Abstract
Cord blood transplantation (CBT) is associated with low risk of leukemia relapse. Mechanisms underlying antileukemia benefit of CBT are not well understood, however a previous study strongly but indirectly implicated cells from the mother of the cord blood (CB) donor. A fetus acquires a small number of maternal cells referred to as maternal microchimerism (MMc) and MMc is sometimes detectable in CB. From a series of 95 patients who underwent double or single CBT at our center, we obtained or generated HLA-genotyping of CB mothers in 68. We employed a technique of highly sensitive HLA-specific quantitative-PCR assays targeting polymorphisms unique to the CB mother to assay CB-MMc in patients post-CBT. After additional exclusion criteria, CB-MMc was evaluated at multiple timepoints in 36 patients (529 specimens). CB-MMc was present in 7 (19.4%) patients in bone marrow, peripheral blood, innate and adaptive immune cell subsets, and was detected up to 1-year post-CBT. Statistical trends to lower relapse, mortality, and treatment failure were observed for patients with vs. without CB-MMc post-CBT. Our study provides proof-of-concept that maternal cells of the CB graft can be tracked in recipients post-CBT, and underscore the importance of further investigating CB-MMc in sustained remission from leukemia following CBT.
INTRODUCTION
As a source of stem cells for allogeneic hematopoietic cell transplantation for the treatment of leukemia, umbilical cord blood (CB) has advantages over other graft sources, both conventional (HLA-matched) and alternative (HLA-mismatched) donors, including reduced rate and severity of graft-vs.-host disease (GVHD) (1). Consequently, CBT allows greater HLA mismatch, increasing graft availability to nearly all patients, in particular those without a fully HLA-matched (or closely matched) alternative donor source (2,3).
Another important advantage is the potent graft-vs.-leukemia (GVL) effect that is observed post-CBT. Leukemia relapse rate after CBT is reduced more than twofold compared to either HLA-matched or HLA-mismatched donors, threefold when minimal residual disease is present at transplantation (4,5). The explanation for this potent GVL effect despite reduced GVHD, is incompletely elucidated (6,7), although T-cell costimulation and HLA expression are likely to be an important aspect (8). Cells from the mother of the CB donor were strongly, but indirectly, implicated in a study that found reduced leukemia relapse when the recipient had a shared HLA allele with the paternally-inherited HLA allele of the CB (referred to as IPA), indicating that the maternal cells, already exposed to the IPA during pregnancy, recognized the same antigen in the patient’s leukemic cells after transplant (9). These results implicated a GVL effect from the naturally occurring maternal microchimerism (MMc) in the CB administered for transplantation (9,10). Microchimerism (Mc) refers to the presence of a small amount of semi-allogeneic cells (or DNA), a common legacy from maternal-fetal exchange during pregnancy (11,12).
Specializing in the research of such rare cells, we recently showed that CB-MMc could persist 6 months post-CBT in a single recipient (13). Here, we sought to evaluate the use of our technology as a tool for the study of CB-MMc post-CBT in correlation with patient outcomes, as a window of insight into a potential contribution to GVL post-CBT.
SUBJECTS AND METHODS
Ethics Statement
The Internal Review Board of the Fred Hutchinson Cancer Research Center approved this study. All participants gave written informed consent, in accordance with institutional guidelines, the Declaration of Helsinki, and Title 45 United States Code of Federal Regulations, Part 46, Protection of Human Subjects.
Patients Inclusion and Transplant Characteristics
We evaluated a series of 95 patients with hematologic malignancies that received CBT between 2008 and 2017 and had peripheral blood and/or bone marrow specimens stored. Of the 95 patients, 45 had acute myeloid leukemia (AML), 38 acute lymphoblastic leukemia (ALL), 5 myelodysplastic syndrome (MDS), 2 myelofibrosis (MYLF), 2 T cell lymphoma (TCL), 1 GATA2 haploinsufficiency, 1 chronic myeloid leukemia (CML), and 1 chronic myelomonocytic leukemia (CMML). CBT conditioning consisted of high-dose total body irradiation (TBI) with 1320 cGy TBI, fludarabine 75 mg/m2, and cyclophosphamide 120 mg/kg; medium-dose TBI with 300 to 400 cGy TBI, fludarabine 125 to 200 mg/m2 (with or without Thiotepa at 10 mg/kg), and cyclophosphamide 50 mg/kg; or low-dose TBI with 200 cGy TBI, treosulfan 42 mg/m2, and fludarabine 150 to 200 mg/m2. Two patients received TBI-free reduced intensity conditioning either busulfan-based (clinical trial NCT02251821) or alemtuzumab/melphalan/thiotepa-based (14). CBT was double (n=70) or single (n=25). CB units were acquired from CB banks across the world (Australia, Singapore, Finland, France, Germany, Italy, the Netherlands, Spain, Sweden, UK, USA, and Canada). An additional non-engrafting non-HLA-matched ex-vivo expanded CB progenitor cell product was infused for 16 double and 5 single CBT patients (15). Double CBT results in one ‘winning’ CB unit as the primary source of hematopoiesis in the majority of recipients, however the non-engrafting ‘losing’ unit is often detectable as Mc when highly sensitive methods are employed (16). Among 70 double CBT recipients, 57 (81%) had a winning and losing CB, 8 (11%) ‘mixed’ CB chimerism, and 5 (7%) had no evidence of engraftment from either unit. Standard GVHD prophylaxis consisted of cyclosporine A and mycophenolate mofetil and when necessary post-CBT systemic steroid-based therapy was administered (beclomethasone [n=62], budesonide [n=48], prednisone [n=54], methylprednisolone [n=36], and/or topical corticosteroids [n=23]) with or without additional immunosuppressive drugs (infliximab [n=3], rapamycin [n=3]). Four patients had severe GVHD despite primary therapy (grade III-IV acute GVHD) and enrolled in mesenchymal stem cell therapy (clinical trials NCT00366145 and NCT02336230). Eighteen patients did not receive GVHD treatment.
