Abstract
The posttranscriptional modifications of tRNA's anticodon stem and loop (ASL) domain represent a third level, a third code, to the accuracy and efficiency of translating mRNA codons into the correct amino acid sequence of proteins. Modifications of tRNA's ASL domain are enzymatically synthesized and site specifically located at the anticodon wobble position-34 and 3′-adjacent to the anticodon at position-37. Degeneracy of the 64 Universal Genetic Codes and the limitation in the number of tRNA species require some tRNAs to decode more than one codon. The specific modification chemistries and their impact on the tRNA's ASL structure and dynamics enable one tRNA to decode cognate and “wobble codons” or to expand recognition to synonymous codons, all the while maintaining the translational reading frame. Some modified nucleosides’ chemistries prestructure tRNA to read the two codons of a specific amino acid that shares a twofold degenerate codon box, and other chemistries allow a different tRNA to respond to all four codons of a fourfold degenerate codon box. Thus, tRNA ASL modifications are critical and mutations in genes for the modification enzymes and tRNA, the consequences of which is a lack of modification, lead to mistranslation and human disease. By optimizing tRNA anticodon chemistries, structure, and dynamics in all organisms, modifications ensure translational fidelity of mRNA transcripts.
Keywords: Modifications prestructure tRNA, Wobble position tRNA modifications, Modifications 3′-adjacent to the anticodon, Modification chemistry and structure, Human disease and tRNA modifications, Codon degeneracy and decoding
1. Introduction
1.1. Universal Genetic Code
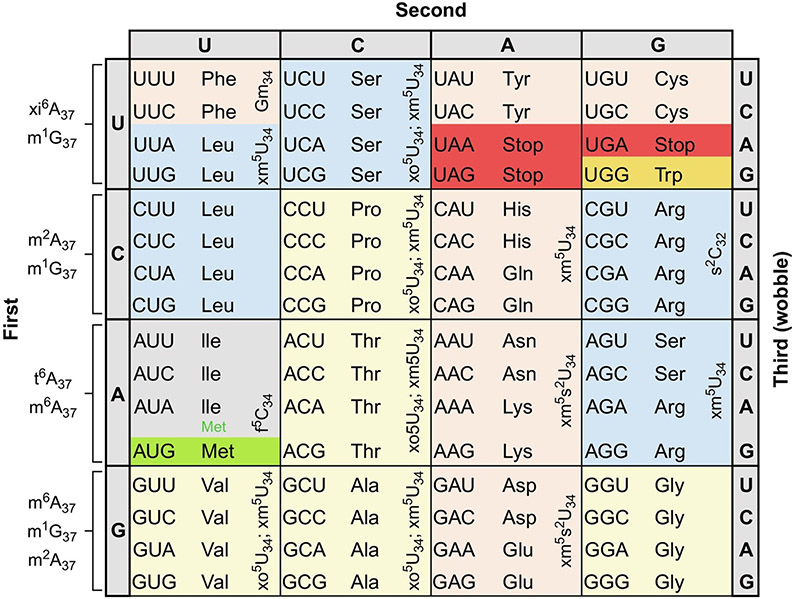
The Universal Genetic Code of 64 triplet codons encoded in DNA and transcribed into each messenger RNA (mRNA) is degenerate (Fig. 1). As a general rule, 61 codons represent the 20 amino acids during translation on the ribosome. Three codons are read by protein factors as translation stops. Transfer RNAs (tRNAs) decode the mRNA codons while bringing the cognate amino acid to the ribosome for incorporation into the growing peptide chain. The ribosome is one of the most complex of enzymes. With the bacterial and eukaryote decoding or aminoacyl site (A-site) and the peptidyl transferase center (PTC) consisting almost completely of RNA [1,2], the ribosome is an RNA-based enzyme with large RNA substrates. Even the mitochondrial ribosome, which is more highly proportioned of proteins, has an RNA-rich decoding and PTC [3]. In humans, the ribosome's substrates include some 25,000 mRNAs for proteins of less than 100 amino acids in length to more than 20,000 amino acids [4]; GTP; a number of protein factors; and some 40 transfer RNAs (tRNA) each aminoacylated with one specific amino acid. The ribosome's enzymatic activity in translating mRNA codons into the amino acid sequences is a “processive” mechanism of action. The ribosome sequentially and covalently links the growing peptide chain to the amino terminus of the incoming amino acid that is bound to its specific tRNA through an ester bond. tRNAs are aminoacylated at their universal 3′-terminal adenosine by amino acid-specific, aminoacyl-tRNA synthetases. The aminoacylated tRNA is brought to the ribosome by a protein, elongation factor, that recognizes that the tRNA is aminoacylated, but is not engaged in codon recognition and specificity.
FIG. 1.
The Universal Genetic Code. Twofold degenerate codons are highlighted in tan; threefold (Ile) in gray, fourfold in yellow, and sixfold in blue. The single codons of Met and Trp are highlighted in green and orange, respectively, and the three stop codons are highlighted in red. The figure is annotated with the abbreviations for those modified nucleosides found in the anticodon domain of tRNAs responding to the codons and discussed in this review. The chemical structures and full names of the modifications are found in Fig. 3.
Codon recognition by aminoacylated tRNA, acceptance and accommodation of the tRNA by the ribosome, peptide bond formation, translocation of the peptidyl-tRNA from the decoding or aminoacyl site (A-site) to the peptidyl-site (P-site), and the movement of unacylated tRNA from the P-site to the exit (E-site) are processive. Without leaving the mRNA template, the ribosome progresses from codon to codon from the mRNA's translational start sequence and first codon AUG for methionine to the 3′- termination codons UAG, UAA, and UGA, and release sequences. The three nucleosides of each aminoacylated tRNA's anticodon recognize a three-nucleotide mRNA codon and bind to the codon in a sequence- and frame-specific manner to deliver the 20 common amino acids for protein synthesis. The aminoacylated tRNA is accepted at the A-site in response to the complementary mRNA codon, and a proofreading mechanism consisting of some nine hydrogen bonds formed between the ribosomal RNA (rRNA), tRNA, and codon ensures a high fidelity in translation. Accurate interaction of the anticodon with codon is very much dependent on the initial Watson–Crick (A●U; G●C) complementarity of the hydrogen bonding of the first two base pairs. Other enzymes having processive mechanisms of action include the DNA- and RNA polymerases. While protein synthesis is accurate to 1 in 10,000 or 20,000 amino acids at a rate of 10–20 peptide bonds formed per second [5,6], DNA- and RNA polymerases have errors of less than 1 in 109 at 50–100s of base pairs per second including proofreading steps [7,8] and 1 in 105 at a rate of 8–85 bases per second [9,10], respectively, in vivo.
tRNA has to be considered one of the most unique of enzyme substrates. The ribosome structure and its processive catalytic mechanism [2] require the tRNA substrates to have a uniformity in chemistry and structure. Nevertheless, some 40 tRNA molecules, 22 within the human mitochondria, possess enough unique chemical diversity and structural malleability to be recognized by 20 aminoacyl-tRNA synthetases and to decode 61 mRNA codons on the ribosome. How do tRNAs achieve uniformity of chemistry and structure and yet a necessary distinctiveness for protein synthesis? Although seemingly contradictory, tRNAs use posttranscriptional modification chemistries to achieve both a stable structure mandated by the ribosome and the originality required for protein recognition determinants and codon reading.
1.2. Posttranscriptional RNA Modification
All RNAs are involved in activities that contribute directly or indirectly to the regulation, accuracy, and efficiency of gene expression that is fulfilled in translating the mRNA codons into the amino acid sequences of proteins. As in translating one language into another, the accuracy of that translation is dependent not only on a literal word-to-word equivalency but also on context, frame, and punctuation. The enzymatically synthesized, site specifically positioned, posttranscriptional modifications of RNA (Fig. 2) could be considered the punctuation marks in the translation of the genetic code. It is difficult to find among RNA species (mRNA; tRNA; ribosomal, rRNA; small nuclear, snRNA; micro, miRNA; long noncoding, lncRNA) one that is not posttranscriptionally modified [11].
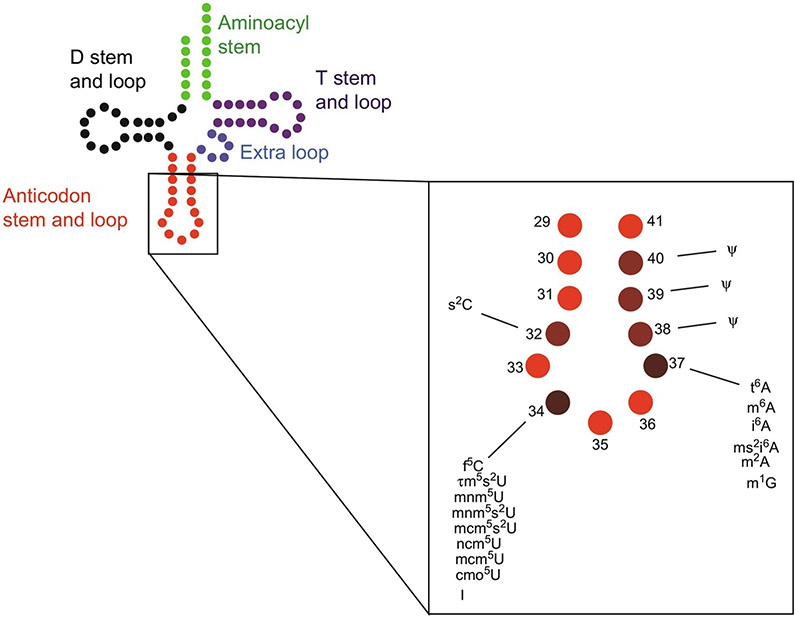
FIG. 2.
tRNA and its anticodon stem and loop (ASL) domain. Left: General cloverleaf structure of a 76-nucleotide tRNA with the aminoacyl-accepting stem in green, dihydrouridine (D) stem and loop in black, ASL in red, extra loop in blue, and ribothymidine (T) stem and loop in plum. Right: An enlargement of the ASL domain, with the abbreviations of the important modifications discussed in this review listed. A darker color hue in plum represents more important modification sites.
The Universal Genetic Code for the 20 amino acids, the first code, and the operational RNA code for the recognition of tRNA by its cognate aminoacyl-tRNA synthetase, the second code [12], did not take into account the posttranscriptional modifications of RNA, for little was known about them at the time, except that they existed. The ubiquitously occurring, conserved modified nucleosides constitute a yet to be fully appreciated third code in regulating translation, its accuracy, and efficiency [13]. Even with more than half a century of research, more than 100 different posttranscriptional modifications of RNAs remain mostly an undetermined regulating factor to translation [14,15]. The posttranscriptional modification of RNA, designated as the epitranscriptome [16], now usually refers exclusively to mRNAs. mRNAs have a half-dozen different types of modifications identified to date [17]. These represent some of the most common of all modifications, the 2′-O methylations of the four major nucleosides adenosine (Am), guanosine (Gm), cytosine (Cm), and uridine (Um); the isomer of uridine with a carbon–carbon glycosidic bond pseudouridine (Ψ); and the base methylations N6-methyladenosine (m6A), 1-methyladenosine (m1A), 7-methylguanosine (m7G), 5-methylcytosine (m5C), and 5-hydroxymethylcytosine (hm5C) [18]. Though these are simple chemical alterations, their influence in regulating translation appears to be truly significant [19-21]. We are only just beginning to understand the relevance of mRNA modifications to decoding [22].
In recent years, the coding regions of mRNA have been found punctuated with posttranscriptional modifications [17]. Early in the study of mRNA biochemistry it was recognized that mRNA had modified nucleosides in noncoding regions, such as the 5′-CAP [23]. Some of the mRNA modifications are transient in that methyltransferases “write” a methylation and demethylases “erase” the methyl group while other proteins, as “readers,” recognize the “mark” and bind the RNA, affecting its structure and function [24]. Some have been proven to affect translation and gene expression [25-28]. The biochemical mechanism by which translation is affected is at present speculative. However, nucleobase modifications of nucleosides within the mRNA coding region will alter recognition of codons by tRNA. Any modification of the Watson–Crick hydrogen bonding face of a nucleoside will change tRNA's anticodon base pairing to the codon. Thus, the methyl of an m6A in an mRNA coding region negates canonical Watson–Crick tRNA base pairing, possibly changing the accuracy and rate of decoding or affecting premature termination, and even the eventual acceptance of the mRNA for translation. The erase mechanism can then functionalize that codon for tRNA reading.
1.3. RNA Modification and the Accuracy and Efficiency of Translation
Accuracy of decoding mRNA at the ribosome's aminoacyl- or A-site depends on tRNA recognition of a complementary mRNA codon. tRNA as the decoder of mRNA is also posttranscriptionally modified for the accuracy and efficiency of reading mRNA codons [11,29-33]. More than 90 different modifications found in tRNAs are site specifically and enzymatically synthesized (Fig. 3). Fully one-quarter of a eukaryotic, cytoplasmic tRNA's nucleosides can be found in a modified form [34,35]. The locations of modifications within tRNA's conserved cloverleaf secondary and “L”-shaped tertiary structures have been known for decades because those sites of modification have been found to be conserved for specific types of modified nucleosides. Modifications outside of the anticodon region are usually of the most common type found in all RNAs, such as 2′-O-methyl, Ψ, m7G, and m1A, but also include modifications that have become synonymous with tRNA structural domains, the dihydrouridine, D, stem and loop and the ribothymidine, T, stem and loop. For the most part, their individual contributions to directing the folding of tRNA into its functioning form have only of late become clearly apparent in physicochemical terms.
FIG. 3.
