SUMMARY
Odors carrying intrinsic values often trigger instinctive aversive or attractive responses. It is not known how innate valence is encoded. An intuitive model suggests that the information is conveyed through specific channels in hardwired circuits along the olfactory pathway, insulated from influences of other odors, to trigger innate responses. Here, we show that in mice, mixing innately aversive or attractive odors with a neutral odor and, surprisingly, mixing two odors with the same valence, abolish the innate behavioral responses. Recordings from the olfactory bulb indicate that odors are not masked at the level of peripheral activation and glomeruli independently encode components in the mixture. In contrast, crosstalk among the mitral and tufted (M/T) cells changes their patterns of activity such that those elicited by the mixtures can no longer be linearly decoded as separate components. The changes in behavioral and M/T cell responses are associated with reduced activation of brain areas linked to odor preferences. Thus, crosstalk among odor channels at the earliest processing stage in the olfactory pathway leads to re-coding of odor identity to abolish valence associated with the odors. These results are inconsistent with insulated labeled lines and support a model of a common mechanism of odor recognition for both innate and learned valence associations.
In brief
Qiu et al. find that odor mixtures abolish innate responses to the component odors. Recordings from the olfactory bulb demonstrate significant crosstalk among mitral/tufted cells to encode odors in a context-dependent manner. These results are inconsistent with insulated labeled-line model and suggest a generalized code for innate odor encoding.
INTRODUCTION
Theories of sensory coding have always seen two major competing views: the specificity and the pattern theories (segregated versus distributed processing). The specificity theory, often referred to as the “labeled-line,” suggests that sensory signals are processed along a fixed, direct line of communication that connects sensory input with a behavior.1 The pattern theory, also termed “across-fiber” and “parallel processing,” stipulates that stimulus information is distributed across different neurons and pathways. Since the mid-19th century, these theories have permeated discussions of sensory coding of all modalities. Whereas the labeled-line theory has been discussed in every sensory modality,2 this type of circuitry is mostly discovered for senses that carry innate valence information and trigger stereotypical responses. For example, the detections of looming threat and pain are thought to be mediated by labeled-line circuits.3–5 In insect and mammalian taste, information about submodalities is suggested to be transmitted by separated pathways.6,7 Complete circuits that link sensory neurons expressing a specific receptor to the cells controlling behavioral output have been delineated in the fly olfactory system.8–10
The mammalian olfactory system is divided into at least four subsystems: the main olfactory epithelium, the vomeronasal organ, the septal organ of Masera, and the Grueneberg ganglion.11 Whereas the functions of the septal organ and the Gruenberg ganglion are debated, the vomeronasal organ is dedicated to detecting intra-species and inter-species cues that elicit innate, stereotypic behaviors and endocrine changes.12,13 Labeled lines have been suggested to link specific pheromone cues to behaviors.14–16 The main olfactory system detects the odor world at large. Individual olfactory sensory neurons (OSNs) express only a single type of odorant receptor. OSNs expressing the same receptor converge their axons into the same glomerulus in the olfactory bulb, where there is a one-to-one connection between glomeruli and the mitral/tufted (M/T) cells.17 Despite the stereotypy in connectivity, studies have largely uncovered a population code for odor identity, given that individual odors generally activate multiple receptor types and each receptor can be tuned to many odors.18–20
Volatile odors also elicit innate responses. Mice are attracted to food and the urine of their own species, but avoid scents from predators, rotten food, and decomposing carcasses. The compound (methylthio)-methanothiol (MTMT) in male urine elicits attraction.21 2-methyl butyric acid (2-MBA), found in spoiled food; 2,3,5-trimethyl-3-thiazoline (TMT) from fox feces; and 2-phenylethylamine (PEA) from bobcat urine trigger avoidance.22–26 Similarly, 2-methylbut-2-enal elicits suckling behavior in neonatal rabbits independent of other milk odors.27 The stereotypical responses to these odorants in different contexts without learning suggest that highly specific inborn pathways transmit the valence information. Indeed, activation of Olfr1019, a receptor for TMT, induces immobility in mice.28 Olfr288 is shown to mediate attraction by urinary odorant (Z)-5-tetradecen-1-ol (Z5–14:OH), but low-affinity activation of other receptors in the absence of Olfr288 leads to aversion.29 The trace-amine-associated receptor 4 (TAAR4) is activated by low-concentration PEA30 and is needed for low-threshold aversive response to PEA.24 These studies provide evidence supporting that dedicated parallel pathways transmit innate valence information. A variant of the specific channel model is that odor valence is topographically encoded. It is hypothesized that the dorsal-ventral axis in the olfactory bulb also corresponds to the aversion-attractive axis in valence coding.21,22 This hypothesis is supported by the finding that genetic ablation of the dorsal olfactory bulb abolishes innate aversive responses to odors without affecting the learning of the same odors in general.22 However, it is challenged by the observation that trimethylamine (TMA) activates the dorsal projecting TAAR5 neurons but elicits attractive responses.31,32
The labeled-line model is incompatible with the population code observed in the main olfactory system. Not only are individual M/T cells tuned to multiple odors17,33,34 in areas traditionally thought to encode odor valence such as the PLCo, a distributed representation for odors with no apparent bias for the aversive odors is found.35 Different ligands for the same TAAR can elicit opposing responses in a context-dependent manner.32 Moreover, except the tracing of TMT and 2-MBA responding M/T cells to the same area in the posterolateral cortical amygdaloid (PLCo), no labeled line has been delineated22,36 (see also Perez-Gómez et. al37). However, it is difficult to demonstrate or refute the labeled line or the population code model experimentally. In the labeled-line model, innately recognized odors are expected to be processed along genetically hardwired circuits, separately from other odors. Disrupting any component in the link is expected to change behavioral responses. On the other hand, all neural circuits require genetic specification to some extent, and population responses depend on the presence of specific set of neurons. Therefore, genetic intervention, optogenetic, or chemogenetic approaches designed to test the labeled-line model can also alter population responses. For example, the loss of innate response after knocking out a specific receptor can be interpreted as the loss of a labeled line, but the knockout might also alter activity patterns in the brain, thereby creating a different perceptual representation based on the population code.
We reason that investigation using odor mixtures can help address this problem. In the labeled-line model, information about an innately recognized odor should be insulated from the background and insensitive to the presence of other odors. In other words, the odor is perceived as elemental—as a separate component in a mixture. The population code model would predict that odor mixtures might be perceived as configural, i.e., as a new odor.38 Indeed, recent evidence indicates that odorants can interact to block attraction or aversion in a mixture without receptor antagonism.32 In this study, we investigate whether the M/T cells that encode innately recognized odors are subject to interference from other odors and whether brain activity patterns and the associated innate behavioral responses are altered when these odors are presented in mixtures. Our evidence points to a model of innate odor coding using a generalized population code that is similar to that for learned odor preference.
RESULTS
Odor mixing abolishes innate valence
An animal’s approach toward an odor source is motivated by novelty-seeking, preference, and odor habituation.39 For an attractive odor, prolonged investigation reflects attraction and novelty-seeking. In contrast, an aversive odor might induce both novelty-seeking (for risk assessment) and avoidance. Odor preference assessed by using place preference by associating odors with their spatial locations can be confounded by intrinsic bias in spatial preference and scent marking.40,41 To avoid intrinsic place bias, we have devised a computerized setup, PROBES, to perform automated single-chamber odor preference assays based on habituation-dishabituation (Figure 1A).40 Despite individual variation in background investigation (Figures 1B–1D), a detailed examination of four episodes of odor presentation revealed that attractive odors such as urine elicited vigorous investigation of the odor port in the first epoch and continued to elicit above-background investigation (Figures 1B, S1A, S1D, S1E, S1G, and S1H). Odors not known to elicit attraction or aversion also triggered investigation above background, but the level was significantly lower than that of the attractive odors (Figures 1C and S1B). They did not elicit investigation above-background level in subsequent epochs (Figures S1D, S1E, and S1H). In contrast, known aversive odors (e.g., coyote urine or PEA) did not elicit investigation above the control level in the first epoch, but triggered a significant depression in investigation in the second one (Figures 1D, S1C, and S1E). We reasoned that investigation of an aversive odor during the first exposure likely reflected the effect of two competing drives: avoidance and risk assessment. During the second exposure, aversion likely became the dominant drive, resulting in a decrease in investigation. Investigations in the third and fourth epochs exhibited a similar trend but were not statistically different (Figures S1A–S1C, S1F, S1I, and S1J). Thus, the high level of investigation during the first epoch indicated attraction, and the depressed investigation in the second epoch indicated aversion. To measure behavioral preference, we combined the two as a single index (Figures 1A, 1E, and S1K). Despite individual variability, the scores within the attractive, neutral, and aversive odor groups can be approximated by normal distribution and are clearly distinguishable (Figures S1L–S1N). Odors or odor mixtures that elicited scores statistically higher than the average were considered as attractive, whereas those with significantly lower scores were considered as aversive.
