Abstract
SARS-CoV-2 are enveloped positive-stranded RNA viruses that replicate in the cytoplasm. It relies on the fusion of their envelope with the host cell membrane to deliver their nucleocapsid into the host cell. The spike glycoprotein (S) mediates virus entry into cells via the human Angiotensin-converting enzyme 2 (hACE2) protein located on many cell types and tissues' outer surface. This study, therefore, aimed to design and synthesize novel pyrazolone-based compounds as potential inhibitors that would interrupt the interaction between the viral spike protein and the host cell receptor to prevent SARS-CoV 2 entrance into the cell. A series of pyrazolone compounds as potential SARS-CoV-2 inhibitors were designed and synthesized. Employing computational techniques, the inhibitory potentials of the designed compounds against both spike protein and hACE2 were evaluated. Results of the binding free energy from the in-silico analysis, showed that three compounds (7i, 7k and 8f) and six compounds (7b, 7h, 7k, 8d, 8g, and 8h) showed higher and better binding high affinity to SARS-CoV-2 Sgp and hACE-2, respectively compared to the standard drugs cefoperazone (CFZ) and MLN-4760. Furthermore, the outcome of the structural analysis of the two proteins upon binding of the inhibitors showed that the two proteins (SARS-CoV-2 Sgp and hACE-2) were stable, and the structural integrity of the proteins was not compromised. This study suggests pyrazolone-based compounds might be potent blockers of the viral entry into the host cells.
Keywords: Sars-CoV-2, Spike glycoprotein, Human angiotensin-converting enzyme 2, Pyrazolone, Computational techniques
Graphical abstract
Introduction
Novel coronavirus disease (COVID-19) is the current and most deadly pandemic the world has faced in more than a century. According to the John Hopkins COVID-19 dashboard, the coronavirus, also known as SARS-CoV-2, has resulted in more than 90.2 million infections and more than 1.9 million deaths worldwide as of January 2021 [1]. Genomic studies have established that SARS-CoV-2 belongs to the beta coronavirus family, including SARS-CoV and MERS-CoV, associated with previous outbreaks of relatively smaller scale [[2], [3], [4]]. Severe acute respiratory syndrome coronavirus 2 is the seventh known coronavirus to infect humans; four of these coronaviruses (HKU1, 229E, NL63 and OC43) only cause slight symptoms of the common cold. The viruses can cause severe symptoms and even death, with SARS-CoV-2 having the highest fatality rate and much more contagious than MERS-CoV or SARS-CoV [5]. The virus emerged in December 2019 and, as of June 29, 2020, 6 months after the first outbreak of pandemics, the global count is rapidly approaching 10 million known cases and 500 000 deaths. Due to its broad clinical scope and high transmissibility, which is responsible for the high death rate, the eradication of SARS-CoV-2, as was achieved with SARS-CoV in 2003, does not seem to be a realistic target in the short term [6]. Since the emergence of the outbreak, many studies and clinical trials launched on SARS-CoV-2 worldwide have started yielding positive results, with vaccines' availability in some parts of Europe and the USA after evidence from randomized clinical trials has shown potential therapy improves patients' recovery.
SARS-CoV-2 is a single-stranded RNA-enveloped virus [7]. An RNA-based metagenomic next-generation sequencing method has been applied to characterize its entire genome, 29,881 bp in length (GenBank no. MN908947), encoding 9860 amino acids [8]. Gene fragments show structural and non-structural proteins. The spike [S]-envelope [E]-membrane [M]-nucleocapsid [N])-poly(T)-3′ genes encode structural proteins, in contrast, to nonstructural proteins, such as 3-chymotrypsin-like protease, papain-like protease, and RNA-dependent RNA polymerase, are encoded by the ORF region [9].
Many glycosylated S proteins cover the surface of SARS-CoV-2 and bind to the host cell receptor angiotensin-converting enzyme 2 (ACE2), mediating viral cell entry [10]. When the S protein binds to the receptor, Transmembrane (TM) protease serine 2 (TMPRSS2), a type 2 TM serine protease located on the host cell membrane, promotes the virus to enter into the cell by activating the S protein [11]. Once the virus gains entrance to the cell, the viral RNA is released, polyproteins are translated from the RNA genome, and replication and transcription of the viral RNA genome occur via protein cleavage and assembly of the replicase–transcriptase complex. Viral RNA is replicated, and structural proteins are synthesized, assembled, and packaged in the host cell, after which viral particles are released [12]. The virion envelope of coronaviruses is typically associated with three viral proteins that include the membrane protein (M), the envelope protein (E), and the spike protein (S). Compared to the three proteins, the spike protein (S) is one of SARS-CoV-II's key biological characteristics and several other viruses because it plays a crucial role in penetrating host cells and initiating infection [13,14]. The S proteins of coronaviruses can be divided into two important functional subunits: The N-terminal S1 subunit, which forms the S protein's spherical head, and the C-terminal S2 region that includes the stalk of the protein, which is directly embedded into the viral envelope. Upon interaction with a potential host cell, the S1 subunit will recognize and bind to the host cell receptors. In contrast, the S2 subunit, which is the most conserved component of the S protein, will fuse the virus's envelope with the host cell membrane [15].
The host cell receptor called Angiotensin-converting enzyme 2 (ACE2) is a protein located on many cell types and tissues' outer surface, including the lungs, heart, blood vessels, kidneys, liver and gastrointestinal tract. It is present in epithelial cells, which line-specific tissues and create protective barriers. It is an enzyme that generates small proteins by cutting up the larger protein angiotensinogen, which then regulates functions in the cell. It is the primary host cell target with which the S protein of SARS-CoV and SARS-CoV-II associates. The SARS-CoV-II virus interacting with the spike-like protein on its surface binds to ACE2 – like a key inserted into a lock – before entry and cell infection. Hence, ACE2 acts as a cellular doorway – a receptor – for the virus that causes COVID- 19. The receptor-binding domain (RBD) of the S1 subunit of these viruses binds to the outer surface of the claw-like structure of ACE2 [[16], [17], [18]].
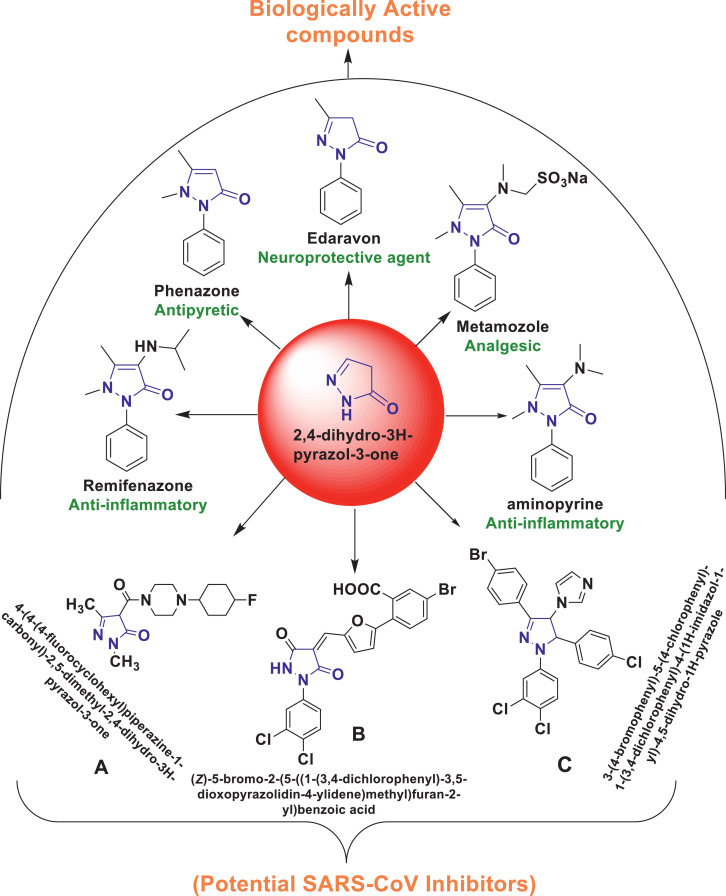
According to literature studies, researchers have found that compounds containing a pyrazole and its related analogues have received significant attention in chemical, medicinal, and pharmaceutical research as a structural scaffold found in various drugs. [19] As shown in Fig. 1, a new pyrazolone compound, edaravone, also known as MCI-186, has been developed as a medical drug for brain ischemia and has also been reported useful for myocardial ischemia [[20], [21], [22]]. Compound (A) has been suggested by Anup Masih and coworkers as a potential lead for therapeutic application to control the inflammatory response in SARS‐CoV‐II infection [23]. Compound (B) 3,5-dioxopyrazolidine has been reported as a SARS-CoV 3CLpro inhibitor [24]. Ramajayam et al., 2010 identified, from high throughput screening, some pyrazolines, particularly those displaying a 1,3,5-triaryl substitution pattern (C), to show activities against SARS-CoV 3CLpro, CoV-229E 3CLpro, CVB3 3Cpro, EV71 3Cpro, and RV14 3Cpro [19].
Fig. 1.
Pyrazolone moieties as drugs or enzyme inhibitors.
This reported antiviral pyrazole activity has motivated this study to design and investigate the novel pyrazolone derivatives' inhibitory potentials synthesized experimentally against the SARS-CoV-2 spike protein human angiotensin-converting enzyme 2 (hACE2) employing in silico methods.
2. Results and discussion
2.1. Chemistry
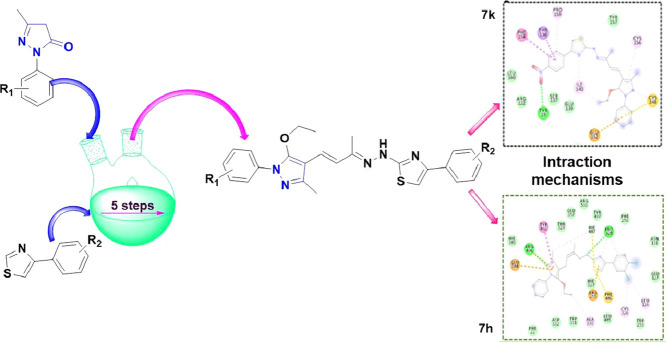
The synthesis of the novel pyrazolone derivatives was achieved through efficient and versatile synthetic routes. The Vilsmeier–Haack reaction of the commercially available starting material 2-(phenyl/substituted phenyl)-5-methyl-2,4-dihydro-3H-pyrazol-3-one (1a-b) lead to chloroformylation to give required 5-chloro-1-(phenyl/substituted phenyl)-3-methyl-1H-pyrazole-4-carbaldehyde (2a–b). Through a nucleophilic substitution reaction, ethoxylation occurs by replacing chloride on the pyrazole ring of compound (2) to yield 1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazole-4-carbaldehyde (3a,b). By simple aldol condensation reaction with acetone in the presence of a base compound (3a,b) transformed to compound (E)-4-(1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-one (4a,b). Furthermore, the compound 4 underwent Schiff base formation with thiosemicarbazide to afford (E)-2-((E)-4-(1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide (5a,b),which were condensed with various commercially available substituted 2-bromo-1-phenylethanones (6b–o). to yield the corresponding final compounds (7b–o and 8a–h; Scheme 1 ). The anticipated structures of the intermediate and final compounds were confirmed with mass and 1H NMR spectral analysis and were further substantiated by IR and 13C NMR, which is summarized in the Supporting Information.
Scheme 1.
Reagents and conditions; (i) (a) POCl3, DMF, 0 °C - rt, 1 h, (b)100 °C, 3 h; (ii), CsCO3, EtOH, Cu(OAc)2, 80 °C, 15 h; (iii) Acetone, NaOH, rt, 4 h; (iv) Thiosemicarbozide, CH3OH, AcOH, reflux, 5 h; (v) methanol, reflux, 5 h.
The 1H NMR spectrum of compound 5a,b exhibited the presence of distinctive singlet, broad singlet and multiple signals at around δ 10.12, 8.15 and 7.66 ppm attributed to the N-H, NH2 and aromatic protons, respectively. The three protons' singlet signals at 2.31 and 2.11 authenticate methyl groups' presence on pyrazole ring and straight-chain. The quartet of two protons and triplet of three protons at 4.01 and 1.21 affirm an ethoxy group's presence.
Also, the appearance of most informative doublet signals around δ 6.65–6.62 ppm (J =16.77 Hz) and 6.82–6.79 (J =16.77 Hz) confirms the presence of olefinic protons.
