Abstract
During neural development, stem and precursor cells can divide either symmetrically or asymmetrically. The transition between symmetric and asymmetric cell divisions is a major determinant of precursor cell expansion and neural differentiation, but the underlying mechanisms that regulate this transition are not well understood. Here, we identify the Sonic hedgehog (Shh) pathway as a critical determinant regulating the mode of division of cerebellar granule cell precursors (GCPs). Using partial gain and loss of function mutations within the Shh pathway, we show that pathway activation determines spindle orientation of GCPs, and that mitotic spindle orientation correlates with the mode of division. Mechanistically, we show that the phosphatase Eya1 is essential for implementing Shh-dependent GCP spindle orientation. We identify atypical protein kinase C (aPKC) as a direct target of Eya1 activity and show that Eya1 dephosphorylates a critical Threonine (T410) in the activation loop. Thus, Eya1 inactivates aPKC, resulting in reduced phosphorylation of Numb and other components that regulate the mode of division. This Eya1-dependent cascade is critical in linking spindle orientation, cell cycle exit and terminal differentiation. Together these findings demonstrate that a Shh-Eya1 regulatory axis selectively promotes symmetric cell divisions during cerebellar development by coordinating spindle orientation and cell fate determinants.
Keywords: Phosphatase, Eya1, Sonic hedgehog, cerebellum, granule cell precursors, mode of division
Introduction
The morphogenic factor Hedgehog (Hh) was initially discovered in Drosophila based on its role in segment polarity [1]. Hh has three vertebrate counterparts, and the closest homolog, Shh, functions as a key developmental regulator by acting as both a morphogen and mitogen in multiple tissues including the central nervous system [2, 3]. Shh has a distinctive role in development of the cerebellum, where it acts as a critical mitogen for granule cell precursors (GCPs), the dividing cells that give rise to cerebellar granule cells [4]. It is now well appreciated that dysregulation of the Shh pathway in GCPs results in one of the most common malignant brain tumors of children, the Shh-subtype medulloblastoma [5].
The cerebellum originates from two distinct progenitor regions, the rostral rhombic lip and the ventricular zone surrounding the IVth ventricle [6]. Early GCPs emerge from the upper rhombic lip during embryonic development and subsequently migrate rostrally along the surface of the cerebellar anlage to form the external granule layer (EGL) [7, 8]. Proliferation of GCPs in the EGL begins toward the end of embryonic development, and continues into the postnatal period, a time period extending from E16 to P21 in mice. The extensive and prolonged proliferation of GCPs is primarily regulated by Shh, which is produced by nearby Purkinje cells and acts directly on GCPs. Beginning at P1, and continuing to P21, GCPs migrate from the EGL to the internal granule cell layer and differentiate into mature granule cells, which are the most prevalent neurons in the brain.
Shh functions as a strong mitogen for GCPs as well as for neocortical progenitors [9]. Moreover, in the developing cortex, Shh signaling can also affect the mode of division. Data from in vitro experiments suggest that Shh signaling promotes symmetric divisions of cerebral cortical progenitors [10], and may also affect the mode of division in motor neuron precursors in the spinal cord [11, 12]. However, it is not yet known how Shh achieves such selectivity in proliferative response.
Changes in phosphorylation state are critical for most growth factor signaling pathways, including Shh signaling [13–15]. Thus kinases including Protein kinase A (PKA), aPKC, as well as phosphatases such as protein phosphatase 2A (PP2A) and Eya1 function as critical regulators of Shh signaling [16–19]. The Eya1 phosphatase is a member of a small gene family originally described in Drosophila, where Eya has a key role in retina determination [20]. The mammalian homologues (Eya1 – 4) [21–23] play critical roles during development as highlighted by the fact that heterozygous loss of function mutations in EYA1, or its partner SIX1, are associated with the congenital disorder branchio-oto-renal syndrome, with ear, kidney and craniofacial defects [24, 25]. In addition, overexpression of Eya family members has been observed in multiple cancers including medulloblastoma [26], breast cancer [27], and glioblastoma [28].
All mammalian Eya proteins contain a highly conserved C-terminal Eya domain, which interacts with Six transcription factors or other members of the retinal determination network, and a less conserved N-terminal Eya domain [29]. Structurally, Eya1–4 belong to the superfamily of haloacid dehalogenases (HAD), which encompasses several different enzyme classes including dehalogenases, ATPases and magnesium-dependent phosphatases [30]. Surprisingly, Eya family members are reported to have dual protein Tyrosine (Tyr) as well as Serine(Ser)/Threonine(Thr) phosphatase activity [31–34]. However, this view has been challenged by a study suggesting that Ser/Thr phosphatase activity of Eya3 might not be intrinsic, but rather arises from association with the PP2A [35]. Moreover, since Eya acts as both a co-transcriptional activator and a phosphatase, it is not clear whether the intrinsic phosphatase activity is required for its actions [36, 37, 27].
In previous studies we found that Eya1 phosphatase activity is essential to promote Shh signaling [19]. While several other studies also suggest that phosphatase activity contributes to Eya-dependent transcriptional output [38], data from the Drosophila system suggest that phosphatase activity of the Eya family members may be dispensable for some of its actions [39–41]. Thus far, four physiological substrates of Eya phosphatase activity have been identified: the histone H2A variant H2AX [42, 43], the estrogen receptor ERβ [44], the proto-oncogene Myc [45] and Notch1 [46], and additional potential substrates have been suggested [47, 48, 34]. It is not clear, which, if any, of these substrates might be important for Shh signaling.
In this study, we address novel roles of Shh signaling and Eya1 activity in regulating GCP development. We show that Shh signaling regulates spindle orientation of mitotic GCPs during cerebellar development and that spindle orientation directly correlates with the mode of division. We further show that Eya1 is required for the effects of Shh signaling on GCP spindle orientation. We find that aPKC is a direct substrate of Eya1, and that de-phosphorylation of aPKC ζ by Eya1 leads to hypo-phosphorylation of the cell fate determinant Numb, a well-known target of aPKC ζ. In this way, Eya1 enables Shh signaling to drive symmetric cell division of cerebellar GCPs during early postnatal development. Together these data provide mechanistic insight into Shh pathway regulation of symmetric cell division and the rapid expansion of the developing GCP population.
Results
Shh regulates spindle orientation of proliferating GCPs
Previous studies suggest that the plane of cell division for GCPs may be developmentally regulated [49, 50], with a progressive increase of mitotic figures with the plane of division oriented horizontally to the pial surface over the course of postnatal development. However, a more recent study instead suggested that vertical divisions are predominant at late postnatal stages of GCP development [51]. Therefore, we first determined the plane of dividing GCPs during the peak of proliferation (postnatal day 1 (P1) to P10) in the murine cerebellum by visualizing anaphase cells using antibodies to phospho-Histone H3 (Fig. S1A). During postnatal development, there is a gradual increase of the proportion of GCP cell divisions with a horizontal mitotic plane (i.e. parallel to the pial surface) (Fig. S1B). Further analyses showed that mitotic orientation did not correlate with the distance from the pial surface, ruling out the possibility that the plane of cleavage might merely be regulated by the actual depth of the dividing cell within the outer external granule cell layer (Fig. S1C). These data demonstrate that GCPs transition from vertical to horizontal divisions during the period of early postnatal development when Shh regulates proliferation [4].
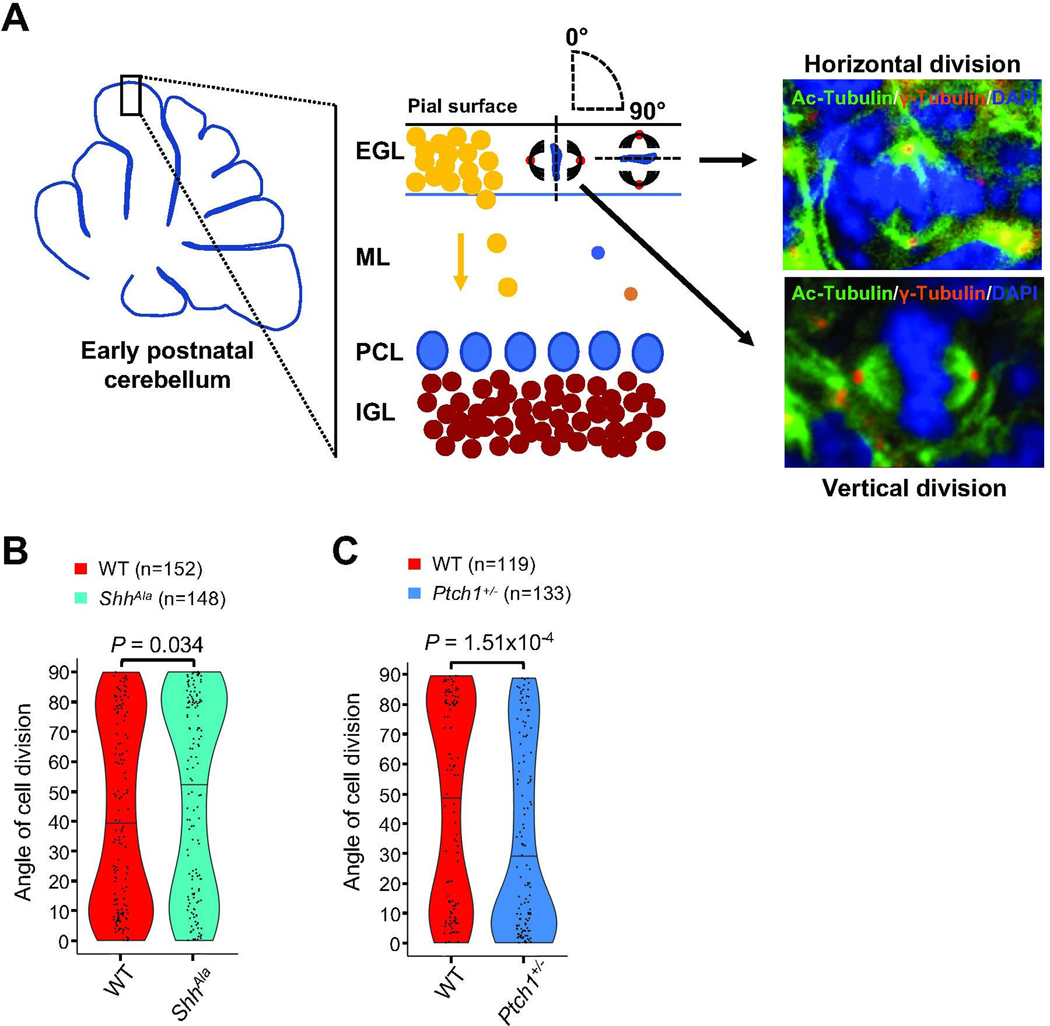
Recent data suggest that Shh signaling can directly promote symmetric divisions of cortical neuroepithelial cells [12]. While there is some evidence from pharmacologic experiments that Shh signaling also affects spindle orientation of GCPs [50, 51], these studies provide conflicting data as to whether Shh activation drives vertical or horizontal spindle orientation of GCPs. We therefore carefully assessed spindle orientation of GCPs in mouse models in which Shh signaling is either reduced (ShhAla) or enhanced (Ptch1+/−). To do so, we used a genetic mouse model expressing a mutant form of Shh (ShhAla) with impaired proteoglycan interactions, which exhibits a reduced Shh output and impaired GCP proliferation [52], and the heterozygous knockout Ptch1+/− mice with enhanced Shh activity and greater proliferation of the GCP population compared to control mice (Fig. S2) [53]. We used antibodies for γ-tubulin and acetylated α-tubulin (Fig. 1A; Movie 1; Table S1), enabling us to determine the angle of cell division of mitotic GCPs relative to the pial surface that lies above the EGL (Fig. S3). The hypomorphic mutation in ShhAla mice alters mitotic spindle orientation, with a decrease in vertical and an increase in horizontal cell divisions of GCPs at P3 (Fig. 1B). In striking contrast to this partial loss of function phenotype, we find that Shh pathway gain of function Ptch1+/− mice show a significant increase in vertical cell divisions in the EGL (Fig. 1C). Together, these gain and loss of function studies suggest that the Shh pathway regulates the orientation of the plane of division of postnatal GCPs in the EGL, in addition to its ability to promote proliferation of these cells [4].
