Abstract
Molecular diagnosis on nasopharyngeal swabs (NPS) is the current standard for COVID-19 diagnosis, but saliva may be an alternative specimen to facilitate access to diagnosis. We compared analytic performances, feasibility and acceptability of NPS, saliva, and oral-self sampling swab for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A prospective, multicenter study was conducted in military hospitals in France among adult outpatients attending COVID-19 diagnosis centers or hospitalized patients. For each patient, all samples were obtained and analyzed simultaneously with RT-PCR or transcription-mediated amplification method. Clinical signs, feasibility, and acceptability for each type of sample were collected. A total of 1220 patients were included, corresponding to 1205 NPS and saliva and 771 OS. Compared to NPS, the sensitivity, specificity, and kappa coefficient for tests performed on saliva were 87.8% (95% CI 83.3–92.3), 97.1% (95% CI 96.1–98.1), and 0.84 (95% CI 0.80–0.88). Analytical performances were better in symptomatic patients. Ct values were significantly lower in NPS than saliva. For OS, sensitivity was estimated to be 61.1% (95% CI 52.7–69.4) and Kappa coefficient to be 0.69 (95% CI 0.62–0.76). OS was the technique preferred by the patients (44.3%) before saliva (42.4%) and NPS (13.4%). Instructions were perceived as simple by patients (> 90%) for saliva and OS. Finally, the painful nature was estimated to be 0.9 for OS, on a scale from 0 to 10, and to be 5.3 for NPS. Performances of OS are not sufficient. Saliva is an acceptable alternative to NPS for symptomatic patient but the process required additional steps to fluidize the sample.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10096-021-04269-4.
Keywords: COVID-19, SARS-CoV-2, Nasopharyngeal swab, Saliva, Oral-self sampling
Introduction
The recent spread of coronavirus disease (COVID-19) caused by SARS-CoV-2 has become an international concern. Rapid and accurate diagnosis is important to control the ongoing COVID-19 pandemic. Nasopharyngeal swabs (NPS), followed by reverse transcription polymerase chain reaction (RT-PCR), are recommended by the World Health Organization, Centers for Disease Control and Prevention, and French Society of Microbiology for the diagnosis of COVID-19 [1–3]. Although the current standard is NPS, collection requires trained medical personnel wearing full personal protective equipment to avoid contamination. Additionally, mass testing requires an increased number of personnel, swabs, and viral transport media causing logistic burden. Furthermore, NPS causes discomfort to the patients [4, 5]. Less invasive methods are needed and collection by the patient could reduce high exposure of the healthcare worker. Previous studies have shown that saliva could be an alternative to NPS. However most studies have a limited number of patients or are monocentric [6–10]. A recent meta-analysis showed a sensitivity of saliva to be 91% (95% CI 80–99) [11]. It highlighted that NPS tests were more sensitive than saliva, but the difference was not statistically significant. Other studies testing symptomatic outpatients [9, 12, 13] showed also that the median cycle threshold (Ct) value was significantly higher in saliva, suggesting lower viral loads in saliva. Finally, some authors reported highest viral loads in posterior oropharyngeal saliva [14–16]. For self-collected oral fluid specimens with swabs, data are scarce in literature and sensitivity varies from 66 to 89.8% [17, 18].
Faced to these heterogeneous results and an uncertainty about the massive screening strategy using saliva in France [19], we carried out a prospective, multicenter study to evaluate the performance, feasibility, and acceptability of saliva samples and by oral self-swabbing.
Methods
Study design, participants, and setting
This prospective, multicenter study was conducted among 1220 volunteer adult patients attending the COVID-19 diagnosis center of 4 military hospitals in France or hospitalized in these hospitals. The inclusion criteria were participants above 18 years, under investigation for a suspected COVID-19 or contact of a confirmed COVID-19 case, and able to obey commands and to give written consent. Patients hospitalized in intensive care unit were excluded. Reason for consultation, clinical signs, feasibility, acceptability, and pain for each type of sample (scale from 0 to 10: 0 no annoyance, 10 extremely unpleasant) were collected.
