Abstract
Microglial surveillance is a key feature of brain physiology and disease. We found that Gi-dependent microglial dynamics prevent neuronal network hyperexcitability. By generating MgPTX mice to genetically inhibit Gi in microglia, we showed that sustained reduction of microglia brain surveillance and directed process motility induced spontaneous seizures and increased hypersynchrony upon physiologically evoked neuronal activity in awake adult mice. Thus, Gi-dependent microglia dynamics may prevent hyperexcitability in neurological diseases.
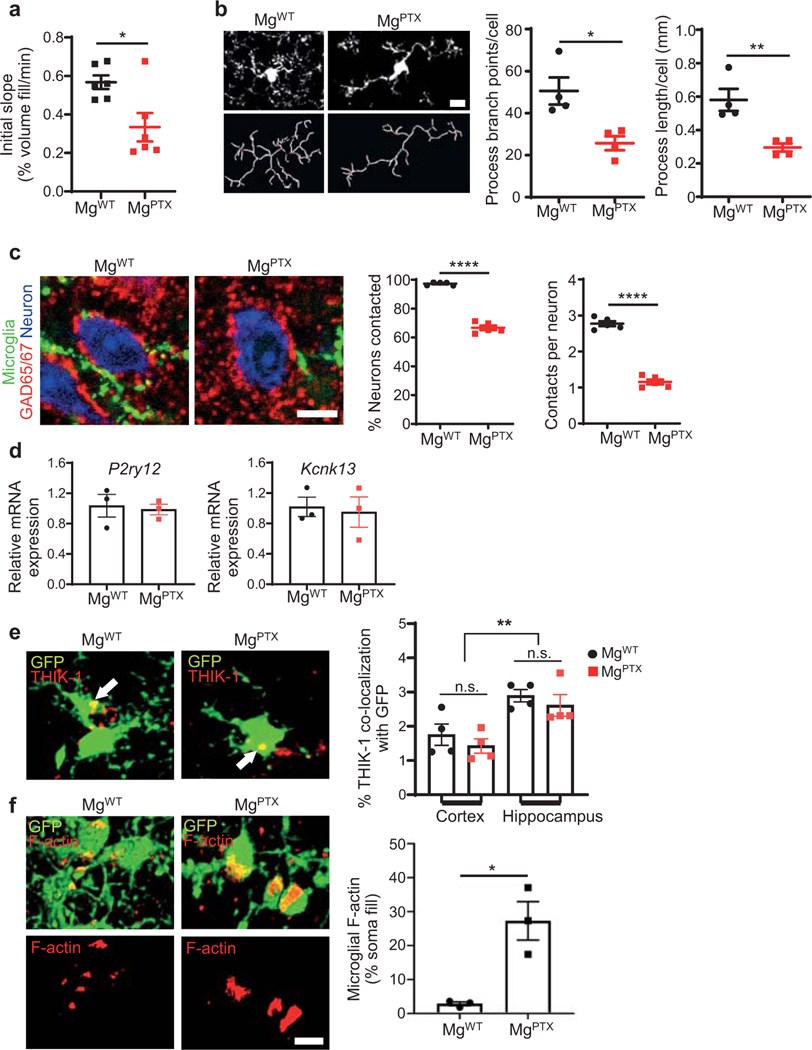
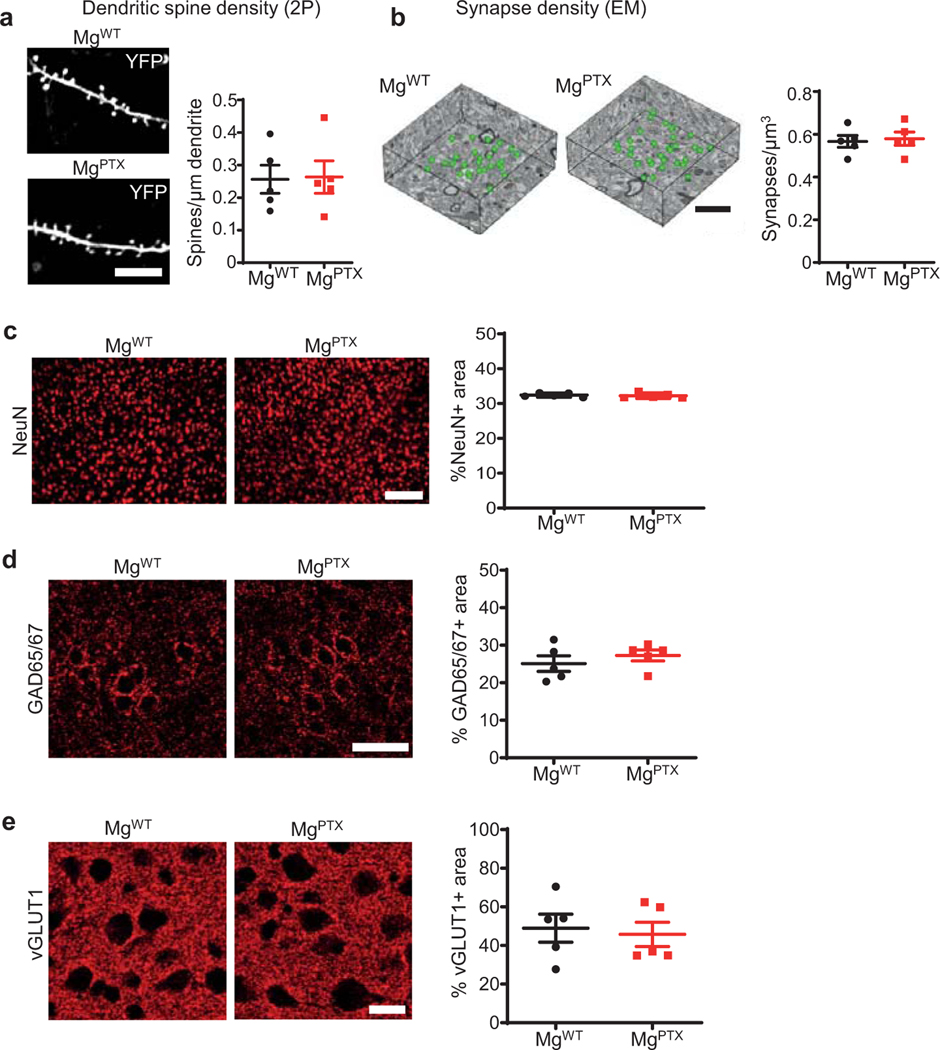
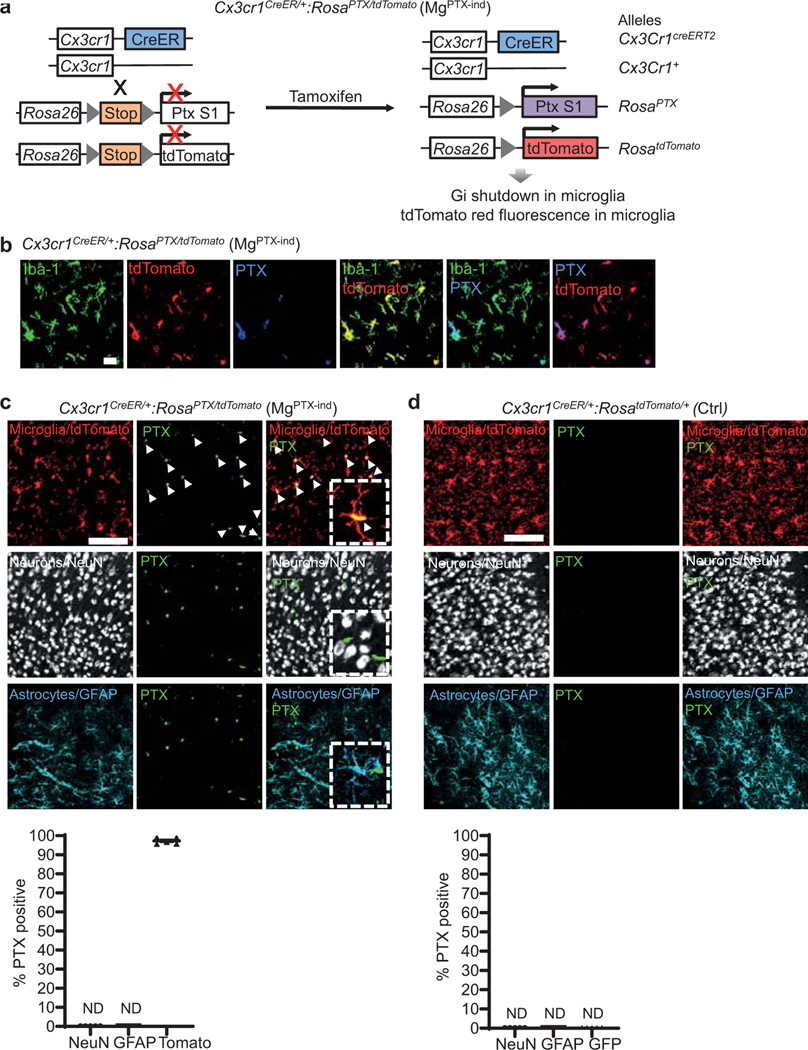
Microglia constantly extend and retract processes to survey the brain, and rapidly respond toward sites of injury1, 2. Microglia process dynamics are perturbed in neuroinflammation and neurodegeneration3, 4. However, the role of microglial surveillance and directed process motility remains poorly understood. Baseline surveillance, but not directed process motility, is regulated by TWIK-related Halothane-inhibited K+ channel (THIK-1), which is gated by the P2Y12 receptor (P2Y12R)5. Genetic depletion of P2Y12R transiently delays microglial directed motility to laser ablation without affecting baseline surveillance potentially due to compensation by other G-coupled protein receptors (GPCRs)6. We hypothesized that inhibiting the Gi pathway in microglia would lead to sustained inhibition of microglia dynamics. We generated Tg(Cx3cr1-Cre):RosaPTX/+ (MgPTX) mice for microglia-specific Gi inhibition (Fig. 1a, Extended Data Fig. 1). In the presence of Cre recombinase, RosaPTX/+ mice express the pertussis toxin (PTX) S1 catalytic subunit, which ADP-ribosylates the α subunits of Gi/o proteins, thereby uncoupling them from upstream GPCRs for cell-autonomous Gi/o inhibition in vivo7. Using in vivo two-photon (2P) time-lapse imaging, we found dramatically reduced microglia surveillance in MgPTX mice compared to littermate control RosaPTX/+ (MgWT) mice (Fig. 1b, Extended Data Fig. 2a and Supplementary Video 1). Lack of P2Y12R delays, but does not abolish, the microglia response to laser ablation6. In contrast, in MgPTX mice, directed process motility was abolished even 2 hours after laser ablation (Fig. 1c and Supplementary Video 2). MgPTX mice had decreased microglial process length and branch points, and fewer microglia contacts onto neuronal somata (Extended Data Fig. 2b, c and Supplementary Video 3). Expression of P2ry12 and Kcnk13 (encoding THIK-1) was similar between genotypes (Extended Data Fig. 2d, e), suggesting that decreased microglial motility in MgPTX mice occurred downstream of Gi. Filamentous actin (F-actin), an indicator of cAMP regulation downstream of Gi in microglia8, was highly decompartmentalized and spread throughout the cytoplasm in MgPTX mice (Extended Data Fig. 2f), indicating increased cAMP upon Gi inhibition. MgPTX mice exhibited normal densities of dendritic spines, synapses, and neurons, as shown by in vivo 2P imaging, serial block-face scanning electron microscopy (SBEM), and immunostaining for NeuN, inhibitory and excitatory presynaptic puncta markers GAD65/67 and vGLUT1, respectively (Extended Data Fig. 3a–e). Thus, inhibition of Gi signaling in microglia impairs both microglia surveillance and lesion-induced directed motility.
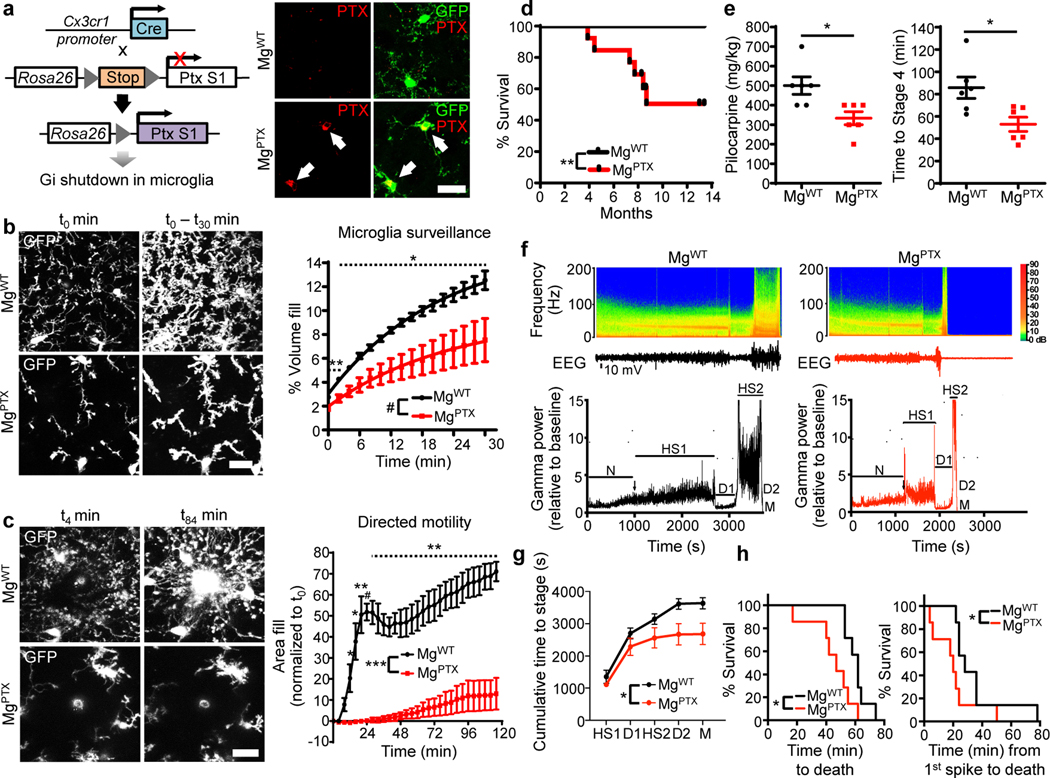
Fig. 1: Effects of Gi inhibition on microglia dynamics and seizures.
a, Generation of MgPTX mice (left); confocal microscopy in cortex of PTX (red) and microglia (green) in MgPTX and MgWT mice. Arrows indicate PTX-positive microglia. Scale bar, 50 μm. 60.5 ± 2.7% (mean ± s.e.m.) of cortical GFP-positive cells express PTX. n = 3 mice. b, In vivo 2P time-lapse imaging of cumulative microglial surveillance. Data are mean ± s.e.m. n = 6 mice per genotype. Overall genotype effect # P = 0.024 by unpaired two-sample t-test of mean AUC. * P < 0.05, ** P < 0.01 at individual time points by permutation test. c, In vivo 2P time-lapse imaging of microglial directed process motility towards laser ablation. Scale bar, 20 μm. Data are mean ± s.e.m. n = 4 mice per genotype. Overall genotype effect *** P = 0.0004 by unpaired two-sample t-test of mean AUC. Dotted line indicates * P < 0.05, ** P < 0.01, # P < 0.001 at individual time points by unpaired two-sample t-test. d, Survival curves of MgPTX and MgWT mice. n = 13 mice per genotype. ** P = 0.0056 by Log-rank test. e, Pilocarpine dose (left) and latency (right) to reach Stage 4 seizures for MgPTX and MgWT mice. Data are mean ± s.e.m. n = 6 mice per genotype. * P = 0.014 and 0.017, respectively, by-unpaired two-tailed t-test. f, Spectrograms (top) and gamma oscillatory power traces (bottom) from EEG recordings during pilocarpine induced-seizures. EEG stages: N, normal; HS1, 1st hypersynchrony; D1, 1st depression; HS2, 2nd hypersynchrony; D2, 2nd depression; M, Mortality. Representative data from n = 7 mice per genotype with similar results. g, Cumulative time to each pilocarpine-induced seizure stage in MgPTX and MgWT mice. Data are mean ± s.e.m. n = 7 mice per genotype. * P = 0.032 for genotype effect by two-way repeated-measures ANOVA. h, Survival curves for pilocarpine-induced time to death (left) and from 1st epileptiform spike to death (right). n = 7 mice per genotype. * P = 0.0127 (time to death) and 0.0333 (time from 1st spike to death) by Gehan-Breslow-Wilcoxon test.