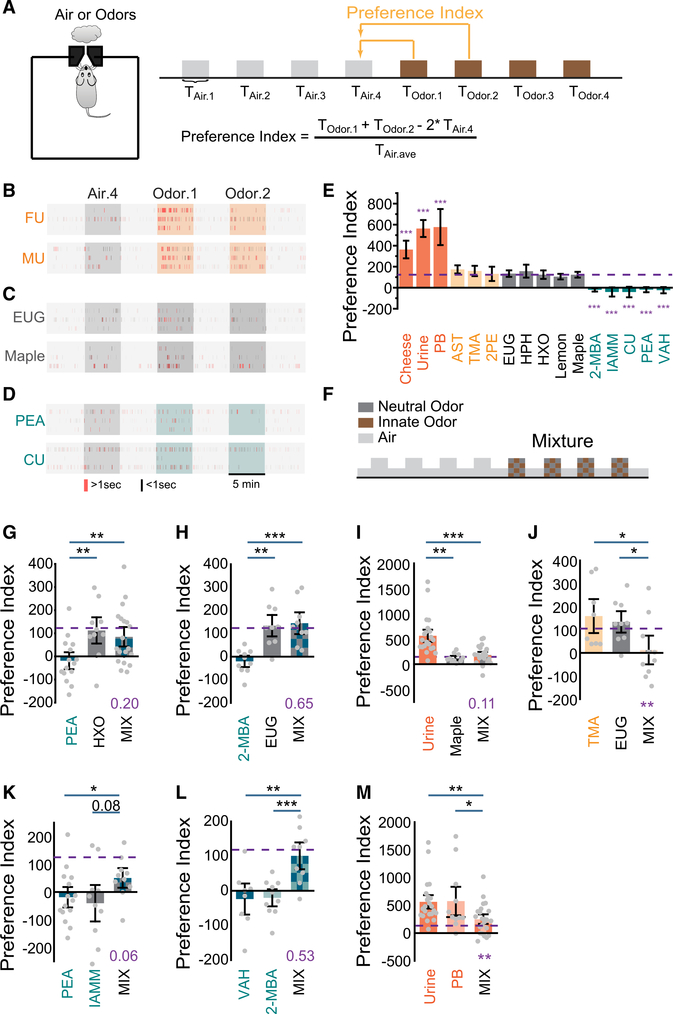
Figure 1. Odor mixtures abolish innate odor preference.
(A) Illustration of the behavior paradigm. Shown on the left is a single-odor port arena. Shown on the right is the odor presentation sequence and the quantification of preference index.
(B) Sample raster plots of odor port investigation of female (FU) and male (MU) urine presentation. Only the fourth air presentation (Air.4, gray box) and the first two odor presentations (Odor.1 and Odor.2, orange boxes) are shown. Each tick represents an investigation event. Investigations longer or shorter than 1 s are marked by red and black ticks, respectively.
(C) Same as (B) but for eugenol (EUG) and maple. Dark-gray-colored boxes indicate presentation of neutral odors.
(D) Same as (B) but for 2-phenylethylamine (PEA) and coyote urine (CU). Olive-colored boxes indicate presentation of aversive odors.
(E) Bar graph showing preference indices for a panel of odors (color coding is as follows: attractive, orange; monomolecular odors previous reported as attractive, light yellow; neutral, gray; aversive, olive). Purple dashed line indicates the averaged level for neutral odors. One-way ANOVA tests were applied to compare individual odors to the average level of neutral odors.
(F) Illustration of odor mixture experiment setup.
(G–J) Bar plots of preference indices measured for individual odor pairs and their mixtures. Urine samples were tested on the opposite sex and grouped together.
(K–M) Bar plots of preference indices measured for individual odor pairs of the same innate valence and their mixtures.
Purple dashed line indicates the average level for neutral odors. One-way ANOVA with Tukey test was applied to determine whether the mixture is attractive or aversive (p value in purple). All bar graph data are shown in mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, probability is shown when p > 0.05.
Notably, several monomolecular odorants that previously had been characterized as attractive, including 2-phenylethanol (2PE), 3-amino-s-triazole (AST), and trimethylamine (Table S1), were classified as neutral in our assay. In the previous studies, these odors evoked more investigation than controls (no odor or water).32,42 In our assay, the responses were also significantly higher than no odor control, but they did not rise to the level of the compound odors known as attractive (Figure 1E).
We performed experiments by using mixtures composed of equal parts of a neutral odor and an odor with innate valence (Figure 1F). Although PEA alone caused pronounced avoidance and 2-hexanone (HXO) did not, mixing PEA with HXO elicited no aversive response (Figure 1G). We observed a similar effect for the mixture of 2-MBA (aversive) and eugenol (EUG) (neutral; Figure 1H). A 2- to 4-fold change in the concentration of neutral odor in the mixtures did not affect this result (Figures S1O and S1P). Urine was attractive to mice of the opposite sex, but maple odor was not (Figures 1E and 1I). A mixture of maple and urine elicited little to no attraction (Figure 1I). In all pairs tested, odor preferences were abolished when innately recognized odors were mixed with neutral odors. We also found that mixing EUG, a neutral odor, with TMA (neutral in our assay but characterized as attractive in others) elicited aversion (Figure 1J).
Mixing odors of same valence abolishes behavioral preference
How could mixing odors abolish innate preference? One possibility is that one odor masks the detection of the other. Alternatively, two odors might activate separate brain regions that drive competing behaviors, resulting in nullification of innate responses. In either scenario, if we mixed two odors of the same valence, the mixture was expected to elicit the same behavioral responses. We, therefore, examined preference tests by using mixtures of two aversive odors, PEA with isoamylamine (IAMM) (Figure 1K).24 Strikingly, the mixture did not elicit aversion. Given that PEA and IAMM were known to activate TAAR-expressing OSNs, we tested 2-MBA together with valeraldehyde (VAH), both were pungent odorants of spoiled foods but not known to activate the TAARs.22 The mixture of the two odors elicited a neutral response (Figure 1L). Similarly, well-mixed, innately attractive odors also reduced their attractiveness (Figure 1M).
Linear decoding of odors from glomeruli responses
These behavioral results cannot be explained by masking or antagonistic interactions between brain areas. They are inconsistent with the labeled-line hypothesis. We hypothesize that mixing alters odor identities such that they are no longer recognizable. Volatile odorants activate multiple receptors in the olfactory epithelium.18–20 Odorants can interact in activating receptor neurons.43,44 Moreover, odors are represented by distributed activities in the olfactory bulb and in the cortices.45–47 Interactions along the pathway might allow odor mixture to create activity patterns that are no longer decoded as individual odors.
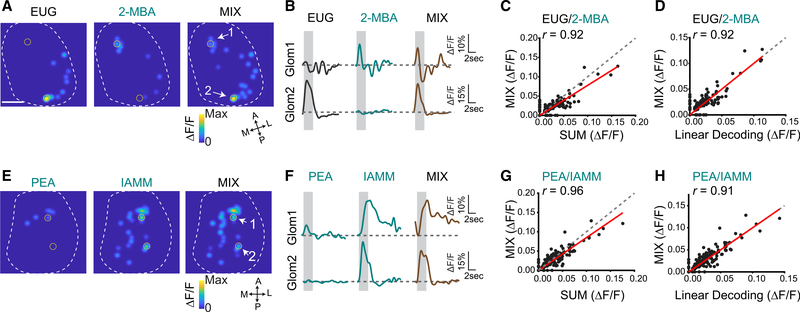
We thus examined neural activities in the olfactory pathway. We first recorded glomerular activation by individual odors and their binary mixtures by using the OMP-IRES-tTA:tetO-GCaMP2 mice.12,47 Aversive odors including PEA, 2-MBA, IAMM, VAH as well as neutral odors, including HXO and EUG, all activated the dorsal bulb. A mixture of EUG and 2-MBA elicited glomerular response patterns similar to the sum of the EUG and 2-MBA alone (Figures 2A and 2B). Response amplitude of individual glomeruli to the mixture was linearly related to the arithmetic sum of the two individual odors along the diagonal line, indicating that the mixture did not cause overt masking at the level of the OSN (Figure 2C). Another pair of aversive/neutral odors, PEA and HXO (Figures S2A–S2C), as well as pairs of innately aversive odors including PEA and IAMM (Figures 2E–2G) and VAH and 2-MBA (Figures S2E–S2G) elicited responses with same characteristics. Even though some responses exhibited sub-linear summation, the mixture responses could be precisely predicted by using linear decoding of the patterns (Figures 2D, 2H, S2D, and S2H). We also extended our studies to the EUG and TMA pair (Figures S3A–S3C) as well as pairs of attractive odors (Figures S3D–S3I). Although neither peanut butter (PB) nor cheese odors evoked strong response in the dorsal bulb, the recorded responses nevertheless exhibited similar linear relationships.
Figure 2. Linear decoding of odors from glomeruli responses.
(A) Glomerular activation patterns elicited by EUG, 2-MBA, and their mixture. Contours of the bulb are outlined. Orientations of the bulb are labeled as follows: A, anterior; P, posterior; M, medial; L, lateral. Scale bar, 500 μm.
(B) Sample response traces from the marked glomeruli in (A). Gray box indicates the odor presentation.
(C) Glomerular response to odor mixture plotted against the sum of response to individual odors. Red line indicates linear fit of the data. Pearson’s correlation coefficient (r) and diagonal line (gray dash) are indicated.
(D) Glomerular response to the odor mixture plotted against the predicted response from linear decoding for odor pair EUG and 2-MBA. Linear decoding is performed by fitting the mixture response as the linear combination of the two component odors (see method details).
(E–H) Same as (A)–(D), but for two aversive odors pair PEA and IAMM.
See also Figures S2 and S3 and Table S1.
These observations showed that odor mixtures containing the innately recognized odors could be represented by linear combination of the glomerular responses to individual odors, similarly to other binary odor mixtures.48 This result indicated that the glomerular representations of the odors were mostly independent of each other. If the insulated labeled-line hypothesis is correct, this type of superposition would allow each odor to activate its distinctive pathway and elicit aversion.
