Abstract
Context
ANGPTL8 (A8) plays a key role in determining the tissue fate of circulating triglycerides (TGs). Plasma A8 levels are associated with several parameters of glucose and TG metabolism, but the causality of these relationships and the contribution of genetic variants to differences in A8 levels have not been explored.
Objective
To characterize the frequency distribution of plasma A8 levels in a diverse population using a newly-developed enzyme-linked immunosorbent assay (ELISA) and to identify genetic factors contributing to differences in plasma A8 levels.
Methods
We studied a population-based sample of Dallas County, comprising individuals in the Dallas Heart Study (DHS-1, n = 3538; DHS-2, n = 3283), including 2131 individuals with repeated measurements 7 to 9 years apart (age 18-85 years; >55% female; 52% Black; 29% White; 17% Hispanic; and 2% other). The main outcome measures were associations of A8 levels with body mass index (BMI), plasma levels of glucose, insulin, lipids, and hepatic TGs, as well as DNA variants identified by exome-wide sequencing.
Results
A8 levels varied over a 150-fold range (2.1-318 ng/mL; median, 13.3 ng/mL) and differed between racial/ethnic groups (Blacks > Hispanics > Whites). A8 levels correlated with BMI, fasting glucose, insulin, and TG levels. A variant in A8, R59W, accounted for 17% of the interindividual variation in A8 levels but was not associated with the metabolic parameters correlated with plasma A8 concentrations.
Conclusions
A8 levels were strongly associated with indices of glucose and TG metabolism, but the lack of association of genetic variants at the A8 locus that impact A8 levels with these parameters indicates that differences in A8 levels are not causally related to the associated metabolic phenotypes.
Keywords: ANGPTL8, ANGPTL3, triglycerides, glucose, insulin, obesity
The angiopoietin-like (ANGPTL) proteins are circulating peptides that play key roles in determining the levels and fate of circulating cholesterol and triglycerides (TGs) in response to food intake. ANGPTL family members contain an unstructured domain at the N-terminus separated by a furin cleavage site from a C-terminal fibrinogen-like domain. The human genome encodes 8 ANGPTLs (A1-A8), 3 of which (A3, A4, and A8) share a sequence motif in their N-terminal regions that binds to lipoprotein lipase (LPL) (1). The last identified family member, A8, differs from A3 and A4 by lacking a C-terminal fibrinogen-like domain. A4 expression rises with fasting and falls with refeeding (2), whereas A8 levels are inversely related: highest in the refed state and lowest in the fasted state (3-5). A3 expression is less influenced by dietary intake relative to A4 or A8 (6).
A3, A4, and A8 influence the tissue destination of circulating cholesterol and TGs by interacting with 2 intravascular lipases: lipoprotein lipase (LPL) (7-9) and endothelial lipase (EL) (10, 11). LPL is synthesized predominantly in parenchymal cells of skeletal muscle, heart, and adipose tissue and is then translocated across the endothelium chaperoned by a glycosylphosphatidylinositol-linked protein, GPIHBP1 (12). GPIHBP1 serves as a scaffold for LPL on the vascular surfaces of endothelial cells (12) where it snags and hydrolyzes circulating TG-rich lipoproteins, thereby delivering fatty acids to adjacent tissues (13). A4 is the most potent inhibitor of LPL (9, 14), while A8 must form a complex with A3 to inhibit intravascular LPL (3, 15). Recently, we showed that hepatic A8 (with A3) acts in an endocrine fashion to inhibit LPL in oxidative tissues, whereas A8 expressed in adipose tissue enhances LPL activity by autocrine/paracrine inhibition of locally produced A4 (16).
Since its discovery, A8 has garnered interest as a potential target for therapeutic reduction of plasma TG levels and potentially improving cardiovascular outcomes (17-19). Findings in genetically-modified mice treated with antibodies that inactivate A8 also suggest that inhibition of A8 may reduce adipocyte mass in addition to lowering plasma TG levels (20). In support of a therapeutic role for A8 inhibition, individuals heterozygous for an inactivating mutation in ANGPTL8 (A8) have reduced levels of plasma TG and increased levels of high-density lipoprotein (HDL)-cholesterol (HDL-C) (21). To date, no individuals who are homozygous for inactivating mutations in A8 have been identified (3).
To examine the relationship between plasma A8 levels and various metabolic risk factors and disorders, including obesity (22–24), type 2 diabetes (23–27), fatty liver disease (28-30), and coronary atherosclerosis (27, 31), several enzyme-linked immunosorbent assays (ELISAs) have been established. However, most of the studies using these ELISAs focused on small, racially/ethnically homogeneous samples, and thus the contribution of A8 to racial/ethnic differences in energy substrate metabolism has not been fully investigated.
To glean further insights into the role of A8 in lipid and glucose homeostasis, we developed a robust, sensitive, and specific sandwich ELISA. We used this assay to measure circulating A8 levels in a large, multiracial/multiethnic population, the Dallas Heart Study (DHS). Here, we report the relationship between circulating levels of A8, genetic variations, and metabolic features in a population-based study.
Methods
Antibodies and Purified Proteins
A list of the names and sources for the reagents, instruments, software, and other materials used in the studies described in this paper is provided in Table 1. Recombinant full-length human A8 was cloned into pCold Trigger Factor to generate an A8 expression construct with a hexa-histidine tag at the C-terminus. The recombinant protein was expressed in bacteria and purified with Ni-NTA agarose. The purified protein was prepared using the Sigma Adjuvant System (50 µg) and injected into New Zealand Black (NZB) mice (n = 2) 7 times. Mouse anti-human A8 monoclonal antibody (mAb) was prepared by fusing SP2-mIL6 mouse myeloma cells with splenic B lymphocytes from the NZB mice. Supernatants from cultured hybridomas were screened by ELISA, and positive clones were confirmed by immunoblotting. Two hybridomas, designated IgG-34B4 (subclass 1, light chain κ) and the other IgG-42C1 (subclass 2a, κ), were subcloned by serial dilution 3 times and purified by gravity-flow affinity chromatography on Protein G Sepharose 4 Fast Flow columns.
Table 1.
Reagents, instruments, software, and other materials used in this study
| Reagents | Source | Catalog # |
|---|---|---|
| Antibodies | ||
| Mouse mAb anti-human A8 | This paper | 34B4 |
| Mouse mAb anti-human A8 | This paper | 42C1 |
| Mouse mAb anti-human A8 (Capture Ab) | Regeneron | H4H15347P-L2 |
| Mouse mAb anti-human A8 (Detection Ab) | Regeneron | H4H15318P-L4 |
| Rabbit pAb anti-fibronectin | Abcam | Ab2413 |
| Goat pAb anti-mouse IgG, light chain specific | Jackson ImmunoResearch | 115-035-174 |
| Chemicals and recombinant proteins | ||
| 3,3′,5,5′-Tetramethylbenzidine (TMB) Liquid Substrate System for ELISA | Sigma | T0440 |
| Alfa Aesar Laemmli SDS Sample Buffer, reducing, 6X | ThermoFisher Scientific | AAJ61337AC |
| BSA Fraction V | Roche | 10735086001 |
| Dulbecco’s Modified Eagle’s Medium (DMEM)–high glucose | Sigma-Aldrich | D6429 |
| Dulbecco’s Phosphate-Buffered Saline Solution 1X | Corning | MT21031CV |
| Fetal Calf Serum (FCS) | Sigma | F2442-500ML |
| FuGENE 6 | Promega | E2692 |
| GelCode Blue Safe Protein Stain | ThermoFisher Scientific | PI24596 |
| OmniPur 10X PBS Liquid Concentrate | Sigma-Aldrich | 6505-OP |
| Penicillin-Streptomycin Solution, 100x | Corning | 30-002-CI |
| Recombinant human A3 | Regeneron | Not applicable |
| Recombinant human A4 | Regeneron | Not applicable |
| Recombinant human A8 | GenScript | U9125BC070801/ P30011604 |
| Horseradish Peroxidase-conjugated Streptavidin (HRP) | Jackson ImmunoResearch | 016-030-084 |
| Sulfuric Acid | Ward’s Science | 470302–858 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | 34577 |
| Trypsin EDTA 1X | Corning | 25-052-CI |
| TWEEN 20 | Sigma | P1379 |
| Cell lines | ||
| Chinese Hamster Ovary (CHO) cells | Not applicable | Not applicable |
| Recombinant DNA | ||
| pCold TF (Trigger Factor) | Addgene | 69741–3 |
| Commercial assay kits | ||
| ANGPTL3 (human) ELISA kit | Adipogen | AG-45A- 0014YEK-KI01 |
| Betatrophin Human ELISA | BioVendor | RD191347200R |
| Betatrophin (139–198) (Human)–EIA Kit | Phoenix | EK-051-55 |
| Human ANGPTL8 ELISA Kit | Cloud-Clone Corp. | SEW803Hu |
| Betatrophin (Human) ELISA Kit | Aviscera | SK00528-06 |
| Human ANGPTL8 Assay Kit | IBL | 27795 |
| Quick Start Bradford Protein Assay kit 2 | BioRad | 5000202EDU |
| Instruments | ||
| 405 TS Microplate Washer | Agilent | 405TSRVSQ |
| Biacore T200 | GE Healthcare | 28979415AE |
| BioTek Epoch 2 Microplate Spectrophotometer | Agilent | UEPOCH2T |
| LI-COR Odyssey Fc | LI-COR Biosciences | 2801-0X |
| Compact Digital MicroPlate Shaker | Thermo Scientific | 88880023 |
| Software | ||
| Image Studio Lite v5.2.5 | LI-COR | Not applicable |
| LocusZoom | http://locuszoom.org (32) | Not applicable |
| PLINK version 1.9b | www.cog-genomics. org/plink/1.9/ (33) | Not applicable |
| Prism 8 version 8.1.1 | GraphPad | Not applicable |
| R statistical package | https://www.R-project.org/ (34) | Not applicable |
| Other | ||
| 2mL Deep Well Plates | Thomas Scientific | 1148Z37 |
| 96-well Clear Polystyrene High Bind Stripwell™ Microplate | Corning | CLS2592-100EA |
| Adhesive Seal Applicator | ThermoFisher Scientific | AB1391 |
| Amersham Protran 0.45 NC supported Western blotting membranes | Cytiva | 10600016 |
| Blue X-Ray films | Phenix Research | F-BX810 |
| Protein G Sepharose 4 Fast Flow | GE Healthcare | 17061805 |
| SealPlate film | Sigma | Z369659 |
| Sigma Adjuvant System | Millipore Sigma | S6322 |
Recombinant hA4 (G26-D148).mFc and hA3 (S17-E460).mmh proteins were cloned into vectors containing an optimized nonnative signal peptide and expressed in Chinese hamster ovary (CHO) cells that were cultured in DMEM-high glucose with 10% FCS and 1% penicillin-streptomycin. Proteins were purified using protein A (hA4.mFc) or immobilized by metal affinity chromatography (hA3.mmh) and dialyzed into phosphate-buffered saline (PBS) containing 5% (v/v) glycerol, pH 7.4.
