored (41.7%) the wild-type phenotype (Fig. 1), while attempts to complement the mgtB and clsA mutants (44D3 and 34F11) using a similar process failed to restore the wild-type phenotype altogether (data not shown).
Microscopy confirmed that the mltD mutant (39G5) experiences a dramatic loss in both viable and nonviable cells.
All of the selected mutants were included in microscopic analysis to assess changes in phenotype despite the failure to complement a number of the mutations. Fluorescence microscopy showed that, after 48 h of incubation of the mutants on polystyrene surfaces in MWB at 30°C, all appeared to show some differences in the structure of their biofilms compared to the wild type (Fig. 2). The biofilm of the mltD mutant (39G5) (Fig. 2d) consisted of a few (mainly individual) cells, which seemed to be elongated and in chains. The lack of visible cells (whether alive or dead) was consistent with the low biofilm formation observed in the initial screen. The biofilm produced by the mgtB mutant (44D3) also contained few live cells, although it did appear to have a cloudy structure under fluorescence (Fig. 2e). Consistent with this, there appeared to be greater numbers of dead cells associated with the biofilm produced by the mgtB mutant. The uvrB mutant (33E11) was a low biofilm former and exhibited only sparse biofilm formation after 48 h (Fig. 2c), which was consistent with the approximately 80% reduction in biofilm formation observed in in vitro assays. The clsA mutant (34F11) showed a biofilm with a high number of living cells but also showed a larger amount of dead, red-stained cells (Fig. 2b).
FIG 2.
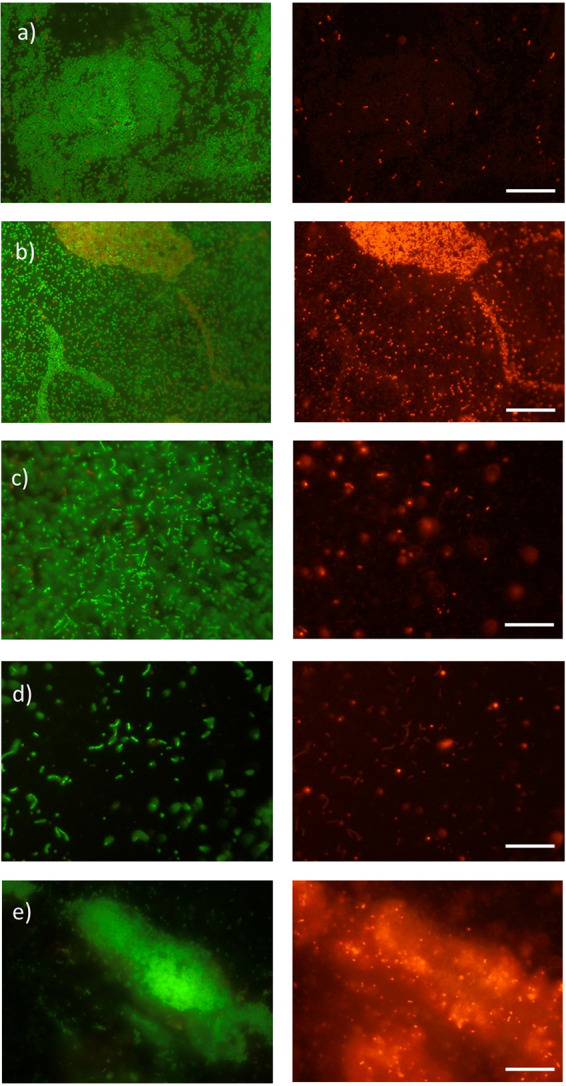
Images of the biofilms produced by L. monocytogenes 15G01 (wild type) (a) and selected transposon mutants with altered biofilm formation (34F11 [b], 33E11 [c], 39G5 [d], and 44D3 [e]) grown on polystyrene surfaces in MWB for 48 h at 30°C. The biofilms were stained with a Live/Dead BacLight bacterial viability kit according to the manufacturer’s instructions (Life Technologies, Thermo Fisher, New Zealand). Living cells were labeled with SYTO9 (green) and dead cells with propidium iodide (red). Scale bars, 20 μm. Mutants: 34F11, clsA mutant; 33E11, uvrB mutant; 39G5, mltD mutant; 44D3, mgtB mutant.
SEM was used to analyze further the structure of the biofilms produced by the mutants after growth of the bacteria on stainless-steel (SS) coupons coated with mussel juice for 7 days at 30°C. An SS coupon coated with mussel juice was used as a control, and organic debris was clearly visible (Fig. 3k and l). Bacterial cells attached and formed biofilms on the coupons preferably where organic debris of the mussel juice was present (Fig. 3i and j).
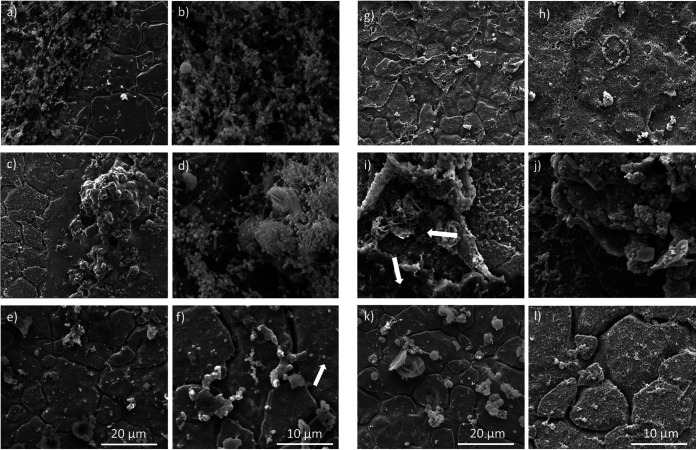
FIG 3.
Images of the coupon coated with mussel juice (k and l) and the biofilms produced by L. monocytogenes 15G01 (wild type) (a and b) and selected transposon mutants with altered biofilm formation (34F11, clsA mutant [c and d]; 33E11, uvrB mutant [e and f]; 39G5, mltD mutant [g and h]; 44D3, mgtB mutant [i and j]) grown on stainless steel coupons coated with mussel juice for 7 days at 30°C. The images were obtained with a scanning electron microscope at 5,000× and 10,000× magnifications. The cracks are features of the stainless-steel surface. The white arrows point at coccoid-shaped bacteria (f) and two different types of biofilm (i).
SEM images of 39G5 confirmed the long-chain phenotype with some elongated cells (Fig. 3g and h). Transposon mutant 33E11 was a low biofilm former and exhibited only sparse biofilm formation on SS coupons after 7 days, although extracellular matrix was present. 33E11 appeared to have a different cell morphology compared to the wild type, with coccoid-shaped rather than rod-shaped cells (Fig. 3e and f). Transposon mutant 34F11, identified as a high biofilm former in both the microtiter plate screening test and under the fluorescence microscope, also exhibited greater biofilm formation on the SS coupons compared to the wild type (Fig. 3c and d). In addition, 34F11 exhibited an extensive thread structured biofilm attached to organic mussel debris (Fig. 3d).
Confocal analysis reveals a unique sandwich structure for the biofilm of the mgtB mutant.
The influence of magnesium on the biofilm structure was investigated using confocal analysis. COMSTAT was used to calculate the biomass, roughness, and maximum and average thickness of the biofilms (Table 3).
TABLE 3.