Specimen collection
For each participant, oral information was given to explain collection procedure and sequence with 3 steps. First, patients should not eat or drink within 30 min before collection, and then, they were asked to take 2 ml saliva with a simple drooling technique [20]. To assist the patient, a container with 2 ml of colored solution was available in the collection room to indicate the correct collection volume. Then, oral self-sampling (OS) was performed by swabbing the internal face of the cheeks, the groove between the cheeks and gums, and then under the tongue for 30 s in order to obtain a good hydratation of the swab and to collect saliva and mucous cells. This swab was then placed in 3 ml of viral medium transport (VMT) (PBS, Virus stabilization tube, Vacuette®). Finally, NPS were collected by experienced and trained nurses and placed in VMT. In this study, NPS was used as reference and this result was communicated to patients.
Molecular analysis for SARS-CoV-2
Different molecular techniques were used in this multicenter study according to the routine method implemented in investigating centers.
The TMA (transcription-mediated amplification) technique was used by centers 1 and 4. This technique (Aptima SARS-CoV-2, Hologic®) that amplifies 2 targets of ORF1ab does not require pretreatment for analysis of nasopharyngeal samples. No pretreatment was carried out on the buccal swabs resuspended in VMT. For saliva samples, pretreatment procedure with 5 μl of proteinase K (PK, 20 mg/mL) into 500 μl of specimen at 65 °C during 10 min was systematically added to avoid invalid results. Swabs used in centers 1 and 4 for NPS and oral swabs were sterile swab with flocked tip (G1022FN, Gongdong Medical Technology®).
The RT-PCR technique was used by centers 2 and 3. Center 2 performed chemical inactivation (PK) before RNA extraction with NucleoMag Pathogen Kit on the Genomic STARlet (Hamilton®) extractor and performed the amplification step with the IDNCOV-2 kit (ID-solutions®) on the CFX-96 system (Biorad®). This RT-PCR amplifies 2 viral targets (N1 and N2 gene of the nucleocapsid) and 1 cellular control.
Center 3 performed chemical (PK) and thermal inactivation, RNA extraction with the Genoxtract Kit and platform (Biocentric®), and then the amplification step with the TaqPathTM COVID-19 CE-IVD RT-PCR Kit on the QuantStudio 5 system (ThermoFisher®). This RT-PCR amplifies 3 viral targets (N gene, S gene and ORF1). Swabs used in centers 2 and 3 for NPS and oral swabs were sterile swab (LYFT001, Lemoine®).
Data processing and analysis
For the categorical variables, the chi-square test was used. Comparisons between quantitative variables were made using Student’s test. Additionally, sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV), Cohen’s kappa (K coefficient), and their 95% confidence intervals (CI) were calculated to assess diagnostic performance. A p value < 0.05 was considered statistically significant. All statistical analyses were performed using R version 3.6.0. Interim analysis was included in the study design in order to stop a sample type in the event of poor performance.
Results
Patient characteristics
In the study between September 15 and October 19, 2020, 1220 subjects were included with 57.2% male and 42.8% female (Supplementary Information Table 1). The mean age was 38.6 ± 14.6 years. Of them 55.4% were symptomatic. Outpatients represented over 99% of the included volunteers.
Comparison of SARS-CoV-2 RNA detection between saliva and nasopharyngeal swab
Out of 1205 analyses conducted in parallel on saliva and NPS, 180 were positive and 971 negative in both tests (Table 1). Saliva samples from 17 patients (1.4%) generated invalid RT-PCR or TMA results due to internal control failure or automated sample processing errors. Invalid rates were, respectively, 1.6%, 1.7%, 0%, and 1.6% for centers 1 (TMA), 2 (RT-PCR), 3 (RT-PCR), and 4 (TMA).
Table 1.