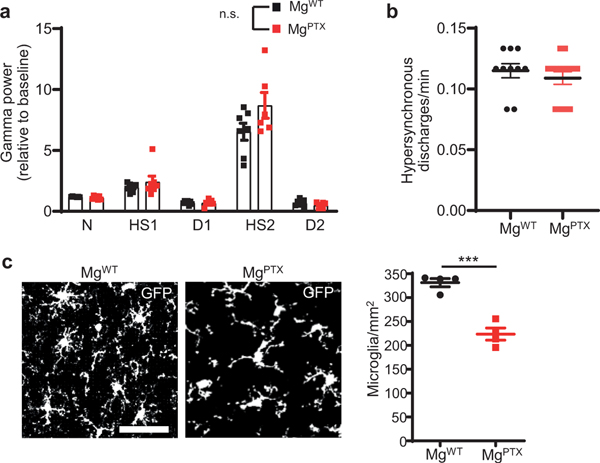
MgPTX mice developed spontaneous seizures that decreased survival (Fig. 1d, Supplementary Video 4). MgPTX mice had a greater susceptibility to pilocarpine-induced seizures both in terms of latency and dose at which they reached tonic-clonic seizures (Racine scale stage 4 ) (Fig. 1e). These results are in accordance with greater susceptibility to kainic acid-induced seizures of mice lacking P2Y12R or CX3CR1—two Gi-coupled receptors expressed on microglia9. Electroencephalography (EEG) recordings during pilocarpine administration revealed distinct consecutive phases of network hypersynchrony and post-ictal depression, particularly in the gamma frequency range (30–90 Hz). Thus, we used gamma power to categorize six stages of pilocarpine-induced aberrant network activity (Fig. 1f). While the gamma power during each network stage was not different between genotypes (Extended Data Fig. 4a), MgPTX mice transitioned between stages significantly faster than controls (Fig. 1g, h). Baseline EEG was similar between genotypes prior to pilocarpine effects (Extended Data Fig. 4b). Microglia numbers were decreased in MgPTX mice (Extended Data Fig. 4c). To overcome any confounding effects of constitutive Gi inhibition during development, we inhibited microglial Gi in the adult by generating Cx3cr1CreER/+:RosaPTX/tdTomato (MgPTX-ind) mice with tamoxifen-induced PTX expression and littermate Cx3cr1CreER/+:RosatdTomato/+ controls (Ctrl) (Extended Data Figs. 5, 6, 7a, b). Similar to MgPTX mice, upon tamoxifen MgPTX-ind mice had deficits in microglia baseline and directed motility and increased susceptibility to pilocarpine-induced seizures compared to control mice (Extended Data Fig. 7a–c and Supplementary Video 5, 6). MgPTX-ind mice had no differences in THIK-1 expression or microglia numbers (Extended Data Fig. 7d, e). These results suggest that microglial dynamics modulate neuronal network hypersynchrony.
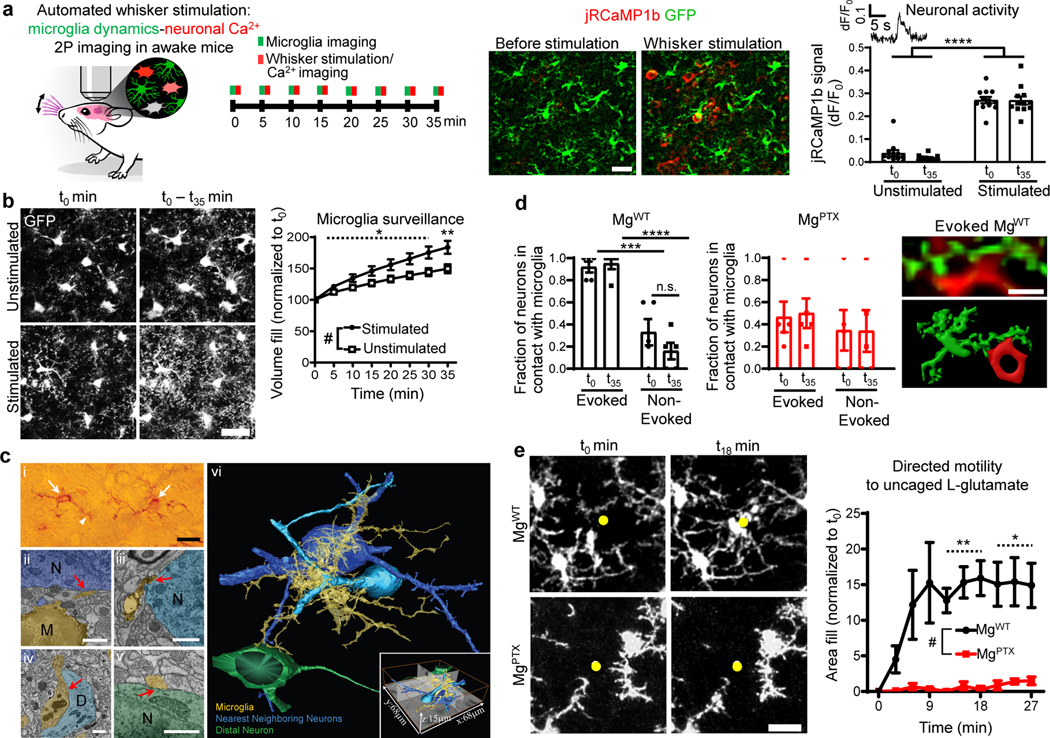
To investigate the role of microglia in hyperexcitability, we developed an automated whisker stimulation model to simultaneously image microglia and evoked neuronal Ca2+ dynamics in the whisker barrel cortex by in vivo 2P imaging in awake mice (Fig. 2a and Supplementary Video 7). Repetitive whisker stimulation causing evoked neuronal Ca2+ transients increased microglia brain surveillance in MgWT mice (Fig. 2b and Supplementary Video 8). We reconstructed in three dimensions (3D) the contacts between P2Y12R-expressing microglia and neurons at the subcellular level using CDeep3M machine learning algorithm for large-scale EM data10. Microglial processes were in contact with the soma of neighboring neurons, while extending processes contacting the soma of distal neurons (Fig 2c, Supplementary Video 9). Whereas in MgWT mice microglia processes contacted neuronal somata with evoked activity at a greater proportion than non-evoked neurons (Fig. 2d, Extended Fig. 8a), Gi-deficient microglia in MgPTX mice lost preferential interaction with evoked neurons (Fig. 2d). Quantification of microglia in the fields of view (FOVs) showed a non-significant reduction in MgPTX mice (Extended Data Fig. 8b), but no correlation between microglia numbers and intraneuronal Ca2+ in either genotype (Extended Data Fig. 8c). Since MgPTX-ind mice phenocopy MgPTX mice without altered microglia numbers and neuronal activity did not correlate with microglia numbers, microglia numbers in MgPTX mice likely do not underlie the effects of microglial-Gi inhibition on neuronal activity. Expression of Grm3, encoding the Gi-coupled glutamate receptor mGluR3, was slightly reduced in MgPTX mice compared to MgWT mice, while gene expression changes in other metabotropic and ionotropic receptors or glutamate transporters were not detected (Extended Data Fig. 8d). In vivo glutamate uncaging induced neuronal activity and microglial directed process motility in MgWT mice, which was abolished in MgPTX mice (Fig. 2e, Extended Data Fig. 8e, and Supplementary Video 10), suggesting that microglia sense and act upon excitatory, glutamate-driven neuronal activity. Together, these results reveal an interdependence between sensory-evoked neuronal activity and microglia surveillance and suggest that microglial Gi signaling regulates microglia-neuron interactions.
Fig. 2: Microglial Gi-dependent dynamics and evoked neuronal activity in awake mice.
a, In vivo 2P imaging of microglia (GFP, green) and neuronal Ca2+ (jRCaMP1b, red) transients in the barrel cortex during whisker stimulation in awake Cx3cr1GFP/+ mice. Scale bar, 50 μm. Data are mean ± s.e.m. n = 12 mice. **** P < 0.0001 by two-way ANOVA. b, In vivo 2P images of cumulative microglial surveillance during whisker stimulation. Scale bar, 20 μm. Data are mean ± s.e.m. n = 12 mice per condition. Overall condition effect # P = 0.021 by unpaired two-sample t-test of mean AUC. Dotted line indicates * P < 0.05 at individual time points by permutation test. c, SBEM and volumetric image analysis of microglial cell contacts with surrounding neurons. (i) Transmitted light microscopy of immunoperoxidase-labeled P2Y12R cell bodies (arrows) and processes (arrowheads). Scale bar, 20 μm. 2D electron micrographs of microglia (yellow) contact sites on nearest neighbor neuron somata (ii, iii; deep and light blue), a dendrite (iv, light blue), a distal neuron soma (v, green). Scale bars, 1 μm. (vi) 3D reconstruction of a microglia (yellow) contacting two nearest neurons (deep and light blues) and a distal neuron (green). SBEM volume dimensions (inset). Representative 3D reconstruction from 4 microglia from n = 3 MgWT mice. d, Evoked and non-evoked neuronal somata upon whisker stimulation in contact with microglia in MgWT and MgPTX mice. Data are mean ± s.e.m. n = 5 mice per genotype. *** P = 0.0003 and **** P < 0.0001 by two-way ANOVA and Tukey’s multiple comparisons test; Right, in vivo 2P image (top) and 3D reconstruction (bottom) of microglia (green) contacting a jRCaMP1b-expressing neuronal soma (red) in MgWT barrel cortex during whisker stimulation. Scale bar, 20 μm. e, In vivo 2P time-lapse of microglial directed motility in response to glutamate uncaging in somatosensory cortex (left); right, microglia area fill around uncaging site (yellow dot). Scale bar, 20 μm. Data are mean ± s.e.m. n = 5 MgPTX and n = 4 MgWT mice. Overall genotype effect # P = 0.015 by unpaired two-sample t-test of mean AUC. * P < 0.05, ** P < 0.01 at individual time points by unpaired two-sample t-test.
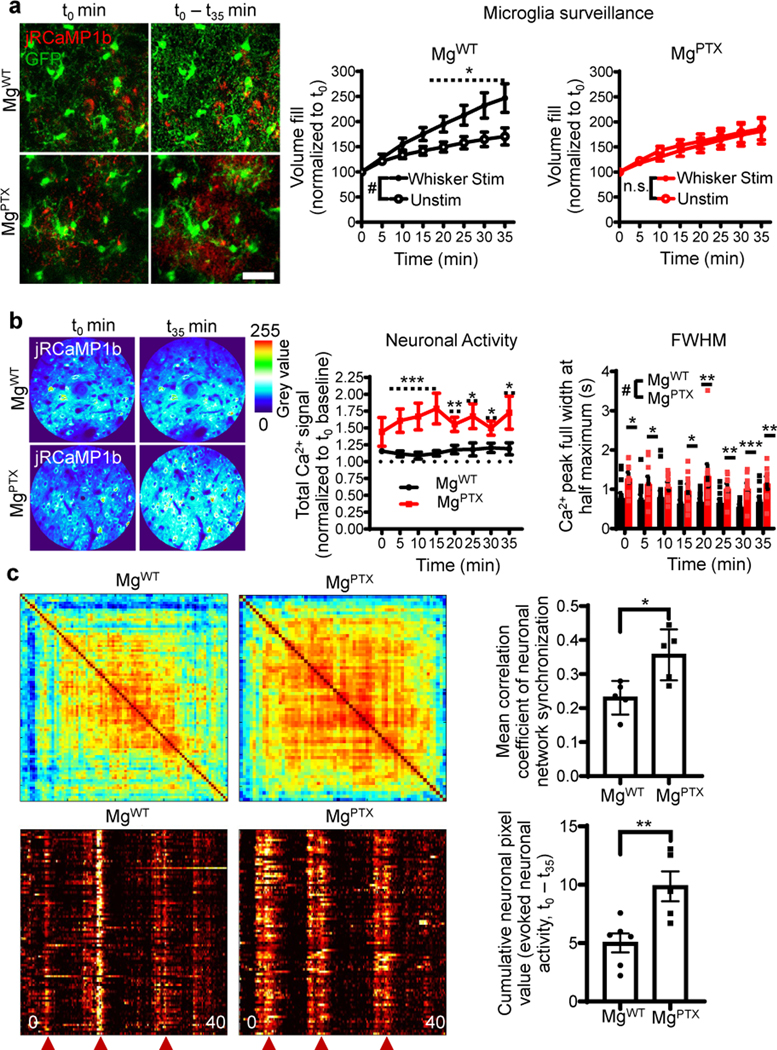
To determine the effects of microglia dynamics on neuronal network hypersynchrony, we measured microglial surveillance and evoked neuronal Ca2+ transients upon whisker stimulation. In response to whisker stimulation-evoked neuronal activity, brain surveillance was increased in MgWT mice, but not in MgPTX mice (Fig. 3a, Supplementary Video 11). Microglia process extension and retraction was reduced in MgPTX mice compared to MgWT mice without differences in process velocity (Extended Data Fig. 9a, b). This lack of microglial response was associated with a cumulative increase in evoked neuronal Ca2+ transients and prolonged decay times of evoked Ca2+ transients (Fig. 3b).Whisker stimulus-induced neuronal activity in MgPTX mice was hypersynchronized as assessed by quantification of synchronized neuronal firing and network burst activity (Fig. 3c). The average amplitude of evoked neuronal Ca2+ transients was similar between genotypes (Extended Data Fig. 9c), indicating similar peak response to whisker stimulation. These results suggest that microglia–neuron interactions prevent network hypersynchrony.