Biomass, roughness coefficient, maximum thickness, and average thickness values for of biofilms formed by WT and mutants 39G5 (mltD mutant) and 44D3 (mgtB mutant) strainsa
| Parameter | Result (SD) |
|||||
|---|---|---|---|---|---|---|
| WT | WT-Mg2+ | 39G5 | 39G5-Mg2+ | 44D3 | 44D3-Mg2+ | |
| Biomass (μm3/μm2) | 1.145 (0.054) | 0.497 (0.409) | 1.813 (1.620) | 0.191 (0.043) | 0.841 (1.130) | 1.626 (0.950) |
| Roughness coefficient (Ra*) | 0.639 (0.074) | 1.515 (0.256) | 0.891 (0.750) | 1.787 (0.009) | 1.284 (0.874) | 0.654 (0.455) |
| Maximum thickness (μm) | 4.784 (1.452) | 4.951 (0.317) | 9.162 (3.767) | 4.280 (0.00) | 10.966 (6.764) | 5.077 (0.262) |
| Avg thickness (entire area) (μm) | 2.295 (0.776) | 0.840 (0.561) | 4.063 (2.399) | 0.320 (0.089) | 3.107 (4.807) | 2.398 (1.340) |
| Avg thickness (biomass) (μm) | 3.357 (1.377) | 3.281 (0.533) | 7.408 (2.243) | 3.026 (0.913) | 4.904 (4.283) | 3.238 (1.082) |
The biomass, roughness coefficient, maximum thickness, and average thickness of biofilms formed by the wild type (WT) and the mutants 39G5 (mltD mutant) and 44D3 (mgtB mutant) in MWB and MWB with a final concentration of 5 mM Mg2+ after incubation at 30°C for 7 days were calculated using COMSTAT. Standard deviations were calculated from three analyzed images taken of each sample.
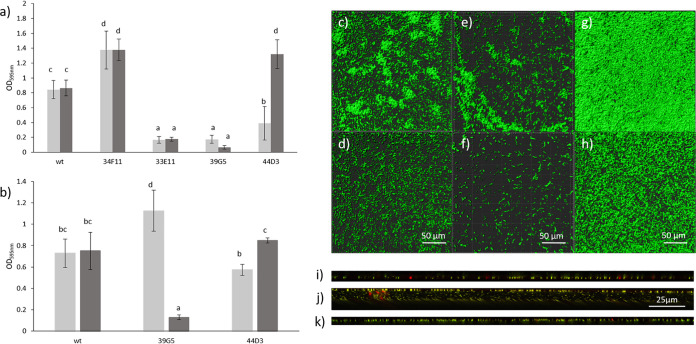
Isosurface images of the biofilms stained with SYTO 9 were generated with Imaris (Bitplane, Zurich, Switzerland). The wild-type control formed microcolonies on glass after 7 days of incubation in MWB at 30°C (Fig. 4g and h). In contrast to the CV assay (Fig. 4a and b), where magnesium addition did not alter biofilm amount significantly, biomass was reduced in the presence of 5 mM Mg2+ for the wild type when analyzed by confocal laser scanning microscopy (CLSM). However, the maximum thickness and the average thickness of the biomass remained the same when analyzed by CLSM. It is possible that magnesium led to increased production or stabilization of extracellular polymeric substance (which is not detected by CLSM) for the wild type. In addition, biofilms were formed on two different surfaces (polystyrene and glass), which could have also contributed to the observed differences.
FIG 4.
Biofilm formation by the wild type and selected mutants in MWB (light gray bars) and MWB with a Mg2+ concentration of 5 mM (dark gray bars) at 30°C (a) and 37°C (b) after 48 h of incubation measured with a CV assay. Error bars represent the SD of three independent experiments (n = 6). Letters in common indicate no significant difference. (c to h) Isosurface images of biofilms of wild-type (c), 39G5 (e), and 44D3 (g) strains formed on glass after 7 days of incubation in MWB (1.67 mM Mg2+) and in MWB with a final Mg2+ concentration of 5 mM (wild type [d], 39G5 [f], and 44D3 [h]). Biofilms were grown at 30°C (wild type, 44D3) or 37°C (39G5) and stained with SYTO9. Orthogonal view of biofilms formed on a glass surface after 7 days at 30°C by the wild type in MWB (i), by the mgtB mutant (44D3) in MWB (j), and by the mgtB mutant (44D3) in the presence of 5 mM Mg2+ (k). Images were taken after removal of media and staining with a Live/Dead BacLight kit with a confocal laser scanning microscope.
Since the differences in biofilm mass in the presence of 5 mM Mg2+ were greatest when the mltD mutant 39G5 was incubated at 37°C (Fig. 4b), this temperature was used for the CLSM analysis. The presence of magnesium led to a decrease in biofilm and attached cells for 39G5 after 7 days of incubation at 37°C (Fig. 4e and f). This was also observed with the CV assays after 48 h (Fig. 4a and b). The maximum thickness and average thickness were more than halved in the presence of 5 mM Mg2+ (46.71 and 40.84%, respectively, of the thicknesses without additional magnesium) (Table 3).
44D3 (mgtB mutant) produced more biofilm mass in the presence of 5 mM Mg2+ compared to 1.67 mM Mg2+ and less biomass than the wild type when grown in MWB (1.67 mM Mg2+) (Table 3), a finding in line with the observations made with the CV assay (Fig. 4a). Calculations with COMSTAT revealed a double maximum thickness for the biofilm of 44D3 compared to biofilms formed by the wild type and 44D3 in the presence of 5 mM Mg2+ (Table 3). This suggests that Mg2+ restored the wild-type phenotype for the mgtB mutant (44D3) (Fig. 4g to k; Table 3). Mutant 44D3 produced a unique sandwich structure for the biofilm (Fig. 4j) with monolayers of bacterial cells at the top and bottom and extrapolymeric substance (EPS) or fluid in between. This structure has, to the best of our knowledge, not been reported for a biofilm before. In the presence of 5 mM Mg2+, 44D3 produced a biofilm similar in structure to the wild type (Fig. 4i and k), further strengthening the hypothesis of restoration of the wild-type phenotype.
The mltD mutant (39G5) was defective in autolysis and motility, as well as in biofilm formation.
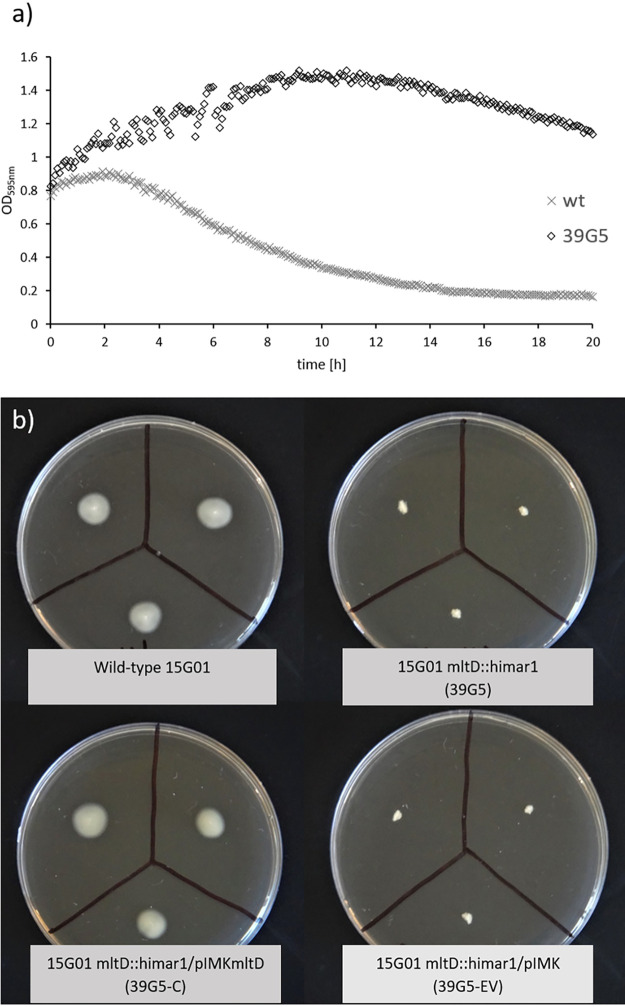
Inactivation of mltD (a murA homologue) has previously been shown to result in loss of motility in L. monocytogenes (27). Consistent with these findings, the mltD mutant created in this study exhibited no motility (Fig. 5b), which was restored upon gene complementation (Fig. 5b). The Triton X-100-induced autolysis rate was reduced for the mltD mutant compared to the wild type (Fig. 5a), confirming the direct involvement in autolysis.
FIG 5.
