Abstract
Numerous human diseases are caused by mutations in genomic sequences. Since amino acid changes affect protein function through mechanisms often predictable from protein structure, the integration of structural and sequence data enables us to estimate with greater accuracy whether and how a given mutation will lead to disease. Publicly available annotated databases enable hypothesis assessment and benchmarking of prediction tools. However, the results are often presented as summary statistics or black box predictors, without providing full descriptive information. We developed a new semi-manually curated human variant database presenting information on the protein contact-map, sequence-to-structure mapping, amino acid identity change, and stability prediction for the popular UniProt database. We found that the profiles of pathogenic and benign missense polymorphisms can be effectively deduced using decision trees and comparative analyses based on the presented dataset. The database is made publicly available through https://zhanglab.ccmb.med.umich.edu/ADDRESS.
Keywords: database, single-nucleotide polymorphism, disease variant, pathogenicity prediction
Graphical Abstract
INTRODUCTION
Mendelian human disease is often the result of a change in a single amino acid within a gene. With the ever-increasing wealth of genomic and structural data, it is possible to quantitatively assess, on a genome-wide scale, why some missense mutations result in disease, while others are benign. Early studies on few proteins revealed changes in protein stability as a causative factor, with buried residues being more common among pathogenic variants [1]. Later studies focused also on the important role of protein and ligand binding interactions, as well as active sites involved in enzymatic function [2–4]. The overall conclusion is that there are multiple routes by which a protein’s function can be deterred within the cellular environment, in which it must maintain a sizable folded fraction [5, 6] and be able to carry out its function, including binding with proteins and other molecules in its interaction network.
Pathogenicity prediction tools often utilize protein evolutionary information, identifying homologous sequences and determining whether mutated residues are well or poorly conserved [7–9]. Such predictors may also utilize information on changes in amino acid sequence identity [10–14]; for instance, a change from a hydrophobic to a charged residue may be expected to have a high likelihood of disease association. In recent programs, the structural neighborhood of the mutation may also be taken into account. Examples include DAMpred [15], which includes many structural contact-related features and builds models of unknown structures, RAPSODY [16, 17], which takes into account structural dynamics, and Missense3D [18], which considers a variety of potentially disruptive structural changes to evaluate whether a given variant is pathogenic. Likewise, predictions of mutation-induced free-energy change (ΔΔG) and changes in binding affinity to functional partners could be expected to carry useful information relevant to potential loss of protein function, as explored in previous database annotations [19]. Efforts to integrate structural information with mutation data in database format include COSMIC [20], which, however, only contains data on somatic mutations in cancer. Other related resources include Swissvar and MSV3d [21], although many servers lack full annotation details and comparison with computational predictions [22, 23] or are no longer maintained.
The UniProt Humsavar database (https://www.uniprot.org/docs/humsavar) contains information on pathogenicity of more than 70,000 human variants and is often used to benchmark tools developed to predict pathogenicity of missense single nucleotide polymorphisms. The majority of variants are annotated either as neutral “Polymorphism” or disease-associated variants (39.4% and 50.0%, respectively), with a small number of unclassified entries (10.6%). While the Humsavar file includes UniProt accession, it does not link directly to experimental structures or provide overall insight into the relationship between structure and disease. Integration with structural information and stability predictions could be useful in assessing which factors are most important to maintaining or disrupting protein functions, as well as facilitating new prediction tools to aid researchers in developing results relevant to clinical practice.
In this study, we present a new semi-manually curated database, ADDRESS (Annotated Database of Disease-RElated Structures and Sequences), mapping mutation sites annotated in UniProt Humsavar to residue numbers in example structural files from the Protein Data Bank (PDB). Next to each structure, we provide the number of contacts in which the mutated residue participates and the predicted ΔΔG of the mutation, according to the EvoEF empirical force field [24, 25]. The database may be searched by the type of variant, by the starting/ending amino acid type, the number of contacts, or predicted ΔΔG. Our database presents an exploratory interface to pathogenicity data, as well as a useful starting point for advanced statistical and machine-learning based method developments.