Results from nasopharyngeal specimens and saliva in same-day matched pairs (n = 1205)
| SARS-CoV-2-RNA | Nasopharyngeal specimens | |||
|---|---|---|---|---|
| Detected | Not detected | Invalid | ||
| Salivary self-sampling | Detected | 180 | 15 | 0 |
| Not detected | 22 | 971 | 0 | |
| Invalid | 3 | 14 | 0 | |
The overall sensitivity and specificity for molecular tests performed on saliva compared to NPS were 87.8% (95% CI 83.3–92.3) and 97.1% (95% CI 96 96.1–98.1), respectively. The positivity rate of NPS was 17% (205/1205) and 16.2% (195/1205) for saliva with no significant difference (p = 0.155). Kappa coefficient was 0.84 (95% CI 0.80–0.88), suggesting excellent agreement.
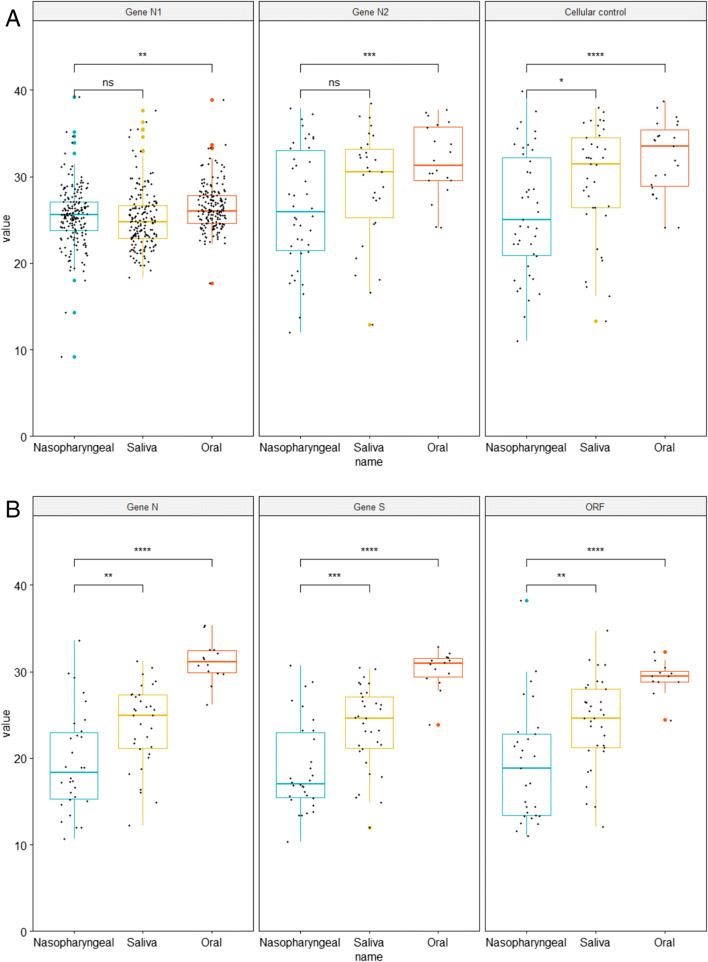
Among the 56 individuals with concordant results in centers using RT-PCR providing Ct values, the Ct values were significantly lower in NPS than saliva for N-gene (19.5 vs 23.8; p = 0.003), S-gene (18.9 vs 23.5; p = 0.001) and ORF1-gene (19.3 vs 24.1; p = 0,003) for center 3 and N2-gene (25.9 vs 29.4 ; p = 0.023) for center 2. In center 2, no difference for N1-gene (26.5 vs 28.9; p = 0.145) and cellular control (25.5 vs 25.1; p = 0.327) was observed (Fig. 1). For centers 1 and 4 using TMA, Ct values and viral load were not estimated due to end point reading technique but intensities of the reaction, reflected by relative light units (RLU) were analyzed. The overall mean RLU were 1195 (95%, CI 1087–1303) for NPS and 1380 (95%, CI 1243–1517) for saliva with no significant difference (p = 0.288).
Fig. 1.
