Abstract
The conceptual basis for a genetic predisposition underlying the risk for developing type 1 diabetes (T1D) predates modern human molecular genetics. Over half of the genetic risk has been attributed to the human leukocyte antigen (HLA) class II gene region and to the insulin (INS) gene locus – both thought to confer direction of autoreactivity and tissue specificity. Notwithstanding, questions still remain regarding the functional contributions of a vast array of minor polygenic risk variants scattered throughout the genome that likely influence disease heterogeneity and clinical outcomes. Herein, we summarize the available literature related to the T1D-associated coding variants defined at the time of this review, for the genes PTPN22, IFIH1, SH2B3, CD226, TYK2, FUT2, SIRPG, CTLA4, CTSH and UBASH3A. Data from genotype-selected human cohorts are summarized, and studies from the non-obese diabetic (NOD) mouse are presented to describe the functional impact of these variants in relation to innate and adaptive immunity as well as to β-cell fragility, with expression profiles in tissues and peripheral blood highlighted. The contribution of each variant to progression through T1D staging, including environmental interactions, are discussed with consideration of how their respective protein products may serve as attractive targets for precision medicine-based therapeutics to prevent or suspend the development of T1D.
Keywords: coding variant, human, precision medicine, risk gene, single nucleotide polymorphism, type 1 diabetes
INTRODUCTION
The development of type 1 diabetes (T1D) occurs as a result of complex genetic, environmental and stochastic determinants interacting to surpass a threshold for disease induction.1,2 The dominant genetic risk for T1D is conferred by the highly polymorphic major histocompatibility complex (MHC) class II region that is responsible for antigen presentation to CD4+ T cells,3 along with the insulin (INS) gene locus.4 Together, these are thought to contribute to tissue-specific T-cell autoimmunity resulting in targeted destruction of insulin-producing β-cells within the pancreatic islets of Langerhans. With the development of large biobanks of peripheral blood from persons with and at-risk for T1D, the improvement in genetic microarray technologies, and the advent of whole genome bioinformatics tools, genome-wide association studies (GWAS) initially identified over 57 additional single nucleotide polymorphisms (SNPs) associated with a minor risk for the disease, with the majority of these loci occurring in the lymphocyte-specific promoter and enhancer regions.5 More recently, with the extension of T1D GWAS subject cohorts from solely European ancestry to include individuals of African and East Asian descent, as well as admixed populations, 36 additional variants with genome-wide significance for T1D were reported, with differential accessibility enriched primarily in effector CD4+ T cells (Teff).6
Although the majority of T1D-associated variants reside in non-coding regions,7 there is considerable interest in the few known coding variants due to the possibility that these may not only impact gene expression in the pancreas and/or immune system (Figure 1), but also, potentially induce loss- or gain-of-function effects. Onengut-Gumuscu et al.5 identified T1D-associated coding variants in seven genes: PTPN22, IFIH1, SH2B3, CD226, TYK2, FUT2 and SIRPG,5 and Robertson et al.6 implicated non-synonymous SNPs (nsSNPs) in three additional genes: CTLA4, CTSH and UBASH3A.6 Here, we describe these ten genes containing T1D-risk coding variants (Table 1), highlighting their relevance in progression through the stages of pre-T1D (Table 2)8 and the potential for precision medicine-based treatment approaches with consideration of possible inter-variant interactions (Figure 2). Notably, many of these variants are implicated in various other T-cell-mediated autoimmune conditions in addition to T1D (Table 1), implying that therapies targeting these specific variants may provide clinical benefit in multiple disease settings.
