Abstract
Stabilities and structure(s) of proteins are directly coupled to their local environment or Gibbs free energy landscape as defined by solvent, temperature, pressure, and concentration. Solution pH, ionic strength, cofactors, chemical chaperones, and osmolytes perturb the chemical potential and induce further changes in structure, stability, and function. At present, no single analytical technique can monitor these effects in a single measurement. Mass spectrometry and ion mobility-mass spectrometry play increasingly essential roles in studies of proteins, protein complexes, and even membrane protein complexes; however, with few exceptions, the effects of the solution temperature on the stability and structure(s) of analytes have not been thoroughly investigated. Here, we describe a new variable-temperature electrospray ionization (vT-ESI) source that utilizes a thermoelectric chip to cool and heat the solution contained within the static ESI emitter. This design allows for solution temperatures to be varied from ~5 to 98 °C with short equilibration times (<2 min) between precisely controlled temperature changes. The performance of the apparatus for vT-ESI-mass spectrometry and vT-ESI-ion mobility-mass spectrometry studies of cold- and heat-folding reactions is demonstrated using ubiquitin and frataxin. Instrument performance for studies on temperature-dependent ligand binding is shown using the chaperonin GroEL.
Graphical Abstract

Since Hopkin’s 1930 report on the temperature-dependent denaturation of ovalbumin,1 the influence of temperature on protein stability, structure, and self-assembly has captivated scientists’ interest. Privalov2 and Ben-Naim3 emphasize in their reviews the importance of the local environment (Gibbs free energy landscape (GEL)) on the stabilities, structures, and folding/denaturation of proteins. While Anfinsen’s hypothesis considers the native state as being a thermodynamic minimum,4 the native state is, in fact, unique to the local environment as defined by temperature, pressure, and concentration of the protein, viz., GEL. While solvent effects are known as determinants for charge state distributions observed by ESI MS, very little is known about how they affect the structure/conformations of gas-phase ions.5-8 Despite the importance of temperature on peptide and protein structure, careful studies aimed at understanding these effects have only been implemented recently.9-19 Here, we describe a variable-temperature electrospray ionization (vT-ESI) source that can be used to study both cold- and heat-induced solution-phase reactions of peptides and proteins at temperatures of ~5–98 °C. vT-ESI-MS is not new; Cong et al. developed a vT-ESI-MS source to study lipid binding of membrane protein complexes,20-22 El-Baba et al. developed a vT-ESI to study heat-induced protein denaturation (ambient to >90 °C),23 Köhler et al. developed a vT-ESI source that operates between 15 and 85 °C,24 and Marchand et al. developed a dual-block vT-ESI capable of temperature-jump mass spectrometry.25 These vT-ESI-MS devices afford an exquisite approach for studies of solution-phase thermochemistry. For example, using this approach, it is possible to directly measure temperature dependence of the formation of specific products by monitoring the masses of the products.23,26-34 Using vT-ESI-IM-MS, which reports the masses and sizes/shapes (rotationally averaged collision cross sections) of products, folding/unfolding and self-assembly reactions can be investigated.35,36 All of the above-mentioned studies illustrate the unique capabilities of vT-ESI MS and vT-ESI IM-MS to directly measure individual peptide/protein conformers that comprise a conformationally heterogeneous population,31 while simultaneously measuring the thermodynamics, viz., ΔG, ΔH, and TΔS, of the chemical reactions.
While different strategies for implementing vT-ESI have been reported,20-25 the present work aims to develop a vT-ESI device compatible with static ESI and capabilities for investigating both cold- and heat-induced folding and self-assembly reactions using mass spectrometry and ion mobility-mass spectrometry. Most importantly, the device needs to be compatible with static nanospray ESI, allowing for small volumes (<10 μL) of protein solutions and measurement of temperature-dependent product profiles between 5 and 98 °C.
EXPERIMENTAL SECTION
Design of the vT-ESI Source.