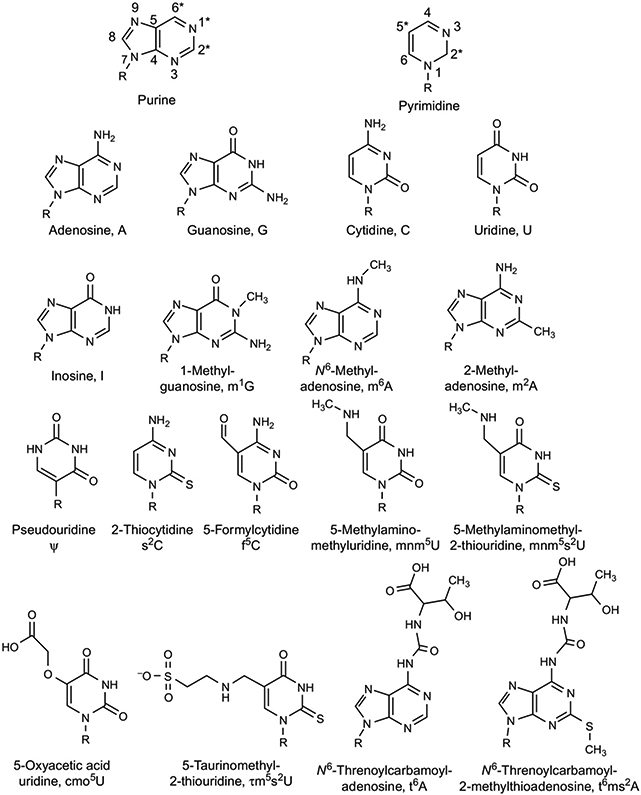
Chemical structures of the major and modified nucleosides of tRNA. Top row: General structures of purine and pyrimidine nucleosides with numbering of the atoms. R represents ribose and the asterisk (*) marks the most important and/or frequent sites of modification. Second row: Conventional RNA nucleosides and their letter abbreviations. Third through fifth rows: The modified nucleosides discussed in this review with their shorthand notions [14,15].
tRNAs are also modified in their anticodons and surrounding sequences. In fact, modifications in the anticodon stem and loop domain of tRNAs (ASL) (Figs. 2 and 3) are the most diverse and the most complex chemically and structurally of all modifications found in RNAs [36]. The ASL domain of tRNAs is critical to accurate and efficient translation, and thus, the modifications found in this domain are equally important. ASL modifications contribute directly to the correct and operationally effective decoding of mRNA codons. After the Universal Genetic Code and the operational recognition of tRNA, they constitute a third regulatory level of translational control no less, and probably more important, than those modified nucleosides found in the coding regions of mRNAs.
The 40 some tRNA species outnumber the 20 amino acids, except for instance in the human mitochondria where tRNAs and their genes number 22. Thus, there are fewer tRNA species than there are codons by a considerable amount in the mitochondrion but also in the cytoplasm of any organism. tRNA recognition of a codon through Watson–Crick canonical base pairing is irrespective of the amino acid attached to the tRNA, depending almost completely on the tRNA's anticodon sequence [37,38]. Naturally occurring mutations that give rise to premature stop codons only to be suppressed by tRNAs prove this point. Suppressor tRNAs in responding to premature stop codons in mRNAs are a naturally occurring example of the introduction of an amino acid in which the tRNA anticodon complements a stop codon, not an amino acid codon [39,40]. tRNA complementation of a stop codon has allowed for natural expansion of the genetic code [41] as well as the introduction of nonstandard amino acids via stop codon recognition by tRNA [42].
The efficiency and accuracy of translation in reading mRNA codons is shaped by the base modifications of tRNA's anticodon, the anticodon loop, and the stem [29]. With the codes being degenerate and with fewer tRNA species than the 61 amino acid codes, some tRNAs need to respond to multiple codons. In reading multiple codons effectively, tRNAs recognize exact Watson–Crick complements of their anticodon, G●C, C●G, A●U, and U●A at the first two base pairs. Degeneracy of the codes and limitation in the number of tRNA species predicates recognition of a “wobble” codon at the third base pair [43]. Wobble codon reading occurs when the nucleoside of the codon is recognized by a nucleoside of the tRNA anticodon other than the complementary base. Such a “wobble” base recognition has been realized for 50 years ever since Crick proposed the wobble hypothesis [43] for the third position of the codes, for instance U●G and G●U, and I●A/U/C [30]. For accurate insertion of the correct amino acid, tRNA discrimination of the third codon base is critically important to “mixed codon boxes” that have more than one amino acid represented. In contrast, some tRNA anticodon nucleoside modifications expand the ability of tRNA species to read as many as all four codons in a fourfold degenerate codon box [44-46]. In this review, we present examples of tRNA anticodon and loop modifications that make possible unique solutions for individual tRNA recognition of a single codon, two synonymous codons, all four codons of a fully degenerate codon box, and the six codons of a single amino acid.
2. Human Mitochondrial tRNAMet Decodes the 1:3 Degenerate Codon Box
2.1. Human Mitochondrial and Cytoplasmic tRNAMet
The mammalian mitochondrial DNA encodes all of the 22 functional tRNAs, one for each of the 18 amino acids and two each for serine and leucine [47] that are required for the synthesis of 13 critical proteins necessary for ATP synthesis and energy production [48,49]. In contrast to cytoplasmic protein synthesis, human mitochondrial tRNA for the amino acid methionine (mt tRNAMetCAU where CAU is the anticodon) is unique in that a single gene codes for the tRNA used for initiation of the translation initiation start codon and for elongation. The universal AUG codon and what is nominally the cytoplasmic isoleucine codon, AUA, are both read as methionine by this one mitochondrial tRNAMetCAU. In the cytoplasm of all cells of all living organisms, two classes of methionine tRNAs decode the only AUG methionine codon: (i) initiator tRNAMetCAU used for initiation of protein synthesis and (ii) elongator tRNAMetCAU that functions to insert methionine into the growing peptide chain. In the cytoplasm, AUA codes for isoleucine and is not used as a methionine codon. However, the AUA codon is present as 20% of the initiator codon in mitochondrial mRNAs and as 80% of internal elongator methionine codons [50,51]. The prevalence of the AUA codon in the mammalian mitochondrial genome indicates that a unique modification exists that allows the recognition of the AUA codon by the single tRNAMetCAU (mt tRNAMetCAU). Although the Crick wobble hypothesis allows for one tRNA to read more than one codon [43], it does not predict the unfavorable base pairing of CAU to AUA due to the C○A mismatch. This is overcome by the presence of a unique posttranscriptional modification of tRNAMetCAU at wobble position-34 transforming cytidine to 5-formylcytidine (f5C34). This modification is critical for the single mt tRNAMet to recognize both the nonuniversal AUA and the universal AUG codons [52,53]. N-terminal formylation of methionine is important for proper initiation of translation by bacterial and eukaryotic organelle tRNAMetCAU[54,55]. In mitochondria, it is catalyzed by the mitochondrial methionyl-tRNAMet transformylase (MTFMT) [56,57]. The formylated fMet-tRNAMet is used as an initiator tRNA, while the nonformylated methionyl-tRNAMet incorporates methionine into the elongating peptide chain.
Although the basic cloverleaf structure of mammalian mitochondrial tRNAs resembles the canonical tRNAs, they have weaker tertiary interactions in their three dimensional structure due to the lack of several conserved nucleotides that facilitate and contribute to the L-shaped tertiary structure. These features in the human mt tRNAMetCAU include (i) a smaller D-loop that lacks G18 and G19 residues which enables interactions with the T-loop in the tertiary structure; (ii) a Mg2+-binding site critical to folding of the full-length tRNA; (iii) a short variable loop lacking G47; (iv) two adjacent pyrimidine:pyrimidine base pairs, U○U and U○Ψ in the T-stem; (v) the presence of only six nucleotides instead of seven in the T-stem; and (vi) the absence of TΨC in the T-loop [58,59]. The loss of these conserved residues indicates that the tertiary interactions occurring in the shortened D- and T-loops of hmtRNAMet are weaker and different from canonical cytoplasmic tRNAs [58] with the highly conserved (Type 0) cloverleaf structure (Fig. 4).
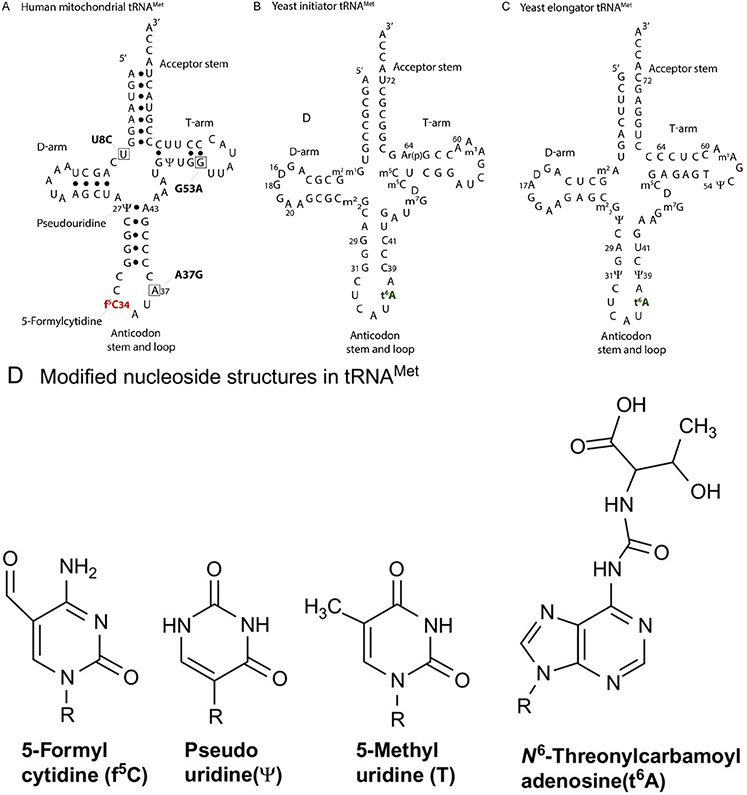
FIG. 4.
Nucleoside sequence and cloverleaf structures of tRNAMet. (A) Human mitochondrial tRNAMet displaying the condensed variable loop and the shortened D- and T-arms. Nucleotides affected by disease-causing point mutations are boxed. (B) Saccharomyces cerevisiae initiator tRNAMet. Ar(p) at position-64 refers to the 2′-O-ribosyl phosphate modification, which is an identity element of initiator tRNAMet in yeast [60]. (C) S. cerevisiae elongator tRNAMet; 5-methyluridine at position-54, indicated by T, is an important determinant of elongator tRNAMet. (D) Chemical structures of the modifications f5C34, Ψ at various noted positions, and t6A37 found in the three methionyl tRNA [61].
2.2. Posttranscriptional Modification of Mitochondrial tRNAMet
Posttranscriptionally modified cytidines have been shown to control switching between the Universal Genetic Code and a deviant code in the isoleucine/methionine codon box. The unique posttranscriptional modification of the wobble position-34 C to f5C34 allows mt tRNAMetCAU to engage in unfavorable base pairing of the CAU anticodon to the AUA codon [52,53]. This facilitates the single human mt tRNAMetCAU to be brought to the P-site of the ribosome for translation initiation as well as to interact with mitochondrial elongation factor Tu (EF-Tumt) and bind to the aminoacyl A-site of the ribosome during peptide elongation. The single modification of f5C34 alters the binding kinetics enabling the ASL to bind the Ile codons AUU and AUC, as well as AUA, in the P-site and to initiate translation in mitochondria [59]. An in vitro reconstituted mitochondrial translation system demonstrated that the modified f5C34 nucleoside of tRNAMetCAU was critical for the recognition of the nonuniversal AUA codon as methionine [62]. The f5C modification is not only found in the mt tRNAMetCAU of bovine and rodents but is thought to be universal in all mammalian mt tRNAMetCAU[53,63,64]. In addition, it is also present in the mt tRNAMetCAU of several other species including nematodes, squids, frogs, chicken, and fruit flies [65-67].
The first reported synthesis and codon binding analysis of the human mt tRNAMet ASL (hmtASLMet-f5C34) compared to the unmodified ASL showed that the f5C34 modification enhanced the motional dynamics of the loop, destabilizing the ASL [68]. The modification decreased the thermodynamic stability of the RNA, observed as a diminished melting temperature. It also reduced base stacking interactions as seen in the decreased ellipticity of the circular dichroism (CD) spectrum at 270nm. NMR and CD studies made apparent that the modification directed the conformational dynamics of the anticodon bases, facilitating binding to the P- and A-sites. A five-base-paired stem consisting of three G●C/C●G base pairs proximal to the anticodon loop also contributes to the thermodynamic stability of RNA stacking in tRNAMet species, increasing the affinity of initiator tRNAMet to binding to the P-site [59,68,69]. The wobble position f5C34 was important for the binding of the hmtASLMetCAU to the AUA codon at the A-site in canonical Watson–Crick geometry, and the rare imino-oxo tautomer of cytidine stabilized the f5C34 base pair with two hydrogen bonds [70] (Fig. 5). This is in contrast to the restrictive modification of 4-acetylcytidine (ac4C34) that prevents the cytoplasmic tRNAMetCAU from reading the AUA codon [71], and of lysidine (k2C) [72] and agmatidine (agm2C) [73,74] from preventing tRNAIleCAU from reading AUG in bacteria and archaea.
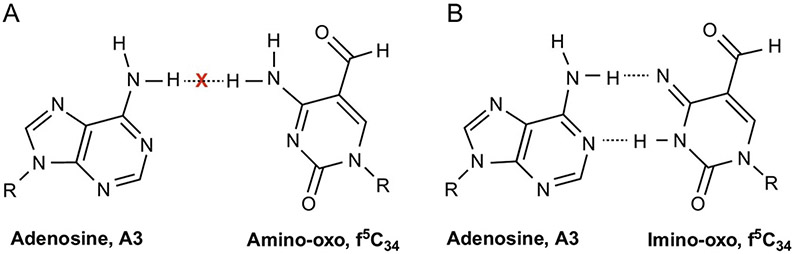
FIG. 5.
The tautomeric forms of 5-formylcytidine, f5C34. Tautomerism of f5C34 allows for the stereochemistry of the Watson–Crick base pairing between tRNA's f5C34 and A3 of the AUA codon to approximate that of a canonical U●A pair. (A) Steric repulsion between the common amino-oxo form of f5C34 N4H and A3N6H marked by an X. (B) Favorable interactions between the imino-oxo form of f5C34 and A3[70]. Dotted lines indicate hydrogen bonding between nucleoside residues [70].
A recent report demonstrated the synthesis of the 5-formyl group of f5C through consecutive reactions involving methylation, hydroxylation, and oxidation [75]. The previously uncharacterized methyltransferase, NOL1/NOP2/Sun domain-containing protein 3 (NSUN-3), catalyzed the methylation of C34 of mt tRNAMet in the presence of S-adenosylmethionine (AdoMet) to form 5-methylcytidine (m5C) in the initial step of f5C generation in human mitochondria. Human embryonic kidney cells lacking NSUN-3 showed reduced mitochondrial protein synthesis and oxygen consumption, causing deficient mitochondrial activity [75]. Mass spectrometric analysis indicated that f5C34 was absent in the NSUN3-KO cells and was replaced by m5C in these cells.
