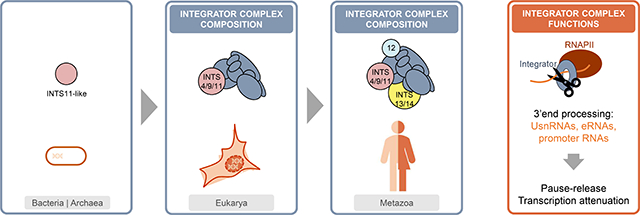
Abstract
Genomic transcription is fundamental to all organisms. In metazoans, the Integrator complex is required for endonucleolytic processing of non-coding RNAs, regulation of RNA polymerase II pause-release, and premature transcription attenuation at coding genes. Recent insights into the structural composition and evolution of Integrator subunits have informed our understanding of its biochemical functionality. Moreover, studies in multiple model organisms point to an essential function of Integrator in signaling response and cellular development, highlighting a key role in neuronal differentiation. Indeed, alterations in Integrator complex subunits have been identified in patients with neurodevelopmental diseases and cancer. Taken together, we propose that Integrator is a central regulator of transcriptional processes and that its evolution reflects genomic complexity in regulatory elements and chromatin architecture.
Graphical Abstract
Introduction
The discovery of the Integrator complex by Baillat et al. in 2005 was a major advance in our understanding of the interplay between RNA Polymerase II (RNAPII)-mediated transcription and the maturation of spliceosome-relevant U-rich small nuclear RNAs (UsnRNAs) [1]. This ~1.5 MDa multimeric protein complex is specific to higher eukaryotes, contains at least 15 subunits including SEM1 (also known as DSS1), and tightly interacts with the RNAPII C-terminal domain (CTD) [1–4]. The pivotal subunit to Integrator function is Integrator subunit (INTS) 11, which contains a metallo-β-lactamase domain with endonucleolytic activity. Together with INTS9 and INTS4, INTS11 forms the core catalytic complex of Integrator (‘cleavage module’) [5]. INTS11/INTS9 are homologous to each other and to members of the cleavage and polyadenylation specificity factor (CPSF) complex CPSF3/CPSF2 (also called CPSF73/CPSF100) [1,6] that are required for maturation and polyadenylation of pre-mRNAs [7].
Since the initial identification of Integrator’s role in UsnRNA processing [1,8,9], Integrator’s function in the regulation of the non-coding transcriptome has been further expanded to the 3’-end maturation of enhancer RNAs (eRNAs) [10], long non-coding RNAs (lncRNAs) [11], including NEAT1 [12], and the human telomerase-associated RNA TERC [13]. Recently, Integrator was reported in the regulation of coding transcription, from transcriptional activation [14,15], the control of RNAPII pause-release [14,16–19], to transcription termination [20]. These various roles of the Integrator complex impact on cellular homeostasis on multiple levels with alterations implicated in neurodevelopmental defects [21–23] and cancer [24,25].
We discuss in this review the evolution of INTS in multicellular organisms, with a particular focus on the homologs INTS11/INTS9 and CPSF3/CPSF2, as well as recent structural studies that have provided insights into subunit interactions and functions. We further outline Integrator’s roles in transcriptional regulation of the non-coding and coding transcriptome and discuss recent key findings that not only advance our knowledge of how Integrator modulates transcriptional homeostasis, but also how INTS alterations mediate pathophysiology.
