Abstract
Mass molecular diagnostic testing for the SARS-CoV-2 pandemic has drawn on laboratory developed tests, commercial assays, and fully-automated platforms to accommodate widespread demand. The Alinity m instrument by Abbott is capable of detecting several clinically relevant pathogens and has recently received FDA emergency use authorization for SARS-CoV-2 molecular testing. The Alinity m performs automatic sample preparation, RT-PCR assembly, amplification, detection, and result calculation in under two hours. Here, we validate the performance characteristics of the Alinity m SARS-CoV-2 assay in comparison with the Roche cobas 6800 and Hologic Panther Fusion platforms. Across 178 positive and 195 negative nasopharyngeal swab specimens (CT range 14.30–38.84), the Alinity m detected one additional positive specimen that was found to be negative on the Roche cobas 6800 (PPA 100%, NPA 99.5%). Across a separate set of 30 positive and 174 negative nasopharyngeal swab specimens (CT range 14.1–38.5), the Alinity m had 100% positive and negative agreement with the Hologic Panther Fusion. Using SeraCare SARS-CoV-2 RNA standards, the assay limit of detection was verified to be two-fold more sensitive than the parameters stated by the SARS-CoV-2 AMP kit package insert, at 50 virus copies/mL. Assay specificity was 100% over 20 specimens positive for other respiratory viruses and intraday precision was 100% concordant with <2% CV. These data illst u illustrate the Abbott Alinity m system's high concordance with reference assays and analyti high analytical for SARS-CoV-2 molecular detection.
Keywords: Alinity m, Abbott, SARS-CoV-2, Limit of Detection, Coronavirus, COVID-19
1. Introduction
As SARS-CoV-2 continues to spread, multiple automated molecular diagnostic platforms have received emergency use authorization (EUA) from the US Food and Drug Administration (FDA) in 2020. The benefits of assay automation and standardization are many, such as improved reproducibility, efficiency, and safety [[1], [2], [3], [4], [5], [6], [7]]. However, automated platforms can take considerable time and resources to implement into clinical workflows [[8], [9], [10]].
The Alinity m SARS-CoV-2 assay is intended for the qualitative detection of nucleic acid from nasal swabs, nasopharyngeal (NP) and oropharyngeal (OP) swabs, or bronchoalveolar lavage fluid (BAL). The assay detects two targets, located in the RdRp and N genes of SARS-CoV-2. A sequence unrelated to SARS-CoV-2 (RNA from the hydroxypyruvate reductase gene of Cucurbita pepo, a pumpkin plant) is introduced into each specimen at the beginning of sample preparation. This unrelated RNA sequence is simultaneously amplified by RT-PCR and serves as an internal control (IC) to demonstrate that the process has proceeded correctly for each sample.
Here, we evaluate the clinical performance of the Alinity m SARS-CoV-2 assay (Abbott Laboratories, Chicago, IL, USA) using clinical NP specimens in comparison with reference assays on the cobas 6800 (Roche, Basel, Switzerland) and Panther Fusion SARS-CoV-2 (Hologic, Marlborough, MA) real-time RT-PCR assays [[11], [12], [13]]. To determine the analytical sensitivity for this assay, we ran initial and confirmatory dilutions of previously quantified SARS-CoV-2 RNA standards. To evaluate specificity, a panel of non-SARS-CoV-2 laboratory-confirmed respiratory infections was run on the Alinity m. Assay precision was evaluated utilizing RNA standards in triplicate with appropriate controls over 72 h for each target concentration. Although the Alinity m SARS-CoV-2 assay is qualitative, here we also assess the linearity and PCR efficiency by serial tenfold dilutions of a remnant clinical NP specimen. Finally, we compare two Alinity m instruments side-by-side using positive clinical specimens over a range of SARS-CoV-2 viral loads.
2. Materials and methods
2.1. Specimen collection and panel selection
NP specimens in viral transport media (VTM) were sent to the University of Washington (UW) Clinical Virology Laboratory for SARS-CoV-2 RT-PCR testing, and de-identified excess sample was used to evaluate platform clinical concordance [14,15]. This work was approved under a consent waiver by the University of Washington institutional review board. 577 remnant clinical NP specimens (208 positives, 369 negatives) were compared to reference SARS-CoV-2 assays on the Roche cobas 6800 and Panther Fusion real-time RT-PCR platforms. Positive cycle thresholds (CTs) from the cobas 6800 ranged from 14.30 to 38.84 while Panther Fusion panel CTs ranged from 14.1 to 38.5.
