Abstract
ORM1-like 3 (ORMDL3) has strong genetic linkage to childhood onset asthma. To determine whether ORMDL3 selective expression in airway smooth muscle (ASM) influences ASM function, we used Cre-loxP techniques to generate transgenic mice (hORMDL3Myh11eGFP-cre), which express human ORMDL3 selectively in smooth muscle cells. In vitro studies of ASM cells isolated from the bronchi of hORMDL3Myh11eGFP-cre mice demonstrated that they developed hypertrophy (quantitated by FACS and image analysis), developed hyperplasia (assessed by BrdU incorporation), and expressed increased levels of tropomysin proteins TPM1 and TPM4. siRNA knockdown of TPM1 or TPM4 demonstrated their importance to ORMDL3-mediated ASM proliferation but not hypertrophy. In addition, ASM derived from hORMDL3Myh11eGFP-cre mice had increased contractility to histamine in vitro, which was associated with increased levels of intracellular Ca2+; increased cell surface membrane Orai1 Ca2+ channels, which mediate influx of Ca2+ into the cytoplasm; and increased expression of ASM contractile genes sarco/endoplasmic reticulum Ca2+ ATPase 2b and smooth muscle 22. In vivo studies of hORMDL3Myh11eGFP-cre mice demonstrated that they had a spontaneous increase in ASM and airway hyperreactivity (AHR). ORMDL3 expression in ASM thus induces changes in ASM (hypertrophy, hyperplasia, increased contractility), which may explain the contribution of ORMDL3 to the development of AHR in childhood onset asthma, which is highly linked to ORMDL3 on chromosome 17q12-21.
Keywords: Muscle Biology, Pulmonology
Keywords: Asthma
Introduction
ORM1-like 3 (ORMDL3), a gene located on chromosome 17q12-21, has been highly linked to childhood onset asthma in genetic association studies (1). ORMDL3 is an ER-localized protein (2) expressed in multiple cell types important to the pathogenesis of asthma, including immune and inflammatory cells [i.e., CD4+ cells (3, 4), eosinophils (5), and macrophages (2)] and lung structural cells [i.e., epithelial cells (2) and airway smooth muscle (ASM) (6)]. At present it is not known which of these or other cells alone or in combination express the key ORMDL3-regulated pathways important to the development of asthma. In terms of immune and inflammatory cells expressing ORMDL3, CD4+ cells expressing SNPs linked to ORMDL3 have enhanced Th2 responses (4), suggesting that ORMDL3-regulated pathways in CD4+ cells may be the key cell implicating ORMDL3 in asthma. However, epidemiologic studies demonstrate that chromosome 17q12-21 is linked to childhood asthma (1) but not to allergic rhinitis (7). As both childhood asthma and allergic rhinitis are associated with increased Th2 responses, one would have expected that ORMDL3 enhancing Th2 responses would result in association of ORMDL3 with both childhood asthma and allergic rhinitis, which is not the case. Thus, ORMDL3 expression in CD4+ cells may be very important for enhancing Th2 responses in childhood asthma but may not be the key cell expressing ORMDL3, implicating it in the onset of asthma in childhood.
One of the other key cells that is present in the lung in asthma, but not present in the upper airway in allergic rhinitis, is ASM, whose expression of ORMDL3 may help explain the epidemiologic association of chromosome 17q21-21 with childhood onset asthma but not allergic rhinitis. To determine the role of selective ORMDL3 expression in ASM, in this study we have used Cre-loxP techniques to generate mice expressing human ORMDL3 (hORMDL3) selectively in smooth muscle cells. These studies demonstrated that ASM cells isolated from the bronchi of hORMDL3Myh11eGFP-cre (myosin heavy chain 11, Myh11) mice spontaneously developed hypertrophy, hyperplasia (mediated by tropomysins TPM1 and TPM4), and increased contractility to histamine in vitro, which was associated with increased levels of intracellular Ca2+, increased cell surface membrane Orai1 Ca2+ channels — which mediate influx of Ca2+ into the cytoplasm — and increased expression of ASM contractile genes, including sarco/endoplasmic reticulum Ca2+ ATPase 2b (Serca2b) and smooth muscle 22 (Sm22). In vivo studies of hORMDL3Myh11eGFP-cre mice demonstrated that they had a spontaneous (in the absence of experimental exposure to allergen or other environmental trigger) increase in ASM, which was associated with a spontaneous increase in airway hyperreactivity (AHR). ORMDL3 expression in ASM thus induces changes in ASM (hypertrophy, hyperplasia, and increased contractility), which may explain the contribution of ORMDL3 to the development of AHR in childhood onset asthma but not to the development of allergic rhinitis.
Results
hORMDL3Myh11eGFP-cre mouse ASM cells for in vitro study.
Mouse ASM cells were obtained from the bronchial airways dissected free of lung tissue of hORMDL3Myh11eGFP-cre and WT mice (Figure 1, A–C). Bronchial airway cells were cultured in mouse smooth muscle cell media (Cell Biologics) for 3 to 4 weeks at which time 90% of the cells were ASM cells as defined by expression of ASM genes (alpha–smooth muscle actin, myh11) (Figure 2, A and B), expression of EGFP (Figure 2, A and B), and morphology.
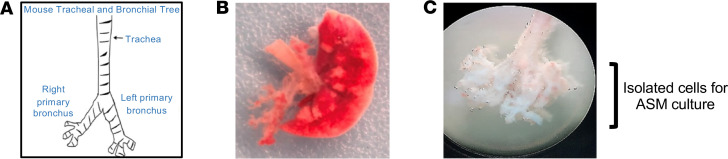
Figure 1. Mouse ASM cells.
(A) The mouse tracheobronchial tree was dissected free from (B) whole lung tissue using (C) a dissecting microscope under 10× objective, and a single-cell suspension of bronchial cells was prepared for culture in mouse smooth muscle cell media.
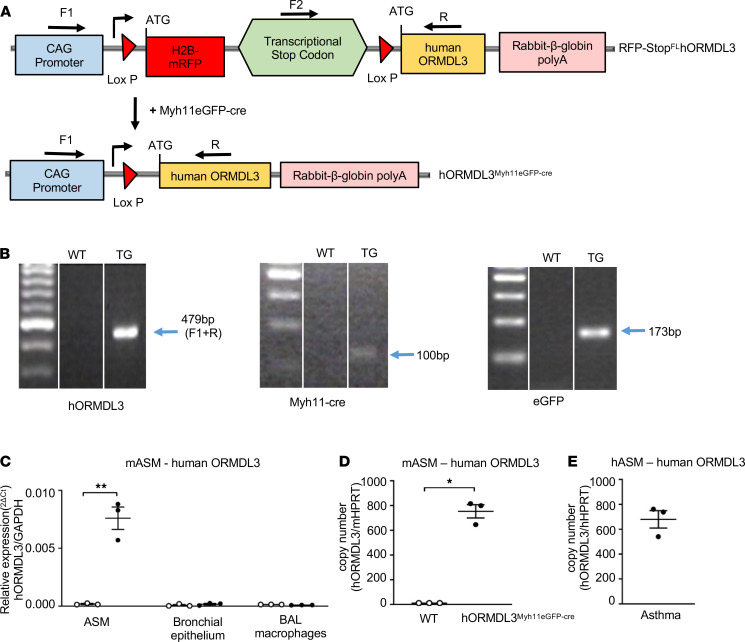
Figure 2. Genotyping and hORMDL3 expression in ASM from mice and asthmatics.
(A) RFP-StopflhORMDL3-Tg mice were crossed with smooth muscle–specific Myh11-creEGFP mice to generate hORMDL3Myh11eGFP-cre (both on C57BL/6 background), which expresses hORMDL3 specifically in smooth muscle cells. (B) WT and hORMDL3Myh11eGFP-cre mice were genotyped to determine expression of human ORMDL3, Myh11-Cre, and EGFP. (C) Human ORMDL3 mRNA expression was quantitated by RT-qPCR in mouse ASM, mouse bronchial epithelium, and mouse bronchoalveolar lavage (BAL) macrophages derived from WT and hORMDL3Myh11eGFP-cre mice (n = 3/group). (D) Human ORMDL3 mRNA copy number was quantitated by dPCR in mouse ASM derived from WT and hORMDL3Myh11eGFP-cre mice (n = 3/group). (E) Human ORMDL3 mRNA copy number was quantitated by digital PCR (dPCR) in human asthma postmortem lung ASM (n = 3) *P < 0.05 compared with WT. **P < 0.01 compared with WT. P values are results of unpaired 2-tailed t tests.
Generation of hORMDL3Myh11eGFP-cre Tg mice.
To generate hORMDL3Myh11eGFP-cre mice, we crossed RFP-StopflhORMDL3-Tg mice (8) (which have a floxed transcriptional stop codon positioned in front of the hORMDL3 transgene, preventing hORMDL3 transcription) with a smooth muscle–specific cre mouse (Myh11-creEGFP) (cre expressed selectively in smooth muscle cells under the smooth muscle myosin heavy chain Myh11 promoter tagged with EGFP) (9) (Figure 2A). Crossing the RFP-StopFLhORMDL3-Tg mice with Myh11-creEGFP mice results in smooth muscle–expressed cre excising the floxed transcriptional stop codon positioned in front of the hORMDL3 transgene, which then allows hORMDL3 expression in smooth muscle cells in hORMDL3Myh11eGFP-cre but not littermates (referred to as WT) (Figure 2A).
Selective expression of hORMDL3 in ASM derived from hORMDL3Myh11eGFP-cre mice.
Primer sets detected predicted sizes of mRNA by reverse transcription quantitative PCR (RT-qPCR) for hORMDL3, Myh11-Cre, and eGFP in progeny of RFP-StopflhORMDL3-Tg mice crossed with Myh11-creeGFP mice (i.e., hORMDL3Myh11eGFP-cre or WT littermate mice) (Figure 2B). The hORMDL3 transgene in hORMDL3Myh11eGFP-cre mice was selectively expressed in smooth muscle cells and was not detected in airway epithelium (Figure 2C), or in BAL macrophages (Figure 2C), as assessed by RT-qPCR. Significant expression of the ORMDL3 transgene was detected in ASM derived from hORMDL3Myh11eGFP-cre mice but not in ASM derived from WT mice (Figure 2C). We also quantitated hORMDL3 mRNA copy number in ASM derived from hORMDL3Myh11eGFP-cre mice compared with WT mice. Copies of hORMDL3 mRNA were readily detected in ASM derived from hORMDL3Myh11eGFP-cre mice but not in ASM derived from WT mice (Figure 2D). Additionally, copies of hORMDL3 mRNA in human ASM from asthmatic airways showed relatively similar ORMDL3 copy number (Figure 2E) as compared with ORMDL3 copy number in ASM derived from hORMDL3Myh11eGFP-cre mice (Figure 2D). Expressing human ORMDL3 in ASM in hORMDL3Myh11eGFP-cre mice did not influence levels of endogenous murine Ormdl3 expression in lung ASM (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136911DS1).
Increased ASM hypertrophy in hORMDL3Myh11eGFP-cre mice.