Subjects
Plasma samples were obtained from participants of the Dallas Heart Study (DHS), a multiracial/multiethnic, population-based probability sample (n = 3557) of Dallas County residents that was collected between 2000 and 2002 (DHS-1) (35). A subset of the DHS-1 participants (n = 2485) were re-sampled between 2007 and 2009 (DHS-2). DHS-2 also included 916 spouses of the DHS-1 participants and other unrelated individuals. The current study included all DHS-1 (and DHS-2) participants who provided blood samples for analysis and in whom A8 was measured. Race/ethnicity was self-reported. DHS-1 included 1828 (51.7%) (DHS-2, 1654 [50.4%]) non-Hispanic Blacks (henceforth “Blacks”), 1038 (29.3%) (DHS-2, 1072 [32.7%]) non-Hispanic Whites (“Whites”), 598 (16.9%) (DHS-2, 466 [14.2%]) Hispanics and 74/2.1% (DHS-2, 91 [2.8%]) individuals of other races/ethnicities. In total among these, 2131 participants had A8 measured in both DHS-1 and DHS-2. Venous blood was obtained from each participant in citrate-EDTA tubes after a 12-hour fast and maintained at 4 °C until plasma was isolated by centrifugation. Plasma was divided into aliquots (100 µL each) in barcoded microfuge tubes that were stored at −80 °C. All participants provided written informed consent. The study protocol was approved by the Institutional Review Board of University of Texas Southwestern Medical Center.
Anti-Human A8 ELISA
Both the Capture and Detection antibodies used in the ELISA were fully human mAbs (IgG4) against human A8 developed using Regeneron’s VelocImmune technology platform (36, 37). The specificity of the A8 ELISA was evaluated by assaying purified A8, A3, A4, and A8 combined with A3. The recombinant purified A8, A3, and A4 proteins were thawed on ice and diluted to a concentration of 2 µg/mL in 400 µL of PBS. Standard curves of A8, A4, and A3 alone and A8 in the presence of A3 (0.88nM or 43.72 ng/mL) were serially diluted 9 times by progressively adding 200 μL to wells containing 200 μL of PBS. In the final dilution, all samples were diluted 1:20 in PBS to generate an 11-point calibration curve. Duplicates of the samples (100 μL each) were added to each plate.
Purified full-length recombinant A8 was prepared according to the specifications of the manufacturer and was used as the standard for the ELISA. The protein was analyzed for purity by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with Coomassie blue staining. Bands were quantified by densitometry, and the protein was estimated to be >95% pure. A8 protein concentration was determined to be 0.06 mg/mL using the Bradford Assay and by comparing the intensity of the Coomassie blue-stained bands to that of serially diluted bovine serum albumin (BSA) after SDS-PAGE (Fig. S1A) (38).
To measure plasma levels of A8, the Capture mAb was diluted in PBS to a concentration of 4 μg/mL, and then 100 µL was added to wells of 96-well flat-bottomed microtiter plates. Plates were sealed and maintained at 4 °C overnight. The following morning, 250 µL of PBST (PBS + 0.1% Tween20) was added to each well using a 405 TS Microplate Washer. The plates were shaken for 10 seconds at 500 rpm and then the PBST was aspirated. The coated plates were washed 3 times prior to addition of 300 µL of Blocking Buffer (PBS + 1% BSA Fraction V). The plates were shaken for 1 hour at room temperature and then washed 3 times in PBST.
An aliquot of A8 standard was thawed on ice and diluted to a concentration of 0.375 µg/mL in 400 µL of Sample Buffer (PBS + 1% BSA Fraction V + 0.1% Tween20). The standard was serially diluted 7 times by progressively adding 200 μL to wells containing 100 μL of Sample Buffer. In the final dilution, all samples were diluted 1:20 in Sample Buffer to generate a 9-point calibration curve. Duplicates of the calibration curve (100 μL/well) were added to each plate. Both DHS plasma samples (10 μL) and 2 control samples diluted 1:10 in Sample Buffer were added in duplicate to each plate (100 μL/well). Four wells contained Sample Buffer alone. Plates were shaken at room temperature for 2 hours and then washed 3 times with PBST as described above. To detect bound A8, wells were incubated with biotinylated Detection Antibody (Ab) diluted to 0.4 µg/mL in Sample Buffer (150 μL/well) and shaken for 1 hour at room temperature. The plates were washed again and incubated for 30 minutes with horseradish peroxidase enzyme diluted 1:50 000 in Sample Buffer (150 μL/well). After a final wash, wells were incubated with 3,3′,5,5′-tetramethylbenzidine (TMB) detection substrate (100 μL/well) for 4 minutes, and the reaction was terminated by adding 1.0 M sulfuric acid (100 µL/well). Plates were shaken for 3 seconds to ensure homogenous mixing. The absorbances at 450 nm and 540 nm wavelengths were measured using an Epoch 2 Microplate Spectrophotometer. To calculate A8 concentrations, the mean value obtained from 2 wells containing Sample Buffer alone was subtracted from the 450 nm absorbance value minus the 540 nm absorbance value. The mean values of the serially diluted purified protein duplicates were used to generate standard curves, which were plotted using a quadratic fit (Fig. S1B) (38). Plasma A8 concentrations were interpolated from the standard curves using Prism 8 version 8.1.1. Values were multiplied by 10 to obtain the plasma concentration of A8. Samples (n = 6) with values exceeding the range of the standard curve were diluted 1:20 and remeasured. To minimize plate-to-plate variation, measurements were normalized by multiplying each individual A8 value by a plate-specific scaling factor, calculated as the ratio of the median value of the control samples across all plates by the median value of the control samples for each plate.
To determine the intra- and interassay variation, 3 plasma samples with high (30.8 ng/μL), medium (15.9 ng/μL), and low (5.0 ng/μL) A8 levels were measured 72 times across 3 plates. For intra-assay variation, coefficients of variability (CV) for each duplicate were calculated using the formula:
Intra-assay variation was determined by taking the median of all duplicate CVs on each plate to account for skewness of low concentration samples. The value reported in Table 2 is the mean of the 3 plate-specific median CVs. The interassay variation was calculated by taking the mean of high, medium, and low concentration values for each plate separately and then using the formula:
Table 2.
Performance of plasma ANGPTL8 ELISA
| Parameter | Characteristics |
|---|---|
| Detection range | 2.1-187.5 ng/mL |
| Calibration curve, R2 | >0.99 |
| Intra-assay variation (% CV) | 5.3% |
| Interassay variation (% CV) | 6.0% |
| Recovery (%) | 74.4%-90.5% |
| Linearity (%) | 91.9%-132.5% |
Formulas used to calculate intra- and interassay coefficients of variations (CV), recovery, and linearity are provided in the “Methods” and in the legends to Tables S1 and S2 (39).
The value reported in Table 2 is the mean of the 3 CVs from each respective concentration group.
A spike-recovery test was performed to rule out any interference from components in human plasma that could affect the measurement of A8 levels. Spikes with known concentrations of purified A8 (117 ng/mL, 77 ng/mL, and 37 ng/mL) were added to samples of human plasma (n = 6) diluted 1:10 or to Sample Buffer alone. The mean values of spiked plasma samples across 3 plates were compared to those obtained from samples in Sample Buffer alone (Table S1) (38). The percent recovery was calculated using the formula:
A range of recoveries was generated from the respective spike concentrations (high, medium, and low) in multiple plasma samples.
The linearity test was performed to validate the precision of the assay across a wider range of A8 levels. Human plasma with a known concentration of A8 was serially diluted 1:2 in Sample Buffer 5 times. The mean values obtained on 3 plates were compared to the expected values, with the expected being 50% of the previously diluted concentration value. The percent linearity was calculated with the formula:
A range of linearities was generated from different dilution steps 1:4 to 1:64 (Table S2) (38).
To assess intra-individual variation of A8, plasma was obtained on 4 occasions (day 0, week 2, week 5, and week 6) from 48 fasted, unrelated individuals (40) (Fig. S1C) (38). The intra-class coefficient (ICC), a measure of similarity between measurements taken in a single individual, was calculated with the formula (41):
The lower limit of detection for the assay was determined by following the Clinical and Laboratory Standards Institute (CLSI) guidelines (39) using 144 repeated measurements of a plasma sample with low A8 concentration (1:10 dilution in Sample Buffer) and of Sample Buffer alone (blank). The limit of blank (LoB) was calculated using the formula: LoB = mean of the blank measurements + 1.645*(standard deviation of the blank measurements). Using the LoB value, the limit of detection (LoD) was calculated as follows: LoD = LoB + 1.645*(standard deviation of sample measurements). The upper limit of detection was determined by taking the highest theoretical concentration on the calibration curve and multiplying the value by 10 to account for the 1:10 plasma dilution.
Immunoblot Analysis
Venous blood was collected from study subjects in citrate-EDTA tubes after a 12-hour fast and collected again 4 hours after a standard meal. Plasma was isolated by centrifugation at 2000g for 10 minutes at 4 °C and stored at −80 °C. Prior to use, plasma samples from all subjects were thawed on ice, diluted 8-fold in 0.9% NaCl and Laemmli SDS reducing sample buffer, and then incubated at 95 °C for 4 minutes. Diluted plasma was size-fractionated by SDS-PAGE (12%) and transferred to a nitrocellulose membrane. Membranes were blocked in PBST with 5% fat-free milk for 60 minutes at room temperature before addition of primary antibodies diluted in PBST with 5% fat-free milk.
After overnight incubation with the primary Ab at 4 °C, membranes were washed 5 times for 5 minutes in PBST and incubated with secondary antibody in PBST with 1.25% fat-free milk (1:10 000 dilution). Following a 60-minute incubation at room temperature, membranes were washed again 5 times with PBST and visualized using SuperSignal-enhanced chemiluminescence substrate. Gel images were captured on Blue X-Ray films and LI-COR Odyssey Fc, and band intensities were analyzed using LI-COR Image Studio Lite version 5.2.5.
Exome-Wide Association Study
Genomic DNA extracted from circulating leukocytes of DHS participants was used to perform whole-exome sequencing at the Regeneron Genetics Center (Tarrytown, New York) as described (42, 43). After excluding variants that failed our quality control criteria (missing rate of > 5%, deviated from Hardy-Weinberg equilibrium with a P < 1.0 × 10−6, were outside of the capture region or had a minor allele count < 5), a total of 346 167 bi-allelic variants were tested for association with circulating A8 levels. Our primary analysis was performed in DHS-1 participants with whole-exome sequencing, plasma A8 levels and covariate data available (n = 3457). Plasma A8 levels were transformed using a rank-based, inverse normal transformation and tested for association with sequence variants using linear regression with adjustment for age, sex, 4 principal components of genetic ancestry, and body mass index (BMI), using PLINK version 1.9b (33). The reported P values were calculated assuming an additive genetic model.
Statistical Methods
Baseline characteristics were compared between racial/ethnic groups using Kruskal-Wallis test (continuous traits) and Fisher’s exact test (categorical variables). The correlations between A8 levels and continuous phenotypes were assessed using Spearman’s rank correlation coefficient. Partial correlations were computed after adjusting both traits for age, sex, self-reported race/ethnicity and BMI, and correlating the residuals. Independent predictors of A8 levels were assessed using a multiple linear regression model adjusted for age, sex and self-reported race/ethnicity. The association of A8 levels with type 2 diabetes was tested using logistic regression, adjusted for age, sex, self-reported race/ethnicity and BMI. The association of rs2278426 A8(R59W) genotype with participant characteristics was assessed using linear and logistic regression models, adjusted for age, sex, 4 principal components of ancestry, and BMI (where appropriate). Partial R2 values, measuring the independent contribution of individual variables to explain variability in A8 levels, were calculated by comparing the residual variation in nested models including and excluding a particular variable, after adjustment for other covariates. All statistical analyses were performed with the R statistical language (34).