RESULTS
Descriptive analysis of residue identities
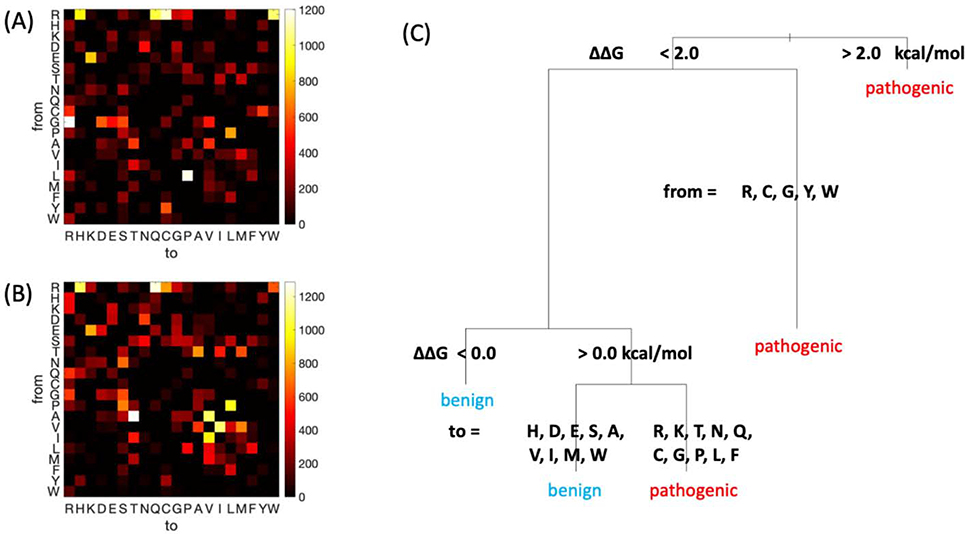
By generating heatmaps displaying the frequency of each possible amino acid change, it is clear that pathogenic and benign mutations have distinct profiles (Figure 1A and 1B, respectively). The arginine to glutamine or histidine mutations are common in both pathogenic and benign cases. Also common in disease are glycine to arginine, leucine to proline, arginine to cysteine, and arginine to tryptophan, representing dramatic changes in terms of physicochemical properties and/or torsional preference. Common benign mutations appear overall more conservative: alanine to threonine, isoleucine to valine, valine to isoleucine, alanine to valine, and proline to leucine. Other mutations, such as cysteine to tyrosine are found much more often in pathogenic cases (or benign, as for threonine to alanine), although they are overall rare. Since the dataset represents a broad range of residue changes, such data can be informative towards predicting pathogenicity. While the heatmaps are largely symmetric, we find statistical significance for several asymmetries, including glycine to arginine being more frequent, in the pathogenic case, than arginine to glycine. A full statistical treatment of amino acid changes upon mutation is presented in the Supplemental Information (Text S1 and Tables S1–S3).
Figure 1.
Summary of sequence and pathogenicity information from the ADDRESS database. A-B) Amino acid identity change represented in a two-dimensional histogram, for pathogenic (A) and benign (B) variants. Brighter colors indicate higher frequencies. 2) A simple decision tree predicting whether a variant is pathogenic or benign. Stability change ΔΔG is the EvoEF predicted value.
Next, we develop a rule-based approach using data on pathogenicity. The decision tree in Figure 1C shows a data-informed process based on the ADDRESS dataset, generated by rpart in R, using only four features: the type of amino acid before and after mutation, the stability change predicted by EvoEF, and the number of contacts formed by the mutated residue. Here, we chose the decision tree over more powerful supervised machine learning algorithms mainly considering the better interpretability of the decision tree results. Nonetheless, the simple decision tree method still achieves an appreciable Mathews Correlation Coefficient (MCC) of 0.34 in the binary classification of pathogenic versus benign mutations.
The first branching of the decision tree is based on the predicted ΔΔG of the mutation, such that changes that lead to a sufficiently large decrease in stability (large positive values) predict pathogenic consequences. However, the value of 2 kcal/mol is still relatively small in comparison to common folding stability values, such that if considering equilibrium thermodynamics alone, in most cases the majority of the protein would still be in the folded state. A similar observation was made previously, where selection for kinetics in a crowded environment was proposed to be the cause of such a low ΔΔG value, for a small number of mutations in a single protein [26]. In the decision tree, when the stability change is small or negative, the mutation is still predicted to be pathogenic in the case of mutation to a subset of relatively “extreme” residues, including mutation from cysteine or tryptophan. For other residues, if the protein is stabilized upon mutation (ΔΔG <0), the mutation is predicted to be benign. Otherwise, for intermediate ΔΔG values, the pathogenic versus benign distinction depends on the type of residue that is mutated to. Such a decision tree supports intuition regarding which changes are likely to be pathogenic, while providing additional insights, such as support for the selection for kinetic stability hypothesis on the scale of thousands of mutations.