For analytical sensitivity, recombinant virus containing SARS-CoV-2 RNA (SeraCare Life Sciences, Milford, MA, USA) was used as a positive control at a stock concentration of 1000 copies/mL. Assay calibrators at 1000 copies/mL, 200 copies/mL, 100 copies/mL, 50 copies/mL, and 25 copies/mL were prepared based on dilutions of the positive control in dH2O and tested in triplicate for initial analytical sensitivity. Confirmatory limit of detection experiments were performed using 20 replicates at concentrations of 100 copies/mL, 50 copies/mL, and 25 copies/mL. Twenty non-SARS-CoV-2 respiratory NP specimens with CTs ranging 17.5–37.6 from an in-house respiratory panel were used to evaluate assay specificity.
For precision, triplicates of the SeraCare RNA standards were run at 1000 copies/mL and 200 copies/mL with He-La cells serving as a negative control. Mean cycle number (CN), standard deviation (SD), and percent coefficient of variation (%CV) were recorded over 3 days at each target concentration. Of note, the Alinity platform's CN is analogous to a standard PCR's CT. Linearity and PCR efficiency was evaluated with a remnant clinical NP specimen serially diluted 10-fold. For instrument comparisons, two separate Alinity m platforms were tested with a panel of remnant clinical NP samples (n = 32) with a CT range of (12.34–37.70). CT shifts at 40 cycles were calculated based on Passing-Bablok linear regression analyses.
3. Results
3.1. Clinical concordance and platform comparison
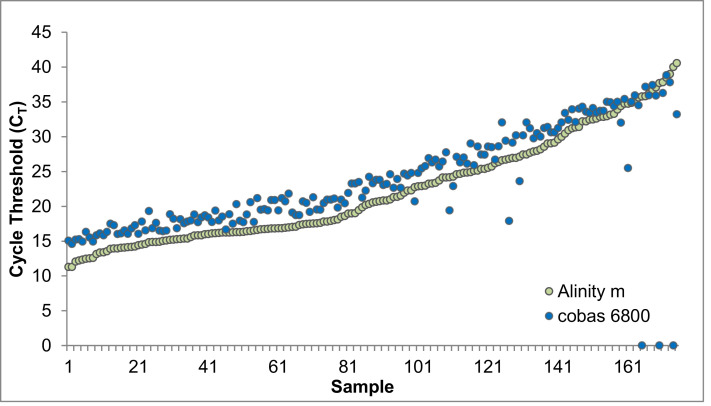
Using 373 remnant clinical NP specimens (178 positive and 195 negative) assayed on the Roche cobas 6800 platform, the clinical performance of the Alinity m SARS-CoV-2 assay was evaluated (Table 1 , Supp. Data 1). The accuracy and percent agreement between the candidate (Alinity m) and reference method (cobas 6800) were calculated as follows: overall percent agreement = 99.7%, PPV = 99%, NPV = 100%, PPA = 100%, and NPA = 99.5%. Individual CTs for all 178 positive specimens run on the cobas 6800 were plotted against mean CTs from the two Alinity m instruments (Fig. 1 ).
Table 1.
Abbott Alinity m Clinical Concordance with Roche cobas 6800 and Hologic Panther Fusion SARS-CoV-2 Assay.
| Roche cobas 6800 | Hologic Panther Fusion | ||||||
| Positive | Negative | Total | Positive | Negative | Total | ||
| Positive | 83 | 0 | 83 | 30 | 0 | 30 | Abbott Alinity m #1 |
| Negative | 0 | 106 | 106 | 0 | 90 | 90 | |
| Total | 83 | 106 | 189 | 30 | 90 | 120 | |
| Positive | Negative | Total | Positive | Negative | Total | ||
| Positive | 94 | 1 | 95 | 30 | 0 | 30 | Abbott Alinity m #2 |
| Negative | 0 | 89 | 89 | 0 | 84 | 84 | |
| Total | 94 | 90 | 184 | 30 | 84 | 114 | |
The positive percent agreement (PPA) and for both Alinity m instruments was reported at 100% compared to the cobas 6800 SARS-CoV-2 assay. The negative percent agreement (NPA) was determine at 100% and 98.9% for instrument #1 and #2, respectively. In comparison to the Panther Fusion SARS-CoV-2 assay, both PPA and NPA were calculated at 100%. Line-item CN values for all samples on each platform are included in Supplemental Data 1.