Results
Development of a Specific and Sensitive Assay to Measure Plasma A8 Levels
First, we compared the relative levels of A8 detected in the plasma using 3 different commercial assays with the results obtained using an immunoblotting assay previously established in our laboratory (3) (Fig. S2) (38). The relative values obtained using the commercial A8 ELISAs correlated poorly with each other and with results obtained using immunoblotting (r = 0.04, 0.09, and 0.02). Moreover, none showed a consistent increase in circulating A8 levels after food intake (3). Therefore, we established a new sandwich ELISA to measure plasma A8 levels.
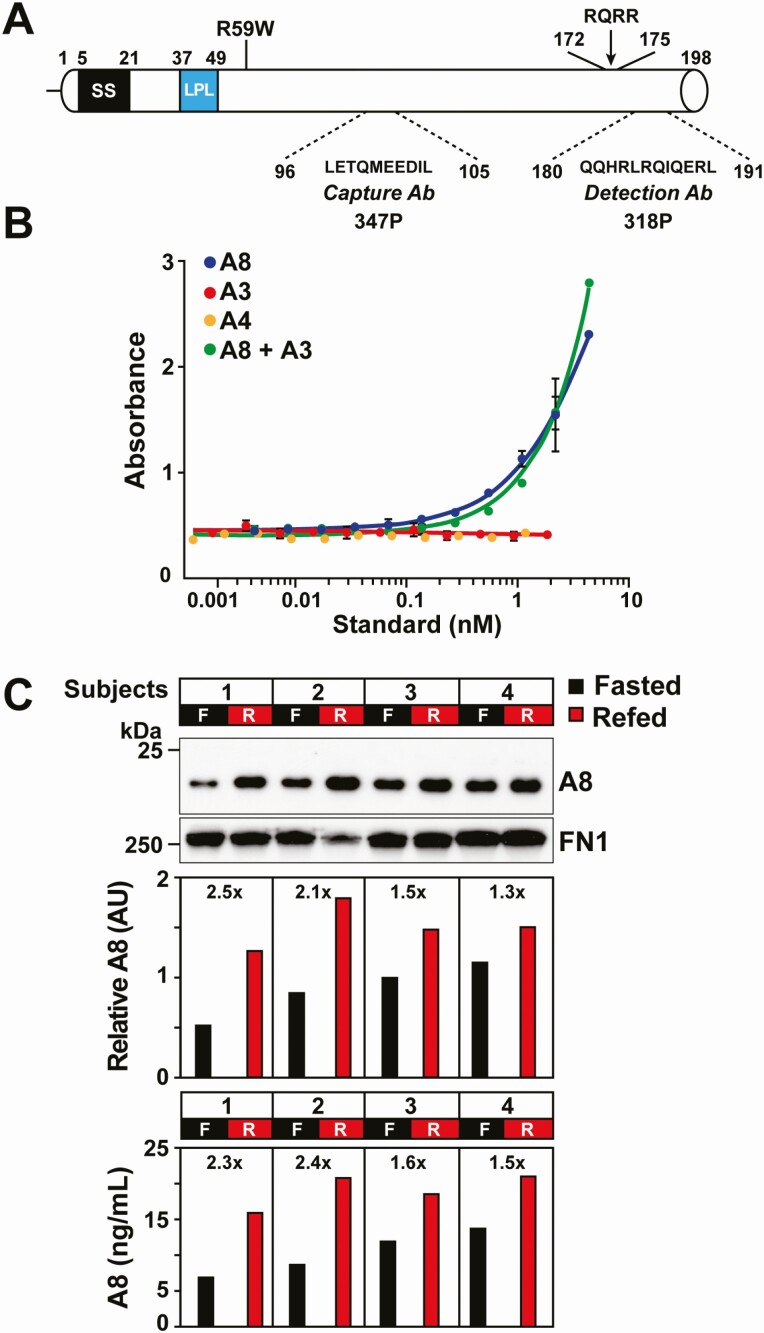
A series of fully humanized anti-human A8 mAb (IgG4) were developed using the VelocImmune technology platform (36, 37). Affinities of the mAbs were tested and compared using a Biacore assay (44), and the Abs with greatest affinities and highest specificities for human A8 were used as Capture and Detection Abs in the ELISA (Fig. 1A). The Capture Ab recognizes a 10-residue motif in the N-terminal domain (LETQMEEDIL), and the Detection Ab recognizes a 12-residue sequence (QQHRLRQIQERL) located 7 residues from the C-terminus of A8.
Figure 1.
Schematic of A8 with locations and specificities of the anti-A8 mAbs used in the ELISA and assay validation. A, A8 is a 198-residue protein that contains at its N-terminus a signal sequence (residues 5-21) and a coiled-coil domain with a lipoprotein lipase (LPL) binding motif. The epitopes for the Capture and Detection mAbs, the putative furin cleavage site, and the A8(R59W) variant are shown. B, The A8 ELISA is specific for human A8 and does not detect human A3 or A4. To evaluate the ELISA specificity, increasing concentrations of human A8, A4, A3, and A8 in the presence of A3 were measured by ELISA as described in the “Methods.” C, Comparison of plasma levels of human A8 obtained by immunoblotting or by ELlSA. Plasma samples were immunoblotted with a mouse A8 mAb (34B4) that recognizes the N-terminal domain. Fibronectin (FN1) was used as loading control. The relative intensity of bands was determined by LI-COR (middle panel) and the A8 levels quantified by ELISA (lower panel). Abbreviations: AU, arbitrary units; F, fasted state; R, refed state.
A8 is an ortholog of A3 (3) and also shares sequence identity with A4 (15). To ensure that the A8 ELISA did not cross-react with A3 or A4, we tested for interference by these ANGPTL family members (Fig. 1B). Addition of human A3 did not interfere with detection of A8. Moreover, A3 and A4 were not detected by the A8 ELISA.
Calibration curves were constructed using purified recombinant human A8 as a standard (0.06 mg/mL) (Fig. S1A) (38). The relationship between levels of A8 and absorbance by the spectrophotometer was best fit using a quadratic curve (R2 ≥ 0.99) (Fig. S1B) (38). Table 2 provides an overview of the assay characteristics. The lower limit of reliable detection for the assay (ie, the lowest plasma concentration that was consistently and accurately greater than zero) was 2.1 ng/mL, and the upper limit was 187.5 ng/mL. Samples with A8 levels greater than 187.5 ng/mL were remeasured at a higher dilution factor (1:20). The intra- and interassay variations were 5.3% and 6.0%, respectively. The levels of A8 obtained using the ELISA were strongly correlated (r = 0.906) with those obtained by immunoblot analysis using a mAb to the N-terminal domain that was developed in-house as described in the “Methods.” The signal intensity of the bands was quantitated using LI-COR Odyssey (Fig. 1C). The intra-individual variation in plasma A8 levels was determined in 48 unrelated fasting individuals during 4 separate visits over a 6-week period (40). The ICC for A8 was 0.69 (95% CI, 0.57-0.79) (Fig. S1C) (38), as calculated in the “Methods.” By comparison, the ICC for low-density lipoprotein-cholesterol (LDL-C) levels in the same samples was 0.65 (95% CI, 0.53-0.77).
Correlation of Circulating A8 Levels With Demographic and Cardiometabolic Variables
We measured circulating A8 levels in DHS, a multiracial/multiethnic, population-based probability sample of Dallas County (35). Measurements were obtained on 3538 participants collected in 2000 to 2002 (DHS-1) and on 3283 participants collected in 2007 to 2009 (DHS-2), including 2131 participants with paired measurements in DHS-1 and DHS-2. Baseline characteristics of the cohorts are summarized in Table 3 (DHS-1) and Table S3 (38) (DHS-2). Several characteristics differed significantly among racial/ethnic groups, as has been reviewed previously (35). In this study, we used data from DHS-1 for our primary analysis. Results from DHS-2 were similar and were used to confirm trends identified in the DHS-1 cohort.
Table 3.
Characteristics of Dallas Heart Study-1 participants
| Characteristic | N | Total | N | Blacks | Whites | Hispanics | P value |
|---|---|---|---|---|---|---|---|
| N (%) | 3538 (100%) | 1828 (51.7%) | 1038 (29.3%) | 598 (16.9%) | - | ||
| Age, years | 3538 | 43.9 ± 10.1 | 3464 | 44.8 ± 10.2 | 44.8 ± 10 | 40.2 ± 9.2 | 1.5E-24 |
| Female, N (%) | 1975 (55.8%) | 1057 (57.8%) | 541 (52.1%) | 347 (58%) | 0.0077 | ||
| BMI (kg/m2) | 3531 | 30.6 ± 7.5 | 3458 | 31.7 ± 8.2 | 29 ± 6.7 | 30.3 ± 6.5 | 1.5E-18 |
| Systolic BP (mmHg) | 3534 | 125 ± 19 | 3461 | 130.1 ± 20 | 119.9 ± 15.3 | 118.7 ± 16.9 | 3.1E-62 |
| Diastolic BP (mmHg) | 3534 | 78.3 ± 10.3 | 3461 | 81 ± 10.6 | 75.9 ± 8.8 | 74.7 ± 9.2 | 7.0E-54 |
| Hypertension, N (%) | 1070 (30.3%) | 758 (41.5%) | 208 (20.1%) | 89 (14.9%) | 1.1E-51 | ||
| Glucose (mg/dL) | 3537 | 92 (85–102) | 3463 | 92 (84–104) | 92 (84–99) | 96 (88–104) | 3.6E-08 |
| Insulin (mIU/L) | 3059 | 12.6 (7.4–20.8) | 2997 | 13.9 (8.2–22.5) | 10.2 (6.2–17.4) | 14 (8.1–21.1) | 3.1E-18 |
| HOMA-IR (mass units) | 2976 | 3 (1.6–5.1) | 2914 | 3.3 (1.8–5.7) | 2.3 (1.4–4.1) | 3.4 (1.8–5.6) | 1.0E-19 |
| Diabetic, N (%) | 408 (11.5%) | 262 (14.3%) | 68 (6.6%) | 71 (11.9%) | 6.6E-10 | ||
| GFR (mL/min/1.73m2) | 3537 | 100.7 ± 24 | 3463 | 103.5 ± 25.8 | 92.2 ± 18.2 | 107.6 ± 23.7 | 4.1E-60 |
| Total cholesterol (mg/dL) | 3537 | 180.3 ± 39.6 | 3463 | 177.9 ± 40.2 | 183.4 ± 38.3 | 182.2 ± 40 | 7.6E-05 |
| LDL-C (mg/dL) | 3536 | 106.3 ± 35.4 | 3462 | 104.9 ± 36.6 | 108.2 ± 34.4 | 107.2 ± 33.2 | 0.010 |
| HDL-C (mg/dL) | 3537 | 49.8 ± 14.7 | 3463 | 52.2 ± 15.3 | 48.3 ± 14.9 | 45.7 ± 11.2 | 3.3E-25 |
| Triglycerides (mg/dL) | 3537 | 96 (67–147) | 3463 | 85 (62–123) | 109 (75–167) | 119 (81–176) | 4.0E-47 |
| A8 (ng/mL) | 3538 | 13.3 (6.8–23.1) | 3464 | 15.8 (8.6–27.1) | 9.6 (4.5–16.5) | 13.5 (7.4–23.1) | 4.6E-49 |
| Men | 1563 | 12.5 (6.3–22.5) | 1519 | 15 (7.7–26.7) | 9.6 (4.3–16.4) | 13.6 (7.6–21.9) | 3.5E-18 |
| Women | 1975 | 13.9 (7.2–23.6) | 1945 | 16.4 (9.5–27.6) | 9.5 (4.9–16.5) | 13.4 (7.4–24.7) | 3.8E-31 |
Data are shown as N (%), mean ± SD, or median (25th-75th percentile). Abbreviations: A8, ANGPTL8; BMI, body mass index; BP, blood pressure; GFR, glomerular filtration rate; HbA1c, glycated hemoglobin A1C; HDL-C, high-density lipoprotein-cholesterol; HOMA-IR, homeostatic model assessment of insulin resistance; LDL-C, low-density lipoprotein-cholesterol.
