Abstract
Recent technological advances have demonstrated the feasibility of achieving high-throughput slide-free three-dimensional (3D) pathology of biopsy and surgical specimens. In comparison to conventional slide-based pathology, 3D pathology has the potential to provide a transformative improvement in diagnostic performance for a number of reasons: (1) vastly greater (multiple log orders) sampling of tissue specimens, (2) volumetric imaging of cell distributions and tissue structures that are prognostic and predictive, (3) nondestructive imaging, which allows valuable biopsy specimens to be used for downstream molecular assays, and (4) a simplified process with cost benefits for pathology laboratories. However, due to the immense size of these feature-rich datasets, new challenges exist in terms of data management and computer-aided interpretation. In this forward-looking Perspective, we first provide a brief overview of the imaging technologies that can enable nondestructive 3D pathology, including computational tools needed to support these 3D methods. We then provide a roadmap for how machine learning, which is already being developed within the context of 2D digital pathology, should be leveraged and refined for 3D pathology. Finally, we discuss future challenges and opportunities for the clinical validation, regulatory approval, and clinical adoption of this new paradigm for precision medicine, including for reducing health disparities across populations. This includes learning from, and integrating with, other diagnostic modalities such as radiology and genomics.
One Sentence Summary:
In this Perspective article, Liu et al. motivate and summarize the optical methods and related computational technologies being developed to enable a new diagnostic paradigm of slide-free nondestructive 3D pathology, as well as a vision for its clinical adoption.
1. Introduction and summary of clinical vision and impact
1.1. Motivation and vision for nondestructive 3D pathology
Disease diagnosis and characterization depend upon the rapid and accurate pathological analysis of biopsies and surgically excised tissues. This careful interrogation of the morphological and molecular characteristics of the tissue plays a key role in determining which treatments are most appropriate for individuals. For many cancers, the biopsy-determined grade of the disease provides the basis for stratifying patients for clinical management, and can result in dramatically different treatment paths 1,2. As an example, for prostate cancer, patients with low-grade cancer (Gleason score < 7) are candidates for active surveillance whereas patients with higher-grade cancer (Gleason score ≥ 7) are candidates for curative therapy (radiation and/or surgery), often in conjunction with neoadjuvant therapy for those with the highest-grade cancer (Gleason scores 8 to 10). Unfortunately, interobserver variance amongst pathologists is high, with kappa values ranging from 0.3 and 0.8 3-6. This ambiguity can, in some cases, result in the overtreatment of certain patients with indolent disease 7,8, which results in unnecessary side effects and financial toxicity to both patients and the healthcare system. Likewise, the undertreatment or nonoptimal treatment of patients with aggressive disease leads to preventable morbidity and mortality 9, along with dramatically increased costs of care for patients with late-stage diseases. For example, largely due to delayed diagnosis and undertreatment of disease, uninsured men are roughly 5 times more likely to have incurable metastatic prostate cancer at initial diagnosis than insured men 10, and have 2.5 times the risk of prostate cancer-specific mortality 11.
Another example to illustrate the limitations of conventional pathology relates to the use of immune-checkpoint inhibitors (targeting PD-1 and PD-L1) as a treatment for patients with non-small-cell lung cancers (NSCLC), which account for 80 – 85% of lung cancers. Compared to standard chemotherapy, these drugs can decrease the risk of progression by up to 60% but are only effective in approximately 20% of patients 12. With treatment costs ranging from $150K to $1M per patient, as well as the possibility of auto-immune side effects, reliable assays are needed to predict which patients will respond to these treatments. Current companion diagnostics and complementary diagnostics for predicting PD-1/PD-L1 treatment response are all based on immunohistochemistry (IHC) of PD-L1 expression, but have been criticized for their unreliability 13,14. A multitude of reasons have been offered to explain the shortcomings of these assays, including the variability in pathologist interpretation of PD-L1 IHC 15, the spatial and temporal heterogeneity of PD-L1 expression within the tumor microenvironment, and the general complexity of the immune response, in which multiple cell types must interact in a coordinated fashion to deliver a durable immunomodulatory response.
One contributing factor for the unreliability in diagnosing and grading malignant tissues, and for developing predictive assays, is that pathology laboratories handle biopsies and surgical specimens via a decades-old analog imaging process that only samples a small fraction of the specimens in 2D. In terms of reliability, imaging a small percentage of a heterogeneous tissue specimen is bound to introduce errors. For example, a 5-micron thick section of a 1-mm thick biopsy represents only 0.5% of the biopsy. This limited sampling is due, in part, to the complexity and time-consuming nature of traditional slide-based pathology, but also by its inherent destructiveness. In an era of molecular medicine where increasing numbers of assays (e.g. genomics and proteomics) are desired for personalized care (i.e. “precision medicine”), there is an ever-increasing need to preserve valuable tissue specimens for such downstream assays 16, further constraining the number of tissue sections that can be analyzed via destructive sectioning. In addition to sampling limitations, the thin slide-mounted tissue sections that are currently viewed by pathologists provide only a 2D view of tissue structures and molecular targets, which can be challenging to interpret accurately and reproducibly (with high interobserver concordance) 17-19. Subjective visual interpretation of these 2D images is another source of uncertainty, which digital pathology with machine learning strives to remedy 20.
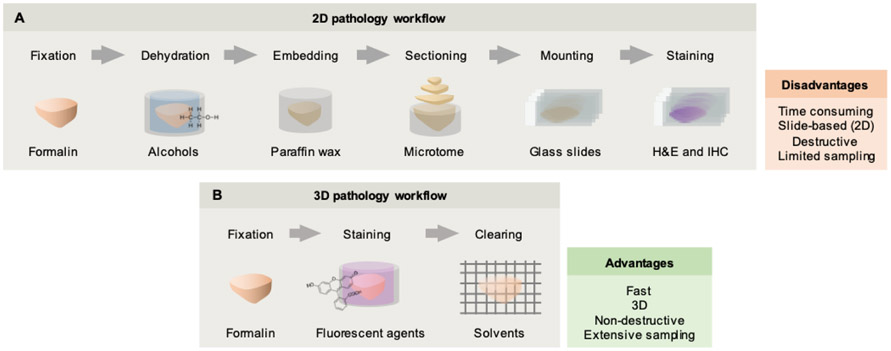
As a complement to traditional slide-based 2D pathology, there has been a re-invigorated interest in nondestructive 3D pathology, especially in light of recent technological advancements in optical clearing, high-throughput microscopy, and computational tools (including artificial intelligence or AI). At the heart of this movement is a belief that nondestructive volumetric microscopy can improve concordance and accuracy (diagnostic, prognostic, and predictive power) in the analysis of tissue specimens, thus resulting in superior patient outcomes. Here, diagnosis refers to the identification of a disease condition, including sub-type, whereas prognosis refers to the anticipated trajectory and outcome of the condition. In the context of this article, the term “predictive” refers to the anticipated effects of a specific treatment. Technical benefits of 3D pathology over traditional pathology include: (1) improved sampling of large volumes of tissue rather than sparse sampling with thin slide-mounted sections; (2) volumetric imaging of diagnostically relevant structures; (3) non-destructive imaging, which allows intact tissue specimens (e.g. core-needle biopsies) to be made fully available for downstream proteomic/genetic assays; (4) a simpler slide-free imaging process, which has the potential to save time and costs. A summary of some of these points is provided in Fig. 1.
Figure 1. Conventional pathology vs. nondestructive 3D pathology.
(A) The conventional histology workflow was developed over a century ago and involves the use of harsh fixatives and dehydration reagents (e.g. xylene) followed by wax embedding, destructive sectioning, and staining of slide-mounted sections with chromogens such as H&E. In addition to being time consuming and destructive, only a small fraction of a clinical specimen is viewed in 2D. (B) Recent advances in optical clearing and fluorescence labeling, along with high-throughput volumetric microscopy, enable entire specimens (e.g. core-needle biopsies) to be imaged in 3D with minimal tissue processing or mounting requirements. This method provides rich 3D structural (and molecular) information of large intact specimens, and preserves valuable clinical specimens for downstream assays (e.g. DNA and RNA sequencing).
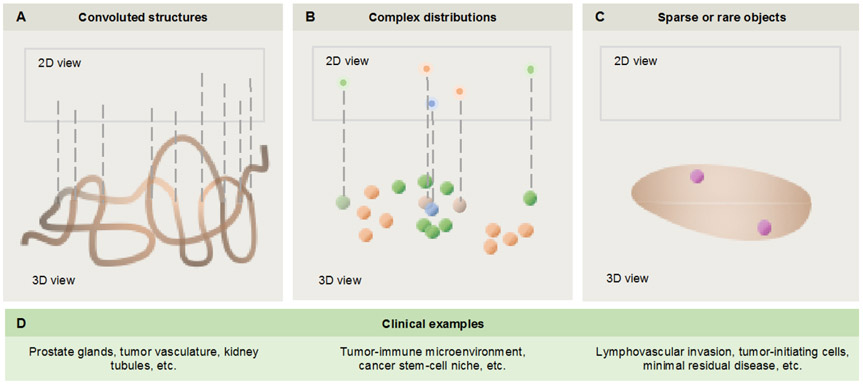
While the full value of 3D pathology data has yet to be determined, three illustrative examples are provided in Fig. 2. The first example deals with convoluted structures, such as the branching-tree vascular and glandular networks known to be disrupted during disease progression. When viewing such complex 3D structures as 2D cross sections on glass slides, artifacts and ambiguities are unavoidable. This is particularly problematic for Gleason grading of prostate cancers, which is currently based on glandular architecture alone. For example, what appears in 2D to be a poorly formed gland (a variant of Gleason pattern 4) might be a tangential section of a well-formed gland (Gleason pattern 3) when viewed in 3D. Consequently, this cancer could be re-categorized from the 2D-determined grade (Gleason score 3+4=7) to a lower-grade Gleason score (3+3=6) when viewed in 3D 21,22. The second example is of complex distributions of cells, such as within the tumor-immune microenvironment. Here, the spatial relationships and interactions between cell types – e.g. the extent and proximity of PD-1 expressing cytotoxic T cells to PD-L1 expressing cancer cells 23, or the balance between immunosuppressive neutrophils and cytotoxic T cells 24,25 – could be more ideally characterized and quantified in 3D. Furthermore, the ability to interrogate larger volumes of tissue would be beneficial in light of the spatial heterogeneity of most diseases at microscopic length scales 26-28. The third example is of rare cells that can play critical roles in the initiation, treatment resistance, and/or dissemination of various diseases. Examples of rare events that are difficult to identify on thin tissue sections include tumor-progenitor cells 29-31, minimal residual disease 32,33, and aggressive tumor subclones that invade the lymphovascular network 34-36 and metastasize 37. Note that the second and third categories mentioned above, of “complex distributions” and “rare events,” are related. In particular, as we begin to understand tumors as hierarchical collections of cells with significant intratumoral heterogeneity, one must recognize the pivotal role of rare cell populations, such as tumor-initiating cells (i.e. cancer stem cells), in some of the largest challenges/opportunities in oncology such as disease progression, treatment resistance, and recurrence 38-40.
Figure 2. Examples in which 3D pathology could outperform 2D pathology.
(A) For convoluted 3D structures, 2D cross sectional views can be misleading. (B) For distributions of cells and other structures, 2D cross sectional views might preclude accurate quantification of complex spatial relationships. (C) Finally, for rare cells and microstructures, 2D sections might not provide adequate sampling to identify and quantify such targets. (D) Clinical examples are provided corresponding to the three categories above.
1.2. Current status of 3D pathology
Three-dimensional microscopy / pathology is a technology that has been embraced and driven by life scientists for a number of decades. This has been catalyzed primarily by the invention of confocal microscopy, followed by multiphoton microscopy and most-recently light-sheet microscopy. In section 2, we will provide a cursory overview of these imaging technologies that are foundational to nondestructive 3D microscopy. In the realm of clinical diagnostics, however, 3D pathology is in its infancy. Much of this is due to technical adoption barriers, in terms of microscopy, tissue preparation, and computational methods (see section 2).