Specimen Collection and Cell Sorting
We studied bone marrow aspirate (BMA) and peripheral blood samples collected at the following days: (mean ± standard deviation [SD]) 33 ±5, 64 ±5, 92 ±11, and 174 ±14 and at up to 405 ±60 days post-CBT for BMA samples. Studies of peripheral blood included an unsorted whole blood (WB) aliquot, as well as aliquots from the major cellular populations. These consisted of peripheral blood mononuclear cells (PBMCs) processed by density-gradient centrifugation and cryopreserved in dimethylsulfoxide 7%, as well as CD66b+ neutrophils after magnetic-activated cell sorting (Miltenyi, Bergisch Gladbach, Germany) with an acceptable purity ≥90% assessed by flow cytometry using CD66abce—PE (BD Biosciences, Franklin Lakes, NJ, USA). Neutrophils constitute 55–75% of white cells in WB and have a short lifespan (about 5 days) (17) thus presence of CB-MMc in this population reflects on the presence of CB-MMc in the corresponding bone marrow progenitor cells. When fluorescence-activated cell sorting (FACS) was needed, it was conducted on PBMC samples to isolate CD3+ T cells, HLA-DR+ CD19+ B cells, CD3−CD56+ and/or CD16+ NK cells, and CD14+ monocytes, as previously described (13). In brief, cryopreserved PBMCs were resuspended, first stained by LIVE/DEAD®-Aqua-Fluorescent fixable dead cell stain (Life Technologies, Carlsbad, CA, USA), followed by staining with a six-color cocktail (CD14—BV711, HLADR—AlexaFluor700, CD19—BUV737, CD16—APC-Cy7, [BD Biosciences, Franklin Lakes, NJ, USA], CD56—BV605 [BioLegend, San Diego, CA, USA], and CD3—PE-TexasRed [Beckman Coulter, Brea, CA, USA]). Gating included ‘alive’ cells and avoided doublets. Purity was assessed by flow cytometry for the sorted cells and the overall median (and interquartile range [IQR]) purity was 95% [92–97%]. Subpopulations were stored as dry pellets at −80 °C for DNA extraction.
Identifying a Non-shared Maternal HLA Polymorphism to Target for CB-MMc Testing
To characterize CB-MMc post-CBT in the recipients, HLA-genotyping of the mothers of CB donors was either obtained from the Cord Blood Banks (class I and/or class II) or conducted in-house after obtaining stored samples, extracting DNA, and using a Luminex-based PCR sequence-specific oligonucleotide probe technique to determine alleles for the class II loci HLA-DRB1, HLA-DQA1, and HLA-DQB1 (One Lambda, Canoga Park, CA, USA). We reviewed HLA-genotyping results for triads (recipient, CB graft, CB mother) for single CBT, for both CB grafts and respective mothers for double CBT, and also non-engrafting non-HLA matched ex-vivo expanded CB progenitor cell products if administered (15). Our goal was to identify a maternal DNA ‘marker’, i.e. a non-shared non-inherited maternal HLA allele (NIMA) unique to the CB mother for testing recipient specimens post-CBT (Supplementary Figure S1).
HLA-Specific Quantitative PCR (qPCR) to Evaluate CB-MMc
We employed a panel of highly sensitive quantitative HLA-specific PCR assays that we developed to quantitatively assess CB-MMc in genomic DNA extracted from patients’ specimens (13,18). Each sampled specimen was assayed for CB-MMc by selecting the qPCR assay specific to the non-shared CB-NIMA. All assays were validated to be highly specific (never amplify unintended alleles) and to have limits of detection ≤ 2.7 per million as previously described (13,18). Real-time qPCR reactions were carried out on ABI Prism® 7700, and on QuantStudio™−5 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA), using the absolute quantification method by standard curves as previously described (19). CB-MMc concentrations were calculated according to the number of cell genome equivalents (gEq) of microchimeric cells (estimated by a target gene marker standard curve) proportional to the number of gEq of total cells tested (estimated by a reference gene marker standard curve). The gEq is defined as the amount of DNA in one human cell and corresponds to ~6.6 pg of DNA (20). Moreover, each measured CB-MMc quantity is associated with a 95% confidence interval (95%CI) of the measured value, derived from the ‘Wilson Score’ without continuity correction (approximating mid-P-exact 95%CI) (21). This 95%CI encapsulates the precision of our measurements, accounting for experimental variability including the total amount of cells tested (i.e. total genomic DNA available) in the assay, by which the Wilson score is affected. For example, there is higher confidence in measuring a Mc value of 0.0 gEq/10 when testing in 100,000 total gEq available (95% CI at [0.0–38.4]) vs. when testing in only 10,000 total gEq (95% CI at [0.0–384.0])
Statistical Analysis
CB-MMc quantities were analyzed as a continuous variable. Mc occurs by definition at low concentrations and approximates a Poisson distribution (data distribution skewed to the right, often with excess of zeros and occasional large outlying values). A negative binomial regression model was used because it was found to best account for the higher level of variability in the data than expected in a Poisson model (22), with the same interpretation of the ‘mean’ as in a Poisson model. The model assesses the association between the Mc gEq count data (dependent variable) and one or more independent variables. The total gEq count data in each sample (the reference gene total) were included as an ‘exposure’ variable (indicating the total number of times Mc event(s) could have happened, and which could differ significantly between a rare and an abundant specimen or subpopulation). CB-MMc measurements were not independent but related per participant and per CB unit within a single participant (in particular those recipients of a double CBT). To account for this, a command ensuring clustering of data-points per CB unit per participant was included (‘cluster’ by CB unit ID); we assumed each CB unit was its own independent entity in the model. The output of this model is a detection rate ratio (DRR), derived from exponentiating the coefficients in the model and interpreted as the fold-change of MMc quantities from one versus another group. Additionally, the Mann-Whitney rank test for 2-group comparisons was used when appropriate. Kaplan-Meier curves were used to assess probability (cumulative incidence) of relapse, overall mortality, and treatment failure post-CBT with long-term follow-up in two groups of patients with detectable CB-MMc in at least one timepoint or specimen vs. undetectable CB-MMc at any timepoint or specimen. To compare the two groups statistically, the log-rank (Mantel-Cox) test was used. Analyses were performed using GraphPad Prism 7 (La Jolla, CA, USA) and STATA-15-SE (College Station, TX, USA).