The 1H NMR spectrum (400 and 600 MHz, DMSO-d6, CDCl3) of the final compounds (7b–o, 8a–h) displayed some distinctive singlet signals at around δ 11.07−10.12 ppm for N−H proton, multiplet at δ 7.68–7.45 for aromatic protons attached to the 1st position of pyrazole ring, singlet signals of 3H at around δ 2.41–2.30 and 2.30–2.05 ppm attributed to the methyl group at 3rd position of pyrazole ring and straight-chain (N=C-CH3), respectively. Additionally, the most informative singlet signal resonated around δ 8.09–7.31 ppm, attributed to the aromatic proton at H-5 of the thiazole ring, thus indicating its formation through the cyclocondensation process. Whereas most characteristic doublet signals around δ 6.86–6.68 ppm (J = 16.91–16.00 Hz, Ar-HC=CH-) and δ 6.74–6.63 ppm (J = 16.84–16.01 Hz, Ar-HC=CH-) indicates the presence of olefinic protons. This observation was found to be in line with previously reported works of a similar type of compound [25]. Furthermore, the unique quartet signal of two protons and triple signal of three protons resonating around δ 4.00–3.99 and 1.36–1.23 ppm indicated the presence of ethoxy (OCH2CH3) group in the final compounds. The various signals appearing as either doublets or multiplets around δ 8.09–6.96 ppm accounted for aromatic protons. These findings were further corroborated from their respective 13C NMR spectra of the title compounds. The characteristic signals resonating at around δ 170.10–158.80 and 149.65–138.10 ppm were assigned to carbons C-2 and C-5 of the thiazole ring. The characteristic carbon signals appearing around δ 71.46–68.70 and 21.52–11.90 ppm indicate methylene and the three different methyl groups in the title compounds, while the various aromatic carbons resonated around δ 140.78–94.33 ppm. The final compounds’ physical properties are given in Table 1 .
Table 1.
Physical properties of the synthesized compounds.
| Comp'd Name | R1 | R2 | Molecular Formula | Molecular Weight (g/mol) | % Yield | m. p. ( °C.) | Exact Weight Calculated | Exact Weight Found |
|---|---|---|---|---|---|---|---|---|
| 5a | H | – | C17H21N5OS | 343.45 | 87 | 150–153 °C | 343.15 | 344 |
| 5b | H | – | C17H20ClN5OS | 377.89 | 76 | 152–155 °C | 377.11 | 378 |
| 7b | H | Br | C25H24BrN5OS | 522.46 | 78 | 177–180 °C | 521.09 | 522 |
| 7c | H | F | C25H24FN5OS | 461.55 | 72 | 124–127 °C | 461.17 | 462 |
| 7d | H | OH | C25H25N5O2S | 459.56 | 89 | 176–179 °C | 459.17 | 460 |
| 7e | H | H | C25H25N5OS | 443.56 | 73 | 154–157 °C | 443.18 | 444 |
| 7f | H | 2,6-OH, OCH3 | C26H27N5O3S | 489.59 | 76 | 132–135 °C | 489.18 | 490 |
| 7 g | H | 2,5-OH, F | C25H24FN5O2S | 477.55 | 87 | 175–179 °C | 477.16 | 478 |
| 7h | H | 2Cl | C25H23Cl2N5OS | 512.45 | 83 | 187–190 °C | 511.10 | 512 |
| 7i | H | 2,5-OH, Br | C25H24BrN5O2S | 538.46 | 74 | 178–181 °C | 537.08 | 538 |
| 7j | H | 2,6–OCH3 | C27H29N5O3S | 503.62 | 76 | 182–185 °C | 503.2 | 505 |
| 7k | H | NO2 | C25H24N6O3S | 488.56 | 75 | 166–169 °C | 488.16 | 489 |
| 7l | H | OCH3 | C26H27N5O2S | 473.59 | 77 | 194–197 °C | 473.19 | 474 |
| 7m | H | Cl | C25H24ClN5OS | 478.01 | 79 | 135–137 °C | 477.14 | 477 |
| 7n | H | CH3 | C26H27N5OS | 457.59 | 63 | 125–128 °C | 457.19 | 458 |
| 7o | H | 3,4–OCH3 | C27H29N5O3S | 503.62 | 68 | 190–193 °C | 503.2 | 504 |
| 8a | Cl | H | C25H24ClN5OS | 478.01 | 69 | 195–198 °C | 477.4 | 480 |
| 8b | Cl | Br | C25H23BrClN5OS | 556.91 | 71 | 198–201 °C | 555.04 | 556 |
| 8c | Cl | F | C25H23ClFN5OS | 496.00 | 81 | 195–198 °C | 495.13 | 496 |
| 8d | Cl | Cl | C25H23Cl2N5OS | 512.45 | 74 | 187–190 °C | 511.10 | 512 |
| 8e | Cl | 2,5-OH, OCH3 | C25H23ClFN5O2S | 512.00 | 66 | 199–202 °C | 511.12 | 512 |
| 8f | Cl | NO2 | C25H23ClN6O3S | 523.01 | 64 | 197–200 °C | 522.12 | 523 |
| 8 g | Cl | CH3 | C26H26ClN5OS | 492.04 | 73 | 200–203 °C | 491.15 | 492 |
| 8h | Cl | 3,4–OCH3 | C27H28ClN5O3S | 538.06 | 68 | 203–206 °C | 537.16 | 538 |
2.2. Thermodynamic Binding free energy of the synthesized compounds and the standard drugs
Several studies have employed the molecular mechanics/generalized born surface area (MMGBSA) computational technique to estimate the binding free energies (ΔGbind) between a ligand and a protein/enzyme [26,27]. In the study, after 100 ns molecular dynamic simulations, the best-docked ligands' binding energies and the reference drugs for the two proteins were estimated ( Table 2 ). Table S1 in the supplementary materials showed the docking results for all the compounds. According to the study's outcome, CFZ, a standard inhibitor of the spike protein (SAR-CoV-2 Sgp), showed binding energy of -32.816 Kcal/mol. However, out of all the synthesized compounds, compounds 7i and 8f exhibited better and higher (higher than CFZ) binding energies of -34.726 Kcal/mol and -32.956 Kcal/mol, respectively. Table S2 (Supplementary materials) showed the binding free energies of all the best-docked compounds. Compound 7k exhibited a relatively close binding energy of -25.898 Kcal/mol. From Table 2, the result showed the binding free energy of the best six molecules and the standard inhibitor, MLN-4760 toward hACE-2, with compounds 8h, 8g and 7k exhibiting the highest binding energies of -49.320 Kcal/mol, -48.229 Kcal/mol, and -46.008 Kcal/mol, respectively.
Table 2.
Average Thermodynamic Binding Free Energy Profiles for the Synthesized compounds and standard drugs towards SAR-CoV-2 Sgp and hACE-2.
| Energy Components (kcal/mol) | |||||
|---|---|---|---|---|---|
| Complex | Δ EvdW | ΔEelec | ΔGgas | ΔGsolv | ΔGbind |
| SAR-CoV-2 Sgp | |||||
| CFZ | -45.828 ± 5.481 | -27.566 ± 7.274 | -73.394±10.076 | 38.577±5.720 | -32.816± 6.070 |
| 7i | -20.002 ± 3.629 | -70.271 ± 9.366 | -90.273±7.63 | 55.547±10.525 | -34.726±8.387 |
| 7k | -34.080 ± 10.437 | 9.760 ± 26.189 | -24.319±35.263 | -1.579±26.365 | -25.898±9.878 |
| 8f | -47.124 ± 3.565 | 43.181 ± 10.061 | -3.942±10.495 | -28.813±9.856 | -32.956±3.091 |
| hACE2 | |||||
| MLN-4760 | -60.100 ± 4.673 | -341.381 ± 21.212 | -461.482±23.262 | 447.923±21.420 | -43.558±4.132 |
| 7b | -58.982 ± 6.227 | -398.646 ± 20.363 | -457.628±25.322 | 414.511±20.383 | -43.116±6.717 |
| 7h | -59.362 ± 4.122 | -407.838±8.108 | -467.200 ± 10.210 | 425.164±7.467 | -42.036±5.542 |
| 7k | -59.985± 5.699 | -427.345±12.415 | -487.330±15.695 | 441.322±11.135 | -46.008±6.552 |
| 8d | -57.574±5.421 | -412.991 ± 15.436 | -470.566±19.570 | 428.832±15.841 | -41.733±5.821 |
| 8 g | -64.115 ± 3.524 | -433.521 ± 11.806 | -497.637±12.474 | 449.407±10.815 | -48.229±5.191 |
| 8h | -64.828 ± 4.295 | -423.331 ± 14.438 | -488.159 ± 17.120 | 438.839 ± 12.930 | -49.320 ± 6.217 |
ΔEele electrostatic energy, ΔEvdW van der Waals energy, ΔGbind total binding free energy, ΔGsol solvation free energy, ΔEgas gas-phase free energy
Compounds 7b, 7 h, and 8d with average binding energies of -43.116 kcal/mol, -42.036 kcal/mol and -41.733 kcal/mol, respectively, might also be potential inhibitors of the human ACE-2 receptor. These binding energy values might suggest that 7i and 8f could inhibit the activities of SAR-CoV-2 spike protein, and compounds, 7b, 7 h, 7k, 8d, 8 g and 8 h might be potent and promising inhibitors of hACE-2, thereby inhibiting the entry of SAR-CoV-2 into the host cells.
2.3. Protein-ligand interaction with SAR-CoV-2 sgp and hACE-2
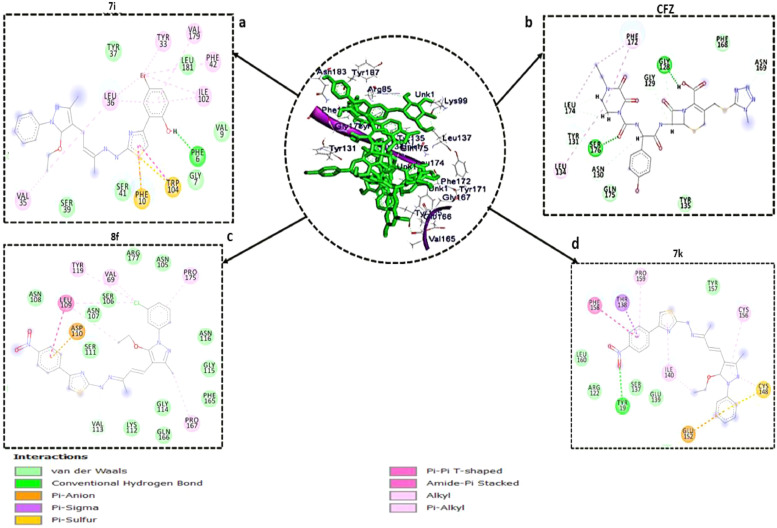
To better understand the interaction better and further between the compounds and the amino residues at the two proteins' binding pockets, plots of ligand-protein interactions were plotted for each compound against the proteins. Studies from Ndagi et al. and Idowu et al. have reportedly used ligand-protein interaction plots to examine the molecular interactions between amino residues at the binding sites of a protein and bound ligands [28,29]. Fig. 2 presented the interaction plots for the SAR-CoV-2 Sgp and hACE-2, respectively. Interactions such as hydrogen bond, π-sigma, π-anion, π-Sulfur, alkyl, π-alkyl, amide-π stacked interaction, π-π T-shaped interactions and Van der Waals (vdW) overlaps observed in the interaction plots. As shown in Fig. 2 (SAR-CoV-2 Sgp bound systems), 7i and 8f have the highest number of interactions of 15 and 19, respectively. Therefore, their higher number of interactions with the amino acid at the binding site of SAR-CoV-2 Sgp justified their higher binding free energies reported in this study. The standard drug, CFZ, and compound 7k had total interactions of 12 and 13, respectively. From this study, the interaction plots showed that the three pyrazolone compounds had π-anion interactions residues Glu152, Phe10, Asp110, and Trp104, with compound 7k having a π-Sulfur with Cys148 residue. The interactions reported in compounds 7i and 8f together with the estimated binding energy suggest they might be promising inhibitors of SAR-CoV-2 Sgp.
Fig. 2.
Interaction types and receptor (SAR-CoV-2 Sgp)-ligand interactions plots of (a) 7i, (b) CFZ, (c) 8f, and (d) 7k.
