Abstract
Small-molecule inhibitors of enzyme function are critical tools for the study of cell biological processes and for treatment of human disease. Identifying inhibitors with suitable specificity and selectivity for single enzymes, however, remains a challenge. In this study we describe our serendipitous discovery that NMS-873, a compound that was previously identified as a highly selective allosteric inhibitor of the ATPase valosin-containing protein (VCP/p97), rapidly induces aerobic fermentation in cultured human and mouse cells. Our further investigation uncovered an unexpected off-target effect of NMS-873 on mitochondrial oxidative phosphorylation, specifically as a dual inhibitor of Complex I and ATP synthase. This work points to the need for caution regarding the interpretation of cell survival data associated with NMS-873 treatment and indicates that cellular toxicity associated with its use may be caused by both VCP/p97-dependent and VCP/p97-independent mechanisms.
Keywords: small-molecule inhibitor, VCP/p97, aerobic fermentation, oxidative phosphorylation, Complex I, ATP synthase
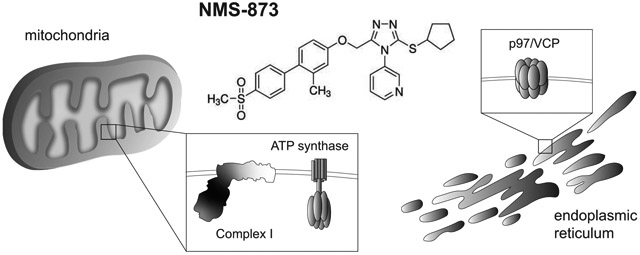
Graphical Abstract
1. Introduction
Small-molecule enzyme inhibitors are an important pharmacologic tool for basic research. Prior to the wide-spread use of RNA interference and genome editing techniques, selective pharmacologic inhibitors were the mainstay for perturbing enzymes as a means to inferring their functions in the cell. The relative stability and ease with which most small-molecule inhibitors can be delivered to cells continues to make them an attractive choice for basic science investigators seeking to selectively and specifically block a given enzyme or cellular process.
Despite their advantages, one of the key problems associated with pharmacologic agents is their potential for off-target effects [1]. Because many classes of enzymes share similar structural motifs within their catalytic domain, it is common for small-molecule inhibitors to bind a broad set of related enzymes at various levels of affinity. This challenge has been well documented for compounds that function as competitive inhibitors, especially those that mimic the structure of generic enzyme substrates such as ATP [2,3]. As such it is often difficult to achieve a high degree of selectivity when screening for drugs that inhibit the activity of ATP-dependent enzymes such as kinases and ATPases.
One means by which the specificity and selectivity of small-molecule inhibitors can be improved is to identify compounds that bind to allosteric regulatory sites on a target enzyme rather than to the catalytic domain [4-6]. Because allosteric sites are usually less conserved than the active site across families of enzymes, it is possible to achieve significant selectivity for a specific enzyme within a broader class. The drug NMS-873, which was identified as a low nanomolar concentration inhibitor of the AAA ATPase VCP/p97, provides a good example of this concept [7]. NMS-873 functions by binding to the interface of two adjacent domains within the active hexameric structure of VCP/p97 and leads to an interruption of its catalytic cycle by stabilizing the ADP-bound state. Unlike competitive inhibitors discovered in the same study, NMS-873 showed a high degree of selectivity for VCP/p97 compared to other AAA ATPases and a panel of 53 kinases. The authors attribute this selectivity to fact that NMS-873 is not an ATP mimetic and binds to a site on VCP/p97 that is unique to its functional quaternary structure.
Since its initial discovery, several studies have employed NMS-873 to infer a role for VCP/p97 in a variety of pathologically relevant processes, most notably in oncogenic transformation and viral replication [8-12]. Given the well-described role of VCP/p97 in in the ERAD and autophagy pathways, its association with diseases that require alteration of cellular proteostasis is not surprising [13,14]. Since VCP/p97 is overexpressed in various cancers and has been shown to facilitate important pro-survival functions, such as DNA repair after ionizing radiation, NMS-873 would seem to be a logical candidate for further therapeutic development [15-17]. Initial testing of the drug against a panel of 38 different cancer cell lines seemed to support its viability for this purpose [7].
While using NMS-873 as a tool to disrupt ERAD in a study related to GLUT transporter trafficking, we consistently observed acidification of culture media every time the drug was employed. This led us to further explore the broader metabolic impact of NMS-873 in both human and mouse cells. Our investigation uncovered an unexpected off-target effect of NMS-873 on mitochondrial OXPHOS in which both Complex I and ATP synthase were inhibited. This observation points out that NMS-873 may not be as selective an inhibitor as previously thought, and that caution should be taken when interpreting the cytotoxic effects of this drug on cultured cells.
2. Materials and Methods
2.1. Chemical reagents
All chemical reagents and small-molecule inhibitors used in this study were obtained from Sigma-Aldrich (St. Louis, MO) except antimycin-A1, atractyloside, trifluoromethoxy-carbonylcyanide-phenylhydrazone (FCCP), nigericin, rotenone, and oligomycin, which were purchased from Cayman Chemical (Ann Arbor, MI). Drugs were dissolved to 250-1000x concentration in the appropriate vehicle solvent (DMSO or water) and stored at −20°C prior to use in assays.
2.2. Cell culture
HK-2 immortalized human kidney cells used in this study were obtained from American Type Culture Collection (ATCC, Manassas, VA). L929 fibroblasts were obtained from the lab of Dr. Larry Louters (Calvin University, Grand Rapids, MI). Both lines were tested in-house for mycoplasma contamination by PCR-based methods and found to be negative. The HK-2 cell line was maintained in standard Roswell Park Memorial Institute 1640 (RPMI-1640) medium containing 2 g/L of glucose, which was supplemented with GlutaMAX (Thermo-Fisher, Waltham, MA) and 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA). The L929 cell line was maintained in low glucose (1 g/L) DMEM supplemented with GlutaMAX (Thermo-Fisher) and 5% fetal bovine serum. Cells were split 2-3 times per week to maintain log-phase growth. Detachment of cells from culture plates was performed with TrypLE Express reagent (ThermoFisher) after washing cells with sterile DPBS (ThermoFisher). For all assays, cells were seeded at the specified densities after counting with Luna II cell counter (Logos Biosystems, Annandale, VA). Nutrient-restricted media formulations were made by supplementing a basic DMEM powder stock (Sigma-Aldrich) with 3.7 grams/liter of sodium bicarbonate and the required amounts of glucose, glutamine, or sodium pyruvate to achieve 10 mM final concentration. Reconstituted media was sterilized by filtration through a 0.22-micron filter prior to use.