Integrator evolution parallels that of genomic complexity
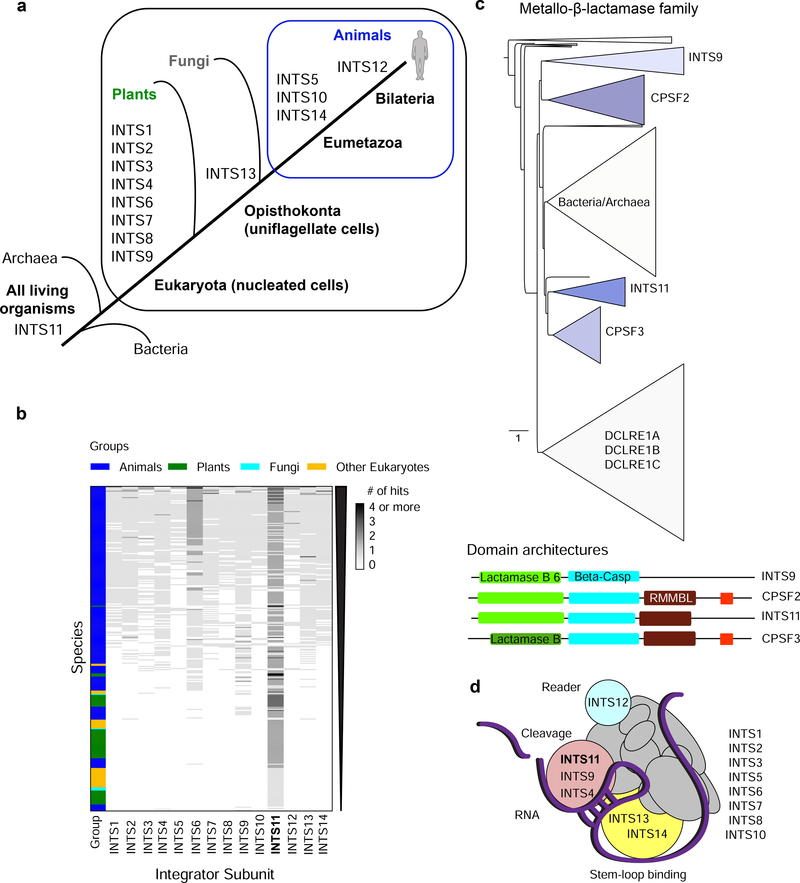
The Integrator complex is confined to multicellular animals (Metazoa) [1,26]. Using gene age estimates based on publicly available ortholog databases revealed the emergence of most INTS at the origin of eukaryotes (nucleated cells, Figure 1a, [27]). Later, full-length INTS13, INTS5, INTS10, and INTS14 evolved with multicellularity at the level of Eumetazoa (animals, except sponges). INTS12 was the latest arrival to the complex in Bilateria (bilateral body symmetry and three germ layers, Figure 1a). Interestingly, this subunit contains a plant homeodomain (PHD) finger, a domain generally referred to as epigenetic reader because of its recognition of histone H3 modifications [28], thus linking Integrator complex to chromatin.
Figure 1. Evolution of the Integrator complex.
a) Overview of the INTS gene ages throughout evolution. Eukarya (nucleated cells), Opisthokonta (uniflagellate cells), Eumetazoa (animals, except sponges), Bilateria (animals with bilateral body symmetry and three germ layers). b) Heatmap of the presence/absence of Integrator-homologous proteins in >300 species (compared to full-length human sequences with the following cut-offs: 30% pair-wise sequence identity, and 50% coverage of the human proteins, in addition to blastp default parameters [29]). Grayscale accounts for the number of hits in one species that aligned to a human INTS. c) Phylogenetic tree of the metallo-β-lactamase superfamily from eggNOG 4.5.1 (COG1236) [33]. Triangle length indicates divergence and triangle height the number of species. Below: domain architecture of human INTS9, CPSF2, INTS11, and CPSF3, as retrieved from Pfam 33.1 [32]. In red CPSF73–100_C domain. d) Model of INTS and their modular interactions. Cleavage module depicted in pink, stem-loop binding module in yellow, and reader module in cyan. Bold: catalytic subunit INTS11.