Fig. 1.
Alinity m and cobas 6800 SARS-CoV-2 Cycle Threshold Comparison of Positive Samples
Alinity m positive cycle threshold (CT) values compared to cobas 6800 positive sample CTs. 178 positive samples were assayed with one discordant sample not detected on the cobas 6800 but was detected on the Alinity m with a CT of 40.02. Two other samples are inconclusive and would be resulted as a positive by the cobas 6800, however they are displayed not detected here as these CT values are plotted from the E-gene target.
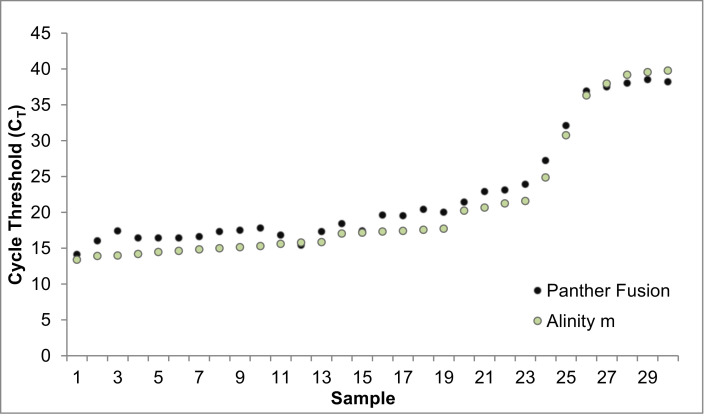
The clinical performance of the Alinity m SARS-CoV-2 assay was also compared to the Hologic Panther Fusion SARS-CoV-2 assay. 204 remnant clinical NP specimens (30 positive and 174 negative) were assayed (Table 1). Overall percent agreement, PPV, NPV, PPA, and NPA were all calculated at 100% accuracy for the candidate method. Individual CTs from the 30 positive specimens assayed on the Panther Fusion were plotted against mean CTs from both Alinity m platforms (Fig. 2 ).
Fig. 2.
Alinity m and Panther Fusion SARS-CoV-2 Cycle Threshold Comparison of Positive Samples
Alinity m positive cycle threshold (CT) values compared to Panther Fusion positive sample CTs. Across 30 positive nasopharyngeal specimens with a range of viral titers assayed on the Panther Fusion, 100% were detected on the Alinity m SARS-CoV-2 assay.
3.2. Analytical sensitivity
The initial estimation of LoD was performed in triplicate at 1000 copies/mL, 200 copies/mL, 100 copies/mL, 50 copies/mL, and 25 copies/mL using SeraCare RNA standards of recombinant virus containing SARS-CoV-2 (Table 2 , Supp. Data 2). Confirmation of the LoD was performed at 100 copies/mL and 50 copies/mL with 20 replicates for each instrument. A total of 110 contrived samples ranging from 50 to 1000 virus copies/mL were run to evaluate the analytical sensitivity of the assay on two separate Alinity m instruments.
Table 2.
Abbott Alinity m SARS-CoV-2 Assay Analytical Sensitivity.
| Analytical Sensitivity | Virus Copies/mL | Replicates | % Positive | Mean CN | Instrument |
| Initial | 1000 | 3 | 100% | 32.11 | Alinity m #1 |
| 200 | 3 | 100% | 34.77 | ||
| 100 | 3 | 100% | 35.36 | ||
| 50 | 3 | 100% | 37.11 | ||
| 25 | 3 | 100% | 38.71 | ||
| Confirmatory | |||||
| 100 | 20 | 100% | 35.80 | ||
| 50 | 20 | 100% | 36.91 | ||
| Initial | 1000 | 3 | 100% | 33.08 | Alinity m #2 |
| 200 | 3 | 100% | 35.40 | ||
| 100 | 3 | 100% | 35.82 | ||
| 50 | 3 | 100% | 37.11 | ||
| 25 | 3 | 67% | 38.17 | ||
| Confirmatory | 100 | 20 | 100% | 36.38 | |
| 50 | 20 | 100% | 37.12 | ||
| 25* | 20 | 60% | 39.18 |
Abbreviations: CN, cycle number.