The past few years have witnessed early signs of a renewed interest in 3D pathology for clinical diagnostics. For example, in 2016, van Royen et al. 41 demonstrated the basic feasibility to image archival formalin-fixed paraffin-embedded (FFPE) and fresh prostate tissues in 3D by employing optical clearing and confocal microscopy. That same year, Olson et al. described the use of multiphoton microscopy with optical clearing to generate 3D histology images of kidney biopsies 42. The following year, Tanaka et al. 43 utilized optical clearing and light-sheet microscopy to examine FFPE specimens (bladder), showing the ability to quantify certain histomorphometric features, such as vascular density and tortuosity. Here, they showed that 3D pathology is superior to 2D pathology for staging cancers and predicting progression-free survival. This same group also imaged and analyzed the lymphatics within bladder tumors in 2018 44. In 2017, Glaser et al. 21 described an open-top light-sheet (OTLS) microscopy system that was specifically designed for high-throughput 3D pathology of large clinical specimens. This report included an anecdotal study demonstrating that 3D pathology could improve the grading of prostate core-needle biopsies by mitigating some of the ambiguities and artifacts encountered when viewing 2D tissue sections of prostate carcinoma. A follow-up study by Reder et al. 22, using the OTLS technology, provided additional insights into the potential value of 3D pathology, including the high variability in glandular morphology throughout an entire core-needle biopsy, which could have a dramatic influence on treatment decisions. More recently, Lee et al. 45 performed 3D confocal microscopy of immune infiltrates in preclinical and human core needle biopsies (head & neck cancer), showing that the spatial distribution of tumor-infiltrating cytotoxic T cells (CTLs) is correlated with other hallmarks of disease progression such as the tumor microvasculature. In addition, Verhoef et al. 46 performed a detailed investigation of the 3D structure of prostate cancers, revealing two major architectural subgroups of growth patterns that could potentially be of prognostic value. Note that the previous examples are not intended to constitute an exhaustive list of 3D pathology studies, but serve to illustrate the growing interest and progress in this field, as well as the many unknowns that remain. In addition, while there is some overlap in technologies, we are not discussing intraoperative applications of optical microscopy, as described in many other articles for rapid ex vivo 21,47-56 and in vivo 57-61 examination of excised tissues and wound cavities to guide surgical decisions.
1.3. Major challenges in nondestructive 3D pathology
A number of challenges will need to be overcome in order to translate nondestructive 3D pathology into mainstream clinical practice:
The ease-of-use, technical capabilities, and throughput of tissue-preparation protocols (clearing and labeling), imaging devices, and data-processing routines, as described in section 2. This includes a discussion of big-data challenges that 3D pathology datasets will introduce.
Computational analysis and visualization techniques to assist clinicians with the interpretation of large 3D pathology datasets within reasonable time frames, as discussed in section 3. This includes developing and applying new classes of computational pathology algorithms for detection, segmentation, visualization and characterization of 3D morphology and histologic primitives.
Strategies for regulatory approval and acceptance by clinicians and patients, including a staged approach for introducing new technologies such as machine learning, and issues related to cost and time (workflow). These issues will be discussed in section 4.
Integration with other diagnostic technologies, such genomics and radiology, for delivering holistic decision support for patients/clinicians across disparate populations. These issues will be discussed in section 5.
2. Enabling technologies for nondestructive 3D pathology
2.1. Tissue-clearing and labeling protocols
In recent decades, numerous protocols have been developed to improve the transparency of excised biological tissues for the purposes of enabling deep optical imaging at high resolution. A number of review articles have been written that describe the principles and popular approaches 62,63. In short, optical clearing seeks to homogenize the refractive index profile within tissues, primarily through dehydration and replacement of the water (n ~ 1.33) with a high-index solvent that is better-matched to the remaining tissue components (mostly proteins and lipids). Certain protocols also perform lipid removal with detergents, in some cases accelerated with active electrophoretic transport, and/or decalcification to enable optical imaging through bone 64,65. Early protocols were often developed for clarification of brain tissue for neuroscience applications with limited optimization for other organs 66-69. Such protocols were often complex, including the use of hydrogel embedding, in order to preserve the structure of delicate brain tissues as well as to maintain the brightness of genetically encoded fluorescent proteins 68,70. Recent efforts have focused on clearing whole organisms and various types of human tissues 64,65,71,72. While an in-depth analysis and comparison of these approaches is beyond the scope of this article, one relatively simple method that has gained popularity for clearing a variety of preclinical and clinical tissues has been iDISCO 71, including a more-recent variant that utilizes a food-grade cinnamon oil (ethyl cinnamate, ECi) 72 for final index matching rather than the corrosive and carcinogenic dibenzylether (DBE) utilized in the original iDISCO protocol. Note that different clearing protocols exhibit varying levels of compatibility with fluorescent labeling approaches, including small-molecule fluorescent probes and large antibodies 62,73.
For clinical applications of 3D pathology, unique requirements and constraints exist for tissue processing in preparation for imaging. First and foremost, until these techniques become the standard of care in future decades, they should ideally not interfere with current methods in histopathology, such as slide-based H&E staining and immunohistochemistry. For most clearing methods, the reagents are relatively gentle in comparison to formalin fixation. In other words, once the tissue is fixed in formalin – as is currently required within strict time frames upon clinical biopsy or resection – subsequent steps for most 3D pathology pipelines are less harsh and damaging to the tissue and its molecular constituents (e.g. proteins, DNA, and RNA). Nonetheless, studies are needed to rigorously demonstrate that 3D pathology methods are compatible with standard pathology techniques. For example, initial studies by us and others have shown that fixed and deparaffinized tissues that are labeled with fluorescent stains and cleared with methods such as iDISCO / ECi can be subsequently embedded in paraffin and subjected to standard H&E and IHC methods with no apparent degradation in quality 22,43,56.
Beyond demonstrating that 3D pathology methods do not interfere with current histopathology methods, the advent of 3D pathology offers the potential to modernize tissue-preservation methods. In particular, as genomics and transcriptomics assays rapidly improve and gain clinical acceptance, a major performance bottleneck is the damaging effects that formalin fixation has on DNA and RNA integrity. Alternative tissue-preservation methods, such as RNAlater 74,75 and PAXgene 76,77, have been developed to maintain the fidelity of nucleic acids. A tissue-preservation protocol, relying upon intra- and intermolecular crosslinking of biomolecules, has also been developed to enable effective optical clearing and fluorescent labeling of molecular targets (including RNA transcripts) 78. The ability to incorporate and standardize new tissue-preservation strategies within a clinical pipeline for 3D pathology, for high-quality volumetric imaging and optimization of nucleic acids, could be transformational for precision medicine. Section 5 provides additional discussions on these forward-looking topics.
2.2. Technical requirements for translation of 3D pathology into clinical practice
While there are many factors to consider when designing any 3D optical imaging system, key attributes to consider include resolution, contrast (signal to background ratio), speed/throughput, ease of use, and cost/complexity. In terms of importance, characteristics such as ease-of-use and speed/throughput are of higher priority for applications in anatomic pathology in comparison to most research applications – while life-science researchers often seek to resolve increasingly finer structures in order to gain novel biological insights, pathologists generally do not image tissues at the high levels of resolution that biologists are accustomed to (e.g. with 1.4 NA oil-immersion objectives). With the exception of a few specialties such as renal pathology, which routinely employs electron microscopy, the vast majority of anatomic pathology cases are viewed at low resolution (standard 5x to 10x objectives, NA ~ 0.1 to 0.3) and occasionally at moderate resolution (standard 20x to 40x objectives, NA ~ 0.4 to 0.8). Rather, for pathologists, the ability to view large areas/volumes of tissue is often of paramount importance, especially in light of the spatial and molecular heterogeneity of neoplastic lesions 28,79. Therefore, in general, the requirements for spatial resolution are relaxed for clinical versus research applications of 3D microscopy.
In terms of achieving high image contrast, which directly impacts imaging depth, a key distinction is that while biologists often desire to visualize living organisms, pathologists tend to examine excised and preserved specimens. With most living tissues, the ability of an optical-sectioning microscope to deliver high contrast (signal to background ratio, SBR) is important for high-quality imaging at reasonable depths (e.g. up to ~0.5 to 1 mm for confocal and multiphoton microscopy in most tissue types). However, recent advances in optical-clearing approaches enable nearly all excised tissues (including bone) to be rendered highly transparent 63,69-71,80-82, thereby allowing for imaging depths of several millimeters, and even centimeters (see section 2.1). This ability to make tissues transparent to light has greatly relaxed the requirements for contrast (rejection of background light) for 3D microscopy of excised specimens.
2.3. Destructive 3D microscopy techniques
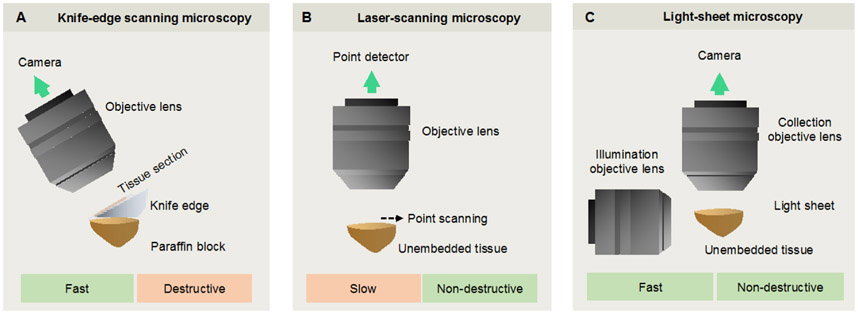
While the concept of 3D pathology is not new, the technologies have evolved considerably over the years. Early studies relied upon destructive serial sectioning techniques 83, which required considerable expense and labor for imaging large numbers of sectioned tissues, and their subsequent 3D reconstruction. Automated serial-sectioning approaches, such as knife-edge scanning (KESM) and micro-optical sectioning tomography (MOST) 84,85, have greatly improved throughput, and have been commercialized for the purposes of delivering 3D pathology data, but are destructive of tissue specimens and introduce sectioning artifacts (Fig. 3A).
Figure 3. A comparison of selected imaging methods for 3D pathology.
(A) Knife-edge scanning microscopy (KESM) and micro-optical sectioning tomography (MOST) are destructive methods in which 2D images are acquired as a specimen is serially sectioned. These stacks of adjacent 2D images are used to reconstruct a 3D image of the specimen. (B) With confocal and multiphoton laser-scanning microscopy, a single point is typically imaged within a thick specimen, and is spatially scanned in three directions to nondestructively generate a 3D image over time. (C) With light-sheet microscopy, a 2D “optical section” within a transparent thick specimen is illuminated. Fluorescence generated within that light sheet is imaged in the orthogonal direction onto a sensitive high-speed camera. Scanning the 2D light sheet through the sample (or vice versa) allows for rapid generation of a 3D image in a plane-by-plane fashion.
2.4. Confocal and multiphoton laser-scanning microscopy
In terms of nondestructive high-resolution 3D imaging of tissues, the traditional work-horse technologies have been confocal and nonlinear/multiphoton microscopy 49,50,86-88. As mentioned previously, several recent studies on 3D pathology have utilized confocal microscopy 41,46,89, and it remains the most prevalent volumetric microscopy technique in academic and industrial labs. Multiphoton microscopy has also been widely adopted in academic research due to the ability to provide enhanced imaging depths (contrast) when imaging nontransparent tissues, and also the ability to achieve label-free imaging by relying upon a number of linear and nonlinear contrast mechanisms such as autofluorescence generation 50,54,87,90-93, second harmonic generation (SHG) 94-96, coherent four-wave mixing (e.g. CARS and SRS) 51,52,87,97,98, and pump-probe methods 99,100. A number of groups have recently explored the use of multiphoton microscopy for clinical applications of anatomic pathology such as surgical guidance and diagnosis 42,50,87,93,101.
While confocal and multiphoton microscopy have become ubiquitous in biomedical research labs, offering exquisite contrast (i.e. rejection of background) and spatial resolution, there are certain challenges to overcome for clinical applications. For example, confocal and nonlinear microscopy typically generate an image in a point-by-point fashion, which requires spatial scanning in all three dimensions to create a volumetric image (Fig. 3B). This tends to add mechanical complexity and is often slow. While methods do exist to accelerate this process, such as the use of spinning disks for confocal microscopy 102,103, and temporal-focusing 104,105 or multifocal methods106-108 for nonlinear microscopy, speed is often still a significant constraint when trying to image large 3D volumes. Furthermore, these technologies, which rely upon complex high-NA optics, high-speed laser scanning, and pulsed lasers (for multiphoton microscopy), are often expensive both in terms of equipment and technical support staff.
2.5. Light-sheet microscopy
Over the past decade, light-sheet microscopy, also known as selective plane illumination microscopy (SPIM), has emerged as the technique of choice for rapid 3D fluorescence microscopy of relatively transparent specimens (e.g. embryos and optically cleared tissues) 109-117. In light-sheet microscopy, a thin excitation beam is incident upon the specimen perpendicular to detection axis such that only one localized focal plane (“optical section”) of interest is excited within the sample (Fig. 3C). Light-sheet systems utilize highly sensitive and fast sCMOS detector arrays to obtain 2D images from a specimen, from which a 3D dataset may be rapidly generated by scanning the specimen (or light sheet). A well-appreciated feature of light-sheet microscopy is its highly efficient geometry that only excites fluorescence within the detection plane, thereby minimizing photobleaching and photodamage compared with alternative 3D microscopy techniques 111,112,114. Thus, light-sheet microscopy has been referred to as a “gentle” form of 3D microscopy 118.