When screening more than 1000 eukaryote reference proteomes [29], we identified over 300 species with homologous proteins to human Integrator subunits within our cut-offs of 30% sequence identity and 50% protein coverage to the full-length human INTS (Figure 1b). The majority (70%) of the retrieved species belonged to the animal kingdom, followed by plants (18%), other eukaryotes (10%), and fungi (2%). Integrator is largely absent in fungi, including Saccharomyces cerevisiae, suggesting that other RNA processing factors function in UsnRNA processing [30]. In plants (Arabidopsis thaliana), the DSP1–4/CPSF73-l complex identified as being responsible for UsnRNA maturation contains subunits homologous to INTS7, INTS3, INTS4, and INTS9, respectively, while the catalytic subunit seems to more closely resemble CPSF3 than INTS11 [31].
Homologues of the catalytic subunit INTS11, containing the three-domain architecture metallo-β-lactamase, β-CASP, and Zn-dependent metallo-hydrolase RNA specificity domain (RMMBL) [32] (Figure 1c), are commonly detected in bacteria and archaea (Figure 1a). The protein phylogeny of the metallo-β-lactamase superfamily revealed at least two duplication events from a common ancestor for the paralog proteins INTS11/CPSF3 and INTS9/CPSF2, suggesting a loss of cleavage activity in the INTS9/CPSF2 branch (Figure 1c, [33]). The two catalytic proteins CPSF3/INTS11 are found most conserved within their orthologous groups, consistent with their functional relevance, while CPSF2 and INTS9 are more divergent (Figure 1c). The heterodimeric interaction between INTS9 and INTS11 is mediated through their conserved C-termini, which represent distinguishing differences between the Integrator complex and the CPSF complex [6].
Novel structural insights indicate Integrator’s modularity
Since INTS are essential for life, most studies utilize RNA interference (RNAi) targeting the panel of INTS to deduce functions of individual Integrator components. However, technical difficulties resulting from knock-down efficiencies in different species and disrupting the integrity of the complex following knock-down provide barriers to assigning function for individual subunits. Therefore, elucidating Integrator’s structure would provide further insights into its mechanism of action and its modular nature. The C-terminal interaction INTS9/INTS11 was the first to be structurally resolved [34]. More recently, the INTS13/INTS14 heterodimer structure was determined and found to form a subcomplex together with INTS10 [35]. Interestingly, all three subunits appear at the origin of Eumetazoa (animals, except sponges). This subcomplex was described to interact with the cleavage module (INTS4/INTS9/INTS11) through INTS13, and has affinity to single-stranded RNA hairpin structures. These observations provoke the model of INTS10/INTS13/INTS14-RNA interactions recruiting the Integrator cleavage module and forming a platform for remaining Integrator subunits for efficient RNA processing ([35] and Figure 1d).
By contrast, specific INTS interacting with RNAPII CTD, as well as requirements for their interaction remain largely elusive. The CTD consists of a number of tandem heptapeptide repeats (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) that varies according to the organism. Its phosphorylation code changes dynamically throughout the transcription cycle, exhibiting distinct patterns for initiation, elongation and termination (reviewed in [36]). For UsnRNA genes, Integrator-CTD interaction seems to strictly depend on the CTD phosphorylation status and requires both Ser2-phosphorylation (Ser2-P, productive elongation) and Ser7-P (promoter located) [2]. Interestingly, interactome studies by mass spectrometry find equally strong Integrator interaction with both hypo-phosphorylated and hyper-phosphorylated CTDs [3], arguing for a general presence of Integrator at RNAPII-engaged genes independent of its phosphorylation status. Recently, Integrator was also found to interact with Tyr1-P CTD, which is required for RNAPII pausing and transcription termination [4].