*The twenty replicates for confirmatory sensitivity at 25 virus copies/mL was run only on a single Alinity m instrument due to reagent/consumable constrictions. Notably, the congruency between both instruments is adequate. The limit of detection for both Alinity m instruments with an observed positivity rate >95% was confirmed at 50 virus copies/mL using SeraCare RNA standards. Confirmatory line-item CN data is described in Supplemental Data 2.
Further assay sensitivity was interrogated after the Alinity platform detected all contrived samples below the manufacturer's claimed LoD at 100 virus copies/mL. Twenty-three additional samples, at 25 virus copies/mL, were assayed to confirm an empirical limit of detection on a single Alinity m due to reagent and consumable constrictions. The mean CN for the manufacturer declared 100 copies/mL concentration was determined at 36.09. However, the Alinity m was able to detect >95% of samples below this threshold, with a confirmed empirical limit of detection of 50 virus copies/mL, with a mean CN of 37.02.
3.3. Assay specificity
Non-SARS-CoV-2 infections from patient NP swabs (n = 20), laboratory-confirmed by UW Virology's laboratory-developed respiratory panel test, were evaluated for assay specificity [[16], [17], [18]]. Specimens included infection with adenovirus (n = 2), bocavirus (n = 2), non-SARS-CoV-2 coronavirus ( n = 5), influenza (n = 3), human metapneumovirus (n = 1), parainfluenza (n = 3), respiratory syncytial virus (n = 2), and rhinovirus (n = 2). Respiratory panel CT ranges for laboratory-confirmed respiratory infections spanned from 17.5 to 37.6. All respiratory infection samples were negative for SARS-CoV-2 when tested on the Alinity m.
3.4. Assay precision
RNA standards were run daily at 1000 copies/mL, 200 copies/mL, and with a negative control - He-La cells - over the course of 3 days. The mean CN, SD, and%CV were calculated at each target concentration (Table 3 A). All positive dilutions were detected with <2% CV and 100% concordance based on qualitative interpretation for both Alinity m instruments. All negative controls did not amplify.
Table 3.
A. Abbott Alinity m SARS-CoV-2 Assay Precision.
| Concentration | Day | CN | Mean CN | SD | %CV | Instrument | ||||
| 1000 virus copies/mL | 1 | 33.48 | 33.0 | 0.5 | 1.50% | Alinity m #1 | ||||
| 2 | 33.10 | |||||||||
| 3 | 32.48 | |||||||||
| 200 virus copies/mL | 1 | 35.18 | 35.4 | 0.2 | 0.60% | |||||
| 2 | 35.48 | |||||||||
| 3 | 35.55 | |||||||||
| Negative | 1 | NDET | N/a | N/a | N/a | |||||
| 2 | NDET | |||||||||
| 3 | NDET | |||||||||
| 1000 virus copies/mL | 1 | 33.41 | 33.1 | 0.3 | 0.80% | Alinity m #2 | ||||
| 2 | 33.07 | |||||||||
| 3 | 32.92 | |||||||||
| 200 virus copies/mL | 1 | 36.22 | 35.5 | 0.7 | 1.90% | |||||
| 2 | 34.99 | |||||||||
| 3 | 35.16 | |||||||||
| Negative | 1 | NDET | N/a | N/a | N/a | |||||
| 2 | NDET | |||||||||
| 3 | NDET | |||||||||
| Table 3B. UW LDT SARS-CoV-2 Assay Precision Comparison | ||||||||||
| Concentration | Day | N1 CN | N2 CN | N1 Mean FCN | N2 Mean FCN | N1 SD | N2 SD | N1%CV | N2%CV | Extraction Instrument |
| 1000 virus copies/mL | 1 | 34.19 | 35.28 | 34.65 | 35.16 | 0.40 | 0.17 | 1.16 | 0.48 | MagNA Pure 96 #1 |
| 2 | 34.95 | 34.97 | ||||||||
| 3 | 34.80 | 35.23 | ||||||||
| 200 virus copies/mL | 1 | 36.81 | 38.02 | 36.36 | 37.34 | 1.82 | 0.96 | 5.00 | 2.57 | |
| 2 | 34.36 | 36.66 | ||||||||
| 3 | 37.91 | NDET | ||||||||
| Negative | 1 | NDET | NDET | N/a | N/a | N/a | N/a | N/a | N/a | |
| 2 | NDET | NDET | ||||||||
| 3 | NDET | NDET | ||||||||
| 1000 virus copies/mL | 1 | 33.76 | 35.79 | 33.89 | 35.44 | 0.32 | 0.51 | 0.95 | 1.44 | MagNA Pure 96 #2 |
| 2 | 33.66 | 35.67 | ||||||||
| 3 | 34.26 | 34.86 | ||||||||
| 200 virus copies/mL | 1 | 36.42 | 36.70 | 37.07 | 37.41 | 0.63 | 0.97 | 1.70 | 2.59 | |
| 2 | 37.11 | 37.02 | ||||||||
| 3 | 37.68 | 38.51 | ||||||||
| Negative | 1 | NDET | NDET | N/a | N/a | N/a | N/a | N/a | N/a | |
| 2 | NDET | NDET | ||||||||
| 3 | NDET | NDET | ||||||||
Abbreviations: CN, cycle number; SD, standard deviation,%CV, percent coefficient of variance, NDET, not detected; N/a, not applicable.