2.3. Other Human Mitochondrial tRNAMet Properties
Base pairing between f5C34 and A3 of the AUA codon was observed through crystallographic studies of the f5C34-containing ASL bound to the 30s subunit of Thermus thermophilus[70]. The authors concluded that the most likely base pairing occurs due to an exceptional shift in the tautomeric equilibrium of f5C34 from the more common amino-oxo form to the less commonly formed imino-oxo tautomer that enables interaction with A3 (Fig. 5) [70]. In addition, this study included a structural investigation of the substitution of cytidine for “invariant” uridine at position-33 (C33) as it occurs in mammalian initiator tRNAMet. The invariant U33 is responsible for the sharp U-turn in the backbone between U33 and the nucleoside in the wobble position, allowing anticodon to codon binding. One characteristic of the U-turn is the hydrogen bonding between U33 and the phosphate of nucleoside 36 (CAU). A weak interaction was seen between C33 and U36, stabilizing the sharp turn of the backbone and base pairing of f5C34. These studies using an induced-fit model not only resulted in the A-site binding conformation but also suggested a favorable canonical U-turn allowing interaction between C33 and U36, permitting it to fit into the P-site in a slightly different conformation [70].
The hmtASLMet-f5C lacks the universal t6A37 (N6-threonylcarbamoyladenosine) modification present in most cytoplasmic methionyl tRNAs and found in almost all tRNAs responding to codons beginning with A (ANN codons) [76]. The A37 modification in elongator tRNAs promotes stacking of the bases that stabilize the first anticodon–codon base pairing. The absence of t6A has been implicated in an increase in +1 and −1 frameshift in cytoplasmic translation, as well as an increase in non-AUG start codons [76]. Since unmodified, hmtASLMet-f5C forms a stable U36●A1 base pair [70], it is possible that the lack of this modification enables efficient binding to the AUA nonstart codon along with the f5C34 modification that enhances the motional dynamics of the ASL [68]. The unusual intraloop base pairing between C32 and the unmodified A37[70] can also promote a more elongator-like conformation; this would expand the codon recognition of f5C34, allowing a single tRNA to perform both as an initiator and an elongator. The human mt tRNAMetCAU is similar to the cytoplasmic elongator tRNAMetCAU in Escherichia coli, whereby they both have a wobble position-34 modification. In E. coli, it is N4-acetylcytidine (ac4C), whereas in humans this modification is 2′-O-methylcytidine (Cm) [59,77].
2.4. Human Mitochondrial tRNA Disease
Mutations of mitochondrial DNA have been associated with numerous mitochondrial diseases caused by pathogenic mutations in mitochondrial tRNA. Mutations have been found in every mitochondrial gene and are associated with several maternally inherited human genetic diseases. More than 250 mt-tRNA mutations [78] represent a large proportion of all reported mutations leading to disease. Mitochondrial dysfunction caused by point mutations in mitochondrial tRNA is associated with most maternally inherited human genetic diseases. Other mutations affecting tRNA modifications are nuclear-encoded point mutations in the genes encoding mitochondrial tRNA modification. They have been linked to mitochondrial dysfunction and contribute to molecular pathogenesis [79,80]. Three diseases caused by mutations in the gene for human mt tRNAMet arise from a single point mutation: (i) the T4409C mutation leads to a change in the conserved U8 to C in the corner of the acceptor and D-stems, causing mitochondrial myopathy that results in dystrophic muscles and intolerance to exercise [81]; (ii) the A4435G mutation resulting in an A37 to G37 change in the anticodon loop acts as a modulator of Leber's hereditary optic neuropathy (LHON), a maternally inherited disorder leading to rapid bilateral loss of central vision [82]; and (iii) the G4450A mutation causes the loss of the third G●C base pair in the T-stem, leading to splenic lymphoma, mainly in lymphocytes resulting in abnormal mitochondria, causing severe disruption of the respiratory chain (Fig. 4A; mt tRNAMet) [83].
The single point mutation of T4409C, resulting in a U8-to-C8 replacement, in the gene for hmtRNAMetCAU causes mitochondrial myopathy [58,81]. Structural probing and molecular reconstitution experiments revealed that the C8 substitution leads to a drastic disruption of the tRNA structure, and the loss of one or more critical Mg2+ binding sites on the tRNA [58]. The U8-to-C8 substitution reduced aminoacylation and resulted in the lack of a formylated mt methionyl-tRNAMetCAU. Thus, correct initiation and the degree of initiation were reduced, which led to the absence of a stable ternary complex formation with eEF1 to participate in chain elongation [58].
The maternally transmitted LHON disease impaired vision of an Asian family of three generations. They had the rare A4435G mutation which is associated with the ND4 G11778A mutation in families of Chinese origin [82]. The A4435G mutation is located in the 3′-end adjacent to the anticodon (A37) of mt tRNAMetCAU. A37 in tRNAMet species is highly conserved from bacteria to human mitochondria, and contributes to high fidelity of codon recognition and structural stabilization of tRNAs. Molecular and genetic analysis of mt DNA identified this novel A37 mutation as contributing to a significant loss of steady-state levels of tRNAMetCAU. The presence of the A4435G mutation increased the expression of the mitochondrial dysfunction associated with the G11778A mutation [82]. In addition, the A4435G mutant has been shown to have reduced m5C34 activity, the first step in f5C34 biosynthesis, indicating a possible lack of NSUN3, and hypomodified f5C34 in these patients [75].
2.5. Summary
Human mitochondria have a unique mechanism to utilize the single hmtRNAMetCAU between initiation of protein synthesis and elongation of the protein chain. The modification 5-formylcytidine at wobble position-34 permits this single tRNAMet to recognize and decode the universal AUG codon as well as the AUA codon as methionine. Codon binding and structural and thermodynamic studies have indicated stable base pairing between the noncanonical CAU and AUA bases at the wobble position, facilitated by the presence of the unique f5C34 modification found in the single tRNA. The rare imino-oxo tautomer of the modified cytidine stabilizes this base pairing and expands the use of the AUA codon in human mitochondria. Substitution of C33 for U33 in hmtASLMetCAU supported a C-turn conformation similar to the U-turn conformation seen with the invariant uridine-33. Importantly, the f5C34 modification contributes a third element of translational control by enabling wobble base pairing to isoleucine codons.
3. tRNA Decoding of the Twofold, 2:2, Degenerate Codon Box
3.1. tRNALys Twofold Degenerate Codons AAA and AAG
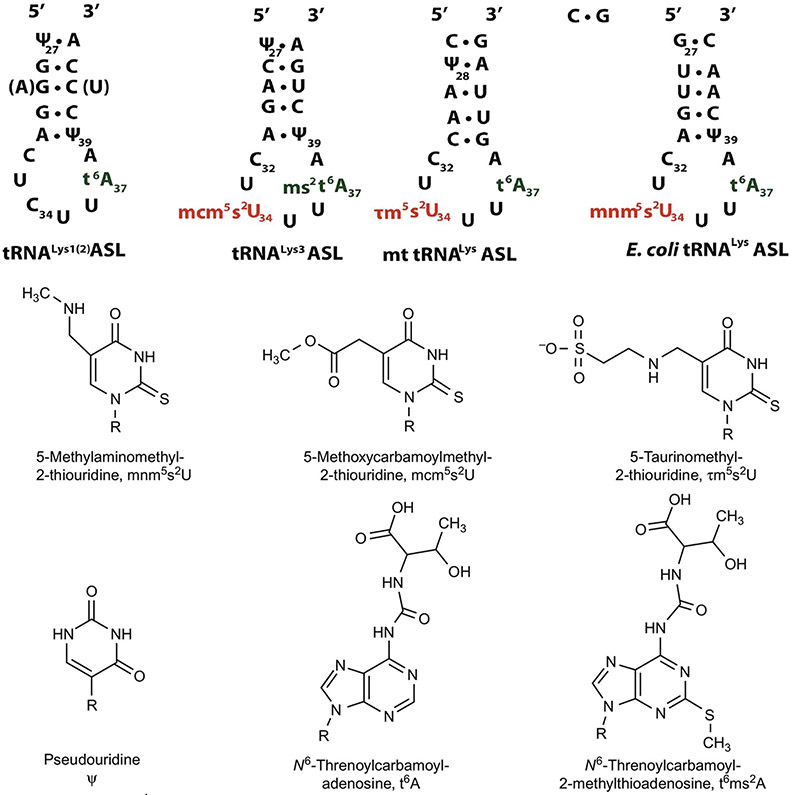
Asparagine and lysine share a twofold degenerate codon box in the Universal Genetic Code (Fig. 1) [29]. Their codons differ only in the last base being a pyrimidine, U or C for asparagine, and an A or G for lysine. Aspartic and glutamic acids, and histidine and glutamine share twofold degenerate codon boxes and similarly have codons differing only in the third base, pyrimidine or purine. Though phenylalanine, tyrosine, and cysteine are encoded by two codons each, phenylalanine shares a twofold degenerate codon box with leucine, which is one of the three amino acids encoded by six codons. Tyrosine's two codons differ from two translational stop codons only in the third base, and cysteine's two codons differ only in the third base from a stop codon and that for tryptophan. Thus, the accurate and efficient reading of mRNA codons by the tRNAs for these amino acids requires discrimination at the third base of the codon. The ability of tRNA's first or wobble anticodon base at position-34 becomes all that more important in distinguishing between codons. The number of tRNAs used for the recognition of these codons varies from amino acid to amino acid and from organism to organism [29,84]. Here, we discuss a twofold codon degeneracy using the AAA and AAG lysine codons as examples. The importance of correctly modified tRNALysUUU is not limited to accurate translation, because it is also important in maintaining the translational reading frame and translocation on the ribosome. Hypomodified or incorrectly modified mt tRNALysUUU has been linked to mitochondrial diseases such as MELAS and MERRF in humans [85], and cytoplasmic tRNALysUUU is the primer for reverse transcriptase of all lentiviruses including HIV. Mutations in modification of human tRNALysUUU have been associated with a possible mistake in proinsulin translation and are a risk factor in Type 2 Diabetes (T2D) [86,87]. The low enthalpy of the three U●A base pairs in the interaction of tRNALysUUU with its cognate codon must be compensated with stabilizing modifications [88]. The modifications present in the ASL are essential for not only codon recognition but additional functions as well [88-90].
E. coli, unlike eukaryotic cells, contains only one tRNA for lysine and it has the anticodon UUU. tRNALysUUU recognizes both AAA and AAG codons [88,90]. Archaea, like prokaryotes, for the most part contain but a single tRNALysUUU which recognizes both AAA and AAG codons [91]. The number of eukaryotic tRNALysUUU species varies among organisms. Mammals such as humans and rabbits contain three species of tRNALys that differ in their anticodons, as well as in nucleoside sequence and modifications. Both tRNALys1CUU and tRNALys2CUU contain the CUU anticodon which recognizes AAG. tRNALys3UUU contains the UUU anticodon capable of recognizing both AAA and AAG with a preference for the former (Fig. 6). In developing tissue, tRNALys2CUU may appear in a hypomodified form, sometimes referred to as tRNALys4CUU, whereas tRNALys5CUU and tRNALys6CUU refer to the various stages in between [92].
FIG. 6.
tRNALys ASL sequences and the chemical structures of their modified. Top: Sequences and secondary structures of the anticodon stem and loop (ASL) domains of human tRNALys1,2CUU, tRNALys3, human mitochondrial tRNALysUUU, and E. coli tRNALysUUU. Nucleosides in parenthesis indicate differences between tRNALys1 and tRNALys2 sequences. Bottom: Chemical structures of the modified nucleosides in the ASLs above and their shorthand notations [92,93].
The smaller genomes of organelles encode fewer tRNAs than the nuclear genome in eukaryotes, approximately 22 in mitochondria and 30 in chloroplasts [94]. Indeed, much like in E. coli, mitochondria utilize a single mt tRNALysUUU for the recognition of both AAA and AAG codons [95,96]. Additionally, it has been shown that it is feasible to design E. coli strains with reduced codon degeneracy, where degenerate codons are replaced with allowed codons [97]. There are some exceptions to the Universal Genetic Code, however. In some species, such as echinoderms and platyhelminthes, the lysine AAA codon in the mitochondria is translated as asparagine, and in starfish mitochondria, mt tRNALysCUU is the sole lysine tRNA [98]. One of the most common coding errors for lysine tRNA is mistranslation as asparagine [92,99]. Similar to mammalian mitochondria, chloroplasts utilize a single tRNALysUUU for translation [100]. Therefore, tRNALysUUU must be able to read both AAA and AAG codons in all organisms where it is utilized. This is made possible by extensive modifications of the ASL (ASLLysUUU) at positions-34 and -37, in particular [29,88,101].
3.2. Wobble Position Uridine-34 Modifications
In humans and other mammals tRNALys1CUU and tRNALys2CUU, position-34 contains an unmodified cytosine; these tRNALysCUU species read only the AAG codon. tRNALys3UUU as well as the bacterial and organelle tRNALysUUU contains the UUU anticodon that is capable of decoding both AAA and AAG codons [92,93]. In order for codons ending in a purine to be efficiently read, cognate tRNAs often contain a modified uridine nucleoside at the wobble position, where 5-methyl-2-thiouridine derivatives (xm5s2U) are commonly found (Fig. 6) [99,102]. In E. coli, the one tRNALysUUU contains 5-methylaminomethyl-2-thiouridine at position-34 (mnm5s2U34). Similarly, human tRNALys3UUU contains 5-methoxycarbonylmethyl-2-thiouridine at position-34 (mcm5s2U34; Fig. 6) [90], and mammalian mt tRNALysUUU has 5-taurinomethyl-2-thiouridine (τm5s2U34; Fig. 6) in the wobble position [96,103,104]. In several bacteria as well as in a murine leukemia cell line, the sulfur in carbon position-2 of uridine-34 can be replaced by selenium in an ATP-dependent process, resulting in a mixture of thiolated and selenated tRNAs for the amino acids glutamate, glutamine, and lysine. The s2U is a required precursor for the reaction to occur, but the mnm5 group is not [92].
3.2.1. Functional Significance of tRNA U34 Modifications
The functional significance of U34 modifications is substantial. Many of these modifications are 5-position derivatives of 5-methyl-2-thiouridine, or xm5s2U (Fig. 6). The xm5s2 modifications restrict the conformational flexibility of the anticodon, thus providing translational fidelity [45,96,105]. A complete knockout of U34 modifications is lethal in Saccharomyces cerevisiae[106]. Hypomodified tRNALysUUU cannot efficiently induce the expected conformational changes in the 30S ribosomal subunit during translation as it cannot form favorable enough interactions with its cognate codon, causing the translation to stall [88,107]. The xm5U modification in bacterial and mt tRNALysUUU (mnm5U, mcm5U, and τm5U) at the wobble position stabilizes the invariant U-turn that occurs between U33 and U34. This in turn stabilizes the anticodon–codon duplex [88,108,109]. In prokaryotes, this is achieved through a hydrogen bond between 2′-OH of U33 and the amino group of mnm5s2U34[109,110]. The xm5U modification is essential for a sure and stable recognition of the AAG wobble codon in addition to the AAA codon. For instance, the mcm5s2U34 shifts from the keto to enol tautomeric form in binding to G3, achieving a stable wobble base pair in the Watson–Crick geometry [111]. Due to its contribution to enhanced structural rigidity and preordering of the anticodon loop, mnm5s2U34 enables wobble base pairing with G, and thus allows recognition of both codons [88,112].