Fig. 3a and b show the ligand-receptor interaction plots for the standard drugs and the six compounds with the best binding energies toward hACE-2. The result showed that the compounds interacted with essential amino residues at the binding pocket of hACE-2 and revealed similar interactions between the synthesized compounds and the standard drug. The synthesized compounds had an average of 20 interactions with the amino residues at the binding pocket of the hACE-2 receptor, which is relatively equal to the 21 interactions observed in MLN-4760. With 7 h having the highest number of interactions 24 and 7k having the lowest total number of interactions of 18. Similar to the standard drug, in addition to the essential hydrogen and Van Der Waal interactions, all the compounds (except 8 g) had either π-cation, π-anion, or π-Sulfur interactions with the amino residues. From the interaction plots, the results showed that pyrazolone-based compounds could be promising inhibitors of hACE-2 and eventually inhibiting the formation of receptor-binding domain-hACE-2 (RBD-ACE-2) complex required for SARS-CoV-2 entry.
Fig. 3.
(a):Interaction types and receptor (hACE-2)-ligand interactions plots of a) MLN-4760 b) 7b c) 7k and d) 7 h. (b):Interaction types and receptor (hACE-2)-ligand interactions plots of a) 8 h, b) 8 g and c) 8d
2.4. Structural dynamics of bound SARS-CoV-2 sgp and hACE-2
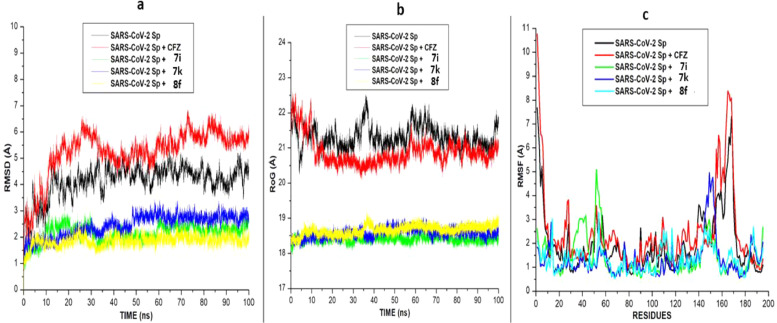
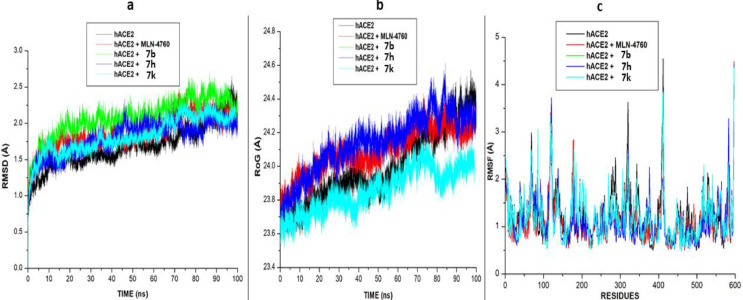
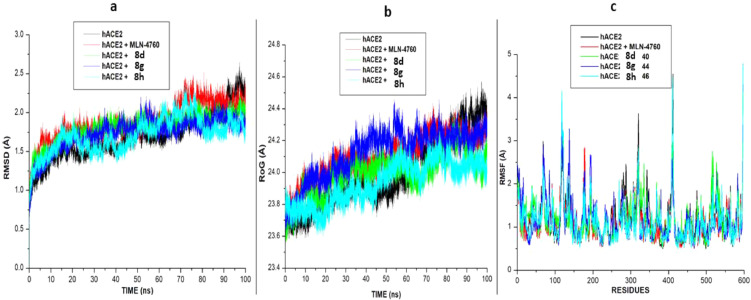
After 100 ns of molecular dynamic (MD) simulation, the root mean square deviation (RMSD), the radius of gyration (RoG) and Root mean square fluctuation (RMSF) of the unbound (apo-enzyme) and ligand-bound trajectories were monitored and calculated to evaluate. The RMSD measured the system's convergence and drift from a reference structural and indirectly the structural stability, and a lowered RMSD values could indicate a more stable system [30]. From the SARS-CoV-2 Sgp systems, all three synthesized compounds achieved convergence in less than 10 ns MD simulation, compared to the CFZ systems, which achieved convergence at approximately 15 ns MD simulation. Signifying their systems reached structural stability and maintained stable conformations earlier than the unbound enzyme (Fig. 4 ). The result also revealed that the binding of the three synthesized compounds (7i, 7k and 8f) with lower RMSD values stabilizes the enzyme more than the standard drug with a relatively higher RMSD value. Unlike the SARS-CoV-2 Sgp systems, the RMSD plots for hACE-2 systems showed that the systems achieved convergence at 100 ns and reached structural stability, as evidence by the lower RMSD values for all the systems ( Fig. 4).
Fig. 4.
Comparative (a). RMSD (b). RoG, and (c). RMSF plots of alpha C atoms of the SARS-CoV-2 Sgp systems calculated throughout 100 ns MD simulations.
The binding of the ligands (the six synthesized compounds and the standard inhibitor) slightly raised the RMSD plots for the bound system above the unbound system (apo-enzyme). However, the plots showed that the binding of the compounds to the two proteins did not affect the proteins' stability and did not alter the protein structural integrity.
In addition to the RMSD, the RoG plots of the two proteins' systems were evaluated (Figs. 5 a). RoG assesses the structural compactness of the proteins upon binding of ligands. The lower the RoG value, the more stable the system. The result of the RoG plots for SARS-CoV-2 Sgp systems correlate with the RMSD plot, showing that the three ligands' binding enhances the structural compactness of the protein and stabilizes the system more than the apo-enzyme and the standard drug. The RoG plots for the hACE-2 also correlate with the RMSD plot. The RoG values for five of the six bound ligands are slightly higher than the RoG plot for the apo-enzyme. The binding of compound 7k lowers the RoG plots slightly below the apo-enzyme, making it more stable and compacted than the other compounds and the standard.
Fig. 5.
(a) Comparative (a). RMSD b). RoG, and (c). RMSF plots of alpha C atoms of the hACE-2 systems calculated throughout 100 ns MD simulations. (b): Comparative (a). RMSD (b). RoG, and (c). RMSF plots of alpha C atoms of the hACE-2 systems calculated throughout 100 ns MD simulations.
RMSF is a measure of the effect of the binding of the ligand on the behavior of the active site residues of a protein. Higher and fewer flexible movements are indicated by high and low RMSF values, respectively. For the SASR-CoV-2 Sgp, as shown in (Fig. 5b), the standard drug and the apo-enzyme showed the highest fluctuation values indicating more flexibility in their residues movement. This result agrees with the high RoG and RMSD plots reported for the apo-enzyme and the standard drug. Lower RMSF values and flexibility were observed in the three synthesized compounds. For the hACE-2 systems, similar fluctuations and flexibilities were observed in all the systems, with higher fluctuations at residues 110–155, 330- 340 and 405–420, respectively.
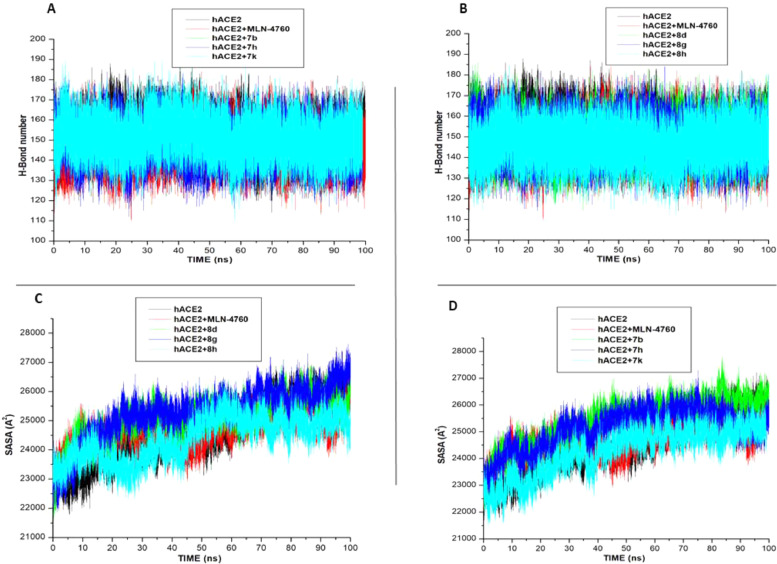
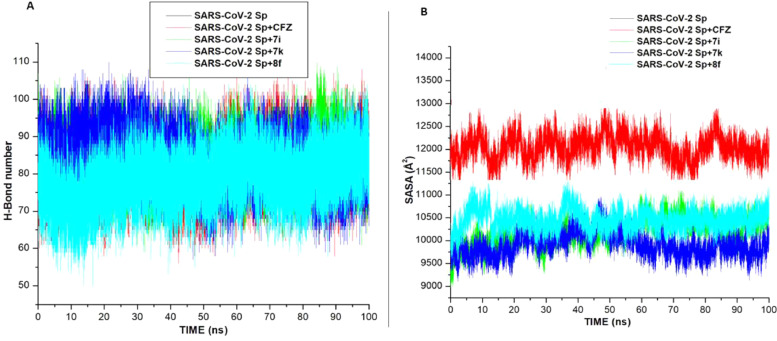
This study also investigated the number of hydrogen bonds (H-bonds) formed before and after ligands bind to the two proteins. H-bonds play vital role in biological system, maintenance of the protein structural integrity, and facilitate protein-ligand binding [29]. Fig. 6 (A,B) and 7A shows H-bonds number formed over the time during the 100 ns simulations of the hACE2 and SARS-CoV-2 Sp respectively. For the hACE2 system, more H-bonds was formed in the Apo-enzyme, MLN-4760, compounds 7b, 7 h, 8d, and 8 g. While the lowest H-bonds formed were observed in compounds 7k and 8 h. In the SARS-CoV-2 Sp systems, higher H-bonds were formed in the Apo-system, CFZ, 7i, 7k than the compound 8f. Reduction in H-bonds observed in compounds 7k and 8 h (in the hACE2), and 8f (in SARS-CoV-2 Sp systems) might lead to structural imbalances and conformational changes that could eventually affect drug binding [31, 32].
Fig. 6.
Comparative of H-bond and SASA plots of alpha C atoms of the hACE-2 systems calculated throughout 100 ns MD simulations.
Solvent accessible surface area (SASA) investigates the effect of the binding of the ligand to a protein, it quantifies the enzyme exposure to solvent molecules [33]. In the hACE2 system (Fig. 6 C,D), the binding of compounds 8d, 8 g, the standard drug (MLN-4760), 7b and 7 h increase solvent accessibility compare to the Apo-enzyme, indicating the binding of the ligand enhanced the accessibility of hACE2 protein to solvent environment. However, the upward increase in solvent accessibility by the compounds showed that hACE2 structural integrity is not altered [27]. Unlike the hACE2 systems, the bind of the compounds 7i, 7k and 8f significantly lowers the exposure SARS-CoV-2 Sp to solvent environment. However, the binding of the standard inhibitor, CFZ, increases the SASA value similar to the Apo-enzyme. Fig. 7 .
Fig. 7.
Comparative of (a). H-bond and (b). SASA plots of alpha C atoms of the SARS-CoV-2 systems calculated throughout 100 ns MD simulations.
2.5. Assessing the drug-likeness of 8 selected phytochemical compounds from the 24 synthesized compounds
As shown in Table 3 , the eight selected phytochemical compounds (7b,7 h, 7i, 7k 8d, 8f, 8 g and 8 h) from the synthesized compounds are poorly soluble in water compared to the two standard drugs (MLN-4760and CFZ) which are soluble in water. And might lower the bioavailability of the eight selected phytochemical compounds.
Table 3.