(a) Triton X-100-induced autolysis of L. monocytogenes 15G01 (wt) and the mltD mutant (39G5) determined by optical density measurement at 595 nm and (b) motility of L. monocytogenes 15G01 (top left), the mltD mutant (39G5 [top right]), and the mltD mutant containing a wild-type copy of the corresponding gene (39G5-C [bottom left]) or the empty vector pIMK (39G5-EV [bottom right]) after 24 h at 30°C.
Furthermore, the mltD mutant, 39G5, produced less biofilm than the wild type at 30°C but more than the wild type at 37°C. At both temperatures, the presence of magnesium (5 mM) reduced biofilm production to a minimum when measured with the CV assay and by CLSM. Attachment studies revealed that the presence of magnesium did not alter the attachment ability of 39G5, suggesting that magnesium influences the biofilm maturation process rather than surface characteristics or initial attachment (see Fig. S5 in the supplemental material).
Observations of both planktonic cells and biofilm cells in the present study using fluorescence microscopy (Fig. 2) and SEM (Fig. 3g and h) showed a high number of elongated cells in long chains for the mltD mutant.
The mltD mutant was found to produce increased cell aggregation, which interfered with optical density measurements for growth behavior. However, cell counts after 24 h growth were similar to the wild-type levels (>9.0 log10 CFU/ml) (see Table S1 in the supplemental material).
DISCUSSION
We identified here 27 genes that are involved in L. monocytogenes 15G01 biofilm formation. Four other research groups have also used Himar1 transposon mutagenesis to identify genes involved in biofilm formation by L. monocytogenes (12–15). In each case, a large number of loci were reportedly associated with biofilm formation. Many of the loci were only identified in a single study, however, probably because different isolates and different assay conditions were used. For example, Piercey et al. (15) performed the assays at 15°C, while other groups used higher temperatures (32, 35, or 37°C). We opted to carry out our analysis at 30 and 37°C, even though food processing plants operate at a lower temperature. These temperatures were chosen for this high-throughput screen to identify potential biofilm associated genes in a timely manner since lower temperatures require longer incubation times due to a lower growth rate.
All studies, including this one, showed that a transposon insertion in lafA (gene ID lmo2555 in L. monocytogenes EGDe), a gene encoding a glycosyltransferase required for membrane development, caused a reduction in biofilm formation (29). Other genes identified by us and other research groups included genes coding for sortase A, a peptidoglycan-linked protein (LPXTG), and the flagellum protein FlaA (13). The detection of common genes associated with changes in biofilm formation under such variable conditions suggests that they are critical to biofilm formation regardless of the environmental conditions the bacterium experiences.
This study revealed the probable involvement of uvrB in biofilm formation. Piercey et al. (15) identified uvrA (located in the same operon as uvrB), which was also associated with biofilm formation. Our research group, as well as Piercey et al. (15), used a serotype 1/2a strain isolated from a processing plant to generate the transposon library.
Ouyang et al. (14) and Alonso et al. (12) both generated a mutant library using L. monocytogenes 10403S, and Chang et al. (13) created a library of L. monocytogenes Scott A mutants. The uvrAB cluster was not identified as involved in biofilm formation in these other studies, suggesting either that the regulatory cascades controlling biofilm formation may be specific to certain environmental triggers or that they may be influenced by genetic variability between strains or serotypes.
Transporter systems are essential in living organisms. The transition from a planktonic to a sessile lifestyle requires changes in metabolism and energy generation as resources within biofilms become scarce (30). Thus, during the screening of the L. monocytogenes 15G01 mutants, it was not surprising to identify seven genes involved in these broad processes that influence biofilm formation.
One transporter system of particular interest is the P-type ATPase, which takes up Mg2+ upon ATP hydrolysis (MgtB). Previous studies have showed that magnesium deprivation triggers biofilm formation (31), and temperature-dependent expression (32) of mgtB is regulated by the PhoP/PhoQ two-component system in Gram-negative bacteria (33, 34). However, knowledge about the function of this P-type ATPase in Gram-positive bacteria, L. monocytogenes in particular, is limited. No data about the involvement of mgtB in biofilm formation are currently available. According to Nielsen et al. (35), mgtB (lmo2689) is regulated by the two-component system CesRK. CesR binding boxes were found upstream of this mgtB gene, which suggests direct control of mgtB. The mgtB gene is part of an operon including genes encoding cell division proteins (i.e., ftsW; see Fig. S3b) which are also regulated by CesR (35). CesRK is associated with virulence (36) and ethanol sensitivity and antibiotic sensitivity (37). The transposon insertion in the mgtB gene (44D3) resulted in the production of a low-biofilm phenotype in this study, suggesting the involvement of mgtB in biofilm formation by L. monocytogenes. The involvement of mgtB in biofilm formation has also been shown for Cronobacter sakazakii (38), wherein a disruption of mgtB led to a 77% reduction in biofilm mass. Other magnesium transporters (MgtE) have also been shown to be involved in biofilm formation or potentially even in virulence (39). Of particular interest in this study was the observed sandwich structure of the biofilm formed by 44D3 with static monolayers at the top and bottom and movement in the fluid in between (Fig. 4). Similarly, Guilbaud et al. (40) also observed movement in the fluid in hollow structures of the biofilm when analyzing it by CLSM. Magnesium has been shown to influence biofilm formation by other bacterial species (31, 41), where high magnesium concentrations (50 mM and higher) led to reduced biofilm formation by Bacillus subtilis and Bacillus cereus (41) without affecting the growth. However, lower concentrations of 5 and 10 mM led to an increase in biofilm formation by B. subtilis in the same study. Other studies showed that Mg2+ presence led to increased attachment of P. fluorescens cells to glass (42), but, on the other hand, Mg2+ limitation resulted in increased biofilm formation by Pseudomonas aeruginosa through repression of the retS gene which is responsible for EPS biosynthesis (31). Although the findings are contradictory, it is clear that magnesium plays a vital role in biofilm formation and should be the focus of further investigations.
Magnesium is not only important for bacterial homeostasis but has also been found to inhibit induced autolysis in E. coli upon its addition (43). One mutant with a disruption of mltD produced low levels of biofilm in this screen. The membrane-bound lytic murein transglycosylase D precursor (mltD) encodes a murein-degrading enzyme (autolysin) and belongs to the class of lytic transglycosylases which are important for cell division and insertion of proteins in the cell envelope and also for the maintenance of bacterial morphology (44). Lytic transglycosylase activity on cell turnover has previously been linked to increased biofilm formation by Gram-negative (45, 46) and Gram-positive (47) bacteria. Cells in a biofilm are protected from exposure to exogenous toxic substances by the surrounding extracellular polymeric substance matrix within a heterogeneous metabolic bacterial population. By altering the cell structure, adhesion to surfaces and the ability of matrix production and anchoring is changed (48). Lamers et al. (49) showed that mltD mutants produced about 70% less biofilm than the wild-type P. aeruginosa. Similar results were observed by Sailer et al. (46) for mltE mutants of E. coli. In our study, mtlD mutants could possibly have suffered from interference with components of the cell membrane, possibly affecting the surface attachment, which is important for the initial steps of biofilm formation. However, we did not investigate the signaling pathway. It has been shown that a double mutation in two lytic transglycolase genes, mltE and mltC, was specifically linked to the regulation of biofilm formation by affecting the expression of the key biofilm gene regulator CsgD (45) in Salmonella enterica serovar Typhimurium. Another possible mechanism was proposed by Artola-Recolons et al. (50). The model involves the maturation of the surrounding peptidoglycan, via lytic transglycosylase, for the proper anchoring and functionality of the flagellar motor, which is required to allow successful colonization of the gastric mucosa by H. pylori.
In line with a previous finding (51), the mltD mutant produced elongated cells when assessed microscopically. The mltD mutant (39G5) was one of the few in the screen that exhibited no motility, which is in agreement with other studies (26, 27). Two other low biofilm formers with insertions in the flaA gene and a gene encoding an unknown protein (44F5) were also motility deficient (see Fig. S6), which might account for their low biofilm production. The successful complementation of the mltD mutant confirmed the gene’s direct involvement in biofilm formation and motility. The reduction in motility and production of long chains with elongated cells might impair the ability of the mutant to move freely and to attach to surfaces, thus resulting in biofilm reduction. The observed lower autolysis rate after exposure of 39G5 to Triton X-100 was also seen in a previous study (51), further confirming the involvement of the mltD gene in autolysis.