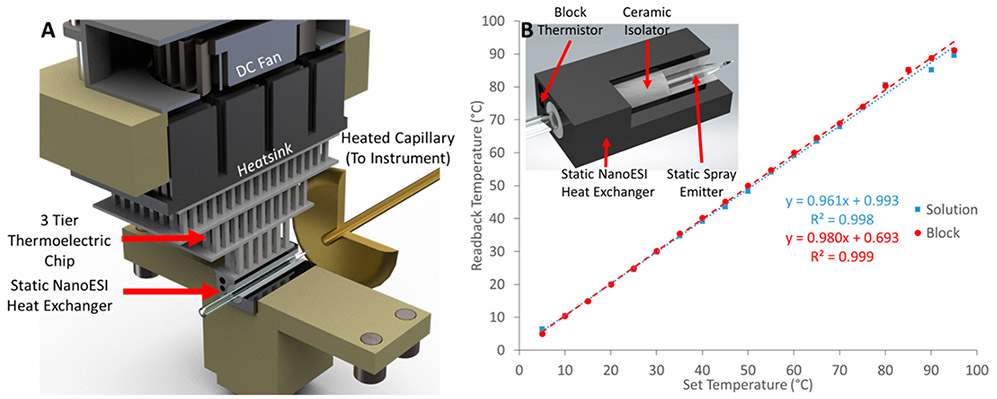
Figure 1A contains a Solidworks rendering of the TEC vT-ESI assembly using static nano ESI emitters. The ESI emitter is positioned inside a ceramic sleeve (Kimball Physics (AL2O3-TU-C-500) and cut to length in-house) that serves as an electrical insulator from an aluminum heat exchanger. The heat exchanger makes direct contact with a three-stage TEC (Peltier chip) (Laird Thermal Systems 9360001-301 three-tier). The TEC is used to maintain the temperature of the heat exchanger and the solution contained in the ESI emitter at the desired temperature, ranging from ~5–98 °C. The vT-ESI capillary temperature was calibrated across the range of temperatures by simultaneously monitoring the static-spray (SS) heat exchanger temperature and the solution contained in the ESI emitter (Figure 1B). The solution temperature was measured using a calibrated T-Type thermocouple (Physitemp Clifton, NJ) paired to a thermocouple (National Instruments USB-TC01) positioned inside the borosilicate pulled glass capillary filled with a 200 mM ammonium acetate solution. The ESI SS capillary was placed inside an alumina silicate ceramic isolator that covered the entire length of the block to ensure the TEC is isolated from ESI voltage (typically between 1.2 and 1.6 kV) applied via a Pt wire (Alfa Aesar, ≥99.9% 0.3048 mm diameter). The block temperature was set to the desired temperature using a temperature controller that utilizes pulse width modulation (TE Technology TC-720); the temperature was monitored for ~2 min to ensure the system had reached equilibrium. The custom power supply developed for the operation of the vT-ESI device is described in the Supporting Information.
Figure 1.
(A) Solidworks rendering of the vT-ESI assembly with labels to identify the significant components. The fan mounted to the top of the device prevents overheating and reduces atmospheric moisture condensation for experiments performed below ~15 °C. The top stage of the thermoelectric chip (TEC) makes direct contact with a 40 mm × 40 mm × 13 mm heatsink (CTS Electronic Components APF40-40-13CB); a 40 mm × 15 mm 24 VDC fan with 14.83 CFM rated airflow (Delta Electronics AFB0424SHB) is used to dissipate the heat off of the heatsink. The vT-ESI assembly uses custom machined PEEK components that mount directly to a commercial Thermo Nanospray Flex source. Additional details about the electronics control system are contained in the Supporting Information. (B) Temperature calibration of the vT-ESI emitter solution is performed using thermocouples inserted into the static spray capillary and the SS heat exchanger (as shown in the inset). The temperature variation ranged from ~±5 °C at the highest and lowest temperature and ~±2 in the range of 5–98 °C.
It is important to note that airflow around the vT-ESI heat exchanger/emitter is essential for reducing condensation at cold temperatures, especially at temperatures below 15 °C, while minimizing air turbulence with deleterious effects for the stability of ionization. We have found that the fan assembly employed here, with controlled fan speed, is essential for both cold and heated operations. The three-tier Peltier TEC provides a much-improved performance over a single-tier design. The temperature ramps obtained using the three-tier design are steeper and allow for reliable operation at much lower temperatures.
Sample Preparation.
Ubiquitin (U6253, Sigma-Aldrich) samples were prepared from a stock solution to have a final concentration of 1 μM in 1% acetic acid, pH of 2.8, which is similar to those used previously by El-Baba et al.23 The A state ubiquitin solutions were prepared by dilution of a 1 mg/mL ubiquitin stock solution to a working concentration of 1 μM in 49:49:2 methanol:water:glacial acetic acid (pH 2).37-39 GroEL40 was expressed in-house as described previously. Frataxin (FXN) was expressed and prepared in-house; a detailed protocol is given in the Supporting Information. Before MS analysis, the sample was biospun in 200 mM ammonium acetate and diluted to a working concentration of 5 μM for FXN and 1 μM for GroEL.
Mass Spectrometry and Ion Mobility-Mass Spectrometry.
The vT-ESI-MS measurements were performed on an Exactive Plus-Extended Mass Range (Thermo Fisher, Bremen, Germany) mass spectrometer. Approximately 7.5 μL of protein solution was loaded into the borosilicate glass capillary (Sutter Instruments, B150-86-10, and P-1000). ESI voltage (~1 kV) was applied by a platinum wire inserted into the solution. The protein solution was equilibrated for 3 min at each temperature prior to each MS acquisition. Data were converted to .txt files using a custom python code and imported into UniDec41 and ProteinMetrics.42 Melting temperatures (TM) were estimated from the inflection point of a four-parameter sigmoidal fit using SigmaPlot version 10.0 from Systat Software, Inc. (San Jose, CA). Additional information on the data processing can be found in the Supporting Information. The IM-MS data were collected using a 1.5 m drift-tube Fourier-transform ion mobility-UHMR Orbitrap, as described previously.34,43-47 A-state ubiquitin IM-MS data were collected on a Waters SYNAPT G2 modified with the newly designed vT-ESI source. CCS calibration was performed using the method described by Ruotolo et al.48
RESULTS AND DISCUSSION
Here, we describe vT-ESI-MS and IM-MS studies using cold- and heat-induced folding reactions for single-domain proteins, viz., ubiquitin and frataxin, and temperature-dependent ligand binding for the chaperonin GroEL. For the purposes of this study, we are interested in comparing and contrasting cold- and heat-induced folding for proteins containing similar secondary structure motifs (helices and β-sheets) similar to ubiquitin.