RESULTS
Of the 95 patients included in the study, CB maternal HLA genotyping could not be obtained, or a unique CB-NIMA target could not be identified, for 28 patients who were excluded from further study. Because Mc can be acquired from other sources (11), we further required that a pre-transplant sample be negative for the CB-MMc target. Conservatively, we further excluded 13 patients with a positive pre-transplant test, although pre-CBT transfusions could potentially occasionally result in a transient positive result (testing was conducted on patient WB and/or PBMC samples collected 19 ±23 days prior to CBT). Additional exclusions included 12 patients who had no pre-CBT or post-CBT samples, 3 patients with graft failure, 1 patient for whom relapse status could not be confirmed post-mortem, and the 2 remaining patients who received mesenchymal stem cell therapy for severe GVHD as it can be an alternative source of allogeneic cells (Supplementary Figure S2).
A total of 36 patients qualified for the study and CB-MMc was assayed in a total of 529 post- and pre-CBT specimens. Clinical characteristics are provided in Table 1. The choice in selecting the type of specimen was on a first level assessing CB-MMc in the bone marrow and peripheral blood; on a second level in neutrophils and PBMCs jointly constituting ~98% of peripheral white blood cells; and on a third level (only if CB-MMc was detectable in any of the previous specimens) in the major constituents of PBMCs (T, B, NK, cells and monocytes). This allowed us to evaluate CB-MMc in the marrow vs. the periphery, in myeloid vs. lymphoid lineages, and in adaptive vs. innate cells. CB-MMc was quantitatively assayed in DNA extracted from BMA, WB, PBMC, and/or neutrophils, at months 1, 2, 3, and/or ≥ 6 post-CBT. The median [and IQR] total number of human gEq tested was 112,259 [51,436–162,149].
Table 1.
Patient characteristics, including detection (yes or no) of cord blood-origin maternal microchimerism (CB-MMc) post-CB transplantation (CBT), degree of HLA matching of CB units to patient and of unit to unit (in double CBT), CB-NIMA target used to detect CB-MMc (n/a = not available or not informative), sustained win-lose (W-L) engraftment status of a double CBT, day of engraftment defined as the first of 3 consecutive days of an absolute neutrophil count of ≥ 0.5×109/L, acute and chronic GVHD, relapse, last contact and death status, as well as pre-transplant minimal residual disease (MRD) assessed by standard multiparameter flow cytometry, cytogenetic and molecular assay below morphologically identifiable disease. Blank entries indicate data not applicable (e.g. a relapse event did not occur in that patient).
| Patient No. | CB-MMc+ (day 30 to ~400) | Sex | Disease | Age diag (yr.) | Age transp (yr.) | HLA match units to patient | HLA match unit to unit | CB-NIMA targeted by qPCR | Unit engr | Engr day | MRD+ | day pre-CBT of MRD test | aGVHD (day 0 to~80) | cGVHD (day 80 to ~365) | Relapse at day | Alive at day | Death at day |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 007 | y | f | AML | 60.4 | 61.0 | 5/6 & 5/6 | 6/6 | DRB1*07 & DRB1*04 | W-L | 16 | n | 35 | II | 0 | 156a | ||
| 017 | y | f | AML | 51.8 | 53.7 | 4/6 & 4/6 | 3/6 | B*13 & DRB1*15 | W-L | 20 | n | 24 | II | 0 | 1916 | ||
| 025 | y | f | ALL | 36.2 | 40.7 | 4/6 & 4/6 | 5/6 | DRB1*15 & DRB1*04 | W-L | 17 | n | 27 | II | mild | 1524 | ||
| 085 | y | f | AML | 28.7 | 31.0 | 4/6 & 4/6 | 2/6 | n/a & DQB1*06 | W-L | 28 | n | 25 | III | moderate | 1240 | ||
| 125 | y | m | AML | 60.4 | 60.9 | 4/6 & 4/6 | 3/6 | DRB1*15 & A*30 | W-L | 28 | n | 32 | 0 | unk. | 917 | ||