As ASM hypertrophy is a feature of asthma (10–12), we investigated whether ORMDL3 expression in ASM influenced hypertrophy by measuring both cell size by FACS and ASM protein to DNA ratio (an index of cell hypertrophy) and by image analysis in ASM derived from hORMDL3Myh11eGFP-cre and WT mice. ASM cells from hORMDL3Myh11eGFP-cre mice were significantly larger in cell size compared with ASM from WT mice (P < 0.05) as quantitated by FACS (Figure 3A). The ratio of protein to DNA content in hORMDL3smgfp-cre (smooth muscle green fluorescent protein, smgfp) ASM cells were also significantly higher compared with WT ASM cells (P < 0.05) (Figure 3B). In addition, ASM cells from hORMDL3Myh11eGFP-cre mice were significantly larger in cell size compared with ASM from WT mice (P < 0.05) as quantitated by image analysis (Figure 3, C and D).
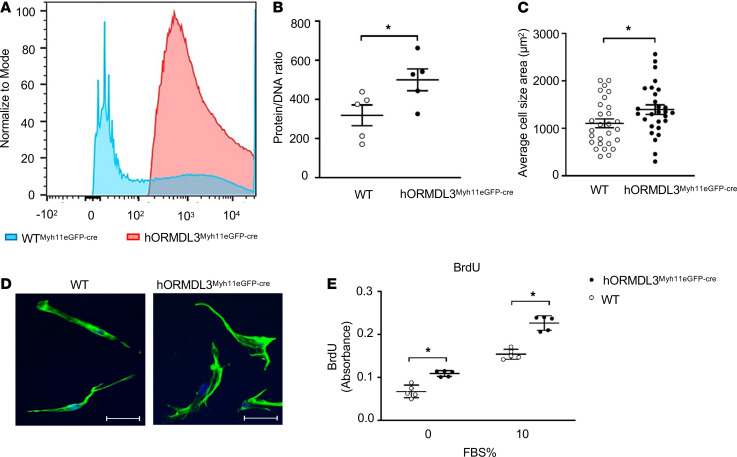
Figure 3. Effect of ORMDL3 on ASM hypertrophy and hyperplasia.
(A) FACS analysis using forward scatter plot of ASM from WT (blue peak) and hORMDL3Myh11eGFP-cre (red peak) mice was used to determine ASM cell size after gating on live ASM cells. (B) Protein/DNA ratio was used as an index of ASM hypertrophy (n = 5/group). (C and D) Microscopic image analysis of ASM quantitated average cell size area following staining with phalloidin tagged with Alexa Fluor 488 (n = 28/group). Scale bar: 50 μm. (E) ASM proliferation was determined by BrdU incorporation at baseline and following 10% FBS stimulation (n = 5/group). *P < 0.05 compared with WT. P values are results of unpaired 2-tailed t tests.
Increased ASM proliferation in hORMDL3Myh11eGFP-cre mice.
As ASM hyperplasia is a feature of asthma (10, 13, 14), we investigated whether increased expression of ORMDL3 in ASM influenced proliferation utilizing a BrdU assay. Unstimulated hORMDL3Myh11eGFP-cre ASM cells were significantly more proliferative compared with unstimulated WT ASM cells (P < 0.05) (Figure 3E). In addition, when stimulated to proliferate with 10% FBS, hORMDL3Myh11eGFP-cre ASM cells were significantly more proliferative compared with 10% FBS–stimulated WT ASM cells (P < 0.05) (Figure 3E).
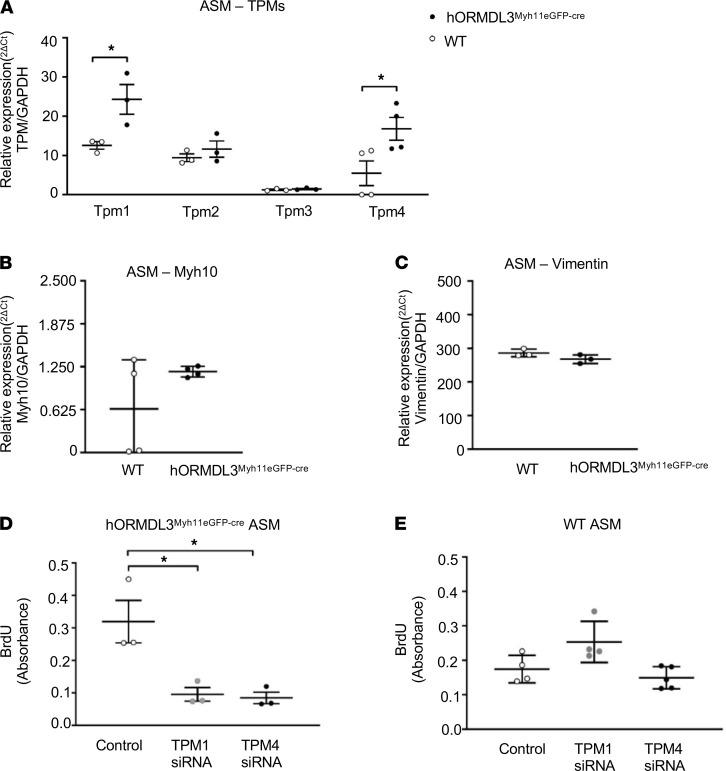
Increased expression of TPM1 and TPM4 in ASM of hORMDL3Myh11eGFP-cre mice.
As ASM derived from hORMDL3Myh11eGFP-cre mice had increased proliferation, we examined whether ORMDL3 expressing ASM had markers associated with a proliferative synthetic ASM phenotype, including tropomysins 1–4, vimentin, and myosin heavy chain 10 (Myh10) (15). These studies demonstrated that ASM from hORMDL3Myh11eGFP-cre mice had significantly higher levels of expression of TPM1 (P < 0.05) and TPM4 (P < 0.05) but not TPM2 or TPM3 mRNA by RT-qPCR compared with WT mice (Figure 4A). There was no increase in Myh10 (Figure 4B), vimentin (Figure 4C), or extracellular matrix proteins including laminin, perlecan, and fibronectin (Supplemental Figure 2A) in ASM from hORMDL3smgfp-cre mice. Thus, ORMDL3 upregulated a specific subset of ASM proliferative synthetic genes, in particular TPM1 and TPM4 (16). ORMDL3 expression in ASM reduced expression of collagen type I and collagen type IV (Supplemental Figure 2A).
Figure 4. Effect of ORMDL3 on ASM TPM expression and ASM proliferation.
(A). Levels of TPM1, TPM2, TPM3, and TPM4 mRNA were quantitated by RT-qPCR in ASM derived from WT and hORMDL3Myh11eGFP-cre mice (n = 3–4/group). (B) Levels of Myh10 and (C) vimentin mRNA were quantitated by RT-qPCR in ASM derived from WT and hORMDL3Myh11eGFP-cre mice (n = 3–4/group). (D) BrdU proliferation assay of ASM from hORMDL3Myh11eGFP-cre (n = 3/group) or (E) ASM from WT mice (n = 3/group) knocked down with TPM1 siRNA, TPM4 siRNA, or control scrambled siRNA. *P < 0.05 compared with control scrambled siRNA. P values are results of unpaired 2-tailed t tests (A–C) or 1-way ANOVA with Tukey’s post hoc test (D and E).
TPM1 and TPM4 mediate ORMDL3-induced ASM proliferation.
As ORMDL3 induced increased levels of TPM1 and TPM4 in ASM (Figure 4A), we investigated whether TPM1 and TPM4 regulate ORMDL3-induced ASM proliferation or hypertrophy by performing knockdown experiments using siRNAs for TPM1 or TPM4 in ASM. In ASM from hORMDL3Myh11eGFP-cre mice siRNA knockdown of either TPM1 or TPM4 significantly reduced ASM proliferation compared with ASM transfected with scramble siRNA as assessed by BrdU incorporation (Figure 4D). In contrast, in WT ASM, which expressed significantly lower levels of TPM1 (P = 0.04) and TPM4 (P = 0.03) compared with ASM from hORMDL3Myh11eGFP-cre mice, siRNA knockdown TPM1 or TPM4 in WT mice did not inhibit ASM proliferation (Figure 4E). Thus TPM1 and TPM4 do not contribute to baseline ASM proliferation in WT mice but do contribute to ORMDL3-induced ASM proliferation. The efficiency of TPM1 and TPM4 siRNA knockdown was approximately 50% and 80%, respectively.
Additionally, we investigated whether siRNA knockdown of TPM1 or TPM4 also regulated ASM hypertrophy by performing cell size measurement. In contrast to TPM1 and TPM4 regulating ORMDL3-induced ASM proliferation, siRNA inhibition of either TPM1 or TMP4 in ASM from hORMDL3Myh11eGFP-cre or WT mice had no effect on ASM hypertrophy as quantitated by cell size using FACS (data not shown). This suggests that TPM1 and TPM4 play a significant role in ORMDL3-induced ASM hyperplasia but do not play a significant role in ORMDL3-induced ASM hypertrophy.
ASM from hORMDL3Myh11eGFP-cre mice have increased contractility to histamine in vitro.
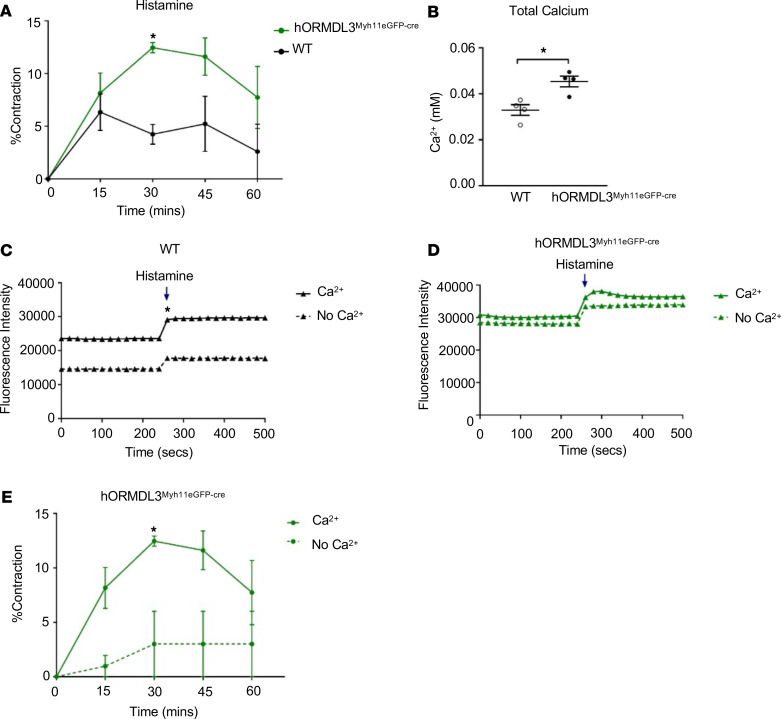
In these in vitro studies using an ASM gel contraction assay, ASM derived from the airways of hORMDL3Myh11eGFP-cre mice had significantly increased contractility to histamine compared with WT ASM cells (P < 0.05) (Figure 5A).
Figure 5. Effect of ORMDL3 on ASM contractility and Ca2+ levels.
(A) ASM contractility to histamine was quantitated in ASM from WT and hORMDL3Myh11eGFP-cre mice (n = 3/group) by measuring the area of ASM-containing collagen gels. (B) Total intracellular Ca2+ levels (n = 4/group) and (C and D) calcium flux following 100 μm histamine stimulation were assessed in ASM from WT and hORMDL3Myh11eGFP-cre mice in the presence or absence of extracellular Ca2+ (n = 3/group). (E) Contractility to histamine was quantitated in ASM-containing collagen gels from hORMDL3Myh11eGFP-cre mice in the presence or absence of extracellular Ca2+ (n = 3/group). *P < 0.05 compared with WT or with no Ca2+. P values are results of unpaired 2-tailed t tests.