Residue contact information and predicted stability change
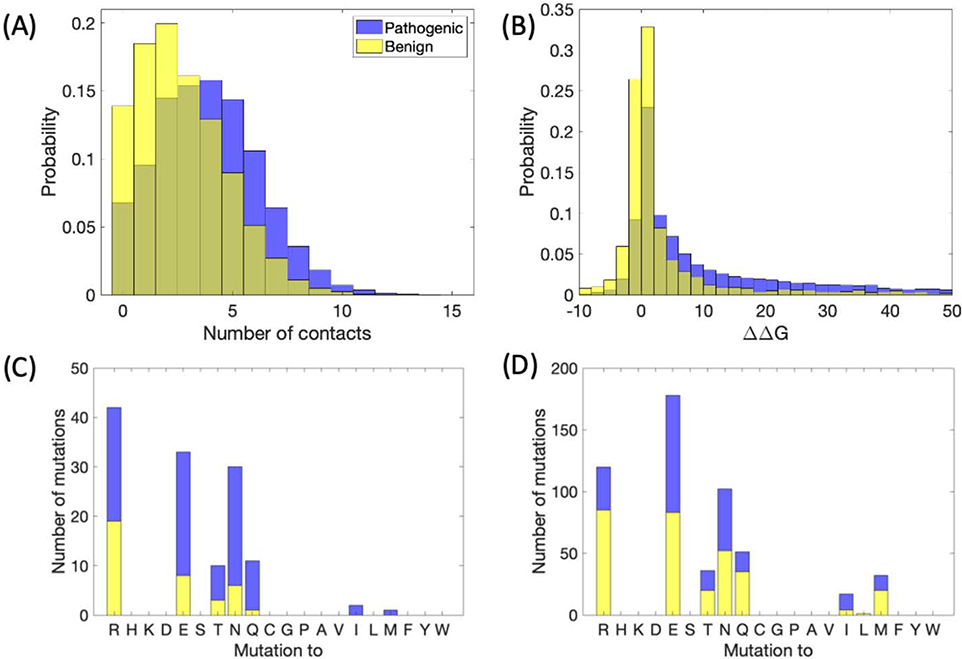
Pathogenic and benign variants show distinct distributions of the number of contacts surrounding the mutated residue (Figure 2A), in line with the observation that local packing density correlates strongly with surface exposure [27] and that solvent exposure in turn is strongly correlated with the site specific rate of mutation [28]. For benign variants, results are more skewed towards smaller numbers, indicating that residues tend to be more buried in the case of disease-causing variants, with a p-value = 3.9×10−295 in Student’s t-test. This is consistent with previous results on a smaller set of proteins showing that pathogenic mutations tend to be located in the protein interior more often than benign ones [4] and with depth and contact information from another former investigation [15]. Overall, the number of contacts for the specified cutoff values peaks around four contacts in the case of pathogenic mutations and two contacts in the case of benign mutations. The mean is 2.7 contacts for benign variants and 3.9 for pathogenic; the median values are 2 and 4, respectively. Likewise, the change in protein stability, ΔΔG, predicted by EvoEF was substantially higher (more positive, with a p-value=2.9×10−253 in Student’s t-test) for pathogenic mutations (mean value 16.13 kcal/mol) than benign ones (mean value 4.92 kcal/mol) (Figure 2B). In comparison, the ΔΔG predicted by another widely used predictor, FoldX [29], shows a somewhat less significant p-value for the difference in pathogenic and benign distributions of 4.7×10−186 (Figure S1), which is part of the reason that our decision tree analysis (Figure 1C) uses EvoEF rather than FoldX. Nevertheless, both EvoEF and FoldX ΔΔG data are listed in the ADDRESS database for comparison.
Figure 2.
Local structural information from missense SNPs. A) Histogram of number of contacts (at least six atom-atom contacts less than five angstroms), for benign and pathogenic variants, with the mutated residue within a known crystal structure. B) ΔΔG in kcal/mol, predicted by EvoEF. C-D) Stacked bar plots indicating the number of benign and pathogenic variants mutated from lysine to the specified residue for C) number of contacts greater than or equal to five, D) number of contacts less than five.
As a case study, we examine in Figures 2C and 2D the frequency of benign and pathogenic variants occurring on Lysine which is an amino acid with both hydrophobic properties and positive charge. Here, we consider differences in the frequency of mutations to different types of residues and their benign or pathogenic state, when the number of contacts in the crystal structure of the original protein is small vs. when it is large. As shown in the plots, mutation to a charged or polar residue is more often benign rather than pathogenic when the number of contacts with the mutated residue is small.