Ct values in nasopharyngeal, saliva, and oral specimens for genes N1, N2 and cellular control for center 2 (A). Ct values in nasopharyngeal, saliva, and oral specimens for genes N, S, and ORF1 center for center 3 (B). ns not significant, * p < 5%, ** p < 1%, *** p < 0.1%, **** p < 0.01%
For symptomatic patients (n = 653), sensitivity, specificity, and kappa coefficient were 89.4 (95% CI 84.9–93.9), 96.2 (95% CI 94.5–97.9), and 0.86 (95% CI 0.82–0.90) (Table 2). In this group, 293 samples were obtained within the first 3 days of symptom onset, 147 between days 4 and 7, and 51 after day 7. Sensitivity was greater than 92% whatever the date of symptom onset. However, performances were excellent between the 4th and 7th day with sensitivity to 95.3% (95% CI 89–100), specificity to 99% (95% CI 97.2–100), and kappa coefficient to 0.95 (95% CI 0.90–1) (Supplementary Information Table 2).
Table 2.
Overall analytical performances for saliva compared to nasopharyngeal specimens and for symptomatic and asymptomatic patients
| Symptomatic and asymptomatic patients | Symptomatic patients | Asymptomatic patients | |
|---|---|---|---|
| Sensitivity (IC95%) | 87.8 (83.3–92.3) | 89.4 (84.9–93.9) | 79.2 (62.9–95.4) |
| Specificity (IC95%) | 97.1 (96.1–98.1) | 96.2 (94.5–97.9) | 98.0 (96.8–99.2) |
| PPV (IC95%) | 92.3 (88.6–96) | 94.7 (91.3–98.1) | 76.0 (59.3–92.7) |
| NPV (IC95%) | 97.8 (96.9–98.7) | 96.4 (94.7–98.1) | 99.2 (98.4–100.0) |
| Kappa (IC95%) | 0.84 (0.80–0.88) | 0.86 (0.82–0.90) | 0.71 (0.57–0.85) |
NP nasopharyngeal; PPV positive predictive value; NPV negative predictive value
For asymptomatic outpatients who represented only 24 NPS positive patients, sensitivity decreased to 79.2% (95% CI 62.9–95.4) and kappa coefficient to 0.71 (95% CI 0.57–0.85).
The number of inclusions was different according to the center and molecular techniques used, with 857 for TMA and 348 for RT-PCR (p = 0.0044). Performance analyses by each center are shown in Table 3. Center 2 obtained a sensitivity to 66.7% (95% CI [51.9–81.5]) and represented 13 of the 22 (59%) discrepant NPS+/saliva- of the study. The 3 other centers found sensitivity greater than 90%. The overall sensitivity, specificity, and kappa coefficient on saliva for these 3 centers (n = 1023) were 92.8% (95% CI 88.8–96.7), 98.7% (95% CI 96–98.2), and 0.87 (95% CI 0.83–0.91), respectively.
Table 3.
Overall analytical performances for saliva compared to nasopharyngeal specimens and by center and prevalence by center.
| All centers | Center 1 | Center 2 | Center 3 | Center 4 | Centers 1 + 3 + 4 |
|
|---|---|---|---|---|---|---|
| Disease prevalence during study | / | 10,1% | 12,5% | 9,1% | 7,4% | / |
| Sensitivity (CI 95%) | 87.8 (83.3–92.3) | 92.2 (85.6–98.8) | 66.7 (51.9–81.5) | 96.8 (90.5–100.0) | 91.5 (85.1–98.0) | 92.8 (88.8–96.7) |
| Specificity (CI 95%) | 97.1 (96.1–98.1) | 97.7 (96.1–99.4) | 97.2 (94.5–99.9) | 94.8 (91.1–98.6) | 97.3 (95.8–98.9) | 97.1 (96.0–98.2) |
| PPV (CI 95%) | 92.3 (88.6–96) | 95.2 (89.8–100.0) | 86.7 (74.5–98.8) | 90.9 (81.1–100.0) | 92.9 (86.8–98.9) | 93.3 (89.5–97.1) |
| NPV (CI 95%) | 97.8 (96.9–98.7) | 99.0 (97.9–100.0) | 91.4 (87.0–95.9) | 99.2 (97.7–100.0) | 98.8 (97.7–99.8) | 98.9 (98.2–99.6) |
| Kappa (CI 95%) | 0.84 (0.80–0.88) | 0.89 (0.83–0.95) | 0.70 (0.56–0.83) | 0.85 (0.76–0.95) | 0.86 (0.80–0.93) | 0.87 (0.83–0.91) |
NP nasopharyngeal; PPV positive predictive value; NPV negative predictive value
Comparison of SARS-CoV-2 RNA detection between oral and nasopharyngeal swabs