Early light-sheet microscopes were constrained to image small non-clinical specimens (e.g. embryos, mouse brains, etc.) that were often embedded in agarose and/or mounted in specialized mounts (e.g. for sample rotation), thus limiting the size/geometry of the specimens and the ease-of-use of the systems 112,114,117,119. More recently, a number of light-sheet microscope systems have been developed to accommodate larger specimens with less physical constraints and simpler mounting requirements 21,115,117,120-130. Many of these systems have utilized an inverted architecture in which one or more tissue samples may be conveniently placed on a platform and imaged from above or below. In particular, imaging from below the specimen(s) allows for “open-top” light-sheet (OTLS) microscopy in which tissues, transparent sample holders, and accessories of arbitrary size can be accommodated 21,123,125,130,131. One downside of OTLS or other forms of inverted light-sheet microscopy is that the imaging depth is limited by the working distance of the angled imaging objectives. Since working distance typically trades off with NA, resolution is usually in the low to moderate range (NA < 0.8) for such inverted systems. However, as mentioned in section 2.2., this level of resolution is adequate for the vast majority of clinical diagnostic applications. In order to maximize imaging depth and resolution, some research groups have explored the use of a single high-NA objective, oriented perpendicular to the sample, which is used to generate both an oblique illumination light sheet, and to image the light-sheet-generated fluorescence signal onto a detector array (camera) 128-130.
In its simplest form, light-sheet microscopy utilizes relatively low-NA illumination such that the light sheet has a long depth of focus at the expense of a thick beam waist. While higher-NA collection ensures that adequate (i.e. micron to sub-micron-scale resolution) is achieved in two dimensions within the plane of the light sheet, most light-sheet systems exhibit anisotropic resolution in which resolution in markedly worse in the direction axial to the light sheet (due to the thick beam waist of the low-NA light sheet). This is similar to conventional slide-based histology, in which paraffin-embedded tissue sections are generally cut at a thickness of 4 to 5 microns, but are imaged in the orthogonal direction with micron- to sub-micron scale resolution. However, with 3D imaging, isotropic resolution can potentially be of importance and several strategies have been developed to achieve this with light-sheet microscopy. A popular approach has been to rapidly swap the imaging and collection paths of a light-sheet microscope such that two volumetric datasets of the specimen are ultimately obtained, but with different low-resolution axes. A “fusion” de-convolution algorithm can then be applied to generate a 3D image with isotropic resolution, albeit with large demands for alignment, calibration, and computational processing 117,132,133. A more-direct approach for achieving isotropic resolution employs axial scanning of a high-NA light sheet (i.e. one in which the thickness of the beam waist matches the lateral resolution of the collection path) 127,134. Synchronizing the beam waist with the “rolling shutter” of a sCMOS camera ensures that an image is generated in a line-by-line fashion from signal generated at the beam waist (isotropic resolution) as it is scanned. Specially engineered illumination strategies, such as “propagation-invariant beams,” have also been employed to achieve isotropic resolution without sacrificing depth of focus, albeit with some trade-offs in terms of image contrast and both hardware and computational complexity 114,135,136. For clinical diagnostic applications of 3D pathology, the value of achieving isotropic resolution is currently unclear and studies are especially needed to determine its value for computer-assisted analysis of datasets.
2.6. Data-handling challenges
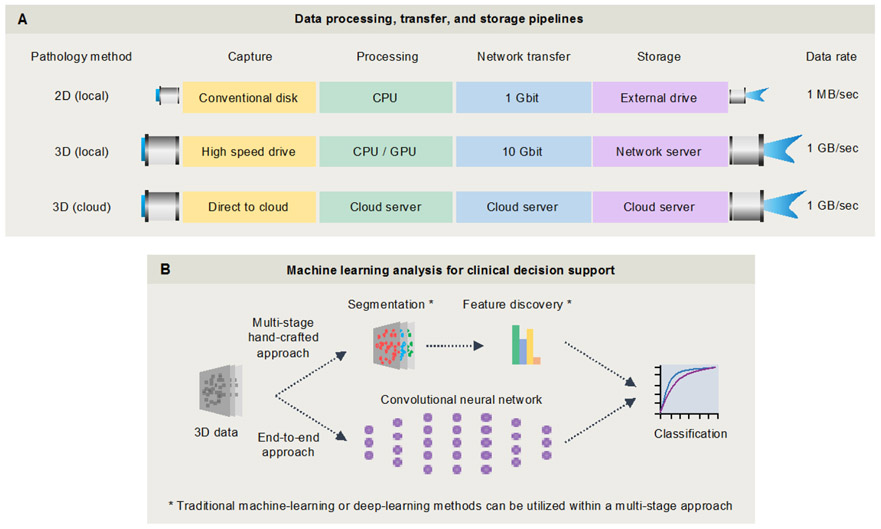
For all of the previously mentioned 3D microscopy techniques, digital reconstruction of large 3D images requires stitching and fusing of large numbers of 2D image tiles. Once generated, these 3D datasets are often on the terabyte (TB) scale, where challenges exist in terms of storage, low-loss compression, quality control, and visualization. In order to streamline progress in the years to come, standard open-source informatics tools and annotated datasets should ideally be established while this field is within a nascent but logarithmic stage of growth. As shown in Fig. 4, 3D microscopy generates vastly more data compared to conventional 2D microscopy approaches. However, data-generation speeds (~800 MB/sec) are similar to current whole-slide imaging (WSI) devices since they utilize similar sCMOS camera technologies – this should facilitate adoption by institutions that are already accommodating a digital pathology workflow. Figure 4 outlines various strategies for processing raw data from 3D microscope systems (e.g. local vs. cloud-based pipelines), as well as general strategies for machine-learning-based analysis of these massive feature-rich datasets.
Figure 4. Example data-processing and image-analysis workflows.
(A) In contrast to conventional microscopes, light-sheet microscopes acquire data at up to 1 GB/sec and require specialized hardware, such as a local 10 Gbit networked server, or a cloud-based storage and analysis solution. (B) Machine-learning tools will be necessary to assist with the analysis of large 3D pathology datasets for clinical decision support. Strategies include a multi-stage “hand-crafted-feature”-based approach, in which intuitive and well-understood microstructures are segmented and quantified as inputs for prognostic and predictive classifiers. Alternatively, an “end-to-end” approach can be used for direct classification based on raw 3D pathology images through a deep-learning model. Note that deep-learning techniques can also be utilized for certain steps within the multi-stage hand-crafted approach, for example to assist with segmentation tasks as described in section 3.2. These topics will be further examined in sections 3 and 4.
2.6.1. 3D image stitching.
A key step in the acquisition pipeline for 3D microscopy/pathology is efficiently assembling large numbers of 2D image tiles into seamless volumetric datasets. A number of commercial software packages, such as Imaris Stitcher and Volocity, have been developed to address this challenge, as well as popular open-source tools, such as TeraStitcher 137,138 and BigStitcher 139. Some software tools, such as BigStitcher 139, are designed to correct for deformation and registration artifacts through affine transformations, including chromatic shifts between wavelength channels. Such operations can be computationally expensive, and should ideally be tailored for specific imaging methods, and parallelized for maximum throughput. For example, for the open-source community, efforts are underway to upgrade the popular hierarchical (multi-resolution) HDF5 file format with a similar format, N5, which will allow for parallel writing (https://github.com/saalfeldlab/n5). For the clinical implementation of 3D pathology, quality-control algorithms will ultimately be needed to ensure the performance of these, and other, image-processing tasks (see section 3.3).
2.6.2. Compression of 3D imaging data.
The detectors of choice for camera-based 3D microscopy approaches, including light-sheet microscopy, are currently 16-bit sCMOS cameras that generate ~800 MB of data per second. Having a large 16-bit dynamic range has practical benefits for avoiding detector saturation when imaging bright tissue regions while also ensuring that bit noise (i.e. digitization noise) is not an issue when imaging dim tissue regions. However, the signal-to-noise ratio (SNR) at any tissue region is generally much lower than 16 bits, due to a combination of detector noise and/or shot noise (signal- and background-induced). This means that a significant level of “lossless” compression can be achieved by windowing the dynamic range of the data to remove noise (at the low end) and unused pixel well capacity (at the high end). For example, unlike lossy compression schemes like JPEG, which down-sample the data in the Fourier or wavelet space, a recently developed B3D compression scheme estimates the noise level of every pixel within an image (based on neighboring pixels) and limits the compression such that pixel deviations remain within that noise level 140. With B3D, fully “lossless” compression can be routinely achieved with a ~10X reduction in file size for the 16-bit imaging data generated by sCMOS-camera-based light-sheet microscopy systems. In terms of speed, B3D is built on the CUDA framework to enable GPU-based compression of imaging data at high speeds (> 1 GB/sec), which surpasses the data rate of standard sCMOS cameras 140. Depending upon the image-analysis task at hand, the use of more advanced 3D compression methods that factor in the inherent redundancy of a 3D dataset could yield significant data-compression results. For optical microscopy, most compression work has been in 2D, even when applied to 3D, as each slice is considered independently. However, other methods such as Fourier or wavelet-based compression are likely to provide superior compression results for 3D datasets with acceptable tradeoffs 141. Additional studies are needed to examine the effects of these various compression schemes on both manual and computerized image-analysis routines.
2.6.3. Visualization.
Human observers may choose to visualize 3D pathology datasets as volume renderings or as a series of 2D cross-sectional views, depending upon the diagnostic problem of interest and the complexity/density of the image. In some cases, it may be necessary to segment (i.e. extract) a subset of tissue structures using machine-learning techniques (see section 3) in order to visualize the 3D structure in an informative way. For example, the ability to visualize a 3D mesh model of a vascular or glandular branching-tree network could yield key insights including tortuosity, branching, and other features that are not easily inferred from 2D images. On the other hand, it is also helpful for pathologists to visualize 2D cross sections of the same vessels or glands within the context of the surrounding cellular and stromal milieu, as is the case with standard H&E histology. In particular, since pathologists are currently trained to interpret 2D cross sections, and a vast body of pathology literature exists for characterizing diseases with 2D images, it is desirable to have both 2D and 3D visualizations. Note that the amount of time required to review large volumes of 3D data is often impractical. For example, in our experience, a prostate biopsy, which can be evaluated in 2D within a few minutes, can require 15-20 minutes for a thorough 3D evaluation 22. Thus, in many cases, the full 3D dataset may only be necessary and practical to visualize in the case of diagnostic ambiguity, where it will have the largest clinical impact. In fact, early clinical implementations of 3D pathology may be as an adjunct to standard 2D pathology in situations when diagnostic ambiguity arise or critical treatment decisions must be made, for example for Gleason 6 – 7 prostate cancer patients needing to decide between active surveillance, surgery, or radiation. In summary, a variety of visualization techniques will likely need to be developed and standardized in order for pathologists to extract maximal utility from 3D pathology datasets (see section 4.4). A number of examples are shown in Fig. 5 of volume-rendered vs. cross-sectional views of 3D pathology.
Figure 5. Examples of nondestructive 3D pathology of clinical specimens.