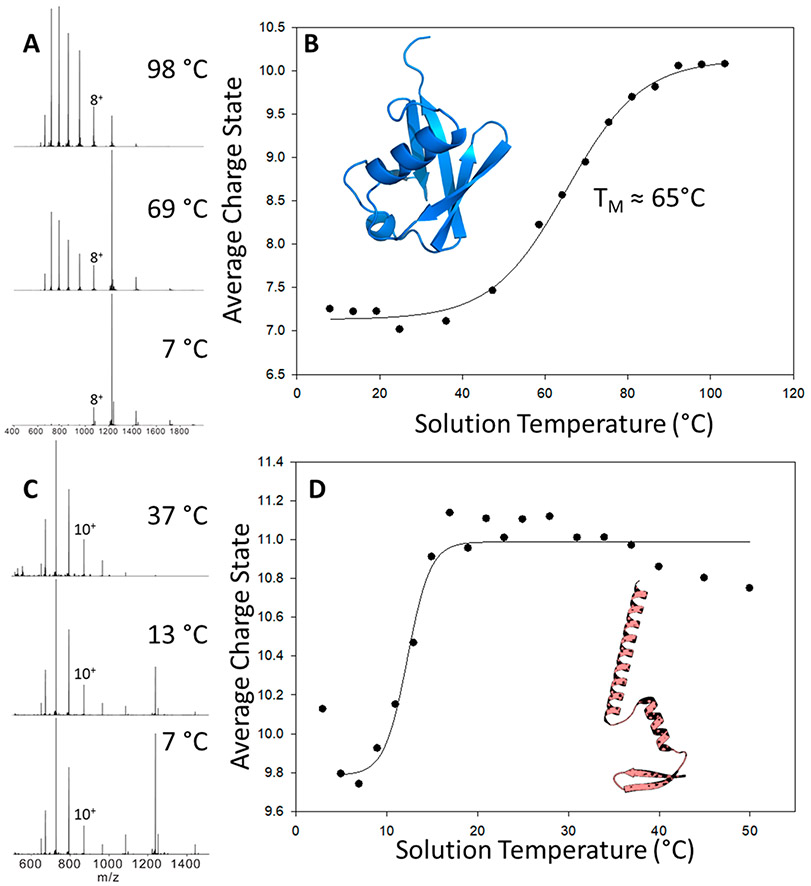
Ubiquitin has been extensively used in variable temperature studies using both MS and IMMS. Thus, it was selected for the initial testing of our vT-ESI device.23,29 The initial experiments employed conditions that preserve native-like, low-charge state ubiquitin.23 Figure 2A contains mass spectra of ubiquitin acquired at different solution temperatures (8 °C, 69 °C, and 98 °C). As expected at these temperatures, changes in the charge state distributions are consistent with temperature-dependent unfolding transitions reported by El-Baba et al.23 However, when the ubiquitin solution conditions were altered to a native buffer system of 200 mM ammonium acetate, no thermal unfolding was observed, indicating the buffer system plays a crucial role in the GEL (data not shown). Figure 2B contains a plot of the average charge state (Zavg) as a function of temperature from 8 to 98 °C; within this range, Zavg changes from 7.25 to 10.07. It is interesting to note that changes in Zavg obtained by reversing the temperature profile (T scans from high- to low-temperature) yield very similar melting profiles.
Figure 2.
(A) Mass spectra of ubiquitin collected at 8 °C, 69 °C, and 98 °C. (B) Plot of the average charge state (Zavg) of ubiquitin (PDB: 1UBQ51) versus temperature of the solution contained in the ESI capillary. Melting temperature (TM) is defined as the midpoint of the sigmoidal curve. (C and D) Plots of the temperature-dependent average charge states (Zavg) of the partially unfolded A-state ubiquitin observed in acidic water/methanol solutions (49:49:2 MeOH:H2O:acetic acid (pH 2.3)). At temperatures above ~50 °C, the signal is attenuated due to the rapid evaporation of the solution. The inset shows the A-state structure adapted from Brutscher et al.38
While the general appearance of the melting curve is in good agreement (within ±0.25 Zavg) with that reported by El-Baba et al.,23 the TM value of 65.2 ± 0.8 °C is lower than their reported value of 71 °C. These differences in TM are attributed to the differences in pH of the solutions. Previous studies reported that the TM values of ubiquitin are pH-dependent; a change in pH from 3.0 to 2.75 shifts TM from 74.1 to 66.3 °C.49 It is also possible that TM values may depend on the size of the ESI emitter owing formation of different droplet sizes. Effects of droplet size on the melting of ubiquitin were noted in experiments using 10.6 μm IR heating of the nanodroplets.29
Prior vT-ESI IMMS studies revealed a complex distribution of intermediate states of ubiquitin using heat-induced melting, including the well-characterized A-state, a partially unfolded conformation consisting of mostly α-helices (structure shown in Figure 2D).23,29,50 Figure 2D contains a plot of the Zavg charge state versus temperature plots obtained using acidic water/methanol solutions (49:49:2 MeOH:H2O:acetic acid (pH 2.3), solution conditions