Abstract
Despite ongoing (macro)pinocytosis of extracellular fluid, the volume of the endocytic pathway remains unchanged. To investigate the underlying mechanism, we used high-resolution video imaging to analyze the fate of macropinosomes formed by macrophages in vitro and in situ. Na+, the primary cationic osmolyte internalized, exited endocytic vacuoles via two pore channels (TPC), accompanied by parallel efflux of Cl− and osmotically-coupled water. The resulting shrinkage caused crenation of the membrane which fostered recruitment of curvature-sensing proteins. These proteins stabilized tubules and promoted their elongation, driving vacuolar remodeling, receptor recycling, and resolution of the organelles. Failure to resolve internalized fluid impairs the tissue surveillance activity of resident macrophages. Thus, osmotically-driven increases in the surface-to-volume ratio of endomembranes promote traffic between compartments and help to ensure tissue homeostasis.
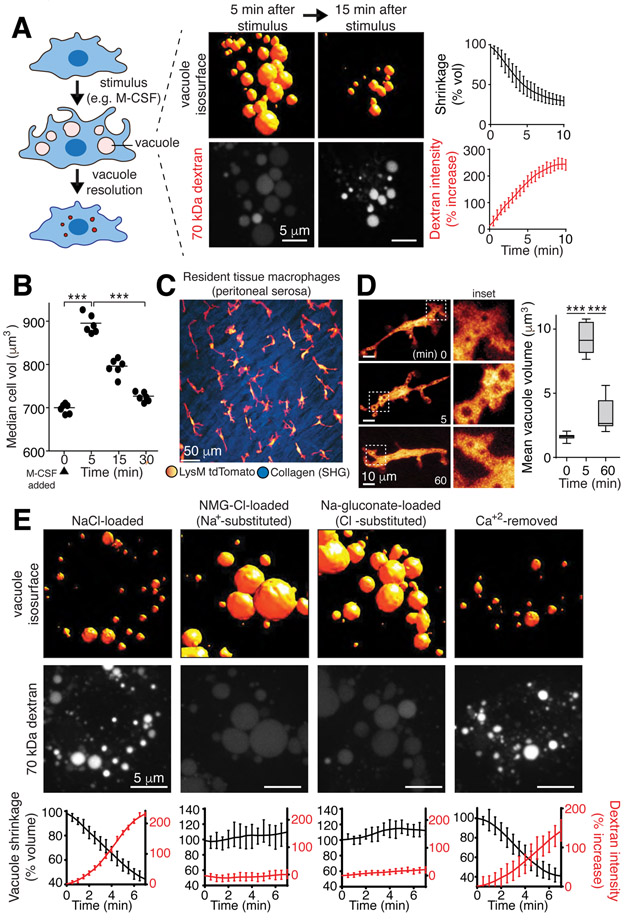
During endocytosis, cells internalize membrane along with extracellular fluid (pinocytosis). The amount of fluid ingested can be substantial: dendritic cells and macrophages take up the equivalent of their entire cellular volume every 4 hours (1). Despite continuous uptake of large amounts of water and osmolytes, the endocytic pathway and the cells as a whole retain their volume and ionic composition over extended periods (1). To investigate endomembrane volume and ionic regulation, we chose macropinosomes, large (up to 5 μm) vacuoles formed by specialized cell types (2, 3) (Figs. 1, S1A-B). Unlike smaller endocytic vesicles, macropinosomes can be resolved by diffraction-limited microscopy (4), enabling detailed assessment of their volume as they mature. Moreover, the volume of medium entrapped by macropinosomes is sufficiently large to alter the overall ionic composition of the cells (Fig. S1C-D). When stimulated by macrophage colony stimulating factor (M-CSF), bone marrow-derived macrophages (BMDM) underwent a large burst of macropinocytosis. Multiple (10-15/cell), large vacuoles (mean volume 7 μm3) form within 5 min (Figs. 1A, S1A-B). The volume of fluid internalized was equivalent to ≈25% of the cell volume, an increase detectable by electronic cell sizing (Fig. 1B). When visualized using rhodamine-dextran (70 kDa), the macropinosomes of BMDM (Fig. 1A), as well as those formed by peritoneal macrophages and human monocyte-derived macrophages (Fig. S1E), resorbed within 30 min and cell volume returned to basal levels (Fig. 1B). Shrinkage of macropinocytic vacuoles was also observed in vivo. Two-photon imaging of live mice revealed that resident tissue macrophages (RTM) of the peritoneal serosa –interstitial, non-migratory cells that constitutively sample the surrounding milieu ((5), Video S1)– formed large macropinosomes that subsequently contracted (Fig. 1C-D and Video S2).
Figure 1. Vacuolar shrinkage requires monovalent ion efflux.
A, Volume and 70 kDa rhodamine-dextran fluorescence intensity changes of macropinosomes induced in bone marrow derived macrophages (BMDM) by macrophage colony-stimulating factor (M-CSF); data are means ± SEM of >100 vacuoles from 3 independent experiments (i.e. n=3). Measurement of vacuole resolution was initiated after a 5-min stimulation with M-CSF in medium containing dextran, followed by an immediate wash. B, Cell (BMDM) volume was measured electronically before and at the indicated times after M-CSF stimulation; >104 cells per point, n=3. p values determined by unpaired, two-sided t-tests. Here and elsewhere *** indicates p<0.001, ** is p<0.01 and * is p<0.05. C, Intravital observation of td-Tomato-labeled resident tissue macrophages (RTM; pseudocolored yellow/red) of the peritoneal serosa and second harmonic imaging (SHG) of collagen (blue). See also Videos S1 and S2. D, Visualization and volume quantification of M-CSF-induced macropinosomes in RTM in vivo; means, upper and lower quartiles (boxes), and distribution (whiskers) are graphed. >50 vacuoles, n=3. p values determined by Mann-Whitney U test. E, Macropinosomes of M-CSF-stimulated BMDM in media containing indicated solutes and dextran. Representative images acquired at 5 min. See also Video S3. Bottom row: mean ± SEM macropinosomal volume and dextran intensity from 3 independent video recordings representing >150 macropinosomes. See also Fig. S1.