Different sample concentrations and a negative control (He-La cells) were run in triplicate over 72 h to evaluate SARS-CoV-2 assay precision between two separate Alinity m instruments.
Abbreviations: UW, University of Washington; LDT, laboratory developed test; CN, cycle number; SD, standard deviation,%CV, percent coefficient of variance, NDET, not detected; N/a, not applicable.
Different sample concentrations and a negative control (He-La cells) were run in triplicate over 72 h to evaluate the Alinity m SARS-CoV-2 assay precision in comparison to UW's LDT.
The Abbott Alinity m SARS-CoV-2 assay precision was then compared to UW Virology's Laboratory Developed Test (LDT which received EUA authorization from the Washington State Department of Health on March 18, 2020). The same RNA standards were diluted to 1000 virus copies/mL and 200 virus copies/mL, run over 3 days on two separate MagNA Pure 96 extraction instruments (Roche, Basel, Switzerland), and amplified on 7500 PCR systems (Applied Biosystems, Foster City, CA, USA) as previously described [14,15]. All positive dilutions of the SeraCare standards were detected by the MagNA Pure 96 extractions except one sample at 200 copies/mL on day 3 for MagNA Pure 96 instrument #1. The N1 target amplified with a CT of 37.91, and the N2 target was not detected, resulting in an inconclusive that would be reported as a positive (Table 3B). All other positive dilutions were detected with <3% CV.
3.5. Linearity and PCR efficiency
Remnant clinical NP swabs were serially diluted 10-fold to evaluate assay linearity and PCR efficiency (Supp. Data 3). The most diluted samples (1:10×105) were omitted from the PCR efficiency calculation. The assay is linear from a CN/CT of approximately 23 - 37 with an R2 value > 0.99. PCR efficiency was calculated at 87.4%, respectively. Notably, the Alinity m assay is qualitative so does not require a quantitative calculation of linearity and PCR efficiency.
3.6. Instrument comparison
To compare the Alinity m instruments #1 and #2, a panel of 32 remnant clinical NP swabs with a range of CTs (12.34–37.70) was tested on each instrument. The CT shift was calculated based on Passing-Bablok Linear Regression analyses where the X-axis comprised CT values from Instrument #1, and the Y-axis corresponds with CT values from Instrument #2 (Supp. Data 4). A Two-tailed test for linearity was performed, and as the computed p-value is greater than the significance level of α = 0.05, the relationship between the two variables can be considered linear. Using the regression analysis equation of Y = A+BX, a comparative CT value [X;40] was calculated as 40 = A +BX. For 32 observations, Y = −0.184 + 0.995B comparative CT values for instrument #1 and #2 are [40.4;40], respectively. In Silico performance analysis determined a 0.4 CT difference between the two instruments at 40 cycles.