Due to the poor performance of OS, this arm of the study was interrupted at mid-term explaining the lower number of these specimens. Among 771 paired OS and NPS, 80 were positive and 633 negative on both tests. Discrepant results were observed for 58 patients: 51 NPS+/OS- and 7 NPS-/OS+. The detection rate for SARS-CoV-2 was higher in NPS than OS (17%, 131/771 vs 11.2%, 87/771, p < 0.001)
These data on 771 inclusions revealed an overall sensitivity for OS of 61.1% (95% CI 52.7–69.4), a specificity of 98.9% (95% CI 52.7–69.4), a PPV of 91.9%, and a NPV of 92.5%. The kappa coefficient was 0.69 (95% CI 0.62–0.76) (Table 4). The Ct values were significantly higher in OS than in NPS for 5 SARS-CoV-2 genes (p < 0.001). For center 2, the mean of Ct was 26.2 in NPS vs 32 in OS and for center 3, 19.2 in NPS vs 30.2 in OS (p < 0.001).
Table 4.
Overall analytical performances for oral specimens compared to nasopharyngeal specimens
| SARS-CoV-2-RNA | Nasopharyngeal specimens | |||
|---|---|---|---|---|
| Detected | Not detected | Invalid | ||
| Oral self-sampling | Detected | 80 | 7 | 0 |
| Not detected | 51 | 633 | 0 | |
| Invalid | 0 | 0 | 0 | |
For symptomatic patients, sensitivity and kappa coefficient were 63.2% (95% CI 54.3–72) and 0.70 (95% CI 0.62–0.78) while they were 46.7 (95% CI 21.4–71.9) and 0.54 (95% CI 0.30–0.78) for asymptomatic patients (Supplementary Information Table 3).
Analytical performance was analyzed according to the date of onset of symptoms and highlighted a sensitivity of 78% (95% CI 65.4–90.7) in patients with symptoms for less than 3 days and 85.7% (95% CI 72.7–98.7) between 4 and 7 days. After 7 days, none of the 28 positive patients on NPS was detected by OS.
Feasibility and acceptability of nasopharyngeal swab, saliva collection, and oral swab
The instructions for the saliva self-sampling were perceived as simple (90%), but 33.6% of patients had difficulty in filling the collection container. The saliva sample was acceptable for 82.6% of the patients.
For the OS, the instructions were simple for 91% of the patients; this type of sampling was acceptable for 89.2% of them. The discomfort caused was estimated at 0.9 ± 1.6 on a scale from 0 to 10.
Regarding the nasopharyngeal swabs, 13% considered it painful, 48.3% rather painful, and the discomfort caused was estimated at 5.3 ± 2.6 on the same scale.
Finally, to the question “among the 3 samples that you have had (salivary, oral, nasopharyngeal) which would you prefer to have for a COVID-19 screening?”, 44.3% preferred OS, 42.4% saliva, and 13.4% NPS.
Discussion
NPS is the present gold standard of sampling for COVID-19 screening, as recommended by most international guidelines. Indeed, NPS showed the best performances in this study regarding positive rate and provided the highest viral loads. However, saliva seems to be a promising alternative that could simplify and accelerate COVID-19 diagnosis in certain circumstances. In study, carried out on 1220 patients, we showed a comparable rate of SARS-CoV-2 detection between NPS and saliva. When the NPS sample collected by a healthcare worker was used as the comparator, the estimated sensitivity of saliva was 87.8% that is consistent with prior literature [11]. The kappa coefficient showed an excellent agreement (> 0.8). For symptomatic patients, sensitivity was 89.4% (95% CI 84.9–93.9), while it was 79.2% (95% CI 62.9–95.4) for asymptomatic patients. Despite the large number of inclusions, only 24 positive patients were asymptomatic, which may explain the low precision in this group.