ASM from hORMDL3Myh11eGFP-cre mice have increased intracellular Ca2+ levels and increased Ca2+ flux response to histamine.
As Ca2+ regulates ASM contraction by regulating the interaction of contractile proteins in ASM (17, 18), we investigated whether selective increased expression of ORMDL3 only in ASM cells would influence not only ASM contractility (Figure 5A) but also ASM Ca2+ levels and flux. Interestingly, these studies demonstrated significantly higher baseline total free intracellular Ca2+ levels in hORMDL3Myh11eGFP-cre ASM cells compared with WT ASM cells (P < 0.01) as assessed with a chromogenic calcium assay (Figure 5B).
To determine whether Ca2+ flux across the ASM cell surface plasma membrane was altered in ASM from hORMDL3Myh11eGFP-cre mice, we measured Ca2+ flux in ASM cells loaded with the Ca2+-sensitive Fluo-4 Direct dye. Baseline levels of intracellular Ca2+ based on this dye indicator method were much higher in ASM derived from hORMDL3Myh11eGFP-cre mice compared with WT ASM cells (P < 0.05) (Figure 5, C and D). However, ASM cells from hORMDL3Myh11eGFP-cre and WT mice had a similar increase in Ca2+ influx following 100 μM histamine stimulation (Figure 5, C and D). Thus, 2 methods of measuring intracellular Ca2+, in which one utilizes a chromogenic assay (Figure 5B) and the second a dye indicator (Figure 5, C and D), both demonstrated that ASM derived from hORMDL3Myh11eGFP-cre mice had increased intracellular Ca2+ levels. Overall, these results demonstrated that increased expression of ORMDL3 in ASM leads to increased ASM contraction (Figure 5A), which is associated with increased baseline intracellular Ca2+ levels (Figure 5, C and D).
ASM from hORMDL3Myh11eGFP-cre mice has reduced requirement for extracellular Ca2+ for Ca2+ flux and contractile response to histamine.
We initially investigated whether removal of extracellular Ca2+ would affect the increased levels of intracellular Ca2+ in ASM from hORMDL3Myh11eGFP-cre mice. Removal of extracellular Ca2+ did not significantly reduce the increased intracellular Ca2+ levels in ASM from hORMDL3Myh11eGFP-cre mice (Figure 5D). In contrast, removal of extracellular Ca2+ significantly reduced the intracellular Ca2+ levels in ASM from WT mice (P < 0.05) (Figure 5C). As removal of extracellular Ca2+ did not significantly reduce the increased intracellular Ca2+ levels in ASM from hORMDL3Myh11eGFP-cre mice (Figure 5D), we assessed whether removal of extracellular Ca2+ would affect the contractility of ASM from hORMDL3Myh11eGFP-cre mice. Interestingly, ASM contraction to histamine in ASM derived from hORMDL3Myh11eGFP-cre mice was significantly reduced in the absence of extracellular Ca2+ (Figure 5E). Taken together this suggests that although increased ORMDL3 in ASM contributes to increased intracellular Ca2+ levels (even in the absence of extracellular Ca2+), extracellular Ca2+ levels play a crucial role in histamine-induced ASM contraction in hORMDL3Myh11eGFP-cre mice.
Does ORMDL3 increase intracellular Ca2+ levels by regulating Ca2+ channels at the ASM cell surface?
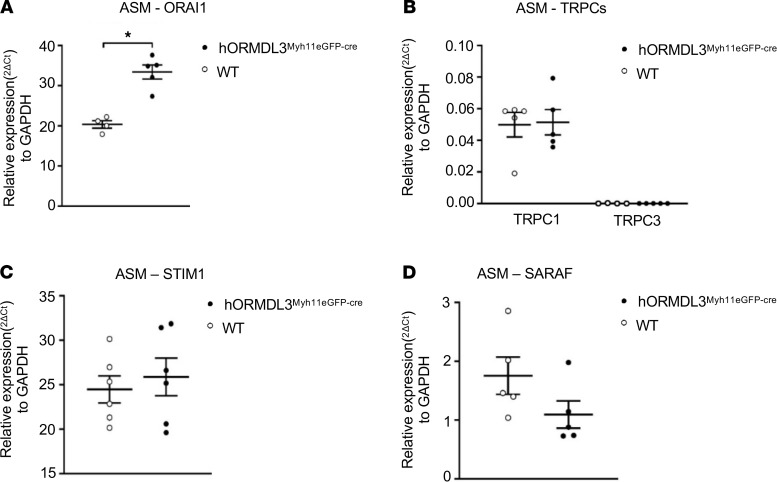
The coordinated interactions of a number of proteins from the plasma and ER membranes control store-operated calcium entry (SOCE) to replenish internal Ca2+ stores and generate intracellular Ca2+ signals (19). As expression of ORMDL3 in ASM increased intracellular Ca2+ levels, we investigated whether ORMDL3 regulated pathways that could increase intracellular Ca2+ levels. Potential pathways linking the ER (where ORMDL3 is localized) (2) to the cell surface plasma membrane to influence cell surface Ca2+ influx include effects of ORMDL3 on cell surface calcium channels, which regulate cell surface Ca2+ influx (includes Orai1 channels, transient receptor potential channels, refs. 20, 21), and/or effects of ORMDL3 on either stromal interaction molecule 1 (STIM1), an ER-resident Ca2+ sensor (22), or store-operated calcium entry associated regulatory factor (SARAF) (23), an ER-resident component of the SOCE, which serves as an important brake on SOCE (Figure 6). We therefore used qPCR to quantitate ASM levels of these cell surface Ca2+ channels (Orai1, transient receptor potential channels [TRPCs]) and the ER-resident Ca2+ sensors STIM1 and SARAF in hORMDL3Myh11eGFP-cre and WT mice. These studies demonstrated that ORMDL3 expression in ASM derived from hORMDL3Myh11eGFP-cre mice had significantly increased levels of the Orai1 cell surface Ca2+ channel, which mediates Ca2+ influx into the cytoplasm (P < 0.05) (Figure 7A). In contrast, ORMDL3 expression in ASM derived from hORMDL3Myh11eGFP-cre mice did not regulate levels of TRPC cell surface Ca2+ channels (TPRC1, TPRC3) (Figure 7B) or levels of either STIM1 (Figure 7C) or SARAF (Figure 7D), which prevents cells from overfilling with Ca2+ (23). Cell surface Orai1 channels are components of 2 different SOCE channels involved in Ca2+ flux at the cell surface, namely calcium release activated calcium (CRAC) channels (composed of Orai1 subunits) (22), and SOC channels (composed of Orai1 subunits as well as TRPC subunits) (19). As ORMDL3 influences only levels of Orai1, but not TRPC, it is more likely to affect levels of CRAC channels (composed of Orai1 subunits) as compared with SOC channels (composed of Orai1 and TRPC subunits). We also found no significant difference in the levels of the Orai1 regulators Septin (24) in ASM from hORMDL3Myh11eGFP-cre and WT mice (Supplemental Figure 2B) to explain the increase in Orai1 channel levels induced by ORMDL3 (Figure 7A). In addition, we found no difference in levels of calcium release activated channel regulator 2A (CRACR2A, a cytosolic Ca2+ sensor that stabilizes CRAC channels) (25) in ASM from hORMDL3Myh11eGFP-cre and WT mice (Supplemental Figure 2B).
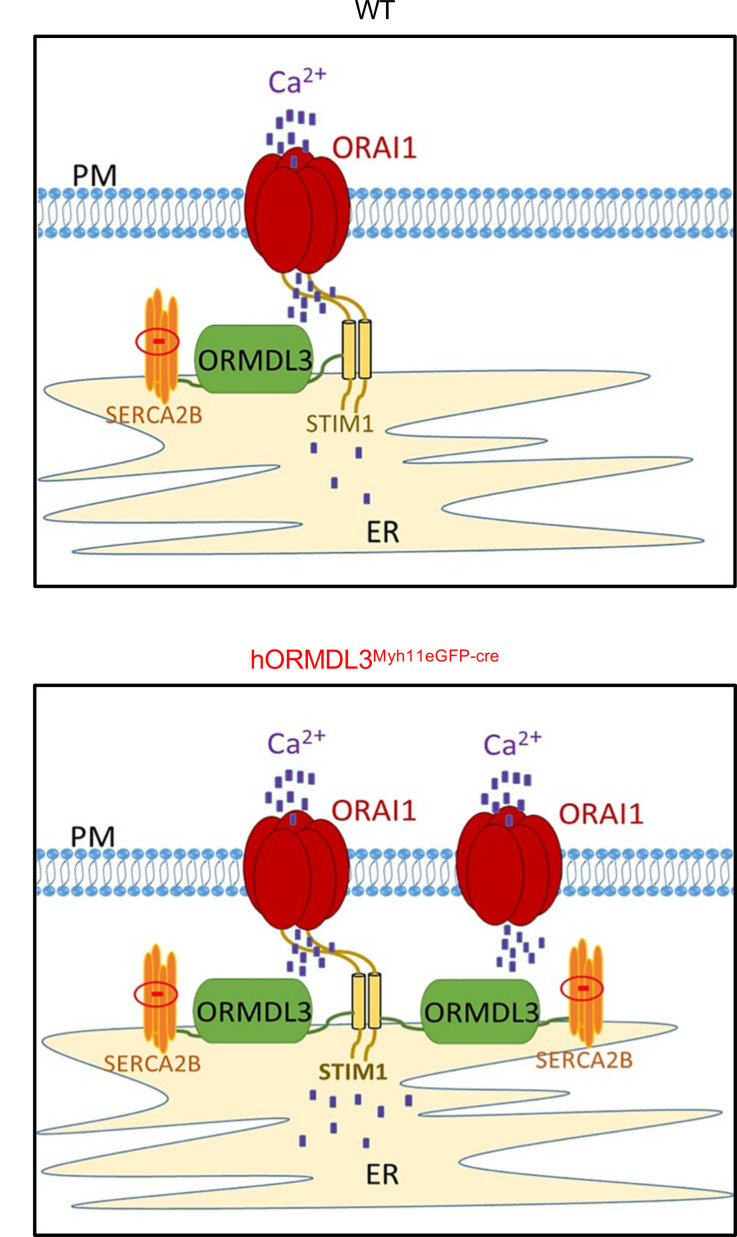
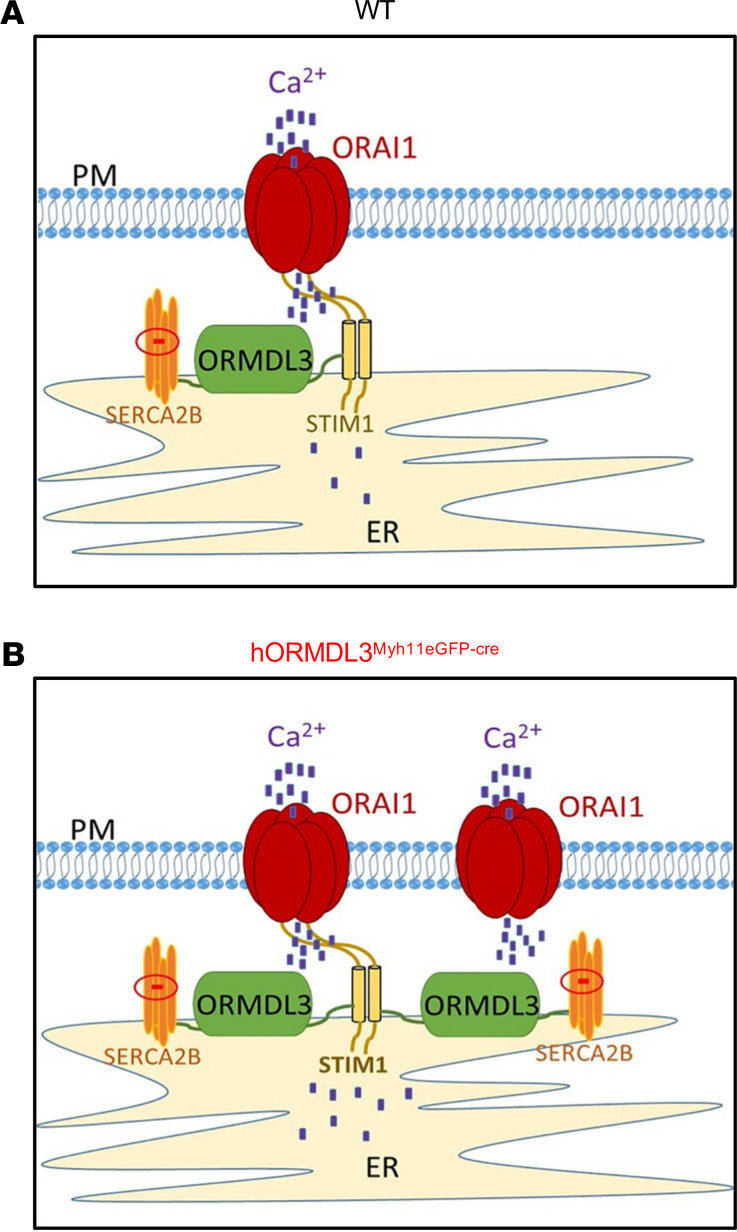
Figure 6. ORMDL3 and regulation of Ca2+ in ASM.