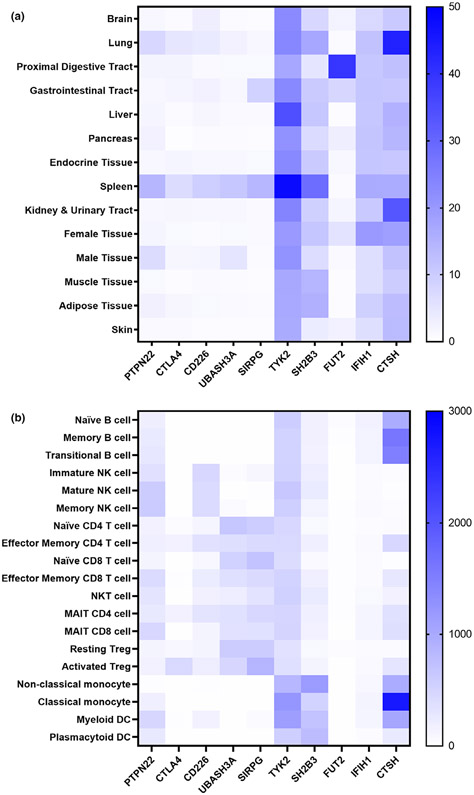
Figure 1.
Expression profiles of genes containing T1D-associated coding variants. (a) Heatmap depicting relative gene expression by organ. Rows are organ classifications utilized in The Human Protein Atlas (HPA), and columns are individual genes containing T1D-associated coding variants. Depicted are the consensus normalized expression (NX) values summarized from HPA, Genotype-Tissue Expression (GTEx), and Functional Annotation of Mammalian Genomes 5 (FANTOM5) transcriptomics datasets.90 (b) Heatmap of gene expression within immune cell subsets. Data are population averaged gene expression from the Human Cell Atlas Census of Immune Cells, an ultra-low-input (ULI) RNA-Seq dataset.91 Blue represents high expression, while white represents low expression levels. Cells indicated as natural killer (NK), mucosal-associated invariant T (MAIT), regulatory T cell (Treg), and dendritic cell (DC).
Table 1.
Coding variants associated with genetic risk for T1D. For each coding SNP, chromosomal region, major and minor alleles, risk allele frequency, odds ratio (OR), amino acid (AA) substitution, prediction of functional effect and association with other autoimmune diseases are presented.
| Gene | Chromosomal region |
T1D associated coding SNP |
Major > minor alleles |
Risk allele |
Risk allele frequency |
T1D OR | Ref for OR |
AA substitution |
SIFT/ PolyPhen predictions |
Association with other autoimmune diseases |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European | African | East Asian |
South Asian |
American | ||||||||||
| PTPN22 | 1p13.2 | rs2476601 | G > A | A | 0.094 | 0.003 | 0.000 | 0.013 | 0.036 | 1.81 | 6 | R620W | Tolerated/benign | RA, CD, JIA, V, MG, UC, CeD |
| CTLA4 | 2q33.2 | rs231775 | A > G | G | 0.359 | 0.388 | 0.637 | 0.310 | 0.463 | 1.20 | 6 | T17A | Tolerated/benign | RA, CD, UC, JIA |
| CD226 | 18q22.2 | rs763361 | C > T | T | 0.473 | 0.737 | 0.388 | 0.513 | 0.452 | 1.10 | 89 | G307S | Tolerated/benign | UC, CD, CeD, MS, RA |
| UBASH3A | 21q22.3 | rs13048049 | G > A | G | 0.949 | 0.973 | 1.000 | 0.990 | 0.981 | 1.19 | 6 | R324Q | Tolerated/possibly damaging | RA, UC, Ps, MS, CeD |
| SIRPG | 20p13 | rs6043409 | G > A | G | 0.660 | 0.825 | 0.848 | 0.858 | 0.782 | 1.14 | 5 | A263V | Tolerated/benign | RA, UC |
| TYK2 | 19p13.2 | rs34536443 | G > C | G | 0.971 | 0.999 | 1.000 | 0.994 | 0.980 | 1.54 | 6 | P1104A | Deleterious/probably damaging | Ps, RA, MS, CeD, UC, Sj |
| rs12720356 | A > C | A | 0.908 | 0.997 | 1.000 | 0.989 | 0.952 | 1.21 | 6 | I684S | Deleterious/possibly damaging | UC, CD, Ps, Sj, CeD, MS, AS, RA | ||
| SH2B3 | 12q24.12 | rs3184504 | C > T | T | 0.464 | 0.019 | 0.003 | 0.069 | 0.254 | 1.27 | 6 | R262W | Tolerated/benign | CeD, CD, Ps, UC, RA, JIA, MS, Sj |