Next, we examined the relationship between plasma A8 levels and other metabolic and cardiovascular parameters (Table S4) (38). A8 levels were associated with BMI (r = 0.254, P = 4.2 × 10−46), as reported previously (27, 45, 46). The association with total fat mass (r = 0.224, P = 5.3 × 10−35) was greater than with lean body mass (r = 0.085, P = 3.3 × 10−6), as determined using dual-energy x-ray absorptiometry. Correlations were also observed between plasma A8 levels and fat depots, including total abdominal (r = 0.253, P = 1.7 × 10−41) and visceral fat (r = 0.199, P = 8.3 × 10−26) that were measured by magnetic resonance imaging (Table S4) (38).
Circulating A8 levels correlated with fasting plasma levels of insulin (ρ = 0.289, P = 4.3 × 10−60), glucose (ρ = 0.101, P = 1.9 × 10−9) and Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) (ρ = 0.290, P = 7.5 × 10−59) which provides an index of insulin sensitivity (47) (Table S4) (38). These relationships were independent of age, sex, race/ethnicity and BMI (Table S5) (38). A8 levels were also positively associated with liver fat content (ρ = 0.235, P = 6.9 × 10−30) and levels of serum liver enzymes. Circulating A8 levels were also positively correlated with plasma levels of TG (ρ = 0.349, P = 6.5 × 10−102) but not with plasma LDL-C levels (ρ = 0.016, P = 0.35). Correlations between plasma A8 levels and other phenotypes assayed in the DHS are provided in Table S4 (38).
Distribution of Plasma A8 Levels in Fasting Plasma Samples From a Population-Based Study
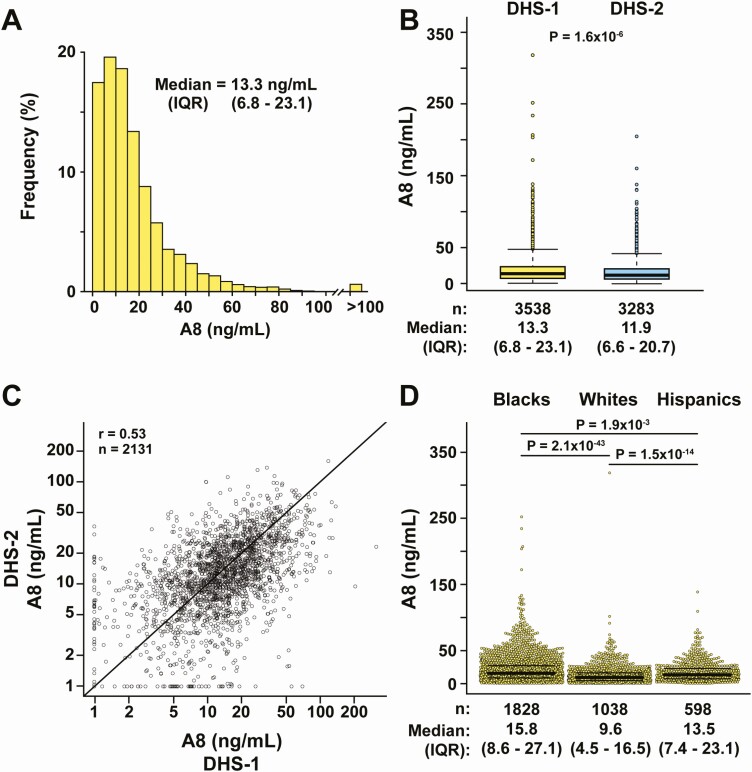
The distribution of plasma A8 levels in DHS-1 (Fig. 2A) was highly skewed to the right, with a median level of 13.3 ng/mL (interquartile range [IQR] = 6.8-23.1 ng/mL). A similar distribution of A8 levels was seen in DHS-2 (Fig. S3A) (38), although the median level was slightly lower at 11.9 ng/mL (IQR = 6.6–20.7 ng/mL) (P = 1.6 × 10−6) (Fig. 2B). Among the 2131 participants with measurements in both DHS-1 and DHS-2, the correlation coefficient was r = 0.53 (Fig. 2C). For comparison, the correlation coefficient between the two studies for other plasma analytes, such as plasma LDL-C levels and plasma TG levels, were 0.47 and 0.60, respectively.
Figure 2.
Fasting plasma A8 levels in DHS-1 and DHS-2. A, Frequency distribution of plasma A8 levels in DHS-1. The median and interquartile range (IQR, 25th–75th percentiles) values are shown. B, Distribution of plasma A8 levels in DHS-1 (yellow, n = 3538) and DHS-2 (blue, n = 3192). Median levels are denoted by a bold horizontal line, and the upper and lower boundaries of the colored boxes denote the IQR. The whiskers extend to the minimum and maximum observations that are within 1.5× the IQR from the box; more extreme observations are plotted as individual dots. C, Correlation between plasma A8 levels in DHS-1 and DHS-2 on log10 scale. The Pearson correlation coefficient is shown. D, Dot plots of plasma A8 levels stratified by race/ethnicity showing median, IQR, and P values.
The A8 levels were similar in men and women (P = 0.71 after accounting for differences in age, race/ethnicity, and BMI) (Table 3). Plasma A8 levels varied significantly among the 3 racial/ethnic groups (Fig. 2D, Fig. S3 (38), and Table 3). Blacks had significantly higher median (IQR) plasma levels of A8 (15.8 [8.6-27.1] ng/mL) than either Hispanics (13.5 [7.4-23.1], P = 0.0019) or Whites (9.6 [4.5-16.5], P = 2.1 × 10−43). The racial/ethnic differences in plasma A8 levels remained significant after adjusting for age, sex, BMI, insulin, plasma TG, and HDL-cholesterol levels (Table 4 and Table S5 (38)). In a multivariable regression model, the variable list in Table 4, when taken together, explained 20.1% of the interindividual variation in A8 levels, of which race/ethnicity accounted for 7.3%.
Table 4.
Predictors of plasma ANGPTL8 levels in DHS-1 in a multivariable regression model (n = 3057)
| Variable | Beta (SE) | P value | Partial R2 (%) |
|---|---|---|---|
| Age (years) | 0.002 (0.012) | 0.19 | 0.06 |
| Female sex | 0.111 (0.587) | 0.0013 | 0.34 |
| Race/ethnicity | 7.30 | ||
| Whites | Ref | - | - |
| Blacks | 0.587 (0.314) | 1.3E-51 | - |
| Hispanics | 0.314 (0.484) | 3.1E-10 | - |
| Other | 0.484 (0.002) | 4.1E-05 | - |
| BMI (kg/m2) | 0.012 (0.011) | 5.9E-07 | 0.82 |
| Insulin (IU) | 0.011 (0.002) | 3.7E-17 | 2.30 |
| TG (mg/dL) | 0.002 (-0.003) | 1.3E-43 | 6.10 |
| HDL-C (mg/dL) | -0.003 (0.001) | 0.012 | 0.21 |
Multiple R-squared (adjusted): 20.1%. Abbreviations: BMI, body mass index; HDL-C, high-density lipoprotein-cholesterol; TG, triglycerides.
We also examined and compared racial/ethnic differences in relative frequency of individuals with A8 levels that fell below the detection limit of the assay, determined according to the Clinical and Laboratory Standards Institute guidelines as described in the “Methods.” Whites had the highest proportion of subjects with A8 levels below the detection limit of the assay (12.2%), compared with both Hispanics (6.0%, P = 3.5 × 10−5) and Blacks (4.3%, P = 1.5 × 10−14).
Immunoblot Analysis of A8 in Individuals With No A8 Detected by ELISA
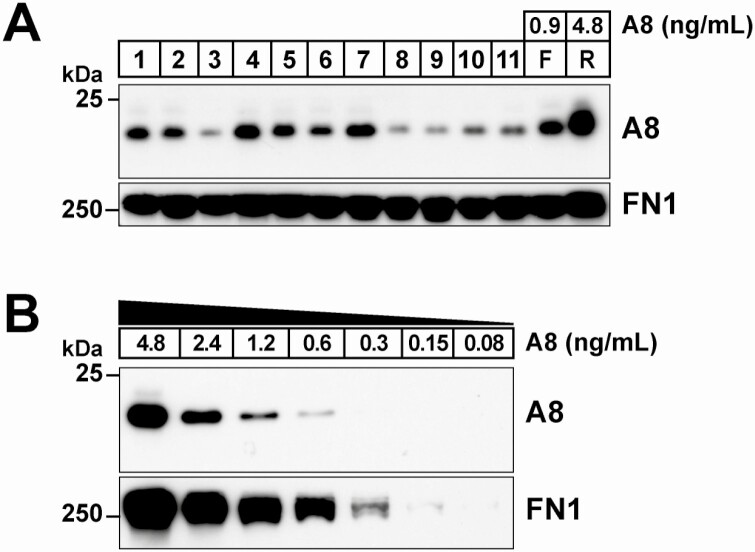
No individuals have yet been described who are homozygous for a loss-of-function mutation in A8. To attempt to identify such an individual and assess the associated phenotype, we focused on all those individuals whose plasma yielded no detectable signal in duplicate ELISA measurements (n = 69 in DHS-1, n = 97 in DHS-2). These subjects comprised 31.6% of those with levels below <2.1 ng/mL. Immunoblot analysis of plasma from 11 of the 166 tested is shown in Fig. 3A. The detection limit of the A8 immunoblotting assay was estimated to be 0.6 ng/mL by analyzing serial dilutions of plasma of known concentration (Fig. 3B). In all 166 subjects, A8 was readily detectable by immunoblotting. Thus, the immunoblot is more sensitive than the ELISA for detecting A8, and no A8 null individuals were identified in this study.
Figure 3.
Immunoblot analysis of plasma from subjects with no detectable A8 in duplicate ELISA measurements. A, Immunoblot analysis of A8 in plasma from a subset of DHS-1 subjects that had no detectable A8 in duplicate assays. Plasma samples were size-fractionated on a 12% agarose gel, and immunoblotting was performed as described in the “Methods” using an anti-A8 mAb (34B4). Fibronectin (FN1) was used as loading control. F and R represent plasma from an individual fasted for 12 hours (fasted) and 4 hours after eating a standard meal (refed). The A8 concentrations (0.9 and 4.8 ng/mL) were calculated from the ELISA values accounting for the dilution factor (1:8) of the immunoblotted sample. B, Plasma from the refed control was serially diluted (1:2) and immunoblotted. The lowest detectable level by immunoblotting was 0.6 ng/mL.