Fig. 1. Shh signaling regulates spindle orientation of mitotic GCPs.
(A) Schematic illustrating the cerebellar cortex and main cell types during early postnatal development. Angles of division for vertically (angle of 0°) and horizontally (angle of 90°) dividing GCPs in the EGL (external granule cell layer) are depicted in the schematic, and representative immunostainings at P3 from sagittal sections in the vermis visualizing both centrioles (γ-Tubulin) and spindles (Ac-Tubulin) of mitotic GCPs in anaphase with the plane of division oriented vertically and horizontally with respect to the pial surface are shown. ML: molecular layer; PCL: Purkinje cell layer; IGL: internal granule cell layer. (B) Violin plots illustrating the distribution of angles of division of mitotic GCPs in the EGL of ShhAla mice at P3 as compared to wild type (WT) mice (n = 5 for each condition, Mann-Whitney test). (C) Violin plots illustrating the distribution of angles of division of mitotic GCPs in Ptch1+/− mice at P3 as compared to WT mice (n = 5 for each condition, Mann-Whitney test).
Spindle orientation correlates with GCP cell fate
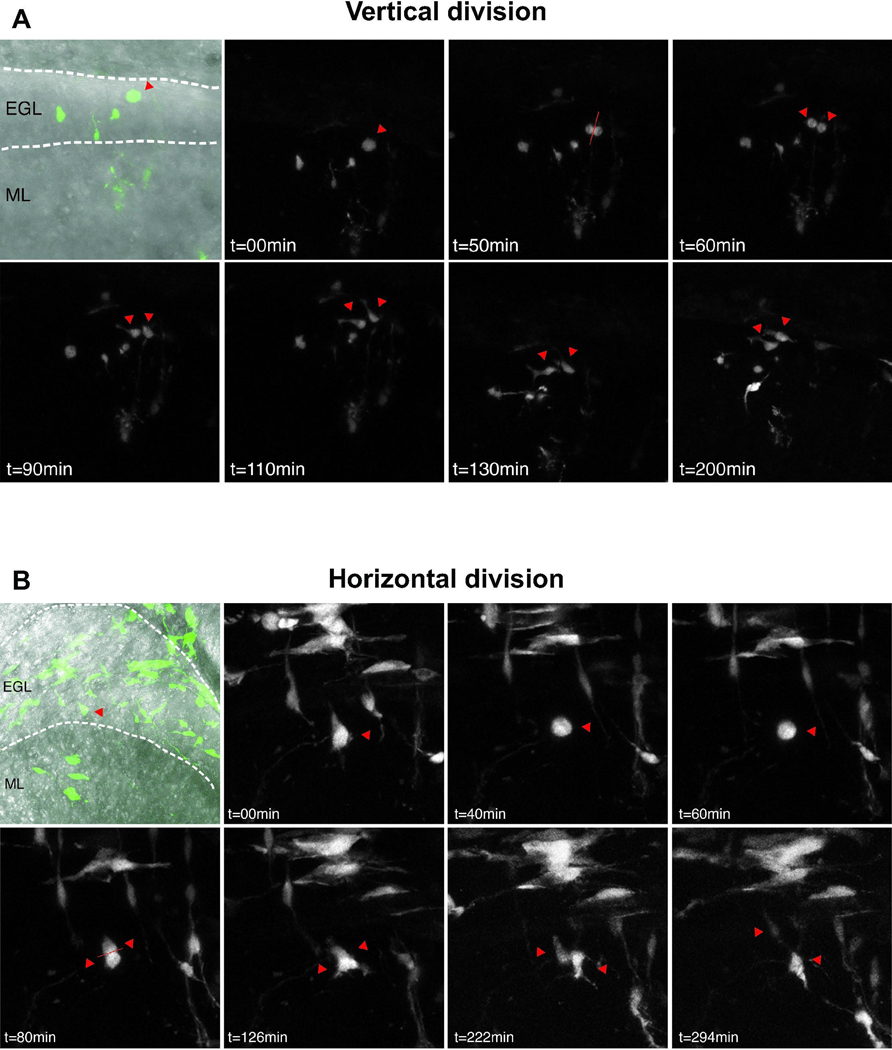
While GCPs uniformly give rise to mature granule cell neurons, the progenitors undergo migration and terminal differentiation over a prolonged time period. Thus, in the early postnatal period, some GCPs migrate from the EGL to the inner granule cell layer (IGL) and differentiate, while other GCPs re-enter the cell cycle, providing two distinct fates for sister cells after each mitotic division. To determine whether mitotic orientation correlates with symmetrical or asymmetrical cell fates of sister cells, we carried out real time analysis of sparsely labeled GCPs in organotypic slice cultures. In an initial experiment, we crossed Atoh1-creER mice that carry an inducible form of the Cre recombinase under control of the Math1 enhancer [7] to a floxed-GFP Cre reporter line. We injected those mice with a low dose of tamoxifen to selectively label a subset of developing GCPs, and prepared organotypic brain slices to monitor individual dividing cells and their corresponding daughter cells over time (Fig. S4A, B). We followed 57 GFP-positive, dividing GCPs from P3 and 31 from P6. For all vertical cell divisions, we find that both daughter cells (34/34 at P3, 10/10 at P6) remain in the EGL (Fig. S4C). In contrast, in many horizontal divisions (6/12 at P3 and 5/18 at P6), one of the daughter cells remains in the EGL while the other one migrates into the IGL, suggesting that the orientation of division might correlate with distinct modes of cell division. In order to more clearly resolve the plane of progenitor cell divisions and the morphology of daughter cells we sought to image GCPs at higher spatial and temporal resolution. To do so we injected a GFP-expressing retrovirus into the EGL of P1 and P4 mice; 48 h later, at P3 and P6, we generated and imaged organotypic cerebellar slices. The proportion of GFP labeled cells in the IGL with the characteristic bipolar morphology of migrating postmitotic granule neurons [54] in P6 slices (54/110; 49%) was increased compared to P3 (25/101; 25%), suggesting a progressive increase in neurogenic divisions by GCPs that were infected at the later postnatal developmental stage (Fig. S4D, E). Consistent with our previous data showing an increase in vertical divisions at P3 compared to P6 (Fig. S4C), we observed a trend of increased vertical divisions at P3 (15/25; 60%) compared to P6 (3/10; 30%). Vertical divisions gave rise to daughter cells that stayed in the EGL and maintained short irregular processes, consistent with GCP morphology (Fig. 2A; Movie 2) [54]. In contrast, the daughter cells of horizontal divisions differed from one another. As shown, one daughter remains in the EGL and maintains the morphology of a GCP, while the other daughter cell adopts newborn granule neuron morphology, including a bipolar shape and a thin trailing process, and migrates into the IGL (Fig 2B; Movies 3 and 4). Taken together our data suggest that vertical GCP divisions correlate with symmetrical fates, while horizontal divisions are associated with asymmetrical fates producing a GCP and one postmitotic neuron.
Fig. 2. Live imaging of retroviral GFP labeled GCPs.
Representative images of individual, GFP-labeled GCPs imaged every 10 minutes for up to 24 hours. At P3, 25 divisions were captured out of 101 cells imaged from 4 slices; at P6, 10 divisions were captured out of 110 cells imaged from 5 slices. Single red arrowheads indicate cells before mitosis and double arrowheads indicate position of daughter cells. Dotted lines indicate boundaries of the EGL and IGL. (A) In vertical divisions, both daughter cells remain in the EGL and maintain GCP morphology with short processes. (B) During horizontal divisions, one daughter cell remains in the EGL while the other one migrates into the IGL and exhibits bipolar migratory neuron morphology.