Interestingly, the two genes influenced by magnesium in this study (mgtB and mltD) are both situated in very close proximity on the genome only 1,282 bp apart (see Fig. S3b). The coding region for mltD is situated on the positive strand and for mgtB on the negative strand. A gene encoding a transcriptional regulator of the TetR family is situated between these two genes. mltD is suggested to be regulated by this gene (52), and TetR is known to be influenced by magnesium (53–55). The mgtB mutant (44D3) showed a low-biofilm phenotype, which was reversed to the wild-type level upon magnesium addition, whereas the mltD mutant (39G5) showed a further reduction in biofilm mass upon magnesium addition. This and the close proximity of the genes suggest a potential common regulatory mechanism, possibly through TetR, although different regulatory systems for the two genes have been suggested (35, 52); however, this needs to be further investigated.
This screen also identified a number of mutants with defects in cell wall and membrane functions that showed changes in biofilm formation. Of particular note, a transposon insertion in the clsA gene (34F11), encoding the cardiolipin synthetase, led to enhanced biofilm formation in the present study. Cardiolipin synthetase catalyzes the formation of cardiolipin from phosphatidylglycerol and is predominantly active in stationary phase (56). Previous research showed that gene disruption of clsA resulted in decreased biofilm formation by other Gram-negative and Gram-positive species (23–25), which indicates that clsA might be differentially regulated in different species, possibly due to differences in membrane composition. The hypothesis of differential regulation in multiple species is supported by studies which showed that changes in the environmental conditions, such as osmotic stress or desiccation, led to activation of clsA in E. coli and Staphylococcus aureus (57, 58), but butanol stress induced the downregulation of clsA in B. subtilis (59). By screening a mutant library of L. monocytogenes for desiccation survival, Hingston et al. (16) found that a transposon insertion in the gene clsA resulted in decreased desiccation survival compared to the wild type in L. monocytogenes.
Changes in environmental conditions not only trigger activation of genes involved in membrane composition to protect cells from damage but also trigger other stress response mechanisms. Many studies have shown that stress response is somehow linked to biofilm formation (11, 60–65). One well-described system is the SOS response which is induced upon replication fork stalling caused by DNA damage through reactive oxygen species (ROS) (66). UvrB is part of an enzyme complex that mediates excision and incision steps of DNA repair, and its expression is induced as part of the SOS response (67). In the present study, a gene disruption of uvrB resulted in a low-biofilm phenotype, although this phenotype could only be partially complemented. Partial complementation of uvrB may have occurred because this gene is part of an operon with uvrA, which may also have been affected by the transposon insertion (see Fig. S3b). The SOS response has been linked to biofilm formation by several bacterial species, including L. monocytogenes, Pseudomonas aeruginosa, and Streptococcus mutans (66–68). Gene expression of uvrB and stress response-associated genes was found to be upregulated in planktonic cells after heat exposure (69). Microscopic analysis of 33E11 showed changes in phenotype and produced coccoid-shaped bacteria (Fig. 3f). The formation of coccoid-shaped bacteria has been reported previously for Listeria cells after exposure to stresses such as starvation due to change from log growth to long-term survival (70, 71). Tremoulet et al. (72) found that the bacterial cells of 7-day-old biofilms of L. monocytogenes were more coccoid-shaped than rod-shaped, which agrees with our findings. The changes in phenotype might be due to maturation of the biofilm since these researchers did not observe this phenotype for biofilm grown for 24 h.
Although complementation was unsuccessful for the clsA and mgtB mutants in this study, their repeated identification in the screen, the different locations of the transposon insertions in these mutants, and in one instance the different orientation of the transposon (clsA) (Table 1) provided substantial evidence for their involvement in biofilm formation.
In addition, both mgtB mutants (30H2 and 44D3; Table 1) identified in this screen behaved similarly in the biofilm formation assay, in growth studies (see Fig. S1 and Table S1) and in motility tests (see Fig. S6). The two clsA mutants (30A9 and 34F11) produced the same amount of biofilm and were also similar in growth behavior; however, they differed in motility. The higher motility for one of the clsA mutants (30A9) could be due to insertion locations producing a partially functional gene (30A9 [insertion after bp 1133] as opposed to bp 729 in 34F11). However, this will require further investigation.
Furthermore, the membrane protein cardiolipin is predominantly found at the cell poles of rod-shaped bacteria (73), and the lack of cardiolipin might affect incorporation or attachment of specific proteins, such as flagella, into the cell poles, resulting in decreased motility. A previous study found that the swimming motility of Rhodobacter sphaeroides was not affected by cardiolipin deficiency (23). However, in contrast to L. monocytogenes, which has four to six peritrichous flagella, R. sphaeroides has just one single flagellum and is usually not situated at the cell pole but medially on the cell body (74). This strengthens the evidence for the involvement of clsA in motility and perhaps indicates this only applies to peritrichous flagella.
The inability to restore the wild-type phenotypes in the mgtB and clsA mutants may have resulted from differential expression of the genes upon site-specific integration of the pIMK vector. This failure may also have been because of polar effects on the expression of downstream genes upon insertion of the transposon (see Fig. S3b). Certainly, the orientation of the transposon in the mgtB mutant suggests that the transposon could influence expression of downstream genes, such as ftsW, which are part of a cell division operon. However, together with literature linking these genes to biofilm formation in other studies (23, 38), their repeated identification provided strong evidence that they are somehow involved in biofilm formation by L. monocytogenes 15G01. A recent publication also found an interesting link between cardiolipin and MgtA (which belongs to the same transporter class as MgtB) in E. coli: both MgtA and cardiolipin were found together in the bacterial membrane. Subramani et al. (75) suggested that the head group of cardiolipin contributes to MgtA activation by possibly acting as a chaperone for MgtA. Whether a similar link is present in Gram-positive bacteria will need further investigation.
To conclude, two genes, clsA and mgtB, were identified as involved in biofilm formation. Both have not, to the best of our knowledge, been previously associated with biofilm formation by L. monocytogenes. The stress responsive gene uvrB is clearly part of an operon involved in biofilm formation, strengthening the link between biofilm formation and stress response. Confocal analysis revealed a unique biofilm structure for the mgtB mutant, which was reversed upon magnesium addition. Further studies analyzing gene regulation are required to assess the exact involvement of the biofilm-associated genes. Ultimately, this level of understanding could then help devise specific intervention technologies that reduce the tendency of these damaging foodborne pathogens to form such persistent biofilms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type strain used in this study was L. monocytogenes 15G01, a representative of the persistent pulsotype 5132 obtained from a New Zealand seafood-processing facility during an extensive sampling program (18). L. monocytogenes 15G01 was kept as glycerol stocks in a −80°C freezer and recovered in a three-step process by (i) growth in tryptic soy broth (TSB) enriched with 0.6% yeast extract (TSBYE; Difco, BD) overnight at 37°C, (ii) plating on tryptic soy agar enriched with 0.6% yeast extract (TSAYE; Difco, BD), and (iii) subculture and storage on Columbian sheep blood agar (Fort Richard, New Zealand) at 4°C. A library of 6,500 mutants of L. monocytogenes 15G01, created by The New Zealand Plant and Food Research Limited using the Himar1 mariner-based transposition system (22) according to a method described previously (76), was kept on 96-well master plates in glycerol at −80°C, subcultured twice before use, and stored on TSAYE plates supplemented with erythromycin (Duchefa, Biochemie, The Netherlands) at a final concentration of 5 ppm. Media and agar plates were supplemented with erythromycin at a final concentration of 5 ppm for the studies with the mutants (the transposon contains the erythromycin resistance gene) and with additional kanamycin (MP Biomedicals, Illkirch, France) at a concentration of 50 ppm for the complemented strains. The optical density measurements for the growth studies were taken at a wavelength of 595 or 600 nm using a microplate reader (Multiskan EX; Thermo Fisher) or an automated microplate reader (SPECTROstar Omega, BMG Labtech).
Biofilm formation assay.