2.3. Lactate quantification assay
Lactate produced by fermentation was measured from conditioned media samples using a L-lactic acid detection kit from Megazyme (Bray, Ireland). The enzyme-coupled reaction was performed as directed by the manufacturer, but decreased in size by ten-fold to accommodate a 96-well plate size for spectrometry. The standard curve for each assay was created by serially diluting a 0.15 mg/mL (1.66 mM) stock solution of sodium lactate into basal media matching the cell type from which samples were collected. Paired measurements were performed on each conditioned media sample to ensure experimental consistency. The absorbance values at 340 nm were obtained with an Eon microplate reader (BioTek, Winooski, VT) and exported to Prism 6 (GraphPad Software, San Diego, CA) for graphing and statistical analysis.
2.4. Quantification of relative cellular abundance
Cells were seeded to black-wall, 96-well culture plates (Greiner, Kremsmunster, Austria) in the indicated culture media at a density of 1.0 x 104 cells/well. At 18-24 hours after plating, media was removed from the plate by inverted shaking and cells were washed twice with DPBS to remove residual media. Media containing the indicated nutrients or drugs was replaced in a volume of 100 μL per well and allowed to incubate on cells for 1-24 hours as indicated for each experiment. The relative abundance of live cells was measured using a Cell Titer Glo assay (Promega, Madison, WI), which produces an ATP-dependent luminescent signal proportional to cell number. Luminescent values were captured with a Synergy H1 plate reader (BioTek, Winooski, WT) and exported to Microsoft Excel (Redmond, WA) for data analysis. Average luminescent values for each triplicate sample were normalized against the average value of control cells treated with vehicle only. Processed data were exported to Prism 6 software for line fitting and calculation of LD50 values.
2.5. Flow cytometry
HK-2 cells were plated to 6-well dishes at a density of 5.0 x 105 cells/well and allowed to adhere for 18-24 hours under normal culture conditions. The following day media was aspirated and cells were treated for 24 hours with fresh media containing DMSO or NMS-873 as indicated. They were then rinsed with PBS and detached from the plate in 1 mL of TrypLE Express (Thermo-Fisher). Cells were subsequently filtered to achieve a single cell suspension and pelleted in polystyrene cytometry tubes (BD Biosciences, San Jose, CA). Cells being assayed for total GLUT1 expression were pelleted, then resuspended in 0.5 mL Cytofix solution (BD Biosciences, San Jose, CA) and incubated at 4°C with constant shaking. After 25 minutes of fixation, the cell suspension was diluted 1:1 with cold PBS and pelleted again. The resulting pellet was resuspended in 0.5 mL Cytoperm solution (BD Biosciences) and incubated at 4°C with constant shaking for 30 minutes to permeabilize and block cells. Cells were pelleted again and then incubated in 100 μL of primary antibody solution composed of Cytoperm buffer with 1:200 dilution of mouse anti-GLUT1 antibody (LS Bio, Seattle, WA) with constant shaking overnight at 4°C. The following day cells were washed with 1 mL of cold Cytoperm solution and pelleted, then resuspended in 100 μL of secondary antibody solution composed of Cytoperm buffer with 1:1,000 dilution of Alexafluor-647-coupled goat anti-mouse antibody (Cell Signaling Technology, Danvers, MA). The cells were incubated in a dark cold room at 4°C with constant shaking for 1 hour, then washed as before and resuspended in 300 μL of Cytoperm solution prior to analysis with a FACScalibur flow cytometer (Becton Dickinson Company, Franklin Lakes, NJ). Laser intensity for the FL4 channel was set to center the negative control cell population (isotype mouse IgG-stained cells) distribution at a mean fluorescent intensity (MFI) of 101 units. No fewer than 2.0 x 104 cells were captured for each sample, and each condition was measured in triplicate to obtain an averaged MFI for each cell population.
2.6. 2-deoxyglucose (2-DG) uptake assay
Cells were plated to 24-well dishes at a density of 8.0 x 104 cells/well and allowed to adhere and equilibrate to culture conditions for 18-24 hours in the specified treatment conditions. Glucose uptake was measured using the radiolabeled glucose analog [1,2-3H]-2-deoxyglucose ([3H]-2-DG). Briefly, the media was replaced with 0.3 mL of glucose-free HEPES buffer [pH 7.4] (140 mM NaCl, 5 mM KCl, 20 mM HEPES, 2.5 mM MgSO4, 1 mM CaCl2, 2 mM sodium pyruvate) supplemented with 1.0 mM (0.3 μCi/mL) [3H]-2-DG. After a 15-minute incubation, cells were washed twice with cold glucose-free HEPES. The cells were digested in 0.25 mL of 0.3 M NaOH prior to measuring the [3H]-2-DG uptake. Quadruplicate [3H]-2-DG uptake values were averaged and tested for significance using student’s T-test.
2.7. ATP quantification assay
Quantification of cellular ATP levels was performed using the ENLITEN ATP Assay System (Promega). Cells were plated to standard 96-well culture plates at 2.0 x 104 cells per well and allowed to adhere and resume growth overnight under standard conditions. The following day they were washed with DPBS and treated for 1 hour with glucose-free DMEM containing 10 mM glutamine, 10 mM pyruvate and the indicated doses of NMS-873 or respiratory chain inhibitors. Following treatment cells were washed with DPBS and lysed with 50 μL of 1% TCA. The resulting lysates were neutralized by addition of 250 μL of cold 50 mM Tris-acetate (pH 7.5) buffer and placed on ice. For each sample, 50 μL of lysate was mixed with an equal volume of reconstituted assay reagent from the kit and then measured for total luminescence using a Synergy H1 plate reader. Luminescence values were converted into ATP concentrations using a standard curve generated by serial dilution of a 0.1 μM stock of ATP with ATP-free water provided with the kit.
2.8. Fluorescent mitochondrial polarization assay
Evaluation of mitochondrial membrane potential was performed using fluorescent microplate assay based on the pH-sensitive cationic dye TMRE (Cayman Chemical). Cells were plated to Corning white-wall, 96-well culture plates (Sigma-Aldrich) at a density of 4.0 x 104 cells per well and allowed to adhere overnight under standard conditions. The following day they were washed with DPBS and treated for 30 minutes with Fluorobright DMEM (Thermo-Fisher) containing the indicated saturating dose of NMS-873 or respiratory chain inhibitors in a volume of 50 μL per well. An equal volume of the same media containing 500 nM TMRE was added to each well and cells were incubated for an additional 30 minutes to allow for dye concentration in the mitochrondrial inner membrane space. After incubation the cells were washed twice with pre-warmed TMRE assay buffer (Cayman Chemical) and then covered with 100 μL per well of the same dye and incubated for 15 minutes at 37°C in the dark. Fluorescent values (excitation/emission = 530/580 nm) were captured with a Synergy H1 plate reader and exported to Microsoft Excel for data analysis.