Macropinosome shrinkage was accompanied by an increase in the intensity of the luminal dextran fluorescence (Fig. 1A), implying that fluid was extracted from the vacuoles. This suggested that the volume loss of the vacuoles is caused by osmotically-driven solvent loss. Na+ and Cl− constitute the majority of the osmolytes in the fluid engulfed during macropinocytosis. Accordingly, inducing macropinocytosis with M-CSF resulted in a 4-fold increase in the total cellular Na+ content (Fig. S1D). Thus, loss of Na+ and Cl−, along with osmotically-coupled water may underlie the rapid shrinkage of macropinosomes. This was validated by ion substitution experiments: replacing Na+ for the impermeant cation, N-methyl-D-glucamine+ (NMG+), virtually eliminated macropinosome resolution (Figs. 1E, S1E-F, and Video S3). Similarly, shrinkage was precluded when substituting the impermeant anion gluconate− for Cl−, implying that electroneutrality needs to be maintained during solute export (Figs. 1E, S1E-F). Preventing monovalent ion efflux from macropinosomes also prevented restoration of the cell volume (Fig. S1G). The absence of luminal Ca2+ did not prevent macropinosome resolution (Fig. 1E).
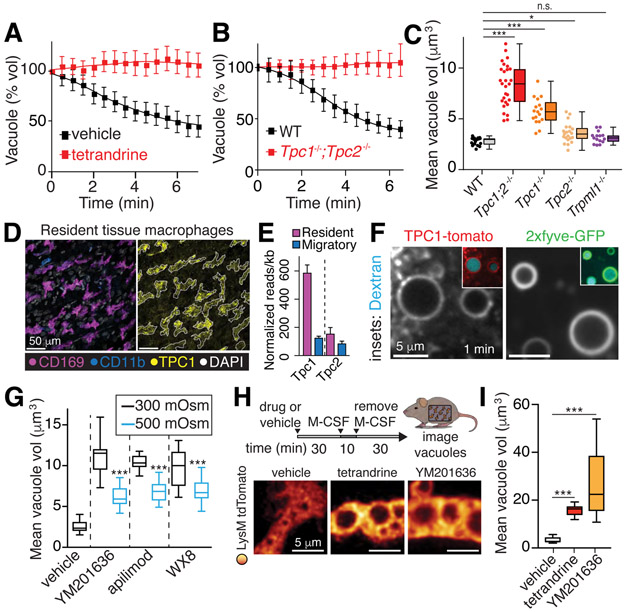
We tested a series of ion transport inhibitors to gain insight into the pathways involved in macropinosome shrinkage. Tetrandrine, a potent inhibitor of two-pore channels (TPC) (6), impaired the volume loss (Figs. 2A, S2A-B; Video S4). Interestingly, the endomembrane isoforms TPC1 and TPC2 are expressed at particularly high levels in myeloid cells, including BMDM (Fig. S6I and (7)). Moreover, TPC1 is highly expressed in the macropinocytic interstitial (Ccr2-negative, CD169-positive) RTM, compared to neighboring stromal or migratory (Ccr2-positive, CD169-negative) myeloid cells that are non-macropinocytic (Fig. 2D-E) (5). While undetectable at the plasma membrane, TPC1 was rapidly (≤1 min) acquired by nascent macropinosomes (Fig. 2F), while TPC2 was recruited later (Fig. S3D). BMDM from Tpc1;Tpc2 double-knockout mice formed large macropinosomes when stimulated by M-CSF, but these did not shrink and resolve during the course of our analyses (Fig. 2B-C). Using single knock-out mice and RNA interference, we discerned this effect to be attributable primarily to TPC1 (Figs. 2C, S3F-G).
Figure 2. Monovalent ion efflux mechanisms.
A, Macropinosome volume changes in presence of 5 μM tetrandrine, measured in BMDM. Measurement of vacuole resolution was initiated once cells were washed after a 5-min stimulation with M-CSF in medium containing dextran; tetrandrine or vehicle were present throughout. Means ± SEM, n=3. See also Fig. S2 and Video S4. B-C, Macropinosome volume changes following stimulation with M-CSF in wt, Tpc1/Tpc2 single and double KO, and Trpml1 KO BMDM. In C, means, upper and lower quartiles (boxes), distribution (whiskers), and observations from fields each containing 3-5 cells (dots) measured 10 min after macropinosome formation; n=3. D, Staining of the peritoneal serosa. Outline of CD169 signal (left) overlaid on TPC1 signal (right). E, RNA-seq. Resident tissue macrophages were Cx3cr1−/Ccr2−. Migratory cells were Cx3cr1−/Ccr2+. F, BMDM expressing TPC1-tomato or 2xfyve-GFP to detect PtdIns(3)P. Dextran shown in cyan. G, BMDM stimulated with M-CSF in the presence and 70 kDa rhodamine-dextran and, where indicated, the PIKfyve inhibitors YM201636, apilimod, or WX8 (all used at 500 nM). Resolution was recorded as in A. 5 min after iso-osmotic recording, a hyperosmotic solution (final 500 mOsm) was added to verify the osmotic responsiveness of the vacuoles. H-I, Visualization and volume quantification of macropinosomes in RTM treated in situ with YM201636 (500 nM) or tetrandrine (5 μM); >15 cells, n=3. All p values determined by Mann-Whitney U tests.
Certain ion channels, including TPCs and TRPMLs, require PtdIns(3,5)P2 for activation (8, 9); this phosphoinositide is generated by phosphorylation of PtdIns[3]P by PIKfyve (10). Consistent with this sequence, PtdIns(3)P and PIKfyve itself were readily detectable on the cytosolic leaflet of nascent macropinosomes (Fig. 2F, S3E). 2xMLN-GFP, a putative probe for PtdIns(3,5)P2 (11) was also found in macropinosomes (Fig. S3E). Macropinosome shrinkage –whether measured directly or assessed indirectly from the overall cell volume gain– was blocked by PIKfyve antagonists (Figs. 2G-I, S1L-N). Inhibiting PIKfyve did not alter the water permeability or pliability of the membrane as indicated by the acute volume loss induced by water abstraction caused by hypertonic medium (Fig. S1K-L). A similar response to hypertonicity was observed in macropinosomes formed in Na+-free solution (Fig. S1J). Despite the fact that TRPML1 channels are recruited to maturing macropinosomes, deletion of the Trpml1 gene was without effect on macropinosome resorption (Figs. 2C, S3D).