that have been shown to promote the formation of A-state ubiquitin. Note that Zavg values for temperatures greater than ~20 °C are higher than that for the “native-like” ubiquitin (see Figure 2A). Note the marked increase in the abundance of 7+ ions that is characteristic of native-like ubiquitin ions; however, the CCS profiles for these 7+ ions (shown in Figure S2) reveal that only a small fraction of the ion population is native-like in terms of the measured CCS 1028 Å2. The CCS profiles obtained under these conditions are very similar to those reported by El Baba et al. at elevated, denaturing conditions, viz., CCS values of 1264 Å2, 1327 Å2, and 1517 Å2.23
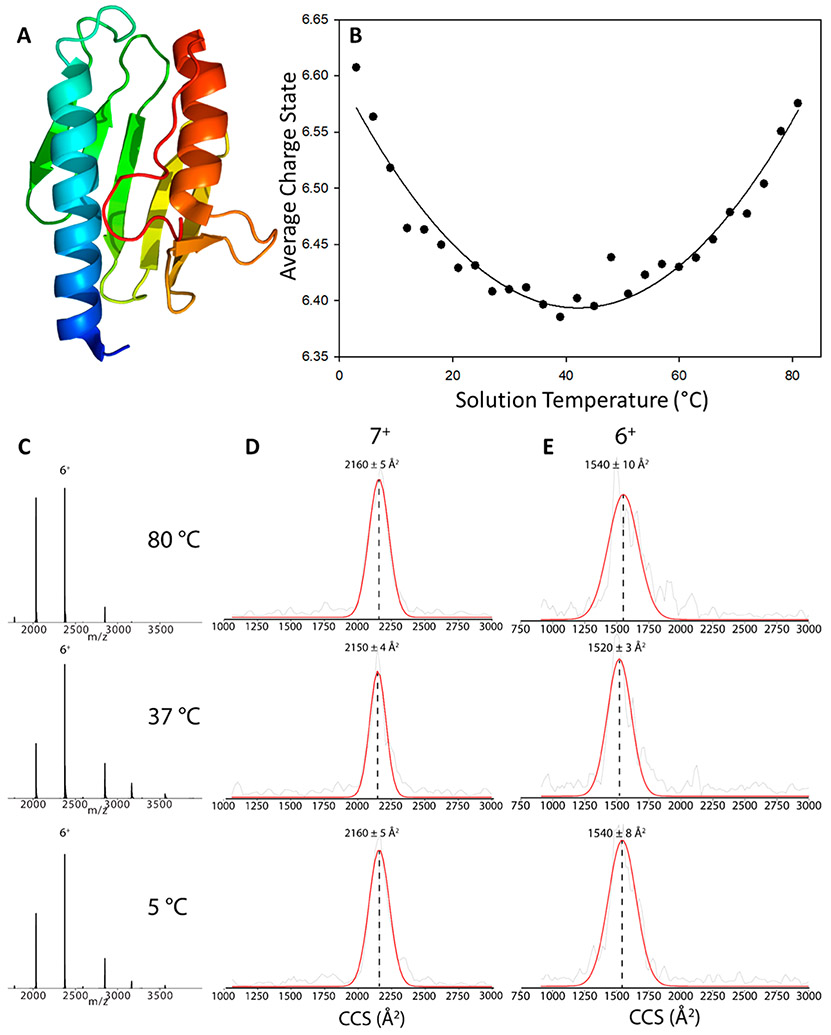
Additional vT-ESI studies were performed using human frataxin (FXN), a 14.237 kDa protein (Uniprot Q16595) allosteric activator of the Fe–S cluster biosynthesis in mitochondria52-56 that has been linked to the neurodegenerative disease Friedreich’s ataxia.57 The vT-ESI data for mature FXN (residues 81–210) are shown over the temperature range of 5–98 °C. Figure 3B contains a T-dependent Zavg plot for FXN. At 3 °C, the Zavg is 6.57, decreases to a minimum of 6.4 at 39 °C, and then increases to 6.57 at 81 °C. The parabolic fit is similar to T-dependent stability reported for yeast frataxin (Yfh1) using CD or NMR spectroscopy.58,59 The changes in the relative abundances of the 7+, 6+, and 5+ FXN charge states (Figure 3C) are consistent with changes in the solvent-accessible surface area (SASA) of the protein. While the collision cross sections (CCSs) (Figures 3D and 3E) for cold- (5 °C) and heat-induced (80 °C) unfolding are similar to that obtained at physiological temperature (37 °C) (see Supporting Information Table S1), the T-dependent CCS differences for the 6+ charge state of FXN are statistically significant (p > 0.05) using an unpaired t test. However, the changes in CCS for the 7+ charge state are not statistically significant.60 More importantly, the peak widths (fwhm) for the CCS profiles are larger for the 6+ ions than for the 7+ ions, which may reflect differences (conformational or dynamics) for the lower charge state ions. For example, the observed differences for cold-induced unfolding may be attributed to differences in the tertiary structure, e.g., differences in the alignment of the helices and or sheet motifs; prior CD studies suggest that the secondary is retained at cold-induced conditions.61 Conversely, the CD spectra suggest that heat-induced unfolding promotes changes in the secondary structure.61 The CD studies were performed using very different buffers and ionic strengths than those used for vt-ESI (20 mM HEPES vs 200 mM ammonium acetate) where solution parameters are known to strongly affect structure and stability.62 Owing to the expected effects of buffers and ionic strengths on cold- and heat-induced folding,2 further investigations of these effects are warranted.