Online database setting
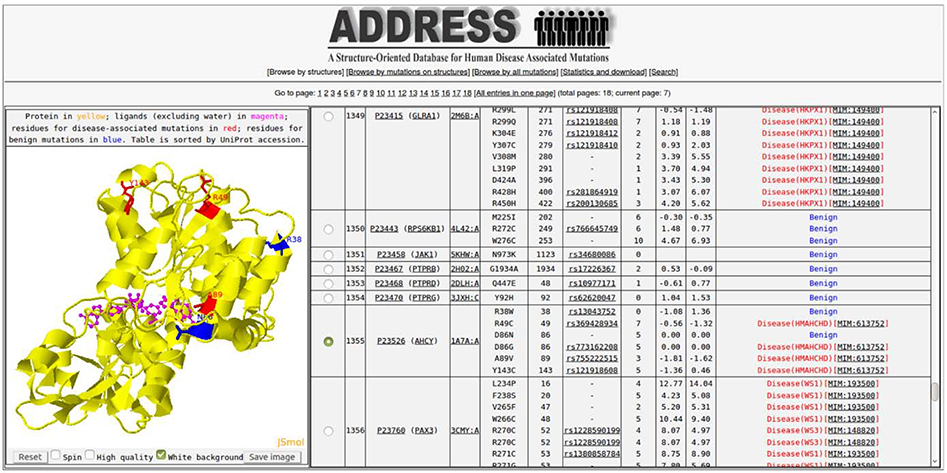
The webserver of ADDRESS mainly consists of three parts: a top banner for view switching, a JSmol [30] applet to display the PDB structure, and a main table that lists the database entries (Figure 3). First, the view switching banner allows the user to select one of the five views: “Browse by structure” lists one PDB chain per row in the main table; “Browse by mutations on structure” lists one mutation mapped to a PDB chain row in the table; “Browse all mutations” displays all mutations, regardless of whether each can be mapped to a structure. Since it is difficult to load all data in a web-browser due to cache size limit, all online tables are split into multiple pages to facilitate browser rendering. If a user would like to view all data contained within ADDRESS, an Excel spreadsheet can be downloaded at the bottom of the “Statistics and download” page, which also includes the data analysis figures (Figure 1 and 2) for general statistics of ADDRESS. Finally, the “Search” page performs database search using PDB IDs, gene names, UniProt accessions, diseases, amino acid types, or range of contact numbers and free-energy change. The database contains information on both the residue number in UniProt, in the column “Mutation on UniProt sequence,” and the residue number mapped to in the PDB structure, under “Residue index in PDB,” which was obtained through sequence alignment as described in the Methods section.
Figure 3.
A screenshot of the ADDRESS online database interface. The interface contains three parts. The view switching banner is the top under the database logo, switched to “Browse by structure” page 1 in this screenshot. A JSmol structure applet is at lower left, displaying PDB structure 1a7a Chain A for human Adenosylhomocysteinase protein AHCY (yellow cartoon) in complex with the NAD and ADC ligands (magenta sticks) and mutation sites (red and blue lines of pathogenic and benign mutations, respectively). The main table at the lower right displays all entries sorted by PDB IDs, where 1a7a Chain A for AHCY is currently selected. At the upper right corner of the database interface is the “Search” button, with which users can search database entries by protein names, PDB IDs, mutations, diseases, and structural features (e.g., contact numbers and free energy changes).
The JSmol applet displays the 3D structure of the PDB chains together with any non-water ligands and the mutation sites, as selected by the first column of the main table. The main table also includes columns for PDB ID (linked to the RCSB PDB website), UniProt accession (linked to the UniProt database) and Gene name (linked to the neXtProt [31]), mutation amino acid types and residue index on the UniProt sequence and on the PDB structure, a link to dbSNP database, number of contacts per residue, EvoEF estimated free energy change upon mutation, and disease association of the mutation. For disease-associated mutations, the disease symbol and disease ID in the OMIM database [32, 33] are displayed in the last column of the table.
CONCLUSION
A better understanding of which mutations lead to disease can be useful in prioritizing experimental study of variants and understanding protein evolution from a theoretical perspective. We have introduced a new database of human variants with each entry enriched with various types of structural bioinformatics information including residue mutation identity, numbers of contacts, and predicted change in protein stability. As an illustration of an application, we have approached data analyses from a descriptive and exploratory perspective, gaining insight into why mutations may be pathogenic or benign. Our results provide data relevant to future prediction tools and permit a variety of comparisons on this sizable dataset.