Altered odor representation by the mitral and tufted cells
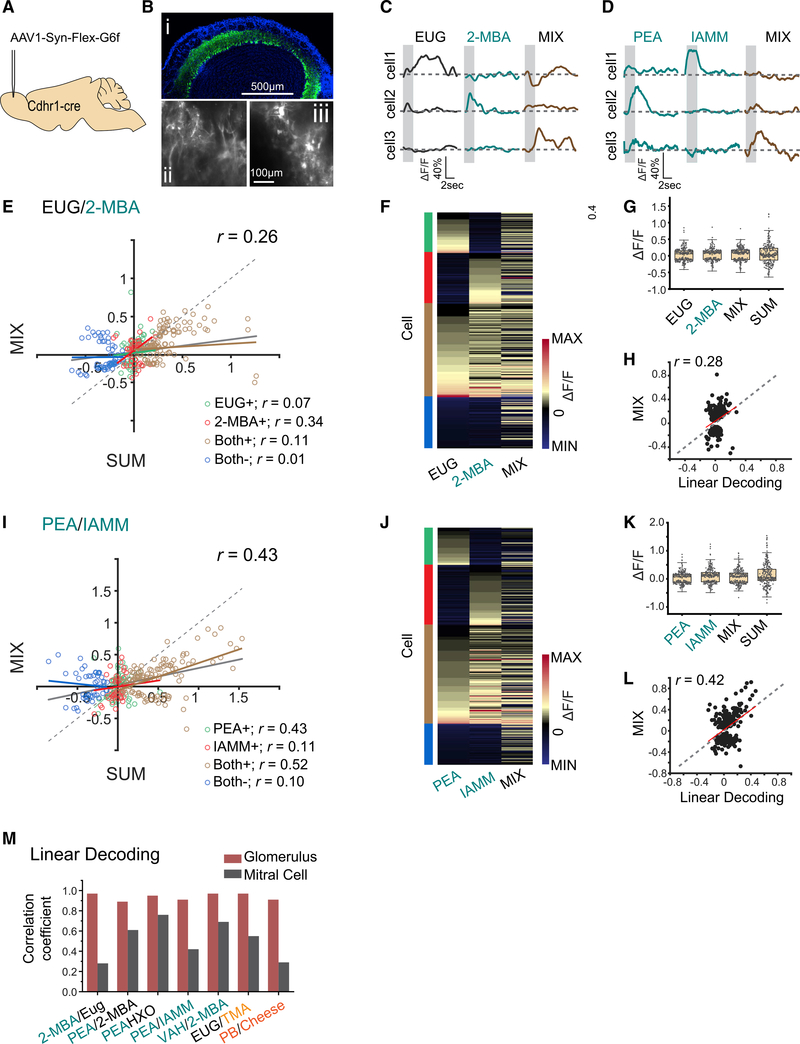
Although M/T cells can sum input from the glomeruli linearly in response to odor mixtures,49 lateral excitation and inhibition mediated by interconnected neuronal networks might transform receptor activation into more complex population activities. To test whether the type of linear separation in the glomerular responses is carried through the olfactory pathways, we performed two-photon calcium imaging to record neural activities of the M/T cells in Cdhr1-Cre mice injected with an adeno-associated virus (AAV) that expresses GCaMP6f in a Cre-dependent manner (pAAV.Syn.Flex.GCaMP6f.WPRE.SV40) (Figure 3A).50 GCaMP6f fluorescence is observed in the cell body and dendrite of the M/T cells (Figure 3B). Although responses could be recorded from both areas, we focused on the cell bodies because these signals more likely reflected activities driven by action potentials. Under awake conditions, we recorded a total of 288 cells in response to PEA, IAMM, 2-MBA, EUG, HXO, and their binary mixtures, and a total of 188 cells in response to VAH, 2-MBA, EUG, TMA, PB, Cheese, and their binary mixtures. The cells exhibited characteristic responses timed to odor stimulations with both increase and decrease in calcium signals (Figures 3C and 3D).
Figure 3. Cross talk among odor channels in the mitral and tufted cell population.
(A) Illustration of AAV-mediated GCaMP6f expression in M/T cells.
(B) GCaMP6f expression in olfactory bulb 3 weeks after virus injection (i). Color coding is as follows: green, GCaMP6; blue, DAPI. Two-photon images showing the GCaMP6f expression in the glomerulus layer (ii) and in cell bodies (iii). Scale bars are indicated.
(C and D) Sample traces of M/T cell response to EUG, 2-MBA, and their mixture (C) or to PEA, IAMM, and their mixture (D). Gray box indicates odor delivery period.
(E) M/T cell response evoked by the mixture plotted against the sum of response to individual odors. They are color coded to indicate positive response (increased calcium signal) to EUG only (EUG+), 2-MBA only (2-MBA+), or both odors (Both+); and negative response (decrease in calcium signal) to both odors (Both−). Gray line indicates the linear fit for all data points. Colored lines indicate linear fit of each group of data. Pearson’s correlation coefficients (r) and diagonal line (gray dash) are indicated.
(F) Heatmap of M/T cell responses to EUG, 2-MBA, and their mixture. Cells are sorted according to their response to EUG only, 2-MBA only, positive response to both odors, and negative response to both odors. Groups are indicated with the same color scheme as in (E).
(G) Boxplot of the distribution of M/T cell response to EUG, 2-MBA, their mixture, and the arithmetic sum of component odors. Boxplot edges indicate the first and third quartiles of the data; whiskers indicate 1.5 interquartile range.
(H) Response of individual M/T cells to the mixture plotted against the predicted response from linear decoding for odor pair EUG/2-MBA. Red line indicates linear-fit of the linear decoding data from the responses of EUG and 2-MBA with activity from mixture. Pearson’s correlation coefficient (r) and diagonal line (gray dash) are indicated.
(I–L) same as (E)–(H) but for two aversive odor pair PEA and IAMM.
(M) Bar graphs shows the correlations coefficients (r) between linearly decoded and the actual responses of the glomeruli (burgundy color) and M/T cells (dark gray) for seven odor pairs.
See also Figures S4 and S5 and Table S1.
Cells responding to only one of the two odors responded to the mixture with either enhanced or diminished amplitudes, and these bidirectional changes were observed for aversive/neutral, aversive/aversive, and attraction/attraction odor pairs, including amine and non-amine odors (Figures 3E, 3F, 3I, 3J, S4A, S4B, S4E, S4F, S4I, S4J, S5A, S5B, S5E, and S5F). For all odor pairs examined, response amplitudes of individual cells to the mixture were smaller than the arithmetic sum of responses to the components for both positive and negative responses. Pairwise comparison between response to the mixture and the sum indicated little correlation between the two for aversive/neutral (Figures 3E and S4A), aversive/aversive (Figures 3I, S4E, and S4I), attractive/neutral (Figures S5A), and attractive/attractive pairs (Figure S5E). The distribution of the mixture response amplitude both positive and negative directions was similar to those of single odors and smaller than the arithmetic sum (Figures 3G, 3K, S4C, S4G, S4K, S5C and S5G). This lack of summation as found in the glomerular response indicated that the overall response was normalized within the cell population.
The patterns of activity that represented the odor mixture were strikingly different from that for individual odors or their sum (Figures 3F, 3J, S5B, S5F, S5J, S5B, and S5F). The mixture responses were poorly correlated with those predicted by linear decoding, indicating that the mixture responses could not be linearly demixed into individual odor patterns (Figures 3H, 3L, 3M, S4D, S4H, S4L, S5D, and S5H). The correlation coefficients between linearly decoded and the actual mixture responses ranged from 0.28 to 0.76, much smaller than those for the glomerular response for all types of mixing we have examined (> 0.9) (Figure 3M). Thus, the population response to the mixture likely represented a new odor identity rather than the combination of two components.
Differential brain activation by aversive odor in mixture
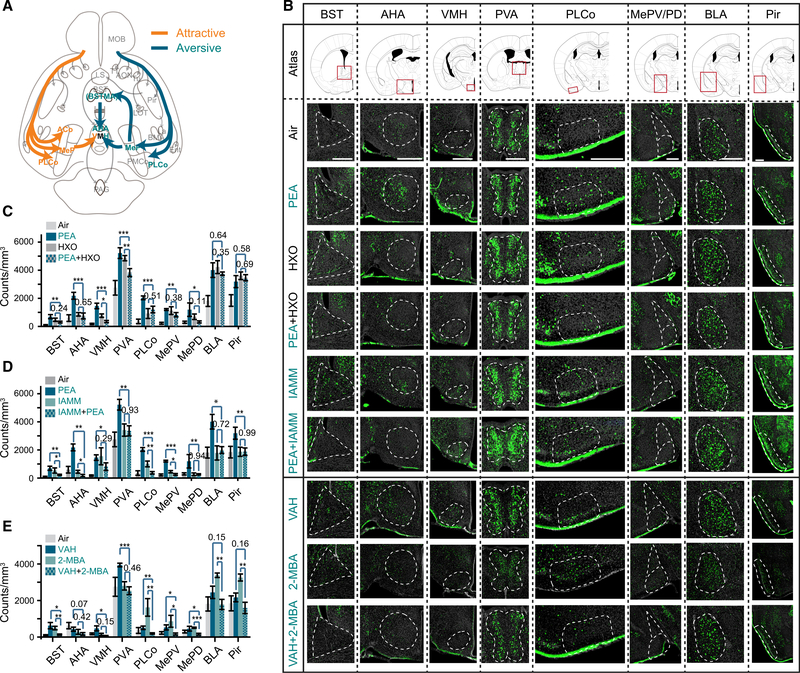
The M/T cells project to at least five cortical areas, including the PLCo and the posterior nuclei of the medial amygdala (MeP), where the pathway appeared to diverge and activate various brain regions associated with different valence (Figure 4A).51–54 Some of the brain regions have been rigorously examined for their involvement of innate aversive or appetitive behaviors. For example, optogenetic intervention has been used to demonstrate that the PLCo mediates both innate aversive and appetitive behaviors in response to odors.42 The ventral medial hypothalamic nucleus (VMH) mediates defensive behaviors including avoidance through activation of the anterior hypothalamic area (AHA),55,56 which is needed to trigger fear responses.57–60 The VMH is also involved in both attraction to mouse urine and aggressive responses.55,61 Optogenetic perturbation of the medial amygdala (MeA), which receives input directly from the olfactory bulb, reveals that it is needed to drive avoidance to predator odor through D1R-expressing cells projecting to the VMH.62 MeA also projects to the bed nucleus of stria terminalis (BST), including the anteriomedial nucleus (BSTMA), to mediate anxiogenic response.59,60 Photo-stimulation of glutamatergic projection from BNST to the ventral tegmental area (VTA) triggers avoidance.63 We, therefore, decided to survey the activation of these brain areas in response to innately recognized odors.