Figure 3.
(A) Structure of human frataxin (PDB: 1EKG)64 and (B) plot of the average charge state (Zavg) of frataxin as a function of solution temperature in the ESI emitter. The ESI signal is unstable at T > 82 °C, presumably owing to decomposition and/or aggregation at higher temperatures. A parabolic fit (Sigmaplot 10.0, solid line) was added to the data. vT-IM-Orbitrap CCS profiles and (C) extract MS of the (D) 7+ charge state and (E) 6+ charge state of FXN at 5 °C, 37 °C, and 80 °C. The red line is a Gaussian function fitted to the raw CCS experimental data (gray). CCS values for the 5+ monomer are not shown due to the overlap with the 10+ dimer.
Previously reported studies suggest different mechanisms for cold- and heat-induced folding for Yfh1, which is directly amendable to vT-IM-MS.58,59,63 The FXN (PDB:1EKG)64 and Yfh1 share a high degree of sequence overlap but differ in terms of the flexibility of the N-terminal residues. The N-2 terminal of FXN is highly dynamic with little to no secondary structure,65,66 whereas Yfh1 has a defined secondary structure.67 The C-terminus length of frataxin orthologues also influences their thermodynamic stability, where FXN has a longer sequence compared to Yfh1.68 Owing to these differences, CD results have shown Yfh1 has unstable secondary structures under cold conditions, while FXN is relatively stable. The observation of FXN cold denaturation by vT-IM-MS in this study highlights the capability of the new device to resolve subtle structural changes that were previously hidden via traditional biophysical techniques.47
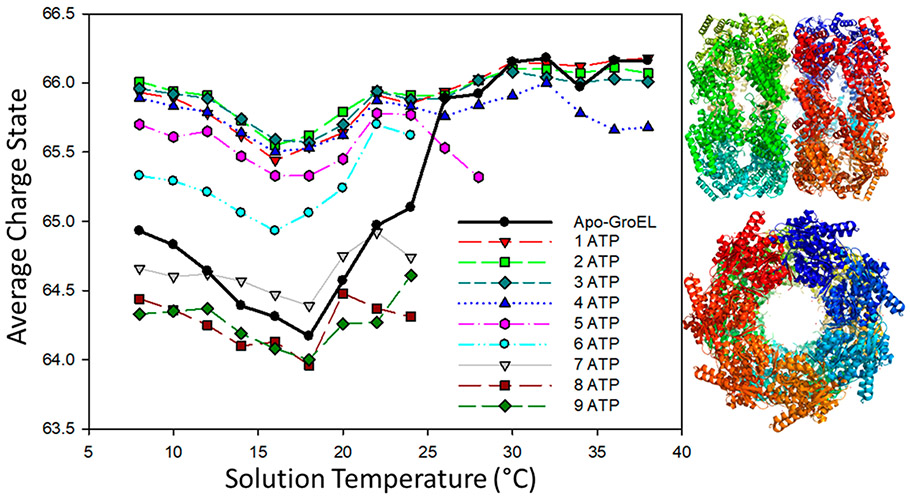
The activities of ion channels and chaperones are highly regulated by conformational (“open” and “closed”) changes that are induced or stabilized by ligand binding that are subject to both enthalpic and entropic barriers.20 GroEL (HSP60, 800 kDa), a 14-mer complex of the HSP10 monomers that are arranged as two stacked heptamers (see Figure 4), forms a large central cavity in which non-native proteins bind via hydrophobic interactions.69,70 The mechanical action GroEL/GroES complex is an example of ATP-dependent movement of the apical domain regulating substrate binding and release.69 Here, vT-ESI is used to investigate the T-dependent ATP binding to GroEL. Figure 4 contains a plot of Zavg for GroEL between 8 and 38 °C, and the products formed upon binding ATP (GroEL(ATP)n); mass spectra are shown in the Supporting Information(Figure S3). Zavg for apo GroEL (solid black data points) decreases from 65 to ~64.5 at temperatures between 8 and 24 °C and then increases to >66 at temperatures above 24 °C. It is interesting to note that significant changes in ATP binding are observed over this same range of temperatures. While the Zavg values obtained for GroEL binding to 1–4 ATPs are mostly similar, a sharp decrease in Zavg is observed for binding the fifth ATP, and there are no detectable signals for binding 6–9 ATPs at T greater than 24 °C. The T-dependent changes in Zavg and ATP binding are interpreted as evidence of changes in the conformation of the architecture of the GroEL complex, as well as altering the entropic barrier. These data were acquired using ammonium acetate (AmA) buffer, and it is interesting to note that such behavior is not observed when using ethylene diammonium diacetate (EDDA) buffer, which produces lower Zavg charge states of GroEL. However, the change in ATP binding occurs in the same range of temperatures for both buffers (manuscript in preparation). For example, at temperatures below 24 °C for GroEL in AmA, bindings of up to 9 ATPs is observed, but at temperatures greater than 24 °C, the maximum number of ATPs bound is 5.
Figure 4.
Temperature dependence of 1 μM GroEL in 200 mM ammonium acetate containing 125 μM ATP (PDB: 1SS8).