Both the non-coding and coding transcriptomes rely on Integrator function
Early work identified a degenerate A/T-rich consensus sequence (GTTTN0–3AAARNNAGA) downstream of the UsnRNA 3’ends termed the 3’box, which is required for efficient UsnRNA processing [37]. Chromatin immunoprecipitation (ChIP) experiments co-localized RNAPII and Integrator at UsnRNA promoter, gene body, and 3’end of U2 snRNA, indicating that Integrator associates with RNAPII constitutively throughout transcription [9]. Furthermore, knock-down of various Integrator subunits leads to UsnRNA extensions beyond the 3’box, suggesting that Integrator recognizes the 3’box and UsnRNA hairpin structure signals for cleavage of the transcripts [1,8]. Moreover, primary miRNA hairpin structures originating from the γ-herpesvirus Herpesvirus saimiri require Integrator for their processing into miRNA precursors using a similar 3’box-like recognition sequence [38].
The scope of Integrator’s action on non-coding RNAs was further expanded by its identification as a crucial factor for eRNA maturation and enhancer-promoter communication. Lai et al. focused on a set of epidermal growth factor (EGF) activated enhancers of immediate early genes (IEGs), and found a requirement for Integrator in the 3’end cleavage and termination of primary eRNA. Additionally, Integrator was deemed essential for EGF-induced chromatin looping, as identified by chromosome conformation capture [10]. Finally, it was shown recently that Integrator displays a global role in eRNA processing at most active enhancers [10,19].
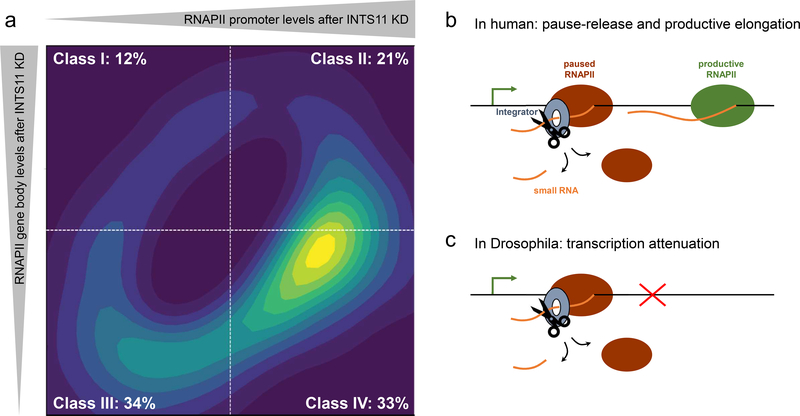
Transcription is regulated at multiple levels to precisely execute gene expression programs critical for cellular homeostasis, development, and response to environmental cues. Besides Integrator’s function in enhancer-promoter looping and subsequent gene activation, it also acts on recruitment of RNAPII to promoters [14,19], premature transcriptional attenuation [17,18,20], and RNAPII pause-release followed by transcriptional elongation [14,16,19]. Advances in nascent RNA sequencing further accelerated our understanding of Integrator’s role in transcription. Two recent studies in Drosophila and human sought to decipher Integrator’s regulatory mechanisms at gene promoters using precision run-on sequencing (PRO-seq), a method that maps actively engaged RNAPII at single nucleotide resolution [17,19]. While in Drosophila, INTS9 depletion leads to activation of a small subset of genes displaying enhancer-like chromatin modifications [17,18], human Integrator plays a more profound and wide-ranging role in the regulation of gene expression [19]. Here, INTS4, INTS9, or INTS11 depletions lead to equal proportions of gene activation and repression, which could be attributed to distinct mechanisms by which Integrator regulates transcriptional initiation and subsequent regulation of transcriptional elongation beyond the +1 nucleosome [19]. While standard PRO-seq analyses approaches like the traveling ratio or pausing index are limited in resolving RNAPII transcriptional dynamics at promoter and gene body, the establishment of the traveling matrix significantly improved our understanding of Integrator in transcriptional regulation (Figure 2a). Separating positional changes in RNAPII at promoters and gene bodies incurred by the loss of INTS11 led to the identification of four gene classes. While Integrator depletion leads to transcriptional de-repression in class I and II genes (33%), 67% of genes (class III and IV) require Integrator for their transcriptional activity (Figure 2a). Additionally, while class I and III genes rely on Integrator for RNAPII recruitment to the promoter, class II and IV genes require INTS11 cleavage activity for proper regulation [19]. Consequently, cleavage of stalled transcripts by INTS11 at these genes leads to premature termination of transcription in the proximity of +1 nucleosome, allowing for productive transcriptional elongation by subsequent rounds of initiation (Figure 2b, [19]). Conversely in Drosophila, Integrator was shown to prematurely terminate transcription at a small subset of genes (~15%) displaying unusual histone modifications of high levels of mono-methyl H3K4 and low levels of tri-methyl H3K4 in the process termed attenuation leading to decreased transcription (Figure 2c, [17]).