Predicted ADME parameters; drug-likeness, pharmacokinetic and physicochemical properties of the 8 synthesized compounds with the highest Molecular Simulation scores and two FDA-approved drugs using SWISSADME server.
| Com'd Name | MolecularFormula | MolecularWeight (g/mol) | Lipophilicity(iLOGP) | WaterSolubility | GITAbsorption | BBBPermeability | BioavailabilityScore | Syntheticaccessibility | DrugLikeness (Lipinski) |
|---|---|---|---|---|---|---|---|---|---|
| 7b | C25H24BrN5OS | 522.46 | 4.53 | Poor | Low | No | 0.17 | 4.15 | No (2) |
| 7h | C25H23Cl2N5OS | 512.45 | 4.70 | Poor | Low | No | 0.17 | 4.11 | No (2) |
| 7i | C25H24BrN5O2S | 538.46 | 4.42 | Poor | Low | No | 0.55 | 4.24 | Yes (1) |
| 7k | C25H24N6O3S | 488.56 | 3.56 | Poor | Low | No | 0.55 | 4.22 | Yes |
| 8d | C25H23Cl2N5OS | 512.45 | 4.70 | Poor | Low | No | 0.17 | 4.15 | No (2) |
| 8f | C25H23ClN6O3S | 523.01 | 4.03 | Poor | Low | No | 0.55 | 4.22 | Yes (1) |
| 8 g | C26H26ClN5OS | 492.04 | 4.93 | Poor | Low | No | 0.55 | 4.26 | Yes (1) |
| 8h | C27H28ClN5O3S | 538.06 | 5.11 | Poor | Low | No | 0.55 | 4.43 | Yes (1) |
| MLN-4760 | C19H23Cl2N3O4 | 428.31 | 2.62 | Soluble | High | No | 0.55 | 3.73 | Yes |
| CFZ | C25H27N9O8S2 | 645.67 | 0.81 | Soluble | Low | No | 0.11 | 5.59 | No (2) |
2.6. Assessing the drug-likeness of 8 selected phytochemical compounds from the synthesized compounds
As shown in Table 3, the eight selected phytochemical compounds (7b,7 h, 7i, 7k 8d, 8f, 8 g and 8 h) from the synthesized compounds are poorly soluble in water compared to the two standard drugs (MLN-4760 and CFZ) which are soluble in water. And might lower the bioavailability of the eight selected phytochemical compounds.
One of the two conventional drugs passes the drug-likeness test (MLN-4760), but only five of the phytochemical compounds (7i, 7k, 8f, 8 g and 8 h) pass the test. These showed that the five phytochemical compounds have good drug properties.
Lipinski's rule is a set of five rules used to assess a compound's drug-likeness with pharmacological or biological activities [34]. The rule says the number of hydrogen bond donors should not be more than 5, hydrogen bond acceptors not more than 10, molecular mass less than 500 daltons and partition coefficient (logP) not higher than 5. From this study, the result revealed one of the conventional drugs (MLN) and two of the phytochemical compounds from the synthesized compounds (7k and 8 g) were predicted to pass the rules. It indicates that the two phytochemical compounds from the synthesized compounds possess the same chemical and physical properties as MLN. CFZ, the conventional drug and six of the synthesized compounds failed a maximum of two rules.
All the eight phytochemical compounds from the synthesized compounds were predicted to have low absorption from the GIT (7b,7 h, 7i, 7k 8d, 8f, 8 g and 8 h). However, they could still eventually attain the required concentration needed for therapeutic effects, just as in the conventional drug, CFZ. The drug bioavailability is a measurement of the degree of absorption and fraction of a given amount of unchanged drug that goes to the systemic circulation [35]. Compared with the two conventional drugs, CFZ has the lowest bioavailability scores of 0.11, followed by the 7b, 7 h and 8d with 0.17. Slightly higher bioavailability scores of 0.55 were predicted for MLN-4760and the other phytochemical compounds from the synthesized compounds.
3. Conclusion
Inhibiting the interaction of SARS-CoV-2 Sgp and hACE-2 is significant for the prevention of SARS-CoV-2 infections. Sequel to this fact, this study has examined and evaluated pyrazolone-based compounds' possibilities as potential inhibitors of both SARS-CoV-2 Sgp and hACE-2 to inhibit the virus entry into the cells. This study employed synthetic chemistry methods to design pyrazolone-based compounds and computational chemistry techniques to identify these compounds' potential inhibitory activity against the spike proteins and hACE-2 receptor. From this study, 24 novel pyrazolone-based compounds were designed and synthesized. According to the in-silico analysis, the results of the binding free energy showed that three compounds (7i, 7k and 8f) and six compounds (7b, 7 h, 7k, 8d, 8 g, and 8 h) showed higher and better binding high affinity to SARS-CoV-2 Sgp and hACE-2, respectively compared to the standard drugs. Furthermore, the outcome of the structural analysis of the two proteins upon binding of the inhibitors showed that the two proteins were stable, and the structural integrity of the proteins was not compromised. The study further revealed that the lead compounds possess similar pharmacokinetic and physicochemical properties to the standard drugs. Therefore, this study suggests pyrazolone-based compounds might be potent inhibitors of the virus entry into the host cells.
4. Experimental section
4.1. Chemistry
4.1.1. General consideration
All the fine chemicals, reagents and solvents were purchased from Sigma Aldrich and Merck and were used without further purification unless otherwise stated. The progress of the reactions and the purity of the compounds was monitored by thin-layer chromatography (TLC) on pre-coated silica gel plates procured from E. Merck and Co. (Darmstadt, Germany) visualized by a UV lamp (254 or 365 nm). Purification was performed by using combi-flash (CombiFlash® NextGen 300+) column chromatography. The synthesized compounds' melting points have been determined and uncorrected using a digital Stuart SMP10 melting point Apparatus. The Fourier transform infrared (FTIR) spectra were recorded in the spectral range of 400–4000 cm−1 on a Bruker Alpha FT-IR spectrometer using the ATR technique. The NMR spectra ( 1H and 13C) were recorded using CDCl3 and DMSO-d6 on Bruker AVANCE III 400 and 600 MHz spectrometers. Chemical shifts were determined relative to internal standard TMS at δ 0.0 parts per million (ppm), and the coupling constants were reported in Hertz. The multiplicities of the NMR resonances were abbreviated as s (singlet), d (doublet), dd (doublet of doublet), (t (triplet), q (quartet), m (multiplet) and brs (broad singlet). Mass spectrometry (MS) was performed using a Bruker ESI-QTOF mass spectrometer in positive and negative ion mode.
4.1.2. General procedure for the synthesis of intermediate (2a,b)
The Vilsmeier-Haack reagent was freshly prepared by the careful addition of POCl3 (7 eq.) in dimethylformamide (3 eq.) at 0 °C with constant stirring. To the reaction mixture maintaining 0 °C was added 2-(Phenyl/substituted phenyl)-5-methyl-2,4-dihydro-3H-pyrazol-3-one (1a-b) (1 Eq.) and stirred initially for 30 min and later slowly brought to rt and stirred for 1 h. Finally, the reaction temperature was raised to 100 °C and allowed to stir for 3 hrs. After cooling to rt, the reaction mixture was poured into a mixture of crushed ice and water, the precipitate formed separated by filtration and washed with ice-cold water. The residue obtained was filtered, dried, and recrystallized from ethanol to afford the title compounds (2a,b).
5-chloro-3-methyl-1-phenyl-1H-pyrazole-4-carbaldehyde (2a): Off white, Yield: 75%; 1H NMR (400 MHz, CDCL3 25 °C) δ 9.98 (s, 1H, -CHO), 7.567.52 (m, 4H), 7.50–7.47 (m, 1H), 2.55 (s, 3H, CH3); 13C NMR (100 MHz, CDCL3 25 °C) δ 183.9, 151.9, 137.0, 133.5, 129.4, 129.3, 125.3, 117.5, 13.9 (CH3)
5-chloro-1-(3-chlorophenyl)-3-methyl-1H-pyrazole-4-carbaldehyde (2b): White Solid, Yield: 70%; 1H NMR (400 MHz, CDCL3 25 °C) δ 9.96 (s, 1H, -CHO), 7.59 (s, 1H), 7.47–7.43 (m, 3H), 2.52 (s, 3H, CH3); 13C NMR (100 MHz, CDCL3 25 °C) δ 183.8, 152.2, 138.0, 135.1, 133.6, 130.3, 129.4, 125.5, 123.2, 117.8, 13.9 (CH3)
4.1.3. General procedure for the synthesis of intermediate (3a,b)
To the solution of 5-chloro-1-(phenyl/substituted phenyl)-3-methyl-1H-pyrazole-4-carbaldehyde (2a,b) (1 eq.) in ethanol was added cesium carbonate (1.5 eq.) followed by copper acetate (0.25 eq.) and refluxed for 15 hrs. The reaction was monitored with TLC (30:70, ethyl acetate; hexane); after the reaction completion, the reaction mixture was cooled to rt, then diluted with ethyl acetate and washed with brine solution. The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated under reduced pressure to get the crude product. The solid crude product was purified by column chromatography using 100–200 mesh silica gel. The desired fractions were collected and concentrated under reduced pressure to afford title products (3a,b).
5-ethoxy-3-methyl-1-phenyl-1H-pyrazole-4-carbaldehyde(3a): Brown Solid, Yield: 68%; .); 1H NMR (400 MHz, CDCl3 25 °C) δ 9.90 (s, 1H, -CHO), 7.64 (d, J = 7.79 Hz, 2H), 7.44 (t, J = 7.91 Hz, 2H), 7.33 (t, J = 7.79 Hz, 1H), 4.41 (q, J = 7.04 Hz, 2H), 2.48 (s, 3H, CH3), 1.33 (t, J = 7.04 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3 25 °C) δ 183.1, 155.6, 151.3, 137.5, 129.1, 127.7, 123.1, 108.1, 72.6 (CH2), 15.3 (CH3), 14.0 (CH3)
1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazole-4-carbaldehyde(3b): Brown Solid, Yield: 71%; .); 1H NMR (400 MHz, CDCl3 25 °C) δ 9.91 (s, 1H, -CHO), 7.91 (s, 1H), 7.64 (d, J = 7.79 Hz, 1H), 7.44 (d, J = 7.91 Hz, 1H), 7.33 (t, J = 7.79 Hz, 1H), 4.40 (q, J = 7.04 Hz, 2H, CH2), 2.48 (s, 3H, CH3), 1.33 (t, J = 7.04 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3 25 °C) δ 183.1, 155.6, 151.3, 147.5, 137.5, 129.1, 127.7, 123.1, 115.6, 108.1, 72.6 (CH2), 15.3 (CH3), 14.0 (CH3)
4.1.4. General procedure for the synthesis of compounds (4a,b)
Compound 1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazole-4-carbaldehyde (3a,b) (1.0 eq.) was dissolved in acetone to which sodium hydroxide solution (7.57 eq.) was slowly added with continues stirring for 2–3 h. The reaction was monitored with TLC (30:70, ethyl acetate; hexane). After completing the reaction, excess acetone was removed under reduced pressure and acidify with 6 N HCl, then extracted with DCM. The organic layer was dried over anhydrous sodium sulfate and concentrated under reduced pressure to obtain the title compounds (4a,b).
(E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-one(4a): Brown Solid, Yield: 91%; 1H NMR (400 MHz, CDCl3 25 °C) δ 7.67–7.65 (m, 2H, ArH), 7.44 (t, J =7.80 Hz, 2H, ArH), 7.40 (s, 1H, Ar-HC=CH-), 7.33–7.29 (m, 1H, ArH), 6.56 (d, J =16.53 Hz, 1H, Ar-HC=CH-), 3.98 (q, J =7.03 Hz, 2H, CH2), 2.39 (s, 3H, CH3), 2.33 (s, 3H, CH3), 1.27 (t, J =7.08 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3 25 °C) δ 198.3, 148.9, 139.0, 133.0, 129.2, 127.3, 124.3, 122.5, 104.2, 71.3 (CH2), 27.6 (CH3), 15.3 (CH3), 14.4 (CH3)
(E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-one(4b): Brown Solid, Yield: 74%; 1H NMR (400 MHz, CDCl3 25 °C) δ 7.66 (d, J =8.28 Hz, 2H, ArH), 7.44 (s, 1H, ArH), 7.40 (s, 1H, Ar-HC=CH-), 7.33–7.29 (m, 1H, ArH), 6.56 (d, J =16.53 Hz, 1H, Ar-HC=CH-), 3.98 (q, J =7.03 Hz, 2H, CH2), 2.39 (s, 3H, CH3), 2.33 (s, 3H, CH3), 1.27 (t, J =J=7.08 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3 25 °C) δ 198.3, 153.0, 148.9, 139.0, 133.0, 129.2, 127.3, 124.3, 122.5, 122.3, 104.2, 71.3 (CH2), 27.6 (CH3), 15.3 (CH3), 14.4 (CH3)
4.1.5. General procedure for the synthesis of compounds (5a-b)
To a constantly stirred solution of (E)-4-(1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-one (4a-b) (1.0 eq.) and thiosemicarbazide (1.1eq.) in anhydrous methanol was added a catalytic amount of glacial acetic acid (0.1 ml) and refluxed for 4–5 h.. After completing the reaction (monitored via TLC), the reaction mass was cooled to rt and concentrated under reduced pressure to give a semi-solid residue. The residue was triturated with 50% diethyl ether and pentane and filtered under suction to afford a pure yellow crystalline solid title compound (5a-b).