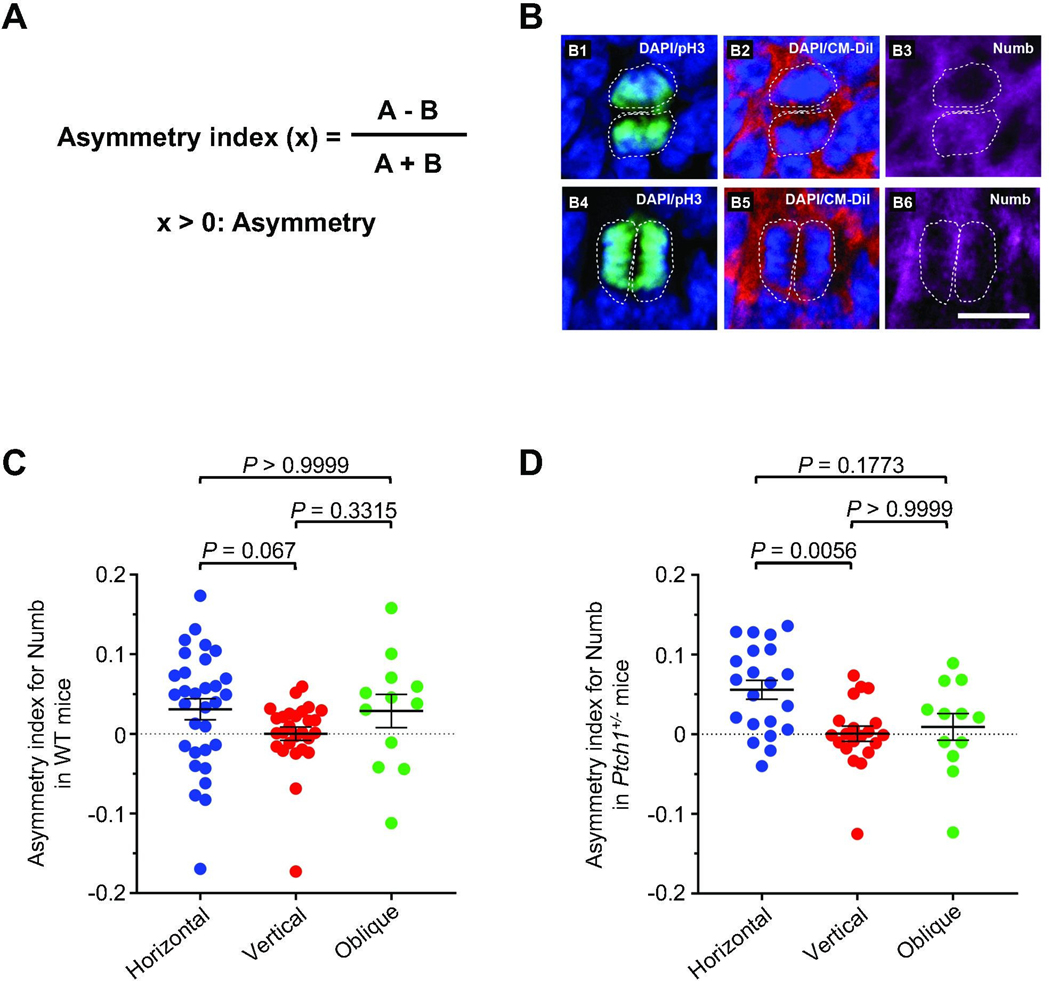
While our data argue for a mechanism that links spindle orientation and mode of division of GCPs, our study does not demonstrate whether daughter cells undergoing horizontal division are distinct from one another at the time of mitosis, or acquire different fates after cell division is complete, perhaps due to changes in the microenvironment [55, 56]. We therefore tested the possibility that unequal distribution of the cell fate determinant Numb might be implicated in the spindle orientation and cell fate of GCPs [57, 58]. We developed a quantitative asymmetry index to analyze the distribution of Numb in dividing, anaphase GCPs in which the mitotic orientation was either vertical, horizontal, or oblique with respect to the pial surface (Fig. 3A, B). While Numb is distributed equally in cells with the plane of division vertical to the pial surface, it is instead distributed unequally in cells with the plane of division horizontal to the pial surface (Fig. 3C). Moreover, Numb expression is higher in the daughter cell further away from the pial surface, consistent with the possibility that Numb facilitates migration and differentiation of these daughter cells. Cells dividing with an oblique orientation exhibit an intermediate asymmetry index. Of note, although vertical divisions are more common in the GCPs from P3 Ptch1+/− than in wild type mice, cells dividing with a horizontal plane exhibit an increased asymmetry index compared to the cells dividing with a vertical plane in both mutant and wild type mice (Fig. 3D). Taken together, these data indicate that mitotic spindle orientation directly correlates with the mode of division of proliferating GCPs, through a mechanism that might involve unequal distribution of the cell fate determinant Numb.
Fig. 3. Spindle orientation of dividing GCPs correlates with the distribution of symmetry markers.
(A) Formula for generating the asymmetry index. A and B, respectively refer to quantified level of immunofluorescent signal of Numb immunochemistry signal from the daughter cell further from, or closer to the pia. (B) Representative immunofluorescent staining for Numb (pink) and phospho-histone H3 (green). CM-DiI (red) and DAPI (blue) were used to demarcate nuclei and outer membrane of individual GCPs in sagittal cerebellar sections from wild type mice at P3 in GCPs dividing horizontally (B1-B3) or vertically (B4-B6) to the pial surface. White dashed lines represent the cell boundaries visualized using CM-Dil. (C, D) Quantitative analyses for the distribution of Numb in GCPs from WT mice (C) and Ptch1+/− mice (D) with vertical (0–30°), horizontal (60–90°), or oblique (30–60°) cell divisions with respect to the pial surface. P=0.0563 for WT mice, P=0.0065 for Ptch1+/− mice, determined using a Kruskal-Wallis test with Dunn`s post hoc test for multiple comparisons. Scale bars, 10 μm.
Eya1 is necessary for Shh-dependent regulation of GCP spindle orientation
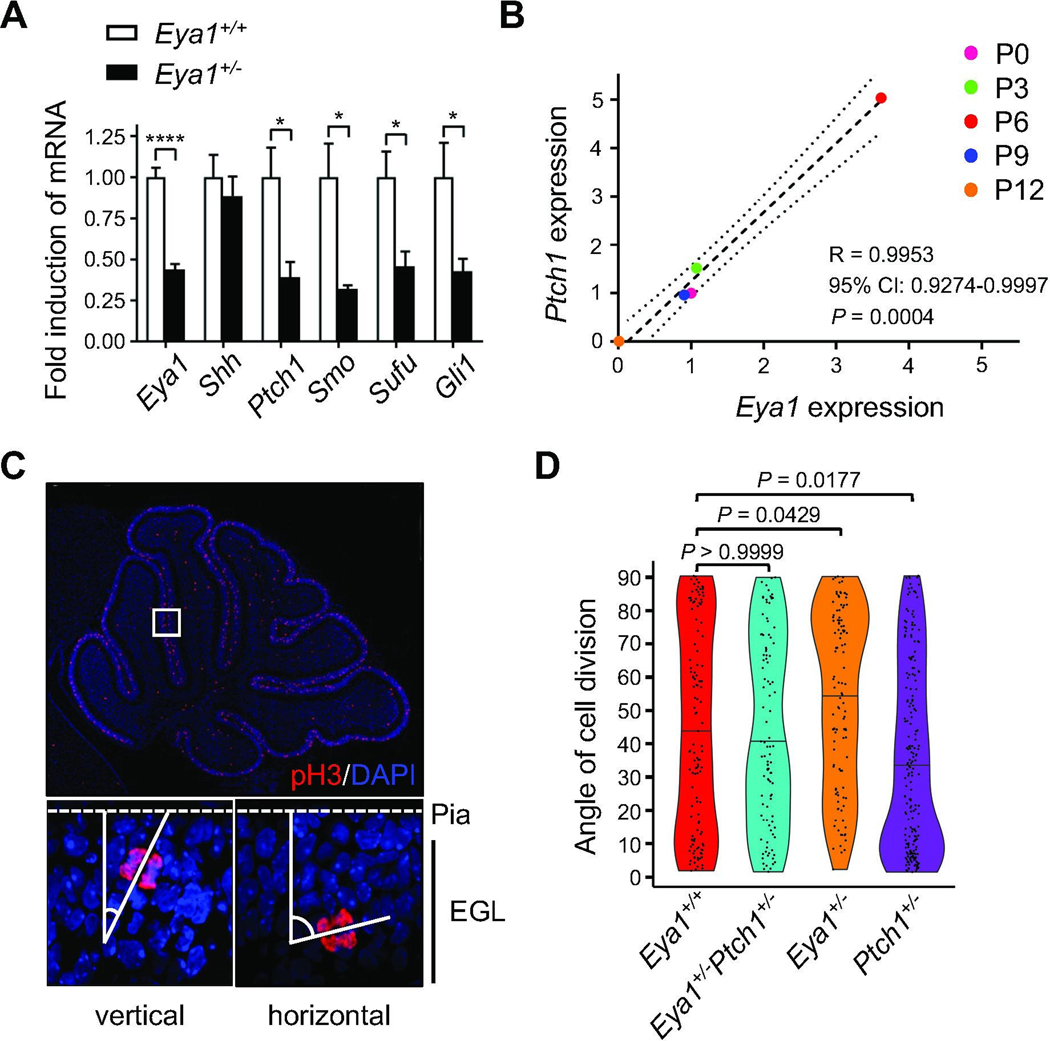
We have previously described the phosphatase Eya1 as a positive regulator of Shh signaling during cerebellar development [19]. Consistent with this role, we find that heterozygous loss of Eya1 in GCPs [59] reduced expression of known Shh pathway members and downstream targets, whereas expression of the Shh ligand itself was not affected (Fig. 4A). Strikingly, expression of Eya1 closely correlates with expression of the major Shh target Ptch1 over the course of cerebellar development (Fig. 4B), with peak levels of expression at postnatal day 6. Together these data indicate that Eya1 regulates the Shh transduction process rather than ligand expression. To determine whether Eya1 enables Shh-dependent orientation of mitotic GCPs, we examined spindle orientation in Eya1+/− mice. We visualized proliferating, anaphase GCPs in the EGL using antibodies against phospho-Histone H3 and assessed the angle of division relative to the pial surface (Fig. 4C). The dividing GCPs in the heterozygotes exhibited a significantly lower proportion of vertical divisions and higher proportion of horizontal divisions as compared to wild type controls (Fig. 4D). Thus Eya1+/− mice phenocopy the changes in GCP spindle orientation observed in hypomorphic ShhAla mice (Fig. 1B). Consistent with our previous data examining the spindle directly, Ptch1+/− mice with a Shh-signaling gain of function phenotype exhibit increased vertical GCP cell divisions compared to wild type animals due to enhanced Shh pathway activation. To identify genetic interactions between the Eya1 and Shh pathway mutations, we generated mice that are both heterozygous for Eya1 and Ptch1. Loss of one copy of Eya1 in the Eya1+/Ptch1+/− mice abrogated the increase of vertical divisions observed in Ptch1+/− mice, and restored the wild type proportions of vertical and horizontal divisions. This genetic interaction suggests that Eya1 is essential for the ability of Shh signaling to promote vertical, symmetric cell divisions of GCPs.