Fig. 3. Hypersynchronized neuronal activity after microglia-specific Gi inhibition.
a, In vivo 2P imaging of cumulative microglia surveillance (green) and neuronal activity (red) in the barrel cortex upon whisker stimulation in awake MgWT and MgPTX mice. Scale bar, 20 μm. Data are mean ± s.e.m. n = 7 mice (unstimulated) and n = 8 mice (whisker stimulation). Overall stimulation effect # P = 0.042 by unpaired two-sample t-test of mean AUC. * P < 0.05 at individual time points by permutation test. n.s., no significant difference for overall effect by-unpaired two-sample t-test of mean AUC. b, Heatmaps of cumulative evoked neuronal Ca2+ and quantification of neuronal activity and peak full width at half maximum (FWHM) during whisker stimulation in awake MgPTX and MgWT mice. Data are mean ± s.e.m. n = 9 MgPTX and n = 14 MgWT mice. Neuronal activity, overall genotype effect # P = 0.016 by unpaired two-sample t-test of mean AUC. * P < 0.05, ** P < 0.01, *** P < 0.001 at individual time points by permutation test. FWHM, overall genotype effect # P = 0.00009 by unpaired two-sample t-test of mean AUC and * P < 0.05, ** P < 0.01, *** P < 0.001 for individual time points by permutation test. c, Clustered correlation matrix of synchronized neuronal network firing (top) and raster plots of synchronized neuronal network burst activity (bottom) during whisker stimulation in MgWT and MgPTX mice. Red arrowheads indicate whisker stimulus. Data are mean ± s.e.m. n = 5 MgPTX and n = 6 MgWT mice. * P = 0.0139, ** P = 0.0092 by unpaired two-tailed t-test.
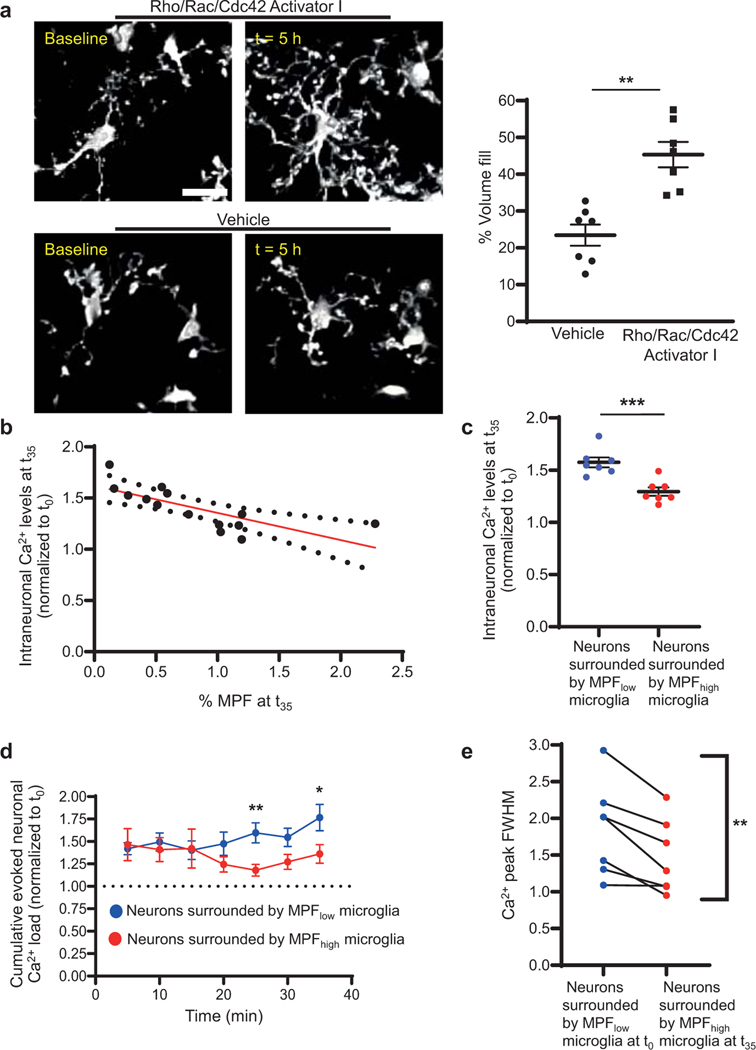
A key PTX-sensitive signaling pathway downstream of Gi is activation of Rho GTPases, which regulate cytoskeletal rearrangement and motility11. We hypothesized that regulation of microglia motility via Gi/Rho GTPases modulates neuronal activity. Cranial bathing with a direct activator of Rho/Rac/Cdc42 GTPases, increased microglia surveillance in MgPTX mice (Extended Data Fig. 10a, Supplementary Video 12), suggesting that constitutive activation of Rho GTPases restores microglia motility in MgPTX mice. Both neuronal Ca2+ load and the decay time of evoked neuronal Ca2+ signals correlated with the degree of microglial process fill in MgPTX mice (Extended Data Fig. 10b, c), further supporting microglia–neuron contact as a major mechanism preventing neuronal hyperexcitability. The aberrant evoked neuronal activity in MgPTX mice was reduced for neurons surrounded by microglia showing increased process extension upon constitutive Rho GTPase activation (Extended Fig. 10d, e). Overall, these data suggest that Gi regulates microglia surveillance and reduces excessive neuronal activity during periods of heightened excitatory neuronal responses.
In summary, we report a microglia–neuron cross-talk mechanism whereby microglia respond to physiologically evoked neuronal activity and prevent dysregulated activity of neuronal circuits. Our data shed new light on the emerging role of microglia in regulating neuronal activity12 by revealing a causal role for Gi-dependent microglia dynamics in preventing hypersynchrony. Although microglia responses to neuronal activity have been shown in zebrafish larvae13, ex vivo brain slices14, spontaneous neuronal activity15, 16, and models of stroke and Alzheimer’s disease (AD)17, 18, we show that microglia process dynamic motility does not merely suppress neuronal activity, but instead maintain brain network synchronization within a physiological range and prevent hyperexcitability. By selective inactivation of the Gi pathway in microglia, we developed MgPTX mice as the first genetic tool for sustained reduction of both microglia baseline surveillance and directed motility. MgPTX mice can be used to study the role of microglia dynamics in pathophysiological conditions with neuronal hyperexcitability19, such as AD, traumatic brain injury, epilepsy, and autism. Our results support a role for Gi/Rho GTPases signaling in the regulation of microglia–neuron interactions and network hypersynchrony. Additional Gi-regulated pathways, such as cytokines released by microglia, may also affect neuronal activity. As Gi controls signaling of purinergic, glutamatergic, and adenosine microglial Gi-coupled receptors, deciphering their role in microglia–neuron interactions will reveal their relative contributions controlling network hyperexcitability9, 20. Thus, strategies to modulate Gi in microglia may hold promise to rein in hyperactivity in neurological and psychiatric disorders with impaired microglial dynamics.
Methods
Animals.
MgPTX [Tg(Cx3cr1-Cre):RosaPTX/+] mice were generated by crossing RosaPTX mice7 with the Tg(Cx3cr1-Cre) (#036395 Tg(Cx3cr1-cre)MW126Gsat/Mmucd MMRRC line21–23) mice. RosaPTX/+ littermates negative for cre (MgWT) were controls for MgPTX mice. RosaPTX/+ mice exMgPTX-ind mice were generated by crossing Cx3cr1CreER/+ line Cx3cr1tm2.1(cre/ERT2)Litt/WganJ (#021160; Jax)24, RosatdTomato/+ line25 (#007914; Jax), and RosaPTX/+ mice to generate Cx3cr1CreER/+:RosaPTX/tdTomato mice. Controls for the Cx3cr1CreER/+:RosaPTX/tdTomato (MgPTX-ind) mice were Cx3cr1CreER/+:RosatdTomato/+ mice (Ctrl). Tg(Cx3cr1-Cre) and Cx3cr1CreER mice were previously tested for microglia-specific cre expression17,21–24 and specific recombination of PTX in microglia using these two lines was experimentally validated in this study. Other constitutive microglia cre lines that were previously generated as Cx3cr1 haploinsufficient23, 26 were not used in this study. For imaging microglia, MgPTX and MgWT mice were bred with Cx3cr1GFP mice (#005582; Jax) to generate Tg(Cx3cr1-Cre):RosaPTX/+:Cx3cr1GFP/+ and RosaPTX/+:Cx3cr1GFP/+ mice, respectively. We utilized the Cx3cr1GFP/+ allele in heterozygosity as a microglia fluorescent reporter, similar to prior studies1–4, 27,28. For dendritic spine imaging, MgPTX and MgWT mice were bred with Thy1-YFP mice (#003782; Jax) to generate Thy1-YFP:Tg(Cx3cr1-Cre):RosaPTX/+ and Thy1-YFP:RosaPTX/+ littermate controls, respectively. All mice were backcrossed more than 8 times to the C57BL/6J background. This strategy ensured that all experimental mice and their littermate controls either had two wild-type Cx3cr1 alleles [Tg(Cx3cr1-Cre):RosaPTX/+, RosaPTX/+, Thy1-YFP: Tg(Cx3cr1-Cre):RosaPTX/+, Thy1-YFP: RosaPTX/+) or one wild-type Cx3cr1 allele and were heterozygous for the Cx3cr1 locus [Tg(Cx3cr1-Cre):RosaPTX/+:Cx3cr1GFP/+, RosaPTX/+:Cx3cr1GFP/+, Cx3cr1GFP/+, Cx3cr1creER/+:RosatdTomato/+ , Cx3cr1creER/+:RosaPTX/tdTomato). Mice used in the study either harbored both wild-type Cx3cr1 alleles or were heterozygous Cx3cr1GFP/+, which do not have neuronal or synapse elimination deficits28, 29. In vivo imaging experiments were performed on both males and females. EEG was performed on male mice. All experiments involving Cx3cr1CreER lines were performed on females. Mice were bred and housed at University of California, San Francisco animal facilities and all animal procedures performed under the guidelines set by Institutional Animal Care and Use Committees at the University of California, San Francisco.
Tamoxifen treatment.
For Cre recombinase induction in inducible Cx3cr1creER/+:RosaPTX/tdTomato (MgPTX-ind) mice used for in vivo imaging 6–7 weeks old females were injected i.p. daily for 10 consecutive days with 2 mg of tamoxifen (Sigma T5648) dissolved in corn oil (Sigma C8267) at 20 mg/ml. Experiments began three weeks after last the tamoxifen injection. For pilocarpine seizure experiments, female pups were injected i.p. with 200 ug of tamoxifen (prepared as above) every other day for 4 days beginning at postnatal day 10. Control Cx3cr1creER/+:RosatdTomato/+ mice (Ctrl) were also injected with tamoxifen.
Cranial window surgery for neuronal activity with microglial surveillance imaging.
Mice 8–22 weeks old were kept at 37°C using a heating pad during all surgical and imaging procedures. Surgery was performed under isoflurane anesthesia (2.0–2.5%) and buprenorphine analgesia (0.1 mg/kg). Following shaving of the head, disinfection of the skin, and local s.c. injection of 2% lidocaine, an incision was made to expose the skull. The skull was dried by removing all bone-attached membrane and fat tissue. Two small self-tapping bone screws (Fine Science Tools, #19010–10) were inserted in the skull for anchoring of the dental cement and steel head bar. Mice were then placed into a stereotactic injection apparatus. A hole was drilled in the skull to inject 1 μl of AAV1.Syn.NES-jRCaMP1b.WPRE.SV40 virus (#AV-1-PV3852, Penn Vector Core) for detection of intraneuronal Ca2+ transients30 into the whisker barrel cortex (−1.7 mm anterior–posterior, 2.65 mm medial–lateral, −0.85 mm dorsal–ventral, relative to bregma) using a beveled 33-gauge Hamilton needle at a rate of 60 nl/min. A craniotomy over the right whisker barrel cortex, including the stereotactic injection site, was prepared immediately afterwards. A circular moat of about 4 mm in diameter was drilled over the whisker barrel cortex until the thinned bone flexed under gentle pressure of a micro scalpel blade (Nordland Blade #6900, Salvin Dental). A custom-designed (eMachineShop, Mahwah, NJ, USA) steel head bar that included an imaging chamber was positioned over the craniotomy and firmly affixed to the skull with cyanoacrylate glue and Metabond® dental cement. A drop of pre-warmed (37°C) artificial cerebrospinal fluid (ACSF, HEPES-based; in mM: 125 NaCl, 10 glucose, 10 HEPES, 3.1 CaCl2, 2.7 KCl, and 1.3 MgCl2; pH 7.4) was applied to the bone island. After ~1 min, the bone island was gently lifted off with a micro scalpel blade (Nordland Blade #6900, Salvin Dental), followed by gentle flushing of the dura mater with pre-warmed ACSF to clean and remove any dural bleedings. A drop of ACSF was placed on top of the dura mater, and the craniotomy was closed and sealed off using a round glass coverslip (3 mm in diameter, type 0; Warner Instruments) which was affixed to the skull and sealed with Flow-It™ ALC™ composite (Pentron). Mice were administered buprenorphine and allowed to recover. Imaging experiments were performed 14–21 days later.
Cranial window surgery for microglial surveillance and process motility imaging.
Surgeries were performed as described above with modifications: Surgery was performed under ketamine/xylazine (KX) anesthesia (140/20 mg/kg body weight) and no AAV was injected. A 2–3 mm cranial window over the motor or somatosensory cortex was made and a dental tool was used to lift off the interior skull flap, revealing the brain surface and keeping the dura mater intact. A drop of 37oC sterile ACSF was used to cover the open brain surface. The brain was either imaged directly or a small cover glass (3–4 mm) was placed on top of the cranial window and affixed to the skull using Flow-It ALC. The skull surrounding the window was attached to a custom-made metal plate using cyanoacrylate allowing for stable head fixation at the microscope. Mice were kept warm with a pumped water-controlled heating blanket set to 37oC during surgery and heated air flow set to 37oC during imaging.