| FUT2 | 19q13.33 | rs601338 | G > A | A | 0.441 | 0.491 | 0.004 | 0.283 | 0.341 | 1.12 | 6 | W154* | Loss of function | CD, MS, Ps |
| IFIH1 | 2q24.2 | rs1990760 | T > C | T | 0.605 | 0.126 | 0.187 | 0.564 | 0.390 | 1.14 | 6 | T946A | Tolerated/benign | UC, Ps, CD, MS, MG |
| rs35667974 | T > C | T | 0.990 | 0.999 | 1.000 | 0.999 | 1.000 | 1.64 | 6 | I923V | Tolerated/probably damaging | UC, Ps, CD | ||
| rs35337543 | C > G | C | 0.981 | 1.000 | 1.000 | 0.998 | 0.990 | 1.58 | 6 | Splice donor variant | Loss of function | UC, Ps | ||
| CTSH | 15q25.1 | rs2289702 | C > T | C | 0.895 | 0.985 | 0.929 | 0.847 | 0.947 | 1.26 | 6 | G11R | Deleterious/benign | - |
Minor allele reported in reference to + strand of DNA from European subjects and risk allele frequencies reported from 1000 Genomes Project phase 3 release V3+. T1D OR shown for risk allele from most recent GWAS including the variant. Predictions of impact on protein structure and function shown from SIFT (Sorting Intolerant From Tolerant) and PolyPhen-2 (Polymorphism Phenotyping v2). The “Association with other autoimmune diseases” column includes conditions in which the risk allele for T1D may carry risk or protection for the noted diseases. Disease associations reported from “Immune system” category of PheWAS from ImmunoBase version 0.4.0 (fcf5738). AS, ankylosing spondylitis; CD, Crohn’s disease; CeD, celiac disease; JIA, juvenile idiopathic arthritis; MG, myasthenia gravis; MS, multiple sclerosis; Ps, psoriasis; RA, rheumatoid arthritis; Sj, Sjogren’s syndrome; SNP: single nucleotide polymorphism; T1D: type 1 diabetes; UC, ulcerative colitis; V, vitiligo.
Table 2.
Coding SNPs associated with progression through the stages of T1D. Presented according to pre-T1D staging from Insel et al.8
| Gene | T1D associated coding SNP(s) | Pre-stage 1: single AAb |
Stage 1: multiple AAb |
Stage 2: glucose intolerance |
Stage 3: clinical diagnosis | References |
|---|---|---|---|---|---|---|
| PTPN22 | rs2476601 | X | - | - | - | 12,26,60 |
| CTLA4 | rs231775 | X | - | X | X | 24,26,27 |
| CD226 | rs763361 | - | - | - | X | 38 |
| UBASH3A | rs13048049 | - | - | - | - | NA |
| SIRPG | rs6043409 | - | - | - | - | NA |
| TYK2 | rs12720356, rs34536443 | - | - | - | - | NA |
| SH2B3 | rs3184504 | X | - | - | - | 60 |
| FUT2 | rs601338 | - | - | X | - | 63,67 |
| IFIH1 | rs35667974, rs1990760, rs35337543 | - | - | - | - | NA |
| CTSH | rs2289702 | - | - | - | - | NA |
X, associated with progression to this stage; - , not previously associated with progression to respective stage; NA, not applicable; SNP, single nucleotide polymorphism; T1D, type 1 diabetes.