Fig. 4. Eya1 contributes to Shh-dependent regulation of spindle orientation.
(A) Quantitative real time PCR analyses for Eya1 and Shh pathway members in P6 cerebellar lysates from wild type controls and mice carrying a heterozygous knockout of Eya1 (5 technical replicates each, unpaired t test). (B) Dot plot illustrating the correlation of expression of Eya1 and Ptch1 over the course of early development (3 technical replicates each). Pearson correlation was computed. Dashed line illustrates result from nonlinear regression, dotted lines show the 95% confidence interval. (C) Immunostainings for phospho-Histone H3 (pH3) of cerebellar sections from P6 wild type mice and representative images for vertically and horizontally dividing GCPs in the EGL. (D) Distribution of angles of division of mitotic GCPs in the EGL of Eya1+/+ (n=126), Eya1+/−Ptch1+/− (n=112), Eya1+/− (n=111), and Ptch1+/− mice (n=211) at P6 (n = 5 for each genotype, Kruskal-Wallis test with Dunn`s post hoc test for multiple comparisons). All data are mean±s.e.m. *P<0.05, ****P<0.0001.
Eya1 dephosphorylates atypical protein kinase C and regulates Numb phosphorylation
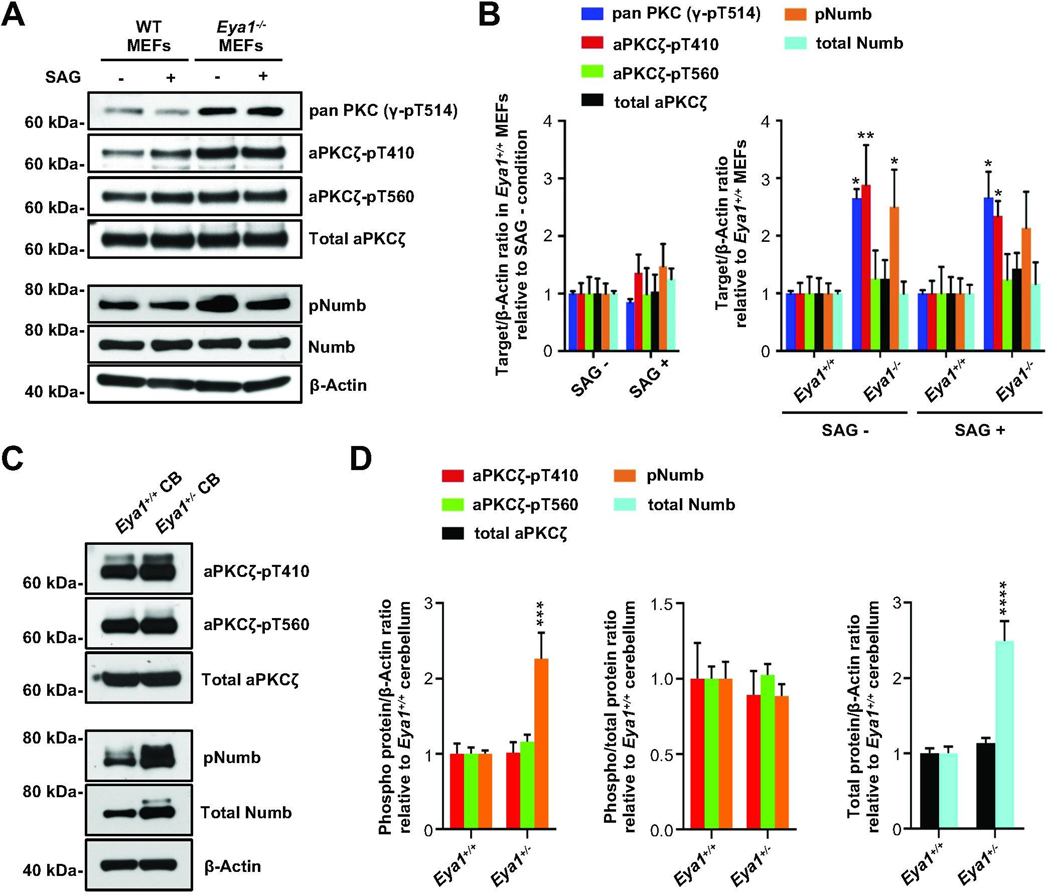
It is possible that the loss of function phenotype in Eya1+/− mice is merely due to an overall impairment of Shh signaling. Alternatively, Eya1 may function directly to regulate mode of division of GCPs. We have shown here that the intracellular distribution of the cell fate determinant Numb [60, 61] correlates with GCP spindle orientation. Unequal distribution of Numb during mitosis is known to be a major regulator of asymmetric division, and data from both fly and vertebrate model systems provide evidence that phosphorylation by the Par complex component aPKC dictates intracellular localization of Numb during asymmetric division [62, 63]. We therefore investigated whether Eya1 might be involved in directly regulating the mode of division of GCPs by altering the aPKC/Numb axis, consistent with previous suggestions [47], in a study that was subsequently retracted [48]. As mice with a homozygous knockout of Eya1 are not viable at postnatal stages [59], we first used mouse embryonic fibroblasts (MEFs) from Eya1−/− mice to study the effects of a complete loss of this phosphatase on both aPKC and Numb phosphorylation. Shh pathway activation in wild type MEFs using the Smoothened agonist SAG (300nM) induces Gli transcription factors (Fig. S5), indicating that MEFs are susceptible to Shh activation. We carried out western blot analyses using an antibody directed against a conserved phosphorylation site within the activation loop of PKC; this antibody recognizes a conserved site that is phosphorylated by pyruvate dehydrogenase kinase 1 in all PKC enzymes. We find that there is an increase in phosphorylation state at this site in Eya1−/− cells compared with wild type MEFs (Fig. 5A, B; Fig. S6A). A more selective antibody that recognizes phosphorylation at the activation loop in one of the atypical PKCs, aPKCζ (pT410), also showed enhanced phosphorylation in Eya1−/− MEFS. In contrast, Eya1 mutations did not alter the phosphorylation state at T560, the turn motif site in aPKCζ [64], suggesting that Eya1 may have a specific effect on the activation loop phosphorylation state of aPKC. While phosphorylation in the activation loop can affect both protein stability and enzymatic activity, we did not detect any changes in total levels of aPKCζ in the Eya1−/− MEFs. Taken together these findings indicate that the phosphatase Eya1 directly, or indirectly, regulates the phosphorylation state of the activation loop in PKC proteins, particularly in atypical PKCs, but these changes do not alter total levels of PKC.
Fig. 5. Eya1 regulates phosphorylation state of aPKCζ and Numb.
(A) Representative western blots of protein lysates from MEF cells generated from wild type or Eya1−/− mice, using indicated phospho-specific antibodies for protein kinase C (PKC) and Numb. (B) Corresponding quantification of relative protein levels from independent experiments (n = 4), β-Actin was used as a loading control. (C) Representative western blots of P6 cerebellar protein lysates (CB) from Eya1+/+ and Eya1+/− mice using phospho-specific antibodies for aPKCζ and Numb. (D) Corresponding quantification of relative protein levels from independent experiments (n = 4), β-Actin was used as a loading control. Analyses for phosphorylated proteins are shown when normalized to total target protein and when normalized to β-Actin. For all statistical comparisons, we use a two-way ANOVA with Bonferroni post hoc test. All data are mean±s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Numb protein is a critical substrate of aPKC that has been implicated in spindle orientation and asymmetric cell division. We found that phosphorylation of Numb at the aPKC site was also significantly increased in the absence of Eya1. Again, these changes in phosphorylation state were not associated with any changes in the overall levels of Numb. Of note, Shh-pathway activation by SAG treatment alone had no significant effect on aPKC and Numb phosphorylation, suggesting that these changes were due to Eya1 function and not due to general changes in Shh signaling.
As Eya1 affects both mitotic orientation and Shh signaling in the developing cerebellum, and both aPKC (Fig. S7) and Numb are expressed in GCPs (Fig. 3B), we analyzed aPKC and Numb phosphorylation in cerebellar lysates from wild type and Eya1+/− mice. Unlike in Eya1−/− MEFs, we could not detect changes in aPKC phosphorylation state in these heterozygotes in vivo (Fig. 5C, D; Fig. S6B), perhaps due to residual Eya1 function. However, we did observe increased phosphorylation of Numb even after heterozygous loss of Eya1, together with an increase in total Numb protein levels. These data provide evidence that Eya1 may contribute to the regulation of the cell fate determinant Numb in the developing cerebellum.