In vivo two-photon brain microscopy.
An Ultima IV 2P microscope (Prairie Technologies/Bruker) equipped with a Mai Tai eHP DeepSee and an Insight X3 Ti:sapphire femtosecond laser (pulse width <120 fs, tuning range 690–1040 nm [Mai Tai] or 680–1300 nm [Insight X3], repetition rate 80 MHz; Spectra-Physics/Newport) was used for all in vivo imaging studies. PrairieView software (v5.4; Bruker) was used for microscope and laser control and image acquisition. The laser excitation wavelength was 910–1100 nm depending on the fluorophore(s). Imaging was performed ~170–200 μm below the dura mater in the whisker barrel cortex for microglial process extension and neuronal activity imaging in awake mice. Imaging was performed ~100–120 μm below the dura mater for dendritic spine imaging or 40–100 μm below the dura mater for microglial surveillance/motility imaging in anesthetized mice. A Nikon 40 × 0.8 NA or an Olympus 25 × 1.05 NA water-immersion lens was used depending on the experiment as specified below. The maximum laser power exiting the objective was ≤ 40 mW during all imaging experiments, except during point scan laser ablations. An IR-blocking filter and 560-nm dichroic were placed in the primary emission beam path before the non-descanned photomultiplier tubes (PMTs). A 660-nm dichroic and a 692/24-nm + 607/45-nm bandpass filter set were used to separate far red (e.g., SiR-actin probe) and red fluorescence emission (e.g., jRCaMP1b), respectively; a 520-nm dichroic and a 542/27-nm + 494/41-nm bandpass filter set separated YFP and GFP fluorescence emission, respectively. A GaAsP PMT (Hamamatsu H7422PA-40) was installed after the 607/45-nm bandpass filter; all other PMTs were of the multi-alkali type (Hamamatsu R3896).
In vivo imaging of microglia in KX-anesthetized mice.
Surveillance: Motor/somatosensory cortex was imaged over 30–120 min, during which a z-stack of at least 30 μm thickness (1 μm z-step) was acquired every 2–3 minutes. Directed process motility toward laser ablation: We ablated a small site approximately 60 μm below the dural surface in the motor/somatosensory cortex with 50–75 pulses of an 880–920 nm laser for 100 ms/pulse. Successful ablation led to rapid development of an autofluorescent spot (average diameter of 6–7 μm). We imaged over 30–120 minutes, acquiring a z-stack of at least 30 μm thickness (1 μm z-step) every 1–6 minutes at 1–3 × optical zoom and 512 × 512 resolution.
In vivo glutamate uncaging.
Uncaging of glutamate was performed as described31, 32 with modifications. The craniotomy/dura mater was bathed with either vehicle alone (ACSF) or 10 mM caged RuBi-Glutamate (Tocris Bioscience) in vehicle for 30 min, shielded from light, followed by closure and sealing of the craniotomy with a glass coverslip. Uncaging was performed under ketamine-xylazine anesthesia in the somatosensory cortex (S1FL/S1HL) at a depth of ~60–80 μm using a Mai Tai eHP DeepSee 2P laser in point scan mode tuned at 800 nm. A total of three 100-ms point scan repetitions at a 1.6-μs pixel dwell time were applied for uncaging. Thirty-minute time-lapse series of microglia within a 60–70 μm z-stack around the uncaging site (30–35 μm above and below the focal plane of uncaging) were acquired in 3-min intervals at 920–940 nm.
In vivo labeling of F-actin.
F-actin in MgPTX and MgWT microglia was visualized with the cell-permeable far-red fluorescent F-actin dye “SiR-actin” that includes the broad spectrum efflux pump inhibitor verapamil (Cytoskeleton, Inc.). SiR-actin and verapamil were diluted with ACSF to 5 and 10 μM, respectively. A craniotomy was prepared as described above. The SiR-actin solution was delivered in the somatosensory cortex (S1FL/S1HL) at a depth of ~100 μm below dura by intracortical micro-injection with a pulled glass electrode using the micro-injection technique33. Thirty min later, the field of view (FOV) as observed with the Olympus 25 × objective at 2 × optical zoom was centered such that the micro-injection site was at the very edge of the region of interest. Subsequently, microglia and SiR-actin fluorescence within a 60–70 μm z-stack were imaged simultaneously by dual wavelength imaging with the Mai Tai eHP DeepSee and Insight X3 laser tuned at 920 nm (GFP excitation) and 1200 nm (SiR-actin excitation), respectively. The degree of microglial Sir-actin labeling was quantified as the percentage of GFP and SiR-actin signal pixels present in the same xyz pixel coordinates by an independent researcher blinded to genotype.
In vivo imaging of neuronal activity and microglial surveillance in awake mice.
On day 11, 12, and 13 post surgery (see “Cranial window surgery for neuronal activity with microglial surveillance imaging”), the mice were trained for 10 min to stay and run head-fixed on a mouse treadmill mounted under the microscope. On day 14–21 post surgery, mice underwent live imaging without whisker stimulation. Mice were head-fixed under the microscope and placed on a treadmill. A z-stack of 65–70 μm was set at 170–200 μm below the dura mater within the whisker barrel cortex. Time-lapse images of microglia in the absence of induced neuronal activity/whisker deflections were acquired every 5 min for a total of 35 min using galvo scan mode at 512 × 512 pixels, 1.5 Hz, and 2 × optical zoom with a 25 × objective (Olympus; 1.05 NA). For live imaging during whisker stimulation, the whiskers at the left side of the snout/contralateral to the cranial window were glued together using cyanoacrylate glue34. The mouse was subsequently head-fixed under the microscope and placed on the treadmill. The glued whiskers were attached to the whisker deflection apparatus consisting of a fine metal rod attached to a servo motor controlled by an Arduino Uno microprocessor under the command of a MATLAB® script. A region within the whisker barrel cortex 170–200 μm below the dura mater was identified that showed neuronal activity evoked by the whisker deflection. The same cortical region or a region close to that imaged in the non-evoked imaging session was identified. Time-lapse images of microglia in the presence of induced neuronal activity/whisker deflections were acquired every 5 min for a total of 35 min using galvo scan mode at 512 × 512 pixels, 1.5 Hz, and 2 × optical zoom with a 25 × objective (Olympus; 1.05 NA) at 965 nm using a Spectra-Physics Mai Tai eHP DeepSee femtosecond 2P laser. Immediately following each 5-min microglial process extension time lapse series, neuronal activity/jRCaMP1b signal was imaged before, between, and during evoked neuronal activity/whisker deflection by acquiring single-plane, 40-s time series images in spiral scan mode at 256 × 256 pixels, 12 Hz and 2 × optical zoom with a 25 × objective (Olympus; 1.05 NA) using dual wave length imaging at 965 nm (Spectra-Physics Ti:sapphire Mai Tai eHP DeepSee) and 1100 nm (Spectra-Physics Ti:sapphire Insight X3) to simultaneously visualize microglia (GFP) and intraneuronal Ca2+ (jRCaMP1b) transients. The evoked neuronal activity/whisker deflection paradigm consisted of five 100-ms, 7-degree angle deflections steered by the whisker deflection apparatus, separated by a 10-second non-stimulus interval repeated three times during each 40-s time series acquisition interval. The start of each image acquisition and each whisker deflection train was controlled by a MATLAB® script that included PrairieLink steering commands (Bruker/Prairie Technologies) to control the microscope and Arduino Support Package steering commands (MATLAB®) to control the Arduino-pulsed servo motor. Two additional whisker deflection sets were performed during the remainder of each 5-min microglia time lapse interval.
To assess synchronized/hypersynchronized neuronal network firing due to whisker stimulation, the same imaging procedure and whisker stimulation paradigm were used but at 1 × optical zoom to allow imaging of a larger FOV and, hence, of a sufficient number of neurons for quantification described in “Quantification of neuronal activity parameters” below.
Activation of Rho GTPases and in vivo imaging of microglial surveillance and neuronal activity.
Mice were surgically prepared and their microglia imaged as described under “In vivo imaging of microglia in KX-anesthetized mice” above, with modifications. A z-stack was acquired under ACSF in a region of interest devoid of large vascular structures (baseline imaging of microglial process morphology). Subsequently, the ACSF in the imaging chamber was removed and replaced with either ACSF or 15 μg/mL Rho/Rac/Cdc42 Activator I35 (Cytoskeleton, Inc.; #CN04) diluted in ACSF. The study was performed double blinded. All researchers involved in the experiments and subsequent analyses were blinded to the treatment (ACSF or Rho/Rac/Cdc42 Activator I); the vials containing the solutions were prepared and labeled with a blinded code by a researcher not involved in the studies. 5 h following the dural bathing with the blinded solution, another z-stack in the very same xyz coordinates used for the baseline z-stack was acquired. Quantification of microglial process morphology and parenchymal fill was performed as described under “Quantification of Rho-induced changes in neuronal activity and microglial surveillance in MgPTX mice” below.
For neuronal activity experiments, MgPTX mice were surgically prepared and trained for imaging under awake conditions as described above. On the day of the experiment, the head bar imaging chamber was filled with pre-warmed ACSF and the glass coverslip sealing the craniotomy was gently removed under light isoflurane anesthesia after which the anesthesia was ended. All subsequent steps were performed in the awake animal. Rho/Rac/Cdc42 Activator I was diluted to 15 μg/mL in ACSF and mixed with 3-kDa rhodamine dextran for visualization purposes. The solution was injected into an area of the whisker barrel cortex that showed neuronal activity evoked by a brief whisker deflection (see description under “In vivo imaging of neuronal activity and microglial surveillance in awake mice” above) by micro-injection with a pulled glass electrode as described previously33. Subsequently, the craniotomy was sealed with a new glass coverslip and the mouse returned to its home cage. 5 h following the Rho/Rac/Cdc42 micro-injection, the mouse was again head-fixed under the microscope and placed on the treadmill. The FOV as observed with the Olympus 25 × objective at 2 × optical zoom was centered such that the micro-injection site was at the very edge of the region of interest and the whisker stimulus paradigm with imaging of neuronal activity in the awake mouse was executed as described above. Quantification of parameters of neuronal activity and microglia surveillance in the focal single-plane time series images as acquired in spiral scan mode at 256 × 256 pixels, 12 Hz and 2 × optical zoom with a 25 × objective (see above) are described under “Quantification of Rho-induced changes in neuronal activity and microglial surveillance in MgPTX mice” below.
Pilocarpine seizure induction.
To induce seizures with pilocarpine using a repeated low-dose treatment36, experimental mice and their littermate controls were first acclimated in fresh individual cages for 1 hour. The experimenter was blinded to mouse genotype. Mice received scopolamine methyl nitrate (1 mg/kg in 0.9% saline) i.p. to inhibit adverse peripheral effects of pilocarpine. Thirty minutes later, mice received i.p. pilocarpine hydrochloride (100 mg/kg in 0.9% saline). Mice additionally received pilocarpine (100 mg/kg) i.p. every 20 minutes until Stage 4 was reached. The seizure activity (latency and severity) was scored by blinded observers according to the following scale: Stage 0, no abnormality; Stage 1, immobility, lying low; Stage 2, appearance of rigid posture, forelimb and/or tail extension, tremor (not continuous), head bobbing; Stage 3, continuous strong tremor and shaking; Stage 4, rearing/hyperexcitability/falling, tonic extension/clonic seizures with loss of posture; Stage 5, status epilepticus (continuous Stage 4 seizures) and/or death. All seizure experiments were performed between 12:00 and 18:00 hours to minimize effects of circadian rhythms. Pilocarpine seizures experiments in MgPTX-ind mice were performed on 20–22-week-old female mice.
EEG implantation, recordings, and analysis.
Tg(Cx3cr1-Cre):RosaPTX/+ mice and their RosaPTX/+ littermate controls were anesthetized with i.p. avertin (tribromoethanol, 250 mg/kg) and implanted for video EEG monitoring as described19. Briefly, teflon-coated silver wire electrodes (0.125-mm diameter) soldered to a multichannel electrical connector were implanted into the subdural space over the left frontal cortex (1 mm lateral and anterior to the bregma) and the left and right parietal cortex (2 mm lateral and posterior to the bregma). The left frontal cortex electrode was used as a reference. All EEG recordings were done at least 10 days after surgery on freely moving mice in circular open-field chambers (30 cm diameter) during the daylight cycle. EEG was recorded with Spike2 (V8) software at 500 Hz sampling rate using the Cambridge Electronics Design (CED) system. Exploratory locomotor activity was tracked using EthoVision (V11.5) software (Noldus).
For spectral analyses, unfiltered EEG recordings (sampling rate 500 sec-1) from freely moving mice were imported and analyzed using Spike2 software (CED). To obtain spectral power, recordings were subjected to a fast Fourier transform (FFT) with a Hanning window of 50% overlap between windows of 500-ms epochs. FFT was done with a 1024-point FFT size to obtain a resolution of 0.1953 Hz. The 30–90-Hz band was used to calculate gamma power and the total signal amplitude or power in V2 in each 500-ms epoch represented the magnitude of gamma oscillatory power. The gamma power values for each 500-ms epoch were further averaged over 1 min of EEG recording. Finally, signals from both hemibrains were averaged to obtain the gamma power values for each mouse. To calculate relative gamma power relative to baseline, absolute gamma power value for each mouse at baseline was used to normalize the gamma power of the pilocarpine treatment session, defined as 1.0 (gamma power relative to baseline). Consecutive stages of pilocarpine-induced hypersynchrony were highly stereotyped and defined as follows: Normal, period with <2.0 gamma relative to baseline; 1st hyper-synchrony (HS1), period with >2.0 gamma relative to baseline; 1st depression (D1), period with <0.75 gamma relative to baseline after HS1; 2nd hyper-synchrony (HS2), period with >2.0 gamma relative to baseline after D1; 2nd depression (D2), period with <0.75 gamma relative to baseline after HS2; death.
Immunohistochemistry/Microscopy.