(E)-2-((E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide(5a) Yield: 87%; mp;150–153 °C; FTIR (ATR, Vmax, cm-1): 3373.89 (N-H Str. of NH2), 3262.82 (N-H Str. of NH), 3.177.48 (Ar C-H Str.), 2973.89 (C-H Str. of CH3), 1597.91 (C = N Str), 1087.70 (C = S); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 10.12 (brs, 1H, NH), 8.15 (brs, 1H, NH2), 7.81 (brs, 1H, NH2), 7.65 (d, J = 7.92 Hz, 2H, ArH), 7.49 (t, J = 8.70 Hz, 2H, ArH), 7.34 (t, J = 8.10 Hz, 1H, ArH), 6.80 (d, J = 16.77 Hz, 1H, Ar-HC=CH-), 6.63 (d, J = 16.77 Hz, 1H, Ar-HC=CH-), 4.00 (q, J = 7.03 Hz, 2H, CH2), 2.30 (s, 3H, CH3), 2.11 (s, 3H, CH3), 1.20 (t, J = 7.76 Hz, 3H, CH3); MS m/z 344.00 (M+H)+.
(E)-2-((E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide(5b) Yield: 76%; mp;152–155 °C; FTIR (ATR, Vmax, cm-1): 366.52 (N-H Str. of NH2), 3258.40 (N-H Str. of NH), 3169.77 (Ar C-H Str.), 2971.39 (C-H Str. of CH3), 1640.27 (C = N Str), 1158.23 (C = S); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 10.12 (brs, 1H), 8.15 (brs, 1H, NH2), 7.77 (brs, 1H, NH2), 7.65 (d, J = 7.90 Hz, 1H), 7.50 (t, J = 7.52 Hz, 2H), 7.34 (d, J = 7.39 Hz, 1H), 6.80 (d, J = 16.30 Hz, 1H Ar-HC=CH-), 6.63 (d, J = 16.98 Hz, 1H, Ar-HC=CH-), 4.01 (q, J = 6.92 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.11 (s, 3H, CH3), 1.21 (t, J = 7.16 Hz, 3H, CH3); m/z 378.00 (M+H)+.
4.1.6. General procedure for the synthesis of final compounds (7b-o and 8a-h)
To a stirred solution of (E)-2-((E)-4-(1-(phenyl/substituted phenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide (5a,b) (1.0 eq.) in anhydrous methanol, was added an appropriate 2-bromo-1-(substituted phenyl)-ethanones ( 6b-o ,1.1 eq.), and the reaction mixture was refluxed for 4–7 h until TLC showed full consumption of starting materials. The reaction mass was cooled to rt and concentrated to remove excess methanol. The obtained reaction mass was diluted with DCM and washed with saturated NaHCO3 solution. The organic layer obtained was dried over anhydrous Na2SO4 and evaporated under reduced pressure to give a solid residue washed with diethyl ether and pentane to afford pure solid title compounds (7b–o and 8a–h).
4.1.6.1. 4-((1E,3E)-3-(2-(4-(4-bromophenyl) cyclopenta-1,3-dien-1-yl) hydrazono) but-1-en-1-yl)-5-ethoxy-3-methyl-1-phenyl-1H-pyrazole (7b); Yellowish Solid, Yield: 78%, mp: 177–180 °C; FTIR (ATR, vmax, cm-1): 3071.06 (Ar-H Str.), 2975.74 (CH Str. of CH3), 1606.33 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.14 (brs, 1H, NH), 7.82 - 7.80 (m, 2H, ArH), 7.68 - 7.65 (m, 2H, ArH), 7.60 -7.57 (m, 2H, ArH), 7.53 -7.48 (m, 2H, ArH), 7.36 (d, J = 8.64 Hz, 1H, ArH), 6.73 (d, J =16.83 Hz, 1H, Ar-HC=CH-), 6.64 (d, J =16.83 Hz, 1H, Ar-HC=CH-), 4.00 (q, J = 7.04 Hz, 2H, CH2), 2.31 (s,3H, CH3), 2.14 (s, 3H, CH3), 1.24 (t, J = 7.05 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 159.9(C-2 of thiazole), 151.5, 149.8, 147.8, 138.2(C-5 of thiazole), 134.6, 132.9, 131.4, 129.1, 127.4, 126.4, 121.8, 121.3, 121.1, 120.4, 112.4, 111.7, 68.5 (CH2), 14.6 (CH3), 13.4 (CH3), 13.0 (CH3). MS m/z 522.00 (M+H)+.
4.1.6.2. 5-ethoxy-4-((1E,3E)-3-(2-(4-(4-fluorophenyl) cyclopenta-1,3-dien-1-yl) hydrazono) but-1-en-1-yl)-3-methyl-1-phenyl-1H-pyrazole (7c); Brownish Solid,Yield:72%,mp:124–127 °C; FTIR (ATR, vmax, cm-1): 3238.79 (N-H Str.), 3112.04 (Ar-H Str.), 2978.96 (CH Str. of CH3), 1549.10 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 40 °C) δ 11.07 (brs, 1H, NH), 789–785 (m, 2H, ArH), 7.66–7.64 (m, 2H, ArH), 7.51–7.47 (m, 2H, ArH), 7.33 (t, J = 7.66 Hz, 1H, ArH), 7.22–7.18 (m, 3H, ArH), 6.68 (d, J =16.91 Hz, 1H, Ar-HC=CH-), 6.63 (d, J =16.66 Hz, 1H, Ar-HC=CH-), 3.98 (q, J = 7.13 Hz, 2H, CH2), 2.29 (s, 3H, CH3), 2.12 (s, 3H, CH3), 1.23 (t, J = 7.24 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.5 (C-2 of thiazole), 160.3, 150.5, 149.4 (C-5 of thiazole), 148.6, 146.8, 137.9, 131.4, 129.2, 127.4 (d, J = 8.08 Hz, 1C), 127.1, 126.6, 121.6, 120.5, 15.3 (d, J = 21.56 Hz, 1C), 104.8, 103.4, 70.5 (CH2), 14.9 (CH3), 13.8 (CH3), 11.9 (CH3). MS m/z 462.00 (M+H) +.
4.1.6.3. 2-(2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl) but-3-en-2-ylidene) hydrazinyl) thiazol-4-yl) phenol (7d); Brownish Solid, Yield:89%, mp:176–179 °C; FTIR (ATR, vmax, cm-1): 3070.84 (Ar-H Str.), 2974.54 (CH Str. of CH3), 1599.88 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.44 (brs, 1H, NH), 11.24 (brs, 1H, OH), 7.80 (d, J=7.94 Hz, 1H, ArH), 7.67 (t, J=8.22 Hz, 2H, ArH), 7.53–7.58 (m, 2H, ArH), 7.36 (t, J=7.33 Hz, 2H, ArH), 7.17–7.13 (m, 1H, ArH), 6.88–6.82 (m, 2H, ArH), 6.77 (d, J=16.67 Hz, 1H, Ar-HC=CH-), 6.66 (d, J=16.71 Hz, 1H, Ar-HC=CH-), 4.02 (q, J=7.04 Hz, 1H, CH2), 2.33 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.26 (t, J=7.06 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 168.5 (C-2 of thiazole), 155, 150.6, 149.3(C-5 of thiazole), 146.9, 137.9, 129.2, 129.1, 128.9, 126.7, 121.8, 121.7, 121.6, 121.2, 119.0, 118.6, 116.9, 116.6, 104.7, 103.6, 70.5(CH2), 14.9 (CH3), 13.8 (CH3), 12.0 (CH3). MS m/z 460.00 (M+H) +.
4.1.6.4.2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2 ylidene)hydrazinyl)-4-phenylthiazole(7e); Light Brownish Solid, Yield:73% mp:154–157 °C; FTIR (ATR, vmax, cm-1): 3070.84 (Ar-H Str.), 2977.30 (CH Str. of CH3), 1614.72 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.11 (brs, 1H, NH), 7.86 (d, J=7.30 Hz, 2H, ArH), 7.67 (d, J=7.68 Hz, 2H, ArH), 7.51 (t, J=7.92 Hz, 2H, ArH), 7.40 (t, J=7.62 Hz, 2H, ArH), 7.35 (t, J=7.34 Hz, 1H, ArH), 7.30 (t, J=7.52 Hz, 2H, ArH), 6.73 (d, J=16.69 Hz, 1H, Ar-HC=CH-), 6.65 (d, J=16.68 Hz, 1H, Ar-HC=CH-)), 4.00 (q, J=7.03 Hz, 2H, CH2), 2.32 (s, 3H, CH3), 2.15 (s, 3H, CH3), 1.25 (t, J=7.01 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.2 (C-2 of thiazole), 159.8, 147.8, 147.0 (C-5 of thiazole), 137.9, 134.6, 133.7, 129.3, 129.1, 128.6 (d, J = 6.86 Hz, 1C), 128.1, 126.8, 126.4, 125.6 (d, J =7.84 Hz, 1C), 121.9, 121.7, 121.4, 112.3, 110.9, 104.8, 68.5 (CH2), 14.7 (CH3), 13.0 (CH3), 12.1 (CH3). MS m/z 444.00 (M+H) +.
4.1.6.5. 2-(2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazol-4-yl)-3-methoxyphenol (7f); Light Brownish Solid,Yield:76%,mp:132–135 °C; FTIR (ATR, vmax, cm-1): 3052.77 (Ar-H Str.), 2978.74 (C-H Str. of CH3), 1600.49 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.27 (brs, 1H, NH), 9.63 (s, 1H, OH), 7.67 (d, J=7.87 Hz, 2H, ArH), 7.57 (d, J=3.13 Hz, 2H, ArH), 7.51 (t, J=7.86 Hz, 2H, ArH), 7.35 (q, J=4.92 Hz, 2H, ArH), 6.81 (d, J=8.96 Hz, 1H, ArH), 6.77 (d, J=16.80 Hz, 1H, 1H, Ar-HC=CH-), 6.66 (d, J=16.65 Hz, 1H, 1H, Ar-HC=CH-), 4.00 (q, J=7.01 Hz, 2H, CH2), 3.72 (s, 3H, OCH3), 2.32 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.25 (t, J=7.03 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 168.3 (C-2 of thiazole), 152.0, 150.6, 149.7, 149.2 (C-5 of thiazole), 146.9, 137.9, 129.2, 126.7–126.6 (d, J = 13.51 Hz, 1C), 124.9, 121.6, 121.4, 118.7, 117.5, 115.5, 111.0, 104.7, 104.4, 70.6(CH2), 55.4(OCH3), 14.9(CH3), 13.8(CH3), 12.0(CH3). MS m/z 490.00 (M+H) +.
4.1.6.6. 2-(2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazol-4-yl)-4-fluorophenol (7 g); Brownish Solid, Yield:87%, mp:175–179 °C; FTIR (ATR, vmax, cm-1): 3070.26 (Ar-H Str.), 2982.37 (C-H Str. of CH3), 1604.59 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.21 (brs, 1H), 9.67 (s, 1H, OH), 7.67–7.63 (m, 3H, ArH), 7.52–7.49 (m, 3H, ArH), 7.35 (t, J = 7.43 Hz, 1H, ArH), 7.01- 6.69 (m, 1H, ArH), 6.90 – 6.86 (m, 1H, ArH), 6.77 (d, J = 16.52 Hz, 1H, Ar-HC=CH-), 6.65 (d, J = 16.43 Hz, 1H, Ar-HC=CH-), 4.00 (q, J = 7.00 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.16 (s, 3H, CH3), 1.24 (t, J = 6.96 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 158.8 (C-2 of thiazole), 154.5, 151.4, 151.2, 150.6, 147.7, 147.0 (C-5 of thiazole), 138.1, 134.4, 129.2–129.1 (d, J = 19.40 Hz, 1c), 128.8, 126.4, 122.2, 120.8 (d, J = 8.00 Hz, 1c), 117.2–117.1 (d, J = 7.67 Hz, 1c), 115.2 -115.1 (d, J = 23.54 Hz, 1c), 114.7, 113.9, 113.7, 112.5, 94.5, 68.7 (CH2), 14.6 (CH3), 13.3 (CH3), 12.9 (CH3). MS m/z 478.00 (M+H) +.