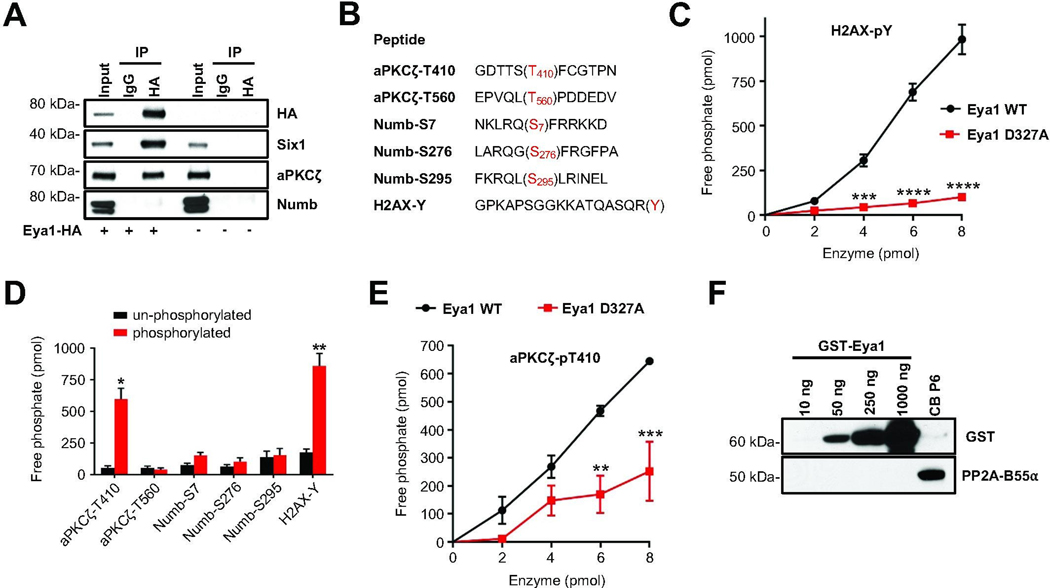
Changes in the phosphorylation state of Numb in Eya1 mutants could reflect direct or indirect effects of the Eya1 phosphatase. As Eya1 binds to its substrates to initiate phosphatase activity, we used HA antibodies to immunoprecipitate protein lysates from HEK293T cells expressing HA-tagged Eya1, and then blotted for aPKCζ and Numb (Fig. 6A; Fig. S6C). Immunoblotting for Six1, a well-known interaction partner of Eya1 phosphatase [65], was included as a positive control. Both Six1 and aPKCζ protein were co-immunoprecipitated with HA-Eya1. In contrast, Numb protein was not present in the co-immunoprecipitates. We next performed in vitro phosphatase assays with purified, full length, recombinant wild type Eya1 protein that was expressed and purified from S2 cells, and various phospho peptides corresponding to sequences from aPKCζ, Numb and H2AX as substrates (Fig. 6B; Table S2). Purified wild type Eya1 catalyzed the dephosphorylation of a peptide corresponding to the known phospho Tyr motif from the Eya1 substrate H2AX [42]. The D327 residue in the Eya domain is required for enzymatic activity, and a D327A substitution results in a phosphatase-dead mutant protein [39, 42, 34]. While wild type Eya1 showed a dose-dependent activity profile in the in vitro assay, Eya1 D327A protein showed essentially no activity even at higher molar concentrations (Fig. 6C). Using this in vitro phosphorylation assay, we tested phospho Thr and phospho Ser peptides corresponding to the phosphorylation sites in aPKC and Numb that are altered in Eya1 mutants in vitro and/or in vivo (Table S2). While Eya1 showed marked activity using the phospho Thr aPKCζ-T410 peptide (Fig. 6D), corresponding to the activation loop of aPKC ζ [66], purified Eya1 did not dephosphorylate the phospho Thr aPKCζ-T560 peptide from the turn motif of the enzyme. Strikingly, Eya1 exhibited little or no activity on peptides corresponding to phospho Ser sites pS7, pS276, and pS295 of Numb [62]. While wild type Eya1 displayed a dose-dependent activity profile for the aPKCζ-T410 peptide, Eya1 D327A protein showed a significant reduction in phosphatase activity (Fig. 6E), consistent with previous studies showing that the C-terminal domain of Eya1 is essential for Thr and Tyr phosphatase activity [34]. These studies suggest that aPKC (T410) is a direct substrate of Eya1, but that Numb is not, and that changes in Numb might instead reflect altered phosphorylation by aPKC.
Fig. 6. aPKCζ is a direct substrate of Eya1 threonine phosphatase activity.
(A) HEK293T cells were transfected with Eya1-HA and protein lysates were precipitated with anti-HA antibody. Immunoprecipitates were blotted with anti-HA, anti-Six1, anti-aPKCζ, and anti-Numb antibodies. Un-transfected HEK293T cells were included as a negative control. (B) Sequences of phospho-peptides used for in vitro Eya1 phosphatase assays. Phosphorylated threonine (T), serine (S) and tyrosine (Y) residues are highlighted in red. (C) Dose-response of phosphatase activity generated by wild type Eya1 enzyme or a mutated D327A form using H2AX-pY phosphorylated peptides as substrate (n = 3, two-way ANOVA with Bonferroni post hoc test). (D) Bar graphs representing the amount of free phosphate generated by Eya1 phosphatase activity using the indicated un-phosphorylated or phosphorylated peptides (n = 4, paired t test). (E) Dose-response of phosphatase activity generated by wild type Eya1 enzyme or a mutated D327A form using aPKC ζ-pT410 phosphorylated peptides as substrate (n = 3, two-way ANOVA with Bonferroni post hoc test). (F) Western blot analysis of GST-tagged Eya1 protein isolated from S2 insect cells. Purified protein was blotted with anti-GST and anti-PP2A-B55α antibodies. Protein lysate (20 μg) from P6 cerebellum was included as a positive control for PP2A-B55α protein. All data are mean±s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
It had been suggested that the Ser/Thr phosphatase activity of Eya3 is not intrinsic but stems from its association with the protein phosphatase 2A (PP2A)-B55α holoenzyme [35]. As we hypothesize that phosphorylation state of aPKC depends on Eya1 activity, we asked whether PP2A-B55α is essential for this function. To do so, we checked for the presence of PP2A-B55α in the recombinant GST-tagged Eya1 prepared from S2 cells. While the Eya1 preparation showed a clear dose-dependent increase in reactivity with the GST-specific antibody, we did not detect any signal for PP2A-B55α protein at any concentration of Eya1 protein tested (Fig. 6F; Fig. S6D). In contrast, we readily detected PP2A-B55α proteins in P6 cerebellar lysates, dorsal root ganglia lysates and insect cells (Fig. 6F and S6E). As PP2A-B55α protein is not present in detectable amounts in our enzymatically active Eya1 protein preparations, the phosphatase activity that dephosphorylates aPKCζ appears to be an intrinsic feature of Eya1 that is altered in D327A mutant. Together, these data suggest that Eya1 directly and specifically dephosphorylates aPKC at residue T410, thereby reducing aPKC-mediated phosphorylation of Numb, altering Numb distribution and regulating spindle orientation and asymmetric cell division in developing GCPs.
Discussion
The results presented here indicate that the Shh pathway and its positive effector Eya1 regulate spindle orientation and the mode of division of GCPs by promoting vertical spindle orientation and symmetric cell divisions. This finding is consistent with previous data showing that active Shh signaling promotes symmetric, self-renewing cell divisions of neuronal progenitors in multiple brain regions [11, 10, 50, 67, 12]. We provide both mechanistic and functional insight into Shh-regulated spindle orientation and symmetric division.
Previous studies suggest that Shh signaling drives symmetric neuroepithelial cell division by a process involving recruitment of protein kinase A to the centrosomes that nucleate the mitotic spindle [12]. Our study adds to these findings by showing that the Eya1 phosphatase promotes Shh pathway-dependent mitotic spindle orientation of GCPs. Given the prominent role of Eya1 and Shh signaling in GCP development, we believe that phenotypes observed in this study can most likely be attributed to cell autonomous changes in the GCP population. However, we cannot rule out the possibility that the actions of Shh and Eya1 within other cell types may also affect the orientation of GCP cell division.
Our data shows that spindle orientation of mitotic GCPs correlates with the mode of division. Using two independent methods for the labeling and timelapse imaging of proliferating GCPs we show that spindle orientation is a strong indicator of cell fate; vertical divisions are associated with symmetrical fates where both daughters remain in the EGL and adopt morphologies consistent with GCPs, while horizontal divisions are associated with asymmetric fates where one daughter cell migrates into the IGL and adopts neuronal morphology while the other one remains in the EGL and maintains GCP morphology. It is worth noting that we did not find a significant association between the distance from the pia and angle of division, suggesting that vertical and horizontal divisions are not being directed by an extrinsic positional cue within the EGL. Finally, the distribution of the cell-fate determinant Numb within the daughter cells further supports this model and provides evidence that GCPs begin to adopt a particular fate during mitosis. Similarly, in other epithelia such as the skin, mitotic orientation and mode of division are tightly linked, and precursors use symmetric and asymmetric division to generate appropriate ratios between proliferation and differentiation [68, 69].
Previous data have implicated Eya in Hh signaling in Drosophila and have provided multiple possible models for the actions of Eya family members in Shh signaling in vertebrates [70, 71, 19]. Here, we show that Eya1 phosphatase activity can remove phosphates from the Thr site within the activation loop of aPKC ζ. aPKC itself is essential for regulation of spindle orientation in a process conserved across species [72]. After genetic loss of Eya1 or developmental downregulation of Eya1 expression, activated aPKC phosphorylates the endocytic component Numb, a change that can facilitate asymmetric cell division [62]. Accordingly, during early development of GCPs, high expression of Eya1 and subsequent inactivation of aPKC might present a mechanism to promote symmetric divisions and preserve canonical Shh signaling.
A recent study indicates that Numb also participates in coordinated endocytosis of the Shh receptors Boc and Ptch1, and thereby enables non-canonical Shh signaling during axon guidance [73]. Interestingly, we have previously shown that aPKC activation and downstream Numb phosphorylation is an important mechanism to promote chemotaxis and migration of GCPs [74], and differentiation of GCPs is tightly linked to their migration out of the EGL [55]. Decreased Eya1 activity in newborn granule neurons entering the IGL could permit Numb to coordinate these later stages of granule neuron differentiation through non-canonical Shh signaling and BDNF-dependent pathways.