| 048 | y | m | ALL | 34.5 | 35.2 | 5/6 | single | DRB1*04 | 21 | n | 14 | II | mild | 1605 | |||
| 108 | y | m | ALL | 4.1 | 4.6 | 5/6 | single | DRB1*01 | 30 | n | 29 | II | moderate | 865 | |||
| 011 | n | f | ALL | 2.3 | 5.4 | 5/6 & 5/6 | 5/6 | DRB1*03 & n/a | W-L | 15 | n | 29 | II | mild | 3360 | ||
| 014 | n | f | AML | 32.3 | 38.7 | 6/6 & 5/6 | 5/6 | DQB1*03 & DRB1*04 | W-L | 31 | n | 26 | II | mild | 1396b | ||
| 015 | n | m | ALL | 32.3 | 32.7 | 5/6 & 4/6 | 4/6 | n/a & DRB1*07 | W-L | 13 | y | 33 | II | 0 | 1218 | ||
| 021 | n | m | TCL | 43.2 | 44.2 | 4/6 & 4/6 | 3/6 | n/a & B*44 | W-L | 31 | n | 21 | II | 0 | 1595 | ||
| 029 | n | f | ALL | 36.4 | 38.9 | 4/6 & 4/6 | 4/6 | DRB1*10 & n/a | mixed | 23 | n | 22 | 0 | 0 | 495a,c | ||
| 032 | n | m | ALL | 44.7 | 45.4 | 6/6 & 4/6 | 4/6 | n/a & DRB1*16 | W-L | 15 | n | 18 | II | mild | 1612 | ||
| 033 | n | m | ALL | 19.4 | 19.9 | 4/6 & 4/6 | 4/6 | A*30 & A*30 | W-L | 26 | y | 35 | 0 | 0 | 135a | ||
| 034 | n | f | AML | 67.9 | 68.5 | 4/6 & 4/6 | 3/6 | n/a & DRB1*03 | W-L | 24 | y | 34 | II | 0 | 208 | 242d | |
| 040 | n | f | ALL | 14.2 | 30.6 | 5/6 & 5/6 | 5/6 | n/a & DRB1*01 | W-L | 21 | n | 34 | II | mild | 1776 | ||
| 053 | n | m | ALL | 58.0 | 58.6 | 4/6 & 4/6 | 2/6 | DRB1*04 & n/a | W-L | 16 | n | 30 | II | 0 | 1077h | ||
| 063 | n | m | AML | 63.9 | 64.3 | 4/6 & 4/6 | 3/6 | DRB1*01 & n/a | W-L | 16 | y | 24 | II | unk. | 26 | 61h | |
| 066 | n | f | GATA2 | 44.6 | 45.0 | 4/6 & 4/6 | 4/6 | DRB1*03 & DRB1*03 | W-L | 23 | n | 32 | III | 0 | 424e | ||
| 068 | n | m | CML | 55.4 | 58.3 | 4/6 & 4/6 | 3/6 | DRB1*07 & n/a | W-L | 19 | y | 42 | II | 0 | 1473 | ||
| 072 | n | f | AML | 16.6 | 17.0 | 4/6 & 4/6 | 3/6 | DRB1*13 & DRB1*15 | W-L | 15 | n | 28 | III | 0 | 760 | 1309 | |
| 087 | n | f | ALL | 30.3 | 30.8 | 4/6 & 4/6 | 2/6 | DRB1*08 & n/a | W-L | 16 | n | 28 | II | 0 | 1075 | ||
| 088 | n | m | MDS | 23.2 | 23.5 | 5/6 & 4/6 | 5/6 | DRB1*15 & DQB1*06 | mixed | 15 | y | 26 | III | severe | 151f | ||
| 093 | n | m | AML | 38.8 | 39.9 | 5/6 & 6/6 | 5/6 | DRB1*08 & n/a | W-L | 9 | n | 36 | I | mild | 480 | 494c | |
| 095 | n | f | ALL | 33.3 | 33.8 | 4/6 & 4/6 | 2/6 | n/a & DRB1*08 | W-L | 17 | n | 25 | II | mild | 458 | 1100 | |
| 096 | n | f | ALL | 20.7 | 27.0 | 4/6 & 4/6 | 2/6 | DRB1*07 & DRB1*07 | W-L | 22 | n | 27 | II | mild | 1214 | ||
| 107 | n | m | ALL | 45.5 | 46.8 | 4/6 & 4/6 | 4/6 | n/a & DRB1*13 | W-L | 23 | y | 36 | III | moderate | 195g | ||
| 113 | n | f | MDS | 33.3 | 34.5 | 6/6 & 4/6 | 4/6 | DRB1*07 & n/a | W-L | 15 | n | 27 | 0 | 0 | 1036 | 1040 | |
| 120 | n | m | ALL | 33.5 | 34.9 | 5/6 & 6/6 | 5/6 | A*30 & n/a | W-L | 20 | y | 26 | 0 | 0 | 279 | 358d | |
| 124 | n | m | AML | 49.9 | 50.4 | 5/6 & 4/6 | 5/6 | n/a & DRB1*15 | W-L | 19 | n | 34 | 0 | unk. | 769 | ||
| 129 | n | f | AML | 27.7 | 28.1 | 4/6 & 4/6 | 4/6 | DRB1*15 & n/a | W-L | 18 | n | 21 | II | 0 | 857 | ||
| 018 | n | m | ALL | 1.5 | 1.9 | 5/6 | single | DRB1*07 | 29 | n | 42 | II | moderate | 1846 | |||
| 073 | n | m | ALL | 14.0 | 14.5 | 4/6 | single | DRB1*01 | 16 | n | 26 | II | 0 | 1114 | |||
| 098 | n | f | AML | 7.7 | 10.5 | 4/6 | single | DRB1*15 | 16 | n | 29 | II | 0 | 1228 | |||
| 112 | n | m | AML | 35.0 | 35.5 | 4/6 | single | DRB1*07 | 19 | y | 21 | II | mild | 872 | |||
| 122 | n | f | AML & ALL | 0.6 | 2.2 | 5/6 | single | DRB1*03 | 12 | n | 25 | II | 0 | 861 | |||
Cause of death:
pneumonia/acute respiratory failure
metastatic squamous cell carcinoma
sepsis/shock
relapse-related
multiple organ dysfunction syndrome
status epilepticus/acute renal failure/thrombotic microangiopathy
acute hemorrhagic stroke
unknown.