CONCLUSIONS
The TEC (Peltier) vT-ESI source was designed for easy implementation on different MS platforms. These include a number of mass spectrometers–the ThermoFisher Orbitraps, including those equipped with REIS and FT-IMS capabilities,22,34,43-47 the Agilent 6560 IM-QTOF and 6545 XT, and the Waters SYNAPT instruments equipped with TWIMS. The performance metrics of the three-stage TEC vT-ESI device make it possible to rapidly acquire reproducible melting curves for static spray capillaries over the temperature range of 5–98 °C. The cold- and heat-folding studies on ubiquitin and frataxin clearly illustrate new applications for vT-ESI-MS. While both ubiquitin and frataxin are monomeric proteins, their respective thermal unfolding curves are significantly different, a sigmodal vs a parabolic curve, respectively. Changes in the Gibbs energy landscape owing to changes in pH, buffers, and/or ligand binding give rise to drastic differences in thermal unfolding curves and may impact the future definition of “native-MS”. The results reported on temperature-dependence of ATP binding to GroEL bode well for future studies on the stabilities and dynamics of protein complexes extending into the mega-dalton molecular weight range. Combining vT-ESI with recent developments in next-generation high-performance IM-MS instrumentation opens new opportunities for studies of intact soluble and membrane protein complexes and other nondenaturing studies.43,47,71,72
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Will Seward and Karl Yeager of the Texas A&M University (TAMU) Machine Shop for fabricating the parts, Greg Matthijetz of the TAMU Laboratory for Biological Mass Spectrometry for assisting in the electrical configuration, and Andrew Roth and Hays Rye of the TAMU Department of Biochemistry and Biophysics for providing the GroEL sample. Funding for this work was provided by the National Science Foundation CHE-1707675 (DHR), National Institutes of Health DP2GM123486 (A.L.), R01GM121751 (D.H.R., A.L., and D.E.C.), R01GM131100 (D.E.C.), P41GM128577 (D.H.R. and V.H.W.), R01GM096100 (D.P.B.), Robert A. Welch Foundation A-1647 (D.P.B.), and endowment funds from the MDS SCIEX Professorship (D.H.R.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c00870.
Description of melting point determination, average charge state, schematic of custom power supply, CCS profile of A-state ubiquitin, and high-resolution vT-ESI MS data of ATP-GroEL binding (PDF)
The authors declare no competing financial interest.
A complete parts list and CAD files of the vT-ESI module for this article are available upon request from the corresponding author.
Contributor Information
Jacob W. McCabe, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Mehdi Shirzadeh, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Thomas E. Walker, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States
Cheng-Wei Lin, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Benjamin J. Jones, Department of Chemistry, Ohio State University, Columbus, Ohio 43210, United States.
Vicki H. Wysocki, Department of Chemistry, Ohio State University, Columbus, Ohio 43210, United States.
David Barondeau, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
David E. Clemmer, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States.
Arthur Laganowsky, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
David H. Russell, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
REFERENCES
- (1).Hopkins FG Nature 1930, 126 (3174), 328–330. [Google Scholar]
- (2).Privalov PL Crit. Rev. Biochem. Mol. Biol 1990, 25 (4), 281–306. [DOI] [PubMed] [Google Scholar]
- (3).Ben-Naim AJ Biomol. Struct. Dyn 2012, 30 (1), 113–124. [DOI] [PubMed] [Google Scholar]
- (4).Anfinsen CB Science 1973, 181 (4096), 223–230. [DOI] [PubMed] [Google Scholar]
- (5).Konermann L; Douglas DJ Biochemistry 1997, 36 (40), 12296–12302. [DOI] [PubMed] [Google Scholar]
- (6).Konermann L; Rosell FI; Mauk AG; Douglas DJ Biochemistry 1997, 36 (21), 6448–6454. [DOI] [PubMed] [Google Scholar]
- (7).Konermann L; Collings BA; Douglas DJ Biochemistry 1997, 36 (18), 5554–9. [DOI] [PubMed] [Google Scholar]
- (8).Wyttenbach T; Bowers MT Annu. Rev. Phys. Chem 2007, 58, 511–33. [DOI] [PubMed] [Google Scholar]
- (9).Clemmer DE; Russell DH; Williams ER Acc. Chem. Res 2017, 50 (3), 556–560. [DOI] [PubMed] [Google Scholar]
- (10).Levy ED; Erba EB; Robinson CV; Teichmann SA Nature 2008, 453 (7199), 1262–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lumry R; Eyring H J. Phys. Chem 1954, 58 (2), 110–120. [Google Scholar]
- (12).Shi L; Holliday AE; Shi H; Zhu F; Ewing MA; Russell DH; Clemmer DE J. Am. Chem. Soc 2014, 136 (36), 12702–12711. [DOI] [PubMed] [Google Scholar]