Mice were anesthetized with avertin and transcardially perfused with 10 mM phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Brains were postfixed in 4% PFA overnight at 4oC and then cryopreserved in 30% sucrose in PBS for 2 days at 4oC. Alternatively, fresh brains were dropfixed in 4% PFA for 2 days at 4oC prior to sucrose cryopreservation. Coronal brain sections (30 μm thickness) were cut on a cryostat. Brain sections were rinsed 3 × with PBS, blocked in 3% normal donkey serum (NDS), 0.3% Triton X-100 in PBS for 1 h at room temperature, and then incubated with primary antibody in 1% NDS, 0.1% Triton X-100 in PBS overnight at 4oC. Primary antibodies used were GAD65/67, Novus #NB100–92033 (1:100); GFP, Aves #GFP-1020 (1:1000); Iba1, Novus #NB100–1028 (1:500); NeuN, Millipore #MAB377 (clone A60; 1:1000); PTX, Abcam #ab188414 (1:4000, pre-adsorbed with wild-type adult cortical tissue); vGlut1, Synaptic Systems #135304 (1:500); THIK-1, Abcam #ab237624 (1:500); rabbit anti-P2Y12R (1:200; kind gift from David J. Julius, UC San Francisco); GFAP, Invitrogen #13–0300 (clone 2.2B10; 1:1000). The next day, sections were rinsed 3 × with PBS and then incubated with AffiniPure fluorescence-conjugated donkey secondary IgG antibodies cross-adsorbed against other species (Jackson ImmunoResearch) diluted at 1:500–1:1000 in 1% NDS, 0.1% Triton X-100 for 1–3 h at room temperature. Sections were rinsed 3 × in PBS and then mounted on slides and covered with ProLong Gold Anti-Fade Mountant (Molecular Probes #P36930) containing DAPI (Molecular Probes #D1306 5 μg/ml) and coverslipped. For GAD65/67 staining, tissue was counterstained with Neurotrace Nissl (Molecular Probes #N-21483) according to manufacturer’s instructions and mounted without 4′,6-diamidino-2-phenylindole (DAPI).
For imaging immunohistochemical staining of mouse brain sections, we used an Olympus Fluoview confocal microscope equipped with a 20 × NA 1.0 objective and a 0.5–1 um z-step or the BIOREVO BZ-9000 inverted fluorescence microscope (Keyence) equipped with a Nikon CFI 60 Series infinite optical system and Keyence BZ-II Analyzer software (v1.0). Epifluorescent images were thresholded to determine positive NeuN signal over background. The average % NeuN-positive area was calculated per mouse. For vGlut1 and GAD65/67 quantification, each confocal z-stack, the z-slice with the maximum % area positive for signal was identified and then used to calculate an average % area positive for signal per mouse. For quantification of microglia-neuron interactions, each confocal z-stack of brain sections triple stained for GFP (microglia), GAD65/67 (inhibitory synapses) and neuronal soma (NeuroTrace), neurons in cortical layer V with GAD65/67-positive puncta surrounding the soma were assessed for microglial contact (only neurons with soma area ≥ 200 μm2 were included in quantification). The average number of microglial contacts per neuronal soma and the % of neurons with at least one microglial contact with the soma was calculated for each mouse.
For quantification of PTX expression in astrocytes, microglia, and neurons, confocal z-stack images with 1 μm z-step were acquired with an Olympus Fluoview confocal microscope equipped with a 20 × NA 1.0 objective and a 0.5–1 um z-step from brain sections triple-stained for GFAP (astrocytes, DyLight 405), NeuN (neuronal somata, Alexa 647), and PTX (Alexa 488 or Cy3). Endogenous GFP (Excitation laser 488 nm) or tdTomato (Excitation laser 543 nm) expression by microglia (MgPTX and MgWT and MgPTX-ind and Ctrl mice, respectively) served to visualize microglia. The same brain regions were imaged as in mice used for in vivo two-photon imaging. All quantifications were performed in non-projected, 1-μm optically sliced z-stacks separated per fluorophore/cell type/fluorescent signal, thus eliminating any potential false-positive signals from cell bodies in close apposition (~1–5 μm). PTX expression/cell type was quantified by merging each fluorophore/cell type channel with the PTX channel. The number of PTX-expressing GFAP-, NeuN-, GFP-, and tdTomato-positive cells was expressed as percentage of the total number counted per cell type. For quantification of tdTomato expression in MgPTX-ind and Ctrl mice with and without induction of tamoxifen, endogenous tdTomato signals were acquired with confocal z-stack imaging at 1 μm z-step in brain sections from each respective genotype immunostained for PTX and counterstained with DAPI. The mean pixel intensity per red fluorescence cell body as appearing in maximum z-projected images was measured and quantified. PTX expression was quantified as described above.
Serial Block-Face Scanning Electron Microscopy (SBEM).
The MgWT mice were anesthetized with ketamine/xylazine and transcardially perfused with Tyrode’s solution followed by 4% formaldehyde/0.1% glutaraldehyde in 0.1M PBS. The brain was dissected and post-fixed with 4% formaldehyde in PBS on ice for 2h and then cut into 100-μm-thick slices. Cortex sections were washed and cryoprotected for 2 h, then freeze-thawed by immersion in liquid nitrogen, with further washes in cryoprotectant and PBS solution. For the immunoperoxidase process37 the slices were blocked by incubating for 2 h in 10% normal goat serum (NGS) (Vector Laboratories), and were incubated overnight in rabbit anti-P2Y12R (1:200, kindly made available by David J. Julius, UCSF). Next, sections were incubated in the biotinylated goat anti-rabbit secondary antibody and avidin–biotin–peroxidase complex (ABC Elite; Vector Laboratories). They were washed in PBS solution and 0.05 M Tris-buffered saline (TBS) solution (Sigma), reacted with diaminobenzidine (DAB free base, Sigma) with subsequent washes in TBS and PBS solutions. Then, DAB-peroxidase labeled tissues were stained for SBEM imaging as previously described38. The tissues were stained in succession with 2% reduced osmium tetroxide, 0.05% thiocarbohydrazide, 2% osmium tetroxide, 2% uranyl acetate, and Walton’s lead aspartate solution, with intervening washes using double distilled water. The specimens were dehydrated with 70%, 90%, 100% EtOH, and dry acetone solutions and then infiltrated and embedded in Durcupan ACM and placed in a 60 °C oven for 48 h. The specimens were imaged on a Zeiss Merlin SEM equipped with a Gatan 3View 2XP system in variable pressure mode controlled by Gatan Microscopy Suite software v2.31.734.0. Stacks were collected with 2.2 kV beam acceleration and 85 pA probe current, with 5.6 nm pixels and 50 nm z-step size.
SBEM Semi-automatic Segmentation and Analysis.
For the microglia-neuron contact 3D reconstruction, CDeep3M10 a deep learning-based image segmentation tool for automated classification of structures in large-scale microscopy images, was applied. A pre-trained model (https://doi.org/10.7295/W9CDEEP3M50692) for plasma membrane detection in SBEM image volumes was applied to the whole SBEM sample. This model was trained on ground truth labels from other image volumes. The resulting cell boundary predictions and image volume were imported to VAST39, where they enabled semi-automatic tracing of microglia using the automated classification of plasma membrane boundaries as a guide to annotate rapid 3D reconstruction of microglia and neurons at the ultrastructural level. For quantification of synapse density, random pixels were chosen to serve as the centers of sub-volumes with dimensions of 5 μm × 5 μm × 2 μm and border zones of 1.25 μm for X and Y and 0.5 μm in Z, while ensuring no overlap between sampled sub-volumes. Synaptic density was estimated by having a blinded expert count all synapses within each sub-volume using the stereology tool in IMOD40, using forbidden and inclusion zones on opposing aspects of each sub-volume.
Fluorescence-activated cell sorting.
Brain tissues were collected from perfused male MgWT and MgPTX mice as previously described41. Single-cell suspensions were prepared from tissue containing cortical and hippocampal regions following the adult brain dissociation (ABD) kit manufacturer’s instructions with modification (Miltenyi Biotec). Briefly, minced tissues were incubated with ABD Mix 1 for 15 min at 34 °C, and then ABD Mix 2 was added for 10 min at 34 °C. Tissues were gently triturated and then incubated for 10 min at 34°C. Homogenized tissue solutions were passed through 70-μm smartstrainer (Miltenyi Biotec), washed with cold Dulbecco’s PBS and centrifuged at 450xg for 7 min at 4°C. Tissue debris was removed following the ABD Kit debris removal step, followed by straining through 30-μm smartstrainer (Miltenyi Biotec) and then centrifuged at 450xg for 7 min at 4°C. Single-cell suspensions were incubated with 1 μM Sytox blue live/dead stain (Thermo Fisher Scientific) for 5 min at 4°C and then cell sorting was performed on an FACSARIA II (BD Biosciences). Live Sytox blue− CX3CR1-GFP+ cells were sorted directly into tubes containing RLT plus lysis buffer (Qiagen) supplemented with 1% 2-mercaptoenthanol (Sigma) and 0.25% reagent DX (Qiagen). Cell lysates were frozen on dry ice and immediately stored at −80°C until use.
Quantitative PCR.
Cell lysates in RLT buffer were lysed using QIAshredder (Qiagen) following manufacturer’s instructions. Total RNA was isolated using the RNAeasy micro kit (Qiagen). cDNA was prepared from total RNA with Superscript IV Vilo kit (ThermoFisher). Quantitative PCR was performed on a Set-One-Plus (Applied Biosystems). Raw Ct values were normalized to Gapdh and data reported as fold change relative to control samples as described42. The following primer sequences were used: Kcnk13: forward, acaacctgagccgtgaagag and reverse, attcttccctcccgttgtgg; P2ry12: forward, gggcgtaccctacagaaaca and reverse, tgttgacaccaggcacatcc; Gapdh: forward, caaggccgagaatgggaag and reverse, ggcctcaccccatttgatgt.
GABA and Glutamate PCR array.
Microglia were isolated from perfused male MgWT and MgPTX mice by FACS41. RNA from purified microglia was analyzed using the RT2 Profiler™ PCR Array Mouse GABA & Glutamate (Qiagen #PAMM-152Z) according to manufacturer’s instructions.
Processing of in vivo imaging data.
To generate images for figures, z-stacks of images were max intensity-projected along the z-axis using ImageJ (NIH) to recreate two-dimensional representations of the imaged volumes. 3D reconstruction and volume rendering were performed using Imaris software (v9.2, Bitplane) or the 3D Viewer plugin with ImageJ (v1.51). Images acquired in awake mice were adjusted for brightness/contrast and background noise with ImageJ using the Subtract Background and/or Remove Outliers plugins. Time-lapse movies were generated using max intensity z-projections of stacks acquired sequentially over time. If drift occurred over the acquisition period, z shifts were corrected manually by taking substacks represented at all time points and lateral shifts were corrected with the PoorMan3DReg, StackReg, or ImageStabilizer plugins. Images acquired for neuronal activity assessments and related microglial dynamics were processed with ImageJ and MATLAB®.
Quantification of microglial surveillance and directed process motility in awake and KX-anesthetized mice.
Quantification of microglial surveillance was determined by the cumulative volume filled by microglia (including their processes) over time using published methods2. We collected a minimum of 20 μm z-stacks every 2–3 minutes for 30–120 minutes and created an ImageJ macro that generated cumulative max projections of the microglial signal from time t and previous time points (t0, t0 + t1, t0 + t1 + t2,…) for every z-slice in a 20-μm sub-stack. The microglial signal was then thresholded and summed for all slices and time points, after which the data were normalized to the total volume of tissue sampled to obtain a % volume surveyed over time. Note that images/movies in figures consist of max intensity projections of stacks made for illustration purposes only. For quantification of microglial surveillance in awake mice, we determined the cumulative volume filled by microglia over 35 minutes from 10 μm z-stacks acquired every 5 minutes. The microglia surveillance curves shown in Fig. 2b were acquired in awake Cx3cr1GFP/+ mice; those in Fig. 3b were acquired in MgWT mice, the controls for the MgPTX mice. There were no statistically significant differences in microglia surveillance in the unstimulated condition between Cx3cr1GFP/+ mice and MgWT mice. Both control groups show significant differences compared to their respective experimental groups. Although all mice underwent the same training for habituation to head-fixed awake imaging, behavioral differences of mice during the awake imaging experiment could have affected the observed differences in microglia motility after whisker stimulation in the control groups. See also the respective notes in the Reporting Summary.
For quantification of directed process motility toward tissue damaged by laser ablation, we generated max projections of z-stacks (40 μm thick) acquired every 4–6 minutes. For every time point, we calculated the area occupied by thresholded microglial signal within a 17.5 μm radius centered at the tissue ablation. The microglial response at each time point t was defined as the microglial signal area at time t minus the microglial signal area at time 0 (stack acquired immediately after ablation), normalized to the circular area examined, and expressed in response units (RU). Due to the initial punctate-like aggregation of the microglial processes around the ablation site/within the circular area examined, the GFP signal showed a transient decrease in intensity. This was followed by the entry of microglial processes from adjacent microglia into the circular area quantified around the ablation site, causing the signal to increase. Thus, the transient decrease in the microglial/GFP signal was not due to decreased directed motility.
To quantify directed process motility toward uncaged glutamate, we generated maximum projections of the 60–70 μm z stacks acquired every 3 minutes. For every time point, we measured the area occupied by thresholded microglial signal within a 17.5 μm radius around the site of glutamate uncaging. The microglial response at each time point t was defined as the microglial signal area at time t minus the microglial signal area at time 0 (stack acquired immediately after uncaging) and expressed as percentage of the area sampled (% area).
The velocity of individual microglial processes was quantified from the in vivo 2P time-lapse images acquired in awake mice under whisker stimulation and non-stimulation conditions by applying a vectorized motion tracking script based on the Lucas-Kanade derivative of Gaussian method (“opticalFlowLKDoG” algorithm) available in the Computer Vision Toolbox within MATLAB®. Data were visualized as quiver/vectorized velocity plots and quantified by extracting and summing the mean of the x and y matrices of velocity (Vx and Vy, respectively) from the “opticalFlow” object. The fraction of microglial process extensions and retractions under whisker stimulation and non-stimulation conditions in awake mice was quantified in MATLAB® from the same in vivo 2P time-lapse images by applying the baseline surveillance index calculation method described previously5.
Quantification of neuronal activity parameters.
From the 2P t-series images acquired at each time interval (t0–t8) during whisker stimulation–microglial surveillance, intensity-over-time data of the jRCaMP1b signals were extracted with ImageJ (NIH). The change in jRCaMP1b fluorescence over time normalized to the jRCaMP1b fluorescence at baseline/before whisker deflection was extracted from the intensity-over-time data with an in-house written MATLAB® script to yield the dF/F0 for each t-series. The amplitude, full-width at half maximum (FWHM), and number of evoked jRCaMP1b/Ca2+ peaks were determined from the dF/F0 datasets using the “findpeaks” function and related “MinPeakProminence” and “MinPeakHeight” subfunctions of the Signal Processing Toolbox within MATLAB®. MinPeakHeight was set at three times the standard deviation of the F0 data to unequivocally distinguish whisker stimulation-evoked jRCaMP1b signals from jRCaMP1b signals due to spontaneous neuronal activity.