2.9. Mitochondrial ATP synthase activity assay
The mitochondrial ATP synthase activity assay was performed using the MitoCheck Complex V assay kit obtained from Cayman Chemical (Ann Arbor, MI). This assay utilizes intact bovine heart mitochondria as a substrate for an enzyme-coupled reaction that terminates in the oxidation of NADH to NAD+, which can be detected by measuring solution absorbance at 340 nm at 30 second intervals for 30 minutes. Measurements were performed on an Eon microplate reader (BioTek, Winooski, WT) and data was exported to Excel for calculation of slopes for each absorbance curve. Relative Complex V ATPase activity in the presence of various concentrations of oligomycin or NMS-873 was normalized to control reactions in which mitochondria were treated with vehicle only (DMSO). All reactions were performed in triplicate to ensure replicability across each concentration of inhibitor.
2.10. Real-time mitochondrial respiration assay
Real-time measurement of ATP production by oxidative phosphorylation was performed with a modified version of a previously published protocol that utilizes ATP-dependent firefly luciferase as a readout for respiratory chain activity [18]. We modified this assay to incorporate the combined luciferase plus D-luciferin (rL/L) reagent obtained from the ENLITEN ATP Assay System (Promega), which was resuspended in 12 mL of fresh Respiratory Medium (RM, 10 mM Tris-HCl [pH 7.4], 10 mM potassium phosphate, 10 mM potassium chloride, 5 mM magnesium chloride,1 mM EGTA, 225 mM sucrose, 0.05% BSA) and stored on ice prior to initiating the assay. HK-2 or L929 cells grown on 10 cm dishes were detached by trypsinization, pelleted at 2,000 rcf and washed in DPBS before counting cell numbers. Cells were then pelleted and resuspended in Permeabilization Buffer (RM with 50 μg/mL digitonin, 1x protease inhibitor cocktail) at a density of 3 x 106 cells/mL. After 1 minute of gentle inversion at room temperature the cells were pelleted and washed twice with cold RM to remove trace digitonin, then stored on ice at a final density of 6 x 106 cells/mL. For each reaction, 1 μL of cell suspension (~6,000 cells) was incubated in 40 mL of Reaction Buffer (RM with 100 mM ADP, 50 mM Ap5A, 1x rL/L reagent, and 5 mM malate or succinate) at room temperature for 10 minutes. Ap5A was added to inhibit mitochondrial adenylate kinase activity. Reaction progress was monitored on a Biotek Synergy H1 plate reader by measuring total luminescence of each well in the white-wall/bottom, 96-well assay plate at 30 second intervals over the course of the 10-minute reaction. Luminescence values were converted into ATP concentrations using a standard curve generated by serial dilution of a 0.1 μM stock of ATP with ATP-free water provided with the ENLITEN kit. Data were exported to Excel and used to generate linear slope values that represent ATP synthesis rate (fmol/sec). Relative ATP synthesis rates at different concentrations of NMS-873 were determined by normalizing the slope values to reactions containing vehicle (DMSO) only. Processed data were exported to Prism 6 software for line fitting and calculation of IC50 values.
2.11. Data fitting and statistical analysis
The results of this study were obtained from exploratory experiments aimed at explaining the unexpected effects of NMS-873 on cellular metabolism. Each experiment was repeated a minimum of three times to ensure that results could be replicated. All data points used for fitting data to LD50 or IC50 curves represent the average of triplicate or quadruplicate values. Regression analysis was carried out in GraphPad Prism 6 software with a 4-parameter, variable-slope dose-response equation to calculate LD50 or IC50 values, which were reported only in cases where the goodness of fit met a minimum threshold of R2 > 0.95. The statistical significance for pairwise comparisons between control and experimental conditions was determined by a two-tailed student’s t-test using Microsoft Excel. Statistical significance is reported for comparisons with p-value < 0.05 as indicated in the figure legends.
3. Results
3.1. NMS-873 induces lactic acid fermentation in human renal tubule cells
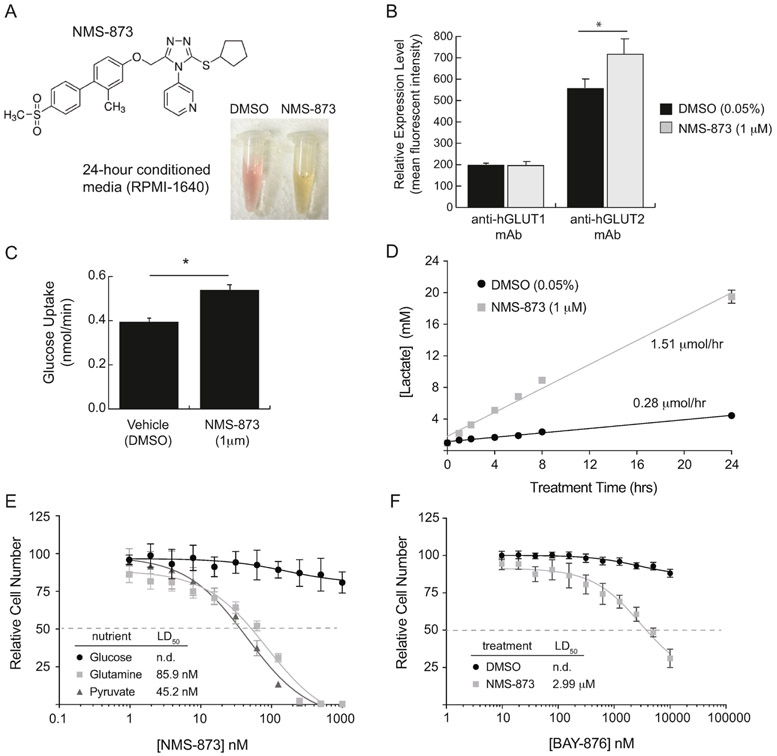
A core project in our laboratory focuses on the trafficking and function of GLUT/SLC2A family glucose transporters in mammalian cells. While studying the impact of glycosylation on GLUT1 trafficking in HK-2 human renal tubule cells, we utilized the drug NMS-873 as a tool compound to inhibit VCP/p97 because of its well-established role in proteolysis of improperly glycosylated membrane proteins (Figure 1A). When used alone, NMS-873 has no impact on GLUT1 expression levels and only modestly increases GLUT2, which is the more abundant glucose transporter found in renal epithelial cells (Figure 1B, Supplemental Figure S1). We observed a mild increase in glucose uptake rate by about 35% when HK-2 cells were treated with NMS-873, which initially seemed to fit with the increase in GLUT2 abundance seen with treatment (Figure 1C, Supplemental Figure S2A).