We anticipate our database to be more useful in generating aggregate statistics and comparisons, rather than predicting results for individual proteins, for which further modeling and predictors with more features may be necessary. With the rapid accumulation and availability of various sequence and structure data, ADDRESS is in active development. Currently, we are working on extending our database to combine literature search and other primary human variance datasets such as the Clinvar [34]. We also plan to provide additional features that are highly discriminatory, such as protein-protein and protein-ligand interaction information, as well as 3D structure models from the start-of-the-art modeling pipelines [35, 36]. We plan to later add information from the ProTherm experimental database on protein stability, for mutants that map to the Humsavar database. However, upon preliminary consideration, we believe that such a task will require substantial additional effort beyond the scope of the initial database (including some manual effort due to labeling errors and inconsistencies in the ProTherm database) and will also cover only a small fraction (about 80) of the proteins in the ADDRESS database. We believe that a high-quality and up-to-date human variance database featured with enriched structural bioinformatics data will have critical importance in facilitating investigations relevant to early diagnosis and treatment of human genetic diseases.
METHODS
Data collection and alignment
The UniProt Humsavar database (version 2020_04 as of this manuscript) was downloaded from Humsavar, where labeled benign “Polymorphism” and pathogenic “Disease” variants were considered. Protein sequences were downloaded from UniProt. For each mutation, the experimental structure containing this mutation in its residue range which had the greatest overall sequence coverage was chosen as a representative structure. The UniProt sequence was aligned with the string of residues contained in the protein structure, using NW-align [37], with adjusted gap penalties to determine the position of the mutated residue in the experimental structure. Cases where the mutated residue aligned with a gap before or after the sequence were discarded. This procedure results in 14,148 pathogenic variants and 7,648 benign variants that are mapped to 3,589 PDB chains.
Feature extraction
Initial and mutated residue identities were extracted from the Humsavar data file. To remove redundant coordinates from a PDB structure, only atoms with the alternative location indicator ‘A’ or ‘ ’ (space) were kept. For the entries with multi-model NMR structures, only the first model was selected. Residues were considered in contact with the residue to be mutated if there were six or more pairwise atomic contacts within five Å.
Folding stability change calculation
The mutation-induced folding stability change, ΔΔG, is estimated by EvoEF [24], which is an empirical force field and has been shown to have a strong correlation with the experimental ΔΔG measurements [25]. Preceding EvoEF calculation, the amino acid types in the PDB are first standardized by removing non-standard amino acids and by mapping selenomethionine (MSE) to methionine (MET), as MSE is commonly engineered to replace MET to facilitate X-ray structure determination. In the case of inconsistent amino acid type between UniProt sequence and PDB structure, the EvoEF BuildMutant subroutine is used to make convert amino acid on a PDB structure to that of the UniProt sequence. EvoEF RepairStructure function is applied to fill in missing atoms such as hydrogens, and EvoEF BuildMutant is performed used to build mutant structure. The folding free energies of wild-type and mutant structures (ΔGwt and ΔGmut, respectively) are estimated by EvoEF ComputeStability function. The stability change is therefore ΔΔG = ΔGmut ―ΔGwt, in the units of kcal/mol.
As FoldX and EvoEF use almost identical function names, ADDRESS also predicts ΔΔG by FoldX following essentially the same protocol as above. The folding free energies of wild-type and mutant structures (ΔGwt and ΔGmut, respectively) are estimated by FoldX Stability function.
Supplementary Material
Highlights.