Analysis according to the date of symptom onset, 0–3rd day, 4th–7th day, > 7th day, showed sensitivity greater than 92% for each group. The best performances were observed between the 4th and the 7th day with sensitivity to 95.3% and kappa coefficient to 0.95 (95% CI 0.90–1). The viral load is described as high in the first week but could be highest in saliva between the 4th and 7th day after symptoms onset [21, 22]. Our results suggest that saliva could be used during the first week after onset of symptoms for SARS-CoV-2 detection. We showed also good performances after the 7th day of illness in this study. In this group of 51 patients, the median time to onset of symptoms was 11 days.
Unlike studies on posterior oropharyngeal saliva [13, 15], our study revealed that Ct values were significantly lower in NPS than saliva suggesting highest viral load in NPS. Posterior oropharyngeal saliva might have a greater viral load than saliva due to debris from nasopharyngeal epithelium in this sample.
Our study revealed discrepancies between NPS and saliva with 22 NPS+/saliva- and 15 NPS-/saliva+.
For NPS+/saliva-, discrepancy rates by center were 4.6% (3/65), 3.2% (1/31), 6.7% (5/75), and 30.9% (13/42). For the latter center, sensitivity was estimated to 66.7%. Specific analysis revealed amplification of cellular control in NPS and saliva, Ct values to 30.6 for N1 gene, and 29.7 for N2 gene in NPS but no amplification in saliva. This low sensitivity on saliva in center 2 does not seem to be related to the limit of detection (LOD) of the RT-PCR used because it was 12.5 copies/reaction while it was 83 copies/reaction for centers 1 and 4 and 10 copies/reaction for center 3. The final dilution of saliva sample in extraction buffer or lysis buffer was 0.5 for centers 2 and 3 and 0.4 for centers 1 and 4. Finally, the prevalence of the disease in the centers during the study period was between 7.4 and 12.5% and does not explain the difference in performance found in center 2 (Table 3). All these data suggest that extraction technique used in center 2 was probably unsuitable for saliva.
Examination of 6 patients confirmed a history of a previous positive RT-PCR from 2 to 28 days earlier. This discrepancy was previously described for patient later in illness [10].
For NPS-/saliva+ discrepancies, cellular control was systematically positive in NPS in one center excluding the hypothesis of a poor quality NPS sample. Further investigations showed low viral load in saliva in few samples and sometimes negative RT-PCR control in saliva.
Additional analysis excluding the center with poor sensitivity revealed excellent results on saliva, on 1023 patients, with an overall sensitivity to 92.8% and Cohen’s kappa to 0.87.
Finally, saliva sampling was perceived as simple for 90% of patients but a third of patients had difficulty filling the collection container.
Despite good feasibility and correct analytical performances for saliva specimen, results seem to be heterogeneous according to the technique and according to the status of the patient (symptomatic/asymptomatic). A major constraint in the management of saliva samples is linked to a systematic physical and chemical pretreatment for fluidification. In our study, 25.6% of saliva samples were not fluid. To our knowledge, this limit was only found in one recent study [23]. This additional processing causes a workflow modification, a prolonged time of process, and ergonomic implications for the clinical laboratory facing increasing number of samples. A solution, proposed in previous studies, could be the addition of VTM in saliva by the laboratory staff before RT-PCR or TMA [13, 24].
The OS was the preferred technique of patients because of simplicity of implementation and its painless character but analytical performances were not optimal.
Sensitivity was estimated to 61.1% (95% CI 52.7–69.4) in our study. Kojima et al. found comparable results in unsupervised self-collection oral fluid specimens (66%) and suggested that coughing may be critical for the detection of SARS-CoV-2 in oral fluid [16].