4.1.6.7. 4-(3,4-dichlorophenyl)-2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (7 h); Brownish solid, Yield:83%, mp:187–190 °C; FTIR (ATR, vmax, cm-1): 3054.80 (Ar-H Str.), 2980.41 (C-H Str. of CH3), 1612.58 (C = N Str.): 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.15 (brs, 1H, NH), 8.10–8.09 (d, J = 2.01 Hz, 1H, ArH), 7.85–7.83 (m, 1H, ArH), 7.67–7.65 (m, 3H, ArH), 7.52–7.48 (m, 3H, ArH), 7.34 (t, J = 7.36 Hz, 1H, ArH), 6.74 (d, J = 16.75 Hz, 1H, Ar-HC=CH-), 6.64 (d, J = 16.75 Hz, 1H, Ar-HC=CH-), 3.99 (q, J = 7.12 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.14 (s, 3H, CH3), 1.24 (t, J = 7.02 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 160.1 (C-2 of thiazole), 151.6, 148.4, 147.8, 138.1 (C-5 of thiazole), 134.5, 134.2, 131.6, 130.6, 130.3, 128.9, 127.5, 127.1, 126.3, 125.3, 122.8, 122.1 -122.0 (d, J = 11.63 Hz, 1C), 121.8, 113.0, 112.5, 94.3, 68.6 (CH2), 14.6 (CH3), 13.3 (CH3), 12.9 (CH3). MS m/z 512.00 (M+H) +.
4.1.6.8. 4-bromo-2-(2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazol-4-yl)phenol (7i); Brownish solid, Yield:74%, mp: 178–181 °C; FTIR (ATR, vmax, cm-1): 3051.84 (Ar-H Str.), 2977.06 (C-H Str. of CH3), 1602.93 (C = N Str.): 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.32 (brs, 1H, NH), 10.26 (s, 1H, OH) 8.04 (d, J=2.41 Hz, 1H, ArH), 7.67 (d, J=7.72 Hz, 2H, ArH), 7.52–7.49 (m, 3H, ArH), 7.35 (t, J=7.40 Hz, 1H, ArH), 7.29 (q, J=3.70 Hz, 1H, ArH), 6.88 (d, J=8.66 Hz, 1H, ArH), 6.78 (d, J=16.58 Hz, 1H, Ar-HC=CH-), 6.66 (d, J=16.67 Hz, 1H, Ar-HC=CH-), 4.01 (q, J=7.01 Hz, 2H, CH2), 2.32 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.25 (t, J=7.02 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 168.7 (C-2 of thiazole), 154.8, 147.4(C-5 of thiazole), 138.3.9, 131.6, 129.9, 129.7, 127.2, 127.0, 122.1, 121.9, 119.3, 110.7, 106.5, 105.2, 71.0 (CH2), 15.4 (CH3), 14.3 (CH3), 12.5 (CH3). MS m/z 538.00 (M+H) +.
4.1.6.9. 4-(2,6-dimethoxyphenyl)-2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (7j); Yellowish Solid, Yield:76%, mp:182–185 °C; FTIR (ATR, vmax, cm-1): 3252.00 (N-H Str.), 3162.33 (Ar-H Str.), 2977.32 (C-H Str. of CH3), 1591.75 (C = N Str); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 10.15 (brs, 1H, NH) 7.66 (d, J=7.83 Hz, 2H, ArH), 7.51 (t, J=7.79 Hz, 2H, ArH), 7.41 (t, J=8.32 Hz, 1H, ArH), 7.36 (t, J=7.28, 1H, ArH), 7.16 (s, 1H, ArH), 7.04 (s, 1H, ArH), 6.93 (d, J=17.79 Hz, 1H, ArH), 6.81 (d, J=16.59 Hz, 1H, Ar-HC=CH-), 6.70 (d, J=16.43 Hz, 1H, Ar-HC=CH-), 4.02 (q, J=6.99 Hz, 2H, CH2), 3.78 (s, 6H, OCH3), 2.33 (s, 3H, CH3), 2.24 (s, 3H, CH3), 1.24 (t, J=6.98 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 158.6 (C-2 of thiazole), 151.5, 147.6, 138.2 (C-5 of thiazole), 132.1, 131.9, 129.7, 127.4, 127.4, 125.6, 122.4, 122.2, 105.1, 104.8, 71.3(CH2), 56.5(OCH3), 15.4 (CH3), 14.4 (CH3), 13.3 (CH3). MS m/z 504.00 (M+H) +.
4.1.6.10. 2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(4-nitrophenyl)thiazole (7k); Brownish Solid, Yield: 75%, mp:166–169 °C;FTIR (ATR, vmax, cm-1): 3069.55 (Ar-H Str.), 2981.67 (C-H Str. of CH3), 1596.47 (C = N Str); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.29 (brs, 1H, NH), 8.27 (d, J = 8.51 Hz, 2H, ArH), 8.11 (d, J = 8.69 Hz, 2H), 7.69–7.66 (m, 3H, ArH), 7.50 (t, J = 7.59 Hz, 2H, ArH), 7.34 (t, 7.32 Hz, 1H, ArH), 6.73 (d, J = 16.60 Hz, 1H, Ar-HC=CH-), 6.64 (d, J = 16.60 Hz, 1H, Ar-HC=CH-), 3.99 (q, J = 7.01 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.15 (s, 3H, CH3), 1.24 (t, J = 6.94 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 170.1 (C-2 of thiazole), 156.4, 151.0, 149.6(C-5 of thiazole), 147.4, 146.6, 141.2 (d, J = 11.1 Hz, 1C), 138.4, 129.6 (d, J = 4.4 Hz, 1C), 127.2 (d, J = 12.2 Hz 1C), 126.7 (d, J =4.5 Hz, 1C), 124.5 (d, J = 16.0 Hz, 1C), 122.1, 121.8, 121.5, 105.2, 71.0 (CH2), 15.4 (CH3), 14.3 (CH3), 12.4 (CH3). MS m/z 489.00 (M+H) +.
4.1.6.11. 2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(4-methoxyphenyl)thiazole (7l); Light Brownish Solid, Yield:77% mp:194–197 °C; FTIR (ATR, vmax, cm-1): 3117.31 (N-H Str.), 3041.74 (Ar-H Str.), 2978.63 (C-H Str. of CH3), 1612.09 (C = N Str.); 1H NMR (400 MHz, CDCl3 25 °C) δ 11.29 (brs, 1H, NH), 7.70–7.66 (m, 4H, ArH), 7.46–7.42 (m, 2H, ArH), 7.33–7.29 (m, 1H, ArH), 7.00–6.69 (m, 2H, ArH), 6.86 (d, J = 16.84 Hz, 1H, Ar-HC=CH-), 6.74 (d, J = 16.84 Hz, 1H, Ar-HC=CH-), 6.58 (s, 2H), 3.98 (q, J = 7.12 Hz, 2H, CH2), 3.84 (s, 3H, OCH3), 2.42 (s, 3H, CH3), 2.33 (s, 3H, CH3), 1.29 (t, J = 7.05 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3 25 °C) δ 169.0 (C-2 of thiazole), 160.9, 155.9, 151.8, 147.9 (C-5 of thiazole), 142.6, 138.2, 129.2, 127.3, 127.1, 125.4, 124.8, 122.5, 122.3, 121.4, 114.9, 105.2, 71.1 (CH2), 55.5 (OCH3), 15.3 (CH3), 14.5 (CH3), 13.3 (CH3). MS m/z 474.00 (M+H) +.
4.1.6.12. 4-(4-chlorophenyl)-2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (7 m); Light Brownish Solid, Yield: 79%, mp: 135–137 °C; FTIR (ATR, vmax, cm-1): 3234.58 (N-H Str.), 3112.25 (Ar-H Str.), 2977.59 (C-H Str. of CH3), 1597.50 (C = N Str); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 10.12 (brs, 1H, NH), 7.85 (d, J = 8.28 Hz, 2H, ArH), 7.65 (d, J = 7.36 Hz, 2H, ArH), 7.50 (t, J = 7.89 Hz, 2H, ArH), 7.44 (d, J = 8.65 Hz, 2H, ArH), 7.34 (t, J = 7.13 Hz, 1H, ArH), 7.28 (s, 1H, ArH), 6.69 (d, J = 16.67 Hz, 1H, Ar-HC=CH-), 6.64 (d, J = 16.67 Hz, 1H, Ar-HC=CH-), 3.98 (q, J = 7.04 Hz, 2H, CH2), 2.30 (s, 3H, CH3), 2.12 (s, 3H, CH3), 1.23 (t, J = 7.13 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.9 (C-2 of thiazole), 150.7, 149.4, 148.9, 147.2(C-5 of thiazole), 138.0, 133.8, 132.0, 129.4, 128.8, 127.4, 127.0, 121.9, 120.7, 105.0, 104.6, 70.7 (CH2), 30.8 (CH3), 15.1 (CH3), 13.9 (CH3), 12.1 (CH3). MS m/z 477.00 (M+H) +.
4.1.6.13.2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(p-tolyl)thiazole(7n); Brownish Solid, Yield:63% mp:125–128 °C; FTIR (ATR, vmax, cm-1): 3306.52 (N-H Str.), 3115.66 (Ar-H Str.), 2977.50 (C-H Str. of CH3), 1592.79 (C = N Str.) 1H NMR (400 MHz, CDCl3, 25 °C), δ 8.71 (s, 1H, NH), 7.69 (q, J=5.63 Hz, 4H, ArH), 7.44 (m, J=3.96 Hz, 2H, ArH), 7.30 (m, J=3.99 Hz, 1H, ArH), 7.19 (d, J=7.92 Hz, 2H, ArH), 6.82 (s, 1H, ArH), 6.79 (d, J=16.72 Hz, 1H, Ar-HC=CH-), 6.64 (d, J=16.72 Hz, 1H, Ar-HC=CH-), 3.99 (q, J=7.06 Hz, 2H, CH2), 2.41 (s, 3H, CH3), 2.36 (s, 3H, CH3), 2.05 (s, 3H, CH3), 1.30 (t, J=7.07 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3) d 169.0 (C-2 of thiazole), 151.3 (d, J=24.27 Hz, 1C), 148.1, 147.6(C-5 of thiazole), 138.4, 137.5, 132.2, 129.27 (d, J=23.33 Hz, 1 C), 126.86 (d, J=12.07 Hz, 1 C), 125.8, 122.2, 121.1, 105.4, 102.9, 70.85 (CH2), 21.31 (CH3), 15.33 (CH3), 14.37 (CH3), 11.01 (CH3). MS m/z 458.00 (M+H) +.
4.1.6.14. 4-(3,4-dimethoxyphenyl)-2-(2-((2E,3E)-4-(5-ethoxy-3-methyl-1-phenyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (7o); Brownish Solid, Yield: 68%, mp: 190–193 °C; FTIR (ATR, vmax, cm-1): 3060.57 (Ar-H Str.), 2902.04 (C-H Str. of CH3), 1613.29 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 10.26 (s, 1H, NH), 7.67 (d, J=7.80 Hz, 2H, ArH), 7.51 (t, J=7.80 Hz, 2H, ArH), 7.38 (m, 3H, ArH), 7.19 (s, 1H, ArH), 6.99 (d, J=8.37 Hz, 1H, ArH), 6.77 (d, J=16.66 Hz, 1H, Ar-HC=CH-), 6.67 (d, J=16.66 Hz, 1H, Ar-HC=CH-), 4.01 (q, J=6.99 Hz, 2H, CH2), 3.79 (d, J=11.77 Hz, 6H, OCH3), 2.32 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.25 (t, J=7.02 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 159.5 (C-2 of thiazole), 151.2 (d, J = 8.80 Hz, 1C), 150.8, 148.8(d, J = 4.37 Hz, 1C), 148.0, 138.2 (C-5 of thiazole), 138.1, 134.3, 129.0, 126.7, 126.3, 121.7, 121.6, 118.0, 112.4, 111.7, 109.2, 109.0, 94.6, 68.7 (CH2), 55.3 (d, J = 24.2 Hz, 1C, OCH3), 14.6 (CH3), 13.3 (CH3), 13.1 (CH3). MS m/z 504.00 (M+H) +.