Figure 1 – NMS-873 promotes lactate fermentation in human renal tubule cells.
(A) Chemical structure of NMS-873 and images of conditioned media from cells treated for 24 hours with vehicle (0.05% DMSO) or NMS-873 (1 μM). (B) Flow cytometry measurement of endogenous human GLUT1 and GLUT2 abundance in HK-2 cells after 24-hour treatment with vehicle or NMS-873. (C) Glucose uptake rates over the course of 30 minutes treatment with vehicle or NMS-873. (D) Time-course measurement of lactate production by HK-2 cells treated with vehicle or NMS-873. (E) HK-2 cell survival after 24 hours of treatment in a two-fold serial dilution of NMS-873. Cells were cultured in serum-free basal media supplemented with 10 mM of the indicated nutrient as the primary source of energy. (F) HK-2 cell survival after 24 hours of treatment in a two-fold serial dilution of the selective GLUT1 inhibitor BAY-876 in combination with vehicle or 1 μM NMS-873. Error bars represent standard deviation of triplicate values for each panel. [*, p-value < 0.05].
More surprisingly, however, we also observed that overnight treatment of HK-2 cells with NMS-873 reproducibly caused the culture media to acidify, as indicated by the color shift of the phenol red indicator dye from pink to yellow (Figure 1A). The simplest explanation for this phenomenon is that NMS-873 induces lactic acid fermentation, which we confirmed by performing a time-course study of lactate concentration in the conditioned media of HK-2 cells treated with the drug. Fitting of the data by linear regression demonstrated that NMS-873 increases lactate secretion 5-fold compared to cells treated with vehicle only (Figure 1D).
Increased aerobic fermentation of glucose by cultured mammalian cells serves as a survival mechanism when mitochondrial oxidative phosphorylation is compromised. To test whether glucose fermentation is necessary for survival of HK-2 cells in the presence of NMS-873, we performed two different experiments. In the first experiment we cultured HK-2 cells in three distinct basal medias in which a single nutrient source is provided for ATP production. Cells grown in media containing 10 mM glucose showed little change in survival over a broad range of NMS-873 concentrations; however, cells grown in media with the non-fermentable substrates glutamine or pyruvate showed significant toxicity at doses of NMS-873 under 100 nM (Figure 1E). In the second experiment we grew cells in complete media containing all three nutrients, but treated them with a high concentration of NMS-873 (1 μM) along with a two-fold serial dilution of the glucose transport inhibitor BAY-876. The combination of these drugs proved to be toxic to HK-2 cells with an IC50 for BAY-876 of about 3 μM, which is consistent with its previously reported IC50 for GLUT2 (Figure 1F) [19]. Together these data demonstrate that HK-2 cells increase glucose transport and lactic acid fermentation as a survival mechanism when NMS-873 is present, suggesting that NMS-873 could be acting as an inhibitor of energy production by OXPHOS.
3.2. NMS-873 induces lactic acid fermentation in mouse fibroblasts
Before investigating the mechanistic aspects of NMS-873, we first wanted to confirm that the drug functioned similarly in mammalian cells of a different tissue and species origin. For this purpose, we repeated the previous experiments in mouse L929 fibroblasts. These cells display a dose-dependent increase in lactate fermentation and glucose uptake after 24 hours of NMS-873 treatment (Figure 2A,B and Supplemental Figure S2B). They also showed significant toxicity in response to NMS-873 treatment in non-fermentable media containing glutamine or pyruvate (Figure 2C). Perhaps even more notable, the sensitivity of L929 cells to BAY-876 (IC50, 6.10 nM) in the presence of NMS-873 was consistent with their expression of only GLUT1, for which BAY-876 has a reported IC50 of 2 nM [19]. These data collectively indicate that both human and mouse cells are sensitive to NMS-873 and that both lines rely upon glucose fermentation for survival when the drug is present at doses higher than about 100 μM.
Figure 2 – NMS-873 promotes glucose uptake and lactate fermentation in mouse L929 fibroblasts.
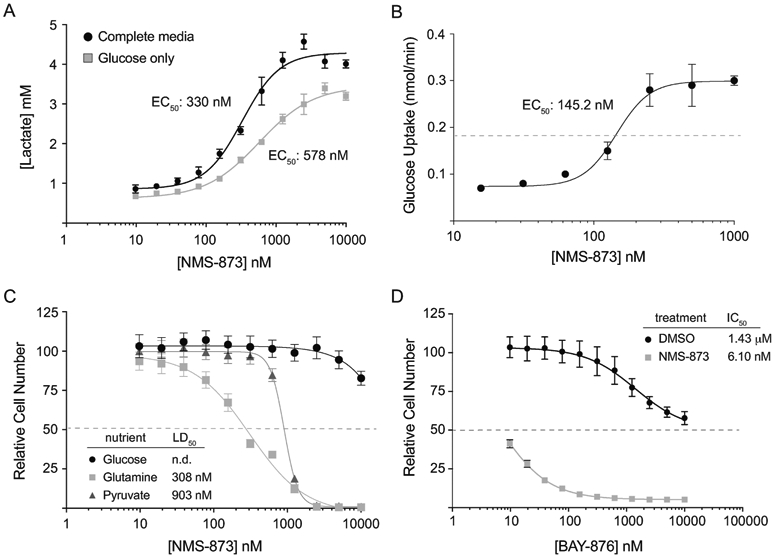
(A) Lactate concentration in conditioned media harvested from L929 cells after 24 hours of treatment with a two-fold serial dilution of NMS-873. Cells were cultured either in complete DMEM media containing glucose (5.5 mM), glutamine (4 mM) and pyruvate (1.25 mM) or in basal DMEM containing only glucose (10 mM). (B) Glucose uptake rates (nmol/min) in L929 cells treated with a two-fold serial dilution of NMS-873. (C) L929 cell survival after 24 hours of treatment in a two-fold serial dilution of NMS-873. Cells were cultured in serum-free basal media supplemented with 10 mM of the indicated nutrient as the primary source of energy. (D) L929 cell survival after 24 hours of treatment in a two-fold serial dilution of the selective GLUT1 inhibitor BAY-876 in combination with vehicle or 1 μM NMS-873. Error bars represent standard deviation of triplicate values for each panel. [*, p-value < 0.05].