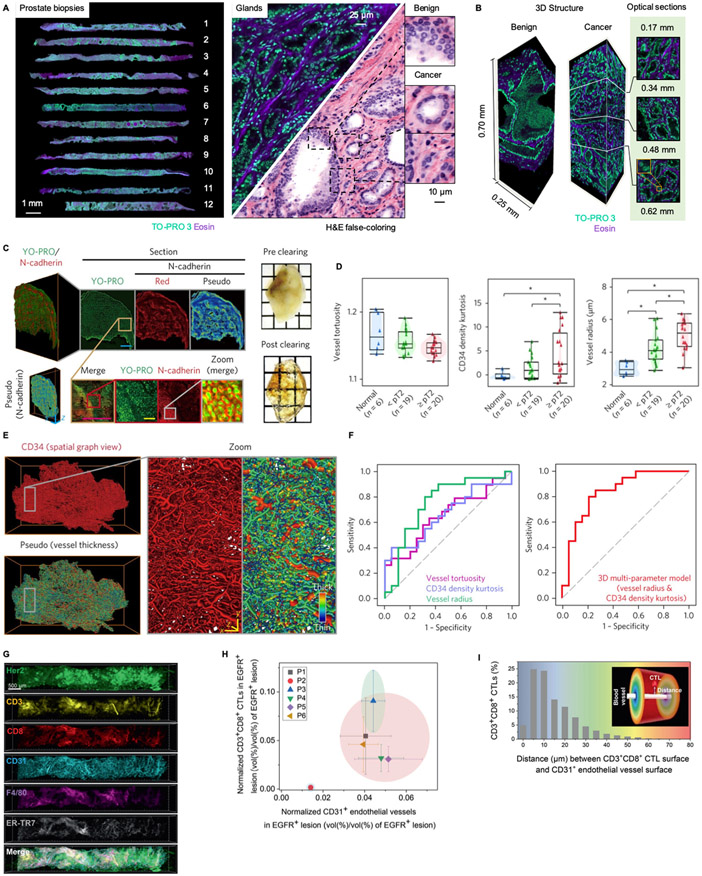
(A) 12 core-needle biopsies from the prostate of a single patient are imaged comprehensively in 3D with an open-top light-sheet (OTLS) microscopy system 123. A fluorescent analog of H&E staining is used to label the specimen, and is false-colored to mimic the appearance of standard H&E histology. (B) Benign and malignant glands are easily identified, with signification variations in appearance as a function of depth, which suggests that 3D pathology may improve diagnosis and grading of prostate carcinoma 21,22. (C) A bladder cancer specimen (FFPE) is deparaffinized, cleared, fluorescently labeled for nuclei and N-cadherin, and then imaged with light-sheet microscopy 43. Scale bars: 80 μm (yellow) and 1,600 μm (cyan). (D) A number of vascular features (tortuosity, kurtosis, and density) are plotted for 45 bladder specimens (human), showing significant differences between normal patients, those with non-muscle-invasive tumor (<pT2) and those with muscle-invasive tumor (>pT2). These quantitative vascular features were obtained after segmenting out the vessel network (E). Scale bars: 80 μm. (F) An ROC analysis was performed for the ability to detect muscle-invasive vs. noninvasive tumor, showing that 3D vascular features outperform 2D features, and that combining all 3D features yields the best performance. (G) Multiplexed 3D immunofluorescence imaging, with confocal microscopy, of intact core-needle biopsies of cancer 45. Scale bar at the top left: 500 μm. (H) Normalized densities of CD3+CD8+ cytotoxic lymphocytes (CTLs), and CD31+ microvasculature in EGFR+ parenchyma, are used to cluster human tumors into inflamed and noninflamed phenotypes. (I) 3D spatial distance mapping of an inflamed patient sample reveals that over 54% of CD3+CD8+ CTLs are located within 10 μm from microvessels.
The ability to render 3D datasets with color palettes that mimic conventional slide-based H&E histology and IHC will likely be important for pathologists to trust and adopt 3D pathology methods in the near future, as well as to validate 3D pathology datasets. While fluorescence images from light-sheet and other forms of 3D microscopy are typically acquired using monochrome cameras (with the appropriate filters in place), it is possible to false-color the datasets to mimic the appearance of standard chromogenic stains that are visualized by standard bright-field pathology microscopes. For example, it is relatively straightforward to utilize the formulae for Beer-Lambert-law absorption of light to convert two-channel images of tissues (labeled with a fluorescent analog of H&E) into H&E-like representations 142. Likewise, 3D immunofluorescence images can be rendered to mimic conventional chromogenic IHC 123. This ability to create images that are familiar to pathologists is crucial for clinical adoption, so that pathologists can continue to rely upon existing disease-classification schemes, while also learning how to improve their diagnostic determinations with the added insights that 3D pathology offers.
2.6.4. Value of open-source tools for data processing, visualization, and analysis.
A key element to utilizing and deploying any imaging method is to identify the optimal informatics and computational workflows. In the life cycle of all image datasets, be it research or clinical, software is used to acquire, visualize, analyze and disseminate results. Currently, in the research imaging world, there are a number of both open- and closed-source software tools. Popular open-source platforms include the Fiji / ImageJ environment 143-145 and related applications such as MicroManager146, BigDataViewer / BigStitcher 139,147, and Napari (https://github.com/napari/napari). Closed-source analysis platforms for 3D microscopy data include Imaris, Amira, Arivis, and Aivia. There is a growing movement in the imaging field advocating for the value and need for open-source workflows to ensure reproducibility, transparency, and broad dissemination 148-151. Since software tools undergo continuous developments over rapid timelines, and obtaining patent protection for software is difficult, many commercial entities have chosen open-source platforms, especially when used to support other technologies and devices that enjoy a stronger IP position. The need for accuracy, transparency and reproducibility is particularly relevant in 3D pathology due its emerging nature, and the large diversity of analysis parameters and metrics that will be developed. Future tools are likely to be developed by many independent researchers and companies with access to well-curated cloud-hosted 3D pathology datasets, in which a common open-source software platform could accelerate progress and standardization.
3. Challenges / opportunities for artificial intelligence in 3D pathology.
3.1. Overview
Recent adoption of whole slide imaging (WSI) scanners by multiple hospitals and health care institutions, which have started digitizing their entire pathology workflows, complemented by rapid increases in computational power, have led to the proliferation of digital pathology approaches in oncology and other areas 20,152-154. The moniker of digital pathology has now become associated with artificial intelligence (AI), including machine-learning approaches to quantitatively examine whole-slide images to address clinical challenges in early detection, diagnosis, prognosis and treatment response. While AI approaches have so far been developed mostly for analysis of 2D pathology images, the advent of 3D pathology techniques has provoked interest in 3D AI and feature-learning approaches. High-quality comprehensive 3D representations of tissue micro-architecture over large regions of interest offer a potential opportunity for AI analyses, due to the large amounts of data that can be generated from each patient specimen in a nondestructive fashion.
AI approaches in oncology primarily aim to develop a machine classifier for clinical decision support, such as identifying patients with aggressive disease who would benefit from aggressive therapies such as chemotherapy, or identifying patients who are likely to respond to specific forms of therapy such as immune-checkpoint blockade. Two commonly used classification approaches include “end-to-end” and “multi-stage” feature-extraction (i.e. “hand-crafted”) approaches 155. The first strategy involves training a deep-learning model to directly classify a lesion/patient based on the imaging data. Such strategies have been shown to be extremely powerful and accurate, but suffer from lack of interpretability in many cases and the need for very large numbers of well-curated patient datasets in order to train a reliable algorithm. Advances are being made on data-efficient techniques as applied to digital pathology, such as multiple instance learning, semi-supervised learning, and transfer learning approaches 156-158. However, the variance on account of pre-analytic factors in the context of pathology often makes it difficult to create generalizable algorithms without sufficiently large and diverse datasets. In addition to being sensitive to minute and often-imperceptible variations in image quality, it is often difficult to spot errors when they occur, or to determine the source of the errors 159. Alternatively, traditional or deep-learning-based image-processing methods can also be used to segment out tissue structures that are already well-known and trusted by pathologists (e.g. cells, glands, collagen, etc.), from which quantitative “hand-crafted features” can be extracted (e.g. density, tortuosity, fractal dimension, angular disorder, etc.). These quantitative histomorphometric features can then be used in a multi-stage approach to train a clinical classifier. An advantage of such an approach is that domain experts (i.e. pathologists) can verify the accuracy of the intermediate segmentation steps of this process, which engenders trust and enables error-checking. Another advantage of a “hand-crafted” approach is that each digital pathology dataset typically contains large numbers of morphological exemplars (i.e. features) that can contribute to the training of an algorithm. Therefore, for low-level tasks (e.g. segmentation of well-conserved micro-architectural features), a modest number of patient specimens or training sets is often sufficient 159-161. The caveat with these approaches, however, is that they require a greater amount of domain-specific information for model training and might be more challenging to train compared to end-to-end approaches. Finally, hybrid approaches are possible, such as using hand-crafted features as intermediate operators within a DL network, and/or in conjunction with DL-derived data-driven features, to develop an optimal classifier 162,163.
3.2. Unique challenges for AI in 3D pathology
While there has been a sharp growth in the use of deep learning (DL)-based approaches for prognostic and predictive classification with digital pathology data, these have been largely confined to 2D pathology images. In training these networks, the images are typically broken up into smaller 2D patches and introduced to a DL network for training. The challenge with 3D pathology images is that traditional 2D networks are not equipped to handle large 3D datasets. This will necessitate 3D CNNs, which have been previously used in the context of 3D radiographic images 164, albeit at much smaller file sizes (low-resolution radiography).
Beyond the computational expense of training 3D CNNs, an additional challenge is definition of the training set and annotating regions or targets of interest. The need for manual annotation of structures and primitives in 3D by a domain expert (i.e. pathologist) will necessitate efficient and user-friendly interactive software tools. These tools will need to be flexible enough to allow the user to navigate through the 3D volume and refine and update annotations of primitives and regions of interest. While tools like Sedeen (Pathcore Inc.)165 and QuPath166 allow for object annotation of 2D pathology images, these toolkits will require substantial modifications to accommodate the annotation of structures in large 3D pathology datasets. Alternatively, in order to bypass the requirements for laborious manual 3D annotations by domain experts, which is somewhat subjective, certain features can be labeled via molecular biomarkers such that traditional intensity- and morphology-based segmentation methods can be employed. Examples include keratin 8 (KRT8) to identify the luminal epithelial cells that line all prostate glands, and CD31 (PECAM-1) to highlight the lymphovasculature. While this molecular-labeling approach is objective and does not rely on manual annotations, the downside is that antibody-based labeling of thick 3D tissues can be very slow and expensive. However, if a DL model can be trained to predict the appearance of 3D immunofluorescence images based on images generated with inexpensive and fast small-molecule fluorophores (e.g. an H&E analog), or even without the use of labels 160,161,167, it could be possible to develop an annotation-free segmentation algorithm based on “synthetic” immunolabeling of specific features. Such a method would be objective (no human annotations), fast (small-molecule labeling of thick tissues), and inexpensive (antibody-free).
3.3. The need for quality control for AI analysis
In addition to data size, one of the most important factors that influences performance of AI approaches in digital pathology is the inherent quality of the data fed into the algorithm. AI analysis of 2D pathology is often limited by pre-analytic sources of variations, such as with staining quality (e.g. hue, saturation, intensity), sectioning artifacts, out-of-focus regions, and other subtle variations due to the different scanners and scanning parameters used 20,168. While the sources of image variability are different for nondestructive 3D pathology approaches, there is a similar need to control such variations when it comes to implementing AI approaches. For example, factors that can influence the quality of 3D pathology include the degree of fixation, cold and warm ischemia times, deparaffinization quality (if FFPE blocks are being used), variations in staining and clearing protocols, fluctuations in laser intensities, optical alignment, and software post-processing routines (e.g. section 2.6.1). Note that with 3D microscopy approaches, images are always “in focus” provided the alignment of the microscope is maintained. Image quality is therefore most influenced by the many tissue-preparation issues mentioned above. As with slide-based histology, the automation of these various steps is of value for reducing variability, but quality-control metrics are still needed to ensure that 3D pathology datasets are of adequate quality to ensure reliable AI-based results.
Recent work has involved building automated and semi-automated quality-control methods for 2D whole-slide images, which can be translated to 3D pathology. These routines aim to automatically detect variations in color, staining, and common artifacts present in histology images. One such tool is HistoQC 169, which interrogates digital tissue slide images and provides a quantitative score of the overall quality of the image. Additionally, the tool enables the precise identification of localized regions that have been compromised by artifacts such as cracks in the glass, hair shafts, tissue folds, and pen markings. Similar tools will need to be extended for quality control of 3D pathology images, in which image artifacts include stitching defects and regions of poor staining/clearing.
Ultimately, some level of variation in image quality is unavoidable for both 2D and 3D pathology, regardless of the degree of automation and standardization of processes. As mentioned in the previous section, a hand-crafted feature-based AI approach towards image interpretation offers potential advantages. First, histomorphometric features can be identified that are relatively insensitive to image-quality variations, as shown in a recent 2D pathology example, where a sensitivity analysis was used to identify a subset of quantitative features that could be integrated into robust clinical classifiers 170. Second, segmentation algorithms (for extraction of quantitative features) can be more-easily trained with diverse datasets such that they are robust against image-quality variations. While this is also possible with an end-to-end classification approach if sufficient numbers of patient datasets are available for training, the use of a hand-crafted approach allows pathologists to visually inspect the results at a critical intermediate step (feature segmentation) and to rely on smaller patient cohorts in which each 3D pathology dataset often contains hundreds to thousands of diverse “exemplars” to accurately train algorithms to segment well-conserved low-level structures (e.g. glands, collagen, broad classes of cells).