There are several limitations in our study. First, patients were essentially outpatients (99%) with mild infection and maybe lower viral loads than severe patients [25]. However, this homogeneity allows to validate our results regarding mass screening population. Furthermore, performances remained good despite patients with low viral loads. Secondly, the number of infected asymptomatic patients was limited (n = 24) so that results in this population are difficult to extrapolate.
Thirdly, the fact that saliva sample was taken systematically before OS could be detrimental for the performance of OS but our data were very closed to that observed in literature and suggest that the change in order of sampling, saliva then OS or OS then saliva, product the same results.
Finally, molecular techniques used were different between centers and could modify study results. Yet, this constraint allows to reflect overall performances in real routine situation and highlights that performances can be impaired with some techniques, which underlines the need for a systematic evaluation of saliva with the local process before routine implementation.
In conclusion, although the oral sample is well accepted by patients, performances are not sufficient. Saliva is an acceptable specimen for the laboratory-based diagnosis of COVID-19 in symptomatic patients during the first week after onset of symptom, but the process requires unfortunately an additional time-consuming step to fluidize or dilute specimen, generating impact on workflow.
Supplementary Information
(DOCX 70 kb)
Acknowledgments
We thank Maryline Vial, Elodie l’Hermitte, Nicolas Yessad, Florence Vilain, Asya Belgrioui, Mireille Moretti, Céline Saliné, Céline Ragot, Christine Bernard, Sarah Bugier, Carine Hejl, Philippe Vest, Rithy Vong, Aurore Rocheteau, Céline Bonnet, Romain Boffy, Giulia Abate, Samuel Lallemand, Landry Bourse, Betty Biteau, Lucie Queval, Nadia Ganouch, Ophélie Brunner, Seta Diakite, Cyrielle Pigeot, David Féry, Carol’Anne Seguin, Delphine Bonnet, Amandine Deroide, Sophie Caron, Paulo Martins, and Mathias David for for their support in performing molecular testing for SARS-CoV-2.
We thank Justine Defrance, Camille Charruau, Celine Tanniou, Amandine Lacaze, Karine Michaud-Gros-Benoit, Damien Ricard, Amandine Steiger, Céline Mizoule, Mona Michaud, Caroline Leiterer, Halima Yssaad, Houria Nait Ammar, Caroline Bedekovic, Julien Blanchard, Audrey Gourden, Carine Malle, Jean-Nicolas Tournier, and Eric Valade for patient’s inclusion, data entry, and methodological support.
Authors’ contributions
Conceptualization: Frédéric Janvier, Catherine Verret, Eric Garnotel, Vincent Foissaud, Audrey Mérens, Julie Plantamura. Methodology: Frédéric Janvier, Catherine Verret, Solenne Martin. Formal analysis : Frédéric Janvier, Solenne Martin, Julie Plantamura, Aurore Bousquet. RT-PCR analysis and investigation: Julie Plantamura, Aurore Bousquet, Marie-Pierre Otto, Christine Bigaillon, Anne-Margaux Legland, Hervé Delacour, Philippe Vest, Hélène Astier, Elodie Valero. Writing—original draft preparation: Frédéric Janvier, Julie Plantamura, Aurore Bousquet, Audrey Mérens. Writing—review and editing: Frédéric Janvier, Audrey Mérens, Vincent Foissaud, Eric Garnotel, Christophe Renard, Olivier Bylicki. Supervision: Frédéric Janvier, Audrey Mérens, Vincent Foissaud, Eric Garnotel, Catherine Verret.
Funding
This work was supported by the French Military Health Service (France).
Data availability
Included in the manuscript.
Code availability
Not applicable.
Declarations
Ethics approval
This study was approved by ethics committee (N° 2020-A02536-33) and referenced on ClinicalTrials (NCT04550390).
Conflict of interest
The authors declare that there is no conflict of interest.