The mnm5U modification in the absence of the s2 group does not appear to restrict a U34●pyrimidine interaction. However, the poor base stacking of the UUU anticodon makes such restriction unnecessary as a U34○U3 interaction would abrogate any base stacking between U34 and U35, leading to a free energy penalty that would make codon recognition unfeasible. Thus, the role of the xm5U modification is to increase translational fidelity by enhancing purine recognition, but not to restrict pyrimidine recognition; this is perhaps predictable from thermodynamic stabilities [88,113]. NMR studies have shown that a modified wobble base is important for the stacking between anticodon positions-34 and -35, since in the absence of modifications noncanonical and closed conformations are highly populated for the UUU anticodon. However, in human tRNALys3UUU the mcm5U modification does not seem to enhance base stacking between U34 and U35 in a significant manner [110]. Therefore, the thio modification is required to compensate for the poor stacking of uridine [110]. The 2-thio group allows the ribose of U34 to adopt C3′-endo puckering augmenting the U-turn and thus facilitates binding with purines and restricting recognition of NNY codons [105,110,114-117]. The thio group stabilizes the U-A base pair, but it is mildly destabilizing for U-G base pairs [29,116].
Hypomodified tRNAs, such as the mammalian tRNALys3UUU lacking the fully modified mcm5s2U at the wobble position, cannot efficiently decode their cognate codons, resulting in codon-specific ribosomal pausing, which in turn effects negative consequences on protein homeostasis [107]. Hypomodified tRNALysUUU is defective in translocation from the A- to P-sites of the ribosome [89]. Interestingly though, the hypomodified bacterial tRNALysUUU is less frequently misread as asparagine than fully modified tRNALysUUU[99,108]. The increased misreading may simply be due to increased efficiency induced by the modifications. The fact that misreading increases for the mnm5U-modified tRNALysUUU compared to unmodified tRNA, but has the opposite effect for tRNAGluUUC, implies that the effect of the modification depends on the structural context as well as the chemical nature of the modification [108]. Thus, hypomodification can lead to mistranslation of alternate codons in twofold degenerate codon boxes of the Universal Genetic Code, or to changes in the translational reading frame leading to aberrant protein synthesis.
3.2.2. Modified Nucleoside-Dependent Synonymous Codon Bias at Uridine-34 and Stress
A mechanism of modified nucleoside-dependent synonymous codon bias regulates gene expression through wobble base pairing at position-34, and is most affected by the uridine-34 modifications. tRNA reading of synonymous codons, those degenerate codons for which the identical amino acid is represented, is “silent” in that the protein is not altered (Fig. 1). Yet, the reading of synonymous codons can lead to different functional outcomes [118-120]. Because tRNA modifications alter tRNA's codon usage or bias, translation can be affected by changes in modifications that support synonymous codon bias in the mRNAs of specific proteins. Variously modified aminoacyl-tRNAs are not necessarily prevalent in equal concentration to codons present, nor is the hypomodified state of the wobble-34 modification most favorable for anticodon–codon binding [119]. Also, optimal reading of codons by fully modified tRNA resulting at what appears to be the most advantageous translation rates is not always the most suitable for a particular cell condition. Thus, particular codons are read by hypomodified tRNA with the result that specific proteins harboring these synonymous codons are expressed. This regulation of translation can occur in response to factors external to cells, such as signal transduction, extracellular RNA, environmental factors and contaminants, illegal and legal drug use. The wobble position-34 uridine modifications have been of particular interest in that their codon biases are altered in response to cellular stress, such as oxidative stress [121-123]. These changes in U34 modification are associated with the translation of specific stress response proteins, the mRNAs of which have a particular codon bias, including the recoding of stop codons [124,125]. In this way, tRNA modifications are “reprogrammed” to support a cell's efficient translation of specialized proteins. Selenated or thiolated uridine-34 may help sense damage caused by irradiation [92]. A lack of the U34 thiolation has also been shown to increase sensitivity to oxidative stress in S. cerevisiae[126], whereas incomplete U34 modifications did not affect a drug-induced stress response in S. cerevisiae compared to controls [107].
3.3. Modifications of Adenosine-37, 3′-Adjacent to the Anticodon
Modifications at position-37 of tRNA are commonly found in a variety of tRNAs, with the type of modification depending on the base found at position-36 [127]. While not essential in all bacteria, the N6-threonylcarbamoyladenosine (t6A) modification at position-37 is found in tRNAs reading ANN codons in all domains of life [88,110,128-130]. It is required for efficient aminoacylation of several tRNAs, tRNA binding to the A-site of the ribosome and codon recognition in ANN codons, efficient translocation, reading frame maintenance, and preventing leaky scanning of initiation codons, and proper recognition of stop codons [88-90,131,132]. The lysine tRNAs of prokaryotes, mammalian mitochondria as well as mammalian tRNALys3 contain the t6A modification [88,110,128]. Adenosine at position-37 can also contain the 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A) or the cyclic-N6-threonylcarbamoyladenosine (ct6A) [131,133,134].
The t6A modification improves the stability of the codon–anticodon duplex through cross-strand stacking with the first base of the codon [88,110]. Furthermore, neither t6A nor ms2t6A modification results in an increase in stability of free tRNA in molecular dynamics simulations, which would be expected for a modification responsible for stabilizing the codon–anticodon interaction in the ribosome [135]. In fact, the unmodified tRNALys3UUU ASL elicits a higher melting temperature and increased stability because of intraloop hydrogen bonding compared to the open loop, t6A37-modified ASLLys3UUU[90]. Pseudouridine (Ψ) at position-39 enhances the stability of both unmodified and t6A37-modified ASLLys3UUU, yet the unmodified ASL with or without Ψ39 is incapable of binding poly-A programmed ribosomes [90,132].
A model of tRNALysUUU with the circular t6 modification, ct6A37, at the ribosomal A-site [133] compared to the structure of tRNALys3UUU containing the t6A modification at the ribosomal A-site [88] predicts the effects of the ct6A modification on codon recognition and ribosomal function. The ct6A37 modification has been shown to enhance the decoding efficiency much like the native t6A37 modification [133]. In both cases, the modification allows the nucleoside to stack with the first adenosine (A1) of the codon and across the anticodon–codon base pair, thus stabilizing the interaction (Fig. 7). A hydrogen bond between N1 and N11 is found in both cases [88,133], enhancing the base stacking ability of t6A37 with A1 as this causes the formation of a third ring that expands the area of the base [88,90]. The C14-OH group in ct6A37 can form a hydrogen bond with N7 of A1, further stabilizing the interaction [133]. The oxazolone ring is also thought to be planar with respect to the adenine [133]. The threonyl group of t6A37 has rotational freedom about the N11-C12 bond [88]. This, due to steric hindrance, prohibits the incorporation of t6A into the helix, which in turn together with its tricyclic structure allows the stacking interaction with the first adenine of the codon [88].
FIG. 7.
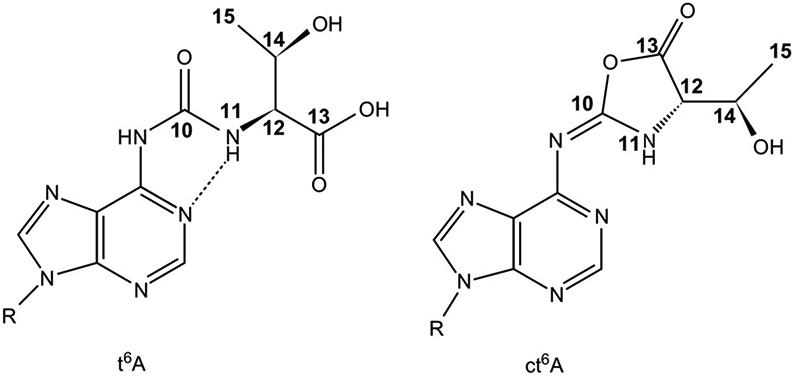
N6-Threonylcarbamoyladenosine and its cyclic derivative, t6A and ct6A. Left: Chemical structure of t6A with the atoms of the modification numbered. The hydrogen bonding that produces a pseudocyclic conformation of the modified nucleoside is shown as a dashed line. Right: The cyclic form of t6A, ct6A, with the corresponding atoms numbered [88,133].
3.3.1. Functional Significance of tRNA A37 Modifications
In a number of organisms, there is but one tRNALysUUU that reads both AAA and AAG codons [88,90,91,94]. In humans and other mammals, tRNALys3UUU decodes AAA and AAG, and is expressed at a ratio of 1:2 with tRNALys1,2CUU that reads only AAG [94]. To enhance the affinity of what is an energetically weak UUU-AAA anticodon–codon interaction, the tRNA is heavily modified with U34 and A37 contributing to binding affinity, translocation ability, and maintenance of the translational reading frame [88,109]. Modifications are necessary in order for tRNALysUUU to read the AAG codon [88,89,109]. The t6A37 modification restores cognate codon binding to unmodified tRNALysUUU, but does not enable translocation of the ASLLysUUU from the A-site to the P-site of the ribosome [89]. The modification mnm5U34 also restores codon binding but not translocation. Interestingly, the modification s2U34 enhanced translocation, but not to the degree of t6A37 and mnm5U34 together.
Together the modifications enabled cognate and wobble codon binding and resulted in a 25-fold increase in translocation in comparison to the unmodified ASLLysUUU[89]. Besides its role in translocation, the modification t6A37 is an essential, strong positive specificity determinant of many but not all prokaryote isoleucyl-tRNA synthetases [129]. The modification may also be a recognition determinant of eukaryote and viral proteins. The viral nucleocapsid protein of HIV, NCp7, recognizes t6A37[136]. Peptides, composed of 15 amino acids and selected to mimic NCp7, not only recognize the modification but are also equally capable of denaturing the modified ASLLys3UUU as does the protein [136,137]. Modification of already modified N6-adenosine-37 (t6A37, i6A37) with the 2-methylthio moiety (ms2) is found in specific tRNA species in almost all organisms [14]. Mutations in the gene for the enzyme, Cdkal1, responsible for ms2-group of ms2t6A37 in human tRNALys3UUU have been shown to be a reliable risk assessment gene for T2D [86,87]. The homozygous recessive mutation of cdkal1 has a 1.50 risk of T2D, comparable to the brca 1 and 2 risks of breast cancer. With an incompletely modified tRNALys3UUU, responsible for inserting Lys88 positioned adjacent to Arg89 at which a crucial protease cleavage separates the insulin A-chain from the C-peptide, the Lys88 codon may not be read, resulting in a proinsulin processing issue, or aberrant protein.
3.4. Summary
The twofold degeneracy within the Universal Genetic Code requires a distinction in decoding between NNA/G and NNU/C. Nature's solution to the twofold degeneracy is to ensure that tRNA anticodons read the appropriate, amino acid-specific codons and not those of the amino acid or stop codons sharing the first and second codon bases. Here again, modifications contribute a third code to controlling translation of mRNAs. In the decoding of twofold degenerate codons, modifications of tRNA's wobble position-34 add precision to the reading of the third codon base. Keto-enol tautomerism enables strong wobble codon reading in a canonical base pair geometry. Modification of the invariant purine at position-37 prestructures and restricts dynamics of the ASL domain to maintain the translational reading frame. The purine-37 modification is all that more important when the first anticodon–codon base pair is a U●A or A●U. The inherent weakness of these base pairs is counterbalanced by the van der Waals forces of the 3′-adjacent modification, creating a hydrophobic platform that is evident for such tRNAs as tRNALysUUU.
4. tRNA Decoding of Codon Fourfold Degeneracy
4.1. Single tRNA Reads All Four Codons of the Prokaryote Fourfold Degenerate Codon Box
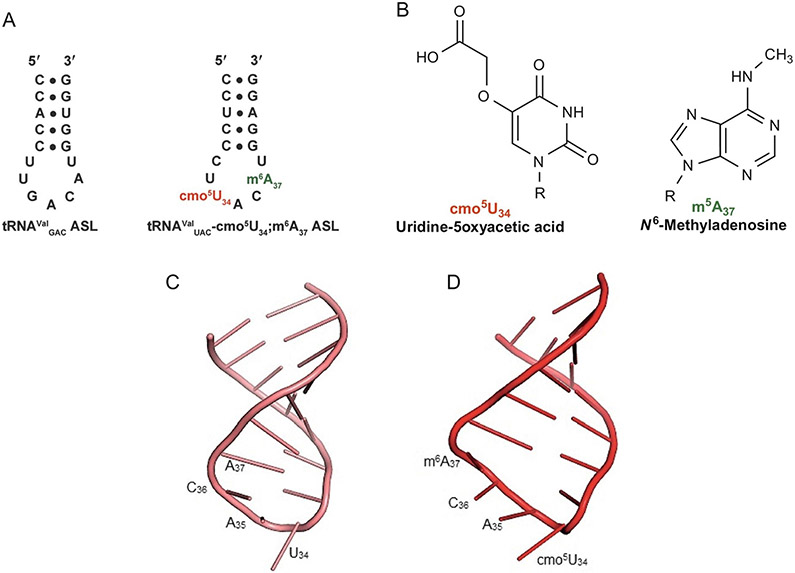
The amino acids alanine, leucine, proline, serine, threonine, and valine each claim, at least, an entire set of four codons, a completely degenerate codon box within the Universal Genetic Code (Fig. 1). Within each set of fourfold degenerate codons, the first two nucleosides of the codons are conserved, while the divergence occurs at the third nucleoside. For each of these amino acids within prokaryotic organisms, there is a single corresponding tRNA that can recognize all four of these codons. In prokaryotes, these tRNAs contain the 5-oxyacetic acid modification at the position-34 uridine (cmo5U34) (Fig. 8) [138]. Although pyrimidine:pyrimidine base pairs were originally thought to be disallowed under the original wobble hypothesis, tRNAs containing cmo5U34 can recognize codons ending in U and even C in addition to codons ending with the canonical A or the traditional wobble G [30,139-142]. The interaction between the cmo5U34 modification and the phosphate backbone has been found to promote the C2′-endo conformation of the ribose ring, which subsequently allows for the U○U base pair that would otherwise be too short (Fig. 9) [142].