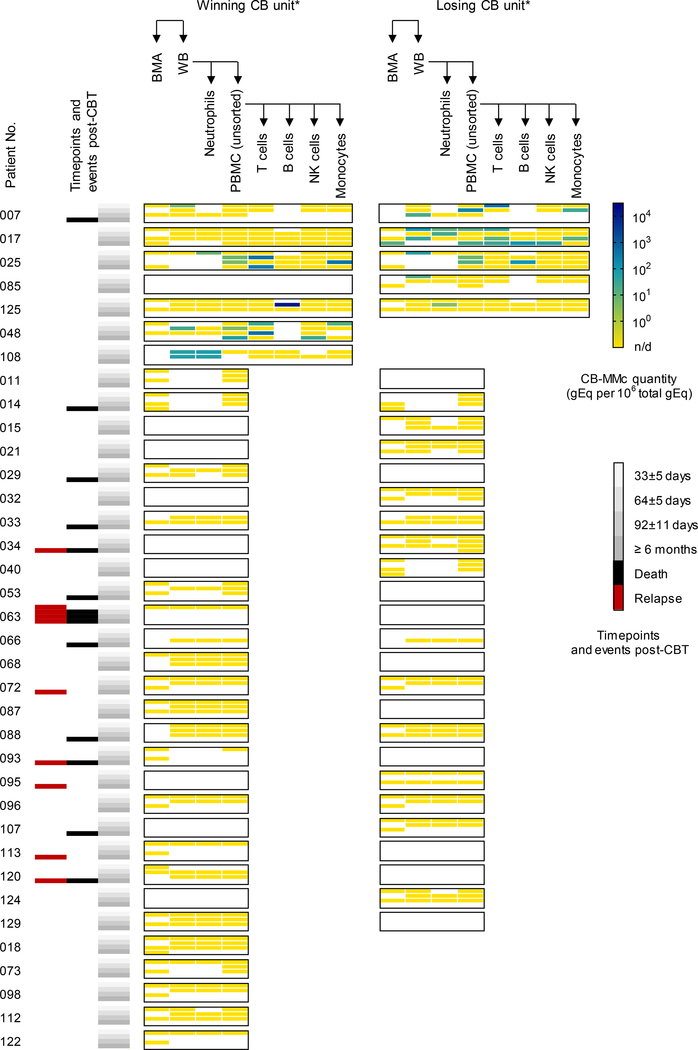
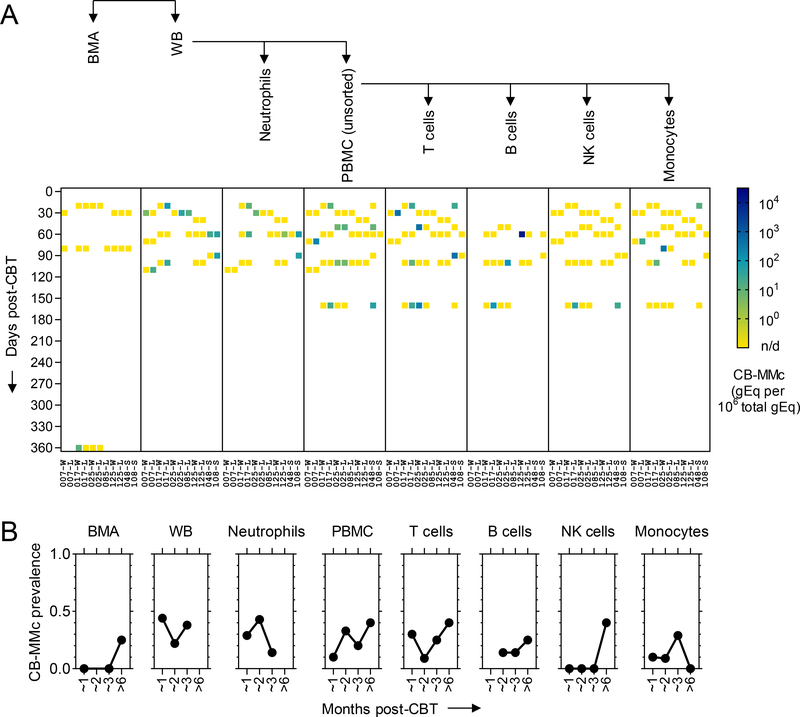
CB-MMc was detected in 19.4% (7/36) of patients in at least one specimen at a timepoint post-CBT (Figure 1). When CB-MMc was present, we conducted FACS and performed additional testing on T cells, B cells, NK cells, and monocytes. For the 7 patients with positive results, CB-MMc was detected across all timepoints and was identified in all cellular subsets. To assess CB-MMc quantitative trends as a function of time since CBT, detection rate ratios (DRR) were calculated, deriving from negative binomial regressions recently described to appropriately model Mc data (22). DRRs are interpreted as the fold-change in expected Mc quantities across the range of a variable. In the tested specimens, CB-MMc post-CBT month-to-month quantitative fold-change appeared unchanging, except in BMA with a late detection of CB-MMc ~1 year post-CBT resulting in a statistically significant increasing trend (Figure 2 and Table 2). Noticeably, CB-MMc quantities remained unchanged across time post-CBT in adaptive immune cells, but tended to begin at lower quantities and increase significantly post-CBT in innate immune cells (Table 2).