Figure 2. Integrator’s main functions on coding genes in human and Drosophila.
a) The travelling matrix separates positional RNAPII changes at promoters and gene bodies into four classes. Graphical depiction of ~3100 significant Integrator-responsive genes [19]. In human, INTS11 depletion predominantly leads to downregulation of actively engaged RNAPII (class III and IV: 67%). Class IV genes are additionally characterized by increased RNAPII pause (33%). b and c) Model representation of Integrator’s functions in human and Drosophila. b) The Integrator complex cleaves promoter-associated small transcripts to allow paused RNAPII eviction and transcriptional elongation by productive RNAPII (class IV). c) Integrator is required for the premature termination (attenuation) of RNAPII transcription.
Interestingly, INTS8 was recently identified to interact with the protein phosphatase 2A (PP2A), which is required for dephosphorylation of RNAPII CTD and pausing factor Spt5 [39], thus inhibiting productive elongation. This study emphasizes the importance of transcriptional regulation mediated by the Integrator complex, albeit independent of its endonucleolytic activity.
What links Integrator malfunctions to neurodevelopmental defects and cancer?
As a result of Integrator’s essential functions, the full disruption of the complex is lethal during embryogenesis [40,41]. Strikingly, individuals harboring biallelic mutations in INTS1 and INTS8 exhibit serious neurodevelopmental defects [21,22]. Integrator was also identified to be crucial for ciliogenesis, the formation of primary cilia responsible for signal transmission [42]. Dysfunctional ciliary proteins are the cause of ciliopathies, which manifest as defects in organogenesis as well as cognitive deficits related to neurodevelopmental disorders. Indeed, INTS6 and INTS10 were identified in a CRISPR based screen as novel ciliopathy genes, linking Integrator and Hedgehog signaling during mammalian ciliogenesis [43].
Multiple recent studies used in vivo or in vitro models to pinpoint the mechanism of Integrator action during embryonic development. Oegema et al. suggest that INTS8 mutations lead to Integrator complex disruption, resulting in UsnRNA misprocessing, and altered splicing patterns. Consequently, resulting gene expression changes lead to impaired neurodevelopment potential [21]. INTS expression peaks during early development, such that INTS RNAi reduces the organism’s differentiation capacity in flatworm Schmidtea mediterranea [44] or brine shrimp Artemia sinica [45]. In mouse, the absence of Integrator leads to neuronal migration defects, an underlying cause of neurological disorders [46]. More interestingly, Van den Berg et al. demonstrate Integrator’s presence at active and poised promoters and enhancers of the relevant gene network, placing Integrator at the junction of signaling and gene expression [46]. Indeed, Integrator-dependent gene regulation has been widely reported as a response to environmental stress [18,45,47] and signaling induction [14,15]. This connection insinuates context-specific interactions of Integrator with transcription factors (TFs), as reported for the fibroblast growth factor downstream TF Esrrb in trophoblast stem cells during self-renewal [48].