FIG. 8.
The primary sequences and nucleoside modifications of the E. coli tRNAVal isoacceptors and the structures of ASLValUAC and ASLValUAC-cmo5U34;m6A37 compared. (A) The anticodon domain primary sequences of the three tRNAVal isoacceptors from E. coli and their modifications: uridine-5-oxyacetic acid at position-34, red and N6-methyladenosine at position-37, green. The two tRNAVal species containing the GAC anticodon have identical ASLs with variations in sequence in other regions of the tRNA molecule. (B) The chemical structures, names, and abbreviations of the modifications that appear in the E. coli tRNAValUAC-cmo5U34;m6A37. (C) Structure of unmodified ASLValUAC, pink. (D) Fully modified ASLValUAC containing the modifications cmo5U34 and m6A37, ASLValUAC-cmo5U34;m6A37, red, showing the base stacking of cmo5U34 and an open loop structure. The structures of the unmodified and modified ASLValUAC were determined by NMR [45] (Protein Data Bank 2JR4 for unmodified ASLVal3UAC and 2JRG for ASLVal3UAC-cmo5U34;m6A37).
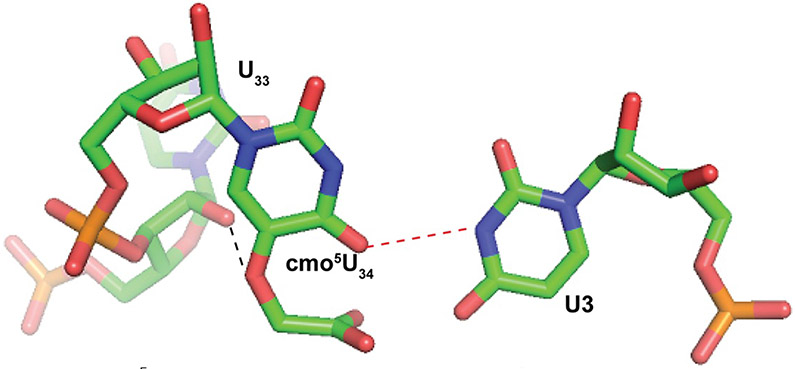
FIG. 9.
cmo5U34○U3 wobble pair. Base pairing between cmo5U34 of the ASLValUAC and U3 of the GUU codon [46] (Protein Data Bank 2UUB). A hydrogen bond occurs between the 2′-OH of the invariant U33 (background) of the ASLValUAC and O5 of cmo5U34 (black dashed line), structuring cmo5U34 for base pairing with U3. The hydrogen bond that allows for the base pair between O2 of cmo5U34 and N2 of U3 (red dashed line).
There are multiple tRNAs for the aforementioned fourfold degenerate amino acid codons, but the tRNA species containing the cmo5U34 modification may be sufficient for viability of prokaryotic organisms. Three tRNAs are responsible for decoding four proline codons in Salmonella enterica: tRNAProCGG, tRNAProGGG, and tRNAProUGG-cmo5U34. When the genes for tRNAProCGG and tRNAProGGG are deleted, the organism is still viable, indicating the ability of tRNAProUGG-cmo5U34 to read all four proline codons [143]. Furthermore, knockout of cmoA and cmoB, two genes implicated in the synthetic pathway of the cmo5U34 modification, in a strain of S. enterica containing only tRNAProUGG-cmo5U34 resulted in reduced decoding efficiency of the four proline codons by the hypomodified tRNA; the cmo5U34 modification, as opposed to the 5-hydroxyuridine 34 (ho5U34) and 5-methoxyuridine 34 (mo5U34) modifications, must be vital for recognition of tRNAProUGG-cmo5U34 by near-cognate codons within the Salmonella system [143].
In E. coli, the cmo5U34-modified tRNAAlaUGC can bind to the GCC codon within the ribosomal A-site, but this binding was found to be seven times less stable than binding to the GCA codon. The rate of GTP hydrolysis during ribosomal translocation with the GCC codon differed from that of the cognate codon only by a factor of 2.5 [144]. In fact, the cmo5U34 modification may be necessary in order to prevent the ribosome from stalling in the presence of an anticodon–codon pair in which a U○U or C○U base pair occurs at the wobble position. Translocation from the A-site to the P-site of the ribosome did not occur when unmodified E. coli tRNAValUAC was bound to the GUU valine codon [89].
4.2. Eukaryote tRNAs That Wobble in Reading Fourfold Degenerate Codons
While the cmo5U34 modification is exclusive to prokaryotic tRNA, eukaryotic tRNA species that decode for the amino acids with fourfold degenerate codons, among others, contain similar modifications at the 5-position carbon of uridine-34 (Fig. 1) [145]. These eukaryotic modifications lack the methoxy group connectivity of cmo5U34 yet include a carboxylic acid derivative. Perhaps this resemblance is enough to enhance recognition at the wobble position in more complex organisms, expanding the wobble capacity of eukaryotic tRNAs as well. In S. cerevisiae, the 5-carbamoylmethyluridine-34 modification (ncm5U34) present in tRNAValUAC, tRNAThrUGU, and tRNASerUGA enhances the efficiency of wobble pairing to the GUG, ACG, and UCG codons, respectively [112]. In fact, fully 25% of the S. cerevisiae tRNA wobble position-34 uridines are modified to be 5-carbamoylmethyluridine, ncm5U34 or mcm5U34 for translational accuracy and efficiency. Synthesis of these modifications requires the Elongator complex associated with elongating RNA polymerase II transcription (see chapter “Structures and activities of the Elongator complex and its co-factors” by Kolaj-Robin and Séraphin). The Elongator complex is composed of six elongator protein subunits and appears conserved among eukaryotes. In addition, three protein subunits of the yeast Kluyveromyces lactis killer toxin (zymocin) insensitivity genes are involved in this modification [146]. tRNAs with the modifications mcm5s2, tRNAGluUUC, tRNALys3UUU, and tRNAGlnUUG are cleaved by zymocin toxin (γ-toxin) at the 3′-side of the modified nucleoside. Resistance to γ-toxin is observed in the modification-deficient mutants [146]. Lack of Elongator activity results in hypomodified tRNA which, in turn, causes defects [147]. ELPC-1 and ELPC-3, the Caenorhabditis elegans homologues to the S. cerevisiae Elongator complex subunits Elp1 and Elp3p, are essential to the synthesis of the ncm5U34 and mcm5U34 modifications within nematode tRNAs [148]. The xm5- and s2 modifications are primary to maintaining the translational reading frame and thus accuracy of protein synthesis. As ELPC-1 and ELPC-3 are expressed in ASE neurons, elpc-1 and elpc-3 mutants demonstrate defective neuronal function, as exhibited through the inability of the mutants to adapt the expected experience-dependent behavioral changes in response to having previously been exposed to 100mM sodium chloride in nutrient-deficient conditions [148].
4.3. Roles of Both Wobble Positions-34 and -37 Modifications in Reading Fourfold Degenerate Codons
In addition to the cmo5U34 modification, the E. coli isoaccepting tRNAValUAC contains another modification on the adenosine adjacent to the anticodon. The N6-methyladenosine modification (m6A37) (Fig. 8), which naturally occurs at position-37 of the tRNAValUAC species with cmo5U34, plays an important role in structurally ordering the ASL for binding to the codon. Modifications at this conserved purine have been proposed to negate intraloop hydrogen bonding, thereby allowing the ASL to attain an open loop structure for binding to the codon, and to simultaneously enhance base stacking (Fig. 8). Base stacking effectively orders the ASL structure [30]. Determination of thermodynamic stability by UV melting of the ASL of E. coli tRNAPhe containing the N6-isopentenyladenosine (i6A37) modification at nucleoside-37 supported the hypothesis that an intraloop hydrogen bond was disrupted [149]. Similar results were determined for the ASL of human tRNALysUUU containing the N6-threonylcarbamoyladenosine (t6A37) modification at position-37 [90].
Differences in thermal stabilities were observed for the fully modified tRNAValUAC with cmo5U34 and m6A37 in comparison to that of the unmodified tRNAValUAC[45]. While the melting temperatures, Tm, did not differ substantially, the hyperchromicity of the modified tRNAValUAC was more than twice that of the unmodified tRNAValUAC[45]. Coupled with the entropic contributions of the modifications, this increased hyperchromicity indicated the increased base stacking, resulting in a more ordered ASL that arose from the presence of the cmo5U34 and m6A37 modifications [45]. The increased ellipticity of the fully modified tRNAValUAC hairpin in comparison to that of the unmodified tRNAValUAC hairpin, obtained by CD spectroscopy, also confirms the increased base stacking within the ASL of the fully modified tRNAValUAC[45].
The fully modified tRNAValUAC with cmo5U34 and m6A37 decodes the GUA, GUG, GUU, and GUC codons [140]. Within the A-site of the 30S ribosomal subunit, the binding affinities of both the fully modified tRNAValUAC and the unmodified tRNAValUAC to each of the four codons were measured [45]. There was a little difference between the dissociation constants for fully modified tRNAValUAC and the unmodified tRNA when binding the cognate GUA codon. Whereas the fully modified tRNAValUAC bound to the GUG and GUU codons (Kd=1.96 and 1.93μM, respectively), the unmodified tRNAValUAC did not have measurable dissociation constants for these three valine codons [45]. The Kd for tRNAValUAC-cmo5U34;m6A37 binding to the GUC codon remains undetermined [45]. Crystal structures have been elucidated for the binding of the fully modified tRNAValUAC on the small subunit of the ribosome in response to mRNA containing each of the valine codons, including the GUC codon [46]. The previously unexpected U○C wobble pairing in the presence of the cmo5U34 modification must occur, despite being a weak interaction.
The cmo5U34 and m6A37 modifications play important roles in structuring the ASL domain of the fully modified tRNAValUAC. The comparatively low root-mean square deviation values obtained from NOESY spectra suggest that the fully modified ASLValUAC is more ordered than unmodified ASLValUAC due to the steric hindrance encountered by the N6-methyl of the m6A37 modification and to the geometry of the cmo5U34 modification brought about by hydrogen bonding between the base and the 5-oxy group [45]. Additionally, both the fully modified and modified ASLs contained some, but not all, defining characteristics of a U-turn at U33, a type of π-turn that confers stability onto the ASL through base-phosphate stacking and hydrogen bonding [45]. With respect to this feature, the ASLValUAC with cmo5U34 and m6A37 contains the shorter hydrogen bond between U33 O2′ and A35 N7 [45].
4.4. Summary
The existence of a single species of tRNA that can decode each codon of a fourfold degenerate set is undoubtedly advantageous. Referring back to S. enterica, the presence of the modified tRNAProUGG with cmo5U34 in the absence of tRNAProCGG and tRNAProGGG allows for survival of the organism, albeit at a reduced growth rate [143]. In some cases, the cmo5U34 modification may prove absolutely essential, even for reading of cognate codons. For instance, the unmodified tRNAAlaUGC cannot recognize the cognate GCA codon or the near-cognate GCG in accordance with traditional base pairing and wobble pairing, respectively, in the ribosomal P-site [150]. In decoding of the wobble codons ending in G, cmo5U34 is crucial [44]. The cmo5U34 is apparently in the enol form and facilitates formation of a Watson–Crick geometry when binding G3. An intramolecular hydrogen bonding stabilizes the conformation of the anticodon for codon binding [46].
5. Three Amino Acids Are Encoded by Six Synonymous Codons Each
5.1. Sixfold Degeneracy and Reading of the Arginine Codons
Within the Universal Genetic Code, three amino acids are coded by six different codons: arginine, leucine, and serine. These six synonymous codons always comprise one whole and an additional half of a twofold degenerate codon box, such that ambiguity exists not only in the identity of the base at the third codon position, but in that of the bases at the first and second positions as well (Fig. 1). This poses a chemical challenge for their set of tRNA isoacceptors, which must together recognize and discriminate among all six degenerate codons while sharing the specific structure and chemistry required for their common recognition by a sole cognate aminoacyl-tRNA synthetase and by the ribosomal A-site. In the case of the E. coli arginine tRNA isoacceptors, a compelling case may be made that a suite of posttranscriptional nucleoside modifications, combined with the chemistry imparted by the identity of the anticodon nucleosides themselves, provides the necessary identity determinants for these whole-and-split codon box tRNA isoacceptors to achieve their function.
In E. coli, the six codons for arginine are decoded by five tRNA isoacceptors [77,151,152]. Usage of these six codons varies widely, from >2% of all codons for the most common CGU and CGC to <0.2% for the very rare AGA and AGG [51,153-155]. Of the four less common codons, three (CGG, AGA, and AGG) are decoded by individual isoacceptors tRNAArg3, tRNAArg4, and tRNAArg5, respectively, while the fourth, CGA, is decoded by the same isoacceptors (tRNAArg1 and tRNAArg2) that also decode the two most common codons CGU and CGC [77]. The primary sequences, and therefore chemical identities, of the E. coli arginine tRNA isoacceptors differ between members of the family; this is particularly true in the region of the anticodon, which must vary in its sequence among isoacceptors to account for the variation in the first and third nucleosides in the six synonymous arginine codons (Fig. 10). Despite these chemical differences, sufficient chemical similarity must exist to permit the single E. coli arginyl-tRNA synthetase (ArgRS) to charge all five isoacceptors [156]. Furthermore, three of the isoacceptors contain bases in the first position of the anticodon that theoretically permit wobble recognition of near-cognate codons, an expansion tRNA decoding capacity that appears to be either regulated or disallowed in vivo [77]. These conflicting chemical requirements—the recognition of a variety of mRNA codons, sufficient similarity to enable charging by ArgRS, and functional regulation of wobble decoding—are facilitated by a suite of isoacceptor-specific posttranscriptional nucleoside modifications.
FIG. 10.
The primary sequences and nucleoside modifications of the E. coli tRNAArg isoacceptors. (A) The anticodon domain primary sequences of the five tRNAArg isoacceptors from E. coli and their modifications: 2-thiocytidine at position-32 (blue); inosine and 5-methylaminomethyluridine at position-34 (red); 2-methyladenosine, N6-threonylcarbamoyladenosine, and 1-methylguanosine at position-37 (green); and pseudouridine at position-40 (orange). (B) The chemical structures, names, and abbreviations of the modifications that appear in the five E. coli tRNAArg isoacceptors.