It has been shown that the highly conserved C-terminal domain (CTD) of Eya family members contains Tyr phosphatase activity [36, 75, 39, 40]. While the less conserved N-terminal domain (NTD) does not contain a sequence motif that shares homology to any other known phosphatase, it has been suggested that the NTD of Eya members harbors Ser/Thr phosphatase activity [32, 76, 45, 34]. A recent study suggests that Ser/Thr phosphatase activity reported for Eya3 reflects a close association between the Eya NTD and the PP2A enzyme [35], rather than direct activity of Eya3 itself. Here, we suggest that the CTD of both murine and human Eya1 is essential for full Thr phosphatase activity, as alterations of the D327 residue within the CTD domain reduce Eya1 activity on Thr-containing peptides, albeit to a lesser extent than its activity on Tyr-containing peptides [34]. Further detailed biochemical and structural understanding of the enzymatic phosphatase activity of Eya1–4 and its substrates will provide greater understanding of the function for this unusual gene family.
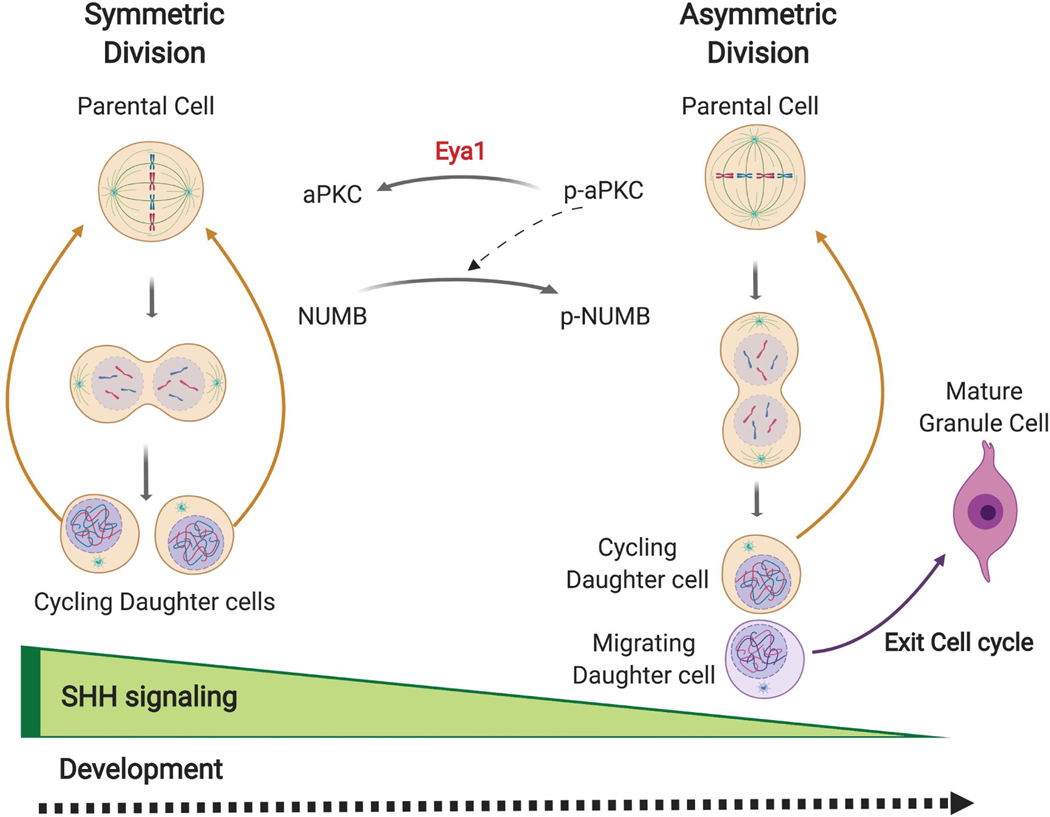
In conclusion, our data indicates that Eya1 promotes active Shh signaling-driven symmetric division of GCPs during early postnatal development (Fig. 7). We suggest that the following sequence of events may provide an underlying mechanism: in early GCPs, Eya1 may dephosphorylate aPKC and so reduce its activity. Atypical PKCs increase the phosphorylation state of Numb protein. Therefore, reduced aPKC activity may lead to decreased phosphorylation and altered intracellular distribution of Numb in dividing GCPs, so that both of the daughter cells remain in the EGL and re-enter the mitotic cycle. However, as development proceeds, decreased levels of Shh and Eya1 (See Figure 4B) result in asymmetric division of GCPs, and two distinct cell fates for the daughters (Fig. 7). As constitutive Shh activation in GCPs enables medulloblastoma, and ectopic activation of Shh pathway has also been implicated in multiple other cancer types, these data suggest that Eya1 may be a developmental regulator that is also an important contributor to tumor initiation and/or tumor growth.
Fig. 7. Eya1 promotes SHH-driven symmetric division in GCPs during cerebellar development.
A model of Eya1 and SHH regulation of symmetric division for GCPs. During development, SHH signaling is highly active, thereby promoting symmetric division. Eya1 de-phosphorylates aPKC, which leads to reduced phosphorylation of Numb and equal distribution of Numb to the two daughter cells. Therefore, the daughter cells both remain in the EGL and re-enter the cell cycle. As development proceeds, Shh and Eya1 expression decrease, and therefore GCPs divide asymmetrically, producing one daughter cell that exits the cell cycle and migrates to the IGL, and one that reenters the mitotic cycle.
Materials and Methods
Animal Studies
All experimental procedures were done in accordance with National Institutes of Health (NIH) guidelines and were approved by the Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee. Eya1−/− mice were obtained from Pin-Xian Xu [59]. Shhala [52] were generated previously and have now been deposited at Jackson Laboratory, Ptch1+/− [53], loxP-Stop-loxP-EGFP (GFPFl/+) [77], and Atoh-1creER [7] mice were obtained from Jackson Laboratories.
Production of Retrovirus
Replication-incompetent GFP-expressing retrovirus was packaged and pseudotyped with the V-SVG glycoprotein using the 293p NIT cell line as previously described [78].
Real-time imaging in organotypic slices
Atoh1-Cre-Estrogen Receptor mice (Atoh1-creER) were crossed with GFPFl/Fl (loxP-Stop-loxP-EGFP) mice to generate Atoh1-creER:GFPFl/+ mice. Neonatal pups (the day when pups were born is designated as P1) were injected intraperitoneally with 10 μl 10 mg/ml tamoxifen. Thirty hours after tamoxifen injection (P3), the pups were dissected to isolate the cerebellum and other regions of the brain were used to check GFP expression by fluorescence microscopy. Cerebella were embedded in 2% low-melting agarose and cut into 200 μm slice. Cerebellar slices were cultured on tissue culture inserts (4 slice per insert) in 6-well plates with phenol-free GCP growth medium at 37°C for 2 hours for recovery before imaging. A Nikon Perfect Focus microscope (Ti Eclipse; Nikon) equipped with a CO2 controlled chamber (5%CO2 and 37°C), a CCD video camera and a far distance 20x objective were used to capture the images on multiple positions with 5 μm-thickness z stacks (1 μm intervals between z-stack) every 45 minutes for the next 72 hours. Time-lapse experiments were analyzed using NLS-Elements software (Nikon). We then analyzed the videos, to identify individual cells that divide within the first 24 hours. We analyzed whether both of the GFP labeled daughter cells remain in the EGL where they will continue to divide (vertical division), or whether one divides and the other migrates to the IGL (horizontal division), or whether both daughters migrate to the IGL.
PNIT-GFP retrovirus (1 – 2 μl) mixed with Fast Green (2.5 mg ml−1) was injected into the cerebellum of Wildtype CD-1 IGS pups 48 hours before imaging to sparsely label GCPs. The cerebella of P3 and P6 pups were dissected in chilled ACSF and checked for successful infection using a fluorescent dissecting microscope. Cerebella with GFP labeled cells were embedded in 4% agarose and vibratome sectioned into 300 μl slices. Slices were cultured on Millipore culture inserts in a 35 mm MatTek glass dish with phenol-free GCP growth medium at 37°C. A Leica SP8X confocal equipped with a Tokai Hit CO2 temperature control unit (5%CO2 and 37°C), HyD detectors, and HC FLUOTAR L 25x/0.95 W VISIR with a long working distance were used to capture images. Images were acquired as 6 μm stacks, 1024 × 1024p, with 10 minute intervals for 24 hours using the Leica X Navigator. Time-lapse movies were rendered using the Leica X software to track the dynamics of labeled cells. We analyzed the morphology and location of cells at time zero to identify and quantify the percentage of neurons from labeled cells. The GCP plane of the division (horizontal or vertical) was determined with respect to the pial surface.
Immunohistochemistry
Mice were perfused, then whole mouse brains or cerebella were fixed in 4% paraformaldehyde overnight at 4°C followed by cryoprotection in 30% sucrose until sinking to the bottom. Sagittal sections of the cerebellar vermis (10 μm) were prepared using a cryostat (Leica Micro-systems). For paraffin sections, mouse brains were fixed in 4% paraformaldehyde overnight, embedded in paraffin using standard protocols, and then cut (5 μm sections). For cerebellar organotypic slices, slices were prepared with a Leica vibratome followed by fixation with ice-cold methanol at - 20°C for overnight. Slices were washed 3 times and rehydrated in PBS overnight. All brain sections were blocked and permeabilized with blocking/permeabilization solution (5% normal goat serum, 0.1% Triton X-100, and 5% BSA in PBS) for 1 hour at room temperature or at 4°C overnight, followed by incubation with the primary antibody (Table S1) at 4°C overnight. After incubation with primary antibodies, sections were washed in PBS, followed by the incubation with appropriate Alexa Fluor 546, 647, or 488-labeled secondary antibodies (Invitrogen, 1:400) and DAPI counterstain. Images were acquired on either a Nikon upright fluorescence microscope and NLS-Element software (Nikon) or confocal microscope (SP5; Leica) using 20X and 40X (Plan Neofluar NA 1.3 oil immersion) objectives and LAS software (Leica). Fiji software was used for image processing (Gaussian blur) and data analysis (spindle orientation measurement).