For the assessment of global cumulative changes in somatic and dendritic jRCaMP1b/Ca2+ signals, summed 8-bit projections of each jRCaMP1b t-series image stack of the entire FOV were made in ImageJ. For each experiment time interval (t0–t8), the t-series image frames acquired before whisker deflection at t0 (baseline), during whisker deflection at t0–t8 (evoked activity), and without whisker deflection at t0–t8 (non-evoked activity) were summed separately. The total jRCaMP1b signal in each set of summed evoked activity image frames at each time interval (t0–t8) and the total jRCaMP1b signal in each set of summed non-evoked activity image frames at each time interval (t0–t8) were extracted with MATLAB®. They were normalized to the total jRCaMP1b signal in the set of summed baseline image frames acquired at t0.
To assess the degree of synchronized/hypersynchronized neuronal network firing due to whisker stimulation, the whisker stimulus neuronal activity time-lapse imaging sequences acquired at 1 × optical zoom (see “In vivo imaging of neuronal activity and microglial surveillance in awake mice” above) were analyzed with the MATLAB®-based toolbox “NeuroCa”43. The “NeuroCa” algorithms enable automated processing and quantitative analysis of neuronal Ca2+ imaging data. Pairwise cross-correlation between two cellular trains were calculated, followed by application to hierarchical clustering to visualize the degree of whisker stimulation-induced neuronal firing synchrony as clustered correlation matrices of the Ca2+/jRCaMP1b signals. The level of red shift within the matrices represents the degree of synchronized neuronal network firing, with each pixel on the x-axis representing the same single neuron as on the inverted y-axis coordinate. The data embedded in the clustered correlation matrices were extracted within the MATLAB® main programming environment and quantified as the mean correlation coefficient of neuronal network firing. Similarly, the cumulative neuronal Ca2+/jRCaMP1b pixel values during the three whisker stimulation events were extracted from the raster plots created by NeuroCa and were used to calculate synchronized neuronal network burst activity.
Quantification of neuronal activity and microglial surveillance in the presence of Rho/Rac/Cdc42 Activator I.
The single-plane images acquired in fast-scanning mode during whisker stimulus-induced neuronal activity were used to perform correlative analyses between the percentage microglia process fill (“MPF”) and cumulative evoked neuronal Ca2+ load as well as the FWHM of evoked Ca2+ transients of the same neurons at t35 of the whisker stimulation paradigm 5 h after micro-injection of Rho/Rac/Cdc42 Activator I. Imaging of microglial process morphology and parenchymal volume fill were performed blinded to treatment (vehicle or Rho Activator) and by two independent researchers, who replicated each other’s results in a double-blinded manner. Quantification was performed by a researcher not involved in the imaging studies who was blinded to the treatments. For each imaged mouse, five microglia and their processes in the baseline z-stack were randomly selected and the same microglia were identified in the post 5-h z-stack. Parenchymal volume fill was quantified for all individual microglia and was expressed as percentage over baseline. Quantification of neuronal activity was performed as described under “Quantification of neuronal activity parameters” above around neurons surrounded by microglia with pronounced MPF increase of 1.0–2.0% at t35 compared to t0 of the whisker stimulation (termed MPFhigh microglia). Neurons surrounded by microglia with a process fill similar to untreated MgPTX mice with minimal MPF change of 0.15–0.75% at both t0 and t35 of the whisker stimulation paradigm (named MPFlow) were quantified as internal control in the same FOV. Data were analyzed blinded to neuron grouping and microglia type.
Quantification of spine density.
Dendritic spine density in Thy1-YFP:RosaPTX and Thy1-YFP:Tg(Cx3cr1-cre):RosaPTX mice was assessed as previously described32 with modifications. YFP-positive dendrites were located in the somatosensory cortex (S1FL/S1HL) of anesthetized mice and subsequently imaged within xyz-stacks of 100 × 100 × 60 μm with a 40 × objective lens at 4 × optical zoom. For each mouse, the number of spines on 10–15 dendrites was counted at all xyz coordinates of the respective dendrites. The length of the dendrites was measured and the number of spines was calculated per μm dendrite length.
Microglial morphology analysis.
Microglia morphology was analyzed using Imaris (Bitplane) software with the FilamentTracer module. Total process length and number of process branch points per microglial cell from three-dimensional confocal images of GFP-positive microglia were quantified in a semi-automated manner. 10–28 cells from 3–6 images per animal were quantified. Only cells whose entire volume was captured in the confocal stack were included.
Statistics.
Statistical analyses were performed with Prism (GraphPad) unless noted. Data distribution was assumed to be normal but this was not formally tested. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications3, 19, 30, 33, 42. Mice were randomly assigned to experimental groups. No data points or animals were excluded from the analysis. If the surgical procedure caused (sub-) dural hemorrhage underneath the cranial window and/or morphological changes in the leptomeningeal vessels (such as swelling, tortuosity, or leakage), mice were not processed for imaging, as these surgical artifacts would cause inflammatory responses unrelated to the experiment. All histopathological analyses, electron microscopy, seizure experiments and EEG recordings, and quantification of neuronal activity and microglia-neuronal contacts were performed in a blinded manner. In vivo imaging and data analysis of pharmacological treatments (Rho/Rac/Cdc42 Activator I or vehicle control) were performed double blinded and were reproduced by two researchers independently. We tested for statistical significance using unpaired t-tests when comparing means of two groups. For in vivo imaging experiments with repeated measures of responses made on mice in two groups at ordered points in time, two kinds of statistical hypotheses were tested using R (v3.5.1; https://www.R-project.org/) in R Studio along with the following packages: Bioconductor (BiocManager), multtest, ggplot2, and reshape2. First, the mean (over the mice) response at each time-point was compared between the two groups. Second, the mean (over the mice) response across time (as captured by the AUC, Area-Under-the-Curve) was compared between the two groups. A permutation test (using 10,000 permutations) was used to estimate the p-value (significance) of differences between the mean responses at each time-point if the number of mice in the control group was greater than 5. A two-sample t-test was used otherwise. A two-sample t-test was also used to estimate the significance of the difference of the mean AUC between the two groups. Two-way repeated-measures ANOVA was used for statistical analysis of EEG stages and Log-rank test for time to death and time from first spike to death. Data are presented as mean ± standard error of the mean (s.e.m.).
Reporting Summary.
Further information on research design is available in the Reporting Summary
Data Availability
The data that support the findings of this study are available from the corresponding author upon request. Source data for figures 1–3 and Extended Data Figures 2–4 and 6–10 are provided with the paper.
Code Availability
The open source machine learning algorithm for large-scale EM data, CDeep3M (National Center for Microscopy and Imaging Research; UCSD, School of Medicine) is freely available at https://github.com/CRBS/cdeep3m2. MATLAB® and R code written for data analysis of this study is available at “Supplementary Software” file.
Extended Data
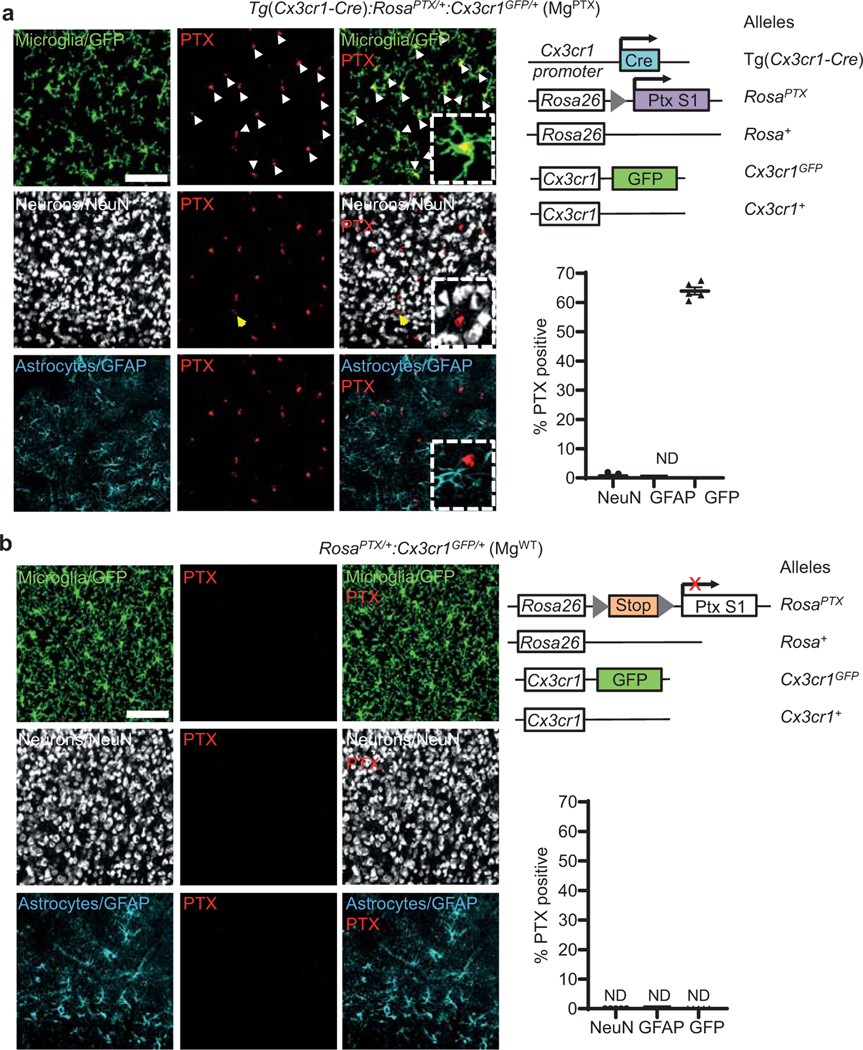
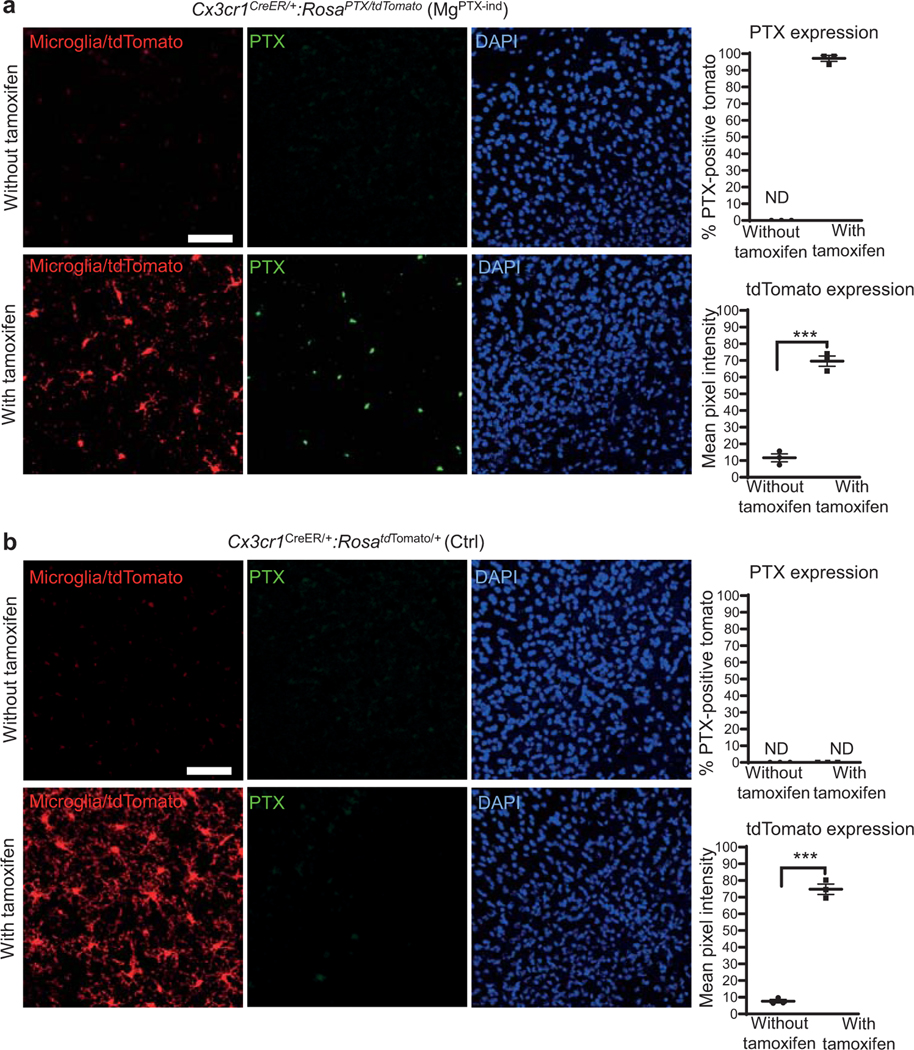
Extended Data Fig. 1. Microglia-specific PTX expression in MgPTX mice.
a, Confocal microscopy of PTX (red), neurons (white), microglia (green), and astrocytes (cyan) in cortex of MgPTX mice (left); right, schematic of MgPTX genetic background. PTX expressed in 63.9 ± 1.3% (mean ± s.e.m.) of GFP-positive microglia and 0.69 ± 0.43% (mean ± s.e.m.) of NeuN-positive neurons. Scale bar, 75 μm. n = 5 mice, 3 brain sections/mouse. ND, not detected. Quantification in an independent mouse cohort of n = 3 mice with similar results is shown in Fig. 1a. PTX expression in microglia (white arrows) and rare occasion of NeuN-positive cell co-localizing with PTX (yellow arrow). b, Confocal microscopy of PTX (red, not detected), neurons (white), microglia (green), and astrocytes (cyan) in cortex of MgWT mice (left); right, schematic of MgWT genetic background. Scale bar, 75 μm. n = 5 MgWT mice, 3 brain sections/mouse. ND, not detected.