4.1.6.15. 2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-phenylthiazole (8a); Brownish Solid, Yield: 69%, mp: 195–198 °C; FTIR (ATR, vmax, cm-1): 3087.50 (Ar-H Str.), 2981.46 (C-H Str. of CH3), 1618.49 (C = N Str.); 1H NMR (400 MHz, CDCl3) d 11.31 (brs, 1H, NH), 7.74 (s, 1H, ArH), 7.70 (d, J=7.09 Hz, 2H, ArH), 7.60 (d, J=8.16 Hz, 1H, ArH), 7.43 (t, J=7.26 Hz, 2H, ArH), 7.37 (m, 2H, ArH), 7.32 (d, J=8.17 Hz, 1H, ArH) 7.24 (s, 1H, ArH), 6.79 (d, J=16.75 Hz, 1H, Ar-HC=CH-), 6.71 (d, J=16.42 Hz, 1H, Ar-HC=CH-),), 4.00 (q, J=7.04 Hz, 2H, CH2), 2.38 (s, 3H, CH3), 2.28 (s, 3H, CH3), 1.30 (t, J=7.06 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.1 (C-2 of thiazole), 150.7, 147.6 (C-5 of thiazole), 139.9, 134.5, 133.5, 131.1, 129.2, 128.5, 127.5, 126.3, 125.5, 124.6, 120.9, 120.5, 119.8, 119.6, 105.2, 103.9, 70.8 (CH2), 14.9 (CH3), 13.8 (CH3), 11.9 (CH3). MS m/z 480.00 (M+H) +.
4.1.6.16. 4-(4-bromophenyl)-2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (8b); Brownish Solid, Yield: 71%, mp: 198–201 °C; FTIR (ATR, vmax, cm-1): 3116.74 (N-H Str.), 2984.47 (Ar-H Str.), 2924.74 (C-H Str. of CH3), 1616.80 (C = N Str.); 1H NMR (400 MHz, DMSO) d 11.40 (brs, 1H, NH), 7.81 (d, J=8.36 Hz, 2H, ArH), 7.73 (s, 1H, ArH), 7.68 (d, J=7.22 Hz, 1H, ArH), 7.59 (d, J=8.38 Hz, 2H, ArH), 7.53 (d, J=8.14 Hz, 1H, ArH), 7.40 (d, J=7.95 Hz, 1H, ArH), 7.35 (d, J=4.91 Hz, 1H, ArH), 6.72 (d, J=16.93 Hz, 1H, Ar-HC=CH-), 6.64 (d, J=16.78 Hz, 1H, Ar-HC=CH-), 4.05 (q, J=7.02 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.14 (s, 3H, CH3), 1.27 (t, J=6.95 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.1 (C-2 of thiazole), 150.6, 148.8, 147.6 (C-5 of thiazole), 139.0, 133.8, 133.4, 131.5, 131.0, 129.1, 127.5, 126.3, 124.5, 123.4, 120.9, 120.5, 120.4, 119.6, 115.1, 105.2, 104.8, 70.8 (CH2), 14.9 (CH3), 13.8 (CH3), 11.9 (CH3). MS m/z 556.00 (M+H) +.
4.1.6.17. 2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(4-fluorophenyl)thiazole (8c); Brownish solid, Yield: 81%, mp: 195–198 °C; FTIR (ATR, vmax, cm-1): 3221.52 (N-H Str.), 3094.49 (Ar-H Str.), 2982.67 (C-H Str. of CH3), 1618.01 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 7.90 (m, J=3.55 Hz, 2H, ArH), 7.73 (t, J=1.93 Hz, 1H, ArH), 7.68 (m, J=1.82 Hz, 1H, ArH), 7.59 (q, J=3.44 Hz, 1H, ArH), 7.41 (q, J=3.03 Hz, 1H, ArH), 7.26 (t, J=4.01 Hz, 2H, ArH), 7.22 (d, J=8.87 Hz, 2H, ArH), 6.73 (d, J=16.60 Hz, 1H, Ar-HC=CH-), 6.65 (d, J=16.70 Hz, 1H, Ar-HC=CH-),), 4.05 (q, J=7.02 Hz, 2H, CH2), 2.32 (s, 3H, CH3), 2.14 (s, 3H, CH3), 1.28 (t, J=7.01 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.2 (C-2 of thiazole), 162.8, 160.3, 150.7, 147.6 (C-5 of thiazole), 139.0, 133.5, 131.2, 130.9 (d, J = 10.96 Hz, 1C), 129.2, 128.3, 127.5 (d, J = 8.13 Hz, 1C), 126.3, 124.8, 124.5, 123.4, 120.7 (d, J = 38.48 Hz, 1C), 119.7 (d, J = 24.30 Hz, 1C), 115.4 (d, J = 21.37 Hz, 1C), 115.1, 105.2, 103.6, 70.8 (CH2), 14.9 (CH3), 13.8 (CH3), 11.9 (CH3). MS m/z 496.00 (M+H) +.
4.1.6.18. 4-(4-chlorophenyl)-2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazole (8d); Light Brownish Solid, Yield: 74% mp: 187–190 °C; FTIR (ATR, vmax, cm-1): 3084.66 (Ar-H Str.), 2982.50 (C-H Str. of CH3), 1616.31 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.29 (brs, 1H, NH), 7.89 (d, J=8.52 Hz, 2H, ArH), 7.75 (t, J=1.91 Hz, 1H, ArH), 7.60 (s, 1H, ArH), 7.55 (t, J=8.13 Hz, 1H, ArH), 7.47 (d, J=8.64 Hz, 2H, ArH), 7.42 (d, J=7.12 Hz, 1H, ArH), 7.38 (s, 1H, ArH), 6.74 (d, J=16.91 Hz, 1H, Ar-HC=CH-), 6.66 (d, J=16.68 Hz, 1H, Ar-HC=CH-), 4.29 (s, 2H, CH2), 2.34 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.30 (t, J=7.01 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.5 (C-2 of thiazole), 150.9, 149.2, 147.9 (C-5 of thiazole), 139.2, 133.7, 133.6, 132.7, 132.1, 131.2, 128.8, 127.4, 126.5, 121.6, 121.3, 121.1, 120.8, 120.1, 119.8, 105.4, 104.9, 71.0 (CH2), 15.1 (CH3), 13.9 (CH3), 12.1(CH3). MS m/z 512.00 (M+H) +.
4.1.6.19. 2-(2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)thiazol-4-yl)-4-fluorophenol (8e); Brownish Solid, Yield: 66%, mp: 199–202 °C; FTIR (ATR, vmax, cm-1): 3073.91 (Ar-H Str.), 2895.89 (C-H Str. of CH3), 1600.42 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.26 (brs, 1H), 10.12 (s, 1H, OH), 7.72 (s,1 H, ArH), 7.67 (d, J=1.96 Hz, 1H, ArH), 7.63 (q, J=4.26 Hz, 2H, ArH), 7.56 (s, 1H), 7.52 (d, J=8.14 Hz, 1H, ArH), 7.48 (d, J=4.35 Hz, 1H, ArH), 7.40 (d, J=7.30 Hz, 1H, ArH), 6.99 (q, J=5.56 Hz, 1H, ArH), 6.89 (t, J=4.43 Hz, 1H, ArH), 6.76 (d, J=16.63 Hz, 1H, Ar-HC=CH-), 6.64 (d, J=16.68 Hz, 1H, Ar-HC=CH-), 4.04 (q, J=6.99 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.15 (s, 3H, CH3), 1.26 (t, J=7.00 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 168.2 (C-2 of thiazole), 154.2, 151.4, 147.7 (C-5 of thiazole), 139.0, 138.5, 133.5, 131.0, 127.1, 124.6, 123.4, 120.9, 119.6, 115.0, 112.8, 105.6, 105.1, 70.8 (CH2), 15.0 (CH3), 13.8 (CH3), 12.0 (CH3). MS m/z 512.00 (M+H) +.
4.1.6.20. 2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(4-nitrophenyl)thiazole (8f); Brownish Solid, Yield: 64%, mp: 197–200 °C; FTIR (ATR, vmax, cm-1): 3087.80 (Ar-H Str.), 2978.29 (C-H Str. of CH3), 1620.73 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.66 (brs, 1H, NH), 8.27 (d, J=8.91 Hz, 2H, ArH), 8.11 (d, J=8.86 Hz, 2H, ArH), 7.72 (t, J=1.87 Hz, 1H, ArH), 7.68–7.66 (m, 2H, ArH), 7.54 (t, J=8.16 Hz, 1H, ArH), 7.40 (d, J=6.87 Hz, 1H, ArH), 6.73 (d, J=16.76 Hz, 1H, Ar-HC=CH-), 6.64 (d, J=16.69 Hz, 1H, Ar-HC=CH-), 4.05 (q, J=7.00 Hz, 2H, CH2), 2.31 (s, 3H, CH3), 2.14 (s, 3H, CH3), 1.27 (t, J=7.00 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 169.8 (C-2 of thiazole), 150.9, 149.3, 148.7, 147.9 (C-5 of thiazole), 146.3, 140.9, 139.2, 133.7, 131.2, 127.5, 126.5, 124.3, 121.1, 120.9, 119.8, 109.0, 105.3, 71.0 (CH2), 15.1 (CH3), 14.0 (CH3), 12.1(CH3). MS m/z 523.00 (M+H) +.
4.1.6.21. 2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(p-tolyl)thiazole (8 g); Brownish Solid, Yield: 73%, mp: 200–203 °C; FTIR (ATR, vmax, cm-1): 3088.16 (Ar-H Str.), 2980.80 (C-H Str. of CH3), 1615.82 (C = N Str.); 1H NMR (400 MHz, CDCl3, 25 °C), δ 11.10 (brs, 1H, NH), 7.76 (s, 1H, ArH), 7.61 (d, J=7.85 Hz, 3H, ArH), 7.56 (d, J=8.07 Hz, 3H, ArH), 7.35 (t, J=8.07 Hz, 1H, ArH), 7.23 (s, 3H, ArH), 6.76 (d, J=16.00 Hz, 1H, Ar-HC=CH-), 6.67 (d, J=16.01 Hz, 1H, Ar-HC=CH-), 4.01 (q, J=7.03 Hz, 2H, CH2), 2.39 (s, 3H, CH3), 2.36 (s, 3H, CH3), 2.30 (s, 3H, CH3), 1.31 (t, J=7.06 Hz, 3H, CH3); 13C NMR (100 MHz, CDCl3, 25 °C), δ 169.0 (C-2 of thiazole), 151.3 (d, J=24.27 Hz, 1C), 148.1, 147.6 (C-5 of thiazole), 138.4, 137.5, 132.2, 129.2 (d, J=23.33 Hz, 1 C), 126.8 (d, J=12.07 Hz, 1 C), 125.8, 122.2, 121.1, 105.4, 102.9, 70.8 (CH2), 21.3 (CH3), 15.3 (CH3), 14.3 (CH3), 11.01 (CH3). MS m/z 492.00 (M+H) +.
4.1.6.22. 2-(2-((2E,3E)-4-(1-(3-chlorophenyl)-5-ethoxy-3-methyl-1H-pyrazol-4-yl)but-3-en-2-ylidene)hydrazinyl)-4-(3,4-dimethoxyphenyl)thiazole (8 h); Brownish Solid, Yield: 68%, mp: 203–206 °C; FTIR (ATR, vmax, cm-1): 3246.20 (N-H Str.), 3077.26 (Ar-H Str.), 2901.07 (C-H Str. of CH3), 1608.77 (C = N Str.); 1H NMR (400 MHz, DMSO-d6 25 °C) δ 11.13 (s, 1H, NH), 7.67 (d, J =7.80 Hz, 2H, ArH), 7.51 (t, J =7.80 Hz, 2H, ArH), 7.38 (m, 3H, ArH), 7.19 (s, 1H, ArH), 6.99 (d, J =8.37 Hz, 1H, ArH), 6.77 (d, J =16.66 Hz, 1H, Ar-HC=CH-), 6.67 (d, J =16.66 Hz, 1H, Ar-HC=CH-), 4.01 (q, J =6.99 Hz, 2H, CH2), 3.79 (d, J =11.77 Hz, 6H, OCH3), 2.32 (s, 3H, CH3), 2.17 (s, 3H, CH3), 1.25 (t, J =7.02 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6 25 °C) δ 162.4 (C-2 of thiazole), 159.5, 151.2 (d, J = 8.80 Hz, 1C), 150.8, 148.8(d, J = 4.37 Hz, 1C), 148.0, 138.2(C-5 of thiazole), 138.1, 134.3, 129.0, 126.7, 126.3, 121.7, 121.6, 118.0, 112.4, 111.7, 109.2, 109.0, 68.7 (CH2), 55.3 (d, J = 24.2 Hz, 1C, OCH3), 14.6 (CH3), 13.3 (CH3), 13.1 (CH3). MS m/z 538.00 (M+H) +.