3.3. The effects of NMS-873 mimic those of known OXPHOS inhibitors
While the effects of NMS-873 to this point were consistent with it being an inhibitor of mitochondrial respiration, we wanted to first confirm that other more well-characterized inhibitors of OXPHOS function in the expected pattern in mouse L929 cells. To this end we obtained a panel of compounds that have distinct inhibitory mechanisms and tested their impact on ATP synthesis capacity in L929 cells (Table 1). As expected, L929 cells show low nanomolar sensitivity to inhibitors targeted against Complex I (rotenone), Complex IIII (antimycin-A1), and ATP synthase (oligomycin) (Figure 3A). They also showed sensitivity to a mitochondrial proton gradient uncoupler (FCCP), albeit at concentrations in the low micromolar range. In contrast, neither of the inner mitochondrial membrane ion transporter inhibitors (atractyloside, mersalyl acid) nor the potassium/proton ionophore nigericin showed significant toxicity at concentrations as high as 20 μM (Figure 3B). Similar to what was shown for NMS-873, the toxicity of all of OXPHOS inhibitors could be rescued by supplementing culture media with glucose (Figure 3C). This rescue was enabled by increased glucose uptake and lactic acid fermentation, both of which increased rapidly upon addition of the inhibitors of L929 cell (Figure 3D,E,F).
Table 1 –
IC50 Values for respiratory complex inhibitors in L929 cells
| Compound | Inhibitory target/mechanism | IC50‡ |
|---|---|---|
| Antimycin A1 | Complex III | 19.5 +/− 0.9 nM |
| Atractyloside | ADP/ATP translocase | n.d. |
| FCCP | H+ gradient uncoupler | 1.13 +/− 0.30 μM |
| Mersalyl acid | H+/PO43- symporter | n.d. |
| Nigericin | K+/H+ exchange ionophore | n.d. |
| Oligomycin | ATP synase | 17.4 +/− 1.3 nM |
| Rotenone | Complex I | 16.1 +/− 1.0 nM |
| NMS-873 | Complex I | 1.30 +/− 0.30 μM |
Figure 3 – NMS-873 mimics the effects of known mitochondrial respiration inhibitors in mouse L929 fibroblasts.
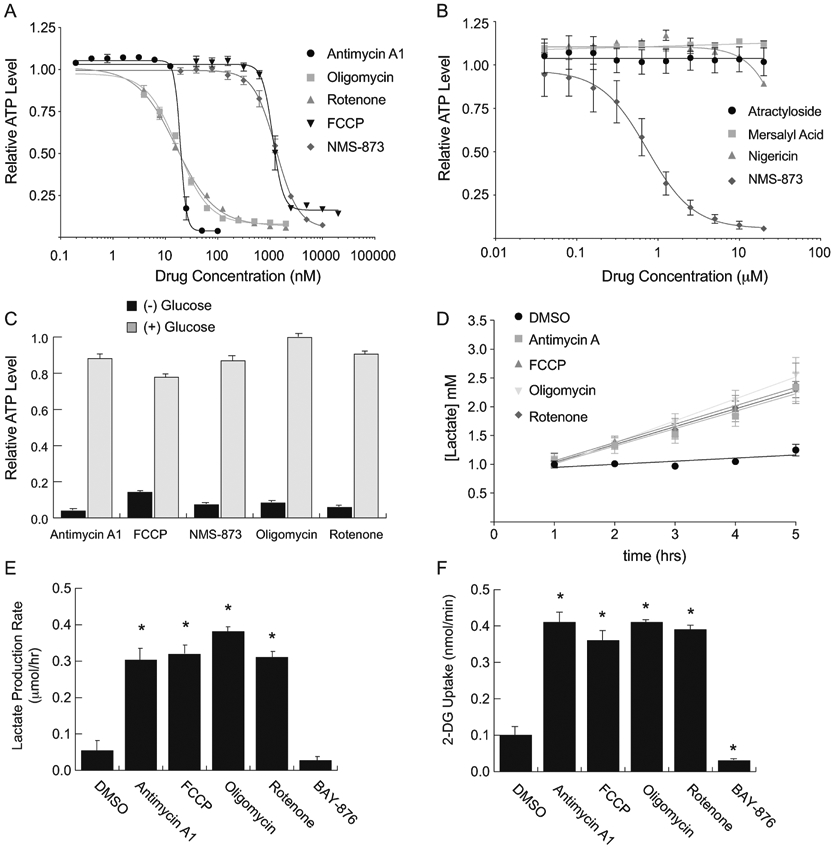
(A and B) Relative ATP concentrations from L929 cells cultured for 1 hour in glucose-free media in the presence of serially-diluted inhibitors of mitochondrial respiration. Values were normalized to cells incubated in vehicle only (0.4% DMSO or water) and fit to standard dose-response inhibition curves to calculate IC50 values for each drug. (C) Relative ATP concentrations from L929 cells cultured for 1 hour in glucose-free media (−) or media containing 10 mM glucose (+). Saturating doses of each inhibitor from panel A were used to block oxidative phosphorylation. [Antimycin A1, 100 nM; FCCP, 20 μM; NMS-873, 1 μM; Oligomycin, 10 μM; Rotenone, 1 μM]. (D) Time-course measurement of lactate production by L929 cells treated with saturating doses of each of the inhibitors from panel A. (E) Lactate production rate [μmol/hr] calculated from the slopes of each line in panel D. (F) Glucose uptake rates from L929 cells treated with saturating doses of each inhibitor from panel A. The selective GLUT1 inhibitor BAY-876 (1 μM) was included in panels E and F to demonstrate that basal lactate production by L929 cells is primarily obtained by glucose fermentation. Error bars represent standard deviation of triplicate values for each panel. [*, p-value < 0.05].
3.4. NMS-873 is a weak inhibitor of ATP Synthase
Having established that NMS-873 displays a similar pattern of toxicity to that of known OXPHOS inhibitors, we sought to determine its mechanistic target in the mitochondria. A relatively simple means that can be used to discriminate among potential inhibitory mechanisms is to measure the impact of a given OXPHOS inhibitor on mitochondrial membrane potential. This can be performed using the pH-sensitive cationic dye TMRE, which spontaneously localizes to mitochondria and produces a fluorescent signal in proportion to the magnitude of the proton gradient across the inner mitochondrial membrane [20]. In this assay, inhibitors that uncouple the proton gradient typically cause a loss of fluorescence, whereas those that inhibit ATP synthase cause an increase in fluorescence.
We performed the mitochondrial polarization assay in both HK-2 and L929 cells and found consistent results for the various OXPHOS inhibitors. Of particular note, the proton gradient uncoupler FCCP caused a significant decrease in polarization, whereas the ATP synthase inhibitor oligomycin and the potassium/proton ionophore nigericin both caused a significant hyperpolarization (Figure 4A). NMS-873 produced an increase in fluorescent signal that was similar to what was observed for oligomycin and nigericin, though this effect was dose-dependent and required relatively high concentrations of the drug (Figure 4B).