Figure 4. Altered representation of innate odors in the brain.
(A) Illustration of pathways processing information about innately recognized odors
(B) Immunofluorescent staining of phospho-S6 (green) of brain sections from animals exposed to air, PEA, HXO, PEA+HXO, IAMM, PEA+IAMM, VAH, 2-MBA, and VAH+2-MBA. Cell nuclei were counter-stained with DAPI (gray). The areas being quantified are marked by dashed lines. Scale bars, 500 μm.
(C–E) Quantification of activated cells in various brain regions in response to control, PEA, HXO, and their mixture (C); PEA, IAMM, and their mixture (D); and VAH, 2-MBA, and their mixture (E).
One-way ANOVA with Tukey test was applied. Data are shown in mean ± SEM, n = 6 half brains for each experiment.
Abbreviations are as follows: AHA, anterior cortical amygdaloid area; AON, anterior olfactory nucleus; BLA, basolateral amygdaloid nucleus, anterior part; BMA, basomedial amygdaloid nucleus, anterior part; BSTMA, bed nucleus of the stria terminalis, medial division, anterior part; Ent, entorhinal cortex; La, lateral amygdaloid nucleus; LOT, lateral olfactory tract; MOB LS, lateral septal nucleus; main olfactory bulb; MePD, medial amygdaloid nucleus, posterodorsal part; MePV, medial amygdaloid nucleus, posteroventral part; VMH, ventromedial hypothalamic nucleus; PAG, periaqueductal gray; PVA, paraventricular thalamic nucleus, anterior part; Pir, piriform cortex; PLCo, posterolateral cortical amygdaloid area; PMCo, posteromedial cortical amygdaloid nucleus; TT, tenia tecta; Tu, olfactory tubercle.
See also Table S1.
We performed immunofluorescent staining against phospho-S6 ribosomal protein (pS6), which stained activated neurons.64 This approach did not have the same temporal resolution as calcium imaging or electrophysiology. Activation of brain areas reflected not only feedforward signals from the olfactory bulb, but also other cortical areas. Nevertheless, these patterns could reveal whether brain activities associated with innate valence were altered by the presence of other odors. A dedicated channel linking a specific odor to these brain areas would lead to their activation regardless of whether the odor was in a mixture. On the other hand, if the activation of these nuclei relied on the proper odor identity encoded by the ensemble activity of M/T cells, then a change of the M/T cell activity in the mixtures was expected to reduce the activation of the corresponding nuclei.
Odor valence might influence approach behavior and lead to reduced odor sampling if the animals avoid an odor. To minimize this influence, we placed the odor vials in the home cage such that the animals were exposed to the odors similarly regardless of odor valence. After odor exposure, we performed serial sectioning through the brain and quantified activated cells in 3D volumes. PEA, but not the mixture with HXO, induced strong signals in BST, AHA, VMH, PVA, PLCo, and MePV (Figures 4B and 4C). We then compared the brain activation patterns elicited by the mixture of two aversive odors with those by individual odors alone. Even though both PEA and IAMM activated the BST, VMH, and PLCo, the PEA/IAMM mixture did not (Figures 4B and 4D). Similar results were found for the aversive odor pairs VAH and 2-MBA, which were not TAAR ligands (Figures 4B and 4E). Thus, areas associated with odor valence were differentially activated by an innately aversive odor depending on whether it was presented alone or in a mixture. The lack of activation of these areas was consistent with the behavioral test results. Areas that received olfactory input but not known to be associated with innate valence, including the basolateral amygdala (BLA) and the piriform cortex (Pir), were activated similarly (Figures 4B–4E).
Distinguishing aversive odors from background
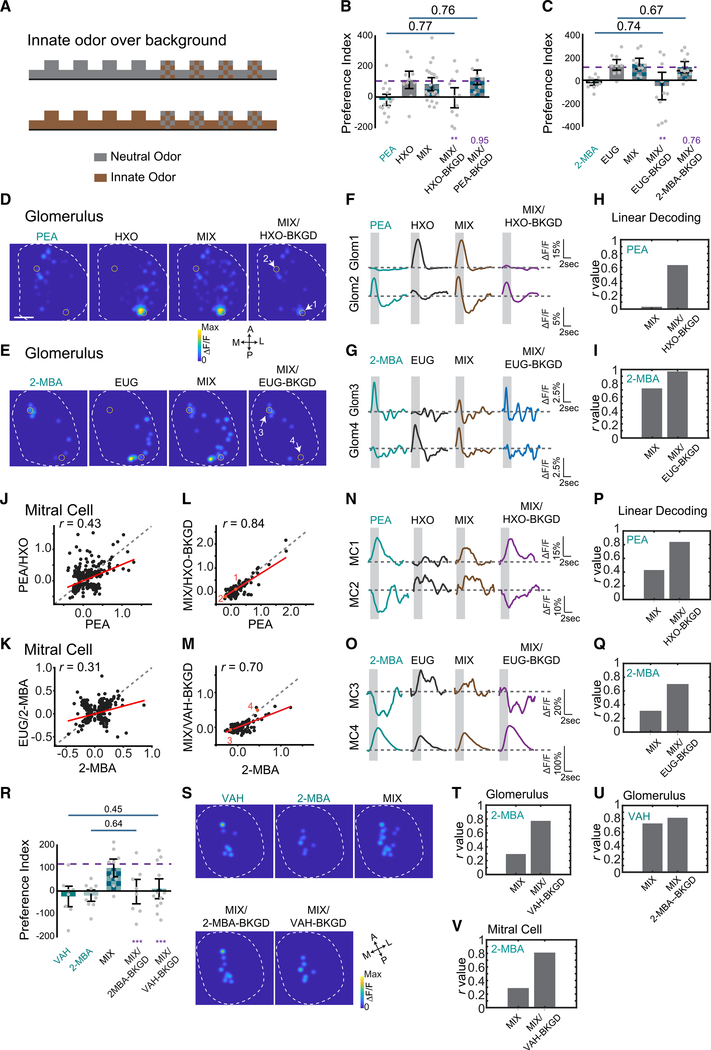
It is counterintuitive that mixing an aversive odor with another odor abolished the innate response. Animals react to predator or food odors in complex odiferous environments, suggesting that they can identify these odors in presence of different background odors. In natural environments, individual odors arrive at the nostril as plumes. A spatial-temporal displacement might allow an odor to be distinguished from other environmental odors and perceived as distinct. In our experiments, two odors were well mixed and presented contemporaneously. We, therefore, tested whether introducing a temporal displacement in odor presentation could allow the aversive odors to be detected as elemental in the mix. We presented one odor as background, followed by the mixture, and scored preference to the mixture (Figure 5A). In experiments using the neutral odor (HXO or EUG) as background and testing with the mixtures with the aversive odor (PEA with HXO, or 2-MBA with EUG), we found that the mixtures elicited the same level of aversion as the aversive odors alone (Figures 5B and 5C). Conversely, with aversive odors as background, the mixtures were perceived as neutral (Figures 5B and 5C). In addition, using either odor from aversive odor pair VAH and 2-MBA as the background, the mixtures also elicited the same level of aversion as the aversive odors alone (Figure 5R). Thus, background presentation of one component odor allowed innate aversive odors within the mixtures to be properly recognized. Temporally displaced presentation of odors was sufficient to allow individual odors to be distinguished.
Figure 5. Background presentation of odors allows segmentation.
(A) Illustration of experimental paradigm. One of the odors (neutral or aversive) is delivered continuously as background. The mixture is delivered during the marked epochs.
(B and C) Bar plots of preference indices measured for individual odors and their mixtures, and the mixture when one of the component odors was presented as background for PEA and HXO (B), and for 2-MBA and EUG (C). Purple dashed line indicates the average level for neutral odors. One-way ANOVA with Tukey test was applied. Statistical significance or the p values are marked by purple labels.
(D and E) Glomerulus activation patterns elicited by PEA and HXO (D) or 2-MBA and EUG (E) mixture after exposed to HXO (D) or EUG (E) as background.
(F and G) Traces of glomerular (Glom) (indicated in D and E) responses to individual odors and the mixture in the absence and presence of background presentation for odor pairs PEA and HXO (F) and 2-MBA and EUG (G).
(H and I) Bar graphs show the Pearson’s correlation coefficients (r) between linearly decoded mixture response and the actual glomerular responses to PEA (H) or 2-MBA (I) with or without background presentation.
(J and K) Scatterplots of M/T cell responses to PEA and HXO mixture against that to PEA (J), and 2-MBA and EUG against 2-MBA (K) without background presentation. Red line indicates linear fit of the data. Pearson’s correlation coefficient (r) and diagonal line (gray dash) are indicated.
(L and M) Same as in (J) and (K) but with neutral odor presented as background.
(N and O) Traces of M/T cells (MCs) (indicated in L and M) responded to individual odors and the mixture with or without background presentation for odor pairs PEA and HXO (N) and 2-MBA and EUG (O), respectively.
(P and Q) Bar graphs show the Pearson’s correlations coefficient (r) between linearly decoded mixture response and the actual M/T cell responses to PEA (P) or 2-MBA (Q) with or without background presentation. Correlation coefficient (r) values are plotted.
(R) Bar plot of preference indices measured for VAH, 2-MBA, and the mixture when one of the component odors is presented as background.
(S) Glomeruli activation patterns elicited by VAH, 2-MBA, and the mixture after exposure to VAH or 2-MBA as background.
(T and U) Bar graphs show the Pearson’s correlation coefficients (r) between linearly decoded mixture response and the actual glomerular responses to 2-MBA (T) or VAH (U) with or without background presentation.
(V) Bar graphs show the Pearson’s correlation coefficients (r) between linearly decoded mixture response and the actual M/T cell responses to 2-MBA with or without VAH as background presentation.
See also Figures S6 and S3, and Table S1.