Overview of potential mechanisms involved in the regulation of baseline intracellular Ca2+ levels in ASM. (A) WT mouse ASM and (B) hORMDL3Myh11eGFP-cre mouse ASM. (B) ASM from hORMDL3Myh11eGFP-cre mice have higher baseline levels of intracellular Ca2+ (depicted as blue dots), more Orai1 channels (through which extracellular Ca2+ enters the cell) at the cell surface plasma membrane (PM), as well as more SERCA2b channels and more ORMDL3 in the ER compared with (A) WT ASM. Levels of the intracellular Ca2+ sensor STIM1 are similar in ASM derived from hORMDL3Myh11eGFP-cre and WT mice.
Figure 7. Effect of ORMDL3 on ASM expression of mRNAs encoding calcium channels and calcium sensors.
(A) ASM cell surface Ca2+ channel expression including (A) Orai1 and (B) TRPC1 and TRPC3 mRNA were assessed by RT-qPCR in WT and hORMDL3Myh11eGFP-cre ASM (n = 4–6/group). (C and D) ER-associated and calcium-sensing STIM1 and SARAF mRNA levels were also assessed via RT-qPCR in WT and hORMDL3Myh11eGFP-cre ASM (n = 4–6/group). *P < 0.05 compared with WT. P values are results of unpaired 2-tailed t tests.
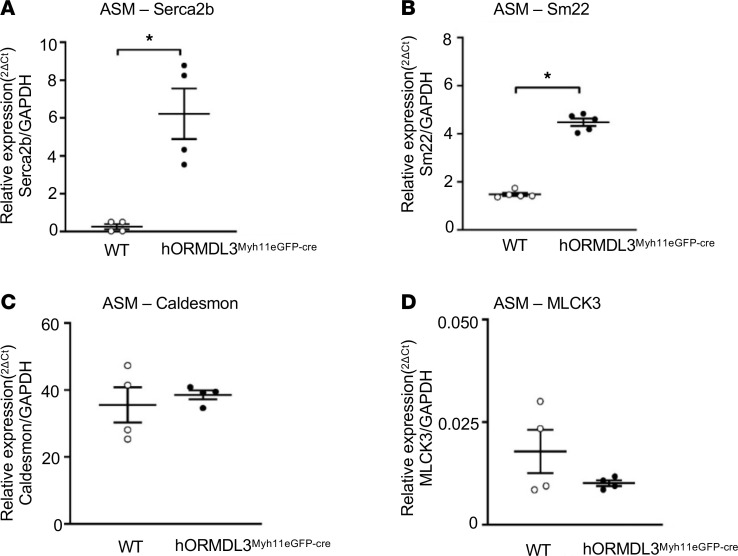
hORMDL3Myh11eGFP-cre mice ASM express increased contractile genes (SERCA2b and SM22, but not Caldesmon or myosin light chain kinase 3).
As ASM derived from hORMDL3smgfp-cre mice had increased contractility and increased intracellular Ca2+ levels, we examined whether ORMDL3-expressing ASM had markers associated with a contractile ASM phenotype, including SERCA2b, SM22, myosin light chain kinase 3 (MLCK3), and Caldesmon (26–29). Levels of expression of ASM contractile genes Serca2b (P < 0.001) (Figure 8A) and Sm22 mRNA (P < 0.05) (Figure 8B), but not Caldesmon (Figure 8C) or MLCK3 (Figure 8D) mRNA, were significantly higher in ASM cells from hORMDL3Myh11eGFP-cre mice compared with WT mice. This suggests that ORMDL3 expression in ASM can lead to altered gene expression of selected genes (SERCA2b, SM22) known to be involved in smooth muscle cell contraction (29, 30), a feature of asthma.
Figure 8. Effect of ORMDL3 on ASM expression of contractile mRNA.
(A) ASM cell expression of (A) SERCA2b, (B) Sm22, (C) Caldesmon, and (D) MLCK3 mRNA were assessed by RT-qPCR in WT and hORMDL3Myh11eGFP-cre ASM (n = 4–5/group). *P < 0.05 compared with WT. P values are results of unpaired 2-tailed t tests.
Previous studies have demonstrated that ORMDL3 is localized to the ER (2) and that the ER plays an important role in regulating cytosolic calcium levels through SERCA pumps, which accumulate Ca2+ in the ER lumen (30). As our studies demonstrate that ORMDL3 upregulated ASM levels of SERCA2b (Figure 8A), a Ca2+ pump localized to the sarcoplasmic endoplasmic reticulum in close proximity to ORMDL3, ORMDL3 can through increased SERCA2b increase accumulation of Ca2+ in the ER lumen from the cytoplasm. However, as this ORMDL3-induced SERCA2b mediates movement of Ca2+ within 2 intracellular compartments (i.e., from the cytoplasm to the ER lumen), it should not increase overall intracellular Ca2+ levels, which we have noted in ASM cells from hORMDL3Myh11eGFP-cre mice.
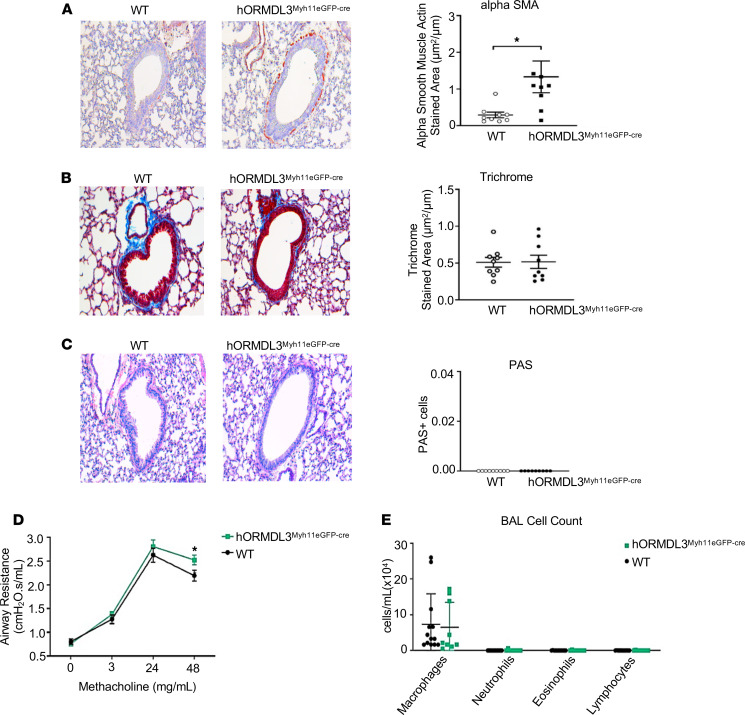
Increased ASM (but not fibrosis or mucus) in airways of hORMDL3Myh11eGFP-cre mice.
To determine whether mice expressing increased ORMDL3 in ASM selectively had increased ASM in vivo, we quantitated levels of ASM in hORMDL3Myh11eGFP-cre and WT mice aged 12 weeks that had not been exposed to any environmental stimulus, such as allergen challenge. Previous studies had demonstrated that at 12-week-old mice universally expressing increased ORMDL3 in all cells spontaneously developed increased ASM, increased peribronchial fibrosis, and increased mucus (8). Selective expression of ORMDL3 in ASM in hORMDL3Myh11eGFP-cre mice resulted in spontaneous significantly increased peribronchial ASM assessed by image analysis quantitation of the area of ASM immunostaining with an antibody against alpha–smooth muscle actin (P < 0.05) (Figure 9A). In contrast to universal ORMDL3-Tg mice, which also exhibited increased peribronchial fibrosis and increased mucus (8), selective expression of ORMDL3 in ASM in hORMDL3Myh11eGFP-cre mice did not show evidence of increased peribronchial fibrosis (Figure 9B) or increased mucus (Figure 9C).
Figure 9. Effect of ORMDL3 on airway remodeling, AHR, and airway inflammation in vivo.
(A) WT and hORMDL3Myh11eGFP-cre mice were sacrificed at 12 weeks of age in the absence of exposure to allergen. Lungs were used to quantitate by image analysis (A) ASM (alpha–smooth muscle actin–stained area) (n = 8–9 mice/group), (B) peribronchial fibrosis (trichrome-stained area) (n = 9 mice/group), and (C) mucus periodic acid–Schiff–positive epithelial cells (n = 8–9 mice/group). (D) Airway resistance following MCh challenge was assessed in intubated mice (n = 9 mice/group). (E) Airway inflammation was determined by quantitation of total BAL cells, macrophages, neutrophils, eosinophils, and lymphocytes in WT and hORMDL3Myh11eGFP-cre mice (n = 9 mice/group). *P < 0.05 compared with WT. P values are results of unpaired 2-tailed t tests.
AHR to methacholine in hORMDL3Myh11eGFP-cre mice.
hORMDL3Myh11eGFP-cre mice not exposed to allergen had a small but statistically significant spontaneous increase in airway responsiveness to methacholine (MCh) 48 mg/mL compared with WT mice at 12 weeks of age (P < 0.05) (Figure 9D). hORMDL3Myh11eGFP-cre mice challenged with house dust mite (HDM) had a significant increase in AHR compared with nonchallenged hORMDL3Myh11eGFP-cre mice (P = 0.0061) (Supplemental Figure 3A). However, there was no increase in AHR in hORMDL3Myh11eGFP-cre mice challenged with HDM compared with WT mice challenged with HDM (Supplemental Figure 3A). The small but statistically significant spontaneous increase in airway responsiveness to MCh noted in hORMDL3Myh11eGFP-cre mice in the initial experiment (Figure 9D) was again noted in the nonchallenged control hORMDL3Myh11eGFP-cre mice group in the acute HDM experiment (P = 0.0253, nonchallenged control hORMDL3Myh11eGFP-cre mice vs. nonchallenged WT) (Supplemental Figure 3A).