Extended Data Fig. 2. Characterization of microglia in MgPTX mice.
a, Initial slope of microglial surveillance (% volume fill/min) in MgPTX and MgWT mice. Data are mean ± s.e.m. n = 6 mice per genotype. * P = 0.0174 by unpaired two-sample t-test. b, Confocal images (top) and filament reconstruction (bottom) of GFP-expressing microglia in MgPTX and MgWT mice and quantification of total process branch points and total process length per cell. Scale bar, 10 μm. Data are mean ± s.e.m. n = 4 mice per genotype. * P = 0.0142 and ** P = 0.0067 by unpaired two-tailed t-test. c, Confocal images of GAD65/67 (red) in MgPTX and MgWT cortex with microglia contacts (GFP, green) onto neuronal somata (NeuN, blue) surrounded by GAD65/67 puncta. Scale bar, 10 μm. Data are mean ± s.e.m. n = 5 mice per genotype. **** P < 0.0001 by unpaired two-tailed t-test. d, Relative expression of P2ry12 and Kcnk13 normalized to Gapdh in FACS-sorted microglia from cortex and hippocampus of MgPTX and MgWT mice. Data are mean ± s.e.m. n = 3 mice per genotype. ns, not significant by unpaired two-tailed t-test. e, Confocal images of THIK-1 (red) and microglial GFP (green) co-localization (yellow, arrows) in MgWT and MgPTX hippocampus. Scale bar, 10 μm. Data are mean ± s.e.m. n = 4 mice per genotype, 3 brain sections/brain region/mouse. **P = 0.0053, ns, not significant by two-way ANOVA. f, In vivo 2P images of microglia (green) and F-actin (red) in MgWT and MgPTX mice. Scale bar, 15 μm. Data are mean ± s.e.m. n = 3 mice per genotype. 5–8 microglia quantified/mouse. * P = 0.0128 by unpaired two-tailed t-test.
Extended Data Fig. 3. Dendritic spine density and synaptic markers in MgPTX mice.
a, 2P in vivo images of cortical dendritic spines (YFP). Scale bar, 10 μm. Data are mean ± s.e.m. n = 5 mice per genotype. No significant difference by unpaired two-tailed t-test. b, 3D reconstruction of excitatory synapses (green spheres) in cortical volumes obtained by SBEM. Scale bar, 2 μm. Data are mean ± s.e.m. n = 5 mice per genotype. No significant difference by unpaired two-tailed t-test. c, Epifluorescent images of NeuN-positive neurons (red) in the cortex. Scale bar, 100 μm. Data are mean ± s.e.m. n = 5 mice per genotype. No significant difference by unpaired two-tailed t-test. d, Confocal images of cortical GAD65/67. Scale bar, 50 μm. Data are mean ± s.e.m. n = 5 mice per genotype. No significant difference by unpaired two-tailed t-test. e, Confocal images of cortical vGLUT1. Scale bar, 20 μm. Data are mean ± s.e.m. n = 5 mice per genotype. No significant difference by unpaired two-tailed t-test.
Extended Data Fig. 4. Effects of microglia-specific Gi inhibition on neural excitability.
a, Gamma oscillatory power for the different EEG stages of pilocarpine-induced network hyper-synchronization in MgPTX and MgWT mice. Data are mean ± s.e.m. n = 7 mice per genotype. N, Normal; HS1, 1st hypersynchrony; D1, 1st depression; HS2, 2nd hypersynchrony; D2, 2nd depression. No significant difference (n.s.) between genotypes for each stage by two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 1. b, Pre-pilocarpine in vivo baseline EEG recordings in MgWT and MgPTX mice. Data are mean ± s.e.m. n = 13 MgPTX and n = 10 MgWT mice. No significant difference by unpaired two-tailed t-test. c, Confocal images of GFP-positive microglia in MgPTX and MgWT cortex. Scale bar, 50 μm. Data are mean ± s.e.m. n = 4 MgPTX mice and n = 4 MgWT mice. *** P = 0.0004 by unpaired two-tailed t-test.
Extended Data Fig. 5. Microglia-specific PTX expression in MgPTX-ind mice.
a, Schematic of Cx3cr1CreER/+:RosaPTX/tdTomato mice (MgPTX-ind) with inducible Gi shutdown and microglia-specific tdTomato expression. b, Confocal images of PTX (blue), Iba1 (green), tdTomato (red), and tdTomato-expressing microglia (yellow, co-localization) in cortex of MgPTX-ind mice 1.5 months after tamoxifen administration. Scale bar, 20 μm. Representative images are shown from n = 3 MgPTX-ind mice. c, Confocal microscopy of PTX (green), neurons (white), microglia (tamoxifen-induced tdTomato expression, red), and astrocytes (cyan) in cortex of MgPTX-ind mice. Exclusive expression of PTX in microglia (white arrows). PTX expressed in 97.3 ± 0.76% (mean ± s.e.m.) of tdTomato-positive microglia. Expression in neurons and astrocytes not detected (ND). Scale bar, 75 μm. Data are mean ± s.e.m. n = 5 mice, 3 brain sections/mouse. d, Confocal microscopy of PTX (green), neurons (white), microglia (tamoxifen-induced tdTomato expression, red), and astrocytes (cyan) in cortex of Ctrl mice. PTX expression not detected (ND). Scale bar, 75 μm. n = 5 mice, 3 brain sections/mouse.
Extended Data Fig. 6. Tamoxifen-dependent microglial PTX expression in MgPTX-ind mice.
a, Confocal images of PTX (green) and nuclei (DAPI, blue) in cortex of MgPTX-ind mice with or without tamoxifen. PTX expression in 97.2 ± 0.62% (mean ± s.e.m.) of tdTomato-expressing microglia after tamoxifen; 69.6 ± 3% (mean ± s.e.m.) tdTomato-positive cells after tamoxifen and 11.67 ± 1.32% (mean ± s.e.m.) tdTomato-positive cells prior to tamoxifen. Scale bar, 75 μm. Data are mean ± s.e.m. n = 3 MgPTX-ind mice per condition, 3 brain sections/mouse. *** P = 0.0001 by unpaired two-tailed t-test. ND, not detected. b, Confocal images of PTX (green) and nuclei (DAPI, blue) in cortex of Ctrl mice with or without tamoxifen (left). PTX expression was not detected; 74.7 ± 1.72% (mean ± s.e.m. tdTomato expression after tamoxifen and 7.65 ± 0.35% (mean ± s.e.m.) tdTomato-positive cells prior to tamoxifen. Scale bar, 75 μm. Data are mean ± s.e.m. n = 3 Ctrl mice per condition, 3 brain sections/mouse. *** P < 0.0001 by unpaired two-tailed t-test. ND, not detected.
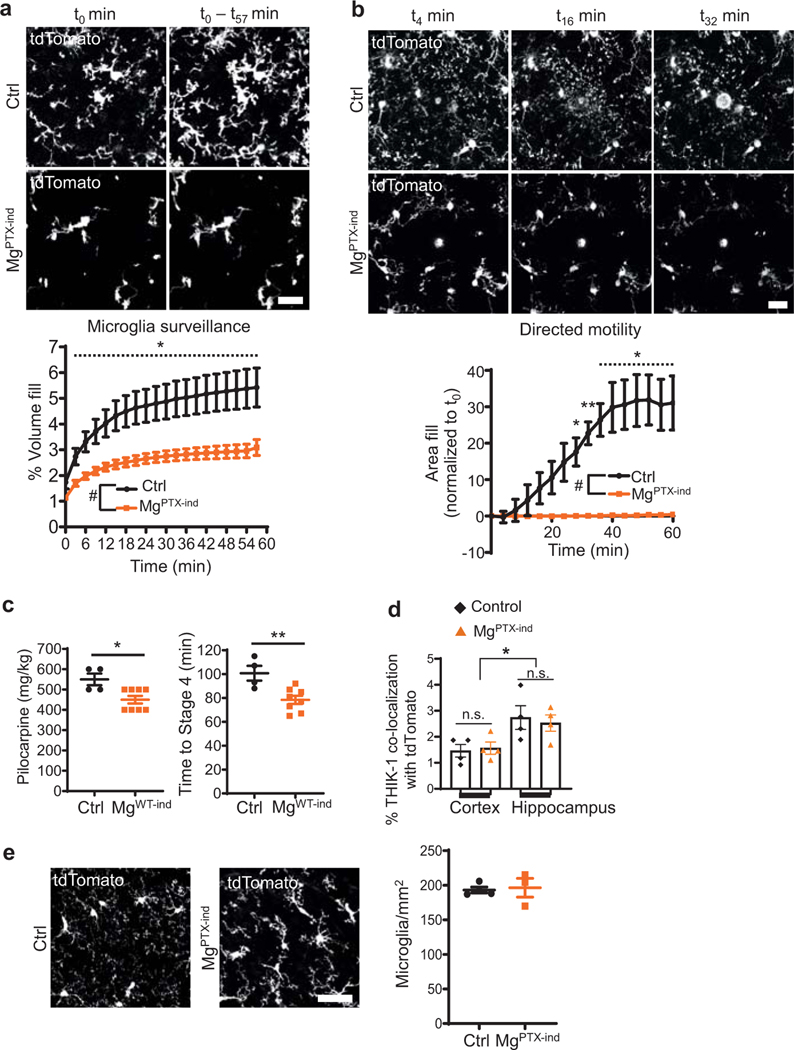
Extended Data Fig. 7. Microglia dynamics and seizures in MgPTX-ind mice.
a, In vivo 2P time-lapse imaging of cumulative microglial surveillance in MgPTX-ind and Ctrl mice after tamoxifen induction. Scale bar, 20 μm. Data are mean ± s.e.m. n = 5 MgPTX-ind and n = 4 Ctrl mice. Overall genotype effect P-value = 0.04 by two-sample t-test of mean AUC. * P < 0.05 for comparison at individual time points by unpaired two-sample t-test. b, In vivo 2P time-lapse imaging of microglial directed process motility toward laser ablation in MgPTX-ind and Ctrl mice after tamoxifen induction. Scale bar, 20 μm. Data are mean ± s.e.m. n = 5 MgPTX-ind and n = 4 Ctrl mice. Overall genotype effect # P = 0.005 by two-sample t-test of mean AUC. * P < 0.05, ** P < 0.01 for comparison at individual time points by two-sample t-test. c, Pilocarpine dose (left) and latency (right) to reach Stage 4 seizures. Data are mean ± s.e.m. n = 8 MgPTX-ind and n = 4 Ctrl mice. * P = 0.0138, ** P = 0.0061, respectively by unpaired two-tailed t-test. d, Percentage of THIK-1 co-localization with tdTomato-expressing microglia in MgPTX-ind and Ctrl mice. Data are mean ± s.e.m. n = 4 mice per genotype, 3 brain sections/brain region/mouse. *P = 0.0105. n.s., not significant by one-way ANOVA. e, Confocal images of cortical Tomato-positive microglia in MgPTX-ind and Ctrl mice 1–2 months after tamoxifen administration. Scale bar, 50 μm. Data are mean ± s.e.m. n = 3 MgPTX-ind mice and n = 4 Ctrl mice. Not significant by unpaired two-tailed t-test. sections/brain region/mouse. *P = 0.0105. n.s., not significant by one-way ANOVA. e, Confocal images of cortical Tomato-positive microglia in MgPTX-ind and Ctrl mice 1–2 months after tamoxifen administration. Scale bar, 50 μm. Data are mean ± s.e.m. n = 3 MgPTX-ind mice and n = 4 Ctrl mice. Not significant by unpaired two-tailed t-test.
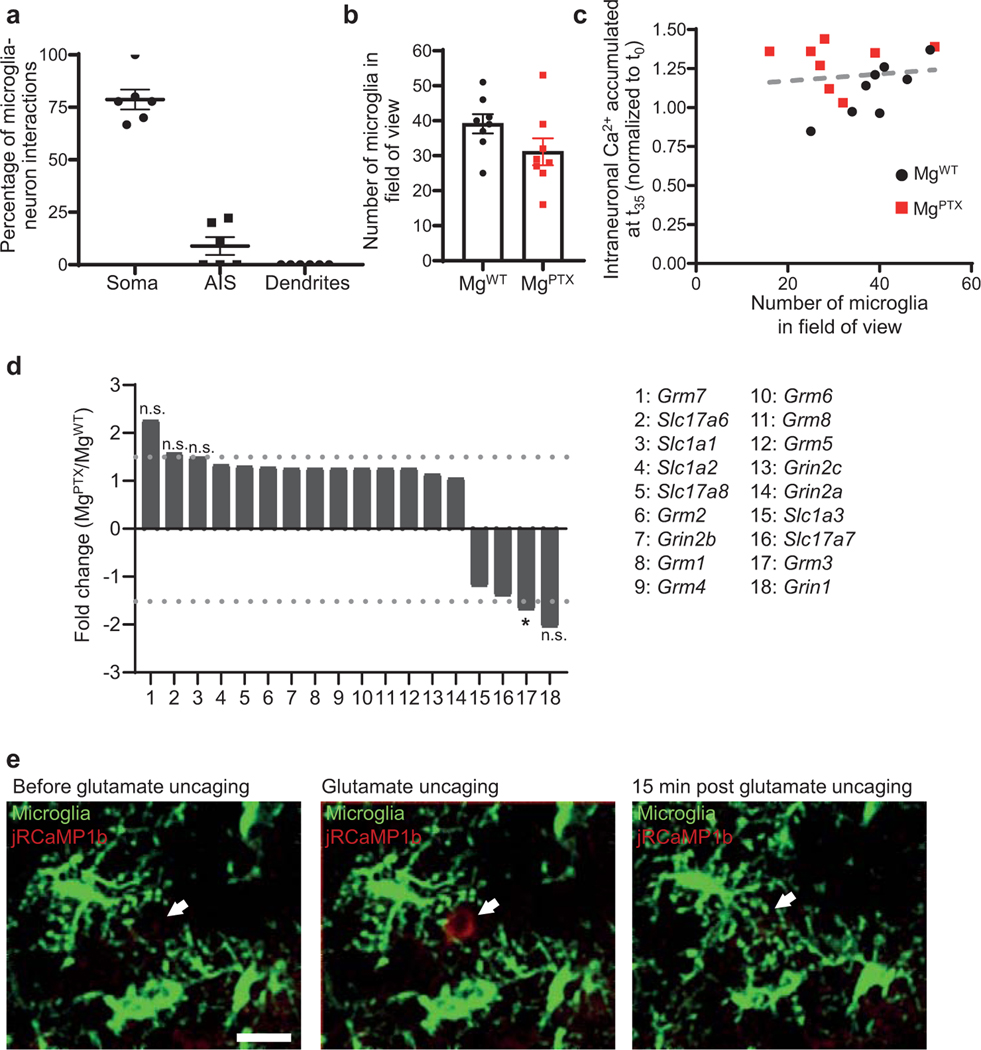
Extended Data Fig. 8. Glutamate-induced microglia–neuron interactions.
a, Location of microglia process contacts onto jRCaMP1b-expressing neurons in the whisker barrel cortex of MgWT mice (images shown in Fig. 2d). Each data point represents the average percentage of all microglia–neuron contacts quantified in an individual mouse. Data are mean ± s.e.m. n = 6 mice. AIS, axonal initial segment. b, Number of microglia in the FOV during in vivo 2P time-lapse imaging of microglial motilities and neuronal activity in awake MgWT and MgPTX mice. Data are mean ± s.e.m. n = 8 mice per genotype. Not significant, (P = 0.114) by two-tailed unpaired t-test. c, Correlation analysis of intraneuronal Ca2+ accumulation and number of microglia in awake MgWT and MgPTX mice. n = 8 mice per genotype. R2 = 0.0059; deviation from zero = not significant (P = 0.856) by linear regression analysis (dotted red line). d, Gene expression of metabotropic (Grm) and ionotropic (Grin) receptors and glutamate transporters (Slc) in MgPTX microglia compared to MgWT microglia. Data from microglia from n = 3 mice/genotype. Dotted lines indicate 1.5-fold change threshold levels. *P = 0.0245; n.s., not significant by unpaired two-tailed t-test. e, In vivo 2P time-lapse imaging of focal (arrows) glutamate uncaging-induced neuronal Ca2+ transients (jRCaMP1b, red) and directed microglia motilities (green) in the cortex of awake MgWT mice. Scale bar, 25 μm. Representative images are shown for n = 3 mice.