Many missense-associated diseases stem from protein structure and stability changes
Development of a new database to associate pathogenic mutations with structures
Quantitative association of human variance with folding free energy changes
Pathogenic mutations are found to lead to lower stabilities and have more contacts
ADDRESS is useful for investigating detailed mechanisms of mutation pathogenicity
ACKNOWLEDGEMENTS
We thank Dr. Jeffrey Brender for helpful discussions. This work is supported in part by the National Institute of General Medical Sciences (GM136422, S10OD026825), the National Institute of Allergy and Infectious Diseases (AI134678), and the National Science Foundation (IIS1901191, DBI2030790, MTM2025426).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–70. [DOI] [PubMed] [Google Scholar]
- [2].Steward RE, MacArthur MW, Laskowski RA, Thornton JM. Molecular basis of inherited diseases: a structural perspective. Trends Genet. 2003;19:505–13. [DOI] [PubMed] [Google Scholar]
- [3].Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. [DOI] [PubMed] [Google Scholar]
- [4].Gao M, Zhou H, Skolnick J. Insights into Disease-Associated Mutations in the Human Proteome through Protein Structural Analysis. Structure. 2015;23:1362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Serohijos AW, Shakhnovich EI. Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics. Curr Opin Struct Biol. 2014;26:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goldstein RA. The evolution and evolutionary consequences of marginal thermostability in proteins. Proteins. 2011;79:1396–407. [DOI] [PubMed] [Google Scholar]
- [7].Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. [DOI] [PubMed] [Google Scholar]
- [8].Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2. [DOI] [PubMed] [Google Scholar]
- [9].Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16 Suppl 8:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–44. [DOI] [PubMed] [Google Scholar]
- [14].Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quan L, Wu H, Lyu Q, Zhang Y. DAMpred: Recognizing Disease-Associated nsSNPs through Bayes-Guided Neural-Network Model Built on Low-Resolution Structure Prediction of Proteins and Protein-Protein Interactions. J Mol Biol. 2019;431:2449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ponzoni L, Bahar I. Structural dynamics is a determinant of the functional significance of missense variants. Proc Natl Acad Sci U S A. 2018;115:4164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ponzoni L, Penaherrera DA, Oltvai ZN, Bahar I. Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics. 2020;36:3084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ittisoponpisan S, Islam SA, Khanna T, Alhuzimi E, David A, Sternberg MJE. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J Mol Biol. 2019;431:2197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–20. [DOI] [PubMed] [Google Scholar]
- [20].Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Luu TD, Rusu AM, Walter V, Ripp R, Moulinier L, Muller J, et al. MSV3d: database of human MisSense Variants mapped to 3D protein structure. Database (Oxford). 2012;2012:bas018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stephenson JD, Laskowski RA, Nightingale A, Hurles ME, Thornton JM. VarMap: a web tool for mapping genomic coordinates to protein sequence and structure and retrieving protein structural annotations. Bioinformatics. 2019;35:4854–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Radusky L, Modenutti C, Delgado J, Bustamante JP, Vishnopolska S, Kiel C, et al. VarQ: A Tool for the Structural and Functional Analysis of Human Protein Variants. Front Genet. 2018;9:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pearce R, Huang X, Setiawan D, Zhang Y. EvoDesign: Designing Protein-Protein Binding Interactions Using Evolutionary Interface Profiles in Conjunction with an Optimized Physical Energy Function. J Mol Biol. 2019;431:2467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang X, Pearce R, Zhang Y. EvoEF2: accurate and fast energy function for computational protein design. Bioinformatics. 2020;36:1135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Godoy-Ruiz R, Ariza F, Rodriguez-Larrea D, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM. Natural selection for kinetic stability is a likely origin of correlations between mutational effects on protein energetics and frequencies of amino acid occurrences in sequence alignments. Journal of Molecular Biology. 2006;362:966–78. [DOI] [PubMed] [Google Scholar]
- [27].Yeh SW, Liu JW, Yu SH, Shih CH, Hwang JK, Echave J. Site-specific structural constraints on protein sequence evolutionary divergence: local packing density versus solvent exposure. Mol Biol Evol. 2014;31:135–9. [DOI] [PubMed] [Google Scholar]
- [28].Franzosa EA, Xia Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol Biol Evol. 2009;26:2387–95. [DOI] [PubMed] [Google Scholar]
- [29].Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. Journal of Molecular Biology. 2002;320:369–87. [DOI] [PubMed] [Google Scholar]
- [30].Hanson RM, Prilusky J, Renjian Z, Nakane T, Sussman JL. JSmol and the next-generation web-based representation of 3D molecular structure as applied to Proteopedia. Israel Journal of Chemistry 2013;53:207–16. [Google Scholar]
- [31].Zahn-Zabal M, Michel PA, Gateau A, Nikitin F, Schaeffer M, Audot E, et al. The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res. 2020;48:D328–D34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McKusick VA. Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. 12th Edition ed. Baltimore: Johns Hopkins University Press; 1998. [Google Scholar]
- [34].Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zheng W, Li Y, Zhang C, Pearce R, Mortuza SM, Zhang Y. Deep-learning contact-map guided protein structure prediction in CASP13. Proteins. 2019;87:1149–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yan R, Xu D, Yang J, Walker S, Zhang Y. A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci Rep. 2013;3:2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.