4. Translational challenges and clinical acceptance
4.1. Clinical studies with archived tissues
In contrast to clinical trials involving therapeutic agents, where new patients must be recruited and followed for response, new technologies in pathology can often be validated using already-established slide repositories or tissue biobanks with detailed follow-up data. Examples include The Cancer Genome Atlas 171, the Prostate Cancer Biorepository Network (http://prostatebiorepository.org/), and many more. These biobanks permit the use of a “prospective-retrospective” study design. For instance, the Oncotype Dx Recurrence Score (Genomic Health), a 21-gene expression assay using FFPE tissue, was validated using archived specimens from NSABP trials B14 and B20 172,173. This randomized clinical trial was completed more than a decade before the validation of Oncotype Dx, enabling the use of 10-year recurrence-free survival as the primary endpoint, thus reinforcing the value of well-characterized archival tissue in translating promising diagnostic technologies to the clinic 174.
Banked tissue is not without its problems. Pre-analytic factors, including formalin fixation time, cold ischemia time, and freezing methodology, all affect the quality of tissue stored in a biobank 175. If the effects of these pre-analytic factors are severe, the assay can be negatively impacted, or samples/patients must be removed from the study, introducing bias. An additional concern with the use of archival tissue is that standard-of-care treatments for the target population could have changed over time, which could potentially confound studies in which the primary endpoints are clinical outcomes rather than biologic measurements.
The gold standard for validation of a diagnostic test is a prospective, randomized trial that is evaluated in a well-defined target population, where evaluation of the diagnostic test is the primary purpose of the trial 176. This type of study design minimizes bias, and is considered the highest level of evidence - LOE 1. To further extend the example of Oncotype Dx Recurrence Score, the assay was further validated by a prospective, randomized trial (TAILORx), which reinforced the value of the test 177. The cumulative body of evidence, including prospective-retrospective studies and prospective, randomized studies, led to the categorization of Oncotype Dx Recurrence Score as the only “preferred” test in the 2018 NCCN breast cancer treatment guidelines for a specific subset of breast cancer patients facing a chemotherapy decision, which is the highest level of clinical acceptance.
4.2. Regulatory strategies
Regulatory approval is a challenging issue that requires close collaboration with relevant regulatory stakeholders (e.g. the FDA in the United States) to develop appropriate strategies for various implementations of 3D pathology. In this section, we provide a high-level overview of potential regulatory scenarios, along with examples from devices and diagnostics that are currently in the market. There are two primary regulatory pathways for diagnostic tests in pathology – FDA approval and laboratory developed tests (LDTs) 178.
Diagnostic tests and kits are classified as medical devices by the FDA, and are subject to the same regulatory processes as other such devices. Medical devices are categorized according to their perceived level risk, from Class I to Class III. Class III devices are of highest risk, including implanted devices and those that are used to sustain life. Class II devices are considered moderate to high risk, and have a predicate device that can be used for comparison. A whole-slide scanner for 2D digital pathology, a technology that was first approved by the FDA in 2017 after an extensive clinical study, is an example of a Class II device 179,180. Class I devices are the lowest risk devices, which include the analog light microscopes used within pathology laboratories. A recent ex vivo tissue microscopy device, the Caliber I.D. Vivascope 2500, has been classified as a Class I device.
An alternative regulatory pathway for many diagnostics tests, if provided as a “medical service” by a single laboratory, is the LDT pathway. Tests designated as LDTs must be performed in a CLIA-certified laboratory with appropriate analytic and clinical validation documentation. The FDA does not currently regulate LDTs, but did express its intentions to issue an oversight framework for LDTs in recent years 181,182.
The regulatory strategy for 3D pathology will be dependent on the paradigm by which the technology is disseminated. If the technology is developed as a device that is marketed to pathology laboratories, FDA approval will be necessary for use of the device, and potentially for decision-support algorithms based on the device. If individual laboratories develop “homebrew” 3D pathology tests, the LDT pathway could be a viable option. Regardless of the regulatory pathway for 3D pathology, a strong base of evidence proving clinical utility will be crucial for adoption.
4.3. Financial and workflow (process time) considerations for clinical integration
The workflows for conventional pathology and 3D pathology are similar for the accessioning, grossing, and initial tissue-processing steps (i.e. fixation and dehydration). The differences begin after the tissue is embedded in paraffin (conventional pathology) or placed in the clearing solution. In the conventional pathology workflow, the tissue must be physically sectioned (by hand), placed on a glass slide, stained, cover-slipped, and scanned using a whole-slide imager to produce a digital 2D dataset. Using a set of 12 prostate biopsies as an example, this process requires ~40 minutes of hands-on histotechnologist time, ~4 hours of total time, and ~$300,000 in capital equipment costs. In the 3D pathology workflow, automated tissue scanning using light-sheet microscopy, with similar resolution to what is provided by a 10X – 20X objective (NA ~ 0.4) in a standard pathology microscope, requires ~30 minutes (10% of sample digitized) to ~4 hours (100% of sample digitized) 123. Since manual sectioning is not required, the 3D pathology workflow can be entirely automated, which means it is not dependent on labor shift timing or histotechnologist availability. The capital equipment costs are likely to be similar, although no FDA-approved devices are currently available for 3D pathology. Thus, the 3D pathology workflow can reduce the labor burden on histotechnologists while nondestructively generating orders-of-magnitude more data than a whole-slide imager.
4.4. A staged approach for clinical translation
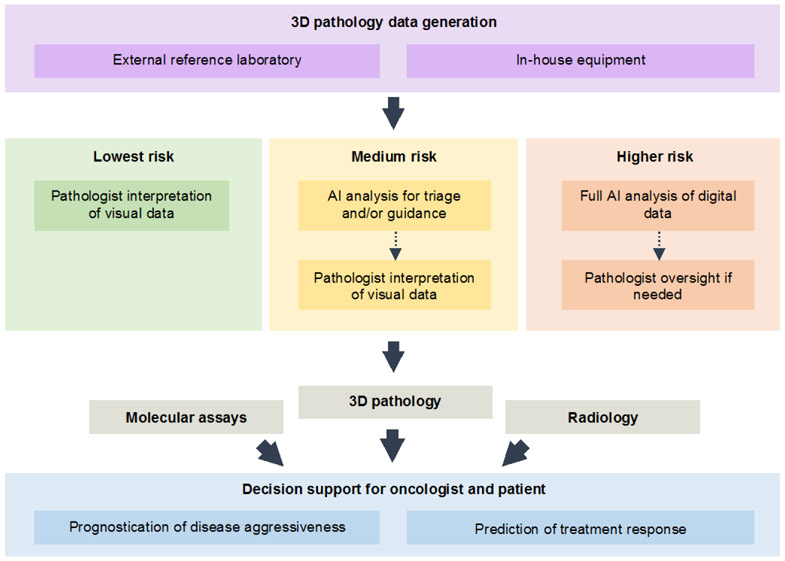
While a fully-automated diagnostic and decision-support workflow could become a reality, the integration of AI will likely occur in a step-wise fashion (Fig. 6). For integration of 3D pathology within clinics, at least three stages can be foreseen. The first stage, which requires the most time and human effort, is also the least risky, in which a pathologist directly interacts with 3D pathology datasets. Datasets would be rendered to mimic the current standards of H&E histology and IHC, and would be viewed much like how 2D whole-slide images are currently viewed, albeit with the ability to scroll through the depth of a specimen. Early studies have shown anecdotally that such a strategy could be highly informative, such as for preventing the over-grading (and over-treatment) of disease 21 and for identifying regions of malignancy that could be missed or misinterpreted with slide-based 2D histology 22. Likewise, the sampling limitations and ambiguity of 2D histology are known to lead to the misdiagnosis and under-grading of certain patients 183-185, which presents an opportunity for 3D pathology to provide significant improvements. Since 3D assessments of tissues (entire biopsies, as an example) are more time-consuming than 2D assessments of sparse 2D sections, a second stage of AI integration would be to automatically triage specimens that are diagnostically unequivocal, and to provide pathologists with only cases that are ambiguous. For example, a recent study, based on 2D digital pathology images, showed that it may be possible to triage up to 75% of cancer cases based on computational analysis, while retaining 100% detection sensitivity 186. Finally, as clinical studies prove their effectiveness, fully-automated computational 3D pathology workflows will emerge. A challenge to achieving these goals will be to integrate 3D pathology seamlessly into laboratory information management systems (LIMS) such that various sources of patient data can be integrated both for clinician- and AI-based treatment decisions.
Figure 6. Staged approach for translation of 3D pathology into clinical practice.
3D pathology datasets, generated by reference labs or in-house pathology labs, may initially provide additional visual information for pathologists as they seek to improve their diagnostic determinations. Early incorporation of AI analysis will likely be for triaging unequivocal cases, in order to reduce pathologist workloads, and to guide their efforts towards regions of ambiguity and/or diagnostic importance. As AI algorithms are increasingly validated and trusted by clinicians, they may eventually be utilized for fully automated analysis of 3D pathology datasets, with pathologist oversight if necessary. The vision for 3D pathology is to provide clinical decision support (prognostication and prediction) to guide treatment decisions, likely in conjunction with other molecular and imaging assays.
5. Future directions
5.1. Integration with lab medicine / molecular assays
It has become increasingly clear that precision medicine will benefit from adopting a multiplexed multi-omics approach that combines diverse diagnostic technologies for patient stratification and clinical decision support. In particular, the complexities of the tumor-immune microenvironment, and various factors that contribute to an individual’s “cancer-immune setpoint,” is becoming well-appreciated by the multitudes working to improve the efficacy and precision delivery of recent immuno-oncology approaches 187. For example, various factors that correlate with response to immune-checkpoint blockade include tumor mutational burden (TMB) 188, microbiome profiles 189,190, patterns of biomarker expression (e.g. PD-1 / PD-L1, CTLA, chemokine receptors, etc.) 14,191,192, and the spatial distribution of tumor and immune cells (e.g. tumor infiltrating lymphocytes) 193.
In order to facilitate the integration of 3D pathology with other clinical assays such as DNA/RNA sequencing, a first step is to ensure that 3D pathology methods do not interfere with standard laboratory methods. Recent studies have indicated that certain clearing and fluorescence labeling methods are relatively gentle, and often utilize reagents that are already part of the standard histology workflow (e.g. xylene, ethanol, etc.). Once tissues have been formalin fixed per standard clinical practice, most clearing reagents are less harsh than the initial fixation step, and standard FFPE processing and histology assays can still be performed (see section 2.1). Furthermore, as described in section 2.1, the advent of nondestructive 3D pathology offers the opportunity to modernize a pathology workflow that dates back nearly a century, for example by developing formalin-free tissue-preservation and labeling/clearing protocols that can maintain RNA integrity.
3D analogs to existing pathology practices, such as laser-capture micro-dissection and manual slide-based macro-dissection of tissues, would also be of value. Such tissue-enrichment techniques could generate orders of magnitude more material for downstream assays than slide-based tissue shavings and laser-captured regions, which could result in a significant leap in sensitivity and accuracy for detecting rare mutations. For rare cell types, 3D micro-aspiration of individual cells or cell contents within thick specimens is also a possibility, as previously demonstrated in the context of neuroscience investigations 194-196. The ability to extract lysates from tens to hundreds of individual rare cells, such as aggressive tumor cells exhibiting lymphovascular invasion, followed by low-input sequencing of those lysates, could enable the discovery of new biomarkers of aggressive disease (for prognostication), druggable targets, and mechanistic insights.
5.2. Radiomics as a model and precedence for the value of 3D imaging
Radiomics or the quantitative interrogation and subsequent mining of pixel-level, sub-visual data from standard medical imaging methods (ultrasound, CT, PET, MRI) has recently been shown to be a non-invasive method to answer clinically relevant questions pertaining to diagnosis, prognosis and treatment response across the oncology spectrum 197,198. Radiomics has traditionally relied on analyzing multiple 2D images across a region of interest, with interpolation being used to extend the analysis in 3D. 3D pathology resembles radiomics, but differs in a few key aspects. Unlike a conventional CT slice which is 512x512 pixels, a whole slide image at 20x magnification is often 40,000 x 40,000 pixels, or 4 orders of magnitude larger. 3D pathology can add an additional 4 orders of magnitude, which introduces a big-data problem but also an opportunity since it provides abundant fuel for data-hungry AI approaches. In addition, unlike reconstructed radiographic images, 3D pathology images contain large numbers of highly-repetitive micro-scale features, such as cells, glands and stromal structures, which can allow for efficient training and use of AI methods (both end-to-end and multi-stage hand-crafted approaches, as discussed in section 3.1). As one example, radiomic and pathomic features from 2D pathology images were used to create a unified predictor of early recurrence in early stage lung cancers 199.