We next recorded glomerular responses to the mixture after presenting the neutral odor as background. We first recorded the responses to component odors and the mixtures separately to provide points for comparison. We then presented the neutral odor as background, followed by the mixture, and recorded the response to the mixture. After background presentation, the mixtures’ elicited responses were dissimilar to those by mixtures alone but similar to the innately aversive odors alone in terms of dynamics and amplitudes (Figures 5D–5G). Linear decoding indicated that the mixtures elicited responses were highly correlated with aversive odors (Figures 5H, 5I, S6C, and S6F). Conversely, presenting the innately aversive odors as background, the mixtures elicited glomerular responses that were similar to the neutral odors alone (Figures S6A, S6B, S6D, and S6E). Similar glomerular response can be found for the aversive VAH and 2-MBA (Figures 5S–5U) and the attractive -peanut butter and female urine (FU) (Figure S3J) odor pairs.
We also imaged M/T the cells to record their responses to the mixture after the neutral odor was presented as a background. Without background odor presentation, the M/T cell activity elicited by the mixture was poorly correlated with the pattern elicited by the aversive odor alone (Figures 5J and 5K). However, when the neutral odor was presented as background, the response to the mixture became highly correlated with the aversive odors (Figures 5L and 5M). For individual cells, the dynamics and amplitude of the responses elicited by the mixture were similar to those elicited by the aversive odors (Figures 5N and 5O). Linear decoding analysis indicated that mixtures presented over background were more likely linearly decoded as the aversive odors, as indicated by the high correlation coefficients, than the mixtures (Figures 5P, 5Q, and 5V). The patterns of activity following adaptation to the background were consistent with the behaviors.
DISCUSSION
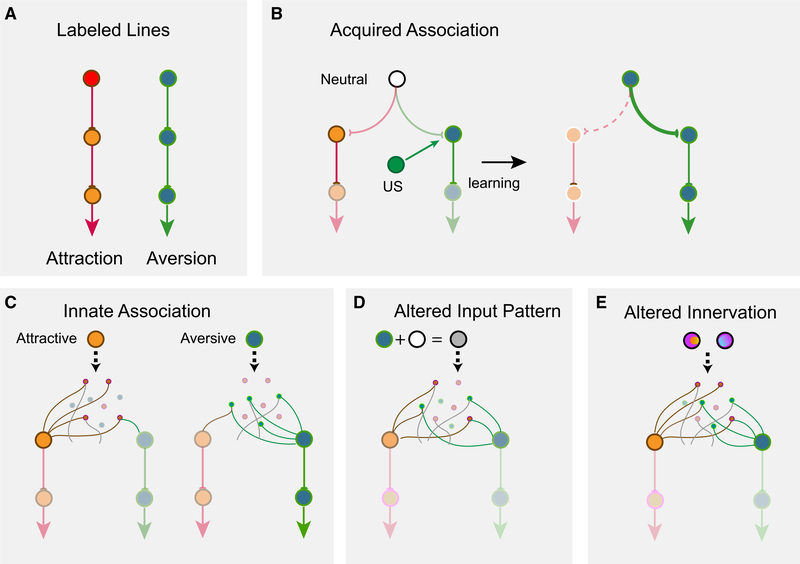
Innate responses to environmental stimuli are shaped by evolution to afford animals with survival advantages in the absence of learning. Information about innately recognized cues is thought to be processed by circuits different from those for learned behaviors. Although labeled lines provide a simple solution by linking sensory cells and behavioral centers (Figure 6A), our results suggest that it does not apply to mammalian olfaction. We find that mixtures abolish innate responses. The most striking discovery is that mixing two odors with the same valence, aversive and attractive alike, also abolishes innate responses. These changes cannot be simply explained by opposing actions of brain centers that drive different behaviors. Consistent with past studies,48,65,66 we find that glomerular responses to odor mixtures can be linearly decoded, indicating that there is no obvious peripheral masking or antagonist interactions of ligands with the receptors.32
Figure 6. Models of encoding innate valence of odors.
(A) The labeled-line model. Receptor activations are directly linked to behavioral outputs.
(B) Acquiring valence through associative learning. A neutral stimulus (white) has an unbiased connection to circuits leaning attraction or aversion. A teaching signal such as an unconditioned stimulus (US) is associated with the neutral stimulus to enhance its connection to aversion, leading to learned response.
(C) A model of innate odor preference. Attractive (orange) or aversive (olive) odors activate sets of cells that have preferential connection to the valence centers. Note that individual cells do not have exclusive connections to one valence center. Activation of a specific set of glomeruli (green) activates a set of cells, which encode the odor identity and are stereotypically connected to brain centers that assign valence.
(D and E) Alterations in the activation of these cells, either through simultaneous activation of additional receptors (D) or through altered connections (e.g., ectopic axon projections into multiple glomeruli) (E), changes the identity of the odor being encoded and leading to changes in valence assignment.
Our imaging study samples 10%–15% of the dorsal olfactory bulb, raising the question of whether the linear coding we have observed for the glomeruli also applies to other part of the bulb. Although it is possible that the dorsal glomeruli might respond differently, it is unlikely given the distributed nature of odor-evoked activities. In addition, the odors we have tested strongly activate the dorsal region. For example, 2-hexanone and EUG have been shown by numerous studies using immediate early gene (IEG) staining and 2-DG uptake to activate primarily the dorsal regions of the bulb.67–70 PEA activates neurons expressing the TAAR4 receptors, which project to a few dorsal glomeruli.24,30
Importantly, there is a contrast between glomerular and M/T cell responses, both recorded from the dorsal bulb. Whereas presynaptic inhibition is mostly intraglomerular, lateral inhibition is observed among the M/T cells.71 The lateral interactions not only normalize M/T cell response, but also redistribute the patterns. M/T cell responses in our recordings clearly show non-linear interactions between odors in the mixture. As such, mixtures elicit M/T cell responses that can no longer be linearly decoded but result in patterns of activities that are likely perceived as novel to the animals. Indeed, behavioral experiments indicate that the mixture creates configural perception.38,72 These results do not fit the classic labeled-line model where straightforward relays of information through the independent lines elicit stereotypical behaviors. Rather, they demonstrate that the activity triggered by innately recognized odors are not insulated from those of others. Interactions within the highly interconnected networks in the mammalian olfactory pathway can change the patterns elicited by the innately recognized odors depending on the presence of other odors. Thus, these odors are encoded similarly as all other odors, through a generalized population code resulting from interactions among the M/T cells. This result differs from a previous study indicating that component mixtures can be linearly decoded by using the M/T cell responses73 and might reflect the differences in the odor set being tested.
However, there is an apparent dilemma because perception of innately recognized odors is usually considered as elemental because they can be recognized in different contexts. How are these odors perceived as independent of the context if they are encoded by a general mechanism? We suggest the following possibilities. First, these compounds are more volatile. Notably, PEA, TMT, MTMT, and 2-MBA are volatile compounds identified from blends. The volatility makes it possible, and likely, for these molecules to diffuse farther and separate from other molecules in the mix (the chromatography effect). Second, animals have evolved more sensitive receptors for these molecules, as exemplified by the extraordinary sensitivity to PEA and MTMT. The sensitivity would allow the single compounds to be detected before others, thereby create the temporal displacement needed for them to be perceived separately from other odorants. Alternatively, but not mutually exclusively, these compounds might be represented at a higher concentration in the source. Our current study has not explored the whole range of odor concentrations. Future study might test these hypotheses. Lastly, the receiving animals might have evolved a mechanism of feature selection, i.e., with specific detection of these molecules while filtering out activity evoked by other compounds.21 These potential mechanisms might have created a situation where an individual component in a mixture is detected as elemental, rather than configural.
If these innately recognized odors are encoded by a general population code, how are their valence assigned? Although the classic labeled-line model (Figure 6A) provides a simple way of understanding intrinsic associated values, it is difficult to envision how it might operate with population activities that are subject to influence by other odors. Odor valence can be assigned through associative learning, presumably by associating the identity of an odor with an unconditioned stimulus (reward or punishment) to assign valence (Figure 6B). We reason that the encoding of innately recognized odor might be similar to those in learned association (Figure 6C). In this model, odor identities are encoded by the population responses starting at the M/T cells, and these identities are associated with cell populations that specify valence. The difference is that for the innate odors this association is intrinsically determined rather than acquired through associative learning. This model allows genetic programs to specify the neuronal connection in a way that generates stereotypic patterns of activated cells to encode odor identities, and these cells have preset connections with the valence circuit to drive approach or aversive behaviors.
This model does not require the M/T cells to establish a labeled-line connection with downstream attractive or aversive centers. The specification of innate connection would simply enable specific patterns of activation to preferentially activate the brain centers. Note that these pathways do not have to be segregated and can be intermingled. In other words, valence per se can also be encoded by distributive sets of cells. Indeed, distributed encoding of odor with different valence can be found in the same brain region.35
This model also does not require neurons that encode innately recognized odors to be specifically tuned, nor does it require the activity encoding odor identities to be insulated from the influence of other odors. When the pattern of activity associated with an innately recognized odor is altered by mixing with another odor, the ensemble activity no longer matches the predetermined pattern. It creates a novel odor identity and abolishes the valence associated with the component odor (Figure 6D). Given that M/T cells have been shown to form divergent patterns of projection in the cortical areas, activity-dependent modification of how olfactory sensory neurons project to the olfactory bulb during development would also alter these predisposed connections and change the valence associated with the odors (Figure 6E).74
This model can also explain the recognition of odors in the presence of background odors. When the odor presentation is temporally or spatially displaced from the background, individual odors in the mixture can be distinguished and their associated valence remain intact.75 This might be achieved through detection of fine temporal fluctuations of odors in turbulent air flow or through target detection by separation of foreground from background.76–78 Regardless of the mechanism, innate detection of ethologically relevant odors is a form of elemental perception that requires their presentation to be independent from the environment. This model, therefore, allows a general coding strategy for all odors alike. It permits a flexible assignment of valence to neutral odors or reassigns valence to those with intrinsic preference.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ron Yu (cry@stowers.org).