5.2. Modification-Dependent tRNA Reading of the Arginine Codons
A number of chemical signatures and naturally occurring posttranscriptional modifications are in fact common to all or nearly all of the E. coli tRNA arginine isoacceptors. A highly conserved adenosine occupies position-38 at the 3′-terminus of the tRNA's ASL domain in all five isoacceptors, and thus, a mismatch base pair between A38 and C32/s2C32 can be formed [77,138]. These mismatches typically feature a stable hydrogen bond between the O2 of C32 and the N6 of A38 and may, together with the invariant uridine at position-33, be important for permitting the ASL to adopt the “U-turn” structural motif for correct tRNA recognition in the ribosomal A-site [110,138,157,158].
A conserved cytidine at position-35, the second position of the anticodon, is also common to all five E. coli tRNA isoacceptors for arginine [138]. C35, positioned to form a Watson–Crick base pair with the invariant guanine that occupies position two of all six arginine codons, is known to provide at least a portion of the common chemical signature that serves as a strong identity determinant for aminoacylation; it is recognized through backbone interactions with ArgRS His22 and His23 and a stacking interaction with Trp569 [159]. Structurally, because C35 must be exposed to the solvent in order to make specific contacts with the ArgRS backbone and/or side chains, but must also accommodate ribosomal binding and mRNA decoding, the ASL domain of arginine tRNA isoacceptors must be significantly deformable. A significant conformational change must occur between binding to the aminoacyl-tRNA synthetase and adopting the canonical U-turn. This deformability is, in fact, visible in a comparison between the ribosome-bound and synthetase-bound structures of the tRNA, which differ considerably in the ASL region [159,160]. Elements of their respective modification schemes are also common among the arginine tRNA isoacceptors. Each has a modified purine at position-37, either a 2-methyladenosine (m2A; tRNAArg1 and tRNAArg2), an N6-threonylcarbamoyladenosine (t6A37; tRNAArg3 and tRNAArg4), or a 1-methylguanosine (m1G; tRNAArg5) [77,138].
The two E. coli isoacceptors responsible for decoding not only the common CGU and CGC codons but also the seldom-used CGA sequence are tRNAArg1ICG and tRNAArg2ICG. These isoacceptors, which have identical primary sequences and differ only in that tRNAArg1ICG lacks the adenosine nucleotide at position-20 (A20) in its dihydrouridine loop and contains the rare modification 2-thiocytidine at position-32 (s2C32), are sometimes referred to jointly as tRNAArg1,2ICG[161,162]. Interestingly, both A20 and s2C32 are present in the other three arginine isoacceptors, tRNAArg3,4,5[77,161,162]. Both tRNAArg1ICG and tRNAArg2ICG also contain posttranscriptional nucleoside modifications at position-34 (inosine, I34) and position-37 (2-methyladenosine, m2A37), which have also been shown to play significant roles in allowing tRNAArg1,2 to achieve its functionality [77,138]. In tRNAArg1, the absence of A20 from the molecule's dihydrouridine loop has been shown to negatively affect aminoacylation efficiency [163]. Mutant tRNAArg2 transcripts in which the expected A20 is either substituted or deleted result in a 370-fold decrease in aminoacylation activity by ArgRS, suggesting that tRNAArg1ICG is constitutively aminoacylated and available to participate in translation at a lower rate of efficiency than tRNAArg2ICG[164].
5.3. The Modification 2-Thiocytidine Modulates Inosine Reading of A, U, and C
The unusual 2-thiocytidine modification also plays a role in differentiating between the properties of the tRNAArg1 and tRNAArg2 isoacceptors. s2C32 is a relatively uncommon modification, present only in a subset of bacterial and archaeal species; in E. coli, it can be found in four of the five arginine tRNA isoacceptors (tRNAArg1,3,4,5) and tRNASerGCU at position-32 in the ASL [14,77]. Functionally, the presence of s2C32 was first demonstrated to promote reading frame maintenance and to reduce the rate of tRNA selection into the ribosomal A-site and was suggested to play a role in improving translational accuracy by reducing the speed of translation, particularly during suboptimal growth conditions [165]. A series of ribosomal A-site binding assays using differentially modified tRNAArg1,2 ASLs later definitively demonstrated that the ASLArg1,2ICG containing either a single s2C32 or m2A37 modification cannot efficiently wobble decode CGA, while the unmodified ASLArg1,2ICG recognizes, per the expectations of the wobble hypothesis, CGC, CGU, and CGA [43,166]. This experiment definitively established the role of s2C32 in tRNAArg1,2 as restricting the ability of the tRNAArg1ICG isoacceptor to decode the rarest of its three wobble codons [166].
The functional difference imparted by the s2C32 modification in tRNAArg1ICG cannot be explained by structural differences in solution; NMR structural analysis of the differentially modified tRNAArg1,2ICG ASL constructs employed in the ribosomal binding assay showed that neither the presence of s2C32 nor of m2A37 altered the solution structure of the ASLArg1,2ICG significantly from the highly stable 5′-U33NCG36-3′ motif observed for the unmodified ASL and found as a tetraloop [166-168]. When a subset of the ASLArg1,2ICG constructs were crystallized bound to their cognate mRNA in the context of the 30S bacterial ribosomal subunit, they adopted the characteristic U-turn structural motif but failed to indicate any significant source of structural difference among them (W. Cantara, personal communication). However, biophysical characterization suggested that s2C32 contributions to stability and thermodynamics properties, reducing the melting temperature of the ASLArg1,2ICG hairpin, increasing the free energy of folding, and decreasing base stacking interactions compared to the unmodified ASL [166]. A computational analysis of the possibly dynamic effect of s2C32 on the properties of tRNAArg1,2ICG in its mRNA-bound context on the ribosome led to the suggestion that the presence of a purine:purine I34-A3 basepair in the A-site wobble position of the unmodified tRNAArg1,2ICG distorts the U-turn from its canonical structural motif and precipitates the formation of a network of water molecules that stabilize the distorted U-turn. Subsequent substitution of the larger, less electronegative sulfur atom for oxygen in the position-32 cytosine disrupts this hydrogen bonding network, such that the unconventional distorted U-turn cannot be simultaneously maintained within the geometric constraints of a large purine:purine I34-A3 base pair (S. Ranganathan and S. Vangaveti, personal communication).
The posttranscriptional modification of some tRNAArg1,2ICG transcripts with s2C32 provides the cell with a regulatory strategy to address the allotment of cellular resources to the translation of the rare codon CGA. Of the two isoacceptors tRNAArg1 and tRNAArg2, only tRNAArg2, bearing an unmodified C32, is capable of decoding CGA [166]. Under typical circumstances, when CGA-containing mRNA transcripts are rare, the tRNAArg1,2 population may be regulated at the level of posttranscriptional modification to favor s2C32-modified tRNAArg1 and the decoding of CGC and CGU only; under stress responses or in other circumstances where a more robust response to CGA is required, the modification of tRNAArg1,2 transcripts can be reduced, favoring tRNAArg2.
Although the tRNAArg1,2ICG isoacceptors have the ability, depending upon modification profile, to decode three codons, the remaining three isoacceptors recognize only one codon each [169,170]. Two of these, tRNAArg3 and tRNAArg5, contain the anticodons CCU and CCG, respectively, and, consistent with the expectations of the wobble hypothesis, do not form non-Watson–Crick wobble base pairs with any nucleosides other than guanosine at the third position of their cognate codons AGG and CGG [43,77]. Both isoacceptors contain a modified purine at position-37 and a 2-thiocytidine modification at position-32 [138]. While the function of s2C32 in these arginine tRNA isoacceptors is not known and cannot be accounted for by a need for the restriction of wobble decoding, the position-37 purine modifications t6A37 and m1G37 have been, respectively, shown to order the ASL domain in an open conformation for the formation of the U-turn motif and to prevent translational frameshifting while enhancing codon– anticodon affinity [90,111,171,172].
5.4. tRNAArg Species Decoding a Twofold Degenerate Codon Box
tRNAArg4UCU, however, provides another example in which nucleoside identity and modification scheme combine to achieve specific functional results. A wide variety of bacterial and archaeal species contain verified tRNAArg species with UCU anticodons for the decoding of the codon AGA; candidate sequences for an equivalent tRNA species in several eukaryotic genomes, while not yet confirmed experimentally, have also been identified [77,173]. The E. coli tRNAArg4UCU, known to be solely responsible for the decoding of the codon AGA in that species, contains four endogenous posttranslational modifications: 2-thiocytidine at position-32 (s2C32); 5-methylaminomethyluridine at wobble position-34 (mnm5U34); t6A37; and Ψ40[77,174]. tRNAArg4UCU shares with the bacterial tRNALysUUU, which decodes both AAA and AAG, a nearly identical ASL sequence and both the mnm5U34 and t6A37 modifications [77]. Although the well-studied human tRNALysUUU does not share s2C32 or Ψ40, its biophysical properties are otherwise so similar to those of E. coli tRNAArg4UCU that it was expected that functional properties of two tRNA species would likewise resemble each other [111]. However, it was found that in tRNAArg4UCU, the mnm5U34 is required for this isoacceptor to recognize the rare codon AGA, but does not allow wobble pairing to AGG. In contrast, mnm5U34 in E. coli tRNALysUUU demonstrably confers the ability to read both AAA and AAG codons [132,175]. The only differences in the loop sequences of E. coli tRNALysUUU and tRNAArg4UCU lie in the identity of the second anticodon nucleoside (C35 for tRNAArg4; A35 for tRNALys) and the s2C32 modification in tRNAArg4UCU[138]; like tRNAArg1,2ICG, it was initially thought that the presence of s2C32 would serve an inhibitory role in AGG recognition [166]. However, A-site codon-specific ribosome binding assays performed on three synthesized tRNAArg4UCU ASL constructs (unmodified ASLArg4UCU, singly modified ASLArg4UCU–s2C32, and triply modified ASLArg4UCU–mnm5U34;t6A37;Ψ40) showed that only the triply modified ASL bound AGA with a physiological dissociation constant (188±21nM), while no construct, with or without modifications, bound AGG (W. Cantara, personal communication).
While the presence of s2C32 surprisingly appears not to restrict the wobble interaction of tRNAArg4 with the AGG codon as it does in tRNAArg1,2, biophysical analysis shows that its modification suite contributes to a reduction in base stacking and overall molecular order, similar to effects observed in the fully modified ASL of ASLLysUUU. Preliminary NMR analysis further indicated that modifications modulate the pKa of A38 to stabilize the noncanonical C32○A38 base pair and enhance cognate decoding by enhancing the base stacking interactions between G31 and s2C32. In this instance, although modifications clearly affect the thermodynamic properties and overall function of tRNAArg4UCU, the chemical determinant for the ability of tRNAArg4UCU to discriminate between A- and G-ending codons lies in the presence of modifications together with the sequence identity of the nucleotide, C35, occupying the second position of the anticodon (W. Cantara, personal communication).
The E. coli tRNA arginine isoacceptors provide an excellent example of a system in which the chemical identity of the nucleotides comprising the RNA, together with their posttranscriptional modifications, operates in tandem to achieve several functional goals for the family of isoacceptors: discrimination between six chemically distinct arginine codon from whole- and split-codon boxes and differing from each other at their first and third nucleotide positions; universal recognition for amino acid charging by a single arginyl aminoacyl-tRNA synthetase; and regulation of wobble decoding to influence the ability of the cell to decode the rare codons CGA, AGA, and AGG. For example, the modification state of tRNAArg1,2ICG regulates whether the isoacceptor decodes all three codons to which it theoretically wobble pairs or only to the two common codons (CGU and CGC) [166]; this regulatory role for modifications counteracts the need for multiple isoacceptors to decode the common and rare codons and be recognizable to the aminoacyl-tRNA synthetase, while also allowing the translation of CGA-containing genes to be regulated by a single modification enzyme under appropriate cellular conditions. It is also clear from the tRNAArg family of isoacceptors that modifications perform different roles depending upon their chemical environment; the rare s2C32 modification, for example, has a clear inhibitory effect on wobble decoding of tRNAArg1,2 but permits and enhances cognate codon binding for tRNAArg4UCU[166]. Such dual functionality may also exist for mnm5U34 in tRNAArg4UCU and tRNALysUUU to explain their respective abilities to inhibit or allow wobble decoding.
5.5. Summary
Sixfold degenerate codons across two codon boxes differ not only in the nucleoside occupying their wobble positions but also in the nucleoside present in their first (and, for serine, second) positions. The tRNA isoacceptors decoding these degenerate codons must simultaneously recognize and discriminate between these divergent codons while remaining universally recognizable to a single aminoacyl-tRNA synthetase and able to bind the ribosomal A-site. In E. coli, posttranscriptional modifications assist the five arginine tRNA isoacceptors in accomplishing distinguishing their cognate and wobble codons. In the tRNAArg1,2ICG isoacceptors, the variable presence of a 2-thiocytidine at position-32 inhibits the ability of the tRNA to wobble decode the rare arginine codon CGA due to steric and hydrogen bonding effects resulting from the replacement of an oxygen with a larger and less electronegative sulfur atom. tRNAArg4UCU, by contrast, is also restricted in its wobble recognition of the near-cognate AGG codon, but depends for this effect not on its s2C32 modification but on the chemical identity of the codon nucleosides together with the presence of four endogenous posttranscriptional modifications. The E. coli arginine tRNA isoacceptors provide an excellent example of the functional specificity conferred by posttranscriptional modifications acting in unique local chemical environments, as well as pointing out how RNA modification-based strategies of cellular regulation might influence the cell's response to codon bias and the translation of rare codons.
6. Posttranscriptional Modification of tRNA Is Required for Accurate and Efficient Decoding
Posttranscriptional modifications evolved and were conserved at tRNA nucleoside positions-34 and -37 to both expand and restrict the capability of isoaccepting tRNAs to read cognate and wobble codons. Though the effects are opposite in function, in both cases the modifications ensure translational fidelity of mRNA transcripts. These site-specific, context-dependent roles for tRNA modifications emphasize their ability to fine-tune tRNA molecular function through chemistry and conformational dynamics in control of gene expression [29,31,45]. This occurs even to the extent that a single modification may perform different functions depending on the sequence and chemical context of the ASL [108].
In their use of modification chemistries, tRNA molecules achieve the simultaneous goals of structural uniformity and ASL sequence variety through the use of a conformational dynamics and chemistries. tRNA structural uniformity is prescribed by the constraints of the ribosomal translational machinery [2,176,177]. Every tRNA species must have ASL domain structures that conform to the spatial limitations and chemistries of the A- and P-sites of the ribosome. To be productively accepted into the decoding site, the backbone chemistries of tRNA's ASL must be structured to engage in stable hydrogen bonding to the rRNAs, bacterial 16 and 23S RNAs, and eukaryote 18 and 28S RNAs. Yet, within the variety of their individual sequence chemistries, each of approximately 40 tRNAs needs to read one or more of the 61 amino acid codons. The individual chemistries, structures, and conformational dynamics required for accurate and efficient decoding of mRNA are provided to the tRNA in the form of prestructuring the ASL domain with modified nucleosides [111].