Genetic Variation Associated With Circulating A8 Levels
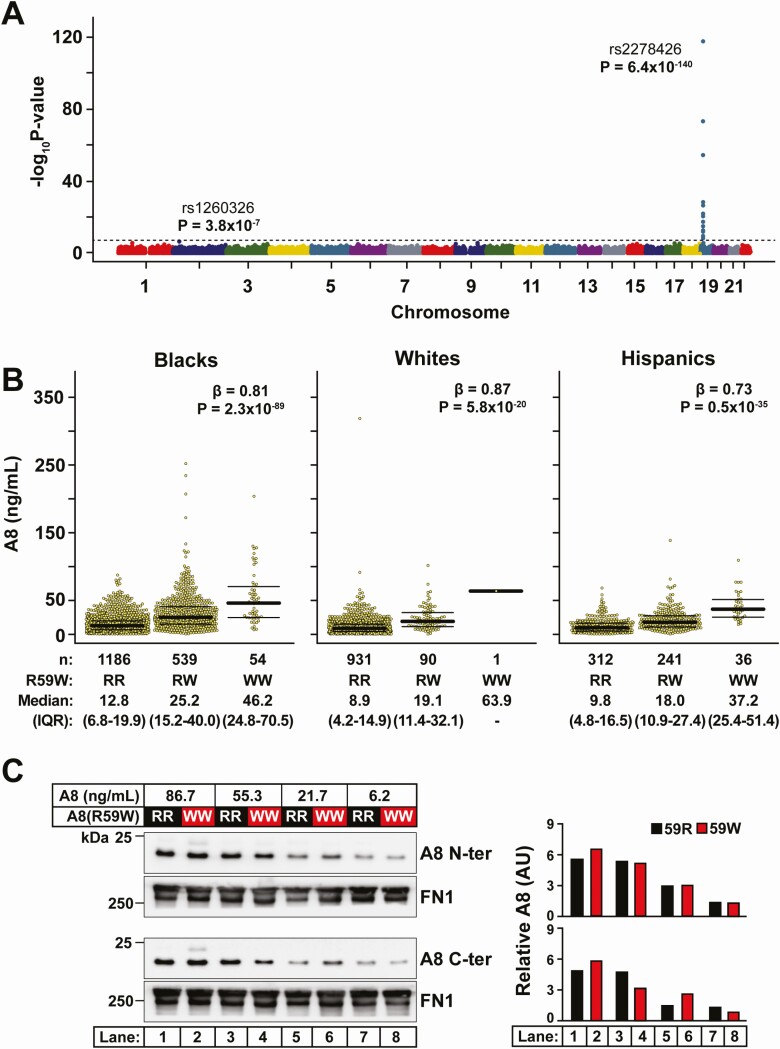
To identify DNA sequence variants associated with plasma levels of A8, we performed an exome-wide association study using whole-exome sequence data in DHS-1. The only locus that exceeded the exome-wide significance threshold (P = 1.44 × 10−7) was a region on chromosome 19 containing the ANGPTL8/DOCK6 locus (Fig. 4A). The lead variant at this locus was rs2278426 (P = 6.4 × 10−140). This variant (c.175C>T) results in substitution of arginine to tryptophan at position 59 (R59W) in A8 (Fig. 1A). We reported previously that this variant is associated with lower plasma levels of LDL-C and HDL-C among both Hispanics and Blacks in DHS-1 (3). A total of 18 other variants at the ANGPTL8/DOCK6 locus also reached exome-wide significance; none of these remained independently associated with A8 levels after conditioning on rs2278426 (Fig. S4) (38). Conditional analysis revealed 3 other variants that were relatively rare (<30 carriers; MAF = 0.7% in Blacks and absent in Whites) and were not in the coding region of ANGPTL8 (Table S6 and Fig. S4 lower panel) (38). As previously reported (3), the frequency of the A8(R59W) variant was highest in Hispanics (26.2%), intermediate in Blacks (17.5%) and lowest in Whites (4.5%).
Figure 4.
Identification of sequence variants associated with A8 levels. A, Manhattan plot of 346 167 biallelic sequence variants were tested for association with circulating A8 levels in DHS-1 (n = 3457). The dashed line denotes the Bonferroni-corrected significance threshold. B, Plasma A8 levels by rs2278426 genotype in Blacks (n = 1779), Whites (n = 1022), and Hispanics (n = 589) in DHS-1 with median and IQR values are provided. Association was tested using linear regression with adjustment for age, sex, ancestry, and BMI. The genotypes (RR, RW, and WW) are indicated. C, Immunoblot analysis of A8 in plasma from individuals homozygous for A8(59R) or A8(59W) who were matched by A8 levels. Plasma samples were immunoblotted with anti-A8 mAb raised against the N-terminal domain (34B4) (top) and the C-terminal domain (Detection mAb 318P in Fig. 1A) (bottom). Fibronectin (FN1) was used as loading control. Band intensities were determined by LI-COR and quantified relative to fibronectin (FN1) levels (right panel).
Next, we examined the relationship between A8(59W) and plasma levels of A8 in subjects stratified by race/ethnicity. Overall, median plasma level of A8 was more than 4-fold higher in homozygotes for the R59W variant than among those who were homozygous for the common allele (median [IQR]: 41.7 [25.3-61.5] vs 10.8 [5.5-17.7] ng/mL) (Table 5). Heterozygotes had intermediate levels of A8 (22.5 [12.9-36.2] ng/mL). We observed that the minor T-allele was associated with increased plasma A8 levels in all 3 racial/ethnic groups (Blacks: β = 0.81, P = 2.3 × 10−89; Whites: β = 0.87, P = 5.8 × 10−20; and Hispanics: β = 0.73, P = 0.5 × 10−35) (Fig. 4B and Fig. S5 (38)).
Table 5.
Characteristics of Dallas Heart Study-1 participants by ANGPTL8 (R59W) genotype
| ANGPTL8 rs2278426 genotype | ||||||
|---|---|---|---|---|---|---|
| Characteristic | N | CC | CT | TT | P value unadjusted | P value adjusted |
| N (%) | 3464 | 2479 (71.6%) | 893 (25.8%) | 92 (2.7%) | - | - |
| Age (years) | 3464 | 44.4 ± 10.1 | 42.8 ± 10 | 43.7 ± 10.1 | 1.5E-04 | NA |
| Female, N (%) | 3464 | 1383 (55.8%) | 500 (56%) | 50 (54.3%) | 0.96 | NA |
| BMI (kg/m2) | 3457 | 30.3 ± 7.5 | 31.1 ± 7.8 | 30.2 ± 5.6 | 0.018 | 0.72 |
| Systolic BP (mmHg) | 3461 | 124.6 ± 18.7 | 125.5 ± 19.2 | 126 ± 20.4 | 0.36 | 0.73 |
| Diastolic BP (mmHg) | 3461 | 78.1 ± 10.2 | 78.7 ± 10.5 | 78.8 ± 10.5 | 0.31 | 0.43 |
| Hypertension, N (%) | 3461 | 717 (28.9%) | 288 (32.3%) | 35 (38.5%) | 0.037 | 0.0021 |
| Glucose (mg/dL) | 3463 | 92 (85–102) | 93 (85–102) | 95.5 (86.8–110.5) | 0.076 | 0.44 |
| Insulin (mIU/L) | 3000 | 12.2 (7.1–20.3) | 14 (7.9–22.1) | 13.8 (8.8–22) | 0.0050 | 0.54 |
| HOMA-IR (mass units) | 2918 | 2.9 (1.6–5) | 3.2 (1.7–5.5) | 3.6 (2.1–5.9) | 0.0068 | 0.50 |
| Type 2 diabetes, N (%) | 3463 | 285 (11.5%) | 98 (11%) | 15 (16.3%) | 0.30 | 0.42 |
| GFR (mL/min/1.73m2) | 3463 | 99 ± 23.6 | 104.6 ± 24.6 | 108.8 ± 26.7 | 5.2E-15 | 0.023 |
| Total cholesterol (mg/dL) | 3463 | 182 ± 39.2 | 176.6 ± 40.4 | 170.9 ± 37.3 | 5.5E-05 | 4.7E-04 |
| LDL-C (mg/dL) | 3463 | 107.6 ± 35.5 | 103.9 ± 34.8 | 96.8 ± 34.8 | 0.0006 | 0.0024 |
| HDL-C (mg/dL) | 3463 | 50.3 ± 14.9 | 48.6 ± 14 | 48.4 ± 17.1 | 0.0026 | 1.9E-04 |
| Triglycerides (mg/dL) | 3463 | 95 (67–148.8) | 96 (68–138) | 108.5 (77.8–180.2) | 0.22 | 0.12 |
| A8 (ng/mL) | 3464 | 10.8 (5.5–17.7) | 22.5 (12.9–36.2) | 41.7 (25.3–61.5) | 3.6E-137 | 6.4E-140 |
Data are shown as N (%), mean ± SD, or median (25th-75th percentile). Abbreviations: A8, ANGPTL8; BMI, body mass index; BP, blood pressure; GFR, glomerular filtration rate; HDL-C, high-density lipoprotein-cholesterol; HOMA-IR, homeostatic model assessment of insulin resistance; LDL-C, low-density lipoprotein-cholesterol; NA, not applicable.
After adjustment for age, sex, race/ethnicity, and BMI, the A8(R59W) variant alone explained ~17% of the interindividual variation in plasma A8 levels, which was more than the contribution of BMI (4.9%) or other factors in the model (<3%) (Table 6). Furthermore, the fraction of variation in A8 levels attributed to self-reported race/ethnicity decreased from 5.4% to 2.7% when the R59W variant was added to the model; thus, the R59W variant accounted for 50% of racial/ethnic differences in A8 levels in DHS-1. Taken together, age, sex, race/ethnicity, BMI, and A8(R59W) explained 26.3% of the variation in A8 levels. In a multivariable model that included R59W but further adjusted for fasting insulin, plasma TG, and HDL-cholesterol levels, all the variables taken together explained 34.4% of interindividual variation in A8 levels. In this model, the R59W variant remained the most important determinant of A8 levels, accounting for 18% of interindividual variation in A8 levels (Table 6).
Table 6.
Effect of A8(R59W) genotype on plasma A8 levels in DHS-1 in multivariable regression models
| Model without genotype | Model with genotype | |||||
|---|---|---|---|---|---|---|
| Variable | Beta (SE) | P value | Partial R-squared (%) | Beta (SE) | P value | Partial R-squared (%) |
| (n = 3457) | ||||||
| Age | 0.004 (0.002) | 0.022 | 0.15 | 0.005 (0.001) | 9.70E-04 | 0.32 |
| Female sex | −0.015 (0.033) | 0.64 | 0.01 | −0.001 (0.03) | 0.97 | 0 |
| Race/Ethnicity | 5.44 | 2.73 | ||||
| White | Ref | - | - | Ref | - | - |
| Black | 0.521 (0.037) | 4.60E-43 | - | 0.305 (0.035) | 4.00E-18 | - |
| Hispanic | 0.382 (0.049) | 1.30E-14 | - | 0.037 (0.047) | 0.42 | - |
| Other | 0.498 (0.114) | 1.40E-05 | - | 0.304 (0.104) | 0.0035 | - |
| BMI | 0.027 (0.002) | 8.60E-34 | 4.17 | 0.026 (0.002) | 2.80E-39 | 4.86 |
| ANGPTL8 rs2278426 | - | - | - | 0.792 (0.029) | 1.60E-144 | 17.31 |
| Multiple R-squared (%) | 10.84 | 26.27 | ||||
| (n = 2998) | ||||||
| Age | 0.002 (0.002) | 0.21 | 0.05 | 0.003 (0.002) | 0.032 | 0.15 |
| Female sex | 0.111 (0.035) | 0.0016 | 0.33 | 0.118 (0.032) | 2.10E-04 | 0.46 |
| Race/Ethnicity | 7.15 | 4.48 | ||||
| White | Ref | - | - | Ref | - | - |
| Black | 0.582 (0.039) | 1.30E-49 | - | 0.363 (0.036) | 1.20E-23 | - |
| Hispanic | 0.316 (0.05) | 3.90E-10 | - | -0.02 (0.047) | 0.68 | - |
| Other | 0.486 (0.118) | 4.10E-05 | - | 0.29 (0.107) | 0.007 | - |
| BMI | 0.012 (0.002) | 8.60E-07 | 0.81 | 0.013 (0.002) | 2.10E-08 | 1.05 |
| Insulin | 0.011 (0.001) | 2.30E-17 | 2.38 | 0.011 (0.001) | 1.90E-21 | 2.98 |
| TG | 0.002 (0) | 1.20E-41 | 5.94 | 0.002 (0) | 3.50E-51 | 7.3 |
| HDL-C | −0.003 (0.001) | 0.017 | 0.19 | −0.001 (0.001) | 0.19 | 0.06 |
| ANGPTL8 rs2278426 | - | - | - | 0.777 (0.03) | 1.00E-131 | 18.1 |
| Multiple R-squared (%) | 19.89 | 34.39 |
Abbreviations: A8, ANGPTL8; BMI, body mass index; HDL-C, high-density lipoprotein-cholesterol; Ref, reference; TG, triglycerides.