Consistent with a fundamental role in modulating gene expression pathways during signal transduction, Integrator mutations were also detected in multiple cancers [25]. Interestingly, lower expression levels of multiple INTS correlate with poorer overall patient survival in various cancer cohort analyses [12]. Additionally, INTS3 was found to be mis-spliced in acute myeloid leukemia [49], resulting in the loss of further INTS and the cells’ differentiation potential, in accordance with observations during development. Finally, a negative selection genetic screen (purifying selection) for mutations in essential genes for cancer identified INTS10 as highest ranked gene, placing Integrator as potential target for cancer treatment [50].
Conclusion and outlook
While the Integrator complex evolved with the appearance of the nucleated organisms, its composition and potential functions in transcription increased with genome complexity, consistent with Integrator’s requirement in development. Additionally, some INTS form smaller subcomplexes [1,35,51], and INTS3 and INTS6 have been also shown to participate in DNA damage response [51]. Similarly, INTS13 represents an independent submodule targeted to poised enhancers [52]. Understanding the structural modularity and assembly of the complex, as well as its interaction with RNAPII (Box 1) will further enhance our understanding of Integrator functions and allow identification of potential interfaces for therapeutic interference.
Box 1: Future perspectives.
To date, many aspects of Integrator’s assembly and functions remain elusive.
Beyond the catalytic module, what are other sub-modules of Integrator complex?
Is Integrator interaction with RNAPII CTD conserved throughout evolution? And which INTS mediate this interaction?
What is the mode of Integrator recruitment to various promoters?
What is the mechanism of Integrator effects on chromatin architecture?
How does Integrator contribute to DNA repair?
What are the determinants of RNA cleavage by INTS11 and how other Integrator subunits contribute to RNA cleavage activity?
Why Integrator mutations in humans are manifested by neurodevelopmental disorders?
Highlights.
The Integrator complex originates in Eukarya and gains additional subunits in parallel to increasing organism/genome complexity
Integrator is required for transcription homeostasis: from 3’end processing of functional small RNAs, to regulation of RNA polymerase II pause-release and premature transcription termination
Alterations in human Integrator subunits contribute to neurodevelopmental defects and cancer
Acknowledgements
We thank the Shiekhattar lab members for discussions and helpful comments. R.S. is supported by the University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center and grants R01 GM078455 and GM105754, and DP1 CA228041 from the National Institute of Health, and the National Cancer Institute of the National Institutes of Health Award Number P30CA240139.
Footnotes
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Baillat D, Hakimi M-A, Näär AM, Shilatifard A, Cooch N, Shiekhattar R: Integrator, a Multiprotein Mediator of Small Nuclear RNA Processing, Associates with the C-Terminal Repeat of RNA Polymerase II. Cell 2005, 123:265–276. [DOI] [PubMed] [Google Scholar]
- [2].Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S: The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. Journal of Biological Chemistry 2010, 285:20564–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ebmeier CC, Erickson B, Allen BL, Allen MA, Kim H, Fong N, Jacobsen JR, Liang K, Shilatifard A, Dowell RD, et al. : Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications. CellReports 2017, 20:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shah N, Maqbool MA, Yahia Y, El Aabidine AZ, Esnault C, Forné I, Decker T-M, Martin D, Schüller R, Krebs S, et al. : Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Molecular cell 2018, 69:48–61.e46. [DOI] [PubMed] [Google Scholar]