- (13).Fuller DR; Glover MS; Pierson NA; Kim D; Russell DH; Clemmer DE J. Am. Soc. Mass Spectrom 2016, 27 (8), 1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shi L; Holliday AE; Khanal N; Russell DH; Clemmer DD J. Am. Chem. Soc 2015, 137 (27), 8680–8683. [DOI] [PubMed] [Google Scholar]
- (15).Fuller DR; Conant CR; El-Baba TJ; Brown CJ; Woodall DW; Russell DH; Clemmer DE J. Am. Chem. Soc 2018, 140 (30), 9357–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Valentine SJ; Counterman AE; Clemmer DE J. Am. Soc. Mass Spectrom 1997, 8 (9), 954–961. [Google Scholar]
- (17).El-Baba TJ; Kim D; Rogers DB; Khan FA; Hales DA; Russell DH; Clemmer DE J. Phys. Chem. B 2016, 120 (47), 12040–12046. [DOI] [PubMed] [Google Scholar]
- (18).Conant CR; Fuller DR; Zhang Z; Woodall DW; Russell DH; Clemmer DE J. Am. Soc. Mass Spectrom 2019, 30 (6), 932–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shi L; Holliday AE; Bohrer BC; Kim D; Servage KA; Russell DH; Clemmer DE J. Am. Soc. Mass Spectrom 2016, 27 (6), 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cong X; Liu Y; Liu W; Liang X; Russell DH; Laganowsky A J. Am. Chem. Soc 2016, 138 (13), 4346–4349. [DOI] [PubMed] [Google Scholar]
- (21).Patrick JW; Laganowsky A Probing Heterogeneous Lipid Interactions with Membrane Proteins Using Mass Spectrometry. In Lipid-Protein Interactions: Methods and Protocols; Kleinschmidt JH, Ed.; Springer New York: New York, NY, 2019; pp 175–190, DOI: 10.1007/978-1-4939-9512-7_9. [DOI] [PubMed] [Google Scholar]
- (22).Lyu J; Liu Y; McCabe JW; Schrecke S; Fang L; Russell DH; Laganowsky A Anal. Chem 2020, 92 (16), 11242–11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).El-Baba TJ; Woodall DW; Raab SA; Fuller DR; Laganowsky A; Russell DH; Clemmer DE J. Am. Chem. Soc 2017, 139 (18), 6306–6309. [DOI] [PubMed] [Google Scholar]
- (24).Köhler M; Marchand A; Hentzen NB; Egli J; Begley AI; Wennemers H; Zenobi R Chem. Sci 2019, 10 (42), 9829–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Marchand A; Czar MF; Eggel EN; Kaeslin J; Zenobi R Nat. Commun 2020, 11 (1), 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Brown CJ; Woodall DW; El-Baba TJ; Clemmer DE J. Am. Soc. Mass Spectrom 2019, 30 (11), 2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Raab SA; El-Baba TJ; Woodall DW; Liu W; Liu Y; Baird Z; Hales DA; Laganowsky A; Russell DH; Clemmer DE J. Am. Chem. Soc 2020, 142 (41), 17372–17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Woodall DW; Brown CJ; Raab SA; El-Baba TJ; Laganowsky A; Russell DH; Clemmer DE Anal. Chem 2020, 92 (4), 3440–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).El-Baba TJ; Fuller DR; Woodall DW; Raab SA; Conant CR; Dilger JM; Toker Y; Williams ER; Russell DH; Clemmer DE Chem. Commun 2018, 54 (26), 3270–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Jakub U; Rosie U; Florian B; Bruno B; Perdita B Protein Unfolding in Freeze Frames: Intermediates of Ubiquitin and Lysozyme Revealed by Variable Temperature Ion Mobility-Mass Spectrometry. ChemRxiv 2020, Preprint. https://chemrxiv.org/articles/preprint/Protein_Unfolding_in_Freeze_Frames_Intermediates_of_Ubiquitin_and_Lysozyme_Revealed_by_Variable_Temperature_Ion_Mobility-Mass_Spectrometry/11888958 (accessed 2021-04-21), DOI: 10.26434/chemrxiv.11888958. [DOI] [Google Scholar]
- (31).El-Baba TJ; Clemmer DE Int. J. Mass Spectrom 2019, 443, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Woodall DW; Henderson LW; Raab SA; Honma K; Clemmer DE J. Am. Soc. Mass Spectrom 2021, 32 (1), 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Woodall DW; El-Baba TJ; Fuller DR; Liu W; Brown CJ; Laganowsky A; Russell DH; Clemmer DE Anal. Chem 2019, 91 (10), 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Poltash ML; McCabe JW; Patrick JW; Laganowsky A; Russell DH J. Am. Soc. Mass Spectrom 2019, 30 (1), 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Shirzadeh M; Poltash ML; Laganowsky A; Russell DH Biochemistry 2020, 59 (9), 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Poltash ML; Shirzadeh M; McCabe JW; Moghadamchargari Z; Laganowsky A; Russell DH Chem. Commun 2019, 55 (28), 4091–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hoerner JK; Xiao H; Kaltashov IA Biochemistry 2005, 44 (33), 11286–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Brutscher B; Brüschweiler R; Ernst RR Biochemistry 1997, 36 (42), 13043–13053. [DOI] [PubMed] [Google Scholar]
- (39).Pan Y; Briggs MS Biochemistry 1992, 31 (46), 11405–12. [DOI] [PubMed] [Google Scholar]