Figure 4 – NMS-873 is a weak inhibitor of ATP Synthase.
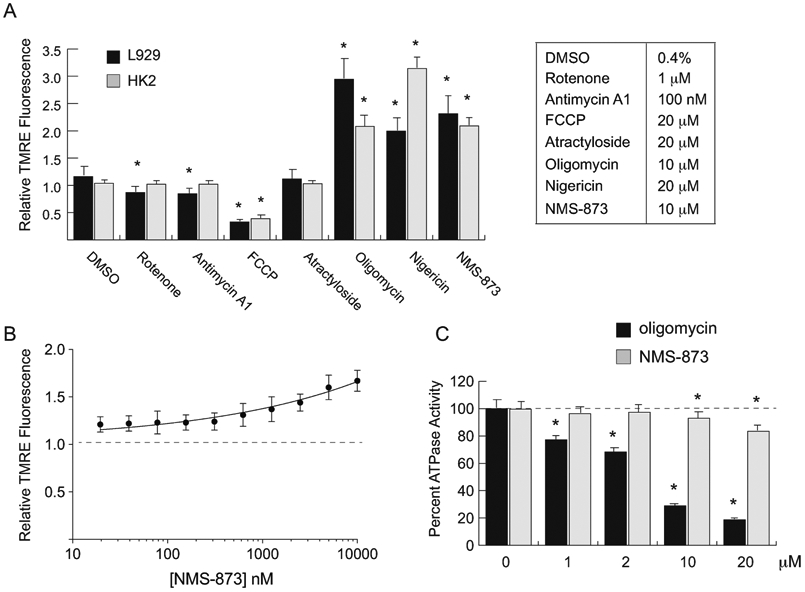
(A) Relative fluorescent intensity of HK-2 or L929 cells stained with the mitochondrial polarization indicator TMRE after treatment with the indicated inhibitors for 1 hour in complete media. Fluorescent signals were normalized to L929 cells incubated in vehicle only (0.4% DMSO or water). Saturating doses of each inhibitor (see table) were used. (B) Relative fluorescent intensity of L929 cells stained with TMRE after treatment with a serial dilution of NMS-873 for 1 hour in complete media. Fluorescent signals were normalized to L929 cells incubated in vehicle only (dashed line). (C) Percent in vitro ATPase activity of bovine mitochondrial ATP synthase in the presence of various doses of the known inhibitor oligomycin or NMS-873. ATP synthase activity over the course of the 30-minute assay was normalized to mitochondria treated with vehicle only (0.1% DMSO). Error bars represent standard deviation of triplicate values for panels A and B and quadruplicate values for panel C. [*, p-value < 0.05].
Because NMS-873 inhibited mitochondrial ATP production and increased mitochondrial polarization in a manner similar to oligomycin, we next tested whether it could directly inhibit the activity of ATP synthase. To accomplish this, we performed an in vitro assay with isolated mitochondria in which the ATPase activity of ATP synthase was tested against various concentrations of NMS-873 and oligomycin, a known inhibitor of ATP synthase (Figure 4C). While oligomycin produced the expected decrease in ATP synthase activity, NMS-873 only weakly inhibited the enzyme at very high concentrations that vastly exceeded its IC50 for ATP production. Though puzzling in view of its stronger impact on ATP synthesis, the weak effect on ATP synthase paralleled the dose-dependency of NMS-873 on membrane polarization. These data suggest that while NMS-873 may be a weak inhibitor of ATP synthase, this activity does not fully explain its potency in the cell-based assays described above.
3.5. NMS-873 is a potent inhibitor of mitochondrial respiratory Complex I
Due to its relatively weak effect on ATP synthase, we next sought to determine whether NMS-873 might also inhibit other complexes in the mitochondrial electron transport chain. One means of distinguishing between the activities of various respiratory complexes is to initiate ATP synthesis at distinct levels in the electron transport chain. This can be accomplished by providing different molecular fuels to isolated mitochondria and measuring their capacity to produce ATP in real-time. To allow our assay to most closely mimic our previous live-cell system, we instead utilized digitonin-permeabilized L929 cells, which are incapable of performing substrate-level phosphorylation due to the evacuation of their cytoplasmic contents [18]. Once isolated and permeabilized, these cells can be used to produce ATP in a defined respiratory medium containing ADP and a molecular fuel to drive OXPHOS. Use of malate as a fuel specifies electron flow through NADH oxidation at Complex I, while use of succinate as a fuel specifies electron flow through FADH2 oxidation at Complex II (Figure 5A,B).
Figure 5 – NMS-873 is a potent inhibitor of respiratory Complex I.
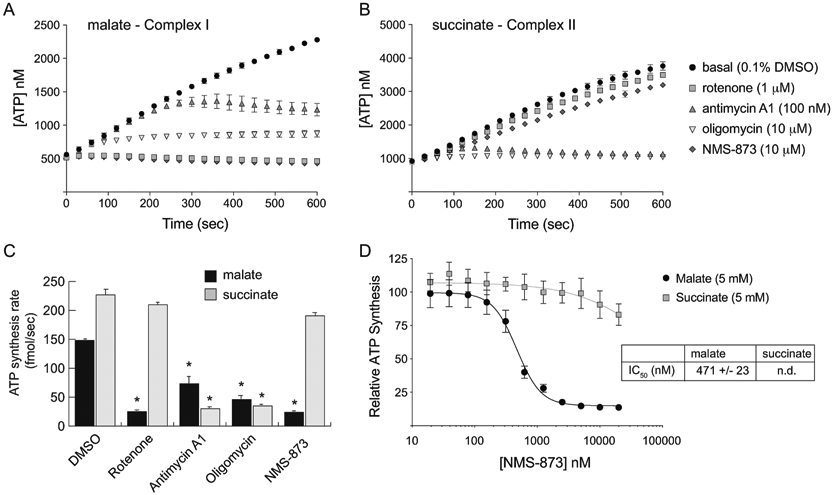
(A and B) Time-course measurement of ATP production by digitonin-permeabilized L929 cells in respiratory medium supplemented with 5 mM malate (A) or 5 mM succinate (B). The cells were treated with the indicated concentration of each respiratory complex inhibitor over the entire course of the experiment. (C) ATP production rate [fmol/sec] per 6,000 cells was calculated from the slopes of each line in panels A and B. (D) The relative ATP synthesis rate was determined as in panel C using a two-fold serial dilution of NMS-873 in each of the respiratory medias (malate and succinate) and fit to standard dose-response inhibition curves to calculate an IC50 value for NMS-873 relative to Complex I. Error bars represent standard deviation of triplicate values. [*, p-value < 0.05].