Figure 1. Maternal microchimerism of the cord blood donor (CB-MMc) in patients who received double or single CB transplantation (CBT).
CB-MMc concentrations are measured in human cell genome equivalent (gEq) of CB-MMc DNA per million gEq of total DNA tested from bone marrow aspirates (BMA), whole peripheral blood (WB), neutrophils, peripheral blood mononuclear cells (PBMC), T, B, NK cells, and monocytes; those last four subsets are tested only if a positive CB-MMc has been detected in BMA, WB, neutrophils, or PBMCs at any timepoint. CB-MMc assays targeted the CB non-shared, non-inherited maternal HLA allele (NIMA) of the winning and/or losing CB units in case of double CBT, or the single unit in case of single CBT. Blank spots are when a specimen at a timepoint was not available or a CB-NIMA-specific assay was not available for testing. Timepoints were grouped into 4 classes, and events of death or relapse are shown. *Patients 048, 108, 018, 073, 098, 112, and 122 were single CBT recipients (i.e. no losing unit), and 029 and 088 had a CBT engraftment that remained mixed (i.e. no winning or losing). n/d= not detected
Figure 2. Dynamics of maternal microchimerism of the cord blood donor (CB-MMc) in the 7 patients who had positive results post-CB transplantation (post-CBT).
(A) CB-MMc concentrations are shown. Measurements are in human cell genome equivalent (gEq) of CB-MMc DNA per million gEq of total DNA tested from bone marrow aspirates (BMA), whole peripheral blood (WB), neutrophils, peripheral blood mononuclear cells (PBMC), T , B, NK cells, and monocytes. Timepoints post-CBT were divided into units of 10 days. Blank spots are when a specimen at a timepoint was not available. Patient numbers (No.) are on the ‘x’ axis and whether the measurement was in the winning (W), losing (L), or single (S) CB unit. (B) Probabilities of having a CB-MMc+ result for each specimen in the 4 major timepoint categories post-CBT, according to data from the 7 patients who had positive results post-CBT. n/d=not detected
Table 2.
Cord blood-origin maternal microchimerism (CB-MMc) quantities according to time since CB transplantation (CBT): month-to-month fold-change estimated by the negative binomial model in the 7 patients with positive results are represented by the detection rate ratios (DRR) accompanied by their 95% confidence intervals (95%CI) and P-values. CB-MMc measurements were not independent and ‘clustered’ by CB unit (assuming each CB as an independent entity in the model). A month post-CBT was equivalent to the number of days post-CBT divided by 30.4375.
| Month-to-month DRR [95%CI] of CB-MMc levels post-CBT | P-value | |
|---|---|---|
| Specimens | ||
| BMA | 2.13 [1.78 – 2.55] | < 0.0001 |
| WB | 0.88 [0.52 – 1.50] | 0.651 |
| Neutrophils | 1.41 [0.57 – 3.48] | 0.454 |
| PBMC (unsorted) | 1.20 [0.76 – 1.88] | 0.431 |
| T cells | 1.04 [0.63 – 1.71] | 0.883 |
| B cells | 0.35 [0.07 – 1.71] | 0.195 |
| NK cells | 2.9E+4 [2.3E−12 – 3.8E+20] | 0.587 |
| Monocytes | 2.16 [0.62 – 7.54] | 0.228 |
| Lineages | ||
| Myeloid (Neutro; Mono) | 1.73 [0.81 – 3.66] | 0.155 |
| Lymphoid (T; B; NK) | 1.31 [0.55 – 3.08] | 0.542 |
| Immune Function | ||
| Adaptive (T; B) | 1.03 [0.48 – 2.20] | 0.943 |
| Innate (Neutro; Mono; NK) | 1.71 [1.13 – 2.60] | 0.011 |
We previously reported persistence of the ‘losing’ (non-engrafting) CB in recipients of double CBT using highly sensitive testing methods (16). In the present study, CB-MMc originated from both the ‘winning’ (primary hematopoiesis source) and ‘losing’ CB units, except in one case where it was detected only from the ‘losing’ unit. In 3 of the 5 double-CB transplant patients with positive results, it was possible to also directly assess chimerism of the ‘losing’ CB unit (thanks to the availability of informative markers). Mc patterns of the losing units were similar to those of the ‘mothers’ of the corresponding losing units (i.e. CB-MMc); only one occurrence in the bone marrow was substantially different with losing-CB chimerism at 990 gEq/106 versus 0 gEq/106 of the losing CB-MMc (Supplementary Table S1).
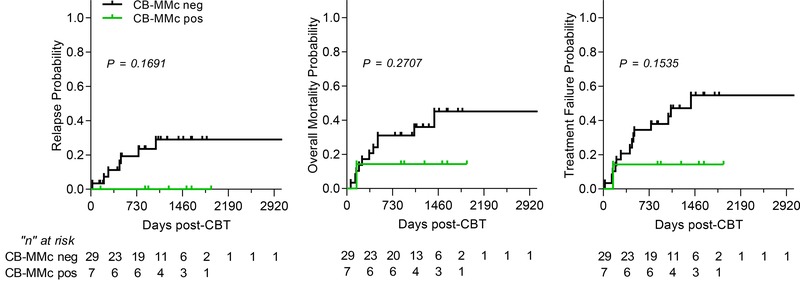
Finally, we examined patient outcomes according to CB-MMc post-CBT. The number of patients in the final study was limited due to stringent inclusion criteria; however, Kaplan-Meier survival curves showed trends towards better outcomes for relapse, mortality, and treatment failure (defined as relapse or death, whichever comes first) when post-CBT CB-MMc was present. Although trends were not statistically significant, we did not observe any case of relapse when CB-MMc was detected (Figure 3). Other outcomes including acute and chronic GVHD, minimal residual disease pre-CBT, engraftment, and HLA matching did not show a correlation with CB-MMc post-transplant and are described in Table 1.