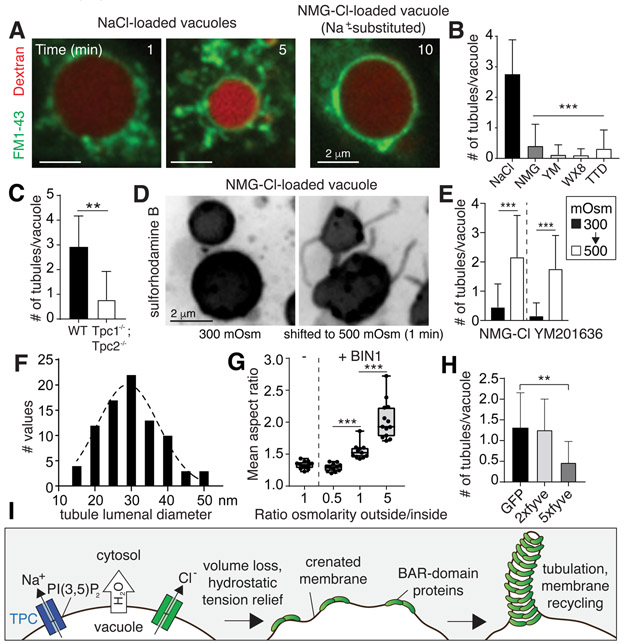
The area of the vacuolar membrane was reduced during shrinkage by emission and severing of tubules and vesicles, which were visualized using FM1-43 (Fig. 3A) or sulforhodamine B (Fig. 3D). Tubule extension accompanies and requires the volume loss that is driven by the export of ions and osmotically-coupled water. Accordingly, substitution of Na+ with NMG+, or blockade of TPC channels by tetrandrine or their inactivation by depleting PtdIns(3,5)P2 impaired tubulation and vesiculation (Fig. 3A-B). Genetic deletion of TPC channels also precluded tubulation (Fig. 3C). Applying a hypertonic solution to macropinosomes that initially failed to shrink because they were loaded with NMG+ or formed in cells treated with a PIKfyve inhibitor revealed that tubulation is a consequence not a cause of volume loss (Figs. 3D-E and Video S5). The tubules emanating from early macropinosomes are remarkably thin, with a modal diameter of ≈30 nm (Fig. 3F, S4C), which likely explains the preferential retention and progressive concentration of large (70 kDa) dextran in the vacuolar lumen. The diameter of the tubules generated during macropinosome resorption is well suited for them to associate with proteins containing BAR domains, concave structures that preferentially bind and stabilize curved membranes of ≈22 nm diameter (12). Indeed, the BAR domain-containing proteins SNX1, SNX2, and SNX5 decorated the tubules emanating from resorbing macropinosomes (Fig. S4A, (13)). Thus, membrane crenation caused by the volume loss potentially generated the necessary curvature to stabilize BAR domains on the membrane and thereby fostered tubulation (Fig. 3I). This notion was tested by generating liposomes and monitoring the effects of hydrostatic tension on the ability of a recombinant BAR-domain protein, BIN1, to induce tubulation. These experiments revealed an exquisite relationship between volume loss and BAR-mediated tubulation: the relief of hydrostatic tension greatly amplified tubulation by BIN1, while swelling the liposomes counteracted it (Figs. 3G, S4D). Given their functional redundancy and ability to form interchangeable heterodimeric complexes, the loss of any one BAR protein is unlikely to prevent tubulation. Because many BAR-domain proteins (including various SNX isoforms) require PtdIns(3)P for optimal binding, we interfered with their association by scavenging the available head-groups of the phosphoinositide by expressing tandem FYVE domains. Indeed, high affinity (multi-copy) FYVE-domain tandems prevented SNX recruitment and precluded tubulation/resolution of the macropinosomes (Figs. 3H, S4B).
Figure 3. Osmotically-driven shrinkage induces tubulation.
A, BMDM were stimulated with M-CSF and the distribution of 70 kDa rhodamine-dextran and FM1-43 imaged at the indicated times after removal of the stimulus. B-C, Mean number of tubules (exceeding 1 μm in length) measured 5 min after stimulation with M-CSF; >100 macropinosomes (n=3) for each condition. D-E, Macropinosomes containing sulforhodamine B and N-methylglucamine chloride or formed in cells treated with YM201636 (500 nM) were recorded 10 min after formation, before and after being subjected to hypertonic solution. See also Video S5. >100 vacuoles, n=6. F, TEM was used to measure the diameter of tubules emerging from macropinosomes; 85 tubules quantified. G, Liposomes formed of whole brain lipid, rhodamine-labelled phosphatidylethanolamine, and PtdIns(4,5)P2 in 20 mOsm solution. The mean aspect ratio for 3-5 fields of liposomes incubated with or without recombinant human BIN1 was quantified by imaging, n=3. H, HT1080 cells expressing GFP, 2xfyve-GFP, or 5xfyve-GFP were pulsed with SRB for 10 min. Mean number of tubules (exceeding 1 μm in length); >100 macropinosomes (n=3) for each condition. All p values determined by unpaired, two-sided t-tests. I, Model of mechanism proposed to underlie macropinosomal tubulation.
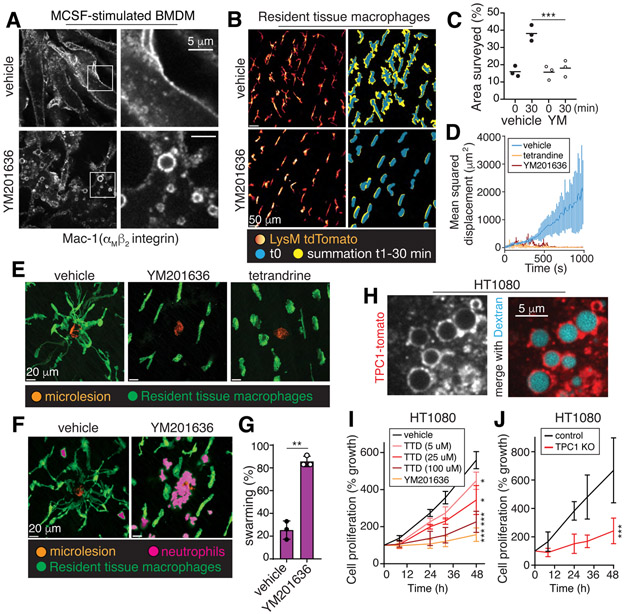
The significance of endomembrane shrinkage driven by efflux of monovalent ions is far-reaching. The resulting tubulation mediates the recycling of plasmalemmal components that are internalized in the course of macropinocytosis and endocytosis. One such example is Mac-1 (αMβ2), an integrin that is key to macrophage adherence, migration and phagocytosis (14). Blocking TPC channels by inhibiting PtdIns(3,5)P2 formation led to a pronounced depletion of plasmalemmal Mac-1, which was instead trapped in endomembrane vacuoles (Fig. 4A, S5C). The effect was phenocopied by simply substituting extracellular Na+ by K+, demonstrating that a Na+ gradient is exploited by the endocytic pathway to direct membrane traffic (Fig. S5A-C). Moreover, non-macropinocytic cells (e.g. fibroblasts) also required Na+ efflux to execute canonical receptor recycling (Fig. S6A-D, G-H) (15, 16). Defective plasmalemmal protein recycling caused by ion substitution had acute functional consequences: the ability of BMDM to bind and ingest complement-coated targets and of fibroblasts to form focal adhesions –processes mediated by integrins– were severely depressed (Figs. S5D-E, S6E-F).