4. Discussion
Here, we validate the Abbott Alinity m SARS-CoV-2 assay and find performance characteristics superior to the parameters stated in the AMP kit package insert. Previous work has evaluated the Alinity m for viral assays such as HCV, HIV, and HPV, though, no research to our knowledge has characterized and validated the Alinity m SARS-CoV-2 assay performance [[19], [20], [21], [22], [23]]. The PPA and NPA for both comparator groups (cobas 6800 and Panther Fusion assays) were calculated at >95% expected correlation as outlined by the FDA EUA minimum requirement for assay validation. Moreover, the PPA compared with both reference assays was calculated at 100%, indicating the Alinity m SARS-CoV-2 assay has equivalent clinical performance to verified and established assays.
Importantly, the Alinity m instruments were able to detect all inconclusive samples from the cobas 6800 platform (n = 7). Inconclusive samples typically correspond with very low viral loads (i.e. late CTs); the mean and median CTs from the seven inconclusive samples were calculated at 36.82 and 37.7, respectively, when detected on the Alinity m (Supp. Data 1). Notably, one sample that was indicated negative for both targets by cobas 6800 was detected by the Alinity m SARS-CoV-2 assay with a CT of 40.02.
According to work done by Zhen et al., the Panther Fusion assay has an LoD of CT 35.6, corresponding with 86 copies/mL (Panther Fusion - https://www.fda.gov/media/136156/download) [24]. The cobas 6800 SARS-CoV-2 assay has an LoD of CT 32.7 and 36.4 for ORF1a and E-gene, or 46 copies/mL for both targets (Cobas 6800 - https://www.fda.gov/media/136049/download). In comparison, Abbott reports the Alinity m SARS-CoV-2 AMP Kit's limit of detection at 100 virus copies/mL according to their package insert (https://www.fda.gov/media/137979/download). However, when quantified with RNA standards, both Alinity m instruments were confirmed to detect >95% of positive specimens at 50 virus copies/mL (with a mean CT 37.02), comparable to other FDA EUA reference assays. At 25 virus copies/mL, 60% of the 20 replicates were detected. It is also worth noting that all samples tested were transported in VTM as phosphate buffered saline is not currently an approved specimen type on the instrument.
Here, the Alinity m SARS-CoV-2 assay by Abbott has been validated to be accurate, specific, and more sensitive than the AMP kit package insert declared, with an empirical limit of detection determined at 50 virus copies/mL. The instrument's sensitivity permits detection of very low viral load specimens that may be missed by other platforms. The clinical concordance to reference assays was greater than 95% NPA and PPA when compared to the cobas 6800 and Panther Fusion. In summary, the Abbott Alinity m SARS-CoV-2 assay performance was superior to the parameters stated in the SARS-CoV-2 AMP kit package insert and equivalent to other FDA EUA molecular assays.
Declaration of Competing Interest
ALG reports contract funding from Abbott and Gilead for testing, research funding from Merck, all outside of the submitted work. Abbott had no role in the design or execution of the study.
Acknowledgments
We would like to thank Jared Castor and Aria Bovell for thoughtful discussion and assistance with clinical data. This research was funded from departmental funds from the Department of Laboratory Medicine and Pathology.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.104869.
Appendix. Supplementary materials
References
- 1.Lippi G., Da Rin G. Advantages and limitations of total laboratory automation: a personal overview. Clin. Chem. Lab. Med. 2019;57:802–811. doi: 10.1515/cclm-2018-1323. (CCLM) [DOI] [PubMed] [Google Scholar]
- 2.Ialongo C., Porzio O., Giambini I., Bernardini S. Total automation for the core laboratory: improving the turnaround time helps to reduce the volume of ordered STAT tests. J. Lab. Autom. 2016;21:451–458. doi: 10.1177/2211068215581488. [DOI] [PubMed] [Google Scholar]
- 3.Dolci A., Giavarina D., Pasqualetti S., Szőke D., Panteghini M. Total laboratory automation: do stat tests still matter? Clin. Biochem. 2017;50:605–611. doi: 10.1016/j.clinbiochem.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Zaninotto M., Plebani M. The “hospital central laboratory”: automation, integration and clinical usefulness. Clin. Chem. Lab. Med. 2010;48 doi: 10.1515/CCLM.2010.192. [DOI] [PubMed] [Google Scholar]