To exclude the possibility that the higher A8 levels associated with the R59W variant were caused by a higher affinity of the Abs used in the ELISA for the A8(59W) isoform, we immunoblotted with the Detection Ab (318P) used in the ELISA, which recognizes an epitope near the C-terminus of A8 (Fig. 1A) and another mAb (34B4) with an epitope in the N-terminal domain (see “Methods”). For this analysis, we selected pairs of subjects of the same race/ethnicity who had similar levels of A8 (within 0.1 ng/mL by ELISA) such that one member of the pair was homozygous for A8(59R) and the other homozygous for the A8(59W) (Fig. 4C). We then compared the amount of A8 between the pairs by immunoblot analysis. No systematic differences were seen in the amount of immunodetectable A8 between the matched pairs. From this analysis, we concluded that the differences in levels of A8 associated with A8(59W) were not an artifact associated with the ELISA.
Higher A3 Levels Associated With A8(59W)
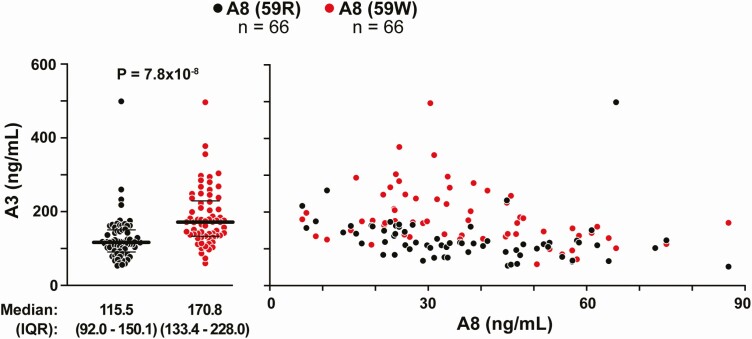
To determine if A8(59W) is associated with differences in plasma A3 levels, we selected 66 pairs of racially/ethnically-matched individuals who were homozygous for either A8(59R) or A8(59W) and had similar levels of A8 (within 1-2 ng/mL by ELISA). Plasma A3 concentrations were measured (1:50 dilution) using a commercial ELISA according to the manufacturer’s instructions (see “Methods”). A3 levels varied over a 10-fold range (from 52.2 to 498.6 ng/mL), and the distribution resembled what has been observed previously (26) (Fig. S6A) (38). Median A3 level was 48% higher in homozygotes for 59W than in 59R homozygotes (170.8 [133.4-228.0] ng/mL vs 115.5 [92.0-150.1] ng/mL, P = 7.76 × 10−8) (Fig. 5, left). The levels of A3 tended to be higher in the A8(59W) homozygotes across the spectrum of A8 levels (Fig. 5, right). In addition, we examined the correlation between plasma levels of A3 and A8 after stratification by genotype (Fig. S6B) (38). Plasma A3 levels were negatively correlated with A8 levels in both groups (r = −0.40 and −0.48 in the 59R and 59W homozygotes, respectively).
Figure 5.
Comparison of plasma A3 levels in individuals homozygous for A8(59R) and A8(59W) who were matched by A8 levels and race/ethnicity. Plasma levels of A3 were measured using a commercial ELISA kit according to manufacturer’s instructions. Dot plots of plasma A3 levels stratified by genotype showing median and IQR values (left). Plasma levels of A3 levels plotted against A8 levels in individuals homozygous for A8(59R) and A8(59W) (right).
Lack of Association Between A8(59W) and Cardiometabolic Parameters
Genetic variants that influence the levels of the encoded protein product can be used to test the hypothesis that circulating protein levels causally affect metabolic health outcomes, an approach referred to as Mendelian randomization (48). Specifically, if increased A8 levels cause a particular condition, then individuals with sequence variants that elevate A8 levels would be expected to be at an increased risk of the condition. To determine if increased A8 levels contribute directly to the adverse cardiometabolic parameters with which A8 levels are associated, we examined the relationship between A8(59W) and these phenotypes (e.g., BMI, insulin levels, glucose levels, HOMA-IR, plasma TG levels). No significant associations were found (Table 5) despite the strong association between A8(59W) and plasma levels of A8 (Fig. 4B) and the even stronger association of plasma levels of A8 and these cardiometabolic variables (Table 4). These data are consistent with the notion that A8 levels are a consequence, not the cause, of the associated metabolic phenotypes.
Variant in GCKR Approaches the Significance Threshold
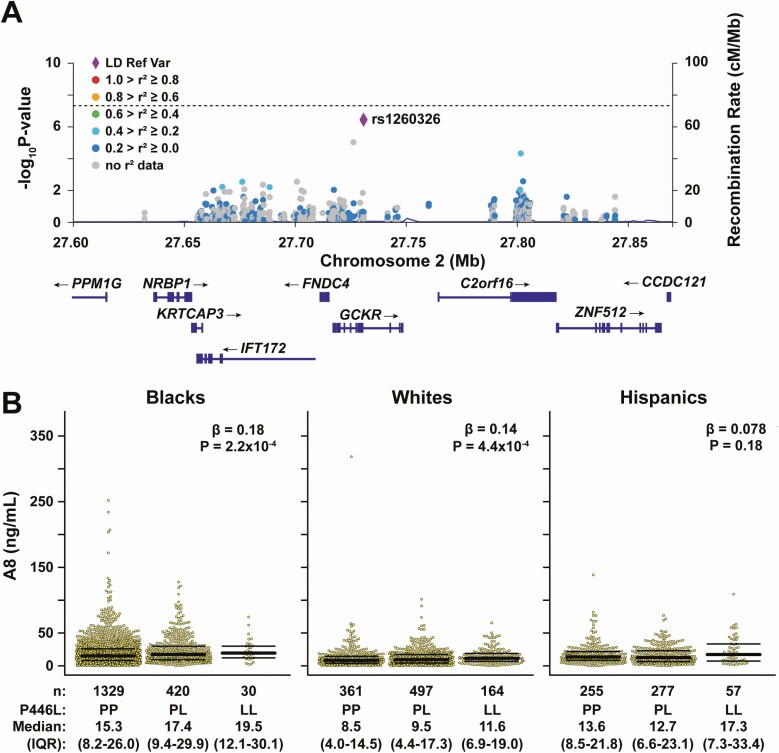
No other genetic locus was significantly associated with A8 levels at the exome-wide significance threshold. Thus, it is unlikely that other common genetic variants have effects on A8 levels that are comparable to those of the A8(59W) variant. However, one variant (rs1260326) in the glucokinase regulator (GCKR) on chromosome 2 approached the significance threshold in DHS-1 (P = 3.8 × 10−7) (Fig. 4A). A similar trend of association with this variant was found among those individuals in DHS-2 who were not in DHS-1, although the association did not reach the nominal significance threshold (n = 1141, β = 0.07, P = 0.12). Therefore, an independent replication will be required to confirm this association.
A regional plot including the GCKR rs1260326 variant and surrounding loci in the Chromosome 2 region is shown in Fig. 6A. This variant (c.1337T>C, p.P446L) results in substitution of proline for leucine at residue 446 of glucokinase regulatory protein (GKRP). GKRP regulates the activity of glucokinase (GCK), a cytoplasmic enzyme that phosphorylates glucose, trapping the monosaccharide after it enters cells.
Figure 6.
Association of GCKR(P446L) variant (rs1260326) and circulating A8 levels in DHS-1. A, Regional plot showing the association of variants with circulating A8 levels in the Chromosome 2 region that includes GCKR, FNDC4, IFT172, KRTCAP3, NRBP1, C2orf16, ZNF512, and CCDC121 loci. The significance threshold is denoted by the dashed line. Diamond in purple represents the rs1260326 variant located in the GCKR locus. B, A8 levels by rs1260326 genotype in Blacks (n = 1779), Whites (n = 1022), and Hispanics (n = 589) in DHS-1 with median and IQR values. Association was tested using linear regression with adjustment for age, sex, principal component analysis of ancestry, and BMI. The number of individuals with each genotype (P/P, P/L, and L/L) is indicated below the corresponding genotype.
The minor T-allele was modestly but consistently associated with increased plasma A8 levels in Blacks (β = 0.18, P = 2.2 × 10−4) and Whites (β = 0.14, P = 4.4 × 10−4), with a directionally concordant effect in Hispanics, although it did not reach nominal statistical significance (β = 0.078, P = 0.18) in this group, likely due to the small sample size (Fig. 6B).
The association of GCKR rs1260326 with plasma A8 levels was only slightly attenuated after additional adjustment for plasma insulin and TG levels (P = 2.77 × 10−6), suggesting that the relationship is not due to the effect of GCKR(446L) on these factors. In joint analysis, both A8(59W) and GCKR(446L) were independently associated with circulating A8 levels (P = 7.7 × 10−139 and P = 2.9 × 10−7, respectively). No interaction between the two variants was detected (P = 0.27). Thus, these two variants appear to act additively.
Discussion
Here we describe a new, sensitive, and specific A8 ELISA that was used to evaluate plasma A8 levels of participants in a multiracial/multiethnic population-based cohort (n = 3538). Levels of A8 varied over a 150-fold range (2.1 ng/mL to 318 ng/mL), and the distribution of levels was strongly skewed to the right with a median of 13.3 ng/mL (Fig. 2A). Levels of A8, measured on 2 occasions separated by 7 to 9 years, were as strongly correlated as plasma levels of LDL-C measured in the same samples (r = 0.53 and 0.47, respectively). No individuals were identified with complete A8 deficiency. Significant racial/ethnic differences in relative plasma A8 levels were observed with the highest levels being in Blacks and the lowest in Whites, with Hispanics being intermediate (Fig. 2D). A missense variant in A8, 59W, was strongly associated with increased levels of plasma A8. Homozygotes for this variant had a 4-fold increase in A8 levels when compared with those homozygous for the common variant (41.7 vs 10.8 ng/mL, respectively) (Table 5). This variant alone accounted for ~17% of the interindividual differences in levels and ~46% of the interracial/ethnic differences in plasma A8 levels. A8(59W) was not associated with any of the metabolic phenotypes found to be strongly correlated with A8 levels, including BMI, plasma glucose, insulin, HOMA-IR, or hepatic TG content. Thus, the strong correlations between plasma levels of A8 and indices of fuel metabolism are not due to causal relationships.