Figure 4. Vacuolar resolution maintains cellular responsiveness and tissue surveillance.
A, BMDM were stimulated for 30 min with M-CSF, with or without YM201636, fixed and immunostained for Mac-1. B-E, Resident tissue macrophages (LysM-tdTomato) with or without YM201636 or tetrandrine were stimulated with M-CSF for 10 min followed by removal of the stimulus for 30 min. Cells were then imaged for 30 min. C, surveillance area measured in the absence (left) or presence (right) of YM201636 (YM) over time, n=3. In D-E, 30 min after M-CSF removal, laser-induced microlesions were generated, marked by the resultant autofluorescence (orange in E). Mean squared displacement of the macrophages is graphed in D; means ± SD, n=3. Representative images in E taken at 15 min post-injury. F-G, Experiments performed as in E. Representative images denoting the presence of neutrophils are shown. In G, the % of lesions with neutrophil swarming was quantified for 6 microlesions per animal, n=3. H, HT1080 cells expressing TPC1-tomato were incubated with 70 kDa dextran for 10 min before imaging. I-J, wildtype and TPC1 KO HT1080 cell growth measured in the absence or presence of YM201636 or tetrandrine (TTD) by cell counting. Means ± SD, n=3. All p values determined by Mann-Whitney U tests.
The need for ion efflux from endocytic compartments for normal cell function was also documented in situ. When Na+ efflux from the endocytic pathway was prevented in vivo, the ability of interstitial RTM to survey their environment was impaired (Fig. 4B-E, Video S6). Blocking β1 and β2 integrins similarly inhibited surveillance (Fig. S7A-C). When microlesions are made by targeted laser ablation to adjacent fibroblasts, RTM normally emit processes to contain the damage (5), preventing the recruitment and activation of neutrophils. When PIKfyve or TPCs were inhibited, the cells failed to resorb vacuoles and were unable to respond to the damage (Fig. 4D-E). As a result, neutrophil swarming ensued (Fig. 4F-G). Thus, vacuole resolution, mediated by lipid-gated Na+ efflux, underpins membrane traffic necessary to maintain cellular responsiveness.
Because macropinocytosis is not restricted to phagocytes, inhibition of vacuolar shrinkage affects also other cell types (17). HT1080 fibrosarcoma cells display vigorous constitutive macropinocytosis (Fig. 4H), express TPC1 at comparatively high levels (Fig. S8A) and localize the channel to macropinosomes (Fig. 4H). HT1080 cells require growth factors for survival and are highly responsive to EGF. Because the EGF receptor is internalized along with the macropinosomal membrane, effective recycling to the cell surface is key. Preventing vacuole shrinkage resulted in the depletion of EGF receptors from the plasmalemma (Fig. S8B) and this was associated with reduced responsiveness to EGF and delayed growth (Fig. 4I-J, S8C).
Solute transport may be involved in the shrinkage and tubulation of other organelles. In this regard, lysosomes are known to undergo swelling in cells treated with PIKfyve antagonists (18). Impaired solute extrusion could account for the volume gain, which is compounded by ongoing membrane fusion that is uncompensated by shrinkage-dependent tubulation and/or vesiculation and scission. Indeed, lysosomes swollen by inhibition of PIKfyve recovered their ability to tubulate when exposed to hypertonic solution (Video S7), and also upon removal of the PIKfyve inhibitor, but failed to do so when tetrandrine was present (Fig. S9A-E). Overexpression of TPC2 alone caused lysosomes to become more tubular, suggesting the channel may be involved in these processes (Fig. S10A-C, Video S8).
In summary, we propose a role for ion fluxes in the endocytic pathway: extrusion of osmotically abundant ions/solutes, accompanied by water, cause vacuolar/vesicular shrinkage leading to crenation of the membrane, forming convex protrusions that stabilize proteins with BAR or similar curvature-sensing domains. These, in turn, foster tubulation and eventual scission that is critical for inter-organellar traffic. In macrophages that continuously survey the extracellular space, an ongoing ion efflux from the endocytic pathway drives the resolution of fluid that is taken up during this process, supporting recycling of receptors to maintain their function. Thus, the resolution of organelles formed during surveillance is necessary to preserve tissue integrity.
Materials and Methods
Cell isolation, culture, and transfection
Murine macrophages were harvested from the marrow of long bones from 6- to 10-week old C57Bl/6 wild-type mice. Cells were washed in PBS before culturing in DMEM containing L-glutamine, 10% heat-inactivated serum, and 10% L929 fibroblast-conditioned medium as a source of macrophage colony-stimulating factor (M-CSF). For primary macrophage transfections, after 5-6 days of culture, cells were lifted with 5 mM EDTA in PBS, centrifuged, and resuspended in T buffer from the Neon™ transfection system (ThermoFisher). 1-5 μg of plasmid DNA or 100 pM siRNA was added to 2x106 cells in 120 μL of T buffer before a single 1500 V, 30 ms pulse was applied through the suspension. For fluorescence microscopy, cells were imaged 4-8 h after transfection. For knockdown experiments, cells were lifted and transfected a second time after 24 h, and then used after 48 h. These approaches routinely resulted in 10-25% transfection efficiency of plasmid DNA as determined by fluorescence microscopy and a 60-90% reduction in mRNA caused by siRNA, as determined by qPCR, respectively.
HT1080 cells from ATCC (CCL-121) were cultured in DMEM containing L-glutamine and 10% heat-inactivated serum. Growth assays were done in medium with low L-glutamine (0.2 mM), 5% FBS, and 2% albumin. Mouse embryonic fibroblasts were from ATCC (SCRC-1008) and cultured in DMEM containing L-glutamine and 5% heat-inactivated serum.
Animal Studies
The following mouse strains were used in this study:
Lyz2Cre/Cre (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; The Jackson Laboratories; 007914).
tdTomfl/fl (B6N.129P2(B6)-Lyz2tm1(cre)Ifo/J; The Jackson Laboratories; 018956).
F1 crossbreed of the above was used for experiments designated “LysM-tomato”.