Materials Availability
There are no new reagents generated in this study.
Data and Code Availability
The original data are available in the Stowers Original Data Repository: https://www.stowers.org/research/publications/ LIBPB-1588_2021
Experimental Model and Subject Details
Animals
The OMP-IRES-tTA (Jackson laboratory), tetO-GCaMP2 (Jackson laboratory), and Cdhr1-Cre (MMRRC) mice were described previously.12,47,79 The C57BL/6J (Jackson laboratory) were used for control group. All the animals were maintained in Lab Animal Services Facility of Stowers Institute at 12:12 h reversed light cycle and provided with food and water ad libitum. All the behavior and functional imaging experiments were conducted during the dark light cycle under red or infrared light illumination. Experimental protocols involving mice were approved by the Institutional Animal Care and Use Committee at Stowers Institute and in compliance with the NIH Guide for Care and Use of Animals. All the experiments were conducted using equal numbers of male and female mice except experiments with mouse urine, in which mice were exposed to the urine of the opposite sex.
Method Details
Odor delivery with olfactometer
Odor delivery was controlled by an automated olfactometer with custom written software developed in the National Instrument Labview programming environment.40 Odorant used are listed Table S1. All single compound chemicals were freshly prepared in mineral oil or water at desired concentration (see below). Mouse urine and natural flavored mixtures were used at original concentration. Odorants were then further diluted in carrier air with a maintained total flow rate (400 mL/min for calcium imaging and 100 mL/min for behavior experiments).
Innate odor preference test
Experiments are the same as previously described.40 2- to 4-month-old mice were used for cross habituation experiments. Each experimental group contained 10–14 animals. Unless otherwise stated, all animals were naive to the testing odors and exposed to the same odor once. Each animal was tested with a total of two separate experiments with at least one week between tests. After being habituated to the testing environment for half an h, the animals were put into a 20 cm x 20 cm chamber for behavioral experiments. Odors from the olfactometer were delivered through a nose cone on one of the side walls, which was 5 cm above the base plate. A vacuum tube connected on the opposite wall of the nose cone provided an air flow to remove residual odors after odor delivery. Pure odorants were diluted into mineral oil at 1:103 (v/v). The odors in the mixture were mixed at either the same concentration as individually (i.e., 100% A + 100% B), 0.5:1 ratio (50% A + 100% B), or at 0.25:1 ratio (25% A + 100% B). To avoid the influence of flow on animal’s investigative behavior, a low flow rate is used for behavior tests. 10 mL/min air flow carried the saturated odor out from the odor vial and was further diluted into a 90 mL/min carrier air to make the final dilution to 1:104 (v/v). Delivery time, concentration, and sequence of odor delivery were controlled by the olfactometer software. Investigation of odor source was registered by infrared beam breaking events and recorded by the same software that controlled the olfactometer.
The sequence of trial sessions is depicted in Figure 1. Odor was delivered for 5 min in each trial. After four trials of air presentations, a testing odor was presented 4 times. In a typical test, mice habituating to the test chambers over the multiple sessions of background air led to decreased TAir. The presentation of an odor elicited an increased TOdor. Repeated presentation of the same odor led to habituation, which was reversed by the test odor if it was perceived as novel. If the odor is attractive to the animal, an increased TOdor1 is expected as the mixed result of novelty and attraction. If the odor is avoided by the animal, a smaller increase or even decrease TOdor1 is expected as the mixed result of novelty seeking (risk assessment) and avoidance, while the TOdor2 is expected as the aversion only because the novelty is habituated quickly while the avoidance persists longer. To use the same index for both behaviors, we define the following preference index:
Complex odors used in innate behavior experiments were as follows: Cheese, FU, MU, CU, PB, maple, and lemon. These odors were delivered without dilution. Single compound odors: PEA, IAMM, 2-MBA, HPH, HXO, EUG, TMA and VAH were diluted at 1:1000 in mineral oil. The concentrations of these odors used were selected based on our previous study imaging glomerular responses in the olfactory glomeruli, which show that at these concentrations odors generally elicited non-saturating responses.47 All the odors were then further diluted in the olfactometer in the air phase for another 10-fold.
Phospho-S6 mapping of odor-evoked activity
For mixture phospho-S6 staining, animals were single housed and habituated in home cages for seven days with a glass vial covered with a plastic cover, which was punched with seven holes for odor evaporation while preventing physical contact with chemicals. For habituation, a small piece of cotton nestlet soaked with 500 μl mineral oil was put inside the vial. Vials were changed every day at one h after light cycle. At day eight, a new glass vial with cotton nestlet soaked with 500 μl 2-MBA, PEA, IAMM, HXO or EUG at 1:103 dilution in mineral oil, or freshly collected male or female urine were added in the home cages. Binary mixtures were prepared with equal mix of two odors. One h after odor stimulation, mice were sacrificed and perfused with 4% PFA in PBS. The mouse brains were dissected and post-fixed with 4% PFA overnight at 4°C.
The phospho-S6 immunochemistry histology was performed based on the published protocol64 with some modifications. The entire brain was cut into 50 μm thick serial sections using a Leica vibratome (VT1000S). Rabbit anti phospho-S6 antibody (Cell Signaling) was diluted 1:1000 in 0.1% PBST (0.1% Triton X-100 in 1X 0.1M PBS) and used to incubate the slices at 4°C overnight. The 3-p peptide (25 nM biotin-QIAKRRRLpSpSLRApSTSKSESSQK, where pS is phosphoserine; synthesized by United Peptide) was also included to reduce background staining.64 Donkey anti-Rabbit Alexa 488 was (Thermo Fisher Scientific) diluted to 1:1000 in 1X PBST for secondary antibody staining overnight. DAPI (1mg/mL, Thermo Fisher Scientific) was used for nuclear staining.
Tiled images were acquired using Olympus VS120 Virtual Slide Microscope or PE Ultraview spinning disk confocal microscope (PerkinElmer), then images were stitched together using the Volocity software (PerkinElmer). Brain nuclei were identified based on the brain atlas (The Mouse Brain Stereotaxic Coordinates, third edition). The pS6 immuno-positive neurons were counted using ImageJ. For quantification, the two sides of the brain were treated independently and the following numbers of sections were used: anterior hypothalamic area, anterior part (AHA), 4 sections at −0.85~−1.05 mm from Bregma; medial amygdaloid nucleus (posterodorsal area, MePD, and posteroventral area, MePV) and ventromedial hypothalamic nucleus (ventrolateral area; VMHvl), 5 sections between −1.35~−1.60 mm from Bregma; hypothalamic nucleus (VMH) in the 2-MBA case, 4 sections between −1.80~−2.00 mm from Bregma; bed nucleus of the stria terminalis (BST), 3 sections between 0.10~0.25 mm from Bregma; posterolateral cortical amygdaloid area (PLCo), 5 sections at −1.35~−1.60 mm from Bregma; paraventricular thalamic nucleus (PVA), 4 sections at −0.25~−0.45 mm from Bregma; piriform cortex (Pir), 5 sections at −1.35~−1.60 mm from Bregma; basolateral amygdaloid nucleus, anterior part (BLA), 5 sections at −1.10~−1.35 mm from Bregma.
Awake head fixed calcium imaging
For calcium imaging of glomerulus, generation of the GCaMP2 mice were described previously.12 Line 12i and 5i mice that exhibited strong fluorescence were used for imaging experiments as described previously.47 One day prior to imaging, mice were anesthetized by intraperitoneal injection of ketamine/xylazine cocktail (100mg/kg, 10mg/kg body weight respectively) for surgery with thinned skull above the olfactory bulb. Animals were head-fixed and free standing on a customized treadmill. Before each experiment, animals were habituated on the experiment apparatus for one h.
For 2-photon mitral/tufted cell imaging, Cdhr1-Cre mice were anesthetized, and two small holes were made above each side of anterior and posterior olfactory bulb for virus injection. At each injection site, 250 nL pAAV.Syn.Flex.GCaMP6f.WPRE.SV40 virus (Addgene, 100833-AAV1, RRID:Addgene_100833) was injected into mitral/tufted cell layer at both 200 μm and 300 μm depths (Chen et al., 2013). After 3–4 weeks, animals were anesthetized to open a craniotomy window over the olfactory bulb and a 3 mm diameter glass coverslip was attached to the skull using dental cement. Animals were used for calcium imaging the next day. Animals were head-fixed and free standing on a customized treadmill. Before each experiment, animals were habituated on the experiment apparatus for one h.
Odorants were diluted in mineral oil (1:102) and delivered at 40 mL/min flow rate. Total flow rate was maintained at 400 mL/min in all experiments. For the mixtures, both odors were delivered at 40 mL/min and mixed inside the olfactometer before being delivered to the mouse nose at the ultimate rate of 400ml/min. The higher flow rate was chosen to allow rapid concentration changes at the nostril. Odors were delivered for 3 s followed by a 27 s interval.
For GCaMP2 glomeruli imaging, responses to odor stimuli were collected on an Olympus BX60WI microscope using 4X air lens (Olympus XLFLUOR4X/340, NA 0.28). The image was collected at 512×512 resolution with 2×2 bin with sampling rate at 8.3 Hz. For mitral/tufted cell 2-photon imaging, the image was collected by an Olympus 2-photon microscope (FLUOVIEW FVMPE-RS) with 940 nm excitation laser using 25X water lens (Olympus XLPLN25XWMP, NA 1.05). A resonate scanner with GaAsP detector was used for image collection at 512×512 resolution with 15 Hz sampling rate.
Custom-written scripts in ImageJ and MATLAB (Mathworks) were used for image processing as described previously[REMOVED HYPERLINK FIELD].47 Briefly, after ROIs were manually defined, the averaged response inside each ROI was extracted by ImageJ. Using customized MATLAB software, a baseline was defined for each ROI and ΔF/F was calculated. To display the response patterns, the peak amplitude of each glomerulus was mapped onto the spatial location of the glomerulus. The value was presented by applying a Gaussian blur with 50 μm standard deviation.