Several other A8 commercial assays have been developed and used to measure plasma A8 levels (25-27, 45, 49, 50). Two new ELISAs were reported while our assay was under development (26, 50). Our results correlated well with those obtained using these 2 assays (Fig. S7) (38). Similar to our findings, A8 levels have been associated with BMI, plasma glucose, insulin, and TG levels (45, 51–53), as well as measures of body fat content including total abdominal and visceral fat (54). Since A8 expression has been reported to be upregulated by insulin (55), it is plausible that the strong correlation observed between plasma A8 and plasma insulin is a reflection of this regulation in the liver. The associations between plasma A8 levels and metabolic parameters have led to speculation that pharmacological inactivation of A8 could have beneficial effects not only by lowering plasma TG levels, but also by reducing body fat content and the other adverse consequences of obesity. Our finding of no association between A8(59W) and these same metabolic phenotypes failed to support the notion that there is a causal basis for the strong associations between plasma A8 levels and cardiometabolic risk. This does not negate the possibility that inactivation of A8, for example with an anti-A8 Ab, might not have therapeutic benefit, as would be predicted based on the marked reductions in plasma TG levels in A8-/- mice (56). It is possible that fasting plasma levels of A8 do not predict the functional activity of A8 in the postprandial period, when the protein is most active. Alternatively, circulating levels of A8 may not reflect the amount of A3/A8 complex that reaches the vascular endothelial surfaces. Finally, it remains possible that variation in these metabolic factors causes changes in plasma levels of A8 (ie, reverse causation), or that a yet-to-be-identified factor acting upstream of A8 causes concomitant changes in plasma levels of A8 as well as in the associated metabolic parameters.
We showed previously that A3 and A8 must form a complex in order to inhibit LPL activity (3, 15). In fasting individuals, the circulating levels of A3 (~5 nM) are notably higher than are those of A8 (0.3nM and 1nM) (26). Chen et al (57) reported that A8 circulates almost entirely as a complex with A3, whereas the major fraction of A3 circulates independently of A8. Taken together, these data are consistent with our hypothesis that the availability of A8 is rate-limiting for the formation of LPL-inhibiting A3/A8 complexes (3). Of note, circulating concentrations of A3 and A8 are 3 orders of magnitude below those of apolipoprotein (Apo) C2 (3-4 μM) (58) and ApoC3 (12 μM) (59), long considered to be two of the primary post-translational regulators of LPL activity. A major difference between these two apolipoproteins and A3/A8 is that ApoC2 and ApoC3 circulate associated with lipoproteins. Further studies will be required to elucidate how A3 and A8 interact in vivo to regulate LPL activity.
Interestingly, the levels of A3 and A8 are inversely correlated (Fig. S5) (38). Those individuals with higher levels in A8 tend to have lower levels of A3. It is possible that the inverse relationship is due to an artifact; the presence of A8 may interfere with the A3 assay. Alternatively, A3 levels may be lower in those with higher A8 levels because of differences in the rates of clearance of A3 and A3/A8 from plasma. The A3 that is bound to A8 may be removed more rapidly from the blood, thus lowering A3 levels. Additional studies will be required to better understand the relationship between levels of A3 and A8.
We failed to identify any individuals in our sample who had no circulating A8. Moreover, loss-of-function mutations in A8 have been rare in all populations examined so far, and no homozygotes or compound heterozygotes for such mutations have been reported (3). This observation raises the possibility that A8 expression is required for viability. Arguing against this notion, A8-/- mice are viable, and offspring of A8-/- homozygotes and heterozygotes are born at the expected Mendelian ratios (56). Nonetheless, we cannot exclude the possibility that A8 is required for viability in humans. If so, this would imply that A8 has a function that is independent of both A3 and LPL, since neither of these proteins are essential for life in humans.
The variant with the greatest impact on A8 levels in a genome-wide association study was a missense variant in the A8 gene: R59W (Fig. 4A). Ironically, A8 was initially identified through the association of this variant with plasma levels of LDL-C and HDL-C (3) rather than plasma levels of TG. Here, we found that R59W had an additive effect on plasma levels of A8. We ruled out the possibility that the increase in A8 levels associated with this variant was due to an artifact associated with the antibodies used in the A8 ELISA (Fig. 4C). We also considered the possibility that the higher levels of A8 associated with A8(59W) were due to reduced cleavage of A8 by furin. A furin cleavage site is present at residue 175 in A8, and the binding epitope of the detection antibody used in the ELISA is C-terminal to this site (Fig. 1A). Unfortunately, we could not test this hypothesis, since we were unable to develop a reliable assay that differentiated the cleaved from the uncleaved form of the protein because the two forms differ by only 23 residues.
Homozygotes for A8(59W) had higher A3 levels than homozygotes for the A8(59R) isoform (Fig. 5). Fasting plasma A3 levels were negatively correlated with A8 levels in both groups: A8(59R) and A8(59W). A more stable interaction between the 59W isoform and A3 may increase the half-life of the complex, and thereby the levels of A8 in the circulation. Additional studies will be required to determine if the higher levels of A8 associated with the A8(59W) variant are due to an increase in the synthesis/secretion of A8 or to a decrease in its clearance.
Heterozygotes for an obligate loss-of-function mutation in A8 (Q121X) have reduced plasma levels of TG, increased HDL-C and little change in LDL-C (21). An essentially identical phenotype is observed in individuals carrying a gain-of-function variant in LPL (60). These findings are consistent with our hypothesis that A8 acts primarily to inhibit LPL and has little effect on endothelial lipase (21). In contrast to the Q121X mutation, the phenotype of A8(59W) cannot be explained by differences in LPL activity. Altered interaction between A8 and A3 may explain the paradoxical phenotype of A8(59W): an association with cholesterol rather than TG levels. A3 inhibition was shown to reduce both HDL-C and LDL-C through endothelial lipase–dependent mechanisms (44, 61). We speculate that the R59W substitution disrupts A3 interaction with endothelial lipase but not with LPL. The higher levels of plasma A8 in the R59W carriers may result in less A3 being unbound to A8 in plasma. The lower concentration of unbound A3 would increase endothelial lipase activity and thereby lower plasma levels of LDL-C and HDL-C (62).
The only other locus in our genome-wide association study that approached genome-wide significance (P = 3.8 × 10−7) was in GCKR (Fig. 6A). The protein product of GCKR, GKRP regulates the activity of GCK and catalyzes the conversion of glucose to glucose-6-phosphate, thus trapping glucose inside cells. In the fasted state, hepatic GKRP forms an inhibitory complex with GCK and sequesters the enzyme in the nucleus (63). In the fed state, fructose-1-phosphate binds to GKRP, causing GCK to dissociate, which results in exposure of a nuclear export signal (64). The missense variant (rs126036) in GCKR that was most strongly associated with A8 levels causes a substitution of proline for leucine at residue 446. This variant was first identified to be associated with lower glucose and higher plasma TG levels (65). GCKR(446L) reduces nuclear sequestration of GCK (66, 67), which would be expected to increase GCK activity and promotes glucose utilization in cells (67). As a consequence, hepatic glycogenesis, glycolytic flux, and fatty acid synthesis are increased (68, 69), as are levels of TG and glucose.
It remains unclear why levels of A8 are higher in subjects with the GCKR(446L) variant. The increase in glucose-6-phosphate levels in hepatocytes or associated changes in insulin signaling may drive A8 secretion, resulting in higher levels of plasma A8. Elucidation of the pathways that control the regulation of A8 and GCKR transcription may provide clues as to the basis for this association. A8 expression is increased in both cultured hepatocytes and 3T3-L1 adipocytes upon addition of glucose and insulin to the medium (55, 70, 71), as is the expression of GCKR in hepatocytes (72). It is possible that A8, like GCKR, is a target gene of ChREBP (Carbohydrate-response element-binding protein), a transcription factor that is upregulated in response to glucose (73, 74). Hepatocyte nuclear factor-1 alpha (HNF1α) has also been implicated in activating A8 expression in primary hepatocytes (66). HNF1α was shown to be required for the insulin-mediated increase in A8 protein expression in primary hepatocytes, but its role in GCKR regulation is not known. Additional studies will be required to determine the molecular basis for the association between A8 and GCKR(446L).
Acknowledgments
We thank Chelsea Burroughs and Fabienne Elwood for assistance with preparation of the manuscript. We also thank Serena Banfi for testing the commercial A8 ELISA kits (Cloud-Clone Corp., Aviscera and Phoenix) and Linda Donnelly for generating the A8 antibodies used for immunoblotting.
Financial Support: J.C.C. and H.H.H. were supported by grants from the National Institutes of Health (5 PO1 HL20948).
Author Contributions: F.O., K.B., J.K., H.H., L.M.S., V.G., J.C.C., and H.H.H. designed the experiments; F.O., K.B., J.K., H.H., L.M.S., and V.G. performed the experiments; L.M.S. and V.G. contributed new reagents/analytic tools; F.O., K.B., J.K., H.H., V.G., J.C.C., and H.H.H. analyzed data; F.O., K.B., J.K., H.H., V.G., J.C.C., and H.H.H. wrote the manuscript.
Abbreviations
- A1-A8
ANGPTL1-ANGPTL8
- Ab
antibody
- ANGPTL
angiopoietin-like
- BSA
bovine serum albumin
- CV
, coefficient of variation
- DHS
Dallas Heart Study
- ELISA
enzyme-linked immunosorbent assay
- GCK
glucokinase
- GKRP
glucokinase regulatory protein
- GPIHBP1
glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1
- HDL
high-density lipoprotein
- HDL-C
high-density lipoprotein cholesterol
- ICC
intra-class coefficient
- IQR
interquartile range
- LDL-C
low-density lipoprotein cholesterol
- LoB
limit of blank
- LoD
limit of detection
- LPL
lipoprotein lipase
- mAb
monoclonal antibody
- PBS
phosphate-buffered saline
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TG
triglyceride.
Additional Information
Disclosures: V.G. and L.M.S. are employees and stockholders of Regeneron Pharmaceuticals, Inc. All other authors have declared that no conflict of interest exists.