Tpc1−/−: The Tpc1 knockout was generated from an ES cell line developed by Velocigene (clone # 13234, purchased from KOMP) to delete the open reading frame. Mice were backcrossed to C57BL/6 (Jax) for more than 10 generations. Genotyping was done using a mixture of three primers with sequences of AGAACACGTTCATCAGTAACGATTATG (forward), AAAGCTGTGTCAGAAGTGAGGGACC (reverse 1) and TCTGTCTGTCCTAGCTTCCTCACTG (reverse 2). The PCR condition was 94°C 2’; (94°C 30”; 55°C 30”; 72 °C 30”) x35; 72°C 5’. Primers amplify the wild-type as 595 bp and the knockout as 381 bp.
Tpc2−/−: The generation of Tpc2 knockout was previously described (1).
Tpc1−/−;Tpc2−/−: The double knockout was generated by crossing the individual knockouts.
Trpml1−/−: The generation of Trpml1 knockout was previously described (2) using crosses of heterozygous mice from S. Slaughenhaupt (Center for Human Genetic Research, Massachusetts General Hospital at Harvard Medical School) and provided by Dr. N. Jones (The Hospital for Sick Children).
C57BL/6J (The Jackson Laboratories; 000664).
Mice used for cell isolation experiments were maintained under a 12:12-h light:dark cycle, and were provided with food and water ad libitum. All animal study protocols were approved by the animal care committee of The Hospital for Sick Children.
Reporter mice for intravital imaging were maintained in specific pathogen-free conditions at an American Association for Laboratory Animal Care–accredited animal facility at the NIAID (National Institute of Allergy and Infectious Diseases, NIH) and were used under a study protocol approved by the NIAID Animal Care and Use Committee (National Institutes of Health, NIH). All imaging experiments were performed with 8-12-week old mice, and both males and females were used at equal ratios for the studies as they displayed similar immune cell dynamics in vivo.
Intravital imaging of the peritoneal serosa
Intravital microscopy was performed as previously described (3). In brief, a midline incision of the abdominal wall of anesthetized mice (Isoflurane; Baxter; FORANE; Cat#1001936040) was performed and the parietal peritoneal serosa was exposed. 10 μL of buffer (pre-warmed HBSS containing 2 mM CaCl2 and 1 mM MgCl2; pH 7.4; 34°C) with inhibitors/vehicle or antibodies was applied topically, before installing a sterilized cover glass (Fisher Scientific; Cat# 12-545-80) and transferring the animal to a heated (36°C) imaging chamber.
To assess endosome resolution in RTM in vivo, YM-201636 (CAS 371942-69-7; Cayman Chemical; dissolved in DMSO; final stock concentration 10 mM), tetrandrine (CAS 518-34-3; Abcam; dissolved in DMSO: final stock concentration 5 mM) or vehicle alone (DMSO) were applied topically and incubated for 30 min. Then, mM-CSF (PeproTech; dissolved in H20; final concentration 100 ng/mL) was added and incubated for another 10 min. To remove M-CSF, the tissue was then carefully washed three times with 1 mL pre-warmed buffer before installing a fresh cover glass with 10 μL buffer. Imaging was started after an additional 30 min incubation time to allow for endosome resolution. The emission of second-harmonic light from non-centrosymmetric structures (matrix, e.g. collagen) was also collected.
To assess integrin function, 1 μg of each anti-CD29 (HMβ1-1; BioLegend; #102235) plus anti-CD11b/β2 integrin (M1/70; BioLegend; #101213) or isotype controls (Armenian hamster IgG plus rat IgG2b) were applied topically, and imaging was started after 30 min incubation time. Tissue staining was done with anti-CD11b and anti-CD169/Siglec-1 (3D6.112; BioLegend; #142407). Imaging was performed on a Zeiss 710 microscope equipped with a Chameleon femtosecond-pulsed laser (Coherent; 930 nm wavelength) and a 20x water immersion objective (NA 1.0; Zeiss). Raw imaging data were processed and analyzed with Imaris 9.2.1 (Bitplane). Macrophage probing dynamics were recorded over 30 min (resolution 512x512; 16 bit; Z- stacks of 12 focal planes each 3 μm apart; frame rate 968 ms; 128 imaging frames in total) and fluorescent intensities from all frames were binarized and summed to visualize and calculate overall tissue surveillance over time using Adobe Photoshop (CC 2019).
To measure individual endosome volumes, still images of macrophages were recorded at indicated time points at higher magnification (4-5x zoom) and resolution (resolution 1024x1024; 16 bit; Z-stacks with 0.5 μm step size). Diameter measurements were performed after deconvolution (Huygens Professional, version 18.04) using the Imaris slice-viewer mode.
Sterile laser damage
Sterile laser damage was performed as previously described (3). In brief, after treating the tissue with inhibitors/vehicle as described above, stromal microlesions (partial-to-unicellular damages) were induced with a brief (< 1 s) two-photon laser pulse using a 20x objective at a 35x zoom.
Macrophage dynamics were recorded immediately after damage induction for 30 min (to assess damage response; “cloaking”) or up to 3 h (to assess endogenous neutrophil response; “swarming”). Spatiotemporal coordinates of individual macrophage pseudopods were used to calculate track displacements in Imaris.
To assess neutrophil damage responses (swarming vs. no swarming), 6 individual microlesions were induced each 1000 μm apart, and static images were recorded every 20 min over 3 h. Neutrophils were identified in LysMgfp/gfp mice by morphology and GFP intensity. Formation of neutrophil clusters at the site of the microlesion was assessed as neutrophil swarming activity.
Intravital imaging experiments were performed using LysMgfp/gfp (NIAID repository, Taconic Farms, Cat#000342) or LysMCre/+ tdTomatofl/+ mice (The Jackson Laboratories; Cat#007914 crossed with Cat#018956) at ages 8-10 weeks, with similar results in males and females.
Generation of TPC1 KO HT1080 by CRISPR/Cas9
A CRISPR/Cas9 and a dual-sgRNA strategy was implemented to inactivate the Tpc1 gene. Two sgRNA sequences (CTAGAAGATGCCTCTAATGG and ATCCTGACCTTGGATGAGGG) flanking the start site of TPC1 within exon 3 were chosen (using the online tool https://benchling.com) and synthesized (Eurofins Genomics). The 2 guides were individually cloned into pX458 vector (Addgene #48538) as described (4) and verified by Sanger sequencing (Eurofins Genomics). The vectors were co-transfected into HT1080 cells and 24 h later the cells were bulk-sorted by FACS based on GFP+ signal. The sorted cells were allowed to recover and grow for 3 days, then diluted and re-plated to achieve well-separated single cells. After 2 weeks in culture, individual colonies were picked by pipette and expanded for ≈1 week in larger wells. Cells were screened by extracting genomic DNA with KAPA Express Extract (KAPA Biosystems, Cat# KK7103) and carrying out PCR with the forward primer 5’AGTATTTGTTCTTGCAGGGTTCC and reverse primer 5’AGGGAAGGCAGCCAAAGTAG to yield a product of approximately 1800 bp in WT cells and 1660 bp in CRISPR-edited cells.