- [5].Albrecht TR, Shevtsov SP, Wu Y, Mascibroda LG, Peart NJ, Huang K-L, Sawyer IA, Tong L, Dundr M, Wagner EJ: Integrator subunit 4 is a ‘Symplekin-like’ scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Research 2018, 46:4241–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Albrecht TR, Wagner EJ: snRNA 3’ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol 2012, 32:1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L: Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature 2006, 444:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ: A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3’-end formation. Molecular and cellular biology 2011, 31:328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S: Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Molecular cell 2012, 45:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lai F, Gardini A, Zhang A, Shiekhattar R: Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, Dujardin G, Dhir A, Murphy S, Proudfoot NJ: Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol Cell 2018, 72:970–984 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barra J, Gaidosh GS, Blumenthal E, Beckedorff F, Tayari MM, Kirstein N, Karakach TK, Jensen TH, Impens F, Gevaert K, et al. : Integrator restrains paraspeckles assembly by promoting isoform switching of the lncRNA NEAT1. Science Advances 2020, 6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rubtsova MP, Vasilkova DP, Moshareva MA, Malyavko AN, Meerson MB, Zatsepin TS, Naraykina YV, Beletsky AV, Ravin NV, Dontsova OA: Integrator is a key component of human telomerase RNA biogenesis. Sci Rep 2019, 9:1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, Shiekhattar R: Integrator Regulates Transcriptional Initiation and Pause Release following Activation. Molecular cell 2014, 56:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yue J, Lai F, Beckedorff F, Zhang A, Pastori C, Shiekhattar R: Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes & Development 2017, 31:1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, et al. : Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 2014, 5:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, Adelman K: The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol Cell 2019, 76:738–752 e737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tatomer DC, Elrod ND, Liang D, Xiao M-S, Jiang JZ, Jonathan M, Huang K-L, Wagner EJ, Cherry S, Wilusz JE: The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes & Development 2019, 33:1525–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Beckedorff F, Blumenthal E, daSilva LF, Aoi Y, Cingaram PR, Yue J, Zhang A, Dokaneheifard S, Valencia MG, Gaidosh G, et al. : The Human Integrator Complex Facilitates Transcriptional Elongation by Endonucleolytic Cleavage of Nascent Transcripts. CellReports 2020, 32:107917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, Washburn MP, Hughes SH, Pagano M: The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res 2015, 25:288–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oegema R, Baillat D, Schot R, van Unen LM, Brooks A, Kia SK, Hoogeboom AJM, Xia Z, Li W, Cesaroni M, et al. : Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS genetics 2017, 13:e1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krall M, Htun S, Schnur RE, Brooks AS, Baker L, de Alba Campomanes A, Lamont RE, Gripp KW, Care 4 Rare Canada C, Schneidman-Duhovny D, et al. : Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur J Hum Genet 2019, 27:582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Y, Koe CT, Tan YS, Ho J, Tan P, Yu F, Sung W-K, Wang H: The Integrator Complex Prevents Dedifferentiation of Intermediate Neural Progenitors back into Neural Stem Cells. CellReports 2019, 27:987–996.e983. [DOI] [PubMed] [Google Scholar]
- [24].Rienzo M, Casamassimi A: Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochimica et biophysica acta 2016, 1859:1269–1280. [DOI] [PubMed] [Google Scholar]
- [25].Federico A, Rienzo M, Abbondanza C, Costa V, Ciccodicola A, Casamassimi A: Pan-Cancer Mutational and Transcriptional Analysis of the Integrator Complex. Int J Mol Sci 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baillat D, Wagner EJ: Integrator: surprisingly diverse functions in gene expression. Trends in biochemical sciences 2015, 40:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Litman T, Stein WD: Obtaining estimates for the ages of all the protein-coding genes and most of the ontology-identified noncoding genes of the human genome, assigned to 19 phylostrata. Semin Oncol 2019, 46:3–9. [DOI] [PubMed] [Google Scholar]
- [28].Sanchez R, Zhou M-M: The PHD finger: a versatile epigenome reader. Trends in biochemical sciences 2011, 36:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 1990, 215:403–410. [DOI] [PubMed] [Google Scholar]