Instances in which a codon box is split between multiple amino acids (Fig. 1) exemplify how modifications prestructure a tRNA to distinguish the correct codon(s) from the incorrect codon(s). As described, mt tRNAMet modifications allow this one tRNA to read AUG and AUA codons as the initial and internal methionine codes. The tRNAValUAC cmo5U34 and m6A37 modifications are crucial for accurate codon recognition of fourfold degenerate codons. Prokaryote and mammalian tRNALysUUU are fully functional in accurate codon recognition, translocation, and maintenance of the reading frame only when fully modified. When the first and second codon bases are shared as they are for lysine and asparagine codons, the introduction of a modification at wobble position-34, such as the 2-thiouridine-34, enables the tRNALysUUU to bind the AAA and AAG lysine codons while hindering recognition of the asparagine AAC and AAU codons [150]. This level of translational regulation through the contribution of modified nucleosides leads to the accurate binding of the anticodon to the cognate and wobble codons within the ribosomal A-site and peptide bond formation. There is still opportunity for the anticodon–codon mismatch within the P-site to trigger early termination of translation [6].
Modified nucleosides are required of tRNAArg species for accurate distinction of sixfold degenerate codons and modulation of wobble position inosine-34 recognition of A, U, and C. The sixfold degenerate codons for leucine claim the entire CUN codon box and share the UUN (where N is A, C, G, or U) mixed codon box with phenylalanine. MiaA, a tRNA isopentenyltransferase implicated in the biosynthesis of the N6-isopentenyl-2-methylthioadenosine-37 (ms2i6A37) modification found in tRNALeuUAA and tRNALeuCAA[178], is required for the accurate translation of the leucine-containing rpoS and iraP gene products in E. coli, whose mRNA transcripts code for leucine using the complementary UUA and UUG codons [179].
Nature rarely discards chemistries that prove successful for one purpose only to be advanced and duplicated for another purpose. In doing so with RNA-modified nucleosides, the value has been conserved. Through site-specific placement and structure/activity relationships, modifications evolved to optimize translational accuracy and efficiency. The ASL was optimized with modifications that prestructures conformation, lowers energy barriers, and negates the need to refashion the chemistry, structure, and dynamics for codon recognition and ribosome binding. The first reportings of modified nucleosides in RNA in general, and in tRNA (s-RNA) in particular, occurred over 50 years ago [180]. The modifications of tRNA's ASL domain are the most studied and understood. Yet, scientists worldwide continue to gain insight into how this encoded third level of translational control within tRNA and other RNAs provide accurate, efficient, and responsive decoding of the Universal Genetic Codes.
Acknowledgment
Our review emphasizes and cites some of the most recent findings regarding the chemistry, structure, and functions of posttranscriptional RNA modifications, focusing on tRNA's anticodon domain. As in any such digest of work, we apologize for inadvertently missing the contributions of some, particularly since interest in RNA modifications has exponentially increased and publications are in a large variety of journals. The summation presented here was only possible through half a century of collaborations that we have had with undergraduate and graduate students, postdoctoral fellows, and colleagues during a long and continuing career supported by grants from NIH, NSF, and DOD, including at present to PFA (NSF, CHE-1407042; DOD, W81XWH-15-PRMRP-DA) and to Manal Swairjo, PI (San Diego State University) and PFA co-PI (NIH, 1R01GM110588).
References
- [1].Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Sun M, Espina N, Madison-Antenucci S, Tong L, Frank J Determination of the ribosome structure to a resolution of 2.5 A by single-particle cryo-EM. Protein Sci. 2017;26(1):82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zimmerman E, Yonath A Biological implications of the ribosome's stunning stereochemistry. ChemBioChem. 2009;10(1):63–72. [DOI] [PubMed] [Google Scholar]
- [3].Greber BJ, Ban N Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem 2016;85:103–132. [DOI] [PubMed] [Google Scholar]
- [4].Brocchieri L, Karlin S Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33(10):3390–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kurland C, Hughes D, Ehrenberg M Limitations of translational accuracy. In: Neidhardt FC, Curtis JL III Ingraham R, Lin ECC, Low JKB, Magasarák B, Reznikoff WS, Riley M, Schaachler M, Umbarger HE, eds. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996:979–1004. [Google Scholar]
- [6].Zaher HS, Green R Quality control by the ribosome following peptide bond formation. Nature. 2009;457(7226):161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McCulloch SD, Kunkel TA The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18(1):148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mechali M Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol 2010;11(10):728–738. [DOI] [PubMed] [Google Scholar]
- [9].Kwak H, Lis JT Control of transcriptional elongation. Annu. Rev. Genet 2013;47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011;12(12):1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Duechler M, Leszczynska G, Sochacka E, Nawrot B Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci 2016;73(16):3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schimmel P, Giegé R, Moras D, Yokoyama S An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. U.S.A 1993;90(19):8763–8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Agris PF The importance of being modified: an unrealized code to RNA structure and function. RNA. 2015;21(4):552–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201 (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267 (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gilbert WV, Bell TA, Schaening C Messenger RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282–285. [DOI] [PubMed] [Google Scholar]
- [19].Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. [DOI] [PubMed] [Google Scholar]
- [20].Maity A, Das B N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. 2016;283(9):1607–1630. [DOI] [PubMed] [Google Scholar]
- [21].Zhao BS, He C Pseudouridine in a new era of RNA modifications. Cell Res. 2015;25(2):153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O'Leary SE, Dominissini D, Rechavi G, Soltis SM, et al. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol 2016;23(2):110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rottman F, Shatkin AJ, Perry RP Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: possible implications for processing. Cell. 1974;3(3):197–199. [DOI] [PubMed] [Google Scholar]
- [24].Cao G, Li H-B, Yin Z, Flavell RA Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6(4):160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19(5):675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schwartz S Cracking the epitranscriptome. RNA. 2016;22(2):169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Agris PF Decoding the genome: a modified view. Nucleic Acids Res. 2004;32(1):223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Agris PF, Vendeix FA, Graham WD tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol 2007;366(1):1–13. [DOI] [PubMed] [Google Scholar]
- [31].Gustilo EM, Vendeix FA, Agris PF tRNA's modifications bring order to gene expression. Curr. Opin. Microbiol 2008;11(2):134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ranjan N, Rodnina MV tRNA wobble modifications and protein homeostasis. Translation. 2016;4(1):e1143076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang R, Luo Z, He K, Delaney MO, Chen D, Sheng J Base pairing and structural insights into the 5-formylcytosine in RNA duplex. Nucleic Acids Res. 2016;44(10):4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jackman JE, Alfonzo JD Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013;4(1):35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014;11(12):1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Agris PF The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol 1996;53:79–129. [DOI] [PubMed] [Google Scholar]
- [37].Chapeville F, Lipmann F, Ehrenstein GV, Weisblum B, Ray WJ, Benzer S On the role of soluble ribonucleic acid in coding for amino acids. Proc. Natl. Acad. Sci. U.S.A 1962;48(6):1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Effraim PR, Wang J, Englander MT, Avins J, Leyh TS, Gonzalez RL Jr., Cornish VW Natural amino acids do not require their native tRNAs for efficient selection by the ribosome. Nat. Chem. Biol 2009;5(12):947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Capecchi MR, Gussin GN Suppression in vitro: identification of a serine-sRNA as a “nonsense” suppressor. Science. 1965;149(3682):417–422. [DOI] [PubMed] [Google Scholar]
- [40].Engelhardt DL, Webster RE, Wilhelm RC, Zinder N In vitro studies on the mechanism of suppression of a nonsense mutation. Proc. Natl. Acad. Sci. U.S.A 1965;54(6):1791–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ambrogelly A, Palioura S, Soll D Natural expansion of the genetic code. Nat. Chem. Biol 2007;3(1):29–35. [DOI] [PubMed] [Google Scholar]
- [42].Wright TH, Vallee MR, Davis BG From chemical mutagenesis to post-expression mutagenesis: a 50 year odyssey. Angew. Chem. Int. Ed. Engl 2016;55(20):5896–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Crick FH Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol 1966;19(2):548–555. [DOI] [PubMed] [Google Scholar]
- [44].Nasvall SJ, Chen P, Bjork GR The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA. 2007;13(12):2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vendeix FA, Dziergowska A, Gustilo EM, Graham WD, Sproat B, Malkiewicz A, Agris PF Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry. 2008;47(23):6117–6129. [DOI] [PubMed] [Google Scholar]
- [46].Weixlbaumer A, Murphy FVT, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol 2007;14(6):498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. [DOI] [PubMed] [Google Scholar]
- [48].Attardi G Animal mitochondrial DNA: an extreme example of genetic economy. Int. Rev. Cytol 1985;93:93–145. [DOI] [PubMed] [Google Scholar]
- [49].Anderson S, de Bruijn MH, Coulson AR, Eperon IC, Sanger F, Young IG Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol 1982;156(4):683–717. [DOI] [PubMed] [Google Scholar]
- [50].Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA. 2008;14(5):862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nakamura Y, Gojobori T, Ikemura T Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bjork GR Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acid Res. Mol. Biol 1995;50:263–338. [DOI] [PubMed] [Google Scholar]
- [53].Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994;33(8):2234–2239. [DOI] [PubMed] [Google Scholar]
- [54].Sinha A, Kohrer C, Weber MH, Masuda I, Mootha VK, Hou YM, RajBhandary UL Biochemical characterization of pathogenic mutations in human mitochondrial methionyl-tRNA formyltransferase. J. Biol. Chem 2014;289(47):32729–32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun 2016;7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli LL, Watanabe K Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J. Biol. Chem 1998;273(24):15085–15090. [DOI] [PubMed] [Google Scholar]
- [57].Takeuchi N, Vial L, Panvert M, Schmitt E, Watanabe K, Mechulam Y, Blanquet S Recognition of tRNAs by methionyl-tRNA transformylase from mammalian mitochondria. J. Biol. Chem 2001;276(23):20064–20068. [DOI] [PubMed] [Google Scholar]
- [58].Jones CN, Jones CI, Graham WD, Agris PF, Spremulli LL A disease-causing point mutation in human mitochondrial tRNAMet results in tRNA misfolding leading to defects in translational initiation and elongation. J. Biol. Chem 2008;283(49):34445–34456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bilbille Y, Gustilo EM, Harris KA, Jones CN, Lusic H, Kaiser RJ, Delaney MO, Spremulli LL, Deiters A, Agris PF The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J. Mol. Biol 2011;406(2):257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Astrom SU, Bystrom AS Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79(3):535–546. [DOI] [PubMed] [Google Scholar]
- [61].Suzuki T, Nagao A, Suzuki T Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet 2011;45:299–329. [DOI] [PubMed] [Google Scholar]
- [62].Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37(5):1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Takemoto C, Koike T, Yokogawa T, Benkowski L, Spremulli LL, Ueda TA, Nishikawa K, Watanabe K The ability of bovine mitochondrial transfer RNAMet to decode AUG and AUA codons. Biochimie. 1995;77(1–2):104–108. [DOI] [PubMed] [Google Scholar]
- [64].Takemoto C, Ueda T, Miura K, Watanabe K Nucleotide sequences of animal mitochondrial tRNAsMet possibly recognizing both AUG and AUA codons. Nucleic Acids Symp. Ser 1999;42:77–78. [DOI] [PubMed] [Google Scholar]
- [65].Tomita K, Ueda T, Watanabe K 5-Formylcytidine (f5C) found at the wobble position of the anticodon of squid mitochondrial tRNAMetCAU. Nucleic Acids Symp. Ser 1997;37:197–198. [PubMed] [Google Scholar]
- [66].Tomita K, Ueda T, Ishiwa S, Crain PF, McCloskey JA, Watanabe K Codon reading patterns in Drosophila melanogaster mitochondria based on their tRNA sequences: a unique wobble rule in animal mitochondria. Nucleic Acids Res. 1999;27(21):4291–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Watanabe Y, Tsurui H, Ueda T, Furushima R, Takamiya S, Kita K, Nishikawa K, Watanabe K Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem 1994;269(36):22902–22906. [PubMed] [Google Scholar]
- [68].Lusic H, Gustilo EM, Vendeix FA, Kaiser R, Delaney MO, Graham WD, Moye VA, Cantara WA, Agris PF, Deiters A Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008;36(20):6548–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lancaster L, Noller HF Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell 2005;20(4):623–632. [DOI] [PubMed] [Google Scholar]
- [70].Cantara WA, Murphy FVT, Demirci H, Agris PF Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. U.S.A 2013;110(27):10964–10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Oashi Z, Murao K, Yahagi T, Von Minden DL, McCloskey JA, Nishimura S Characterization of C+ located in the first position of the anticodon of Escherichia coli tRNAMet as N4-acetylcytidine. Biochim. Biophys. Acta 1972;262(2):209–213. [PubMed] [Google Scholar]
- [72].Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J. Biol. Chem 1988;263(19):9261–9267. [DOI] [PubMed] [Google Scholar]
- [73].Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL Agmatidine, a modified cytidine in the anticodon of archaeal tRNAIle, base pairs with adenosine but not with guanosine. Proc. Natl. Acad. Sci. U.S.A 2010;107(7):2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Voorhees RM, Mandal D, Neubauer C, Kohrer C, RajBhandary UL, Ramakrishnan V The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat. Struct. Mol. Biol 2013;20(5):641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol 2016;12(7):546–551. [DOI] [PubMed] [Google Scholar]
- [76].El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37(9):2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162 (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC mtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics 2013;44:1.23.21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW The genetics and pathology of mitochondrial disease. J. Pathol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].de Almeida RA, Fraczek MG, Parker S, Delneri D, O'Keefe RT Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem. Soc. Trans 2016;44(4):1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vissing J, Salamon MB, Arlien-Soborg P, Norby S, Manta P, DiMauro S, Schmalbruch H A new mitochondrial tRNAMet gene mutation in a patient with dystrophic muscle and exercise intolerance. Neurology. 1998;50(6):1875–1878. [DOI] [PubMed] [Google Scholar]
- [82].Qu J, Li R, Zhou X, Tong Y, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest. Ophthalmol. Vis. Sci 2006;47(2):475–483. [DOI] [PubMed] [Google Scholar]