Cell sorting and RNA sequencing
RNA sequencing protocol has been published previously (3) and the RNAseq data pooled from three independent biological replicates have been deposited online (NCBI GEO: CGSE119870). In brief, Cx3cr1 GFP/+ Ccr2 RFP/+ mice received an i.v. injection of Dextran-Cascade Blue (Invitrogen; #D1976) to label pinocytosing cells in the interstitium of the peritoneal serosa. Intravascular leukocytes were labeled by i.v. injection of anti-CD45 (BioLegend; clone 30-F11) 3 min prior to euthanasia. The peritoneal serosa was harvested and digested for 20 min at 37°C in RPMI (GIBCO; Cat#11875093) with 1% FBS (Gemini; Lot#A20G001; Cat#100-106), 250 μg/ml Liberase TL (Sigma-Aldrich; Cat#5401020001), 100 μg/mL DNase II (Sigma-Aldrich; Cat# D8764), 25 μM Z-VAD-FMK (Tocris; Cat#2163) and 10 mM HEPES (GIBCO; Cat#15630080). Cells were resuspended in PBS (GIBCO; Cat#10010-23; Lot#1943335) with 2% FBS, 2 mM EDTA (Quality Biological; Cat#51-027-101) and 25 μM Z-VAD-FMK and stained for CD11b (BioLegend; clone M1/70), I-A/I-E (BioLegend; clone M5/114.15.2) and Ly6G (BioLegend; clone 1A8). CD45− CD11b+ Ly6G− cells were sorted into Dextran+ MHCII+/− CCR2− CX3CR1+/− (sessile RTM) and Dextran− MHCII+ CCR2+ CX3CR1− (migratory monocytes), directly into 0.5 mL TRIZOL (Invitrogen; Cat#15596026) using a FACS-Aria cell sorter (BD Bioscience).
A Nugen Ovation SoLo kit (NuGEN; Cat#0501-32) was used to generate RNAseq libraries with an input of ≈1 ng total RNA. Sequencing was performed using Illumina NextSeq V2 reagents (Illumina; Cat#FC-404-2005) with pair end 75 pair reads at a sequencing depth of ~ 20 million reads. Reads were mapped to Mus musculus mm10 genome build (UCSC annotation) and Salmon-estimated transcript-per-million values were summarized at the gene level using tximport (v 1.4.0) and the org.Mm.eg.db database.
Additional reagents and plasmids
YM201636 (Cayman Chemical, 371942-69-7) was used at 500 nM. Apilimod (Santa Cruz, sc-40051) was used at 500 nM. WX8 (a generous gift from Dr. M. DePamphilis, NIH) was used at 500 nM (5). Ouabain octahydrate (Sigma Aldrich, O3125) was used at 5 mM. Furosemide (Tocris, 3109) was used at 5 μM. Bumetanide (Tocris, 3108) was used at 5 μM. 4,4’-diisothiocyanatostilbene-2-2’-disulfonic acid disodium salt (DIDS, Sigma Aldrich, D3514) was used at 10 μM. CFTRinh-172 (Tocris, 3420) was used at 1 μM. NPPB (Tocris, 0593) was used at 10 μM. Tetrandrine (Abcam, ab142464) was used at the indicated concentrations. Nocodazole (Sigma, 487928) was used at 5 μM.
mTPCN1 stealth siRNA (cat #MSS217431), murine and human TPCN1/TPCN2/ABT1 TaqMan® probes (Mm00455326_m1, Hs01552053_g1, Hs00330542_m1, Mm00803824_m1, Hs00706003_s1), TaqMan® RT-PCR master mix (4444557) were from ThermoFisher. The generation of plasmids used for the expression of LAMP-1-RFP and tubulin-GFP (6), SNX2-GFP (7), and PX-GFP (8) is described in the references provided. hTPC1wt-tomato was a generous gift from Dr. N. Klugbauer (9). EGFR-GFP was from Dr. A. Sorkin (Addgene plasmid #32751). pEGFP-N3-hTPC2wt was from Dr. S. Di Pietro (Addgene plasmid #80153). PIKfyve-GFP was from Dr. G. van den Bogaart (Addgene plasmid #121148). TRPML1-YFP was from Dr. C. Montell (Addgene plasmid #18826). 2xfyve-GFP, 2xfyve-mCherry, and 5xfyve-GFP were from Dr. L. Cantley (Beth Israel Deaconess Medical Center, Boston, Mass.). Mutagenic primers for TPCN2.K321A were: fw 5'-cggggctacctgatgGCGtctctccagacctcg-3', rv 5'-cgaggtctggagagaCGCcatcaggtagccccg-3'. Sequencing confirmed with the primer 5'-tctggtggactggaccgt-3'. SNX1-GFP was generated by PCR amplification of human SNX1 cDNA using the primers 5' GATCTCGAGCTCAAGCTTCGATGGCGTCGGGTGGTGGTG and 5' GTACCGTCGACTGCAGAATTGGGGAGATGGCCTTTGCC and performing Gibson assembly (New England Biolabs, Cat# E2611) with an EcoRI-digested pEGFP-N1 vector.
Anti-Mac-1 antibody was from BD Bioscience (550282). Anti-β-tubulin was from Sigma (T6199). Anti-EEA1 antibody was from Santa Cruz (sc-6415). Anti-paxillin was from BD Bioscience (610051). Anti-β1-integrin (TS2/16) (303002), anti-CD11b (M1/70) (101201), and CD29 (HMβ1) (102201) were from BioLegend.
All lipids were from Avanti Polar Lipids Inc.: total porcine brain extract (13110P), PE-rhodamine (810150P), PtdIns(4,5)P2 (850185). Alexa568-Tfn was from ThermoFisher (T23365).
Macropinocytosis
Macrophages were seeded onto glass coverslips for 24-48 h at high confluency. The medium was then removed and replaced with solutions warmed to 37°C of either PBS with Ca2+ and Mg2+, or buffers containing 150 mM of either NaCl, N-methylglucamine-chloride, or Na-gluconate, plus 1 mM MgCl2, 1 mM CaCl2, 5 mM KCl, 20 mM HEPES, pH 7.2. The osmolarity of the solutions was equivalent (≈300 mOsm) as measured using a model 5004 automatic osmometer (Precision Systems; 2094). Each solution also contained 70,000 MW tetramethylrhodamine- or Oregon Green-dextran (ThermoFisher, D1818 or D7173) to identify the newly formed vacuoles, and 100 ng/mL recombinant murine M-CSF (Peprotech, cat. # 315-02) to stimulate macropinocytosis.