- [30].Peart N, Sataluri A, Baillat D, Wagner EJ: Non-mRNA 3’ end formation: how the other half lives. Wiley Interdiscip Rev RNA 2013, 4:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu Y, Li S, Chen Y, Kimberlin AN, Cahoon EB, Yu B: snRNA 3’ End Processing by a CPSF73-Containing Complex Essential for Development in Arabidopsis. PLoS Biol 2016, 14:e1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. : The Pfam protein families database in 2019. Nucleic Acids Res 2019, 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huerta-Cepas J, Szklarczyk D, Heller D, Hernandez-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. : eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 2019, 47:D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wu Y, Albrecht TR, Baillat D, Wagner EJ, Tong L: Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proceedings of the National Academy of Sciences of the United States of America 2017, 114:4394–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sabath K, Staubli ML, Marti S, Leitner A, Moes M, Jonas S: INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module. Nat Commun 2020, 11:3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harlen KM, Churchman LS: The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nature reviews. Molecular cell biology 2017, 18:263–273. [DOI] [PubMed] [Google Scholar]
- [37].Hernandez N: Formation of the 3’ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J 1985, 4:1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xie M, Zhang W, Shu MD, Xu A, Lenis DA, DiMaio D, Steitz JA: The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3’ ends. Genes Dev 2015, 29:1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang KL, Jee D, Stein CB, Elrod ND, Henriques T, Mascibroda LG, Baillat D, Russell WK, Adelman K, Wagner EJ: Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. Mol Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hata T, Nakayama M: Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochimica et biophysica acta 2007, 1773:1039–1051. [DOI] [PubMed] [Google Scholar]
- [41].Gomez-Orte E, Saenz-Narciso B, Zheleva A, Ezcurra B, de Toro M, Lopez R, Gastaca I, Nilsen H, Sacristan MP, Schnabel R, et al. : Disruption of the Caenorhabditis elegans Integrator complex triggers a non-conventional transcriptional mechanism beyond snRNA genes. PLoS Genet 2019, 15:e1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jodoin JN, Shboul M, Albrecht TR, Lee E, Wagner EJ, Reversade B, Lee LA: The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol Open 2013, 2:1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, Kennedy MC, Han K, Li A, Hess GT, Bassik MC, et al. : A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet 2018, 50:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schmidt D, Reuter H, Huttner K, Ruhe L, Rabert F, Seebeck F, Irimia M, Solana J, Bartscherer K: The Integrator complex regulates differential snRNA processing and fate of adult stem cells in the highly regenerative planarian Schmidtea mediterranea. PLoS Genet 2018, 14:e1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huang H, Liu J, Yao F, Li X, Wang Y, Shao Y, Wang X, Kong J, Zhang X, Jiang T, et al. : The integrator complex subunit 11 is involved in the post-diapaused embryonic development and stress response of Artemia sinica. Gene 2020, 741:144548. [DOI] [PubMed] [Google Scholar]
- [46].van den Berg DLC, Azzarelli R, Oishi K, Martynoga B, Urban N, Dekkers DHW, Demmers JA, Guillemot F: Nipbl Interacts with Zfp609 and the Integrator Complex to Regulate Cortical Neuron Migration. Neuron 2017, 93:348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu CW, Wimberly K, Pietras A, Dodd W, Atlas MB, Choe KP: RNA processing errors triggered by cadmium and integrator complex disruption are signals for environmental stress. BMC Biol 2019, 17:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Latos PA, Goncalves A, Oxley D, Mohammed H, Turro E, Hemberger M: Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat Commun 2015, 6:7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yoshimi A, Lin K-T, Wiseman DH, Rahman MA, Pastore A, Wang B, Lee SC-W, Micol J-B, Zhang XJ, de Botton S, et al. : Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 2019, 574:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Van den Eynden J, Basu S, Larsson E: Somatic Mutation Patterns in Hemizygous Genomic Regions Unveil Purifying Selection during Tumor Evolution. PLoS Genet 2016, 12:e1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang F, Ma T, Yu X: A core hSSB1-INTS complex participates in the DNA damage response. Journal of Cell Science 2013, 126:4850–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Barbieri E, Trizzino M, Welsh SA, Owens TA, Calabretta B, Carroll M, Sarma K, Gardini A: Targeted Enhancer Activation by a Subunit of the Integrator Complex. Molecular cell 2018, 71:103–116.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]