A biofilm formation assay was performed according to the method described by Djordjevic et al. (77) with some modifications. Briefly, overnight cultures were grown at 37°C in TSBYE in a sterile 96-well plate (polystyrene, U-bottom; Interlab, New Zealand) and transferred to new 96-well plates with a 96-well replicator, each well containing 200 μl of MWB (Himedia, India). The cultures were incubated for 48 h at 30°C and then washed three times with 200 μl of double-distilled H2O (ddH2O) using a microplate strip washer (ELx50; BioTek). After air-drying at ambient temperature for 30 min, 150 μl of a 1% aqueous crystal violet (CV) solution was added to the plates. After 45 min of incubation at 30°C, the CV solution was removed, and the cultures were washed six times with 150 μl of ddH2O. After drying for 30 min at 30°C, 150 μl of 96% ethanol was added to each well to destain the CV-stained cells. The optical density was measured after 1 h at 595 nm with a microplate reader (Multiskan EX, Thermo Fisher). The OD595 values obtained were corrected by subtracting the OD595 value of sterile media. To screen the library of transposon mutants, mutants were stored in 96-well master-plates and subcultured twice before tests for biofilm formation. Biofilm formation for each mutant was measured three times and compared to the wild-type strain. Statistical analysis (two sample t test; P ≤ 0.05) was performed to select mutants of interest. The OD595 values of the mutants selected for further analysis were at least two SD above or below that of the wild-type strain. To eliminate variability caused by growth deficiency, biofilm formation and turbidity measurements were repeated for 120 selected mutants in two independent experiments with eight replicates each. Turbidity was measured at 595 nm with a microplate reader before the washing and staining process. The growth of the mutants was compared to the growth of the wild type in MWB to confirm that the altered biofilm formation observed in the initial screen was not due to differences in the ability of the mutants to grow in this media (see Table S1 and Fig. S1).
To assess the influence of magnesium on biofilm formation, the CV assay was carried out in MWB with a final Mg2+ concentration of 1.67 or 5 mM, respectively. Biofilm formation was measured at 30 and 37°C for 48 h.
Identification of transposon insertion sites in selected mutants.
To locate the transposon insertion sites in the genomes of the mutants of interest, a nested arbitrary PCR was performed using one transposon specific primer and one arbitrary primer (Table 4) to amplify the regions flanking the transposon from the right and the left end. The PCR was performed in two steps with the Mastercycler gradient (Eppendorf, Germany). BioMix Red (Bioline, UK) was used as the mastermix in the first round and the second round was run using AccuPrime Hifi Taq polymerase (Invitrogen, USA) as described previously (22). The annealing temperature was adjusted for each mutant, if necessary, to minimize nonspecific annealing or to increase annealing. A final concentration of 3 mM Mg2+ was used for all PCRs. A portion (5 μl) of each amplified product was visualized on a 1.5% agarose gel (settings, 100 V for 30 min) using Redsafe (Intron Biotechnology, South Korea) under UV light. The PCR products obtained were subsequently purified and sequenced by Macrogen, Ltd. (South Korea) and analyzed using the NCBI BLAST program, version 2.2.30 (available from https://www.ncbi.nlm.nih.gov), and the Geneious 7 program (78). The reference strain L. monocytogenes EGD (accession HG421741) (79) was used to identify the coordinates at the point of the transposon insertion site and the orientation of the transposon in the chromosome. This reference strain was chosen since it is the same serotype as the L. monocytogenes 15G01 strain (1/2a).
TABLE 4.
Primers used in this study
| Usage and primer | Primer nucleotide sequence (5′–3′)a | Product size (bp) | Source or reference |
|---|---|---|---|
| Complementation of 33E11 | |||
| uvrB_Fwd | CAACTGCAGCCTTCAATTAAATCCACATCTGGT (PstI) | 2,528 | This study |
| uvrB_Rev | AACGGATCCTGTGCTTGCAACGTATATGCT (BamHI) | This study | |
| Complementation of 39G5 | |||
| mltD_Fwd | CAACTGCAGTTGACGTAGAAACACCTTAGCAC (PstI) | 2,683 | This study |
| mltD_ Rev | AACGGATCCAAAGGCAATTTCGGTGCGAC (BamHI) | This study | |
| Identification of L. monocytogenes | |||
| 16S_Fwd | CAGCAGCCGCGGTAATAC | 938 | 87 |
| 16S_Rev | CTCCATAAAGGTGACCCT | 87 | |
| Arbitrary PCR | |||
| Marq254 | CGTGGAATACGGGTTTGCTAAAAG | 76 | |
| Marq255 | CAGTACAATCTGCTCTGATGCCGCATAGTT | 76 | |
| Marq206 | TGTCAGACATATGGGCACACGAAAAACAAGT | 76 | |
| Marq207 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGTAAT | 76 | |
| Marq208 | GGCCACGCGTCGACTAGTAC | 76 | |
| Marq257 | CTTACAGACAAGCTGTGACCGTCT | 76 | |
| Marq270 | TGTGAAATACCGCACAGATGCGAAGGGCGA | 76 | |
| Marq271 | GGGAATCATTTGAAGGTTGGTACT | 76 | |
| Presence of vector pIMK | |||
| T7promoter | TAATACGACTCACTATAGGG | Macrogen, Inc. (South Korea) |
Italics represent restriction enzyme sites; the restriction enzymes used are indicated in parentheses. N = A, C, G, or T.
Complementation of selected mutants.
The site-specific integrative vector pIMK (80) was used for the genetic complementation of selected mutants. The vector pIMK is a derivative of pPL2 (1 kb smaller) and facilitates the insertion at the tRNAArg locus (81). Genetic complementation constructs using this plasmid were constructed by amplifying the target genes from wild-type 15G01 using gene-specific primers in PCR (Table 4). The gene-specific PCR products and the pIMK vector were digested with PstI and BamHI and ligated to one another using the LigaFast rapid DNA ligation system (Promega), following the manufacturer’s instructions, in a molar ratio 3:1 (vector to insert). A 1-μl portion of the recombinant plasmid was then introduced into chemically competent E. coli S17 cells by heat shock. Transformants were selected on Luria-Bertani (LB) agar plates supplemented with kanamycin. A colony was picked from the agar plate with a sterile toothpick and dipped in the PCR mix. Colony PCR was performed with one gene-specific primer and T7 (T7 binding site present in the pIMK plasmid) to confirm successful transformation. Recombinant plasmids were extracted from colony PCR-positive cultures, and the gene inserts were sequenced to confirm the authenticity of the constructs. Authentic transformants were then used for the genetic complementation of L. monocytogenes 15G01 mutants. Conjugation was performed according to Azizoglu et al. (82) with some modifications. Single colonies of the donor (E. coli transformants containing the construct pIMK:gene or a control containing only the pIMK vector) were resuspended overnight in LB broth containing kanamycin and incubated at 30°C at 100 rpm to an OD595 of ∼0.55. At the same time, a colony of the recipient (L. monocytogenes 15G01 transposon mutant) was resuspended in brain-heart infusion (BHI; Difco, BD) medium and incubated overnight at 37°C with shaking. The donor culture (3 ml) and a prewarmed (45°C; 10 min) recipient culture (1.5 ml) were mixed and centrifuged at 2,050 × g for 8 min; the bacterial pellet was then washed with 10 ml of BHI broth and centrifuged again under the same conditions. After a washing step, the pellet was resuspended in 500 μl of fresh BHI broth, deposited in the center of a BHI agar plate, and incubated overnight at 37°C. The drop was then resuspended in 2 ml of BHI broth, and a 100-μl aliquot was spread plated on BHI agar plates containing kanamycin and nalidixic acid (Fort Richard, New Zealand) (20 μg/ml). L. monocytogenes strains are naturally resistant against nalidixic acid, and therefore it was used for counterselection. The plates were incubated at 30°C for 2 to 3 days. The authenticity of transconjugants was confirmed by colony PCR using the corresponding gene-specific primers. Their identity as L. monocytogenes was also confirmed using 16S-rRNA specific primers for L. monocytogenes (Table 4). To confirm that the empty vector had no effect on the phenotype, the parent plasmid was transformed in each mutant, as well as in the wild-type strain 15G01.