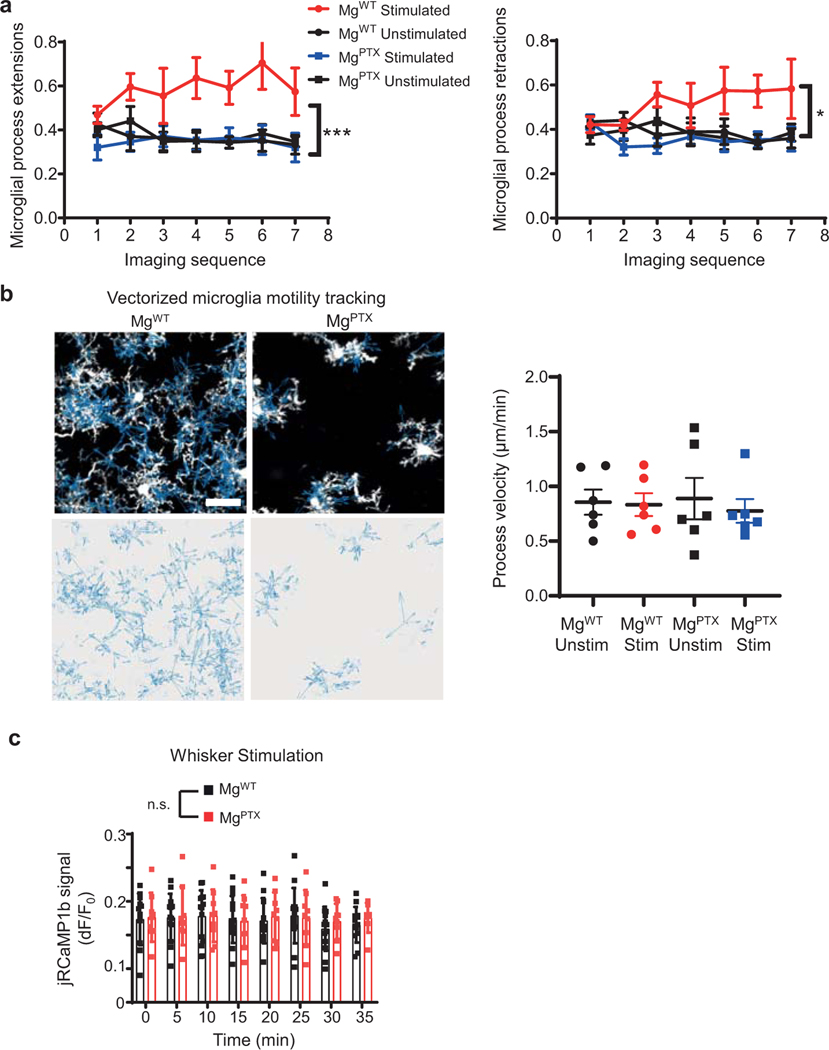
Extended Data Fig. 9. Impaired microglia dynamics in MgPTX mice.
a, In vivo 2P time-lapse imaging of individual microglial process extensions and retractions in awake MgWT and MgPTX mice during whisker stimulation (“Stim”) and non-stimulation (“Unstim”) conditions. Data are mean ± s.e.m. n = 6 mice per genotype. *P = 0.018 (MgWT Stimulated vs. all other conditions) and ***P = 0.0083 (MgWT vs. MgPTX mice) by two-way ANOVA. b, Velocity (blue vectors) of individual microglia process dynamics (white) in MgWT and MgPTX mice. Scale bar, 25 μm. Data are mean ± s.e.m. n = 6 mice per genotype. No significant difference by two-way ANOVA with post hoc Tukey’s multiple comparison test. c, Ca2+ peak amplitudes of whisker stimulation-induced neuronal Ca2+ transients in MgPTX and MgWT mice. Data are mean ± s.e.m. n = 14 MgWT and n = 9 MgPTX mice. No significant difference by-two-sample t-test of mean AUC and Permutation test.
Extended Data Fig. 10. Rho GTPase effects on microglia and neuronal activity in MgPTX mice.
a, In vivo 2P images of microglial process extensions at baseline and after Rho/Rac/Cdc42 Activator I or vehicle (ACSF) in MgPTX mice. Scale bar, 25 μm. Data are mean ± s.e.m. n = 7 mice per condition, 5–10 microglia/mouse/condition. ** P = 0.0054 by paired two-tailed t test. b, Correlation between microglia process fill (MPF) and cumulative whisker stimulus-induced neuronal Ca2+ levels 5 h after Rho/Rac/Cdc42 Activator I administration. Data are mean ± s.e.m. n = 7 MgPTX mice. Deviation from zero = significant (P = 0.0023); R2 = 0.553 (linear regression analysis [red line]); 95% confidence intervals (dotted black lines). c, Cumulative Ca2+ levels in neurons surrounded by microglia with MPF increase at t35 compared to t0 of the whisker stimulation (MPFhigh microglia) 5 h after administration of Rho/Rac/Cdc42 Activator I. Data are mean ± s.e.m. n = 7 MgPTX mice. *** P = 0.0007 for overall treatment effect by unpaired two-tailed t test. d, Total Ca2+ signal in neurons surrounded by either MPFlow or MPFhigh microglia in the FOVs described in (b) and (c). Data are mean ± s.e.m. n = 7 MgPTX mice. * P = 0.01073 and ** P = 0.00672 at individual time points (multiple unpaired t tests, Holm-Sidak). e, Intraneuronal Ca2+ peak FWHM of neurons surrounded by MPFlow microglia at t0 and by MPFhigh microglia at t35. Data are mean ± s.e.m. n = 7 MgPTX mice. ** P = 0.0057 for overall treatment effect (paired two-tailed t test).
Supplementary Material
Acknowledgements
We thank D.J. Julius for P2Y12R antibody, J. Heller Brown and L. Mucke for discussions, G. Maki for graphics, K. Claiborn and F. Chanut for editorial assistance, and the Gladstone Histology and Microscopy Core for Imaris. The EM studies were in part supported by grants for shared infrastructure from NIGMS P41 GM10341, R24GM137200, S10OD021784 (M.H.E.) and Gladstone animal facility by NCRR RR18928. This research was supported by the Berkelhammer Award for Excellence in Neuroscience (V.A.R. and A.S.M.), the UCSF Immunology NIAID T32 AI733426, AHA Postdoctoral Fellowship, and NINDS F32 NS096920 (V.A.R.), NIA K99 AG062776 (K.M.), NMSS Postdoctoral Fellowships FG-1708–28925 and FG-1944-A-1 (A.S.M. and S.B.), NINDS K02 NS110973 (M.A.P.), CTSI grant 5TL1TR001871 and NIGMS Molecular and Cellular Immunology Program Grant T32 AI00733432 (E.G.S.), NIA RF1 AG064926 (K.A., J.J.P, and M.H.E.), NIA AG047313 and AG062234 (J.J.P.), and the Simon Family Trust, the Dagmar Dolby Family Fund, Edward and Pearl Fein, the Conrad N. Hilton Foundation (17348), and NINDS R35 NS097976 (K.A.).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y. et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8, 752–758 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn A, Kirchhoff F. & Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Davalos D, Kyu Ryu J, Merlini M, Baeten KM, Le Moan N. et al. Fibrinogen-induced perivascular microglial clustering is required for the development of axonal damage in neuroinflammation. Nat Commun 3, 1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condello C, Yuan P, Schain A. & Grutzendler J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Abeta42 hotspots around plaques. Nat Commun 6, 6176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madry C, Kyrargyri V, Arancibia-Carcamo IL, Jolivet R, Kohsaka S. et al. Microglial ramification, surveillance, and interleukin-1beta release are regulated by the two-pore domain K(+) channel THIK-1. Neuron 97, 299–312 e296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Regard JB, Kataoka H, Cano DA, Camerer E, Yin L. et al. Probing cell type-specific functions of Gi in vivo identifies GPCR regulators of insulin secretion. J Clin Invest 117, 4034–4043 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernier LP, Bohlen CJ, York EM, Choi HB, Kamyabi A. et al. Nanoscale surveillance of the brain by microglia via cAMP-regulated filopodia. Cell Rep 27, 2895–2908 e2894 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Eyo UB, Murugan M. & Wu LJ Microglia-neuron communication in epilepsy. Glia 65, 5–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haberl MG, Wong W, Penticoff S, Je J, Madany M. et al. CDeep3M-Preview: Online segmentation using the deep neural network model zoo. bioRxiv 10.1101/2020.03.26.010660 (2020). [DOI] [Google Scholar]
- 11.Hall A. Rho GTPases and the actin cytoskeleton. Science 279, 509–514 (1998). [DOI] [PubMed] [Google Scholar]
- 12.York EM, Bernier LP & MacVicar BA Microglial modulation of neuronal activity in the healthy brain. Dev Neurobiol 78, 593–603 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Du XF, Liu CS, Wen ZL & Du JL Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev Cell 23, 1189–1202 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Cserep C, Posfai B, Lenart N, Fekete R, Laszlo ZI et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science 367, 528–537 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Liu YU, Ying Y, Li Y, Eyo UB, Chen T. et al. Neuronal network activity controls microglial process surveillance in awake mice via norepinephrine signaling. Nat Neurosci 22, 1771–1781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stowell RD, Sipe GO, Dawes RP, Batchelor HN, Lordy KA et al. Noradrenergic signaling in the wakeful state inhibits microglial surveillance and synaptic plasticity in the mouse visual cortex. Nat Neurosci 22, 1782–1792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F. et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szalay G, Martinecz B, Lenart N, Kornyei Z, Orsolits B. et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat Commun 7, 11499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verret L, Mann EO, Hang GB, Barth AM, Cobos I. et al. Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell 149, 708–721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A. et al. Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Nemeth DP, McKim DB, Zhu L, DiSabato DJ et al. Cell-type-specific interleukin 1 receptor 1 signaling in the brain regulates distinct neuroimmune activities. Immunity 50, 317–333 e316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold TD, Lizama CO, Cautivo KM, Santander N, Lin L. et al. Impaired alphaVbeta8 and TGFbeta signaling lead to microglial dysmaturation and neuromotor dysfunction. J Exp Med 216, 900–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao XF, Alam MM, Liao Y, Huang T, Mathur R. et al. Targeting microglia using Cx3cr1-Cre Lines: Revisiting the specificity. eNeuro 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yona S, Kim K-W, Wolf Y, Mildner A, Varol D. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyoneva S, Davalos D, Biswas D, Swanger SA, Garnier-Amblard E. et al. Systemic inflammation regulates microglial responses to tissue damage in vivo. Glia 62, 1345–1360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunner G, Cheadle L, Johnson KM, Ayata P, Badimon A. et al. Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat Neurosci 22, 1075–1088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao F, Xu JM & Jiang XH CX3 chemokine receptor 1 deficiency leads to reduced dendritic complexity and delayed maturation of newborn neurons in the adult mouse hippocampus. Neural Regen Res 10, 772–777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dana H, Mohar B, Sun Y, Narayan S, Gordus A. et al. Sensitive red protein calcium indicators for imaging neural activity. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noguchi J, Nagaoka A, Watanabe S, Ellis-Davies GC, Kitamura K. et al. In vivo two-photon uncaging of glutamate revealing the structure-function relationships of dendritic spines in the neocortex of adult mice. J Physiol 589, 2447–2457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fino E. & Yuste R. Dense inhibitory connectivity in neocortex. Neuron 69, 1188–1203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merlini M, Rafalski VA, Rios Coronado PE, Gill MT, Ellisman M. et al. Fibrinogen induces microglia-mediated spine elimination and cognitive impairment in Alzheimer’s Disease. Neuron 101, 1099–1108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocamora N, Welker E, Pascual M. & Soriano E. Upregulation of BDNF mRNA expression in the barrel cortex of adult mice after sensory stimulation. J Neurosci 16, 4411–4419 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M. et al. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725–729 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Muller CJ, Groticke I, Hoffmann K, Schughart K. & Loscher W. Differences in sensitivity to the convulsant pilocarpine in substrains and sublines of C57BL/6 mice. Genes Brain Behav 8, 481–492 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Moss J, Gebara E, Bushong EA, Sanchez-Pascual I, O’Laoi R. et al. Fine processes of Nestin-GFP-positive radial glia-like stem cells in the adult dentate gyrus ensheathe local synapses and vasculature. Proc Natl Acad Sci U S A 113, E2536–2545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilke SA, Antonios JK, Bushong EA, Badkoobehi A, Malek E. et al. Deconstructing complexity: serial block-face electron microscopic analysis of the hippocampal mossy fiber synapse. J Neurosci 33, 507–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger DR, Seung HS & Lichtman JW VAST (Volume Annotation and Segmentation Tool): Efficient manual and semi-automatic labeling of large 3D image stacks. Front Neural Circuits 12, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Mendiola AS, Ryu JK, Bardehle S, Meyer-Franke A, Ang KK et al. Transcriptional profiling and therapeutic targeting of oxidative stress in neuroinflammation. Nat Immunol 21, 513–524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu JK, Rafalski VA, Meyer-Franke A, Adams RA, Poda SB et al. Fibrin-targeting immunotherapy protects against neuroinflammation and neurodegeneration. Nat Immunol 19, 1212–1223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang MJ & Nam Y. NeuroCa: integrated framework for systematic analysis of spatiotemporal neuronal activity patterns from large-scale optical recording data. Neurophotonics 2, 035003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Source data for figures 1–3 and Extended Data Figures 2–4 and 6–10 are provided with the paper.