Figure 3. Probability (cumulative incidence) of relapse, overall mortality, and treatment failure post-cord blood transplantation (CBT), with a follow-up of up to 8+ years (2920+ days).
Treatment failure represents an event of relapse or death, whichever comes first (inverse of disease-free survival). P-values are from the log-rank (Mantel-Cox) test. Black lines represent patients who had no CB-MMc detectable post-CBT and green lines are patients with detectable CB-MMc post-CBT. Ticks represent censored individuals and a table below represents the number of individuals at risk.
DISCUSSION
We present studies that, for the first time, track serial blood and bone marrow samples in CBT recipients for donor CB-MMc. We identify CB-MMc post-CBT in multiple immunophenotypes in almost one fifth of leukemia patients. We achieved this after having developed a highly sensitive and specific technique capable of identifying the mother of the CB donor even when multiple donors were involved. Interestingly, when CB-MMc was present relapse was not observed, and treatment failure and overall mortality rates trended favorably.
The major limitation of our study is the modest sample size, due largely from stringent inclusion criteria. Accordingly, power was reduced in statistical analyses for association with patients’ outcomes including relapse rates which are already low post-CBT. The sample size was further limited because most patients received double CBT necessitating evaluation of genotypes across six different directions to identify a unique CB-MMc ‘marker’. Transfusions potentially could confound some test results. Leukemia patients receive multiple red blood cell and platelet transfusions both pre- and post-CBT as part of their standard treatment protocols (up to a total of 342 in the most transfused patient in our cohort). However, transfusion history and patterns were not significantly different between patients positive vs. negative for CB-MMc (Supplementary Figure S3). Moreover, CB-MMc was observed in many cell populations and consistently at multiple timepoints post-CBT. Based on our study design, sharing of the specific HLA targeted is not likely, and results overall cannot be explained by transient leukodepleted gamma-irradiated transfusion products.
Although we previously found that the ‘losing’ CB unit is frequently detected post-CBT with our highly sensitive technology (16), in the current study it was an unexpected result that we could also detect MMc from the losing CB. Why/how losing CB MMc might affect transplant outcome is unclear; possibilities include benefit conferred due simply to retention of another HLA-disparate cell population, potential for epitope spreading, or indirectly reflecting a patient who more readily accepts low levels of allogeneic cells for which HLA-disparity provides a slight advantage of immunosurveillance against pre-malignant or malignant cells.
CB-MMc was dynamically present in innate and adaptive immune function cell lineages, a phenomenon previously described for maternal immune subsets naturally present at birth (in CB) and in adults (13,23,24). The presence of maternal cells in the fetus as early as the second (25) and third gestational trimesters (26), and their persistence in her progeny into adult life (27) implies their ability to cross the fetal-maternal barrier. Their identification within the short-lived neutrophil compartment (as previously described (28) and as our results show) is suggestive for the presence of an active microchimeric progenitor cell niche (29), leading to the apparent replenishment of immune competent cells (across the timepoints post-CBT) and potentially contributing to the proposed antileukemia benefit. MMc in CB could act directly, augmenting activity against minimal residual disease immediately post-CBT. Alternatively, the fetal immune system is influenced by the mother as it develops and detecting CB-MMc could be a marker for a greater impact of the CB mother in an instructional role to the primary cell population in the CB graft, the fetal cell population. ‘Licensing to kill’ has been described in other settings (30,31) and MMc could be contributory in a similar role acting on fetal cells that become the transplanted graft licensing to kill aberrant cells (instruction that could also occur during gestation).
Our results provide ‘proof-of-concept’ for the study of persisting CB-MMc, a phenomenon that is not uncommon post-CBT, and support previously reported indirect evidence implicating CB-MMc in decreased leukemia relapse rate after CBT. Overall, our study brings to attention the importance of investigating CB-MMc and its correlation with sustained remission of leukemia when CB is the donor transplant source.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health grant R01HL117737. The authors are grateful to the patients and families who consented to participate in the study.
CONFLICTS OF INTEREST
SBK and JLN are co-founders of Chimerocyte, Inc. that develops highly-sensitive chimerism analysis technologies. Chimerocyte, Inc. had no role in funding this research project.