Quantitation of airway inflammation in BAL in hORMDL3Myh11eGFP-cre mice.
hORMDL3Myh11eGFP-cre mice did not develop any spontaneous evidence of airway inflammation at 12 weeks of age (the time point they had spontaneous increased ASM) (Figure 9A) as assessed by quantitation of total cells, macrophages, lymphocytes, eosinophils, and neutrophils in BAL (Figure 9E). Levels of BAL eosinophils were similar in acute HDM–challenged hORMDL3Myh11eGFP-cre mice and HDM-challenged WT mice (Supplemental Figure 3B).
Discussion
In this study we provide potentially novel evidence that ORMDL3 expression selectively in ASM can induce increased ASM hypertrophy, proliferation, and contractility, which together may help explain the epidemiologic linkage of chromosome 17q12-21 to the development of childhood onset asthma (1) (where ASM can express ORMDL3 in the lower airway) but not to the development of allergic rhinitis (7) (where there are no ASM cells in the nasal mucosa). In addition to demonstrating that ORMDL3 expression in ASM induced functional changes (i.e., increased ASM hypertrophy, proliferation, and contractility), we identified potentially novel pathways through which ORMDL3 may contribute to these ASM changes characteristic of asthma. These pathways we identified to be ORMDL3 regulated in ASM include (a) TPM1 and TPM4 mediated ASM hyperplasia, (b) ORMDL3 regulated Ca2+ signaling (increased levels of intracellular Ca2+, increased cell surface membrane Orai1 Ca2+ channels, which mediate influx of Ca2+ into the cytoplasm), and (c) ORMDL3 upregulated expression of ASM contractile genes (Serca2b and Sm22). In addition, the potential importance of ORMDL3-induced increases in Serca2b in ASM to ORMDL3-induced ASM hypertrophy is suggested from studies of the heart, in which mice expressing increased levels of Serca2b exhibited cardiac muscle hypertrophy and increased contractility (30, 31). Interestingly, in our study mice selectively expressing human ORMDL3 in ASM developed increased expression of Serca2b, ASM hypertrophy, and increased contractility, suggesting a potential link between Serca2b and ASM hypertrophy and contractility not only in cardiac muscle (30, 31) but also in ASM, in which levels of ORMDL3 are increased. Thus, childhood asthmatics with SNPs on chromosome 17q12-21 linked to increased ORMDL3 expression may have increased amounts of peribronchial smooth muscle due to ASM hypertrophy and hyperplasia. In addition, it is also likely that ORMDL3 expression in cells other than ASM, such as CD4+ cells, results in enhanced Th2 responses (4), further increasing AHR through effects of Th2 cytokines such as IL-13 on ASM (32) to increase AHR.
The increased thickness of the ASM layer is a characteristic feature of asthma and is due to both ASM hypertrophy (10–12) and hyperplasia (10, 13, 14), without a change in the proportion of extracellular matrix, all of which are related to asthma severity (33). In this study we have demonstrated that ORMDL3 expression in ASM induces both ASM hypertrophy and hyperplasia, without a change in expression of extracellular matrix proteins, recapitulating many of the findings noted in asthmatics. While many studies in mice use tracheal smooth muscle, an advantage of this study is that we used mouse bronchial smooth muscle (of greater relevance to asthma than tracheal smooth muscle), which expressed copy numbers of human ORMDL3 similar to that noted in ASM in human patients with asthma. Evidence that ORMDL3 regulated ASM hypertrophy was noted in studies using FACS, protein/DNA ratio, and microscopic image analysis. While many cytokines/mediators or mechanical stretch signals arising extrinsic to ASM have been identified to modulate ASM hypertrophy and hyperplasia (17, 34, 35), this study makes the likely novel observation of the importance of intrinsic expression in ASM of a gene (i.e., ORMDL3) on chromosome 17q12-21 to the development of ASM hypertrophy and hyperplasia. Thus, children with SNPs linked to chromosome 17q12-21 and increased expression of ORMDL3 in ASM may develop a childhood onset increased thickness of the ASM layer, predisposing to the development of childhood onset asthma.
This study also demonstrated the importance of TPM (16) to ORMDL3-induced ASM proliferation, which can manifest as ASM hyperplasia in the airway in asthma. TPM has not previously been linked to ORMDL3 or to ASM proliferation, underscoring the novelty of this finding. In addition, only 2 of the 4 TPM genes (i.e., TPM1 and TPM4 but not TPM2 or TPM3) were regulated by ORMDL3. TPM is a coiled-coil dimer protein that lies end to end in the actin groove and plays an important role in regulating muscle contraction (16). Thus, ORMDL3 upregulation of TPM1 and TPM4 could regulate not only ASM proliferation but also ASM contractility. ASM contraction results from the interaction of myosin with F-actin, which is controlled by calcium ions via regulatory proteins including TPM associated with F-actin (16). Therefore, the ability of ORMDL3 to regulate TPM1, TPM4, and Ca2+ may have important implications for ASM contractility. Support for the importance of TPMs to muscle contraction is also derived from studies of cardiac muscle in which TPM1 mutations have been associated with systolic hypercontractility in hypertrophic obstructive cardiomyopathy (36).
In addition, we identified that ORMDL3 regulated increased expression of ASM contractile genes (Serca2b and Sm22). Serca2b is an intracellular ATP-driven Ca2+ pump that transports Ca2+ from the cytoplasm into the sarco-endoplasmic reticulum (26, 37). Based on studies of increased expression of Serca2b in cardiac muscle (30, 31), increased expression of Serca2b in ASM may be the downstream pathway by which ORMDL3 influences ASM Ca2+ flux, contractility, and hypertrophy. For example, studies of mice expressing increased levels of Serca2b in the heart had increased cardiac contractility with increased Ca2+ flux in cardiomyocytes and increased cardiac muscle hypertrophy (30, 31). Interestingly, knockdown of SERCA2b in ASM reduces ASM contractility in vitro (6), defining similarities in SERCA2b function as it relates to contractility in both the heart (30, 31) and in ASM (6). While a study has reported that decreased expression of SERCA in ASM from asthmatics (who were not genotyped for chromosome 17q12-21 SNPs or enrolled based on history of childhood onset asthma) contributes to airway remodeling (38), we hypothesize that genotyping childhood asthmatics for chromosome 17q12-21 SNPs may identify a subset of asthmatics with increased ORMDL3 and SERCA2b expression. In addition to ORMDL3 regulating SERCA2b, we also noted that ORMDL3 regulated increased levels of SM22a (transgelin), an actin filament–associated protein in smooth muscle (29). Studies of endobronchial biopsies from asthmatics have shown by qPCR and immunohistochemistry that levels of SM22 are increased in ASM in asthma compared with nonasthmatics (39). Although SM22 is highly expressed in ASM and binds to filaments of actin in smooth muscle cells in vitro and in vivo (29), its function in ASM requires further study in asthma using allergen or viral challenge models because it is not essential for constitutive smooth muscle development as demonstrated in Sm22-KO mice (40). Thus, ORMDL3 expression in ASM is associated with increased expression of selected ASM contractile proteins (Serca2b, SM22a, but not MLCK3) and increased contractility.
In this study we also made the potentially novel observation that ASM from hORMDL3Myh11eGFP-cre mice has increased basal total intracellular Ca2+ levels, which are associated with increased basal ASM contractility. When cytosolic Ca2+ levels are increased, the combination of Ca2+ and calmodulin activate myosin light chain (MLC) kinase, which phosphorylates MLC, leading to enhanced ASM contraction (17, 41). In previous studies we have demonstrated that lung slices from universal ORMDL3-Tg mice exhibit increased Ca2+ oscillations when stimulated with acetylcholine (6), but prior studies have not examined effects of selective ORMDL3 expression in ASM on total intracellular Ca2+ levels or ORMDL3 effects on Ca2+-replenishing pathways in ASM (Serca2b, STIM1, Orai1, or regulators of Orai1). Ca2+ oscillations occur as the Ca2+ is taken up into sarcoplasmic reticulum (SR) stores via SERCA. ASM Ca2+ stores are then replenished via SERCA and by the oligomerization of the SR-localized STIM1 (a Ca2+ sensor), which then associates with Orai1 in the plasma membrane, causing it to form channels, which mediate SOCE (42). In this study, we advance our knowledge of how ORMDL3 regulates intracellular Ca2+ in ASM to demonstrate that ORMDL3 increases total intracellular Ca2+ levels and that this is associated with increased Serca2b (which is able to replenish SR Ca2+ stores), as well as increased Orai1, which mediates cell surface SOCE. Thus, increased total intracellular Ca2+ in ASM may be mediated by ORMDL3 located in the ER regulating a cell surface filling mechanism for Ca2+ entry mediated by Orai1. This ORMDL3-regulated Orai1 cell surface filling mechanism for Ca2+ entry would be controlled by the filling state of intracellular Ca2+ and by SOCE, which has been shown to depend on the cell surface transmembrane protein Orai1 and its interaction with STIM1, an ER Ca2+ sensor (43). Ca2+ influx due to low ER stores gives rise to the Ca2+ released activated Ca2+ current (ICRAC), which then activates Orai1 to open in order to replenish cytosolic Ca2+ concentration. Interestingly, allergen-challenged mice have increased levels of Orai1 in ASM (44).
In contrast to our study demonstrating that ORMDL3 upregulates Ca2+ levels and Orai1 levels in primary mouse ASM cells, studies in a human Jurkat T cell leukemia cell line transfected with ORMDL3 have shown that ORMDL3 negatively modulates SOCE by inhibiting Ca2+ influx and inhibits STIM1 in Jurkat cells (45, 46). It should be noted that in our studies the ORMDL3 mRNA copy number in mouse ASM we studied, which was derived from hORMDL3Myh11eGFP-cre mice, was similar to the hORMDL3 copy number detected in ASM from human patients with asthma. The differences in the results from the 2 studies may relate to the difference in cell types studied (primary mouse ASM cells vs. leukemic T cell line), methods of generating increased ORMDL3 (transgenic mice vs. transfection), methods used (intracellular Ca2+ dye Fura2 vs. Fluo-4, cresolphthalein), outcomes studied (PCR for Stim1), and/or other factors.
While previous studies of universal ORMDL3-expressing transgenic mice (8) and their precision-cut lung slices (6) have demonstrated that universal ORMDL3 expression in all cells results in spontaneous increased AHR to MCh, this study markedly extends these observations to demonstrate that selective ORMDL3 expression in ASM only (rather than in any other lung, immune, or inflammatory cell in universal ORMDL3-Tg mice) (6, 8) is inducing increased ASM contractility as well as ASM hypertrophy, which were not studied in universal ORMDL3-Tg mice. Studies of hORMDL3Myh11eGFP-cre mice also demonstrated that they had spontaneous increased levels of ASM but not spontaneous increased amounts of peribronchial fibrosis or mucus, which were noted in universal ORMDL3-Tg mice (8). Thus, cells expressing ORMDL3 other than ASM are likely to contribute to spontaneous peribronchial fibrosis and mucus. In addition, this study demonstrated that selective ORMDL3 expression in ASM regulates likely novel pathways not studied in universal ORMDL3-Tg mice, including ORMDL3 regulation of ASM hypertrophy, TPM proteins involved in ASM hyperplasia, increased expression of ASM contractile genes (Serca2b and Sm22), increased Orai1 Ca2+ channels, and increased intracellular Ca2+ levels, providing additional pathways through which ORMDL3 may regulate ASM contractility, which may contribute to the development of AHR in childhood onset asthma, which is highly linked to ORMDL3 on chromosome 17q12-21.