Microscopic analysis.
Fluorescence microscopy, scanning electron microscopy (SEM), and confocal laser scanning microscopy were used to visualize biofilm formation by bacterial strains on polystyrene, stainless steel, and glass, respectively. For visualization under fluorescence, a fluorescent Live/Dead BacLight bacterial viability kit (Life Technologies, Thermo Fisher, New Zealand) was used to label living cells green (SYTO9, membrane-permeable stain) and dead cells red (propidium iodide, a non-membrane-permeable stain). Six-well plates (tissue treated; Greiner, Germany) were filled with 2.97 ml of MWB, inoculated with 30 μl of an overnight bacterial culture grown in TSBYE at 37°C, and incubated at 30°C for 48 h. The biofilms were then washed twice with 0.8% NaCl to remove loosely attached cells and stained with 1 ml of fluorescent stain prepared according to the manufacturer’s manual. A fluorescence microscope (Olympus, BX51 fitted with the XC30 digital camera) was used at 10 × 100 magnification to take images.
SEM was performed on stainless steel coupons (5 × 5 mm, food grade) as described previously (83). The coupons were coated with cooked mussel juice (CMJ) produced as described previously (19), but with some modifications. Briefly, Greenshell mussels obtained from the local supermarket were stored for 24 h at 10°C in a fridge and then boiled in a wok closed with a lid without the addition of water. When all mussels opened and released the intervalvular juice, the liquid was collected and autoclaved at 121°C for 15 min. The coupons were pretreated with alkali detergent for 2 h at 45°C, rinsed with ddH2O, and autoclaved in deionized water. The coupons were coated by immersion in CMJ in a 6-well plate (tissue treated, Greiner, Germany) for 4 h at 60°C (or until dried). The coupons were then placed into a fresh 6-well plate containing MWB (2.97 ml) and inoculated with 30 μl of an overnight culture of the bacterium (107.5 to 108 CFU). After incubation for 7 days at 30°C, phosphate-buffered saline (PBS; pH 7.2) was used to remove loosely attached cells on the coupons. After a rinse with 100 mM cacodylate buffer (pH 7.2; Acros Organics, NJ), the coupons were fixed overnight at 4°C in 2% glutaraldehyde (Acros Organics, NJ) and 0.1% ruthenium red solution (Acros Organics) in 100 mM cacodylate buffer. The next morning, coupons were rinsed to remove unbound dye and then dehydrated in serial dilutions of ethanol for 10 min each (30, 50, 60, 70, and 90% [vol/vol]) with three final 10-min rinses in absolute ethanol. The coupons were then critical point dried (BalTec CPD030; BalTec AG, Balzers, Liechtenstein) and sputter coated with gold (Leica EM ACE200; Leica Microscopy Systems Ltd., Heerbrugg, Switzerland) for visualization using a scanning electron microscope (FEI Quanta 250 SEM; Fei Company, Hillsboro, OR).
For confocal laser scanning microscopy (CLSM) analysis, the biofilms were grown on glass-bottom dishes (35-mm petri dish, 10-mm Microwell no. 0 coverglass; MatTek Corporation). First, single colonies picked from an agar plate (TSAYE) were used to inoculate TSBYE and then incubated overnight at 37°C. The glass-bottom dishes were filled with 2.97 ml of MWB (Mg2+ concentration 1.67 or 5 mM) and inoculated with 30 μl of the overnight culture. After 7 days of incubation at 30°C (or 37°C for 39G5), the medium was carefully removed, and the biofilm on the plates washed twice with 0.8% NaCl solution. The fluorescent Live/Dead BacLight bacterial viability kit was used to stain the biofilms according to the manufacturer’s instructions. Three images (246.03 × 246.03 μm) per sample were taken with a Leica DM6000B scanning confocal microscope running LAS AF software version 2.7.3.9723. Excitation and emission levels were as follows: (i) SYTO9 stain, excitation at 488 nm (argon laser) and emission collection at 498 to 550 nm, and (ii) propidium iodide stain, excitation at 561 nm (DPSS 561 laser) and emission collection at 571 to 700 nm.
Images were analyzed using ImageJ software and/or Imaris (Bitplane, Zurich). COMSTAT (www.comstat.dk) was used to calculate the biomass, roughness, and maximum and average thicknesses of the biofilms (84, 85).
Motility assay.
A motility assay was performed according to the method of Knudsen et al. (86). Briefly, semisolid agar plates (TSB plus 0.25% agar [Difco, BD]) were inoculated with L. monocytogenes 15G01 or the mutant (39G5) using a sterile pick and then incubated at 30 or 37°C for 48 h. The diameter of the halo formed around the colony was then measured and compared to the halo surrounding the wild-type strain (15G01). Three independent experiments were performed with each treatment repeated in triplicate.
Autolysis assay.
The assay was performed according to Huang et al. (10) with minor modifications. Briefly, single colonies of the wild type and the selected mutant (39G5) were picked from the TSAYE plate and grown in BHI at 37°C overnight. OD595 was measured in a microplate reader (SPECTROstar Omega; BMG Labtech) and adjusted to 0.6 ± 0.05 for each culture. Each culture (1.5 ml) was transferred to 2-ml microtubes and centrifuged at 4°C at 4,500 × g for 10 min. The supernatant was discarded, and the cell pellet washed twice with ice-cold ddH2O and then resuspended in the same volume of Tris-HCl (pH 7.2) containing 0.05% Triton X-100. Solutions with cells were incubated at 30°C in a 96-well plate, and the OD595 was measured for 20 h in 5-min intervals using an automated microplate reader (SPECTROstar Omega).
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request (sandra.visnovsky@plantandfood.co.nz).
Supplementary Material
ACKNOWLEDGMENTS
We thank Erik Rikkerink and Falk Kalamorz for valuable feedback on the manuscript, Duncan Hedderley for help with the statistical analysis, and Ian Hallett and Paul Sutherland for help with the SEM.
This research was funded through The New Zealand Ministry of Business, Innovation, and Employment (MBIE) funding under contract CAWX1801.