Pathology is often utilized to validate or follow up on an observation that is first seen with radiology (e.g. mammography leading to breast biopsy). However, except for a few specialized multiscale animal studies 200,201, there have been few efforts to gain a multiscale spatial understanding of tissue composition in disease. Modern 3D pathology, when deployed with the type of computational tools that radiography has pioneered, could provide a cell-to-organ view not possible with any other currently available clinical imaging strategy. This need for both microscopic and “mesoscopic” imaging (over larger fields of view) is starting to be appreciated by researchers 202. As an example, light-sheet microscopes have now been developed for highly diverse applications ranging from single-cell investigations with advanced high-resolution systems to large-volume studies of intact organs (e.g. mouse brains) 126,203-205.
5.3. Co-registration of multi-scale imaging data (pathology and radiology)
Extensive research has been carried out to co-register conventional 2D pathology with medical images for improved characterization of diseases over a large range of spatial scales 206-209. A driving motivation has been to use the gold standard method of pathology to elucidate changes seen at more macroscopic tissue scales with standard medical imaging methods 210. For example, in current clinical practice, radiology techniques are often used for the early detection of various diseases, where histopathology is subsequently relied upon to provide diagnostic, prognostic, and predictive insights. Pathology and radiology approaches are also often used for cross validation. For example, numerous recent studies have correlated PET imaging results with histologically determined metrics such as proliferative index (Ki-67 expression), micro-vessel density, and immune phenotype (e.g. CD8 expression) 211-213. However, these correlative studies would benefit from increased standardization between medical imaging and histopathology. Prior efforts to standardize the co-registration of radiology and pathology approaches include placing fiducial markers to guide ex vivo mold-based tissue slicing 214. 3D reconstructions of 2D pathology data have also been generated to improve registration accuracy 207. The emergence of nondestructive 3D pathology, which can enable visualization of large tissue volumes, has the potential to greatly facilitate and improve co-registration with radiographic images in addition to improving genomics assays. In addition, intermediate imaging modalities such as preclinical micro-CT/MRI/PET, may be helpful to bridge the large gap in spatial resolution and volumetric field between microscopy and whole-body imaging techniques 202.
5.4. Opportunity to study and mitigate health disparities across populations
Recent work has suggested that 2D computational pathology features of cancer from H&E-stained whole slide images exhibit population-level differences. In particular, Bhargava et al 215 showed that stromal features of prostate cancer differed significantly between African American (AA) and Caucasian American (CA) men, in which a computational prognostic model trained with these stromal features was strongly associated with risk of recurrence in two validation datasets of AA men. Interestingly, this prognostic model for prostate cancer, which was trained with data from AA men alone, was nearly twice as accurate compared to a model trained with a combination of AA and CA men. Considering that significant morphometric differences have been identified in the stroma between AA and CA men with prostate cancer, there is optimism that the computational interrogation of 3D stromal and epithelial morphology can further improve the treatment of disparate populations. This includes those who have traditionally been underserved, such as AA men with prostate cancer, whose mortality rate is nearly 2.5 times higher than CA men 216.
5.5. Developing holistic decision-support algorithms (imaging, sequencing, health records, etc.)
Recent research has expanded from interrogating a single source of information (e.g. radiology, pathology, genomics, metabolomics) towards multi-modality approaches to improve prognostication and prediction. This is especially attractive when it comes to AI-based methods that thrive on large amounts of orthogonal and complementary data. AI approaches integrating these multiple data types have already been used in cancers of the lung 199,217, breast 218,219, brain 220, and prostate 221, as well as for cardiovascular 222 and neurological diseases 223. These prior examples foreshadow the use of holistic decision-support systems (Fig. 6) that combine data from multiple diagnostic modalities and patient records to build accurate models for diagnosis, prognosis and prediction of therapeutic response.
6. Summary and outlook
Unlike radiology, in which many of the imaging technologies have been developed and refined over the last half-century, the field of pathology is rooted in over a century of tradition. This extensive history behind current pathology practices will make change difficult. However, the fact that pathology is still regarded as the “gold standard” for clinical diagnosis attests to the wealth of insight that microscopy offers, even when obtained from small numbers of thin 2D sections. The recent FDA approval of digital pathology solutions indicates that pathology is entering into a phase of modernization and change that will likely evolve over the next half century. In many ways, this recent advent of 2D slide-based digital pathology will pave the way for nondestructive 3D pathology, for example by establishing the clinical IT infrastructure and resources needed to support a digital 3D pathology workflow. An optimistic viewpoint is that nondestructive 3D pathology would represent the ultimate fulfillment of the vision for digital pathology that is currently being pursued with 2D whole-slide imaging (WSI). For example, a significant factor in the success of digital X-ray imaging at the end of the 20th century was the economic benefits of replacing analog films with reusable X-ray panels 224. Likewise, nondestructive 3D pathology offers the potential to bypass glass slides, as well as associated tissue-sectioning processes, in favor of a simple digital approach that requires less human labor and consumables.
As with any diagnostic method, clinical value must be demonstrated through large-scale randomized studies. The ability to perform studies on 3D pathology with archived FFPE tissue specimens will facilitate and accelerate this process, but studies with prospective patient specimens will be of value as well. Convenience and cost savings for pathologists may not be sufficient to drive rapid adoption of 3D pathology, but studies showing clear advantages for improving clinical outcomes will cause oncologists and patients to demand these new services. As discussed in this article (Fig. 2), initial studies will likely focus on the ability of 3D pathology to improve disease prognostication and prediction of treatment response through: (1) the characterization of complex structures (e.g. vessels, glands, collagen); (2) the quantification of complex spatial relationships (e.g. the tumor-immune microenvironment); and (3) the analysis of rare cells that are challenging to identify on 2D sections. A more-concrete example of the first scenario would be to show that 3D characterization of glandular architecture in prostate biopsies provides superior classification of indolent vs. aggressive prostate cancers in comparison to traditional Gleason grading (2D pathology). For moderate-risk patients, this could help urologists and prostate oncologists to determine which patients to place on active surveillance (watchful waiting) versus curative treatments (surgery or radiation), the latter of which carries risks for significant side effects (e.g. incontinence and impotence). An example of the second scenario would be to show that 3D quantification of the tumor-immune microenvironment (e.g. lymphocytes, neutrophils, tumor cells expressing specific checkpoint receptors, etc.) within large volumes of biopsy or surgical tissue is superior to traditional 2D histology and IHC for predicting patient response to various immuno-oncology regimens, especially when combined with complementary assays such as genomics. Again, the goal is to assign the right patients (likely responders) to appropriate pharmacologic treatments while sparing others (including insurers) from the financial costs and side effects of ineffective treatments. Finally, an example of the third scenario would be to show that identification and quantification of rare events such as lymphovascular invasion (LVI) in large surgical specimens (e.g. prostatectomies or breast lumpectomies) is feasible with high-throughput 3D pathology methods – due to orders of magnitude increased sampling compared to traditional 2D pathology – and is of value for prognostication and guiding adjuvant therapies.
In this Perspective article, we have reviewed some of the key technologies that will underlie the emergence of nondestructive 3D pathology as a clinical diagnostic method in the decades to come. Rapid advances in high-throughput 3D microscopy, data-processing, and AI-based image interpretation are currently being coordinated, along with clinical-validation studies to demonstrate the value of this diagnostic paradigm. We have discussed how 3D pathology has the potential to form a bridge between anatomic pathology and other diagnostic disciplines such as genomics and radiology, all of which should ideally be integrated to support accurate clinical decision-making.
Acknowledgements
We acknowledge research grants from the Department of Defense (DoD) Prostate Cancer Research Program through: W81XWH-18-10358 (JTCL and LDT), W81XWH-18-PC180686 (NPR), and W81XWH-15-1-0558 (AM); DoD Breast Cancer Research Program W81XWH-19-1-0668 (AM); DoD Lung Cancer Research Program W81XWH-18-1-0440 (AM); DoD Peer Reviewed Cancer Research Program W81XWH-16-1-0329 (AM); Ohio Third Frontier Technology Validation Fund (AM); Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and The Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University (AM); Prostate Cancer Foundation (NPR); National Science Foundation 1934292 HDR: I-DIRSE-FW (JTCL); the NIH / NCI through: K99CA240681 (AKG), R01CA175391 (JTCL), R01CA199996 (KWE); U24CA199374 (AM), R01CA202752 (AM), R01CA208236 (AM), R01CA216579 (AM), R01CA220581 (AM), U01CA239055 (AM); the NIH / NIBIB through R43EB028736 (AM); the NIH / NIGMS through P41GM135019 (KWE); the NIH / NCRR through C06RR12463 (AM); US Dept. of Veterans Affairs IBX004121A (AM); Institute for Prostate Cancer Research (IPCR) at the University of Washington (LDT), NCI Breast Cancer SPORE / Safeway Foundation at the Fred Hutchinson Cancer Research Center (JTCL).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Competing interests
JTCL, AKG, NPR, and LDT are co-founders and shareholders of Lightspeed Microscopy Inc., of which JTCL and NPR are board members and NPR is the CEO. Technology developed by JTCL, AKG, NPR, and LDT at the University of Washington has been licensed by Lightspeed Microscopy Inc. KWE is a co-founder and shareholder of OnLume Inc. KWE is a scientific advisory consultant for Bruker Corporation. AM is an equity holder in Elucid Bioimaging and Inspirata Inc. AM has previously served as a scientific advisory board member for Inspirata Inc, AstraZeneca, Bristol Meyers-Squibb and Merck. AM has also had sponsored research agreements with Philips and Inspirata Inc. AM’s technology has been licensed to Elucid Bioimaging and Inspirata Inc. AM is involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
References
- 1.Carroll PR, et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw 14, 509–519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gradishar WJ, et al. NCCN Guidelines Insights Breast Cancer, Version 1.2016. J Natl Compr Canc Netw 13, 1475–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 3.McKenney JK, et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. in J Urol, Vol. 186 465–469 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Shah RB, et al. Diagnosis of Gleason pattern 5 prostate adenocarcinoma on core needle biopsy: an interobserver reproducibility study among urologic pathologists. in Am J Surg Pathol, Vol. 39 1242–1249 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Zhou M, et al. Diagnosis of "Poorly Formed Glands" Gleason Pattern 4 Prostatic Adenocarcinoma on Needle Biopsy: An Interobserver Reproducibility Study Among Urologic Pathologists With Recommendations. in Am J Surg Pathol, Vol. 39 1331–1339 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kweldam CF, et al. Gleason grade 4 prostate adenocarcinoma patterns: an interobserver agreement study among genitourinary pathologists. in Histopathology, Vol. 69 441–449 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Epstein JI, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. in Eur Urol, Vol. 69 428–435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch HG & Black WC Overdiagnosis in cancer. in J Natl Cancer Inst, Vol. 102 605–613 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Haffner MC, De Marzo AM, Yegnasubramanian S, Epstein JI & Carter HB Diagnostic challenges of clonal heterogeneity in prostate cancer. in J Clin Oncol, Vol. 33 e38–40 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Miller DC, et al. Prostate cancer severity among low income, uninsured men. J Urol 181, 579–583- discussion 583-574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahal BA, et al. The association between insurance status and prostate cancer outcomes: implications for the Affordable Care Act. Prostate cancer and prostatic diseases 17, 273–279 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Meyers DE, Bryan PM, Banerji S & Morris DG Targeting the PD-1/PD-L1 axis for the treatment of non-small-cell lung cancer. Current oncology (Toronto, Ont.) 25, e324–e334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hersom M & Jørgensen JT Companion and Complementary Diagnostics-Focus on PD-L1 Expression Assays for PD-1/PD-L1 Checkpoint Inhibitors in Non-Small Cell Lung Cancer. Therapeutic drug monitoring 40, 9–16 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Patel SP & Kurzrock R PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Molecular Cancer Therapeutics 14, 847–856 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Brunnström H, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod Pathol 30, 1411–1421 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Makhlouf H, et al. Toward Improving Practices for Submission of Diagnostic Tissue Blocks for National Cancer Institute Clinical Trials. Am J Clin Pathol 153, 149–155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson E, Levene MJ & Torres R Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. in Biomed Opt Express, Vol. 7 3089–3096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul D, Cowan AE, Ge S & Pachter JS Novel 3D analysis of Claudin-5 reveals significant endothelial heterogeneity among CNS microvessels. in Microvasc Res, Vol. 86 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres R, et al. Three-Dimensional Morphology by Multiphoton Microscopy with Clearing in a Model of Cisplatin-Induced CKD. in J Am Soc Nephrol, Vol. 27 1102–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bera K, Schalper KA, Rimm DL, Velcheti V & Madabhushi A Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nature Reviews Clinical Oncology, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser AK, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. biomed. eng 1, 0084–0010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reder NP, et al. Open-Top Light-Sheet Microscopy Image Atlas of Prostate Core Needle Biopsies. Arch Pathol Lab Med, arpa.2018-0466-OA (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DB, et al. Quantitative Spatial Profiling of PD-1/PD-L1 Interaction and HLA-DR/IDO-1 Predicts Improved Outcomes of Anti-PD-1 Therapies in Metastatic Melanoma. Clin Cancer Res 24, 5250–5260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kargl J, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun 8, 14381–14311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He G, et al. Peritumoural neutrophils negatively regulate adaptive immunity via the PD-L1/PD-1 signalling pathway in hepatocellular carcinoma. Journal of experimental & clinical cancer research : CR 34, 141–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y Spatial Heterogeneity in the Tumor Microenvironment. Cold Spring Harbor perspectives in medicine 6, a026583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masugi Y, et al. Characterization of spatial distribution of tumor-infiltrating CD8+ T cells refines their prognostic utility for pancreatic cancer survival. Mod Pathol 32, 1495–1507 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Heindl A, Nawaz S & Yuan Y Mapping spatial heterogeneity in the tumor microenvironment: a new era for digital pathology. Laboratory Investigation 95, 377–384 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Plaks V, Kong N & Werb Z The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell stem cell 16, 225–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo W, et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 453, 529–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM & Strasser A Is tumor growth sustained by rare cancer stem cells or dominant clones? Cancer Research 68, 4018–4021 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Cavé H, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer--Childhood Leukemia Cooperative Group. N Engl J Med 339, 591–598 (1998). [DOI] [PubMed] [Google Scholar]