Our studies demonstrated that nonchallenged hORMDL3Myh11eGFP-cre mice had a modest, but reproducible, statistically significant spontaneous increase in AHR compared with nonchallenged WT mice in 2 different experiments. However, there was no increase in AHR in hORMDL3Myh11eGFP-cre mice challenged with HDM compared with WT mice challenged with HDM. This result may reflect that there is only a small but reproducible increase in AHR in nonchallenged hORMDL3Myh11eGFP-cre mice compared with nonchallenged WT mice. In addition, the allergen stimulus we used, HDM, induces a robust increase in AHR, which may mask the contribution to AHR provided by ORMDL3 in ASM. It is also likely that ORMDL3 expression in cells other than ASM, such as CD4+ cells, results in enhanced Th2 responses (4), further increasing AHR through effects of Th2 cytokines such as IL-13 on ASM (32) to increase AHR. In support of this effect of ORMDL3 expressed in CD4+ cells on AHR, prior studies of universal ORMDL3-Tg mice (8) demonstrated a greater increase in AHR compared with our studies of selective expression of ORMDL3 in ASM in hORMDL3Myh11eGFP-cre mice, suggesting that the combination of ORMDL3 expression in ASM and other cell types (possibly CD4+ cells) is needed for significant enhancement in AHR. At present no studies have reported the effect of the selective expression of increased hORMDL3 in mouse CD4+ cells on AHR.
In this study we utilized a mouse model in which we expressed increased levels of the human ORMDL3 gene, as studies of human SNPs linked to ORMDL3 in asthma demonstrate increased levels of ORMDL3 (47). ORMDL3 exhibits 96% identity between mouse and human (48), suggesting that mouse Ormdl3 and human ORMDL3 have similar functions. Although we expressed human ORMDL3 in the presence of the endogenous mouse Ormdl3 in hORMDL3Myh11eGFP-cre mice, we do not think the endogenous mouse Ormdl3 influenced our results, as the control WT mice, like the hORMDL3Myh11eGFP-cre mice, expressed similar levels of the endogenous mouse Ormdl3. Thus, the only difference in ORMDL3 between the hORMDL3Myh11eGFP-cre mice and WT mice was the level of human ORMDL3, which was the focus of this study.
In summary, our studies demonstrate that ORMDL3 expression selectively in ASM can induce spontaneous (in the absence of allergen or other environmental trigger) increased ASM hypertrophy, ASM proliferation, and contractility, which together could contribute to the development of AHR and childhood onset asthma. In addition to demonstrating that ORMDL3 expression in ASM induced functional changes, we identified potentially novel pathways regulated by ORMDL3 in ASM, including TPM1 and TPM 4 mediated ASM hyperplasia, and ORMDL3 induced increased ASM contractility to histamine in vitro. These ORMDL3-induced pathways in ASM were associated with increased levels of intracellular Ca2+, increased cell surface membrane Orai1 Ca2+ channels — which mediate influx of Ca2+ into the cytoplasm — and increased expression of ASM contractile genes, including Serca2b and Sm22. These ORMDL3-induced changes in ASM may contribute to the development of AHR in childhood onset asthma, which is highly linked to ORMDL3 on chromosome 17q12-21.
Methods
RFP-StopflhORMDL3-Tg mice
The generation of hORMDL3-Tg floxed mice (RFP-StopflhORMDL3-Tg) on a C57BL/6 background in this laboratory has been previously described (8).
Myh11-creEGFP mice
Myh11-creEGFP mice (smooth muscle–specific cre) on a C57BL/6 background were acquired from The Jackson Laboratory (9).
hORMDL3Myh11eGFP-cre mouse ASM cells for in vitro study
Mouse ASM cells were obtained from the bronchial airways dissected free of lung tissue of hORMDL3Myh11eGFP-cre and WT mice (Figure 1, A–C). Mice were euthanized and the tracheobronchial tree free of lung tissue was isolated up to the third generation by dissecting and removing the surrounding lung parenchyma and vasculature from major bronchi under a dissecting microscope (Figure 1, B and C) (Leica DMLS, Leica Microsystems). The trachea was detached from the bronchial tree, and only the bronchial tree was digested in a cocktail of collagenases (Roche Diagnostics) for 30–60 minutes at 37°C to generate a single-cell suspension of bronchial cells. Cells were then cultured in mouse smooth muscle cell media (Cell Biologics) for 3 to 4 weeks at which time 90% of the cells were ASM cells as defined by expression of ASM genes (alpha–smooth muscle actin), expression of EGFP, and morphology.
Generation of hORMDL3Myh11eGFP-cre Tg mice
To generate hORMDL3Myh11eGFP-cre mice, we crossed RFP-StopflhORMDL3-Tg mice (which have a floxed transcriptional stop codon positioned in front of the hORMDL3 transgene preventing hORMDL3 transcription) with a smooth muscle–specific cre mouse (Myh11-creEGFP) (cre expressed selectively in smooth muscle cells under the smooth muscle myosin heavy chain Myh11 promoter tagged with EGFP) (Figure 2A). Crossing the RFP-StopflhORMDL3-Tg mice with Myh11-creEGFP mice results in smooth muscle–expressed cre excising the floxed transcriptional stop codon positioned in front of the hORMDL3 transgene, which then allows hORMDL3 expression in smooth muscle cells. hORMDL3Myh11eGFP-cre and littermate (referred to as WT) mice were genotyped by PCR as previously described (8) to detect (a) hORMDL3: F1, 5′-GCAACGTGCTGGTTATTGTG-3′; F2, 5′-CCCCCTGAACCTGAAACATA-3′; R, 5′-TACAGCACGATGGGTGTGAT-3′; (b) Myh11Cre: F, GCGGTCTGGCAGTAAAAACTATC; R, GTGAAACAGCATTGCTGTCACTT; and (c) EGFP: F, AAGTTCATCTGCACCACCG; R, TCCTTGAAGAAGATGGTGCG.
The expected sizes of hORMDL3, Myh11Cre, and EGFP PCR products are 479 bp, 100 bp, and 173 bp, respectively (Figure 2B).
Human ASM cells
Postmortem human lungs from asthmatics were procured by the Arkansas Regional Organ Recovery Agency and by the National Disease Research Interchange and delivered to the Lung Cell Biology Laboratory at the Arkansas Children’s Research Institute and processed for studies of the expression of ORMDL3 mRNA copy number in ASM at UC San Diego as described (49). To obtain human ASM from postmortem asthma lungs, surrounding lung tissue was removed from bronchial airways, which were cut into small pieces (4–5 strips per airway) and transferred to a 6-well tissue culture dish for ASM attachment and outgrowth. After 7 days in culture, ASM outgrowth was present and the bronchial strips were removed. Human ASM cells were then maintained with smooth muscle cell (SMC) media (ScienCell Research Laboratories) for up to 5 passages at which time more than 90% of the cells were ASM cells as defined by expression of ASM genes (alpha–smooth muscle actin) and morphology.
hORMDL3 copy number in mouse hORMDL3Myh11eGFP-cre and human ASM cells
Mouse ASM cells were derived from hORMDL3Myh11eGFP-cre mice, and human ASM cells were derived from postmortem asthma lungs (49) as described above. hORMDL3 mRNA copy number values were quantitated in mouse and human ASM by using a dPCR method as previously described in this lab (50). In brief, total RNA was extracted from WT and hORMDL3Myh11eGFP-cre mouse ASM cells, as well as from human ASM cells, with RNA-STAT-60 (Tel-Test) and reverse-transcribed with the Oligo-dT and SuperScript II Kit (Life Technologies, Thermo Fisher Scientific). hORMDL3 mRNA copy number values were quantitated by using a dPCR method, in which the reaction mixture contained 500 ng of target cDNA, Supermix (Bio-Rad Laboratories), as well as hORMDL3-specific primers and probes (Applied Biosystems, Thermo Fisher Scientific). The reaction mixture was loaded into the sample well of a disposable droplet generator cartridge (Bio-Rad Laboratories). After droplet generation, the mixture was transferred into a 96-well PCR plate and then amplified on the Bio-Rad T100 Thermal Cycler. The plate was analyzed with a QX200 Droplet Reader, and the dPCR data were processed by using QuantaSoft software (Bio-Rad Laboratories). Results are expressed as hORMDL3 mRNA copy number normalized to mouse or human HPRT.
Selective expression of hORMDL3 transgene in ASM in hORMDL3Myh11eGFP-cre mice
To determine smooth muscle–specific expression of the hORMDL3 transgene in hORMDL3Myh11eGFP-cre mice, we obtained purified populations of ASM, bronchial epithelial cells, and BAL macrophages. Bronchial epithelial cells were obtained using a bronchial brushing technique previously described in this laboratory (51). BAL macrophages were obtained by inserting a 20-gauge catheter into the trachea and lavaging the lungs with 1 mL of PBS/EDTA, which was repeated on 5 occasions (52). Expression of the hORMDL3 transgene in ASM, bronchial epithelial cells, and BAL macrophages from hORMDL3Myh11eGFP-cre and WT mice was assessed via RT-qPCR. We also used RT-qPCR to quantitate endogenous levels of mouse Ormdl3 in ASM as described (2).
ASM cell size in hORMDL3Myh11eGFP-cre mice
We used 3 methods (FACS, image analysis, and protein/DNA ratio) to quantitate ASM cell size.
FACS.
ASM cell size was compared in hORMDL3Myh11eGFP-cre and WT ASM cells by FACS analysis using measurements of forward scatter after gating on live cells with a Novocyte cytometer (Acea Biosciences, Inc).
Image analysis.
To compare ASM cell area (as an index of hypertrophy) in ASM derived from hORMDL3Myh11eGFP-cre with that of WT mice, ASM cells were cultured on cell culture chamber slides for 24 hours. Adapting image analysis methods previously used for quantitation of cardiac SMC size (53), ASM cells were fixed with paraformaldehyde and stained with phalloidin-488 (Thermo Fisher Scientific), which stains the F-actin filament of ASM (53). Images of individual ASM cells were captured in 10 random fields, and ASM cell area was measured using the ImageJ software (NIH). Results are expressed as ASM cell area in square micrometers.
Protein/DNA ratio.
We also compared protein/DNA ratio in ASM derived from hORMDL3Myh11eGFP-cre with that of WT mice as an index of ASM hypertrophy as described (54). Protein/DNA ratio has been used as an indicator of cellular hypertrophy, as with cell size increase (i.e., hypertrophy), more protein synthesis occurs (relative to DNA synthesis) to meet the increased architectural and functional demands of the growing cell (55). To determine ASM hypertrophy, total protein to DNA content was assessed in WT and hORMDL3Myh11eGFP-cre ASM cells seeded at a density of 2 × 105 cells/well for 2 days. Protein and DNA were then isolated using Qiagen AllPrep kit, and total protein and DNA content was quantified using a NanoDrop Spectrophotometer (Thermo Fisher Scientific).