Quantitative analyses of asymmetric protein distribution
Cryosections from postnatal day 3, wild type cerebella were stained with Vybrant® CM-DiI (Invitrogen) for 20 minutes at room temperature to mark the cell boundary, then post-fixed with 4% PFA. After washing with PBS, CM-DiI-labeling sections were blocked and permeabilized with blocking solution (5% NGS, 1% BSA, 0.1% Triton X-100 in PBS) for 1 hour and followed by co-immunostaining with antibodies to phospho-histone H3 (Table S1), to recognize mitotic cells and visualize mitotic orientation, and with an antibody to Numb, followed by DAPI counterstaining. Images were captured by confocal microscopy (Leica SP5). We examined all phospho-histone H3 positive cells in anaphase or telophase and defined the boundaries of individual GCPs in the crowded EGL using CM-DiI. One individual used phospho-histone H3 and DAPI to define the mitotic plane and then integrated the fluorescence signal for Numb across the Z-axis of the two incipient daughter cells. The Asymmetry Index is the value of signal in Daughter A minus signal value in Daughter B/Signal in Daughter A plus signal in Daughter B. Complete symmetry gives an index of 0, while complete asymmetry gives an index of 1. We analyzed integrated and mean measurements of Numb fluorescence within each cell, and these different analyses did not affect the asymmetry index. To reduce any unconscious bias, a second individual determined whether the mitosis is vertical or horizontal in nature based on the orientation of the chromosomes separating in anaphase or telophase (visualized with phospho-Histone H3 and with DAPI stain of DNA). Orientation was readily apparent in anaphase/telophase cells. We also assessed a third category, of indeterminate or intermediate orientation, which is included as “oblique orientation”. Similar analysis was done for GCPs in postnatal day 3 Ptch1+/− cerebellum.
Measurement of mitotic spindle orientation
For staining using phospho-Histone H3, a line was drawn perpendicular to the mitotic spindle to best represent the plane of division. A second line perpendicular to the pial surface was drawn, and the angle between those lines was measured. For all stainings using acetylated α-tubulin and γ-Tubulin, a line was drawn between two centrosomes to mark the mitotic spindle. A second line between two points of the pial surface on the both ends of the fissure was drawn to act as a reference line for scoring the spindle orientation. For organotypic slices, we defined x, y, and z coordinates of the two centrosomes and five points on the pial surface of the 3D-rendered cerebellar slices. These five points were used to determine the best-fitting plane by orthogonal distance regression. Then, we calculated the angle between the line connecting two centrosomes and line for the best-fitting plane. The angle α of mitotic spindle orientation is calculated as 90° plus the angle. To estimate the uncertainty of each spindle orientation angle, we repeated this calculation for all possible combinations using only four out of the five points within the plane and determine the angular SD of the resulting normal lines. All angles were scaled to correspond to a range between 0° (vertical cell division) and 90° (horizontal cell division). Three pairs of control littler mates and mutants were analyzed. Comparable locations (midpoint of primary, secondary and tertiary fissures within sagittal sections of the cerebellar vermis) were assessed.
Cell culture and constructs
Mouse embryonic fibroblast (MEF) cultures from Eya1−/− and wild type littermates were generated and cultured as previously described [79]. For Shh activation, MEF cells were cultured for 24 hours in MEF media with reduced serum (0.5%) in the presence of 300 nM SAG. HEK293T cells were obtained from American Type Culture Collection and cultured according to their recommendations.
Peptides and plasmids
All peptides were custom synthesized by the Tufts University Core Facility for Peptide Synthesis; sequences are in Table S2. All peptides were purified by high-performance liquid chromatography, and all purified products were analyzed by mass spectrometry. The purity of each peptide was greater than 95%. The coding sequence of full-length Eya1 was cloned into pcDNA3.1(+)-KozakHAHA as previously described [19] for transfection into HEK293T cells. For protein expression in S2 cells, full-length Eya1 was cloned into a custom-modified version of the pFastBac vector HT A (Thermo Fisher) carrying the sequences for His and GST tags. For the construction of the D327A Eya1 mutant, we generated a single nucleotide exchange using the QuikChange II Site-Directed Mutagenesis Kit (Agilent).
Protein expression and purification
Full-length Eya1 wild type and mutant D328A protein in a GST-fusion format were generated using the Bac-to-Bac Baculovirus Expression system (ThermoFisher Scientific) and expressed in Sf9 insect cells. Cells were harvested 65–72 hours post-infection and resuspended with lysis buffer (1X PBS, 10% Glycerol, 1 mM TCEP). Resuspended cell pellets were disrupted by sonication. After sonication, lysed cells were spun down at 40000 rpm for 2 hours. Supernatants were applied to GST sepharose beads (GE Lifescience). After washing with PBS, bound proteins were eluted with GST Elution Buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 10 mM Glutathione, 2 mM TCEP, 10 mM MgCl2). To remove GST tag, eluents were incubated with Tobacco etch virus protease for 6 hours on ice. After cleavage, eluents were applied to a HiTrap Q column (GE Lifescience) and washed with Buffer A (Buffer A. 50 mM Tris-HCl pH8.0, 2 mM TCEP, 10 mM MgCl2). The protein was then eluted with a 0 – 25% gradient of Buffer B (Buffer B. 50 mM Tris-HCl pH8.0, 2 M NaCl, 2 mM TCEP, 10 mM MgCl2). Eya1 protein was concentrated to approximately 2 mg/ml using an Amicon Ultra concentrator (30 MWCO, Millipore) and further purified by size-exclusion chromatography on a Superdex 200 Increase 10/300 column (GE Lifescience) in Buffer C (Buffer C. 40 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM TCEP). Purified Eya1 protein was concentrated to 1mg/ml and frozen by liquid nitrogen for long-term storage.
In vitro phosphatase assay
For peptide assays in half-area 96 well plates, 400 ng (unless specified otherwise) of purified Eya1 protein was incubated in a final volume of 20 μl with 1 mM peptide in a buffer containing 60 mM HEPES (pH7.5), 75 mM NaCl, 75 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 1 mM DTT at 37°C for 2 hours. The released phosphate was quantified using the Malachite Green Phosphate Assay Kit (Sigma-Aldrich). Duplicates values were background corrected and compared to the internal phosphate standard.
Quantitative real-time RT-PCR
RNA was isolated using the RNeasy Mini Kit (Qiagen). Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer’s specifications. qRT-PCR was performed using Taqman gene expression assays with the following Taqman probes: Eya1 Mm01239746_m1, Shh Mm00436527_m1, Ptch1 Mm00970977_m1, Smo Mm01162710_m1, Sufu Mm00489385_m1, and Gli1 Mm00494645_m1. Values were normalied to Gapdh (Mm99999915_g1) levels.
Co-immunoprecipitations and western blotting
For all assays, protein was extracted using RIPA buffer (150mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 25mM Tris pH7.4). For co-immunoprecipitations, we used Dynabeads Protein G (Invitrogen) according to manufacturer’s protocol and optimized antibody concentrations. For western blots, protein lysates were separated electrophoretically in NuPAGE 4 – 20 % Bis-Tris gels (Thermo Fisher) and blotted on a PVDF membrane (Millipore) using standard procedures. Optimal concentrations for all antibodies were evaluated empirically. Protein bands were visualized using horse radish peroxidase (HRP)-coupled secondary antibodies (BioRad) and SuperSignal West Dura solution as substrate (Thermofisher). Image acquisition was done HyBlot CL autoradiography film (Denville Scientific).
Statistical Analysis
Significance threshold for P value was 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). For comparison of two groups, we used a Student t test or a Mann-Whitney test. We used a two-way ANOVA with Bonferroni correction for multiple comparisons to compare data from several groups. For statistical comparisons of angle distributions from several conditions, we used a Kruskal-Wallis test with Dunn`s post hoc test for multiple comparisons For comparison of fractions, Fisher’s exact test was used. All statistical analyses were done in Excel, Prism, or in the R statistical environment.
Supplementary Material
Acknowledgements
We thank CD Stiles and all members of the Segal and Harwell labs for helpful suggestions.
Funding
German Cancer Aid (Mildred Scheel postdoctoral fellowship), and Deutsche Forschungsgemeinschaft (ZUK 63 to DJM); Pussycat Foundation Helen Gurley Brown Fellowship (to GH), HHMI Gilliam Fellowship (CMR); Alex’s Lemonade Stand Foundation and Swifty Foundation (RAS) and support from the NIH (R01CA205255 to RAS and R01MH119156 and R01NS102228 to CCH).
Footnotes
Statement of Ethics
All mouse procedures were approved by the Dana-Farber and Harvard Medical School Institutional Animal Care & Use Committee (IACUC).
Competing Interests
We declare potential conflict of interest with Allergan Pharmaceuticals, Decibel Therapeutics and Amgen (RAS).