- (40).Weaver J; Jiang M; Roth A; Puchalla J; Zhang J; Rye HS Nat. Commun 2017, 8 (1), 15934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Marty MT; Baldwin AJ; Marklund EG; Hochberg GKA; Benesch JLP; Robinson CV Anal. Chem 2015, 87 (8), 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bern M; Caval T; Kil YJ; Tang W; Becker C; Carlson E; Kletter D; Sen KI; Galy N; Hagemans D; Franc V; Heck AJR J. Proteome Res 2018, 17 (3), 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).McCabe JW; Mallis CS; Kocurek KI; Poltash ML; Shirzadeh M; Hebert MJ; Fan L; Walker TE; Zheng X; Jiang T; Dong S; Lin C-W; Laganowsky A; Russell DH Anal. Chem 2020, 92 (16), 11155–11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Clowers BH; Russell DH Anal. Chem 2018, 90 (17), 10472–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).McCabe JW; Hebert MJ; Shirzadeh M; Mallis CS; Denton JK; Walker TE; Russell DH Mass Spectrom. Rev 2021, 40, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lin C-W; McCabe JW; Russell DH; Barondeau DP J. Am. Chem. Soc 2020, 142 (13), 6018–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Russell DH TrAC, Trends Anal. Chem 2020, 124, 115533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ruotolo BT; Benesch JLP; Sandercock AM; Hyung S-J; Robinson CV Nat. Protoc 2008, 3 (7), 1139–1152. [DOI] [PubMed] [Google Scholar]
- (49).Wintrode PL; Makhatadze GI; Privalov PL Proteins: Struct., Funct., Genet 1994, 18 (3), 246–253 [DOI] [PubMed] [Google Scholar]
- (50).Shi H; Pierson NA; Valentine SJ; Clemmer DE J. Phys. Chem. B 2012, 116 (10), 3344–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Vijay-Kumar S; Bugg CE; Cook WJ J. Mol. Biol 1987, 194 (3), 531–44. [DOI] [PubMed] [Google Scholar]
- (52).Bridwell-Rabb J; Fox NG; Tsai CL; Winn AM; Barondeau DP Biochemistry 2014, 53 (30), 4904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Patra S; Barondeau DP Proc. Natl. Acad. Sci. U. S. A 2019, 116 (39), 19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Prischi F; Konarev PV; Iannuzzi C; Pastore C; Adinolfi S; Martin SR; Svergun DI; Pastore A Nat. Commun 2010, 1, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Fox NG; Yu X; Feng X; Bailey HJ; Martelli A; Nabhan JF; Strain-Damerell C; Bulawa C; Yue WW; Han S Nat. Commun 2019, 10 (1), 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Tsai C-L; Barondeau DP Biochemistry 2010, 49 (43), 9132–9139. [DOI] [PubMed] [Google Scholar]
- (57).Campuzano V; Montermini L; Moltò MD; Pianese L; Cossée M; Cavalcanti F; Monros E; Rodius F; Duclos F; Monticelli A; Zara F; Cañizares J; Koutnikova H; Bidichandani SI; Gellera C; Brice A; Trouillas P; De Michele G; Filla A; De Frutos R; Palau F; Patel PI; Di Donato S; Mandel J-L; Cocozza S; Koenig M; Pandolfo M Science 1996, 271 (5254), 1423. [DOI] [PubMed] [Google Scholar]
- (58).Sanfelice D; Morandi E; Pastore A; Niccolai N; Temussi PA ChemPhysChem 2015, 16 (17), 3599–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pastore A; Martin SR; Politou A; Kondapalli KC; Stemmler T; Temussi PA J. Am. Chem. Soc 2007, 129 (17), 5374–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Krzywinski M; Altman N Nat. Methods 2013, 10 (11), 1041–1042. [DOI] [PubMed] [Google Scholar]
- (61).Wallace BA Curr. Opin. Struct. Biol 2019, 58, 191–196. [DOI] [PubMed] [Google Scholar]
- (62).Xia Z; DeGrandchamp JB; Williams ER Analyst 2019, 144 (8), 2565–2573. [DOI] [PubMed] [Google Scholar]
- (63).Bonetti D; Toto A; Giri R; Morrone A; Sanfelice D; Pastore A; Temussi P; Gianni S; Brunori M Phys. Chem. Chem. Phys 2014, 16 (14), 6391–6397. [DOI] [PubMed] [Google Scholar]
- (64).Dhe-Paganon S; Shigeta R; Chi YI; Ristow M; Shoelson SE J. Biol. Chem 2000, 275 (40), 30753–6. [DOI] [PubMed] [Google Scholar]
- (65).Musco G; Stier G; Kolmerer B; Adinolfi S; Martin S; Frenkiel T; Gibson T; Pastore A Structure 2000, 8 (7), 695–707. [DOI] [PubMed] [Google Scholar]
- (66).Prischi F; Giannini C; Adinolfi S; Pastore A FEBS J. 2009, 276 (22), 6669–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Pastore A; Puccio H J. Neurochem. 2013, 126 (s1), 43–52. [DOI] [PubMed] [Google Scholar]
- (68).Adinolfi S; Nair M; Politou A; Bayer E; Martin S; Temussi P; Pastore A Biochemistry 2004, 43 (21), 6511–8. [DOI] [PubMed] [Google Scholar]
- (69).Lin Z; Rye HS Crit. Rev. Biochem. Mol. Biol 2006, 41 (4), 211–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Bukau B; Horwich AL Cell 1998, 92 (3), 351–366. [DOI] [PubMed] [Google Scholar]
- (71).Mallis CS; Zheng X; Qiu X; McCabe JW; Shirzadeh M; Lyu J; Laganowsky A; Russell DH Int. J. Mass Spectrom 2020, 458, 116451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M Anal. Chem 2019, 91 (13), 8564–8573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.