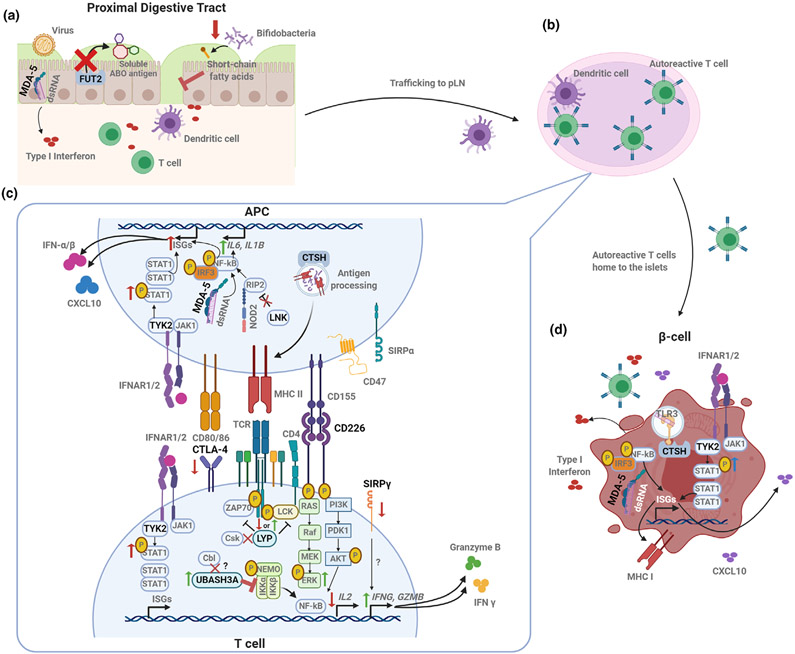
Figure 2.
Coding gene variants converge on signaling and activation pathways to impact autoimmunity. (a) Disruption of FUT2 function and the resulting lack of ABO blood group antigen secretion in the intestinal mucosa can result in impaired barrier function and immunity by increasing susceptibility to some viral infections and altering microbiome composition as well as the microbial metabolites (such as short-chain fatty acids). IFIH1 variants may augment innate responses against enteroviruses. (b) Antiviral or other pro-inflammatory responses in the gut may result in trafficking of APCs to the pancreatic lymph node (pLN), where autoantigen-specific T cells are activated. (c) Activation and function of autoreactive T cells may be exacerbated by T1D-risk variants expressed either in the APC or in the T cell itself. The risk variant of PTPN22 (LYP) abolishes CSK binding, which could result in decreased phosphatase activity and increased TCR signaling. The CTLA4 risk variant results in reduced CTLA4 expression on the T-cell surface, reducing regulation of T-cell activation. Variants in co-stimulatory molecules CD226 and SIRPγ may contribute to pro-inflammatory T-cell skewing by promoting activation of the MAPK/ERK pathway and enhancing the production of inflammatory and cytotoxic molecules, respectively. Treg function may be negatively impacted by a UBASH3A variant, which inhibits NFκB activation and downstream IL-2 production. APCs may contribute to the generation of this pro-inflammatory milieu, as altered LNK function results in enhanced NOD2 signaling, activation of NFκB and production of IL-6 and IL-1β. APC peptide repertoire for presentation may be influenced by a CTSH variant. Variants in TYK2 and IFIH1 may also contribute to innate inflammation through increased induction of IFN stimulated genes (ISGs). (d) Genetic variants may impact β-cell susceptibility to death via sensing of viral or self dsRNA in the case of IFIH1, while a variant within CTSH seems to play a role in TLR3 activation, downstream IFN production and susceptibility to cytokine-induced damage. T1D-associated TYK2 variants may enhance β-cell expression of ISGs, including MHC I and CXCL10. Created with BioRender.