- 33.van Dongen JJ, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 352, 1731–1738 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Herman CM, Wilcox GE, Kattan MW, Scardino PT & Wheeler TM Lymphovascular invasion as a predictor of disease progression in prostate cancer. Am J Surg Pathol 24, 859–863 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Mohammed RAA, et al. Improved methods of detection of lymphovascular invasion demonstrate that it is the predominant method of vascular invasion in breast cancer and has important clinical consequences. Am J Surg Pathol 31, 1825–1833 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Song YJ, et al. The role of lymphovascular invasion as a prognostic factor in patients with lymph node-positive operable invasive breast cancer. Journal of breast cancer 14, 198–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haffner MC, et al. Tracking the clonal origin of lethal prostate cancer. The Journal of clinical investigation 123, 4918–4922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pribluda A, de la Cruz CC & Jackson EL Intratumoral Heterogeneity: From Diversity Comes Resistance. Clin Cancer Res 21, 2916–2923 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Eyler CE & Rich JN Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol 26, 2839–2845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks MD, Burness ML & Wicha MS Therapeutic Implications of Cellular Heterogeneity and Plasticity in Breast Cancer. Cell stem cell 17, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Royen ME, et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 69, 985–992 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Olson E, Levene MJ & Torres R Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express 7, 3089–3088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka N, et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. biomed. eng, 1–14 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Tanaka N, et al. Mapping of the three-dimensional lymphatic microvasculature in bladder tumours using light-sheet microscopy. Br J Cancer 118, 995–999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SS-Y, Bindokas VP, Lingen MW & Kron SJ Nondestructive, multiplex three-dimensional mapping of immune infiltrates in core needle biopsy. Laboratory Investigation, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhoef EI, et al. Three-dimensional analysis reveals two major architectural subgroups of prostate cancer growth patterns. Mod Pathol, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce MC, Javier DJ & Richards-Kortum R Optical contrast agents and imaging systems for detection and diagnosis of cancer. Int J Cancer 123, 1979–1990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abeytunge S, Li Y, Larson B, Toledo-Crow R & Rajadhyaksha M Rapid confocal imaging of large areas of excised tissue with strip mosaicing. J Biomed Opt 16, 050504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abeytunge S, et al. Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue. J Biomed Opt 18, 61227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao YK, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci USA 111, 15304–15309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji M, et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med 7, 309ra163–309ra163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orringer DA, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. biomed. eng 1, 0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abeytunge S, et al. Evaluation of breast tissue with confocal strip-mosaicking microscopy: a test approach emulating pathology-like examination. J Biomed Opt 22, 034002–034012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boppart SA, et al. Label-free optical imaging technologies for rapid translation and use during intraoperative surgical and tumor margin assessment. J Biomed Opt 23, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshitake T, et al. Rapid histopathological imaging of skin and breast cancer surgical specimens using immersion microscopy with ultraviolet surface excitation. Scientific Reports, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, et al. Rapid pathology of lumpectomy margins with open-top light-sheet (OTLS) microscopy. Biomed Opt Express 10, 1257–1216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu JTC, et al. Micromirror-scanned dual-axis confocal microscope utilizing a gradient-index relay lens for image guidance during brain surgery. J Biomed Opt 15, 026029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanai N, et al. Intraoperative confocal microscopy in the visualization of 5-aminolevulinic acid fluorescence in low-grade gliomas. Journal of Neurosurgery 115, 740–748 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Nguyen QT & Tsien RY Fluorescence-guided surgery with live molecular navigation--a new cutting edge. Nat Rev Cancer 13, 653–662 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei L, Roberts DW, Sanai N & Liu JTC Visualization technologies for 5-ALA-based fluorescence-guided surgeries. J Neurooncol 141, 495–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei L, Fujita Y, Sanai N & Liu JTC Toward Quantitative Neurosurgical Guidance With High-Resolution Microscopy of 5-Aminolevulinic Acid-Induced Protoporphyrin IX. Frontiers in Oncology 9, v1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson DS & Lichtman JW Clarifying Tissue Clearing. Cell 162, 246–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Azaripour A, et al. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Progress in Histochemistry and Cytochemistry 51, 9–23 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Berke IM, Miola JP, David MA, Smith MK & Price C Seeing through Musculoskeletal Tissues: Improving In Situ Imaging of Bone and the Lacunar Canalicular System through Optical Clearing. PloS one 11, e0150268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jing D, et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell research 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hama H, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Ke M-T, Fujimoto S & Imai T SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154–1161 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Kim S-Y, et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci USA 112, E6274–6283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renier N, et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Klingberg A, et al. Fully Automated Evaluation of Total Glomerular Number and Capillary Tuft Size in Nephritic Kidneys Using Lightsheet Microscopy. Journal of the American Society of Nephrology : JASN 28, 452–459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silvestri L, Costantini I, Sacconi L & Pavone FS Clearing of fixed tissue: a review from a microscopist's perspective. J Biomed Opt 21, 081205 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Chowdary D, et al. Prognostic gene expression signatures can be measured in tissues collected in RNAlater preservative. J Mol Diagn 8, 31–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mutter GL, et al. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics 5, 88 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ergin B, et al. Proteomic analysis of PAXgene-fixed tissues. Journal of proteome research 9, 5188–5196 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Urban C, et al. PAXgene fixation enables comprehensive metabolomic and proteomic analyses of tissue specimens by MALDI MSI. Biochimica et biophysica acta. General subjects 1862, 51–60 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Park Y-G, et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol 37, 73–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.González-García I, Solé RV & Costa J Metapopulation dynamics and spatial heterogeneity in cancer. Proceedings of the National Academy of Sciences of the United States of America 99, 13085–13089 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung K & Deisseroth K CLARITY for mapping the nervous system. Nat Methods 10, 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Susaki EA, et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc 10, 1709–1727 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Tainaka K, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Humphrey PA Complete histologic serial sectioning of a prostate gland with adenocarcinoma. Am J Surg Pathol 17, 468–472 (1993). [DOI] [PubMed] [Google Scholar]
- 84.McCormick BH, et al. Construction of anatomically correct models of mouse brain networks. Neurocomputing 58-60, 379–386 (2004). [Google Scholar]
- 85.Li A, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Yoshitake T, et al. Direct comparison between confocal and multiphoton microscopy for rapid histopathological evaluation of unfixed human breast tissue. J Biomed Opt 21, 126021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tu H, et al. Stain-free histopathology by programmable supercontinuum pulses. Nature Photonics, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Helmchen F & Denk W Deep tissue two-photon microscopy. Nat Methods 2, 932–940 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Uhlén P & Tanaka N Improved Pathological Examination of Tumors with 3D Light-Sheet Microscopy. Trends in Cancer, 1–4 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Pouli D, et al. Imaging mitochondrial dynamics in human skin reveals depth-dependent hypoxia and malignant potential for diagnosis. Sci Transl Med 8, 367ra169–367ra169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baugh LM, et al. Non-destructive two-photon excited fluorescence imaging identifies early nodules in calcific aortic-valve disease. Nat. biomed. eng 1, 914–924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Skala MC, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proceedings of the National Academy of Sciences of the United States of America 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.You S, et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proceedings of the National Academy of Sciences of the United States of America 3, 201909243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xylas J, Alt-Holland A, Garlick J, Hunter M & Georgakoudi I Intrinsic optical biomarkers associated with the invasive potential of tumor cells in engineered tissue models. Biomed Opt Express 1, 1387–1400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conklin MW, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol 178, 1221–1232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campagnola PJ & Loew LM Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol 21, 1356–1360 (2003). [DOI] [PubMed] [Google Scholar]
- 97.Freudiger CW, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saar BG, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 330, 1368–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fischer MC, Wilson JW, Robles FE & Warren WS Invited Review Article: Pump-probe microscopy. The Review of scientific instruments 87, 031101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matthews TE, Piletic IR, Selim MA, Simpson MJ & Warren WS Pump-probe imaging differentiates melanoma from melanocytic nevi. Sci Transl Med 3, 71ra15–71ra15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Giacomelli MG, et al. Multiscale nonlinear microscopy and widefield white light imaging enables rapid histological imaging of surgical specimen margins. Biomed Opt Express 9, 2457–2475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakano A Spinning-disk confocal microscopy -- a cutting-edge tool for imaging of membrane traffic. Cell structure and function 27, 349–355 (2002). [DOI] [PubMed] [Google Scholar]
- 103.Tanaami T, et al. High-speed 1-frame/ms scanning confocal microscope with a microlens and Nipkow disks. Appl Opt 41, 4704–4708 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Cheng L-C, et al. Spatiotemporal focusing-based widefield multiphoton microscopy for fast optical sectioning. Opt Express 20, 8939–8948 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Oron D, Tal E & Silberberg Y Scanningless depth-resolved microscopy. Opt Express 13, 1468–1476 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Zhang T, et al. Kilohertz two-photon brain imaging in awake mice. Nat Methods 16, 1119–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bewersdorf J, Pick R & Hell SW Multifocal multiphoton microscopy. Opt Lett 23, 655–657 (1998). [DOI] [PubMed] [Google Scholar]
- 108.Bahlmann K, et al. Multifocal multiphoton microscopy (MMM) at a frame rate beyond 600 Hz. Opt Express 15, 10991–10998 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Dodt H-U, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 110.Fahrbach FO, Simon P & Rohrbach A Microscopy with self-reconstructing beams. Nature Photonics, 1–6 (2010). [Google Scholar]
- 111.Huisken J, Swoger J, Del Bene F, Wittbrodt J & Stelzer EHK Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 112.Keller PJ, et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nature Methods 7, 637–642 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keller PJ, Schmidt AD, Wittbrodt J & Stelzer EHK Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Planchon TA, et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods 8, 417–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Power RM & Huisken J A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Meth 14, 360–373 (2017). [DOI] [PubMed] [Google Scholar]
- 116.Tomer R, et al. SPED Light Sheet Microscopy: Fast Mapping of Biological System Structure and Function. Cell 163, 1796–1806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu Y, et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scherf N & Huisken J The smart and gentle microscope. Nat Biotechnol 33, 815–818 (2015). [DOI] [PubMed] [Google Scholar]
- 119.Fahrbach FO, Gurchenkov V, Alessandri K, Nassoy P & Rohrbach A Light-sheet microscopy in thick media using scanned Bessel beams and two-photon fluorescence excitation. Opt Express 21, 13824 (2013). [DOI] [PubMed] [Google Scholar]