Quantification and statistical analysis
All the statistics are conducted in MATLAB or OriginPro. Data were expressed as means ± SEMs in figures and text. Group differences were analyzed using one-way ANOVA with Tukey test. Significance was defined as: * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, p value labeled when > 0.05. MATLAB build in function ‘linsolve’ was used for the linear decoding analysis. It solves the linear system A*X = B using LU factorization with partial pivoting when A is square and QR factorization with column pivoting otherwise. In terms of the mixture case here, the formula is essentially the equation x1*A1+x2*A2 = B. Here A1, A2 and B are the response vectors to the two component odors and the mixture, respectively. Each vector is the peak responses of N glomeruli (mitral/tufted cells). The linear system is expressed as matrix operation A*X = B, where A is a Nx2 matrix representing the component responses, B is a Nx1 vector, and X is 2×1 vector that is to be solved. A lower correlation coefficient indicates poor linear decoding. All the correlation coefficients were analyzed using linear Pearson Correlation in MATLAB.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: Cdhr1-Cre | MMRRC | 030952-UCD; RRID: MMRRC_030952-UCD |
| Mouse: tetO-GCaMP2 | Yu Lab The Jackson Laboratory |
JAX: 017755; RRID: IMSR_JAX:017755 |
| Mouse: OMP-IRES-tTA | The Jackson Laboratory | JAX: 017754; RRID: IMSR_JAX:017754 |
|
Odorants | ||
| 2-Hexanone | MilliporeSigma | CAS# 591–78–6; Abbr: HXO |
| Heptanal | MilliporeSigma | CAS# 591–78–6; Abbr: HPH |
| Eugenol | MilliporeSigma | CAS# 97–53–0; Abbr: EUG |
| 2-Methylbutyric acid | MilliporeSigma | CAS# 116–53–0; Abbr: 2-MBA |
| 2-Phenylethylamine | MilliporeSigma | CAS# 64–04–0; Abbr: PEA |
| Isoamylamine | MilliporeSigma | CAS# 107–85–7; Abbr: IAMM |
| Valeraldehyde | MilliporeSigma | CAS# 110–62–3; Abbr: VAH |
| Trimethylamine | MilliporeSigma | CAS# 75–50–3; Abbr:TMA |
| 2-Phenylethanol | MilliporeSigma | CAS# 60–12–8; Abbr: 2PE |
| 3-Amino-1,2,4-triazole | MilliporeSigma | CAS# 61–82–5; Abbr: AST |
| Female mouse urine | MilliporeSigma | Strain: c57bl/6; Abbr: FU |
| Male mouse urine | Fresh collected | Strain: c57bl/6; Abbr: MU |
| Coyote urine | Harmon scents | Cat# CCHCY4; Abbr: CU |
| Peanut butter | Jif Extra Crunchy | Abbr: PB |
| Shredded Mild Cheddar Cheese | Happy Farms by ALDI | Abbr: Cheese |
| Lemon flavor | Frontier natural products co-op | Cat# 23071 |
| Maple flavor | Frontier natural products co-op | Cat# 23081 |
|
Antibodies | ||
| Donkey anti-Rabbit Alexa 488 | Thermo Fisher Scientific | Cat# R37118; RRID: AB_2556546 |
| Rabbit anti-Phospho-S6 (Ser235/236) | Cell Signaling | Cat# 4854; RRID: AB_390782 |
| DAPI | Thermo Fisher Scientific | Cat# D1306; RRID: AB_2629482 |
|
Virus Strains | ||
| AAV1.Syn.Flex.GCaMP6f.WPRE.SV40 | Addgene | Cat# 100833-AAV1; RRID: Addgene 100833 |
|
Dental cement, Peptides and anesthetics | ||
| C&B Metabond Quick Adhesive Cement System | Parkell | Cat# UN1247 |
| 3-p peptide | United Peptide | biotin-QIAKRRRLpSpSLRApSTSKSESSQK |
| Isoflurane | Patterson Veterinary | Cat# 07–893–1389; CAS# 26675–46–7; |
| Xylazine | Patterson Veterinary | Cat# 07–893–2121 |
| Ketamine | Vedco | Cat# VINV-KETA-0VED |
|
Software and Algorithms | ||
| OriginPro | Origin Lab | https://www.originlab.com/Origin; RRID: SCR_014212 |
| MATLAB | Mathworks | https://www.mathworks.com; RRID: SCR_001622 |
| Custom MATLAB scripts | Yu Lab | N/A |
| ImageJ (Fiji) software | NIH | RRID: SCR_002285 |
Highlights.
Odor mixture abolishes innate behavioral response to component odors
Change of odor preference is correlated with altered brain activity
Glomerular, but not mitral/tufted cell responses can be linearly decoded
Temporally displaced presentation allows odor segmentation
ACKNOWLEDGMENTS
We thank A. Moran and members of the Lab Animal Services at the Stowers Institute for technical assistance. We are also thankful for valuable input from members of the Yu laboratory. The work is supported by funding from Stowers Institute for Medical Research and the National Institutes of Health (R01 DC008003, R01 DC014701, and R01 DC016696).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2021.01.094.
REFERENCES
- 1.Squire L, Berg D, Bloom FE, Du Lac S, Ghosh A, and Spitzer NC (2012). Fundamental neuroscience, Fourth Edition (Academic Press; ). [Google Scholar]
- 2.Erickson RP (1978). Common properties of sensory systems. In Sensory Integration (Springer; ), pp. 73–90. [Google Scholar]
- 3.Klapoetke NC, Nern A, Peek MY, Rogers EM, Breads P, Rubin GM, Reiser MB, and Card GM (2017). Ultra-selective looming detection from radial motion opponency. Nature 551, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig AD (2003). Pain mechanisms: labeled lines versus convergence in central processing. Annu. Rev. Neurosci 26, 1–30. [DOI] [PubMed] [Google Scholar]
- 5.Ma Q (2010). Labeled lines meet and talk: population coding of somatic sensations. J. Clin. Invest 120, 3773–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carleton A, Accolla R, and Simon SA (2010). Coding in the mammalian gustatory system. Trends Neurosci. 33, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Gabitto M, Peng Y, Ryba NJ, and Zuker CS (2011). A gustotopic map of taste qualities in the mammalian brain. Science 333, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, and Liberles SD (2015). Aversion and attraction through olfaction. Curr. Biol 25, R120–R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su CY, Menuz K, and Carlson JR (2009). Olfactory perception: receptors, cells, and circuits. Cell 139, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeholzer LF, Seppo M, Stern DL, and Ruta V (2018). Evolution of a central neural circuit underlies Drosophila mate preferences. Nature 559, 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munger SD, Leinders-Zufall T, and Zufall F (2009). Subsystem organization of the mammalian sense of smell. Annu. Rev. Physiol 71, 115–140. [DOI] [PubMed] [Google Scholar]
- 12.He J, Ma L, Kim S, Nakai J, and Yu CR (2008). Encoding gender and individual information in the mouse vomeronasal organ. Science 320, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, and Dulac C (2011). Molecular organization of vomeronasal chemoreception. Nature 478, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii KK, Osakada T, Mori H, Miyasaka N, Yoshihara Y, Miyamichi K, and Touhara K (2017). A Labeled-Line Neural Circuit for Pheromone-Mediated Sexual Behaviors in Mice. Neuron 95, 123–137.e8. [DOI] [PubMed] [Google Scholar]
- 15.Kimoto H, Haga S, Sato K, and Touhara K (2005). Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature 437, 898–901. [DOI] [PubMed] [Google Scholar]
- 16.Haga S, Hattori T, Sato T, Sato K, Matsuda S, Kobayakawa R, Sakano H, Yoshihara Y, Kikusui T, and Touhara K (2010). The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 466, 118–122. [DOI] [PubMed] [Google Scholar]
- 17.Mori K, Nagao H, and Yoshihara Y (1999). The olfactory bulb: coding and processing of odor molecule information. Science 286, 711–715. [DOI] [PubMed] [Google Scholar]
- 18.Araneda RC, Kini AD, and Firestein S (2000). The molecular receptive range of an odorant receptor. Nat. Neurosci 3, 1248–1255. [DOI] [PubMed] [Google Scholar]
- 19.Malnic B, Hirono J, Sato T, and Buck LB (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 20.Nara K, Saraiva LR, Ye X, and Buck LB (2011). A large-scale analysis of odor coding in the olfactory epithelium. J. Neurosci 31, 9179–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin DY, Zhang SZ, Block E, and Katz LC (2005). Encoding social signals in the mouse main olfactory bulb. Nature 434, 470–477. [DOI] [PubMed] [Google Scholar]
- 22.Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. (2007). Innate versus learned odour processing in the mouse olfactory bulb. Nature 450, 503–508. [DOI] [PubMed] [Google Scholar]
- 23.Fendt M, Endres T, Lowry CA, Apfelbach R, and McGregor IS (2005). TMT-induced autonomic and behavioral changes and the neural basis of its processing. Neurosci. Biobehav. Rev 29, 1145–1156. [DOI] [PubMed] [Google Scholar]
- 24.Dewan A, Pacifico R, Zhan R, Rinberg D, and Bozza T (2013). Non-redundant coding of aversive odours in the main olfactory pathway. Nature 497, 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberles SD, and Buck LB (2006). A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650. [DOI] [PubMed] [Google Scholar]
- 26.Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, and Liberles SD (2011). Detection and avoidance of a carnivore odor by prey. Proc. Natl. Acad. Sci. USA 108, 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaal B, Coureaud G, Langlois D, Giniès C, Sémon E, and Perrier G (2003). Chemical and behavioural characterization of the rabbit mammary pheromone. Nature 424, 68–72. [DOI] [PubMed] [Google Scholar]
- 28.Saito H, Nishizumi H, Suzuki S, Matsumoto H, Ieki N, Abe T, Kiyonari H, Morita M, Yokota H, Hirayama N, et al. (2017). Immobility responses are induced by photoactivation of single glomerular species responsive to fox odour TMT. Nat. Commun 8, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horio N, Murata K, Yoshikawa K, Yoshihara Y, and Touhara K (2019). Contribution of individual olfactory receptors to odor-induced attractive or aversive behavior in mice. Nat. Commun 10, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Pacifico R, Cawley D, Feinstein P, and Bozza T (2013). Ultrasensitive detection of amines by a trace amine-associated receptor. J. Neurosci 33, 3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Korzan WJ, Ferrero DM, Chang RB, Roy DS, Buchi M, Lemon JK, Kaur AW, Stowers L, Fendt M, and Liberles SD (2013). Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr. Biol 23, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saraiva LR, Kondoh K, Ye X, Yoon KH, Hernandez M, and Buck LB (2016). Combinatorial effects of odorants on mouse behavior. Proc. Natl. Acad. Sci. USA 113, E3300–E3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurent G (1999). A systems perspective on early olfactory coding. Science 286, 723–728. [DOI] [PubMed] [Google Scholar]