The authors declare that there are no conflicts of interest and that the research does not involve human participants and/or animals.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rocourt J, Jacquet C, Reilly A. 2000. Epidemiology of human listeriosis and seafoods. Int J Food Microbiol 62:197–209. 10.1016/S0168-1605(00)00336-6. [DOI] [PubMed] [Google Scholar]
- 2.Lecuit M. 2005. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin Microbiol Infect 11:430–436. 10.1111/j.1469-0691.2005.01146.x. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher G, Rogers M, Wong R. 1994. Survey of Listeria monocytogenes in New Zealand seafood. J Aquat Food Prod 3:13–24. 10.1300/J030v03n02_03. [DOI] [Google Scholar]
- 4.Gudbjörnsdóttir B, Suihko ML, Gustavsson P, Thorkelsson G, Salo S, Sjöberg AM, Niclasen O, Bredholt S. 2004. The incidence of Listeria monocytogenes in meat, poultry and seafood plants in the Nordic countries. Food Microbiol 21:217–225. 10.1016/S0740-0020(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 5.Srey S, Jahid IK, Ha S-D. 2013. Biofilm formation in food industries: a food safety concern. Food Control 31:572–585. 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]
- 6.Lou Y, Yousef AE. 1999. Characteristics of Listeria monocytogenes important to food processors, p 131–224. In Ryser ET, Marth EH (ed), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, New York, NY. [Google Scholar]
- 7.Carpentier B, Cerf O. 2011. Review: persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Cruz CD, Fletcher GC. 2012. Assessing manufacturers’ recommended concentrations of commercial sanitizers on inactivation of Listeria monocytogenes. Food Control 26:194–199. 10.1016/j.foodcont.2012.01.041. [DOI] [Google Scholar]
- 9.Van Houdt R, Michiels CW. 2010. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol 109:1117–1131. 10.1111/j.1365-2672.2010.04756.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, Shi C, Yu S, Li K, Shi X. 2012. A putative MerR family regulator involved in biofilm formation in Listeria monocytogenes 4b G. Foodborne Pathog Dis 9:767–772. 10.1089/fpd.2011.1101. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Suo Y, Shi C, Szlavik J, Shi XM, Knochel S. 2013. Mutations in gltB and gltC reduce oxidative stress tolerance and biofilm formation in Listeria monocytogenes 4b G. Int J Food Microbiol 163:223–230. 10.1016/j.ijfoodmicro.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Alonso AN, Perry KJ, Regeimbal JM, Regan PM, Higgins DE. 2014. Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS One 9:e113696. 10.1371/journal.pone.0113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y, Gu W, Fischer N, McLandsborough L. 2012. Identification of genes involved in Listeria monocytogenes biofilm formation by mariner-based transposon mutagenesis. Appl Microbiol Biotechnol 93:2051–2062. 10.1007/s00253-011-3719-z. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang Y, Li J, Dong Y, Blakely LV, Cao M. 2012. Genome-wide screening of genes required for Listeria monocytogenes biofilm formation. J Biotech Res 4:13–25. [Google Scholar]
- 15.Piercey MJ, Hingston PA, Truelstrup Hansen L. 2016. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15°C. Int J Food Microbiol 223:63–74. 10.1016/j.ijfoodmicro.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Hingston PA, Piercey MJ, Truelstrup Hansen L. 2015. Genes associated with desiccation and osmotic stress in Listeria monocytogenes as revealed by insertional mutagenesis. Appl Environ Microbiol 81:5350–5362. 10.1128/AEM.01134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins B, Joyce S, Hill C, Cotter PD, Ross RP. 2010. TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob Agents Chemother 54:4658–4663. 10.1128/AAC.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz CD, Fletcher GC. 2011. Prevalence and biofilm-forming ability of Listeria monocytogenes in New Zealand mussel (Perna canaliculus) processing plants. Food Microbiol 28:1387–1393. 10.1016/j.fm.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Nowak J, Cruz CD, Palmer J, Fletcher GC, Flint S. 2015. Biofilm formation of the L. monocytogenes strain 15G01 is influenced by changes in environmental conditions. J Microbiol Methods 119:189–195. 10.1016/j.mimet.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Nowak J, Cruz CD, Tempelaars M, Abee T, van Vliet AHM, Fletcher GC, Hedderley D, Palmer J, Flint S. 2017. Persistent Listeria monocytogenes strains isolated from mussel production facilities form more biofilm but are not linked to specific genetic markers. Int J Food Microbiol 256:45–53. 10.1016/j.ijfoodmicro.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Cruz CD, Pitman AR, Harrow SA, Fletcher GC. 2014. Listeria monocytogenes associated with New Zealand seafood production and clinical cases: unique sequence types, truncated InlA, and attenuated invasiveness. Appl Environ Microbiol 80:1489–1497. 10.1128/AEM.03305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak J, Visnovsky S, Cruz CD, Fletcher G, van Vliet AHM, Hedderley D, Butler R, Flint S, Palmer J, Pitman A. 2021. Inactivation of the gene encoding the cationic antimicrobial peptide resistance factor MprF increases biofilm formation but reduces invasiveness of Listeria monocytogenes. J Appl Microbiol 130:464–467. 10.1111/jam.14790. [DOI] [PubMed] [Google Scholar]
- 23.Lin TY, Santos TM, Kontur WS, Donohue TJ, Weibel DB. 2015. A cardiolipin-deficient mutant of Rhodobacter sphaeroides has an altered cell shape and is impaired in biofilm formation. J Bacteriol 197:3446–3455. 10.1128/JB.00420-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz-Elias EJ, Marcano J, Camilli A. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun 76:5049–5061. 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puttamreddy S, Cornick NA, Minion FC. 2010. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect Immun 78:2377–2384. 10.1128/IAI.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machata S, Hain T, Rohde M, Chakraborty T. 2005. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol 187:8385–8394. 10.1128/JB.187.24.8385-8394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machata S. 2008. Molecular investigations of peptidoglycan-binding proteins in Listeria monocytogenes. PhD thesis. Justus-Liebig-Universitaet Giessen, Giessen, Germany. [Google Scholar]
- 28.Bao Y, Zhang X, Jiang Q, Xue T, Sun B. 2015. Pfs promotes autolysis-dependent release of eDNA and biofilm formation in Staphylococcus aureus. Med Microbiol Immunol 204:215–226. 10.1007/s00430-014-0357-y. [DOI] [PubMed] [Google Scholar]
- 29.Webb AJ, Karatsa-Dodgson M, Grundling A. 2009. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol 74:299–314. 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek MR, Fuqua C. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J Bacteriol 186:4427–4440. 10.1128/JB.186.14.4427-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulcahy H, Lewenza S. 2011. Magnesium limitation is an environmental trigger of the Pseudomonas aeruginosa biofilm lifestyle. PLoS One 6:e23307. 10.1371/journal.pone.0023307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehres DG, Maguire ME. 2003. The unusual nature of magnesium transporters, p 347–360. In Microbial transport systems. Wiley-VCH Verlag GmbH & Co., Berlin, Germany. [Google Scholar]
- 33.Chamnongpol S, Groisman EA. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol Microbiol 44:561–571. 10.1046/j.1365-2958.2002.02917.x. [DOI] [PubMed] [Google Scholar]
- 34.Groisman EA, Cromie MJ, Shi Y, Latifi T. 2006. A Mg2+-responding RNA that controls the expression of a Mg2+ transporter. Cold Spring Harbor Symp Quant Biol 71:251–258. 10.1101/sqb.2006.71.005. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen PK, Andersen AZ, Mols M, van der Veen S, Abee T, Kallipolitis BH. 2012. Genome-wide transcriptional profiling of the cell envelope stress response and the role of LisRK and CesRK in Listeria monocytogenes. Microbiology (Reading) 158:963–974. 10.1099/mic.0.055467-0. [DOI] [PubMed] [Google Scholar]
- 36.Kallipolitis BH, Ingmer H, Gahan CG, Hill C, Søgaard-Andersen L. 2003. CesRK, a two-component signal transduction system in Listeria monocytogenes, responds to the presence of cell wall-acting antibiotics and affects β-lactam resistance. Antimicrob Agents Chemother 47:3421–3429. 10.1128/AAC.47.11.3421-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottschalk S, Bygebjerg-Hove I, Bonde M, Nielsen PK, Nguyen TH, Gravesen A, Kallipolitis BH. 2008. The two-component system CesRK controls the transcriptional induction of cell envelope-related genes in Listeria monocytogenes in response to cell wall-acting antibiotics. J Bacteriol 190:4772–4776. 10.1128/JB.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartmann I, Carranza P, Lehner A, Stephan R, Eberl L, Riedel K. 2010. Genes involved in Cronobacter sakazakii biofilm formation. Appl Environ Microbiol 76:2251–2261. 10.1128/AEM.00930-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coffey BM, Akhand SS, Anderson GG. 2014. MgtE is a dual-function protein in Pseudomonas aeruginosa. Microbiology (Reading) 160:1200–1213. 10.1099/mic.0.075275-0. [DOI] [PubMed] [Google Scholar]