REFERENCES
- 1.Rocha V, Wagner JE, Sobocinski KA, Klein JP, Zhang MJ, Horowitz MM, et al. Graft-versus-host disease in children who have received a cord-blood or bone marrow transplant from an HLA-identical sibling. Eurocord and International Bone Marrow Transplant Registry Working Committee on Alternative Donor and Stem Cell Sources. N Engl J Med. 2000. June 22;342(25):1846–54. [DOI] [PubMed] [Google Scholar]
- 2.Gragert L, Eapen M, Williams E, Freeman J, Spellman S, Baitty R, et al. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014. July 24;371(4):339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruggeri A, Paviglianiti A, Gluckman E, Rocha V. Impact of HLA in cord blood transplantation outcomes. HLA. 2016. June 1;87(6):413–21. [DOI] [PubMed] [Google Scholar]
- 4.Brunstein CG, Gutman JA, Weisdorf DJ, Woolfrey AE, Defor TE, Gooley TA, et al. Allogeneic hematopoietic cell transplantation for hematologic malignancy: relative risks and benefits of double umbilical cord blood. Blood. 2010. November 25;116(22):4693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milano F, Gooley T, Wood B, Woolfrey A, Flowers ME, Doney K, et al. Cord-Blood Transplantation in Patients with Minimal Residual Disease. N Engl J Med. 2016. September 8;375(10):944–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker J, Hanash A. Cord blood T cells are “completely different.” Blood. 2015. December 24;126(26):2778–9. [DOI] [PubMed] [Google Scholar]
- 7.Hiwarkar P, Qasim W, Ricciardelli I, Gilmour K, Quezada S, Saudemont A, et al. Cord blood T cells mediate enhanced antitumor effects compared with adult peripheral blood T cells. Blood. 2015. December 24;126(26):2882–91. [DOI] [PubMed] [Google Scholar]
- 8.Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25(4):603–11. [DOI] [PubMed] [Google Scholar]
- 9.van Rood JJ, Scaradavou A, Stevens CE. Indirect evidence that maternal microchimerism in cord blood mediates a graft-versus-leukemia effect in cord blood transplantation. Proc Natl Acad Sci USA. 2012. February 14;109(7):2509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milano F, Nelson JL, Delaney C. Fetal maternal immunity and antileukemia activity in cord-blood transplant recipients. Bone Marrow Transplant. 2013. March;48(3):321–2. [DOI] [PubMed] [Google Scholar]
- 11.Nelson JL. The otherness of self: microchimerism in health and disease. Trends Immunol. 2012. August;33(8):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinder JM, Stelzer IA, Arck PC, Way SS. Immunological implications of pregnancy-induced microchimerism. Nature Reviews Immunology. 2017. May 8;17(8):483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanaan SB, Gammill HS, Harrington WE, Rosa SCD, Stevenson PA, Forsyth AM, et al. Maternal microchimerism is prevalent in cord blood in memory T cells and other cell subsets, and persists post-transplant. OncoImmunology. 2017. May 4;6(5):e1311436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parikh SH, Mendizabal A, Benjamin CL, Komanduri KV, Antony J, Petrovic A, et al. A novel reduced-intensity conditioning regimen for unrelated umbilical cord blood transplantation in children with nonmalignant diseases. Biol Blood Marrow Transplant. 2014. March;20(3):326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney C, Milano F, Cicconi L, Othus M, Becker PS, Sandhu V, et al. Infusion of a non-HLA-matched ex-vivo expanded cord blood progenitor cell product after intensive acute myeloid leukaemia chemotherapy: a phase 1 trial. Lancet Haematol. 2016. July;3(7):e330–339. [DOI] [PubMed] [Google Scholar]
- 16.Milano F, Gammill H, Oliver DC, Kanaan SB, Nelson JL, Delaney C. Persistence of the losing cord blood unit following double cord blood transplantation: finding the unseen. Blood. 2017. September 21;130(12):1480–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JAM, et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010. July 29;116(4):625–7. [DOI] [PubMed] [Google Scholar]
- 18.Kanaan SB, Sensoy O, Yan Z, Gadi VK, Richardson ML, Nelson JL. Immunogenicity of a rheumatoid arthritis protective sequence when acquired through microchimerism. Proc Natl Acad Sci USA. 2019. September 6;201904779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, et al. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis and Rheumatism. 2004;50(3):906–14. [DOI] [PubMed] [Google Scholar]
- 20.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988. January 29;239(4839):487–91. [DOI] [PubMed] [Google Scholar]
- 21.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998. April 30;17(8):857–72. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie KA, Gammill HS, Kamper-Jørgensen M, Tjønneland A, Gadi VK, Nelson JL, et al. Statistical Methods for Unusual Count Data: Examples From Studies of Microchimerism. Am J Epidemiol. 2016. November 15;184(10):779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loubiere LS, Lambert NC, Flinn LJ, Erickson TD, Yan Z, Guthrie KA, et al. Maternal microchimerism in healthy adults in lymphocytes, monocyte//macrophages and NK cells. Lab Invest. 2006. September 11;86(11):1185–92. [DOI] [PubMed] [Google Scholar]
- 24.Mold JE, Michaëlsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008. December 5;322(5907):1562–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo ES, Lo YM, Hjelm NM, Thilaganathan B. Transfer of nucleated maternal cells into fetal circulation during the second trimester of pregnancy. Br J Haematol. 1998. March;100(3):605–6. [DOI] [PubMed] [Google Scholar]
- 26.Petit T, Dommergues M, Socié G, Dumez Y, Gluckman E, Brison O. Detection of maternal cells in human fetal blood during the third trimester of pregnancy using allele-specific PCR amplification. Br J Haematol. 1997. September;98(3):767–71. [DOI] [PubMed] [Google Scholar]
- 27.Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, et al. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104(1):41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunku Cuddapah C, Gadi V, deLavaldeLacoste B, Guthrie K, Nelson J. Maternal and fetal microchimerism in granulocytes. Chimerism. 2010;1(1):11–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995. October 1;86(7):2829–32. [PubMed] [Google Scholar]
- 30.Melief CJM. “License to kill” reflects joint action of CD4 and CD8 T cells. Clin Cancer Res. 2013. August 15;19(16):4295–6. [DOI] [PubMed] [Google Scholar]
- 31.Bracamonte-Baran W, Florentin J, Zhou Y, Jankowska-Gan E, Haynes WJ, Zhong W, et al. Modification of host dendritic cells by microchimerism-derived extracellular vesicles generates split tolerance. Proc Natl Acad Sci USA. 2017. 31;114(5):1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.