- 34.Davison IG, and Katz LC (2007). Sparse and selective odor coding by mitral/tufted neurons in the main olfactory bulb. J. Neurosci 27, 2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iurilli G, and Datta SR (2017). Population Coding in an Innately Relevant Olfactory Area. Neuron 93, 1180–1197.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igarishi Kei, et al. (2012). Parallel Mitral and Tufted Cell Pathways Route Distinct Odor Information to Different Targets in the Olfactory Cortex. The Journal of Neuroscience 32 (23), 7970–7985. 10.1523/JNEUROSCI.0154-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Gómez A, Bleymehl K, Stein B, Pyrski M, Birnbaumer L, Munger SD, Leinders-Zufall T, Zufall F, and Chamero P (2015). Innate Predator Odor Aversion Driven by Parallel Olfactory Subsystems that Converge in the Ventromedial Hypothalamus. Curr. Biol 25, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottfried JA (2010). Central mechanisms of odour object perception. Nat. Rev. Neurosci 11, 628–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheal ML, Klestzick J, and Domesick VB (1982). Attention and habituation: odor preferences, long-term memory, and multiple sensory cues of novel stimuli. J. Comp. Physiol. Psychol 96, 47–60. [DOI] [PubMed] [Google Scholar]
- 40.Qiu Q, Scott A, Scheerer H, Sapkota N, Lee DK, Ma L, and Yu CR (2014). Automated analyses of innate olfactory behaviors in rodents. PLoS ONE 9, e93468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han W, Tellez LA, Perkins MH, Perez IO, Qu T, Ferreira J, Ferreira TL, Quinn D, Liu Z-W, Gao X-B, et al. (2018). A Neural Circuit for Gut-Induced Reward. Cell 175, 665–678.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Root CM, Denny CA, Hen R, and Axel R (2014). The participation of cortical amygdala in innate, odour-driven behaviour. Nature 515, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Li W, Voleti V, Zou D-J, Hillman EMC, and Firestein S (2020). Widespread receptor-driven modulation in peripheral olfactory coding. Science 368, eaaz5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfister P, Smith BC, Evans BJ, Brann JH, Trimmer C, Sheikh M, Arroyave R, Reddy G, Jeong H-Y, Raps DA, et al. (2020). Odorant Receptor Inhibition Is Fundamental to Odor Encoding. Curr. Biol 30, 2574–2587.e6. [DOI] [PubMed] [Google Scholar]
- 45.Stettler DD, and Axel R (2009). Representations of odor in the piriform cortex. Neuron 63, 854–864. [DOI] [PubMed] [Google Scholar]
- 46.Poo C, and Isaacson JS (2009). Odor representations in olfactory cortex: “sparse” coding, global inhibition, and oscillations. Neuron 62, 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma L, Qiu Q, Gradwohl S, Scott A, Yu EQ, Alexander R, Wiegraebe W, and Yu CR (2012). Distributed representation of chemical features and tunotopic organization of glomeruli in the mouse olfactory bulb. Proc. Natl. Acad. Sci. USA 109, 5481–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGann JP, Pírez N, Gainey MA, Muratore C, Elias AS, and Wachowiak M (2005). Odorant representations are modulated by intrabut not interglomerular presynaptic inhibition of olfactory sensory neurons. Neuron 48, 1039–1053. [DOI] [PubMed] [Google Scholar]
- 49.Gupta P, Albeanu DF, and Bhalla US (2015). Olfactory bulb coding of odors, mixtures and sniffs is a linear sum of odor time profiles. Nat. Neurosci 18, 272–281. [DOI] [PubMed] [Google Scholar]
- 50.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennan PA, and Kendrick KM (2006). Mammalian social odours: attraction and individual recognition. Philos. Trans. R. Soc. Lond. B Biol. Sci 361, 2061–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon H, Enquist LW, and Dulac C (2005). Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123, 669–682. [DOI] [PubMed] [Google Scholar]
- 53.Kang N, Baum MJ, and Cherry JA (2009). A direct main olfactory bulb projection to the ‘vomeronasal’ amygdala in female mice selectively responds to volatile pheromones from males. Eur. J. Neurosci 29, 624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boehm U, Zou Z, and Buck LB (2005). Feedback loops link odor and pheromone signaling with reproduction. Cell 123, 683–695. [DOI] [PubMed] [Google Scholar]
- 55.Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, and Anderson DJ (2011). Functional identification of an aggression locus in the mouse hypothalamus. Nature 470, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Chen IZ, and Lin D (2015). Collateral pathways from the ventromedial hypothalamus mediate defensive behaviors. Neuron 85, 1344–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gross CT, and Canteras NS (2012). The many paths to fear. Nat. Rev. Neurosci 13, 651–658. [DOI] [PubMed] [Google Scholar]
- 58.Canteras NS, Ribeiro-Barbosa ER, and Comoli E (2001). Tracing from the dorsal premammillary nucleus prosencephalic systems involved in the organization of innate fear responses. Neurosci. Biobehav. Rev 25, 661–668. [DOI] [PubMed] [Google Scholar]
- 59.Duvarci S, Bauer EP, and Paré D (2009). The bed nucleus of the stria terminalis mediates inter-individual variations in anxiety and fear. J. Neurosci 29, 10357–10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim SY, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, et al. (2013). Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature 496, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, Unger EK, Wells JA, and Shah NM (2013). Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller SM, Marcotulli D, Shen A, and Zweifel LS (2019). Divergent medial amygdala projections regulate approach-avoidance conflict behavior. Nat. Neurosci 22, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jennings JH, Sparta DR, Stamatakis AM, Ung RL, Pleil KE, Kash TL, and Stuber GD (2013). Distinct extended amygdala circuits for divergent motivational states. Nature 496, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, and Friedman JM (2012). Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murphy GJ, Darcy DP, and Isaacson JS (2005). Intraglomerular inhibition: signaling mechanisms of an olfactory microcircuit. Nat. Neurosci 8, 354–364. [DOI] [PubMed] [Google Scholar]
- 66.Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, and Albeanu DF (2010). Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nat. Neurosci 13, 1404–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson BA, and Leon M (2000). Odorant molecular length: one aspect of the olfactory code. J. Comp. Neurol 426, 330–338. [DOI] [PubMed] [Google Scholar]
- 68.Johnson BA, Farahbod H, and Leon M (2005). Interactions between odorant functional group and hydrocarbon structure influence activity in glomerular response modules in the rat olfactory bulb. J. Comp. Neurol 483, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inaki K, Takahashi YK, Nagayama S, and Mori K (2002). Molecular-feature domains with posterodorsal-anteroventral polarity in the symmetrical sensory maps of the mouse olfactory bulb: mapping of odourant-induced Zif268 expression. Eur. J. Neurosci 15, 1563–1574. [DOI] [PubMed] [Google Scholar]
- 70.Johnson BA, and Leon M (2007). Chemotopic odorant coding in a mammalian olfactory system. J. Comp. Neurol 503, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Economo MN, Hansen KR, and Wachowiak M (2016). Control of Mitral/Tufted Cell Output by Selective Inhibition among Olfactory Bulb Glomeruli. Neuron 91, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frederick DE, Barlas L, Ievins A, and Kay LM (2009). A critical test of the overlap hypothesis for odor mixture perception. Behav. Neurosci 123, 430–437. [DOI] [PubMed] [Google Scholar]
- 73.Fletcher ML (2011). Analytical processing of binary mixture information by olfactory bulb glomeruli. PLoS ONE 6, e29360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu Q, Wu Y, Ma L, and Yu CR (2020). Acquisition of Innate Odor Preference Depends on Spontaneous and Experiential Activities During Critical Period. bioRxiv, 2020.2001.2028.923722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gschwend O, Abraham NM, Lagier S, Begnaud F, Rodriguez I, and Carleton A (2015). Neuronal pattern separation in the olfactory bulb improves odor discrimination learning. Nat. Neurosci 18, 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hopfield JJ (1991). Olfactory computation and object perception. Proc. Natl. Acad. Sci. USA 88, 6462–6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erskine A, Ackels T, Dasgupta D, Fukunaga I, and Schaefer AT (2019). Mammalian olfaction is a high temporal bandwidth sense. bioRxiv, 570689. [Google Scholar]
- 78.Rokni D, Hemmelder V, Kapoor V, and Murthy VN (2014). An olfactory cocktail party: figure-ground segregation of odorants in rodents. Nat. Neurosci 17, 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagai Y, Sano H, and Yokoi M (2005). Transgenic expression of Cre recombinase in mitral/tufted cells of the olfactory bulb. genesis 43, 12–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data are available in the Stowers Original Data Repository: https://www.stowers.org/research/publications/ LIBPB-1588_2021