- 5.Dharavath B., Yadav N., Desai S., Sunder R., Mishra R., Ketkar M., Bhanshe P., Gupta A., Redhu A.K., Patkar N., Dutt S., Gupta S., Dutt A. A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2. Heliyon. 2020;6:e04405. doi: 10.1016/j.heliyon.2020.e04405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann S. Lab automation in the microbiology lab: an ongoing journey, not a tale? J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02592-20. JCM02592-20, jcm;JCM.02592-20v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culbreath K., Piwonka H., Korver J., Noorbash M. Benefits derived from full laboratory automation in microbiology: a tale of four laboratories. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01969-20. JCM.01969-20, jcm;JCM.01969-20v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell S.L., George K St., Rhoads D.D., Butler-Wu S.M., Dharmarha V., McNult P., Miller M.B. Understanding, verifying, and implementing emergency use authorization molecular diagnostics for the detection of SARS-CoV-2 RNA. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00796-20. e00796-20 jcm/58/8/JCM.00796-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 10.Archetti C., Montanelli A., Finazzi D., Caimi L., Garrafa E. Clinical laboratory automation: a case study. J. Public Health Res. 2017;6(1):881. doi: 10.4081/jphr.2017.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro. Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., KR Jerome, Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00821-20. e00821-20jcm/58/8/JCM.00821-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. Detection of SARS-CoV-2 by use of the cepheid xpert xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00772-20. e00772-20, jcm/58/8/JCM.00772-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalla A.K., Casto A.M., Huang M.L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., KR Jerome, Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00557-20. JCM00557-20, jcm;JCM.00557-20v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perchetti G.A., Nalla A.K., Huang M.L., Zhu H., Wei Y., Stensland L., Loprieno M.A., Jerome K.R., Greninger A.L. Validation of SARS-CoV-2 detection across multiple specimen types. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuypers J., Wright N., Corey L., Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J. Clin. Virol. 2005;33:299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuypers J., Wright N., Ferrenberg J., Huang M.L., Cent A., Corey L., Morrow R. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuypers J., Martin E.T., Heugel J., Wright N., Morrow R., Englund J.A. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119 doi: 10.1542/peds.2006-1406. e70-e76. [DOI] [PubMed] [Google Scholar]
- 19.Maree L., Krügel M., Reinhardt B., Glass A.J. Evaluation of the Alinity m HIV-1 assay for the quantification of HIV-1 RNA plasma viral load in a high-throughput molecular laboratory in South Africa. J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104644. [DOI] [PubMed] [Google Scholar]
- 20.Braun P., Glass A., Maree L., Krügel M., Pacenti M., Onelia F., Gunson R., Goldstein E., Martínez-García L., Galán J.C., Vilas A., D'costa J., Sameer R., Ehret R., Knechten H., Naeth G., Bouvier-Alias M., Marlowe N., Palm M.J., Joseph A.M., Dhein J., Reinhardt B., Pfeifer K., Lucic D., Obermeier M. Multicenter clinical comparative evaluation of Alinity m HIV-1 assay performance. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104530. [DOI] [PubMed] [Google Scholar]
- 21.Chevaliez S., Onelia F., Pacenti M., Goldstein E., Galán J.C., Martínez-García L., Vilas A., Glass A., Maree L., Krügel M., Ehret R., Knechten H., Braun P., Naeth G., Bonanzinga S., Jackson K., Abravaya K., Dhein J., Huang S., Joseph A.M., Lucic D., Marlowe N., Palm M.J., Pfeifer K., Toolsie D., Reinhardt B., Obermeier M., Gunson R. Multicenter clinical evaluation of Alinity m HCV assay performance. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104531. [DOI] [PubMed] [Google Scholar]
- 22.Oštrbenk Valenčak A., Šterbenc A., Seme K., Poljak M. Alinity m HR HPV assay fulfills criteria for human papillomavirus test requirements in cervical cancer screening settings. J. Clin. Microbiol. 2019;58 doi: 10.1128/JCM.01120-19. e01120-19, jcm/58/1/JCM.01120-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouna L., Pallier C., Proust S., Prégermain C., Roque-Afonso A.M. Comparison of the Abbott Alinity m and m2000 assays for the quantification of HIV-1, HCV and HBV in clinical samples. J. Clin. Virol. 2020;126 doi: 10.1016/j.jcv.2020.104331. [DOI] [PubMed] [Google Scholar]
- 24.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00743-20. e00743-20, jcm/58/8/JCM.00743-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.