- [83].Lombes A, Bories D, Girodon E, Frachon P, Ngo MM, Breton-Gorius J, Tulliez M, Goossens M The first pathogenic mitochondrial methionine tRNA point mutation is discovered in splenic lymphoma. Hum. Mutat 1998;11(Suppl 1):S175–S183. [DOI] [PubMed] [Google Scholar]
- [84].Grosjean H, Westhof E An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016;44(17):8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Suzuki TS, Wada T, Saigo T, Watanabe K, Kimitsuna W Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002;21(23):6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. [DOI] [PubMed] [Google Scholar]
- [87].Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet 2007;39(6):770–775. [DOI] [PubMed] [Google Scholar]
- [88].Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF The role of modifications in codon discrimination by tRNALysUUU. Nat. Struct. Mol. Biol 2004;11(12):1186–1191. [DOI] [PubMed] [Google Scholar]
- [89].Phelps SS, Malkiewicz A, Agris PF, Joseph S Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol 2004;338(3):439–444. [DOI] [PubMed] [Google Scholar]
- [90].Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39(44):13396–13404. [DOI] [PubMed] [Google Scholar]
- [91].Mallick B, Chakrabarti J, Sahoo S, Ghosh Z, Das S Identity elements of archaeal tRNA. DNA Res. 2005;12(4):235–246. [DOI] [PubMed] [Google Scholar]
- [92].Björk GR Chapter 11: Biosynthesis and function of modified nucleosides. In: Söll D, RajBhandary U, eds. tRNA: Structure, Biosynthesis, and Function. Washington, DC: American Society for Microbiology; 1995:165–205. [Google Scholar]
- [93].Raba M, Limburg K, Burghagen M, Katze JR, Simsek M, Heckman JE, Rajbhandary UL, Gross HJ Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur. J. Biochem 1979;97(1):305–318. [DOI] [PubMed] [Google Scholar]
- [94].Agris PF Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie. 1991;73:1345–1349. [DOI] [PubMed] [Google Scholar]
- [95].Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem 2004;279(13):12363–12368. [DOI] [PubMed] [Google Scholar]
- [96].Suzuki T, Suzuki T A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353(6301):819–822. [DOI] [PubMed] [Google Scholar]
- [98].Bezerra AR, Guimaraes AR, Santos MA Non-standard genetic codes define new concepts for protein engineering. Life (Basel). 2015;5(4):1610–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hagervall TG, Pomerantz SC, McCloskey JA Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol 1998;284(1):33–42. [DOI] [PubMed] [Google Scholar]
- [100].Sugita M, Shinozaki K, Sugiura M Tobacco chloroplast tRNALys(UUU) gene contains a 2.5-kilobase-pair intron: an open reading frame and a conserved boundary sequence in the intron. Proc. Natl. Acad. Sci. U.S.A 1985;82(11):3557–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fuller W, Hodgson A Conformation of the anticodon loop in tRNA. Nature. 1967;215(5103):817–821. [DOI] [PubMed] [Google Scholar]
- [102].Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem 2008;283(27):18801–18811. [DOI] [PubMed] [Google Scholar]
- [103].Watanabe K, Yokobori S tRNA modification and genetic code variations in animal mitochondria. J. Nucleic Acids 2011;2011:623095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kamble AS, Sambhare SB, Fandilolu PM, Sonawane KD Structural significance of modified nucleoside 5-taurinomethyl-2-thiouridine, τm5s2U, found at ‘wobble’ position in anticodon loop of human mitochondrial tRNALys. Struct. Chem 2016;27(3):839–854. [Google Scholar]
- [105].Agris PF, Sierzputowska-Gracz H, Smith W, Malkiewicz A, Sochacka E, Nawrot B Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc 1992;114(7):2652–2656. [Google Scholar]
- [106].Björk GR, Huang B, Persson OP, Bystrom AS A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13(8):1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Nedialkova DD, Leidel SA Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161(7):1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Manickam N, Joshi K, Bhatt MJ, Farabaugh PJ Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 2016;44(4):1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun 2016;7:10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44(22):8078–8089. [DOI] [PubMed] [Google Scholar]
- [111].Vendeix FA, Murphy FVT, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol 2012;416(4):467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol 2008;28(10):3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Serra MJ, Turner DH Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. [DOI] [PubMed] [Google Scholar]
- [114].Rodriguez-Hernandez A, Spears JL, Gaston KW, Limbach PA, Gamper H, Hou YM, Kaiser R, Agris PF, Perona JJ Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J. Mol. Biol 2013;425(20):3888–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Numata T, Ikeuchi Y, Fukai S, Suzuki T, Nureki O Snapshots of tRNA sulphuration via an adenylated intermediate. Nature. 2006;442(7101):419–424. [DOI] [PubMed] [Google Scholar]
- [116].Sheng J, Larsen A, Heuberger BD, Blain JC, Szostak JW Crystal structure studies of RNA duplexes containing s2U:A and s2U:U base pairs. J. Am. Chem. Soc 2014;136(39):13916–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ashraf SS, Ansari G, Guenther R, Sochacka E, Malkiewicz A, Agris PF The uridine in “U-turn”: contributions to tRNA-ribosomal binding. RNA. 1999;5(4):503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Jacobson GN, Clark PL Quality over quantity: optimizing co-translational protein folding with non-‘optimal’ synonymous codons. Curr. Opin. Struct. Biol 2016;38:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Quax TE, Claassens NJ, Soll D, van der Oost J Codon bias as a means to fine-tune gene expression. Mol. Cell 2015;59(2):149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Supek F. The code of silence: widespread associations between synonymous codon biases and gene function. J. Mol. Evol 2016;82(1):65–73. [DOI] [PubMed] [Google Scholar]
- [121].Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 2007;28(5):860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Chan CT, Deng W, Li F, DeMott MS, Babu IR, Begley TJ, Dedon PC Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem. Res. Toxicol 2015;28(5):978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun 2012;3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Endres L, Dedon PC, Begley TJ Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12(6):603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Katz MJ, Gandara L, De Lella Ezcurra AL, Wappner P Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon. Cell. Mol. Life Sci 2016;73(9):1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Fernandez-Vazquez J, Vargas-Perez I, Sanso M, Buhne K, Carmona M, Paulo E, Hermand D, Rodriguez-Gabriel M, Ayte J, Leidel S, et al. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013;9(7):e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Anton BP, Russell SP, Vertrees J, Kasif S, Raleigh EA, Limbach PA, Roberts RJ Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis. Nucleic Acids Res. 2010;38(18):6195–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Harris KA, Bobay BG, Sarachan KL, Sims AF, Bilbille Y, Deutsch C, Iwata-Reuyl D, Agris PF NMR-based structural analysis of threonylcarbamoyl-AMP synthase and its substrate interactions. J. Biol. Chem 2015;290(33):20032–20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Thiaville PC, El Yacoubi B, Kohrer C, Thiaville JJ, Deutsch C, Iwata-Reuyl D, Bacusmo JM, Armengaud J, Bessho Y, Wetzel C, et al. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol 2015;98(6):1199–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Thiaville PC, Legendre R, Rojas-Benitez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crecy-Lagard V Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 2016;3(1):29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Matuszewski M, Sochacka E Stability studies on the newly discovered cyclic form of tRNA N6-threonylcarbamoyladenosine (ct6A). Bioorg. Med. Chem. Lett 2014;24(12):2703–2706. [DOI] [PubMed] [Google Scholar]
- [132].Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39(44):13390–13395. [DOI] [PubMed] [Google Scholar]
- [133].Miyauchi K, Kimura S, Suzuki T A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol 2013;9(2):105–111. [DOI] [PubMed] [Google Scholar]
- [134].Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest 2011;121(9):3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Xu Y, MacKerell AD Jr., Nilsson L Structural effects of modified ribonucleotides and magnesium in transfer RNAs. Bioorg. Med. Chem 2016;24(20):4826–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Graham WD, Barley-Maloney L, Stark CJ, Kaur A, Stolarchuk C, Sproat B, Leszczynska G, Malkiewicz A, Safwat N, Mucha P, et al. Functional recognition of the modified human tRNALys3UUU anticodon domain by HIV's nucleocapsid protein and a peptide mimic. J. Mol. Biol 2011;410(4):698–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Spears JL, Xiao X, Hall CK, Agris PF Amino acid signature enables proteins to recognize modified tRNA. Biochemistry. 2014;53(7):1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sprinzl M, Vassilenko KS Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33(Database issue):D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Ishikura H, Yamada Y, Nishimura S Structure of serine tRNA from Escherichia coli. I. Purification of serine tRNA's with different codon responses. Biochim. Biophys. Acta 1971;228(2):471–481. [DOI] [PubMed] [Google Scholar]
- [140].Mitra SK, Lustig F, Akesson B, Axberg T, Elias P, Lagerkvist U Relative efficiency of anticodons in reading the valine codons during protein synthesis in vitro. J. Biol. Chem 1979;254(14):6397–6401. [PubMed] [Google Scholar]
- [141].Sorensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Bjork GR, Ehrenberg M Over expression of a tRNALeu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J. Mol. Biol 2005;354(1):16–24. [DOI] [PubMed] [Google Scholar]
- [142].Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. U.S.A 1985;82:4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Nasvall SJ, Chen P, Bjork GR The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAProcmo5UGG promotes reading of all four proline codons in vivo. RNA. 2004;10(10):1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kothe U, Rodnina MV Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell 2007;25(1):167–174. [DOI] [PubMed] [Google Scholar]
- [145].Kimura S, Suzuki T Iron-sulfur proteins responsible for RNA modifications. Biochim. Biophys. Acta 2015;1853(6):1272–1283. [DOI] [PubMed] [Google Scholar]
- [146].Huang B, Johansson MJ, Bystrom AS An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11(4):424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Esberg A, Huang B, Johansson MJ, Bystrom AS Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 2006;24(1):139–148. [DOI] [PubMed] [Google Scholar]
- [148].Chen C, Tuck S, Bystrom AS Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5(7):e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cabello-Villegas J, Winkler ME, Nikonowicz EP Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol 2002;319(5):1015–1034. [DOI] [PubMed] [Google Scholar]
- [150].Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem 2002;277(19):16391–16395. [DOI] [PubMed] [Google Scholar]
- [151].Soll D, Cherayil JD, Bock RM Studies on polynucleotides. LXXV. Specificity of tRNA for codon recognition as studied by the ribosomal binding technique. J. Mol. Biol 1967;29(1):97–112. [DOI] [PubMed] [Google Scholar]
- [152].Soll D, RajBhandary UL Studies on polynucleotides. LXXVI. Specificity of transfer RNA for codon recognition as studied by amino acid incorporation. J. Mol. Biol 1967;29(1):113–124. [DOI] [PubMed] [Google Scholar]
- [153].Ikemura T Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol 1985;2(1):13–34. [DOI] [PubMed] [Google Scholar]
- [154].Wada K, Wada Y, Ishibashi F, Gojobori T, Ikemura T Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992;20(Suppl):2111–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Maloy SR, Stewart VJ, Taylor RK Genetic Analysis of Pathogenic Bacteria: A Laboratory Manual. Plainville, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- [156].Martinis SA, Plateau P, Cavarelli J, Florentz C Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie. 1999;81(7):683–700. [DOI] [PubMed] [Google Scholar]
- [157].Auffinger P, Westhof E Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol 1999;292(3):467–483. [DOI] [PubMed] [Google Scholar]
- [158].Auffinger P, Louise-May S, Westhof E Molecular dynamics simulations of solvated yeast tRNAAsp. Biophys. J 1999;76(1 Pt. 1):50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Delagoutte B, Moras D, Cavarelli J tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000;19(21):5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Murphy FVT, Ramakrishnan V Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol 2004;11(12):1251–1252. [DOI] [PubMed] [Google Scholar]
- [161].Chakraburtty K Primary structure of tRNAArgII of E. coli B. Nucleic Acids Res. 1975;2(10):1787–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Murao K, Tanabe T, Ishii F, Namiki M, Nishimura S Primary sequence of arginine transfer RNA from Escherichia coli. Biochem. Biophys. Res. Commun 1972;47(6):1332–1337. [DOI] [PubMed] [Google Scholar]
- [163].Sissler M, Giege R, Florentz C Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J. 1996;15(18):5069–5076. [PMC free article] [PubMed] [Google Scholar]
- [164].Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M In vitro study of E. coli tRNAArg and tRNALys identity elements. Nucleic Acids Res. 1992;20(9):2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Jager G, Leipuviene R, Pollard MG, Qian Q, Bjork GR The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol 2004;186(3):750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Cantara WA, Bilbille Y, Kim J, Kaiser R, Leszczynska G, Malkiewicz A, Agris PF Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNAArg1,2. J. Mol. Biol 2012;416(4):579–597. [DOI] [PubMed] [Google Scholar]
- [167].Cheong C, Varani G, Tinoco I Jr. Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990;346(6285):680–682. [DOI] [PubMed] [Google Scholar]
- [168].Moore PB Structural motifs in RNA. Annu. Rev. Biochem 1999;68:287–300. [DOI] [PubMed] [Google Scholar]
- [169].Sauge-Merle S, Falconet D, Fontecave M An active ribonucleotide reductase from Arabidopsis thaliana. Eur. J. Biochem 1999;266(1):62–69. [DOI] [PubMed] [Google Scholar]
- [170].Soll D, Jones DS, Ohtsuka E, Faulkner RD, Lohrmann R, Hayatsu H, Khorana HG Specificity of sRNA for recognition of codons as studied by the ribosomal binding technique. J. Mol. Biol 1966;19(2):556–573. [DOI] [PubMed] [Google Scholar]
- [171].Ashraf SS, Guenther RH, Ansari G, Malkiewicz A, Sochacka E, Agris PF Role of modified nucleosides of yeast tRNAPhe in ribosomal binding. Cell Biochem. Biophys 2000;33(3):241–252. [DOI] [PubMed] [Google Scholar]
- [172].Stuart JW, Koshlap KM, Guenther R, Agris PF Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J. Mol. Biol 2003;334(5):901–918. [DOI] [PubMed] [Google Scholar]
- [173].Lowe TM, Eddy SR tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Garcia GM, Mar PK, Mullin DA, Walker JR, Prather NE The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986;45(3):453–459. [DOI] [PubMed] [Google Scholar]
- [175].Spanjaard RA, Chen K, Walker JR, van Duin J Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res. 1990;18(17):5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Ramakrishnan V The ribosome emerges from a black box. Cell. 2014;159(5):979–984. [DOI] [PubMed] [Google Scholar]
- [177].Steitz TA A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol 2008;9(3):242–253. [DOI] [PubMed] [Google Scholar]
- [178].Thompson KM, Gottesman S The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J. Bacteriol 2014;196(4):754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Aubee JI, Olu M, Thompson KM The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA. 2016;22(5):729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Dunn DB The occurrence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta 1961;46:198–200. [DOI] [PubMed] [Google Scholar]