PTPN22
PTPN22 encodes the protein tyrosine phosphatase non-receptor type 22 known as lymphoid tyrosine phosphatase (LYP).9,10 The rs2476601 coding variant in exon 14 of PTPN22 results in an arginine to tryptophan at amino acid 620 (R620W) and is significantly associated with T1D, carrying the highest risk after HLA Class II and INS (Table 1).9,10 In T cells, LYP acts as a negative regulator of T-cell receptor (TCR) signaling, dephosphorylating multiple proximal signaling proteins, including Src family tyrosine kinase (LCK) and the zeta chain of T-cell receptor associated protein kinase 70 (ZAP-70) (Figures 1, 2).10,11 The R620W variant impairs interactions between a proline-rich motif of LYP and CSK, a tyrosine kinase that associates with T-cell receptor scaffolding to modulate signaling.
The R620W mutation has been linked to insulin autoantibodies (IAA) appearing first in children with high-risk HLA or a first-degree relative with T1D (Table 2), potentially driving an earlier age-at-onset.12 However, uncertainty surrounding the impact of R620W on LYP function complicates the investigation of targeted therapeutics for T1D and necessitates further study. While it is widely accepted that rs2476601 alters T-cell activation,10 there is considerable debate concerning whether R620W represents a gain- or loss-of-function variant.9,13 One potential mechanism by which the gain-of-function could predispose to autoimmunity involves reduced activation of regulatory T cells (Tregs), which are required for the suppression of autoreactivity.10,14 Loss-of-function models propose impaired localization of R620W to T-cell receptor signaling machinery, resulting in less efficient dephosphorylation of signaling proteins and increased Teff activation.9,11 Our own studies modulating PTPN22 in the context of CD4+ T-cell activation support the notion of R620W functioning as a hypomorph (unpublished data). Furthermore, LYP modulates several other pathways in multiple hematopoietic lineages, including B cell receptor, toll-like receptor, NLRP3 and integrin signaling.15-21 Ultimately, therapeutic strategies should also consider impacts in these pathways and impacts on shared phosphatases.
CTLA4
CTLA4 encodes cytotoxic T-lymphocyte associated protein 4, an inhibitory checkpoint molecule on T cells, which competes with its activating co-stimulatory counterpart CD28 for binding B-7 molecules (CD80/CD86) on antigen-presenting cells (APCs) (Figures 1, 2). Previous T1D GWAS have identified one missense, one intronic and four intergenic CTLA4-associated variants.6 rs231775 is an A > G missense mutation that results in a T17A substitution (Table 1) within the signal peptide region and has been suggested to impair protein glycosylation, in turn, inhibiting trafficking of CTLA4 to the cell surface.22 Indeed, in a Chinese Han cohort of healthy subjects without an autoimmune diagnosis, heterozygosity or homozygosity for the T1D-risk genotype (G) at rs231775 was associated with reduced CTLA4 expression on circulating naïve and activated Treg subsets.23
The constitutive expression of CTLA4 on Tregs suggests that the rs231775 autoimmune-associated variant may negatively impact Treg function, potentially allowing for uncontrolled Teff activation. In support of this notion, T1D donors of Chilean origin possessing the rs231775 homozygous risk genotype displayed higher serum levels of the pro-inflammatory cytokines, TNFα and IFNγ, compared with donors with the homozygous protective genotype.24 While the rs231775 risk allele has been associated with increased immunoregulatory cytokine production by primary human peripheral blood mononuclear cell (PBMCs) in response to enterovirus, a speculated trigger of T1D,25 additional studies are required to examine the role of this SNP in modulating β-cell specific responses, particularly in combination with other T1D risk alleles. The Environmental Determinants of Diabetes in the Young (TEDDY) study recently identified that the T1D susceptible allele of rs231775 was associated with risk of developing glutamic acid decarboxylase autoantibodies (GADA) at initial seroconversion (Table 2),26 potentially implicating that this variant may associate with a later age-at-onset of T1D.