- 120.Tomer R, Ye L, Hsueh B & Deisseroth K Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9, 1682–1697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marx V Microscopy: seeing through tissue. Nat Methods 11, 1209–1214 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Wu Y, et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci USA 108, 17708–17713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Glaser AK, et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat Commun 10, 2781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Migliori B, et al. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC biology 16, 57–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mcgorty R, et al. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt Express 23, 16142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Voigt FF, et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat Methods 16, 1105–1108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chakraborty T, et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat Methods 16, 1109–1113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bouchard MB, et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nature Photonics 9, 113–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Voleti V, et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat Methods 16, 1054–1062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang B, et al. Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat Methods 16, 501–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Strnad P, et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nature Methods 13, 139–142 (2016). [DOI] [PubMed] [Google Scholar]
- 132.Verveer PJ, et al. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods 4, 311–313 (2007). [DOI] [PubMed] [Google Scholar]
- 133.Swoger J, Verveer P, Greger K, Huisken J & Stelzer EHK Multi-view image fusion improves resolution in three-dimensional microscopy. Opt Express 15, 8029–8042 (2007). [DOI] [PubMed] [Google Scholar]
- 134.Dean KM, Roudot P, Welf ES, Danuser G & Fiolka R Deconvolution-free Subcellular Imaging with Axially Swept Light Sheet Microscopy. Biophysj 108, 2807–2815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Remacha E, Fahrbach FO, Vermot J & Friedrich L How to define and optimize axial resolution in light-sheet microscopy: a simulation-based approach. Biomed Opt Express 11, 8–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vettenburg T, et al. Light-sheet microscopy using an Airy beam. Nature Methods (2014). [DOI] [PubMed] [Google Scholar]
- 137.Bria A, Bernaschi M, Guarrasi M & Iannello G Exploiting Multi-Level Parallelism for Stitching Very Large Microscopy Images. Frontiers in neuroinformatics 13, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bria A & Iannello G TeraStitcher - a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC bioinformatics 13, 316–315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hörl D, et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat Methods 16, 870–874 (2019). [DOI] [PubMed] [Google Scholar]
- 140.Balazs B, Deschamps J, Albert M, Ries J & Hufnagel L A real-time compression library for microscopy images. bioRxiv, 164624 (2017). [Google Scholar]
- 141.Stefansson HN, et al. Wavelet compression of three-dimensional time-lapse biological image data. Microsc Microanal 11, 9–17 (2005). [DOI] [PubMed] [Google Scholar]
- 142.Giacomelli MG, et al. Virtual Hematoxylin and Eosin Transillumination Microscopy Using Epi-Fluorescence Imaging. PloS one 11, e0159337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rueden CT, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC bioinformatics 18, 529–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Edelstein A, Amodaj N, Hoover K, Vale R & Stuurman N Computer control of microscopes using manager. Current Protocols in Molecular Biology 92, Unit14.20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pietzsch T, Saalfeld S, Preibisch S & Tomancak P BigDataViewer: visualization and processing for large image data sets. Nat Methods 12, 481–483 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Pitrone PG, et al. OpenSPIM: an open-access light-sheet microscopy platform. Nat Methods 10, 598–599 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marx V Microscopy: OpenSPIM 2.0. Nat Methods 13, 979–982 (2016). [DOI] [PubMed] [Google Scholar]
- 150.Carpenter AE, Kamentsky L & Eliceiri KW A call for bioimaging software usability. Nat Methods 9, 666–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cardona A & Tomancak P Current challenges in open-source bioimage informatics. Nat Methods 9, 661–665 (2012). [DOI] [PubMed] [Google Scholar]
- 152.Ghaznavi F, Evans A, Madabhushi A & Feldman M Digital imaging in pathology: whole-slide imaging and beyond. Annual review of pathology 8, 331–359 (2013). [DOI] [PubMed] [Google Scholar]
- 153.Niazi MKK, Parwani AV & Gurcan MN Digital pathology and artificial intelligence. Lancet Oncol 20, e253–e261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pantanowitz L, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform 2, 36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Madabhushi A & Lee G Image analysis and machine learning in digital pathology: Challenges and opportunities. 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cheplygina V, de Bruijne M & Pluim JPW Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal 54, 280–296 (2019). [DOI] [PubMed] [Google Scholar]
- 157.Tajbakhsh N, et al. Embracing Imperfect Datasets: A Review of Deep Learning Solutions for Medical Image Segmentation. arXiv.org eess.IV(2019). [DOI] [PubMed] [Google Scholar]
- 158.He K, Fan H, Wu Y, Xie S & Girshick R Momentum Contrast for Unsupervised Visual Representation Learning. arXiv.org cs.CV(2019). [Google Scholar]
- 159.Belthangady C & Royer LA Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat Methods, 1–13 (2019). [DOI] [PubMed] [Google Scholar]
- 160.Christiansen EM, et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell 173, 792–803.e719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ounkomol C, Seshamani S, Maleckar MM, Collman F & Johnson GR Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat Methods 15, 917–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maier AK, et al. Learning with Known Operators reduces Maximum Training Error Bounds. Nature machine intelligence 1, 373–380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maier A, Syben C, Lasser T & Riess C A gentle introduction to deep learning in medical image processing. Zeitschrift fur medizinische Physik 29, 86–101 (2019). [DOI] [PubMed] [Google Scholar]
- 164.Dou Q, et al. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal 41, 40–54 (2017). [DOI] [PubMed] [Google Scholar]
- 165.Martel AL, et al. An Image Analysis Resource for Cancer Research: PIIP-Pathology Image Informatics Platform for Visualization, Analysis, and Management. Cancer Research 77, e83–e86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Scientific Reports 7, 16878–16877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rivenson Y, et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. biomed. eng 3, 466–477 (2019). [DOI] [PubMed] [Google Scholar]
- 168.Khan AM, Rajpoot N, Treanor D & Magee D A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng 61, 1729–1738 (2014). [DOI] [PubMed] [Google Scholar]
- 169.Janowczyk A, Zuo R, Gilmore H, Feldman M & Madabhushi A HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides. JCO clinical cancer informatics 3, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Leo P, et al. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: a multi- site study. Scientific Reports, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu J, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416.e411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Paik S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24, 3726–3734 (2006). [DOI] [PubMed] [Google Scholar]
- 173.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- 174.Bast RC & Hortobagyi GN Individualized care for patients with cancer - a work in progress. N Engl J Med 351, 2865–2867 (2004). [DOI] [PubMed] [Google Scholar]
- 175.Engel KB, Vaught J & Moore HM National Cancer Institute Biospecimen Evidence-Based Practices: a novel approach to pre-analytical standardization. Biopreservation and biobanking 12, 148–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goossens N, Nakagawa S, Sun X & Hoshida Y Cancer biomarker discovery and validation. Translational cancer research 4, 256–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sparano JA, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 373, 2005–2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Allen TC Food and Drug Administration approval of laboratory tests. Arch Pathol Lab Med 137, 13–18 (2013). [DOI] [PubMed] [Google Scholar]
- 179.Evans AJ, et al. US Food and Drug Administration Approval of Whole Slide Imaging for Primary Diagnosis: A Key Milestone Is Reached and New Questions Are Raised. Arch Pathol Lab Med 142, 1383–1387 (2018). [DOI] [PubMed] [Google Scholar]
- 180.Mukhopadhyay S, et al. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study). Am J Surg Pathol 42, 39–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Joly Y, et al. Regulatory approval for new pharmacogenomic tests: a comparative overview. Food and drug law journal 66, 1–24- i (2011). [PubMed] [Google Scholar]
- 182.DAngelo R, et al. Facing the Inevitable: Being Prepared for Regulatory Requirements for Laboratory Developed Tests. Am J Clin Pathol 149, 484–498 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Madabhushi A, Feldman MD & Leo P Deep-learning approaches for Gleason grading of prostate biopsies. Lancet Oncol 21, 187–189 (2020). [DOI] [PubMed] [Google Scholar]
- 184.King CR & Long JP Prostate biopsy grading errors: a sampling problem? Int J Cancer 90, 326–330 (2000). [DOI] [PubMed] [Google Scholar]
- 185.Ruijter E, van Leenders G, Miller G, Debruyne F & van de Kaa C Errors in histological grading by prostatic needle biopsy specimens: frequency and predisposing factors. The Journal of pathology 192, 229–233 (2000). [DOI] [PubMed] [Google Scholar]
- 186.Campanella G, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nature Medicine 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen DS & Mellman I Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 188.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gajewski TF, Schreiber H & Fu Y-X Innate and adaptive immune cells in the tumor microenvironment. Nature immunology 14, 1014–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Corredor G, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res, clincanres.2013.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Annecchino LA, et al. Robotic Automation of In Vivo Two-Photon Targeted Whole-Cell Patch-Clamp Electrophysiology. Neuron 95, 1048–1055.e1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Long B, Li L, Knoblich U, Zeng H & Peng H 3D Image-Guided Automatic Pipette Positioning for Single Cell Experiments in vivo. Scientific Reports, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Suk H-J, et al. Closed-Loop Real-Time Imaging Enables Fully Automated Cell-Targeted Patch-Clamp Neural Recording In Vivo. Neuron 95, 1037–1047.e1010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Thawani R, et al. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung cancer (Amsterdam, Netherlands) 115, 34–41 (2018). [DOI] [PubMed] [Google Scholar]
- 198.Lambin P, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nature Reviews Clinical Oncology 14, 749–762 (2017). [DOI] [PubMed] [Google Scholar]
- 199.Vaidya P, et al. RaPtomics: integrating radiomic and pathomic features for predicting recurrence in early stage lung cancer. Vol. 10581 (eds. Gurcan MN & Tomaszewski JE) 105810M (International Society for Optics and Photonics, 2018). [Google Scholar]
- 200.Reusch LM, et al. Nonlinear optical microscopy and ultrasound imaging of human cervical structure. J Biomed Opt 18, 031110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pinkert MA, et al. Review of quantitative multiscale imaging of breast cancer. Journal of medical imaging (Bellingham, Wash.) 5, 010901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mori K From macro-scale to micro-scale computational anatomy: a perspective on the next 20 years. Med Image Anal 33, 159–164 (2016). [DOI] [PubMed] [Google Scholar]
- 203.Gao R, et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hoyer P, et al. Breaking the diffraction limit of light-sheet fluorescence microscopy by RESOLFT. Proceedings of the National Academy of Sciences of the United States of America 113, 3442–3446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen B-C, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Li L, et al. Co-Registration of ex vivo Surgical Histopathology and in vivo T2 weighted MRI of the Prostate via multi-scale spectral embedding representation. Scientific Reports 7, 8717–8712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rusu M, et al. Co-registration of pre-operative CT with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof-of-concept study. Eur Radiol 27, 4209–4217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rusu M, et al. Prostatome: a combined anatomical and disease based MRI atlas of the prostate. Medical physics 41, 072301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Antunes J, et al. Coregistration of Preoperative MRI with Ex Vivo Mesorectal Pathology Specimens to Spatially Map Post-treatment Changes in Rectal Cancer Onto In Vivo Imaging: Preliminary Findings. Academic radiology 25, 833–841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schillaci O, et al. Combining Diagnostic Imaging and Pathology for Improving Diagnosis and Prognosis of Cancer. Contrast media & molecular imaging 2019, 9429761–9429710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Grönroos TJ, et al. Hypoxia, blood flow and metabolism in squamous-cell carcinoma of the head and neck: correlations between multiple immunohistochemical parameters and PET. BMC Cancer 14, 876–811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Surov A, Meyer HJ & Wienke A Standardized Uptake Values Derived from 18F-FDG PET May Predict Lung Cancer Microvessel Density and Expression of KI 67, VEGF, and HIF-1α but Not Expression of Cyclin D1, PCNA, EGFR, PD L1, and p53. Contrast media & molecular imaging 2018, 9257929–9257910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bensch F, et al. 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nature Medicine, 1–13 (2018). [DOI] [PubMed] [Google Scholar]
- 214.Gibson E, et al. Registration of prostate histology images to ex vivo MR images via strand-shaped fiducials. Journal of Magnetic Resonance Imaging 36, 1402–1412 (2012). [DOI] [PubMed] [Google Scholar]
- 215.Bhargava HK, et al. Computationally Derived Image Signature of Stromal Morphology Is Prognostic of Prostate Cancer Recurrence Following Prostatectomy in African American Patients. Clin Cancer Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.OKeefe EB, Meltzer JP & Bethea TN Health disparities and cancer: racial disparities in cancer mortality in the United States, 2000-2010. Frontiers in public health 3, 51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhou M, et al. Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 286, 307–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Savage RS & Yuan Y Predicting chemoinsensitivity in breast cancer with 'omics/digital pathology data fusion. Royal Society open science 3, 140501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pinker K, Chin J, Melsaether AN, Morris EA & Moy L Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology 287, 732–747 (2018). [DOI] [PubMed] [Google Scholar]
- 220.Mobadersany P, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proceedings of the National Academy of Sciences of the United States of America 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Penzias G, et al. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings. PloS one 13, e0200730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dainis AM & Ashley EA Cardiovascular Precision Medicine in the Genomics Era. JACC. Basic to translational science 3, 313–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kai C, Uchiyama Y, Shiraishi J, Fujita H & Doi K Computer-aided diagnosis with radiogenomics: analysis of the relationship between genotype and morphological changes of the brain magnetic resonance images. Radiological physics and technology 11, 265–273 (2018). [DOI] [PubMed] [Google Scholar]
- 224.Montalto MC An industry perspective: An update on the adoption of whole slide imaging. J Pathol Inform 7, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]