Data Availability
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Supplementary Data: All supplemental figures and tables are located in a digital research data repository. (38)
References
- 1. Korn ED, Quigley TW Jr. Studies on lipoprotein lipase of rat heart and adipose tissue. Biochim Biophys Acta. 1955;18(1):143-145. [DOI] [PubMed] [Google Scholar]
- 2. Kersten S, Mandard S, Tan NS, et al. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J Biol Chem. 2000;275(37):28488-28493. [DOI] [PubMed] [Google Scholar]
- 3. Quagliarini F, Wang Y, Kozlitina J, et al. Atypical angiopoietin-like protein that regulates ANGPTL3. Proc Natl Acad Sci U S A. 2012;109(48):19751-19756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Z, Yao F, Abou-Samra AB, Zhang R. Lipasin, thermoregulated in brown fat, is a novel but atypical member of the angiopoietin-like protein family. Biochem Biophys Res Commun. 2013;430(3):1126-1131. [DOI] [PubMed] [Google Scholar]
- 5. Zhang R. Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochem Biophys Res Commun. 2012;424(4):786-792. [DOI] [PubMed] [Google Scholar]
- 6. Kersten S. Angiopoietin-like 3 in lipoprotein metabolism. Nat Rev Endocrinol. 2017;13(12):731-739. [DOI] [PubMed] [Google Scholar]
- 7. Mattijssen F, Kersten S. Regulation of triglyceride metabolism by Angiopoietin-like proteins. Biochim Biophys Acta. 2012;1821(5):782-789. [DOI] [PubMed] [Google Scholar]
- 8. Kersten S. Regulation of lipid metabolism via angiopoietin-like proteins. Biochem Soc Trans. 2005;33(Pt 5):1059-1062. [DOI] [PubMed] [Google Scholar]
- 9. Shan L, Yu XC, Liu Z, et al. The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. J Biol Chem. 2009;284(3):1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shimamura M, Matsuda M, Yasumo H, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27(2):366-372. [DOI] [PubMed] [Google Scholar]
- 11. Wu L, Soundarapandian MM, Castoreno AB, Millar JS, Rader DJ. LDL-cholesterol reduction by ANGPTL3 inhibition in mice is dependent on endothelial lipase. Circ Res. 2020;127(8):1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young SG, Davies BS, Voss CV, et al. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J Lipid Res. 2011;52(11):1869-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goulbourne CN, Gin P, Tatar A, et al. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014;19(5):849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee EC, Desai U, Gololobov G, et al. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL). J Biol Chem. 2009;284(20):13735-13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haller JF, Mintah IJ, Shihanian LM, et al. ANGPTL8 requires ANGPTL3 to inhibit lipoprotein lipase and plasma triglyceride clearance. J Lipid Res. 2017;58(6):1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oldoni F, Cheng H, Banfi S, Gusarova V, Cohen JC, Hobbs HH. ANGPTL8 has both endocrine and autocrine effects on substrate utilization. JCI Insight. 2020;5(17):e138777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17..Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371(1):32-41. [DOI] [PubMed] [Google Scholar]
- 18. Dewey FE, Gusarova V, Dunbar RL, et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med. 2017;377(3):211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dewey FE, Gusarova V, O’Dushlaine C, et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016;374(12):1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gusarova V, Banfi S, Alexa-Braun CA, et al. ANGPTL8 blockade with a monoclonal antibody promotes triglyceride clearance, energy expenditure, and weight loss in mice. Endocrinology. 2017;158(5):1252-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peloso GM, Auer PL, Bis JC, et al. ; NHLBI GO Exome Sequencing Project . Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94(2):223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abu-Farha M, Sriraman D, Cherian P, et al. Circulating ANGPTL8/betatrophin is increased in obesity and reduced after exercise training. Plos One. 2016;11(1):e0147367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abu-Farha M, Al-Khairi I, Cherian P, et al. Increased ANGPTL3, 4 and ANGPTL8/betatrophin expression levels in obesity and T2D. Lipids Health Dis. 2016;15(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al-Shawaf E, Al-Ozairi E, Al-Asfar F, et al. Biphasic changes in angiopoietin-like 8 level after laparoscopic sleeve gastrectomy and type 2 diabetes remission during a 1-year follow-up. Surg Obes Relat Dis. 2018;14(9):1284-1294. [DOI] [PubMed] [Google Scholar]
- 25. Ebert T, Kralisch S, Hoffmann A, et al. Circulating angiopoietin-like protein 8 is independently associated with fasting plasma glucose and type 2 diabetes mellitus. J Clin Endocrinol Metab. 2014;99(12):E2510-E2517. [DOI] [PubMed] [Google Scholar]
- 26. Morinaga J, Zhao J, Endo M, et al. Association of circulating ANGPTL 3, 4, and 8 levels with medical status in a population undergoing routine medical checkups: A cross-sectional study. Plos One. 2018;13(3):e0193731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abu-Farha M, Abubaker J, Al-Khairi I, et al. Higher plasma betatrophin/ANGPTL8 level in Type 2 Diabetes subjects does not correlate with blood glucose or insulin resistance. Sci Rep. 2015;5:10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee YH, Lee SG, Lee CJ, et al. Association between betatrophin/ANGPTL8 and non-alcoholic fatty liver disease: animal and human studies. Sci Rep. 2016;6:24013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cengiz M, Ozenirler S, Kocabiyik M. Serum β-trophin level as a new marker for noninvasive assessment of nonalcoholic fatty liver disease and liver fibrosis. Eur J Gastroenterol Hepatol. 2016;28(1):57-63. [DOI] [PubMed] [Google Scholar]
- 30. Hu W, Shao X, Guo D, et al. Relationship of serum betatrophin with nonalcoholic fatty liver in a Chinese population. Plos One. 2017;12(1):e0170758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fadaei R, Shateri H, DiStefano JK, et al. Higher circulating levels of ANGPTL8 are associated with body mass index, triglycerides, and endothelial dysfunction in patients with coronary artery disease. Mol Cell Biochem. 2020;469(1-2):29-39. [DOI] [PubMed] [Google Scholar]
- 32. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. R: a language and environment for statistical computing. R Foundation for Statistical Computing. 2020. https://www.R-project.org/ [Google Scholar]
- 35. Victor RG, Haley RW, Willett DL, et al. ; Dallas Heart Study Investigators . The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93(12):1473-1480. [DOI] [PubMed] [Google Scholar]
- 36. Murphy AJ, Macdonald LE, Stevens S, et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci U S A. 2014;111(14):5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macdonald LE, Karow M, Stevens S, et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc Natl Acad Sci U S A. 2014;111(14):5147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oldoni F, Bass K, Kozlitina J, et al. Genetic and metabolic determinants of plasma levels of ANGPTL8. Supplemental data. figshare. Deposited February 2, 2021. 10.6084/m9.figshare.13691893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29 Suppl 1:S49-S52. [PMC free article] [PubMed] [Google Scholar]
- 40. Lakoski SG, Xu F, Vega GL, et al. Indices of cholesterol metabolism and relative responsiveness to ezetimibe and simvastatin. J Clin Endocrinol Metab. 2010;95(2):800-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86(2):420-428. [DOI] [PubMed] [Google Scholar]
- 42. Dewey FE, Murray MF, Overton JD, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354(6319):aaf6814. [DOI] [PubMed] [Google Scholar]
- 43. Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gusarova V, Alexa CA, Wang Y, et al. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J Lipid Res. 2015;56(7):1308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu Z, Berhane F, Fite A, Seyoum B, Abou-Samra AB, Zhang R. Elevated circulating lipasin/betatrophin in human type 2 diabetes and obesity. Sci Rep. 2014;4:5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trends in ischemic heart disease death rates for blacks and whites-- United States, 1981-1995. 1998. [PubMed]
- 47. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412-419. [DOI] [PubMed] [Google Scholar]
- 48. Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1(8479):507-508. [DOI] [PubMed] [Google Scholar]
- 49. Chen X, Lu P, He W, et al. Circulating betatrophin levels are increased in patients with type 2 diabetes and associated with insulin resistance. J Clin Endocrinol Metab. 2015;100(1):E96-100. [DOI] [PubMed] [Google Scholar]
- 50. Leiherer A, Ebner J, Muendlein A, et al. High betatrophin in coronary patients protects from cardiovascular events. Atherosclerosis. 2020;293:62-68. [DOI] [PubMed] [Google Scholar]
- 51. Gao T, Jin K, Chen P, et al. Circulating betatrophin correlates with triglycerides and postprandial glucose among different glucose tolerance statuses-a case-control study. Plos One. 2015;10(8):e0133640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gómez-Ambrosi J, Pascual E, Catalán V, et al. Circulating betatrophin concentrations are decreased in human obesity and type 2 diabetes. J Clin Endocrinol Metab. 2014;99(10):E2004-E2009. [DOI] [PubMed] [Google Scholar]
- 53. Guo K, Lu J, Yu H, et al. Serum betatrophin concentrations are significantly increased in overweight but not in obese or type 2 diabetic individuals. Obesity (Silver Spring). 2015;23(4):793-797. [DOI] [PubMed] [Google Scholar]
- 54. Zheng J, Liu J, Hong BS, Ke W, Huang M, Li Y. Circulating betatrophin/ANGPTL8 levels correlate with body fat distribution in individuals with normal glucose tolerance but not those with glucose disorders. BMC Endocr Disord. 2020;20(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ren G, Kim JY, Smas CM. Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. Am J Physiol Endocrinol Metab. 2012;303(3):E334-E351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Y, Quagliarini F, Gusarova V, et al. Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proc Natl Acad Sci U S A. 2013;110(40):16109-16114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen YQ, Pottanat TG, Siegel RW, et al. Angiopoietin-like protein 8 differentially regulates ANGPTL3 and ANGPTL4 during postprandial partitioning of fatty acids. J Lipid Res. 2020;61(8):1203-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barr SI, Kottke BA, Chang JY, Mao SJ. Immunochemistry of human plasma apolipoprotein C-II as studied by radioimmunoassay. Biochim Biophys Acta. 1981;663(2):491-505. [DOI] [PubMed] [Google Scholar]
- 59. Qamar A, Khetarpal SA, Khera AV, Qasim A, Rader DJ, Reilly MP. Plasma apolipoprotein C-III levels, triglycerides, and coronary artery calcification in type 2 diabetics. Arterioscler Thromb Vasc Biol. 2015;35(8):1880-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Groenemeijer BE, Hallman MD, Reymer PW, et al. Genetic variant showing a positive interaction with beta-blocking agents with a beneficial influence on lipoprotein lipase activity, HDL cholesterol, and triglyceride levels in coronary artery disease patients. The Ser447-stop substitution in the lipoprotein lipase gene. REGRESS Study Group. Circulation. 1997;95(12):2628-2635. [DOI] [PubMed] [Google Scholar]
- 61. Adam RC, Mintah IJ, Alexa-Braun CA, et al. Angiopoietin-like protein 3 governs LDL-cholesterol levels through endothelial lipase-dependent VLDL clearance. J Lipid Res. 2020;61(9):1271-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jaye M, Lynch KJ, Krawiec J, et al. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21(4):424-428. [DOI] [PubMed] [Google Scholar]
- 63. Toyoda Y, Miwa I, Satake S, Anai M, Oka Y. Nuclear location of the regulatory protein of glucokinase in rat liver and translocation of the regulator to the cytoplasm in response to high glucose. Biochem Biophys Res Commun. 1995;215(2):467-473. [DOI] [PubMed] [Google Scholar]
- 64. Shiota C, Coffey J, Grimsby J, Grippo JF, Magnuson MA. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J Biol Chem. 1999;274(52):37125-37130. [DOI] [PubMed] [Google Scholar]
- 65. Orho-Melander M, Melander O, Guiducci C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57(11):3112-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Watanabe T, Ozawa A, Masuda S, et al. Transcriptional Regulation of the Angptl8 Gene by Hepatocyte Nuclear Factor-1 in the Murine Liver. Sci Rep. 2020;10(1):9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rees MG, Wincovitch S, Schultz J, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55(1):114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beer NL, Tribble ND, McCulloch LJ, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18(21):4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santoro N, Caprio S, Pierpont B, Van Name M, Savoye M, Parks EJ. Hepatic de novo lipogenesis in obese youth is modulated by a common variant in the GCKR gene. J Clin Endocrinol Metab. 2015;100(8):E1125-E1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nidhina Haridas PA, Soronen J, Sädevirta S, et al. Regulation of angiopoietin-like proteins (ANGPTLs) 3 and 8 by insulin. J Clin Endocrinol Metab. 2015;100(10):E1299-E1307. [DOI] [PubMed] [Google Scholar]
- 71. Zhang L, Shannon CE, Bakewell TM, Abdul-Ghani MA, Fourcaudot M, Norton L. Regulation of ANGPTL8 in liver and adipose tissue by nutritional and hormonal signals and its effect on glucose homeostasis in mice. Am J Physiol Endocrinol Metab. 2020;318(5):E613-E624. [DOI] [PubMed] [Google Scholar]
- 72. Arden C, Petrie JL, Tudhope SJ, et al. Elevated glucose represses liver glucokinase and induces its regulatory protein to safeguard hepatic phosphate homeostasis. Diabetes. 2011;60(12):3110-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Poungvarin N, Chang B, Imamura M, et al. Genome-wide analysis of ChREBP binding sites on male mouse liver and white adipose chromatin. Endocrinology. 2015;156(6):1982-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yamashita H, Takenoshita M, Sakurai M, et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A. 2001;98(16):9116-9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Supplementary Data: All supplemental figures and tables are located in a digital research data repository. (38)