In some cases, FM1-43 dye (ThermoFisher, T35356) was added to label the membrane, or high molecular weight dextran was substituted with sulforhodamine B (Sigma-Aldrich, 230162). Where specified, inhibitors were added to the macrophages at the indicated concentrations 5 min before the addition of M-CSF. After 5 min of M-CSF stimulation, cells were washed and vacuole resolution was monitored by spinning-disc microscopy as described previously (10) in PBS with or without inhibitors. The mean macropinosomal volume and dextran intensity were determined using Volocity software (PerkinElmer), excluding objects below 0.5 μm3.
In experiments where macropinosomes were treated with PIKfyve inhibitors or loaded with NMG+ and sulforhodamine B, then induced to tubulate by elevating the tonicity of the media, the sulforhodamine B was loaded at 1 μg/mL. After 10 min post-macropinosome sealing, an 800 mOsm, pH 7.2 solution of NaCl was used at 1:1 to adjust the tonicity to 500 mOsm overall. In experiments where hypotonic solutions were used, the medium was adjusted using 4:1 solution 100 mOsm NaCl, pH 7.2.
Cell volume measurements
Macrophages seeded on non-treated Petri dishes were lifted mechanically. The median cell diameter for >5000 cells was determined using the Z2 Coulter particle count and size analyzer (Beckman Coulter) and converted to volume by assuming that the suspended cells were spherical.
Atomic absorption spectrometry
Differentiated BMDM were seeded onto 6-well plastic TC dishes and cultured for an additional 24 h. Cells were counted by imaging 3 fields by phase microscopy. After stimulation, dishes were washed x3 by gentle, brief submersion of the dishes in Na+-free solutions to remove contaminating ions. Cells were lysed with 1 mL H2O containing 1% HNO3 and 0.1% LiCl. Lysis was done by scraping and vigorous pipetting. Samples were analyzed using a PinAAcle 900T Atomic Absorption Spectrometer (PerkinElmer).
STED microscopy
STED microscopy was performed as previously described (11). Briefly, using a Leica TCS SP8 STED 3X microscope, images were acquired using HyD detectors and a 90X/1.4 glycerol objective. Samples were labeled with Cy3-conjugated secondary antibodies and excited at 554 nm using a white-light laser. Emissions were time-gated 0.5-6 ns. A 1.5 W depletion laser at 660 nm was used for the red channel, at 45% of maximal power. Images were acquired using Leica LAS X software with a xy pixel size of 31 nm and z step of 200 nm and then deconvolved with Leica Lightning software (Scientific Volume Imaging, The Netherlands).
Liposome generation and tubulation
Liposomes were made as previously described (12). Briefly, whole brain lipid (porcine) at 94% of the total lipid concentration, PtdIns(4,5)P2 at 5% and PE-rhodamine at 1% were mixed in chloroform and the solvent was removed under a stream of nitrogen. Liposomes were generated by vortexing into 20 mM Tris-HCl and sonication in an ice-bath, then incubated with 20 μg/mL recombinant human BIN1 protein (Abcam, 98238) on ice for 30 min. Liposomes were then seeded onto acid-washed glass coverslips coated with a 1:1000 solution of poly-L-lysine in 20 mM Tris-HCl (Sigma, P4707) for 10 min and imaged by spinning-disc microscopy. The osmolarity of the solution was then adjusted with Tris-HCl containing NaCl or water and liposomes were again imaged 30 sec later. Objects were identified and the mean aspect ratio, equal to the major axis/minor axis of each liposome with a fitted ellipse, was determined using ImageJ software.
Immunoblotting
HT1080 cells were serum-starved for 2 h in DMEM, then given 100 ng/mL human recombinant EGF (Sigma Aldrich, E9644) for the indicated times and lysed in RIPA buffer containing protease and phosphatase inhibitors. Laemmli sample buffer with 2-mercaptoethanol was added to the cell lysates, samples were boiled for 5 min, then run on 10% polyacrylamide gels. Blots were incubated with rabbit p-Ser473-Akt and mouse total-Akt antibodies (Cell Signaling Technology, 736E11 and 40D4) at 1:2000 in TBST overnight. Secondary antibodies conjugated with LI-COR-compatible fluorophores were used for detection on an Odyssey FC imaging system (LI-COR Biosciences).
Particle Binding
Sheep red blood cells (MP Biomedicals, 55876) were opsonized with rabbit-anti sheep red blood cell IgM, washed, then incubated with C5a-deficient serum (Accurate Chemical & Scientific Corp., AIAM6840). Prior to adding phagocytic targets, the primary murine macrophages were treated with vehicle control, YM201636 at 500 nM, or 10 μM tetrandrine along with M-CSF for 30 min. Binding was determined post-fixation, washing, and permeabilization of the cells by staining the red cells with goat anti-rabbit IgM conjugated with DyLight488 (Abcam, ab96979).
Transmission electron microscopy
Monolayers of BMDMs grown on glass coverslips were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, rinsed in buffer, post-fixed in 1% osmium tetroxide in buffer, dehydrated in a graded ethanol series followed by propylene oxide, and embedded in Quetol-Spurr resin. Sections (90 nm thick) were cut on a Leica Ultracut ultramicrotome, stained with uranyl acetate and lead citrate and viewed in an FEI Tecnai 20 TEM.
Lysosomal motility
TPC2-GFP-expressing compartments were recorded over 10-30 sec at 10 frames per sec on a Zeiss Axiovert 200M microscope equipped with a 100x, 1.45 N.A. oil objective, a custom 2.4x magnification lens, and a back-thinned EM-CCD camera (Hamamatsu). The motion type for individual lysosomes was determined using an MSS analysis, previously described (13). Directed motion was defined as those lysosomes having linear or super-diffusive trajectories, indicative of motor-based movement of the organelle.
Supplementary Material
Video S1. Resident tissue macrophages (LysM-tomato) of the peritoneal serosa continuously probe their local microenvironment. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S2. The stimulation of resident tissue macrophages (LysM-tomato) with M-CSF causes the formation of large macropinosomes as the cells continue to probe. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S3. Na+ efflux is required for macropinosome shrinkage. BMDM were stimulated with M-CSF in isosmotic medium containing either 150 mM NaCl or N-methylglucamine chloride as primary solutes, as indicated. Solutions also contained 1 mM CaCl2, 1 mM MgCl2, 5 mM KCl, 20 mM HEPES, plus 70 kDa tetramethylrhodamine dextran to identify the newly formed vacuoles. Fields of 5-10 cells were imaged every 30 s. Scale in lower right indicates time in minutes.