ASM hyperplasia in hORMDL3Myh11eGFP-cre mice
To compare ASM proliferation (as an index of ASM hyperplasia) in vitro in ASM derived from hORMDL3Myh11eGFP-cre and WT mice, we measured BrdU incorporation in ASM cells using a BrdU cell proliferation ELISA (Exalpha Biologicals Inc). In brief, ASM cells were seeded at 2 × 104 cells/well overnight in a 96-well plate. BrdU was incubated for 18 hours with ASM cells cultured in the presence or absence of 10% FBS (FBS stimulates ASM proliferation) (56). BrdU incorporation was measured by ELISA according to the manufacturer’s instructions.
hORMDL3Myh11eGFP-cre mouse expression of ASM cell synthetic, contractile, and Ca2+ flux genes
We utilized RT-qPCR to quantitate levels of expression of synthetic genes including tropomysins (TPM1, TPM2, TPM3, TPM4) (16), Myh10 (57), and vimentin (58); contractile genes including Serca2b (26), MLCK3 (27), Caldesmon (28), and SM22 (29); Ca2+ flux-regulating genes including STIM1 (22), Orai1 (20), TRPC1 and TRPC3 (21), and SARAF (23); regulators of Orai1 channels including CRACR2A (25) and SEPTIN (24); as well as extracellular matrix genes expressed by ASM (59) including laminin, perlecan, collagen, and fibronectin in ASM derived from hORMDL3Myh11eGFP-cre and WT mice. RT-qPCR was performed as previously described in this laboratory (8). In brief, total RNA was extracted with RNA-STAT-60 (Tel-Test) and reverse-transcribed with Clontech cDNA Synthesis Kit. RT-qPCR was performed with TaqMan PCR Master Mix, and primers were obtained from Life Technologies (Thermo Fisher Scientific). The relative amounts of transcripts were normalized to those of housekeeping gene (GAPDH) mRNA and compared between the different genes by the double delta Ct (ΔΔ) cycle threshold method as previously described in this laboratory (8).
Effect of siRNA knockdown of TPM1 or TPM4 in ASM on proliferation and cell size
Mouse ASM cultures were grown in 6-well plates using SMC media with smooth muscle growth supplement (Cell Biologics). ASM cultures were transfected with 20 nM TPM1 siRNA, TPM4 siRNA, or control siRNA by using transfection reagents siTran1.0 (Origene) and Opti-MEM (Thermo Fisher Scientific) according to the manufacturers’ instructions. The transfected ASM cells were used 24 hours after the transfection in all experiments including quantitation of ASM proliferation, ASM cell size, and expression of TPM1 and TPM4 mRNA. Detection of expression of TPM1 and TPM4 mRNA by RT-qPCR was performed with TaqMan PCR Master Mix and mouse TPM1 or TPM4 primers (Life Technologies, Thermo Fisher Scientific). The relative amounts of transcripts were normalized to those of housekeeping gene (GAPDH) mRNA and compared between control siRNA–transfected samples and TPM1 siRNA– or TPM4 siRNA–transfected samples by the ΔΔ cycle threshold method as previously described in this laboratory (2, 8).
hORMDL3Myh11eGFP-cre mouse ASM cell contraction assay in vitro
Mouse ASM (passage 3) from hORMDL3Myh11eGFP-cre and WT mice were used in an in vitro smooth muscle gel contraction assay previously described in this laboratory (6). In brief, ASM cells were seeded at a density of 2 × 105 cells/well in collagen gels free of LPS (Advanced BioMatrix). After a 3-day incubation in collagen gels containing smooth muscle complete media (ScienCell Research Laboratories) containing 1.8 mM Ca2+ or similar media containing no calcium, hORMDL3Myh11eGFP-cre or WT ASM cells were cultured in the presence or absence of an agonist (100 μM histamine) or media control for 0, 15, 30, 45, and 60 minutes. With agonist-induced ASM contraction, the area of the gel decreases significantly (6). The area of the gels was quantitated by using a Bio-Rad ImageDR transilluminator and Versadoc scanner (Bio-Rad Laboratories) with an accompanying image capture and analysis program to generate area in square millimeters. Results are expressed as the percentage of contracted gel area divided by the area at time 0 minutes.
Levels of total intracellular Ca2+ in ASM in hORMDL3Myh11eGFP-cre mice
Levels of total intracellular Ca2+ were measured in isolated ASM cells from WT and hORMDL3Myh11eGFP-cre mice. Briefly, ASM cells were seeded at 2 × 105 cells/well overnight in a 96-well plate with smooth muscle complete media (ScienCell Research Laboratories) containing 1.8 mM Ca2+. ASM cells were then collected and homogenized, and total calcium concentration was measured using a chromogenic assay. In this assay a complex is formed between calcium ions and 0-cresolphthalein and measured at optical density of 575 nm according to manufacturer’s instructions (Abcam).
Calcium flux in ASM in hORMDL3Myh11eGFP-cre mice
To determine calcium flux, 2 × 105 ASM cells/well from WT and hORMDL3Myh11eGFP-cre mice were seeded overnight in a 96-well plate in media containing 1 mM extracellular Ca2+. Calcium flux experiments were performed in minimum essential medium in the presence or absence of 1 mM extracellular Ca2+. Twenty-four hours later, the ASM cells were loaded with the calcium-sensitive Fluo-4 Direct dye reagent and incubated at 37°C for 60 minutes according to manufacturer’s instruction (Invitrogen, Thermo Fisher Scientific). Fluorescence intensity of cultured ASM cells was then measured to determine both the baseline unstimulated level and the level following histamine stimulation (100 μM) with spectrophotometry setting for excitation at 494 nm and emission at 516 nm. Fluorescence intensity at baseline and following histamine stimulation were both measured every 20 seconds for 4 minutes.
Quantitation of ASM, peribronchial fibrosis, and mucus in lungs of hORMDL3Myh11eGFP-cre mice
Lungs from naive hORMDL3Myh11eGFP-cre and naive WT mice aged 12 weeks (n = 12 mice/group) were processed for immunohistology (paraffin-embedded lung sections) as previously described in this laboratory (8). As naive universal hORMDL3-Tg mice develop spontaneous increased ASM, peribronchial fibrosis, mucus, and AHR in the absence of airway inflammation by 12 weeks of age (8), this same 12-week time point was chosen to study naive mice selectively expressing hORMDL3 in ASM. For paraffin-embedded sections, the left lung was inflated with an intratracheal injection of the same volume of 4% paraformaldehyde solution (Sigma Chemicals) to preserve the pulmonary architecture. Additionally, right lung lobes were initially snap-frozen in liquid nitrogen and stored at –80°C for later processing for protein and RNA extraction.
Quantitation of ASM.
Lung sections were immunostained with an anti–alpha–smooth muscle actin primary antibody (Abcam; catalog number ab7817; clone 1A4) to detect peribronchial smooth muscle. Quantification of the positively stained peribronchial area in paraffin-embedded lung sections was outlined and measured using a light microscope (Leica DMLS, Leica Microsystems) attached to an image analysis system (Image-Pro Plus, Media Cybernetics) as previously described (8). Results are expressed as the area of peribronchial alpha–smooth muscle actin staining per micrometer length of basement membrane of bronchioles 150–200 μm internal diameter.
Quantitation of peribronchial fibrosis.
The area of peribronchial trichrome staining in paraffin-embedded lungs was outlined and quantified under a light microscope (Leica DMLS, Leica Microsystems) attached to an image analysis system (Image-Pro Plus). Results are expressed as the area of trichrome staining per micrometer length of basement membrane of bronchioles 150–200 μm.
Mucus.
To determine the level of mucus expression in the airway, the numbers of periodic acid–Schiff–positive (PAS-positive) and PAS-negative epithelial cells in individual bronchioles were counted as previously described in this laboratory (60).
Quantitation of airway inflammation in BAL in hORMDL3Myh11eGFP-cre mice
BAL fluid was collected from 12-week-old naive hORMDL3Myh11eGFP-cre and naive WT mice by lavaging the lung with 1 mL PBS via a tracheal catheter as previously described (8). BAL fluid was centrifuged and the cell pellets were resuspended with 1 mL of PBS to make Wright Giemsa–stained cytospin slides for total and differential cell counts.
AHR to MCh in hORMDL3Myh11eGFP-cre mice
To determine whether expression of hORMDL3 in ASM influenced spontaneous AHR to MCh, AHR was assessed in naive hORMDL3Myh11eGFP-cre and naive WT mice at 12 weeks of age (n = 12 mice/group). Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) i.p., intubated, and ventilated (FlexiVent ventilator; Scireq) as previously described (8). The dynamic airway resistance and elastance were determined using Scireq software in mice exposed to nebulized PBS and methacholine doses at 0, 3, 24, and 48 mg/mL. Ventilator settings include tidal volume set at 10 mL/kg, frequency at 150/min, and positive end-expository pressure at 3 cmH20.
Acute HDM challenge
WT and hORMDL3Myh11eGFP-cre mice aged 12 weeks were administered intranasal Dermatophagoides pteronyssinus extract (Greer Laboratories) at 100 μg on days 0, 7, 14, and 21 as previously described (61). On day 24 mice had AHR to MCh assessed and were sacrificed (61). Control groups of WT and hORMDL3Myh11eGFP-cre mice aged 12 weeks were not administered HDM.
Statistics
All results are presented as mean ± SEM. A statistical software package (GraphPad Prism) was used for the analysis. A 2-tailed t test was used for analysis of 2 groups. One-way ANOVA was used when more than 2 groups were compared. P values of less than 0.05 were considered statistically significant.
Study approval
The acquisition of deceased human lung donor tissue was reviewed by the University of Arkansas for Medical Sciences Institutional Review Board and determined not to be human subject research. The mouse studies were approved by the University of California, San Diego, Institutional Animal Care and Use Committee.
Author contributions
AKP designed and performed the experiments, analyzed the data, and prepared the figures. MM, PR, SD, NW, and SJ assisted AKP in performing experiments. JB and TAD assisted AKP in performing the FACS studies. RCK provided postmortem asthma lungs for ASM studies and provided feedback on the manuscript. BO reviewed ASM studies and provided feedback on the manuscript. DHB conceived the study, obtained funding for the study, designed experiments, supervised the work, and wrote the manuscript with AKP.
Supplementary Material
Acknowledgments
This study was supported by NIH grants AI 070535, AI 124236, and AI 107779 (to DHB). AKP and NW were supported by NIH T32 AI 007469 and TAD by Veterans Affairs Biomedical Laboratory Research and Development Award BX005073 and NIH grants AI 114585 and AI 070535.
Version 1. 03/04/2021
In-Press Preview
Version 2. 04/08/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, Pham et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2021;6(7):e136911.https://doi.org/10.1172/jci.insight.136911.
Contributor Information
Alexa K. Pham, Email: akpham@health.ucsd.edu.
Marina Miller, Email: mamiller@ucsd.edu.
Peter Rosenthal, Email: peter.simon.rosenthal@gmail.com.
Sudipta Das, Email: sudiptadasphd@gmail.com.
Ning Weng, Email: ningweng@gmail.com.
Sunghoon Jang, Email: sam04010@gmail.com.
Richard C. Kurten, Email: KurtenRichardC@uams.edu.
Jana Badrani, Email: jhbadrani@gmail.com.
Taylor A. Doherty, Email: tdoherty@ucsd.edu.
David H. Broide, Email: dbroide@ucsd.edu.