There is substantial interest in interventions targeting CTLA4 to augment T-cell regulation and thereby prevent or delay autoimmunity.27,28 Treatment with Abatacept (CTLA4-Ig) initially delayed the decline of β-cell function in recent-onset T1D subjects, though 1 year later, the rate of decline mirrored the placebo group.27 Trials are currently approved to evaluate Abatacept alone and in combination with Rituximab (anti-CD20) for the ability to prevent the progression of T1D in participants with Stage 1 disease (i.e. positive for two or more islet autoantibodies with normal glucose tolerance; NCT03929601 and NCT01773707), expected to initiate after SARS-CoV-2-related holds.
CD226
CD226 (also known as DNAX accessory molecule-1; DNAM-1) is a co-stimulatory molecule in the immunoglobulin (Ig) superfamily known to augment effector/memory T-cell and natural killer (NK) cell activation (Figures 1, 2).29 Similar to the CD28/CTLA4 axis, CD226 competes with its immunoregulatory counterpoint, TIGIT, to bind CD155 expressed on APCs. rs763361 is a C > T missense mutation in CD226 (Table 1)30 with the resulting G307S located in the cytoplasmic tail of CD226 near two intracellular phosphorylation sites, Tyr322 and Ser329.31 This introduction of an additional serine (S) has been hypothesized to provide an extra phosphorylation site, thereby amplifying downstream PI3K/AKT and MAPK/ERK signaling (Figure 2). Indeed, in vitro anti-CD226 and anti-CD3 co-activation of genotype-selected human primary CD4+ T cells showed enhanced p-ERK induction in subjects carrying the rs763361 T1D-risk allele, although p-AKT induction was not rs763361 genotype-dependent.32 In addition to potential modulation of CD226 signaling, our group has shown that CD226 protein expression is augmented in NK cells of subjects with T1D.33
The ERK pathway regulates T-cell activation and skewing, and accordingly, the rs763361 risk allele was associated with increased in vitro skewing of Th17 and Th1/17 populations,32 with the latter in particular being widely considered diabetogenic. There is a need for additional studies to determine the impact of rs763361 on CD8+ T cell, Treg and NK cell function specifically. In support of this notion, our group has characterized a novel Cd226 knockout (KO) non-obese diabetic (NOD) mouse strain, observing attenuated T1D onset with reduced peripheral CD8+ T-cell activation and avidity of the immunodominant islet-reactive CD8+ T-cell specificity, islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP).34 Our studies of the impact of CD226 on human Treg function showed that CD226+TIGIT− Tregs lack Helios expression, have reduced suppressive capacity and increased effector cytokine production,35 suggesting that the deletion of Cd226 may also provide disease protection via enforcement of Treg lineage stability. Further evaluation of Treg-specific Cd226 KO NOD mice and gene-edited human PBMC subsets are currently ongoing.
The T1D risk-associated T allele of rs763361, in combination with the tightly linked G allele of rs727088, has been associated with an increased risk of severe influenza A (H1N1) infection,36 representing a potential viral trigger of islet autoimmunity.37 Notably, patients with T1D of less than 2 years duration who were homozygous for the rs763361 risk allele showed lower fasting serum C-peptide levels compared with individuals heterozygous or homozygous for the protective allele (Table 2).38 While replication of these observations will be necessary, these data support evaluation of precision medicine efforts with CD226 blocking antibodies or TIGIT-Ig fusion proteins to block CD155 interactions in strategies aimed at disease prevention or the maintenance of residual C-peptide post-onset of T1D.