As expected, ATP synthesis from malate and succinate are both blocked by inhibitors that function downstream of Complex II, such as antimycin A1 and oligomycin (Figure 5A-C). In contrast, the Complex I inhibitor rotenone only blocks ATP synthesis when malate is used as a fuel of OXPHOS and has virtually no effect when succinate is used. Remarkably, NMS-873 functions in a similar fashion to rotenone when used at 10 mM concentration, suggesting that it too acts as a Complex I inhibitor (Figure 5C). To ensure that this inhibitory effect on Complex I more closely matches the dose required to kill live cells and inhibit OXPHOS, we repeated the assay across a broad range of NMS-873 using both malate and succinate as fuels (Figure 5D). Curve-fitting of the data when malate is used as a substrate provided an IC50 of 471 nM, which is much closer to the doses of NMS-873 required for cellular toxicity (Figure 2C) and inhibition of OXPHOS (Figure 3A,B) seen in live L929 cells. It is notable that at very high concentrations of NMS-873 (≥10 μM), a 10-15% inhibition of ATP synthesis is also observed when succinate is used as a fuel of OXPHOS. We attribute this effect to the weak inhibitory activity of NMS-873 on ATP synthase, which—like oligomycin—would be expected to inhibit ATP synthesis regardless of the organic fuel used to drive OXPHOS.
4. Discussion and conclusions
Since the advent of chemotherapy in the early 1940s, the field of clinical oncology has continuously sought to identify new compounds with increased potency toward cancer cells and decreased toxicity toward normal tissues. Much of this work has relied upon well-known functional distinctions between transformed and normal cells, such as differences in growth rate, metabolism, or DNA repair capacity. Drug screening strategies that rely upon these functional categories of distinction, however, often suffer from a lack of specificity because they target entire processes rather than specific molecular differences that emerge during the process of oncogenic transformation. This realization has led to increasing demand for molecular biomarkers of drug efficacy and for specific enzymatic targets whose expression and activity are distinctly altered in cancer cells relative to their normal counterparts.
One of the more recently identified targets for chemotherapy is the proteasome, which is required for a wide variety of proteolytic functions in both normal and cancerous cells [21]. Changes to protein metabolism that are induced by oncogenic transformation or chemotherapeutic stress often necessitate increased proteolytic flux through the proteasome, which has been exploited for combinational therapy with the clinically approved proteasome inhibitor bortezomib [22,23]. While this approach has found considerable success in a variety of cancer types including multiple myeloma and mantle cell lymphoma, it comes as the cost of toxicity to the peripheral nervous system and other tissues. As such there remains continued interest in identifying enzymatic targets that mediate selective aspects of proteosome function in cancer cells.
The AAA ATPase VCP/p97 serves as an important adaptor for proteosomal function based on its ability to segregate specific protein substrates for degradation [14]. Several prior studies have shown that VCP/p97 expression and function are increased in various types of cancer and that cancer cells rely upon this alteration for survival and proliferation [13,15,16]. Although substrates of VCP/p97 are certainly more restricted than those of the proteasome at large, it is unlikely that any single substrate or pathway can account for the importance of its activity across various cancer types. Instead, it seems likely that the role of VCP/p97 in cancer is pleiotropic and context-dependent rather than pathway-specific [14].
While this observation does not necessarily diminish the potential utility of VCP/p97 as a chemotherapeutic target, it does present a significant challenge in the context of drug discovery due to the lack of a specific biomarker for activity. Careful pairing of RNA interference or CRISPR/cas9-mediated gene deletion along with drug screening is therefore necessary to verify that the impact of a putative VCP/p97 inhibitor matches genetic loss-of-function phenotypes. This carefully controlled approach was properly employed in HCT116 cells to allow for the initial identification of NMS-873 as an allosteric inhibitor of VCP/p97, which provided relatively strong confidence that the cytotoxic effects of NMS-873 on these cells was a result of target engagement rather than other off-target effects [7]. Subsequent work aimed at identifying suppressor mutations in VCP/p97 have clearly shown that cytotoxicity of NMS-873 in full-nutrient media—such as McCoy’s 5A formulation—can be attributed to its specific inhibition of VCP/p97 [24,25].
In this study we have demonstrated that NMS-873 also functions as a sub-micromolar inhibitor of mitochondrial respiration. This effect was initially observed in the absence of visible toxicity to both human renal cells and to mouse fibroblasts, which we attribute to the presence of glucose in the growth media. Under these conditions both HK-2 and L929 cell lines were able to undergo aerobic fermentation to compensate for the loss of OXPHOS activity. When glucose was removed from the media or its uptake was inhibited pharmacologically, both cell lines showed significant toxicity to NMS-873, consistent with its function as a respiratory inhibitor. Further investigation demonstrated that it specifically functions as a dual inhibitor of Complex I and ATP synthase.
While NMS-873 remains an excellent tool compound for probing VCP/p97 function in cellular proteostasis, inferences regarding its cytotoxicity should be made with caution. In the absence of careful genetic controls, it should not be assumed that decreases in cellular survival or proliferation in response to NMS-873 can be explained simply in terms of VCP/p97 inhibition [7,8,11,26]. Though sensitivity to NMS-873 might well be indicative of reliance on VCP/p97 function, it is equally possible that sensitivity points to a requirement for mitochondrial respiration above the capacity of glycolysis. Similar inferences regarding the viral replication cycle should also be made with great caution due to the energy-intensive nature of this process that could be impacted by OXPHOS inhibition [9,10,12]. As such, suggestions that NMS-873 or its derivatives could be of therapeutic value for various diseases are premature [8,9]. Given the known toxicity of Complex I inhibitors such as rotenone at the level of whole organisms, it seems unlikely that NMS-873 would be sufficiently safe for use in vivo.
Supplementary Material
Highlights.
Off-target effects of pharmacologic inhibitors limit their value in cellular assays
The allosteric inhibitor NMS-873 produces unexpected metabolic effects in vitro
A real-time luminescent assay was utilized to monitor mitochondrial respiration
NMS-873 directly inhibits mitochondrial respiratory Complex I and ATP synthase
The impact of NMS-873 on viability and other cellular functions is multifactorial
Acknowledgements
We wish to thank Lori Keen and David Ross (Calvin University) for their help procuring reagents and support work as lab managers of the Biology and Chemistry Departments, respectively. This research was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases [Grant 1-R15-DK081931].