References
- 1.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980. October 30;287(5785):795–801. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011. June;12(6):393–406. [DOI] [PubMed] [Google Scholar]
- 3.Pak E, Segal RA. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell. 2016. August 22;38(4):333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–14. [DOI] [PubMed] [Google Scholar]
- 5.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012. April;123(4):465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altmann J, Bayer SA. Development of the Cerebellar System in Relation to its Evolution, Structure and Functions. New York: CRC Press; 1997. [Google Scholar]
- 7.Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. [DOI] [PubMed] [Google Scholar]
- 8.Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Hou S, Han YG. Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat Neurosci. 2016. July;19(7):888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araujo GL, Araujo JA, Schroeder T, Tort AB, Costa MR. Sonic hedgehog signaling regulates mode of cell division of early cerebral cortex progenitors and increases astrogliogenesis. Front Cell Neurosci. 2014;8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saade M, Gutierrez-Vallejo I, Le Dreau G, Rabadan MA, Miguez DG, Buceta J, et al. Sonic hedgehog signaling switches the mode of division in the developing nervous system. Cell Rep. 2013. August 15;4(3):492–503. [DOI] [PubMed] [Google Scholar]
- 12.Saade M, Gonzalez-Gobartt E, Escalona R, Usieto S, Marti E. Shh-mediated centrosomal recruitment of PKA promotes symmetric proliferative neuroepithelial cell division. Nat Cell Biol. 2017. April 27;19(5):493–503. [DOI] [PubMed] [Google Scholar]
- 13.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005. December;37(12):1323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillman RT, Feng BY, Ni J, Woo WM, Milenkovic L, Hayden Gephart MG, et al. Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev. 2011. November 15;25(22):2333–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purzner T, Purzner J, Buckstaff T, Cozza G, Gholamin S, Rusert JM, et al. Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci Signal. 2018. September 11;11(547). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999. November 1;13(21):2828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia H, Liu Y, Yan W, Jia J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development. 2009. January;136(2):307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013. February 28;494(7438):484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisner A, Pazyra-Murphy MF, Durresi E, Zhou P, Zhao X, Chadwick EC, et al. The Eya1 phosphatase promotes Shh signaling during hindbrain development and oncogenesis. Dev Cell. 2015. April 06;33(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993. February 12;72(3):379–95. [DOI] [PubMed] [Google Scholar]
- 21.Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997. January;124(1):219–31. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman JE, Bui QT, Steingrimsson E, Nagle DL, Fu W, Genin A, et al. Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res. 1997. February;7(2):128–41. [DOI] [PubMed] [Google Scholar]
- 23.Borsani G, DeGrandi A, Ballabio A, Bulfone A, Bernard L, Banfi S, et al. EYA4, a novel vertebrate gene related to Drosophila eyes absent. Hum Mol Genet. 1999. January;8(1):11–23. [DOI] [PubMed] [Google Scholar]
- 24.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997. December;6(13):2247–55. [DOI] [PubMed] [Google Scholar]
- 25.Vincent C, Kalatzis V, Abdelhak S, Chaib H, Compain S, Helias J, et al. BOR and BO syndromes are allelic defects of EYA1. Eur J Hum Genet. 1997. Jul-Aug;5(4):242–6. [PubMed] [Google Scholar]
- 26.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017. June 12;31(6):737–54 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu K, Li Z, Cai S, Tian L, Chen K, Wang J, et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013. July 15;73(14):4488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, et al. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 2013. June 27;3(6):2127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 2013. June;70(11):1897–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravind L, Galperin MY, Koonin EV. The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biochem Sci. 1998. April;23(4):127–9. [DOI] [PubMed] [Google Scholar]
- 31.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004. June 15;101(24):9085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009. July 23;460(7254):520–4. [DOI] [PubMed] [Google Scholar]
- 33.Sano T, Nagata S. Characterization of the threonine-phosphatase of mouse eyes absent 3. FEBS Lett. 2011. September 2;585(17):2714–9. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Rodriguez Y, Cheng C, Zeng L, Wong EY, Xu CY, et al. EYA1’s Conformation Specificity in Dephosphorylating Phosphothreonine in Myc and Its Activity on Myc Stabilization in Breast Cancer. Mol Cell Biol. 2017. January 01;37(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Zhou H, Li X, Vartuli RL, Rowse M, Xing Y, et al. Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat Commun. 2018. March 13;9(1):1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003. November 20;426(6964):247–54. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012. February 14;22(2):377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis TL, Hoi CSL, Rebay I. Mutations that impair Eyes absent tyrosine phosphatase activity in vitro reduce robustness of retinal determination gene network output in Drosophila. PLoS One. 2017;12(11):e0187546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003. November 20;426(6964):299–302. [DOI] [PubMed] [Google Scholar]
- 40.Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007. October 15;310(2):416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin M, Jusiak B, Bai Z, Mardon G. Eyes absent tyrosine phosphatase activity is not required for Drosophila development or survival. PLoS One. 2013;8(3):e58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009. April 2;458(7238):591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, et al. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009. June 12;284(24):16066–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan B, Cheng L, Chiang HC, Xu X, Han Y, Su H, et al. A phosphotyrosine switch determines the antitumor activity of ERbeta. J Clin Invest. 2014. August;124(8):3378–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Wong EY, Cheng C, Li J, Sharkar MT, Xu CY, et al. Eya1 interacts with Six2 and Myc to regulate expansion of the nephron progenitor pool during nephrogenesis. Dev Cell. 2014. November 24;31(4):434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Wang L, Wong EYM, Tsang SL, Xu PX, Lendahl U, et al. An Eya1-Notch axis specifies bipotential epibranchial differentiation in mammalian craniofacial morphogenesis. Elife. 2017. November 15;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Hashash AH, Turcatel G, Al Alam D, Buckley S, Tokumitsu H, Bellusci S, et al. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011. April;138(7):1395–407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.El-Hashash AH, Turcatel G, Al Alam D, Buckley S, Tokumitsu H, Bellusci S, et al. Retraction: Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development doi: 10.1242/dev.058479. 2017. October 15;144(20):3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zagon IS, McLaughlin PJ. The location and orientation of mitotic figures during histogenesis of the rat cerebellar cortex. Brain Res Bull. 1987. March;18(3):325–36. [DOI] [PubMed] [Google Scholar]
- 50.Haldipur P, Sivaprakasam I, Periasamy V, Govindan S, Mani S. Asymmetric cell division of granule neuron progenitors in the external granule layer of the mouse cerebellum. Biol Open. 2015. May 15;4(7):865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyashita S, Adachi T, Yamashita M, Sota T, Hoshino M. Dynamics of the cell division orientation of granule cell precursors during cerebellar development. Mech Dev. 2017. October;147:1–7. [DOI] [PubMed] [Google Scholar]
- 52.Chan JA, Balasubramanian S, Witt RM, Nazemi KJ, Choi Y, Pazyra-Murphy MF, et al. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009. April;12(4):409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. [DOI] [PubMed] [Google Scholar]
- 54.Hanzel M, Rook V, Wingate RJT. Mitotic granule cell precursors undergo highly dynamic morphological transitions throughout the external germinal layer of the chick cerebellum. Sci Rep. 2019. October 23;9(1):15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi Y, Borghesani PR, Chan JA, Segal RA. Migration from a mitogenic niche promotes cell-cycle exit. Journal of Neuroscience. 2005;25(45):10437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leto K, Arancillo M, Becker EB, Buffo A, Chiang C, Ding B, et al. Consensus Paper: Cerebellar Development. Cerebellum. 2016. December;15(6):789–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994. February 11;76(3):477–91. [DOI] [PubMed] [Google Scholar]
- 58.Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995. October 19;377(6550):624–7. [DOI] [PubMed] [Google Scholar]
- 59.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999. September;23(1):113–7. [DOI] [PubMed] [Google Scholar]
- 60.Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004. February 3;14(3):R129–38. [PubMed] [Google Scholar]
- 61.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005. April;132(8):1751–62. [DOI] [PubMed] [Google Scholar]
- 62.Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007. January 24;26(2):468–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008. October 3;135(1):161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chou MM, Hou W, Johnson J, Graham LK, Lee MH, Chen CS, et al. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr Biol. 1998. September 24;8(19):1069–77. [DOI] [PubMed] [Google Scholar]
- 65.Buller C, Xu X, Marquis V, Schwanke R, Xu PX. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet. 2001. November 15;10(24):2775–81. [DOI] [PubMed] [Google Scholar]
- 66.Parker PJ, Parkinson SJ. AGC protein kinase phosphorylation and protein kinase C. Biochem Soc Trans. 2001. November;29(Pt 6):860–3. [DOI] [PubMed] [Google Scholar]
- 67.Yang R, Wang M, Wang J, Huang X, Yang R, Gao WQ. Cell Division Mode Change Mediates the Regulation of Cerebellar Granule Neurogenesis Controlled by the Sonic Hedgehog Signaling. Stem Cell Reports. 2015. November 10;5(5):816–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005. September 8;437(7056):275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011. February 17;470(7334):353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pappu KS, Chen R, Middlebrooks BW, Woo C, Heberlein U, Mardon G. Mechanism of hedgehog signaling during Drosophila eye development. Development. 2003. July;130(13):3053–62. [DOI] [PubMed] [Google Scholar]
- 71.Lu K, Reddy R, Berika M, Warburton D, El-Hashash AH. Abrogation of Eya1/Six1 disrupts the saccular phase of lung morphogenesis and causes remodeling. Dev Biol. 2013. October 1;382(1):110–23. [DOI] [PubMed] [Google Scholar]
- 72.Guilgur LG, Prudencio P, Ferreira T, Pimenta-Marques AR, Martinho RG. Drosophila aPKC is required for mitotic spindle orientation during symmetric division of epithelial cells. Development. 2012. February;139(3):503–13. [DOI] [PubMed] [Google Scholar]
- 73.Ferent J, Giguere F, Jolicoeur C, Morin S, Michaud JF, Makihara S, et al. Boc Acts via Numb as a Shh-Dependent Endocytic Platform for Ptch1 Internalization and Shh-Mediated Axon Guidance. Neuron. 2019. April 24. [DOI] [PubMed] [Google Scholar]
- 74.Zhou P, Alfaro J, Chang EH, Zhao X, Porcionatto M, Segal RA. Numb links extracellular cues to intracellular polarity machinery to promote chemotaxis. Dev Cell. 2011. May 17;20(5):610–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, et al. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003. November 20;426(6964):295–8. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Sano T, Guan Y, Nagata S, Hoffmann JA, Fukuyama H. Drosophila EYA regulates the immune response against DNA through an evolutionarily conserved threonine phosphatase motif. PLoS One. 2012;7(8):e42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001. January 1;97(1):324–6. [DOI] [PubMed] [Google Scholar]
- 78.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–20. [DOI] [PubMed] [Google Scholar]
- 79.Jozefczuk J, Drews K, Adjaye J. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J Vis Exp. 2012. June 21(64). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.