References
- 1.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Miller M, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109(41):16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmiedel BJ, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun. 2016;7:13426. doi: 10.1038/ncomms13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schedel M, et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in TH2 cytokine levels. J Allergy Clin Immunol. 2015;136(4):893–903. doi: 10.1016/j.jaci.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Ha SG, et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, et al. Orosomucoid-like 3 (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca(2+) oscillations in asthma. J Allergy Clin Immunol. 2018;142(1):207–218. doi: 10.1016/j.jaci.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andiappan AK, et al. Functional variants of 17q12-21 are associated with allergic asthma but not allergic rhinitis. J Allergy Clin Immunol. 2016;137(3):758–766. doi: 10.1016/j.jaci.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 8.Miller M, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regan CP, et al. Development of a smooth muscle-targeted cre recombinase mouse reveals novel insights regarding smooth muscle myosin heavy chain promoter regulation. Circ Res. 2000;87(5):363–369. doi: 10.1161/01.RES.87.5.363. [DOI] [PubMed] [Google Scholar]
- 10.James AL, et al. Airway smooth muscle hypertrophy and hyperplasia in asthma. Am J Respir Crit Care Med. 2012;185(10):1058–1064. doi: 10.1164/rccm.201110-1849OC. [DOI] [PubMed] [Google Scholar]
- 11.Ebina M, et al. Hyperreactive site in the airway tree of asthmatic patients revealed by thickening of bronchial muscles. A morphometric study. Am Rev Respir Dis. 1990;141(5 pt 1):1327–1332. doi: 10.1164/ajrccm/141.5_Pt_1.1327. [DOI] [PubMed] [Google Scholar]
- 12.Regamey N, et al. Increased airway smooth muscle mass in children with asthma, cystic fibrosis, and non-cystic fibrosis bronchiectasis. Am J Respir Crit Care Med. 2008;177(8):837–843. doi: 10.1164/rccm.200707-977OC. [DOI] [PubMed] [Google Scholar]
- 13.Heard BE, Hossain S. Hyperplasia of bronchial muscle in asthma. J Pathol. 1973;110(4):319–331. doi: 10.1002/path.1711100406. [DOI] [PubMed] [Google Scholar]
- 14.Woodruff PG, et al. Hyperplasia of smooth muscle in mild to moderate asthma without changes in cell size or gene expression. Am J Resp Crit Care Med. 2004;169(9):1001–1006. doi: 10.1164/rccm.200311-1529OC. [DOI] [PubMed] [Google Scholar]
- 15.Beamish JA, et al. Molecular regulation of contractile smooth muscle cell phenotype: implications for vascular tissue engineering. Tissue Eng Part B Rev. 2010;16(5):467–491. doi: 10.1089/ten.teb.2009.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marston S, El-Mezgueldi M. Role of tropomyosin in the regulation of contraction in smooth muscle. Adv Exp Med Biol. 2008;644:110–123. doi: 10.1007/978-0-387-85766-4_9. [DOI] [PubMed] [Google Scholar]
- 17.Prakash YS. Emerging concepts in smooth muscle contributions to airway structure and function: implications for health and disease. Am J Physiol Lung Cell Mol Physiol. 2016;311(6):L1113–L1140. doi: 10.1152/ajplung.00370.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koziol-White CJ, Panettieri RA. Modulation of bronchomotor tone pathways in airway smooth muscle function and bronchomotor tone in asthma. Clin Chest Med. 2019;40(1):51–57. doi: 10.1016/j.ccm.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez JJ, et al. Molecular basis and regulation of store-operated calcium entry. Adv Exp Med Biol. 2020;1131:445–469. doi: 10.1007/978-3-030-12457-1_17. [DOI] [PubMed] [Google Scholar]
- 20.Spinelli AM, Trebak M. Orai channel-mediated Ca2+ signals in vascular and airway smooth muscle. Am J Physiol Cell Physiol. 2016;310(6):C402–C413. doi: 10.1152/ajpcell.00355.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong HL, et al. Dynamic assembly of TRPC1-STIM1-Orai1 ternary complex is involved in store-operated calcium influx. Evidence for similarities in store-operated and calcium release-activated calcium channel components. J Biol Chem. 2007;282(12):9105–9116. doi: 10.1074/jbc.M608942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437(7060):902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palty R, et al. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 2012;149(2):425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 24.Katz ZB, et al. Septins organize endoplasmic reticulum-plasma membrane junctions for STIM1-Orai1 calcium signaling. Sci Rep. 2019;9(1):10839. doi: 10.1038/s41598-019-46862-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srikanth S, et al. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12(5):436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wuytack F, et al. Molecular physiology of the SERCA and SPCA pumps. Cell Calcium. 2002;32(5–6):279–305. doi: 10.1016/s0143416002001847. [DOI] [PubMed] [Google Scholar]
- 27.Hong F, et al. Biochemistry of smooth muscle myosin light chain kinase. Arch Biochem Biophys. 2011;510(2):135–146. doi: 10.1016/j.abb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CL. Caldesmon and smooth-muscle regulation. Cell Biochem Biophys. 2001;35(3):275–288. doi: 10.1385/CBB:35:3:275. [DOI] [PubMed] [Google Scholar]
- 29.Rattan S, Ali M. Role of SM22 in the differential regulation of phasic vs. tonic smooth muscle. Am J Physiol Gastrointest Liver Physiol. 2015;308(7):G605–G612. doi: 10.1152/ajpgi.00360.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene AL, et al. Overexpression of SERCA2b in the heart leads to an increase in sarcoplasmic reticulum calcium transport function and increased cardiac contractility. J Biol Chem. 2000;275(32):24722–24727. doi: 10.1074/jbc.M001783200. [DOI] [PubMed] [Google Scholar]
- 31.Ver Heyen M, et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca(2+)-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction-relaxation of the heart. Circ Res. 2001;89(9):838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- 32.Chiba Y, et al. Interleukin-13 augments bronchial smooth muscle contractility with an up-regulation of RhoA protein. Am J Respir Cell Mol Biol. 2009;40(2):159–167. doi: 10.1165/rcmb.2008-0162OC. [DOI] [PubMed] [Google Scholar]
- 33.James AL, et al. Airway smooth muscle thickness in asthma is related to severity but not duration of asthma. Eur Respir J. 2009;34(5):1040–1045. doi: 10.1183/09031936.00181608. [DOI] [PubMed] [Google Scholar]
- 34.Goldsmith AM, et al. Transforming growth factor-beta induces airway smooth muscle hypertrophy. Am J Respir Cell Mol Biol. 2006;34(2):247–254. doi: 10.1165/rcmb.2005-0166OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erle DJ, Sheppard D. The cell biology of asthma. J Cell Biol. 2014;205(5):621–631. doi: 10.1083/jcb.201401050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redwood C, Robinson P. Alpha-tropomyosin mutations in inherited cardiomyopathies. J Muscle Res Cell Motil. 2013;34(3–4):285–294. doi: 10.1007/s10974-013-9358-5. [DOI] [PubMed] [Google Scholar]
- 37.Vandecaetsbeek I, et al. The Ca2+ pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3(5):1–24. doi: 10.1101/cshperspect.a004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahn K, et al. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci U S A. 2009;106(26):10775–10780. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leguillette R, et al. Myosin, transgelin, and myosin light chain kinase: expression and function in asthma. Am J Respir Crit Care Med. 2009;179(3):194–204. doi: 10.1164/rccm.200609-1367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang JC, et al. Analysis of SM22alpha-deficient mice reveals unanticipated insights into smooth muscle cell differentiation and function. Mol Cell Biol. 2001;21(4):1336–1344. doi: 10.1128/MCB.2001.21.4.1336-1344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam M, et al. Regulation of airway smooth muscle contraction in health and disease. Adv Exp Med Biol. 2019;1124:381–422. doi: 10.1007/978-981-13-5895-1_16. [DOI] [PubMed] [Google Scholar]
- 42.Zou JJ, et al. Role of STIM1/Orai1-mediated store-operated Ca2+ entry in airway smooth muscle cell proliferation. J Appl Physiol (1985) 2011;110(5):1256–1263. doi: 10.1152/japplphysiol.01124.2010. [DOI] [PubMed] [Google Scholar]
- 43.Soboloff J, et al. Orai1 and STIM reconstitute store-operated calcium channel function. J Biol Chem. 2006;281(30):20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 44.Spinelli AM, et al. Airway smooth muscle STIM1 and Orai1 are upregulated in asthmatic mice and mediate PDGF-activated SOCE, CRAC currents, proliferation, and migration. Pflugers Arch. 2012;464(5):481–492. doi: 10.1007/s00424-012-1160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carreras-Sureda A, et al. ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum Mol Genet. 2013;22(3):519–530. doi: 10.1093/hmg/dds450. [DOI] [PubMed] [Google Scholar]
- 46.Cantero-Recasens G, et al. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19(1):111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 47.Verlaan DJ, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85(3):377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hjelmqvist L, et al. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3(6):research0027.1. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das S, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci U S A. 2016;113(46):13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller M, et al. Does reduced zona pellucida binding protein 2 (ZPBP2) expression on chromosome 17q21 protect against asthma? J Allergy Clin Immunol. 2018;142(2):706–709. doi: 10.1016/j.jaci.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doherty TA, et al. Alternaria induces STAT6-dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J Immunol. 2012;188(6):2622–2629. doi: 10.4049/jimmunol.1101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Churg A, et al. Alpha1-antitrypsin suppresses TNF-alpha and MMP-12 production by cigarette smoke-stimulated macrophages. Am J Respir Cell Mol Biol. 2007;37(2):144–151. doi: 10.1165/rcmb.2006-0345OC. [DOI] [PubMed] [Google Scholar]
- 53.Watkins SJ, et al. Angiotensin II-induced cardiomyocyte hypertrophy in vitro is TAK1-dependent and Smad2/3-independent. Hypertens Res. 2012;35(4):393–398. doi: 10.1038/hr.2011.196. [DOI] [PubMed] [Google Scholar]
- 54.McKay S, et al. Angiotensin II induces hypertrophy of human airway smooth muscle cells: expression of transcription factors and transforming growth factor-beta1. Am J Respir Cell Mol Biol. 1998;18(6):823–833. doi: 10.1165/ajrcmb.18.6.2924. [DOI] [PubMed] [Google Scholar]
- 55.Marguerat S, Bahler J. Coordinating genome expression with cell size. Trends Genet. 2012;28(11):560–565. doi: 10.1016/j.tig.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Hawker KM, et al. Interleukin-4 inhibits mitogen-induced proliferation of human airway smooth muscle cells in culture. Am J Physiol. 1998;275(3):L469–L477. doi: 10.1152/ajplung.1998.275.3.L469. [DOI] [PubMed] [Google Scholar]
- 57.Leal A, et al. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/S0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- 58.Li J, et al. Polo-like Kinase 1 regulates vimentin phosphorylation at Ser-56 and contraction in smooth muscle. J Biol Chem. 2016;291(45):23693–23703. doi: 10.1074/jbc.M116.749341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson PR, et al. The production of extracellular matrix proteins by human passively sensitized airway smooth-muscle cells in culture: the effect of beclomethasone. Am J Respir Crit Care Med. 2000;162(6):2145–2151. doi: 10.1164/ajrccm.162.6.9909111. [DOI] [PubMed] [Google Scholar]
- 60.Cho JY, et al. Inhibition of airway remodeling in IL-5-deficient mice. J Clin Invest. 2004;113(4):551–560. doi: 10.1172/JCI19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doherty TA, et al. The tumor necrosis factor family member LIGHT is a target for asthmatic airway remodeling. Nat Med. 2011;17(5):596–603. doi: 10.1038/nm.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.