Video S4. Tetrandrine, a cation channel inhibitor, blocks vacuole shrinkage. BMDM treated with vehicle control or tetrandrine (1 μM) were simultaneously stimulated with M-CSF in medium containing 70 kDa tetramethylrhodamine-dextran to identify the newly formed vacuoles. Fields of 5-10 cells were imaged every 30 s. Scale in lower left indicates time in minutes.
Video S5. Osmotically-induced shrinkage of macropinosomes promptly causes tubulation. Macropinosomes loaded with sulforhodamine B and N-methylglucamine chloride and imaging started 10 min after their initial formation. After acquisition of the first frame, cells were exposed to a medium made hypertonic (500 mOsm final) by addition of sucrose. Images were acquired every 10 s.
Video S6. Preventing vacuole resolution by inhibition of PIKfyve abrogates tissue surveillance. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S7. Lysosome swelling upon inhibition of PIKfyve is reversed by exposure to hyperosmotic solution. HeLa cells expressing LAMP1-RFP were treated with 500 nM of the PIKfyve inhibitor YM201636 for 1 h, causing swelling of the vesicles to the extent that their lumen could be resolved by confocal microscopy. The vesicles were monitored at 1 frame/10 s for the indicated times before and after addition of sucrose (second frame) to bring the medium osmolarity to 500 mOsm.
Video S8. TPC2 expression level controls lysosome tubulation in a PtdIns(3,5)P2-dependent manner. HeLa cells expressing TPC2wt-GFP or TPC2K321A-GFP were recorded at 10 Hz for 30 s.
Acknowledgements
We thank Kimberley Aranda and Zhengjiang Liu for tissue preparation and genotyping. We are grateful to the Transgenic and Chimeric Mouse Core of the University of Pennsylvania (supported by NIH center grants P30DK050306, P30DK019525, and P30CA016520) for the generation of the TPC1 line. Supported by grant FDN-143202 from CIHR to S.G. This work was also supported in part by the Intramural Research Program of the NIH to R.G. and S.U. and by NIH grants R01 HL147379 and R01 GM133172 to D.R.
References and Notes
- 1.Steinman RM, Brodie SE, Cohn ZA, Membrane flow during pinocytosis. A stereologic analysis. J Cell Biol 68, 665–687 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swanson JA, Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol 9, 639–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson JA, Watts C, Macropinocytosis. Trends Cell Biol 5, 424–428 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Yoshida S, Hoppe AD, Araki N, Swanson JA, Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J Cell Sci 122, 3250–3261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uderhardt S, Martins AJ, Tsang JS, Lammermann T, Germain RN, Resident Macrophages Cloak Tissue Microlesions to Prevent Neutrophil-Driven Inflammatory Damage. Cell 177, 541–555 e517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakurai Y et al. , Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347, 995–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lattin JE et al. , Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4, 5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.She J et al. , Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 556, 130–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X et al. , TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove SK et al. , Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390, 187–192 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Li X et al. , Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proceedings of the National Academy of Sciences of the United States of America 110, 21165–21170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peter BJ et al. , BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Kerr MC et al. , Visualisation of macropinosome maturation by the recruitment of sorting nexins. J Cell Sci 119, 3967–3980 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Dustin ML, Complement Receptors in Myeloid Cell Adhesion and Phagocytosis. Microbiol Spectr 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castonguay J et al. , The two-pore channel TPC1 is required for efficient protein processing through early and recycling endosomes. Sci Rep 7, 10038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm C et al. , High susceptibility to fatty liver disease in two-pore channel 2-deficient mice. Nature communications 5, 4699 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Commisso C et al. , Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferies HB et al. , A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep 9, 164–170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capurro MI et al. , VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat Microbiol 4, 1411–1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran FA et al. , Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma G et al. , A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy 15, 1694–1718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherer NM et al. , Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4, 785–801 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Carlton JG et al. , Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J Cell Sci 118, 4527–4539 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanai F et al. , The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol 3, 675–678 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Canton J et al. , Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat Commun 7, 11284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman SA et al. , Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 172, 305–317 e310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung T et al. , Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Jaqaman K et al. , Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Resident tissue macrophages (LysM-tomato) of the peritoneal serosa continuously probe their local microenvironment. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S2. The stimulation of resident tissue macrophages (LysM-tomato) with M-CSF causes the formation of large macropinosomes as the cells continue to probe. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S3. Na+ efflux is required for macropinosome shrinkage. BMDM were stimulated with M-CSF in isosmotic medium containing either 150 mM NaCl or N-methylglucamine chloride as primary solutes, as indicated. Solutions also contained 1 mM CaCl2, 1 mM MgCl2, 5 mM KCl, 20 mM HEPES, plus 70 kDa tetramethylrhodamine dextran to identify the newly formed vacuoles. Fields of 5-10 cells were imaged every 30 s. Scale in lower right indicates time in minutes.
Video S4. Tetrandrine, a cation channel inhibitor, blocks vacuole shrinkage. BMDM treated with vehicle control or tetrandrine (1 μM) were simultaneously stimulated with M-CSF in medium containing 70 kDa tetramethylrhodamine-dextran to identify the newly formed vacuoles. Fields of 5-10 cells were imaged every 30 s. Scale in lower left indicates time in minutes.
Video S5. Osmotically-induced shrinkage of macropinosomes promptly causes tubulation. Macropinosomes loaded with sulforhodamine B and N-methylglucamine chloride and imaging started 10 min after their initial formation. After acquisition of the first frame, cells were exposed to a medium made hypertonic (500 mOsm final) by addition of sucrose. Images were acquired every 10 s.
Video S6. Preventing vacuole resolution by inhibition of PIKfyve abrogates tissue surveillance. Intravital microscopy. Scale in lower right indicates time in minutes.
Video S7. Lysosome swelling upon inhibition of PIKfyve is reversed by exposure to hyperosmotic solution. HeLa cells expressing LAMP1-RFP were treated with 500 nM of the PIKfyve inhibitor YM201636 for 1 h, causing swelling of the vesicles to the extent that their lumen could be resolved by confocal microscopy. The vesicles were monitored at 1 frame/10 s for the indicated times before and after addition of sucrose (second frame) to bring the medium osmolarity to 500 mOsm.
Video S8. TPC2 expression level controls lysosome tubulation in a PtdIns(3,5)P2-dependent manner. HeLa cells expressing TPC2wt-GFP or TPC2K321A-GFP were recorded at 10 Hz for 30 s.