Abbreviations
- Ap5A
P1,P5-di(adenosine-5’)-pentaphosphate
- CRISPR
clustered regularly interspaced short palindromic repeats
- DMEM
Dulbecco’s Modified Eagle Medium
- DPBS
Dulbecco’s phosphate buffered saline
- DMSO
dimethyl sulfoxide
- ERAD
endoplasmic reticulum associated degradation
- FCCP
trifluoromethoxy-carbonylcyanide-phenylhydrazone
- GLUT
glucose transporter
- OXPHOS
oxidative phosphorylation
- PBS
phosphate buffered saline
- rcf
relative centrifugal forces
- RPMI-1640
Roswell Park Memorial Institute 1640
- SLC2A
solute carrier family 2A
- TCA
trichloroacetic acid
- TMRE
tetramethylrhodamine ethyl ester
- VCP/p97
valosin-containing protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
No conflicts of interest or disclosures are declared by any author involved in this work.
References
- 1.Lynch JJ 3rd, Van Vleet TR, Mittelstadt SW, Blomme EAG. Potential functional and pathological side effects related to off-target pharmacological activity. J Pharmacol Toxicol Methods 2017;87:108–26 doi 10.1016/j.vascn.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 2.Badrinarayan P, Sastry GN. Rational approaches towards lead optimization of kinase inhibitors: the issue of specificity. Curr Pharm Des 2013;19(26):4714–38. [DOI] [PubMed] [Google Scholar]
- 3.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol 2005;12(6):621–37 doi 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Groebe DR. Screening for positive allosteric modulators of biological targets. Drug Discov Today 2006;11 (13-14):632–9 doi 10.1016/j.drudis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Lazareno S, Birdsall NJ. Detection, quantitation, and verification of allosteric interactions of agents with labeled and unlabeled ligands at G protein-coupled receptors: interactions of strychnine and acetylcholine at muscarinic receptors. Mol Pharmacol 1995;48(2):362–78. [PubMed] [Google Scholar]
- 6.Soudijn W, Van Wijngaarden I, AP IJ. Allosteric modulation of G protein-coupled receptors: perspectives and recent developments. Drug Discov Today 2004;9(17):752–8 doi 10.1016/S1359-6446(04)03220-9. [DOI] [PubMed] [Google Scholar]
- 7.Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol 2013;9(9):548–56 doi 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 8.Duscharla D, Reddy Kami Reddy K, Dasari C, Bhukya S, Ummanni R. Interleukin-6 induced overexpression of valosin-containing protein (VCP)/p97 is associated with androgen-independent prostate cancer (AIPC) progression. J Cell Physiol 2018;233(10):7148–64 doi 10.1002/jcp.26639. [DOI] [PubMed] [Google Scholar]
- 9.Lin YT, Prendergast J, Grey F. The host ubiquitin-dependent segregase VCP/p97 is required for the onset of human cytomegalovirus replication. PLoS Pathog 2017;13(5):e1006329 doi 10.1371/journal.ppat.1006329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyupina YV, Erokhov PA, Kravchuk OI, Finoshin AD, Abaturova SB, Orlova OV, et al. Essential function of VCP/p97 in infection cycle of the nucleopolyhedrovirus AcMNPV in Spodoptera frugiperda Sf9 cells. Virus Res 2018;253:68–76 doi 10.1016/j.virusres.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Walworth K, Bodas M, Campbell RJ, Swanson D, Sharma A, Vij N. Dendrimer-Based Selective Proteostasis-Inhibition Strategy to Control NSCLC Growth and Progression. PLoS One 2016;11(7):e0158507 doi 10.1371/journal.pone.0158507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Hu Y, Hau R, Musharrafieh R, Ma C, Zhou X, et al. Identification of NMS-873, an allosteric and specific p97 inhibitor, as a broad antiviral against both influenza A and B viruses. Eur J Pharm Sci 2019;133:86–94 doi 10.1016/j.ejps.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan B, Chai S, Wang P, Wang K. VCP/p97/Cdc48, A Linking of Protein Homeostasis and Cancer Therapy. Curr Mol Med 2017;17(9):608–18 doi 10.2174/1566524018666180308111238. [DOI] [PubMed] [Google Scholar]
- 14.van den Boom J, Meyer H. VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 2018;69(2):182–94 doi 10.1016/j.molcel.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Tsujimoto Y, Tomita Y, Hoshida Y, Kono T, Oka T, Yamamoto S, et al. Elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer. Clin Cancer Res 2004;10(9):3007–12. [DOI] [PubMed] [Google Scholar]
- 16.Valle CW, Min T, Bodas M, Mazur S, Begum S, Tang D, et al. Critical role of VCP/p97 in the pathogenesis and progression of non-small cell lung carcinoma. PLoS One 2011;6(12):e29073 doi 10.1371/journal.pone.0029073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 2011; 13(11):1376–82 doi 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- 18.Fujikawa M, Yoshida M. A sensitive, simple assay of mitochondrial ATP synthesis of cultured mammalian cells suitable for high-throughput analysis. Biochem Biophys Res Commun 2010;401(4):538–43 doi 10.1016/j.bbrc.2010.09.089. [DOI] [PubMed] [Google Scholar]
- 19.Siebeneicher H, Cleve A, Rehwinkel H, Neuhaus R, Heisler I, Muller T, et al. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016;11(20):2261–71 doi 10.1002/cmdc.201600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scaduto RC Jr., Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J 1999;76(1 Pt 1):469–77 doi 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thibaudeau TA, Smith DM. A Practical Review of Proteasome Pharmacology. Pharmacol Rev 2019;71(2):170–97 doi 10.1124/pr.117.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med 2003;348(26):2609–17 doi 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 23.Teicher BA, Ara G, Herbst R, Palombella VJ, Adams J. The proteasome inhibitor PS-341 in cancer therapy. Clin Cancer Res 1999;5(9):2638–45. [PubMed] [Google Scholar]
- 24.Her NG, Toth JI, Ma CT, Wei Y, Motamedchaboki K, Sergienko E, et al. p97 Composition Changes Caused by Allosteric Inhibition Are Suppressed by an On-Target Mechanism that Increases the Enzyme's ATPase Activity. Cell Chem Biol 2016;23(4):517–28 doi 10.1016/j.chembiol.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Toth JI, Blanco GA, Bobkov AA, Petroski MD. Adapted ATPase domain communication overcomes the cytotoxicity of p97 inhibitors. J Biol Chem 2018;293(52):20169–80 doi 10.1074/jbc.RA118.004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeo SK, French R, Spada F, Clarkson R. Opposing roles of Nfkb2 gene products p100 and p52 in the regulation of breast cancer stem cells. Breast Cancer Res Treat 2017;162(3):465–77 doi 10.1007/s10549-017-4149-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.