4.2. Molecular docking and simulation
4.2.1. Proteins (COVID-19 sgp and hACE2) acquisition and preparation
The X-ray crystal structures of the SARS-CoV-2 Sgp (PDB code: 6LZG) and hACE2 (PDB code: 1R4l) were obtained from the RSCB Protein Data Bank [36,37]. The two proteins' structures were then prepared on the UCSF Chimera software package [38]. The structures of the proteins were prepared by removing water molecules, nonstandard naming, protein residue connectivity. The missing atoms of sidechains and protein backbone were added to the protein structure before the molecular docking. The standard drugs cefoperazone (CFZ) and MLN-4760 were accessed from PubChem [39]. The 3-D structures of the pyrazolone compounds and the standard drugs were prepared on the Avogadro software package [40].
4.2.2. Molecular docking
Autodock available on Chimera was used for molecular docking [41] with default docking parameters. Before docking, Gasteiger charges were added to the molecules, and the non-polar hydrogen atoms were merged into carbon atoms. The molecules were then docked into the proteins' binding pocket, COVID-19 Sgp and hACE2, by defining the grid box with a spacing of 1 Å each and size and (64 × 108 × 46) and (78 × 102 × 90) pointing in x, y and z directions, respectively. A duplicate docking and of the The standard drug systems and all molecules with the two best docking scores (for duplication) were then subjected to molecular dynamics simulations.
4.2.3. Molecular dynamic (MD) simulations
The MD simulation was performed as described by Idowu et al. [29]. The simulations were performed using the GPU version provided with the AMBER package (AMBER 18), in which the FF18SB variant of the AMBER force field (Nair and Miners, 2014) was used to describe the systems.
ANTECHAMBER was used to generate atomic partial charges for the ligand by utilizing the Restrained Electrostatic Potential (RESP) and the General Amber Force Field (GAFF) procedures. The Leap module of AMBER 18 allowed for the addition of hydrogen atoms and Cl− and Na+ counter ions to COVID-19 Sgp and hACE2, respectively, to neutralize all systems. The systems were then suspended implicitly within an orthorhombic box of TIP3P water molecules such that all atoms were within 8 Å of any box edge [42].
An initial minimization of 2000 steps was carried out with an applied restraint potential of 500 kcal/mol for both solutes. They were performed for 1000 steps using the steepest descent method followed by 1000 steps of conJugate gradients. An additional full minimization of 1000 steps was further carried out using the conJugate gradient algorithm without restraint. A gradual heating MD simulation from 0 K to 300 K was executed for 50 ps, such that the systems maintained a fixed number of atoms and fixed volume. The systems' solutes were imposed with a potential harmonic restraint of 10 kcal/mol and collision frequency of 1.0 ps. Following heating, an equilibration estimating 500 ps of each system was conducted; the operating temperature was kept constant at 300 K. Additional features such as several atoms and pressure were also held constant, mimicking an isobaric-isothermal ensemble. The system's pressure was maintained at 1 bar using the Berendsen barostat (Gonnet, 2007; Basconi and Shirts, 2013).
The total time for the MD simulations conducted was 100 ns. In each simulation, the SHAKE algorithm was employed to constrict hydrogen atoms' bonds [43]. The step size of each simulation was 2fs, and an SPFP precision model was used. The simulations coincided with the isobaric-isothermal ensemble (NPT), with randomized seeding, the constant pressure of 1 bar maintained by the Berendsen barostat a pressure-coupling constant of 2 ps, a temperature of 300 K and Langevin thermostat with a collision frequency of 1.0 ps [42–44].
4.2.4. Post-Dynamic analysis
Analysis of root mean square deviation (RMSD), root means square fluctuation (RMSF) Solvent accessible surface area (SASA), Hydrogen bond (Hbond) and radius of gyration (RoG) was done using the CPPTRAJ module employed in the AMBER 18 suit. All average raw data plots were generated using the Origin data analysis software [45].
4.2.5. Binding free energy calculations
To estimate and compare the systems' binding affinity, the free binding energy was calculated using the Molecular Mechanics/GB Surface Area method (MM/GBSA) (Ylilauri and Pentikäinen, 2013). Binding free energy was averaged over 100,000 snapshots extracted from the 100 ns trajectory. The free binding energy (ΔG) computed by this method for each molecular species (complex, ligand, and receptor) can be represented as:
Egas denotes the gas-phase energy, which consists of the internal energy Eint, Coulomb energy Eele and the van der Waals energies Evdw. The Egas was directly estimated from the FF14SB force field terms. Solvation free energy, Gsol, was estimated from the energy contribution from the polar states, GGB, and non-polar states, G. The non-polar solvation energy, SA. GSA was determined from the solvent-accessible surface area (SASA), using a water probe radius of 1.4 Å. In contrast, the polar solvation, GGB, the contribution was estimated by solving the GB equation. S and T denote the total entropy of the solute and temperature, respectively.
4.2.6. Pharmacokinetic properties analysis
For the prediction of the lead compounds' pharmacokinetic properties and the standard drugs, the SwissADME server was employed [44].The server predicts the target of small molecules.
Credit author statement
Vincent A. Obakachi and Narva Deshwar Kushwaha synthesized the compounds and drafted the manuscript. Babita Kushwaha contributed to the data analysis and experimental writing. Idowu Kehinde performed in-silico analysis and assisted with manuscript drafting. Mavela Cleopus Mahlalela and Suraj Raosaheb Shinde contributed to the data analysis and manuscript drafting. Rajshekhar Karpoormath designed the experiments and supervised the project.
CRediT authorship contribution statement
Vincent A. Obakachi: Conceptualization, Investigation, Methodology, Writing - original draft. Narva Deshwar Kushwaha: Conceptualization, Investigation, Methodology, Writing - original draft. Babita Kushwaha: Visualization, Investigation, Data curation. Mavela Cleopus Mahlalela: Writing - review & editing. Suraj Raosaheb Shinde: Writing - review & editing. Idowu Kehinde: Writing - original draft, Software. Rajshekhar Karpoormath: Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are thankful to the Discipline of Pharmaceutical Sciences, College of Health Sciences, University of KwaZulu-Natal (UKZN), Durban, South Africa, for providing all the necessary facilities. R.K. gratefully acknowledges National Research Foundation-South Africa (NRF-SA) for funding this project (Grant Nos. 103728 and 112079). The authors would also like to acknowledge fellow UKZN colleagues Dr. Vuyisa Mzozoyana (NMR spectroscopy).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.130665.
Appendix. Supplementary materials
References
- 1.Bigelow B.F., Tang O., Barshick B., Peters M., Sisson S.D., Peairs K.S., Katz M.J. Outcomes of universal COVID-19 testing following detection of incident cases in 11 long-term care facilities. JAMA Intern. Med. 2021;181:127–129. doi: 10.1001/jamainternmed.2020.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Huang, C. Yang, X.F. Xu, W. Xu, S.W.J.A.P.S. Liu, Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19, 41 (2020) 1141–1149. [DOI] [PMC free article] [PubMed]
- 6.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 With SARS-CoV and Influenza pandemics. The Lancet Infectious Diseases. 2020;20:238–244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Zhan P., Liu X. Targeting the entry step of SARS-CoV-2: a promising therapeutic approach. Signal Transduct. Target. Ther. 2020;5:1–2. doi: 10.1038/s41392-020-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16:1718. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. J. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan Y., Graham R., Baric R., Li F. An analysis based on decade-long structural studies of SARS 3, JVI Accepted Manuscript Posted Online 29 January 2020. J. Virol. 2020;94:1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorg. Med. Chem. 2010;18:7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe T., Yuki S., Egawa M., Nishi H. Protective effects of MCI-186 on cerebral ischemia: possible involvement of free radical scavenging and antioxidant actions. J. Pharmacol. Exp. Ther. 1994;268:1597–1604. [PubMed] [Google Scholar]
- 21.Kawai H., Nakai H., Suga M., Yuki S., Watanabe T., Saito K.I. Effects of a novel free radical scavenger, MCI-186, on ischemic brain damage in the rat distal middle cerebral artery occlusion model. J. Pharmacol. Exp. Ther. 1997;281:921–927. [PubMed] [Google Scholar]
- 22.Wu T.W., Zeng L.H., Wu J., Fung K.P. Myocardial protection of MCI-186 in rabbit ischemia–reperfusion. Life Sci. 2002;71:2249–2255. doi: 10.1016/s0024-3205(02)01965-3. [DOI] [PubMed] [Google Scholar]
- 23.Masih A., Agnihotri A.K., Srivastava J.K., Pandey N., Bhat H.R., Singh U.P. Discovery of novel pyrazole derivatives as a potent anti-inflammatory agent in RAW264. 7 cells via inhibition of NF-KB for possible benefit against SARS-CoV-2. J. Biochem. Mol. Toxicol. 2020;35:e22656. doi: 10.1002/jbt.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;11:915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampannavar G.A., Karpoormath R., Palkar M.B., Shaikh M.S., Chandrasekaran B. Dehydrozingerone inspired styryl hydrazine thiazole hybrids as promising class of antimycobacterial agents. ACS Med. Chem. Lett. 2016;7:686–691. doi: 10.1021/acsmedchemlett.6b00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehinde I., Ramharack P., Nlooto M., Gordon M. Dynamics, molecular dynamic mechanism (s) of inhibition of bioactive antiviral phytochemical compounds targeting cytochrome P450 3A4 and P-glycoprotein. J. Biomol. Struct. 2020:1–11. doi: 10.1080/07391102.2020.1821780. [DOI] [PubMed] [Google Scholar]
- 27.Farrokhzadeh A., Akher F.B., Soliman M.E. Probing the dynamic mechanism of uncommon allosteric inhibitors optimized to enhance drug selectivity of SHP2 with therapeutic potential for cancer treatment. Appl. Biochem. Biotechnol. 2019;188:260–281. doi: 10.1007/s12010-018-2914-0. [DOI] [PubMed] [Google Scholar]
- 28.Ndagi U., Mhlongo N.N., Soliman M.E. The impact of Thr91 mutation on c-Src resistance to UM-164: molecular dynamics study revealed a new opportunity for drug design. Mol. Biosyst. 2017;13:1157–1171. doi: 10.1039/c6mb00848h. [DOI] [PubMed] [Google Scholar]
- 29.Kehinde I., Ramharack P., Nlooto M., Gordon M. The pharmacokinetic properties of HIV-1 protease inhibitors: a computational perspective on herbal phytochemicals. Heliyon. 2019;5:e02565. doi: 10.1016/j.heliyon.2019.e02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess B. Convergence of sampling in protein simulations. Phys. Rev. E. 2002;65 doi: 10.1103/PhysRevE.65.031910. [DOI] [PubMed] [Google Scholar]
- 31.Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ndagi U., Mhlongo N.N., Soliman M.E. Re-emergence of an orphan therapeutic target for the treatment of resistant prostate cancer–a thorough conformational and binding analysis for ROR-γ protein. J. Biomol. Struct. Dyn. 2018;36:335–350. doi: 10.1080/07391102.2016.1277555. [DOI] [PubMed] [Google Scholar]
- 33.Boyce S.E., Tirunagari N., Niedziela-Majka A., Perry J., Wong M., Kan E., Lagpacan L., Barauskas O., Hung M., Fenaux M. Structural and regulatory elements of HCV NS5B polymerase–β-loop and C-terminal tail–are required for activity of allosteric thumb site II inhibitors. PLoS One. 2014;9:e84808. doi: 10.1371/journal.pone.0084808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Heaney R.P. Factors influencing the measurement of bioavailability, taking calcium as a model. J. Nutr. 2001;131:1344S–1348S. doi: 10.1093/jn/131.4.1344S. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C.C., Pettersen E.F., Goddard T.D., Meng E.C., Sali A., Ferrin T.E., Chimera UCSF. Modeller, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosdidier A., Zoete V., Michielin O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 42.Ryckaert J.P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 43.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 44.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifert E. OriginPro 9.1: Scientific Data Analysis and Graphing Software Software Review. J. Chem. Inf. Model. 2014;54:1552. doi: 10.1021/ci500161d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.