- 40.Guilbaud M, Piveteau P, Desvaux M, Brisse S, Briandet R. 2015. Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl Environ Microbiol 81:1813–1819. 10.1128/AEM.03173-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oknin H, Steinberg D, Shemesh M. 2015. Magnesium ions mitigate biofilm formation of Bacillus species via downregulation of matrix genes expression. Front Microbiol 6:907. 10.3389/fmicb.2015.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song B, Leff LG. 2006. Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol Res 161:355–361. 10.1016/j.micres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Leduc M, Kasra R, van Heijenoort J. 1982. Induction and control of the autolytic system of Escherichia coli. J Bacteriol 152:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheurwater E, Reid CW, Clarke AJ. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol 40:586–591. 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Monteiro C, Fang X, Ahmad I, Gomelsky M, Römling U. 2011. Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J Bacteriol 193:6443–6451. 10.1128/JB.00425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sailer FC, Meberg BM, Young KD. 2003. β-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol Lett 226:245–249. 10.1016/S0378-1097(03)00616-5. [DOI] [PubMed] [Google Scholar]
- 47.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J, Elasri MO, Shaw LN. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology (Reading) 157:2206–2219. 10.1099/mic.0.049692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucher T, Oppenheimer-Shaanan Y, Savidor A, Bloom-Ackermann Z, Kolodkin A, Gal I. 2015. Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ Microbiol Rep 7:990–1004. 10.1111/1758-2229.12346. [DOI] [PubMed] [Google Scholar]
- 49.Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RN, Burrows LL. 2015. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. Microbiologyopen 4:879–895. 10.1002/mbo3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Artola-Recolons C, Lee M, Bernardo-García N, Blázquez B, Hesek D, Bartual SG, Mahasenan KV, Lastochkin E, Pi H, Boggess B, Meindl K, Usón I, Fisher JF, Mobashery S, Hermoso JA. 2014. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem Biol 9:2058–2066. 10.1021/cb500439c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carroll SA, Hain T, Technow U, Darji A, Pashalidis P, Joseph SW, Chakraborty T. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J Bacteriol 185:6801–6808. 10.1128/JB.185.23.6801-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74:1323–1338. 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leypold CF, Marian DT, Roman C, Schneider S, Schubert P, Scholz O, Hillen W, Clark T, Lanig H. 2004. How does Mg2+ affect the binding of anhydrotetracycline in the TetR protein? Photochem Photobiol Sci 3:109–119. 10.1039/b303431n. [DOI] [PubMed] [Google Scholar]
- 54.Scholz O, Schubert P, Kintrup M, Hillen W. 2000. Tet repressor induction without Mg2+. Biochem 39:10914–10920. 10.1021/bi001018p. [DOI] [PubMed] [Google Scholar]
- 55.Werten S, Dalm D, Palm GJ, Grimm CC, Hinrichs W. 2014. Tetracycline repressor allostery does not depend on divalent metal recognition. Biochemistry 53:7990–7998. 10.1021/bi5012805. [DOI] [PubMed] [Google Scholar]
- 56.Koprivnjak T, Zhang D, Ernst CM, Peschel A, Nauseef WM, Weiss JP. 2011. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol 193:4134–4142. 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maudsdotter L, Imai S, Ohniwa RL, Saito S, Morikawa K. 2015. Staphylococcus aureus dry stress survivors have a heritable fitness advantage in subsequent dry exposure. Microbes Infect 17:456–461. 10.1016/j.micinf.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Romantsov T, Guan Z, Wood JM. 2009. Cardiolipin and the osmotic stress responses of bacteria. Biochim Biophys Acta 1788:2092–2100. 10.1016/j.bbamem.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinayavekhin N, Mahipant G, Vangnai AS, Sangvanich P. 2015. Untargeted metabolomics analysis revealed changes in the composition of glycerolipids and phospholipids in Bacillus subtilis under 1-butanol stress. Appl Microbiol Biotechnol 99:5971–5983. 10.1007/s00253-015-6692-0. [DOI] [PubMed] [Google Scholar]
- 60.Suo Y, Huang Y, Liu Y, Shi C, Shi X. 2012. The expression of superoxide dismutase (SOD) and a putative ABC transporter permease is inversely correlated during biofilm formation in Listeria monocytogenes 4b G. PLoS One 7:e48467. 10.1371/journal.pone.0048467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, Andrew PW, Roberts IS. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol 184:621–628. 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Veen S, Abee T. 2010. Dependence of continuous-flow biofilm formation by Listeria monocytogenes EGD-e on SOS response factor YneA. Appl Environ Microbiol 76:1992–1995. 10.1128/AEM.02680-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Veen S, Abee T. 2010. HrcA and DnaK are important for static and continuous-flow biofilm formation and disinfectant resistance in Listeria monocytogenes. Microbiology (Reading) 156:3782–3790. 10.1099/mic.0.043000-0. [DOI] [PubMed] [Google Scholar]
- 64.van der Veen S, Abee T. 2010. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl Environ Microbiol 76:7854–7860. 10.1128/AEM.01519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Veen S, Abee T. 2011. Generation of variants in Listeria monocytogenes continuous-flow biofilms is dependent on radical-induced DNA damage and RecA-mediated repair. PLoS One 6:e28590. 10.1371/journal.pone.0028590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Veen S, Abee T. 2011. Bacterial SOS response: a food safety perspective. Curr Opin Biotechnol 22:136–142. 10.1016/j.copbio.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 67.van der Veen S, van Schalkwijk S, Molenaar D, de Vos WM, Abee T, Wells-Bennik MH. 2010. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology (Reading) 156:374–384. 10.1099/mic.0.035196-0. [DOI] [PubMed] [Google Scholar]
- 68.Inagaki S, Matsumoto-Nakano M, Fujita K, Nagayama K, Funao J, Ooshima T. 2009. Effects of recombinase A deficiency on biofilm formation by Streptococcus mutans. Oral Microbiol Immunol 24:104–108. 10.1111/j.1399-302X.2008.00480.x. [DOI] [PubMed] [Google Scholar]
- 69.van der Veen S, Hain T, Wouters JA, Hossain H, de Vos WM, Abee T, Chakraborty T, Wells-Bennik MH. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology (Reading) 153:3593–3607. 10.1099/mic.0.2007/006361-0. [DOI] [PubMed] [Google Scholar]
- 70.Doijad SP, Barbuddhe SB, Garg S, Poharkar KV, Kalorey DR, Kurkure NV, Rawool DB, Chakraborty T. 2015. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS One 10:e0137046. 10.1371/journal.pone.0137046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen J, Anantheswaran RC, Knabel SJ. 2009. Changes in barotolerance, thermotolerance, and cellular morphology throughout the life cycle of Listeria monocytogenes. Appl Environ Microbiol 75:1581–1588. 10.1128/AEM.01942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tremoulet F, Duche O, Namane A, Martinie B, Labadie JC, European Listeria Genome Consortium. 2002. Comparison of protein patterns of Listeria monocytogenes grown in biofilm or in planktonic mode by proteomic analysis. FEMS Microbiol Lett 210:25–31. 10.1016/S0378-1097(02)00571-2. [DOI] [PubMed] [Google Scholar]
- 73.Dworkin J. 2009. Cellular polarity in prokaryotic organisms. Cold Spring Harb Perspect Biol 1:a003368. 10.1101/cshperspect.a003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol 169:514–518. 10.1128/JB.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramani S, Perdreau-Dahl H, Morth JP. 2016. The magnesium transporter A is activated by cardiolipin and is highly sensitive to free magnesium in vitro. Elife 5:e11407. 10.7554/eLife.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao M, Bitar AP, Marquis H. 2007. A mariner-based transposition system for Listeria monocytogenes. Appl Environ Microbiol 73:2758–2761. 10.1128/AEM.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68:2950–2958. 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kuhbacher A, Brisse S, Pucciarelli MG, Garcia-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerda J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5:e00969-14. 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184:4177–4186. 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Azizoglu RO, Elhanafi D, Kathariou S. 2014. Mutant construction and integration vector-mediated gene complementation in Listeria monocytogenes. Methods Mol Biol 1157:201–211. 10.1007/978-1-4939-0703-8_17. [DOI] [PubMed] [Google Scholar]
- 83.Borucki MK, Peppin JD, White D, Loge F, Call DR. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69:7336–7342. 10.1128/AEM.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiol (Reading) 146:2395–2407. 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 85.Vorregaard M. 2008. Comstat2: a modern 3D image analysis environment for biofilms. Technical University of Denmark, Copenhagen, Denmark. [Google Scholar]
- 86.Knudsen GM, Olsen JE, Dons L. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett 240:171–179. 10.1016/j.femsle.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 87.Park S, Jung J, Choi S, Oh Y, Lee J, Chae H, Ryu S, Jung H, Park G, Choi S. 2012. Molecular characterization of Listeria monocytogenes based on the PFGE and RAPD in Korea. Adv Microbiol 2:26214. 10.4236/aim.2012.24079. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request (sandra.visnovsky@plantandfood.co.nz).