Disclaimer
The views expressed in this article reflect the results of research conducted by the authors and do not necessarily reflect the official policy or position of the French Military Health Service
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (2020) Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/331329
- 2.Centers for Disease Control and Prevention (2020) Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19), 14 April 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
- 3.Fiche de gestion des prélèvements biologiques d’un patient suspect ou confirmé de la COVID-19, Version 6, Septembre 2020. Available at: https://www.sfm-microbiologie.org/wp-content/uploads/2020/09/Fiche-COVID-19_V6.0.pdf
- 4.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19) : current status and future perspectives. Int J Antimicrob Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marty FM, Chen K, Verill KA. How to obtain a nasopharyngeal swab specimen. N Engl J Med. 2020;382(22):e76. doi: 10.1056/NEJMvcm2010260. [DOI] [PubMed] [Google Scholar]
- 6.To KK. Tsang OT, Yip C, Chan KH, Wu TC, Chan JM, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matic N, Stefanovic A, Leung V, Lawson T, Ritchie G, Li Y et al (2020) Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur J Clin Microbiol Infect Dis. 10.1007/s10096-020-04090-5 [DOI] [PMC free article] [PubMed]
- 8.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283e6. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skolimowska K, Rayment M, Jones R, Madona P, Moore L, Randell P. Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect. 2020;26:1711–1713. doi: 10.1016/j.cmi.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li A, Paterson A, et al (2020) Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. Clin Infect Dis ciaa848. 10.1093/cid/ciaa848 [DOI] [PMC free article] [PubMed]
- 11.Czumbel LM, Kiss S, Farkas N, Mandel I, Hegyi A, Nagy A, et al. Saliva as a Candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. 2020;7:465. doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams E, Bond K, Zhang B, Putland M, Williamson D. Saliva as a noninvasive Specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58:e00776–e00720. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W et al (2020) Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 10.1016/j.cmi.2020.05.001 [DOI] [PMC free article] [PubMed]
- 14.Cheuk S, Wong Y, Tse H, Siu HK, Kwong TS, Chu MY et al (2020) Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis. 10.1093/cid/ciaa797 [DOI] [PMC free article] [PubMed]
- 15.To KK. Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:656–674. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto M-P, Darles C, Valero E, Benner P, Dutasta F, Janvier F (2020) Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis. 10.1093/cid/ciaa1181 [DOI] [PMC free article] [PubMed]
- 17.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S et al (2020) Self-Collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. Clin Infect Dis. 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed]
- 18.Tu Y, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020;383:494–496. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avis n° 2020.047/AC/SEAP du 18 septembre 2020 du collège de la Haute Autorité de santé relatif à l’inscription sur la liste des actes et prestations mentionnée à l’article L. 162-1-7 du code de la sécurité sociale, de la détection du génome du virus SARS-CoV-2 par technique de transcription inverse suivie d’une amplification (RT-PCR) sur prélèvement salivaire. https://www.has-sante.fr/jcms/p_3202319/fr/avis-n-2020-047/ac/seap-du-18-septembre-2020-du-college-de-la-haute-autorite-de-sante-relatif-a-l-inscription-sur-la-liste-des-actes-et-prestations-mentionnee-a-l-article-l-162-1-7-du-code-de-la-securite-sociale-de-la-detection-du-genome-du-virus-sars-cov-2-par-technique-de-transcription-inverse-suivie-d-une-amplification-rt-pcr-sur-prelevement-salivaire
- 20.Lee R, Herigon J, Benedetti A, Pollock N, Denkinger C (2020) Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: A systematic review and meta-analysis. J Clin Microbiol. 10.1128/JCM.02881-20 [DOI] [PMC free article] [PubMed]
- 21.Wölfel R, Corman V, Guggemos W, Seilmaier M, Zange S, Müller M, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson K, Barker A, Hillyard D, Gilmore N, Barrett J, Orlandi R, et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020;58:e01824–e01820. doi: 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berenger B, Conly J, Fonseca K, Hu J, Louie T, Schneider A et al (2020) Saliva collected in universal transport media is an effective, simple and high-volume amenable method to detect SARS-CoV-2. Clin Microbiol Infect. 10.1016/j.cmi.2020.10.035 [DOI] [PMC free article] [PubMed]
- 25.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral and dynamics in mild and severe cases of COVID-19. The Lancet. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 70 kb)
Data Availability Statement
Included in the manuscript.
Not applicable.