UBASH3A
UBASH3A encodes the ubiquitin-associated and SH3 domain-containing A protein, which serves as a negative regulator of TCR signaling.39 Downstream of the TCR, UBASH3A inhibits activation of the IKK complex, thereby restricting NF-κB signaling and downstream IL-2 production (Figure 2), as shown using the Jurkat T-cell line.40 The majority of credible T1D-associated variants in UBASH3A thus far are intronic, with evidence suggesting that the risk variants increase the expression of UBASH3A within TCR-activated primary CD4+ T cells, resulting in decreased IL-2 production.40,41 Moreover, the Diabetes Autoimmunity Study in the Young (DAISY) observed that non-coding UBASH3A risk variants were associated with progression from islet autoimmunity to overt disease, and children possessing these variants were more likely to develop T1D by the age of 15.42 Despite decreased UBASH3A expression being associated with T1D protection in humans, murine studies have shown complete Ubash3a deficiency to accelerate T1D43 and to protect from systemic Candida albicans infection,44 suggesting that UBASH3A levels require fine-tuning to simultaneously prevent infections and to avoid autoimmunity.
Recently, a UBASH3A coding variant, rs13048049, which is a G > A missense variant resulting in an arginine to glutamine substitution at position 324 (R324Q), was associated with T1D protection (Table 1). This mutation is located within the SH3 domain, which has previously been shown to interact with the ubiquitin ligase Cbl and dynamin to facilitate downmodulation of the TCR-CD3 complex, thus negatively regulating TCR signaling.45 Mutating a nearby residue (W279L) in the SH3 domain has been shown to impair binding to dynamin;45,46 however, the impact of R324Q on similar protein–protein interactions and ultimately, T-cell signaling remains unknown. Importantly, previous studies demonstrating the impact of non-coding variants on IL-2 expression warrant investigation into the influence of rs13048049 on Treg function, as IL-2 signaling is critical for the maintenance of Treg stability.47
SIRPG
SIRPG, which encodes signal-regulatory protein gamma (SIRPγ), has recently been associated with T1D.48-50 SIRPγ is found predominantly on T cells and NK cells and as a member of the SIRP family, includes conserved extracellular regions containing three Ig-like domains (D1–D3).51,52 While it has no cytoplasmic signaling domain, SIRPγ can engage and promote signaling downstream of its binding partner, integrin-associated protein (IAP; CD47) (Figures 1, 2).48,49,51,52 CD47 is an Ig superfamily protein, which provides a “don’t-eat-me” signal to phagocytes, promoting survival of the SIRP family expressing T and NK cells and regulating their activation.48,49,51,52
Two SNPs in SIRPG have been observed in T1D GWAS: rs2281808 (C > T; intronic) and rs6043409 (G > A; A263V) (Table 1).48,51,52 Decreased SIRPγ expression on CD8+ T cells has been associated with a lowered activation threshold and enhanced cytotoxic potential,48 suggesting SIRPγ would function as a negative regulator of activation. The V263A mutation, rs6043409, is located in the extracellular D3 domain, the function of which is currently unknown. The proximity of D3 to the D1 and D2 domains, which bind CD47 and mediate SIRPγ dimerization, suggests that V263A may inhibit either of these processes and thereby promote CD8+ T-cell activation.49
Compared with SIRPγ, more is known about the function of SIRPα, which competes to bind CD47. SIRPα is expressed mainly on macrophages and dendritic cells (DCs) and inhibits phagocytosis of CD47+ cells.48,49,51,52 In the absence of functional CD47-SIRPα signaling, T cells have been shown to exhibit a more activated phenotype.53 Although this study did not examine the effects of SIRPγ expression on T-cell immunoregulation, the binding of SIRPγ to CD47 has been hypothesized to induce similar effects to SIRPα-CD47.48,49,51,52 The paucity of evidence concerning the role of SIRPγ in T1D indicates a need for additional studies to determine the mechanisms by which the associated variants promote disease.
TYK2
TYK2, a Janus tyrosine kinase (JAK) family member, is an essential regulator of cytokine and type I interferon (TI-IFN) signaling (Figure 2).54 In the context of T1D, TYK2 enhances antigen presentation by stimulating MHC class I expression and promotes CXCL10 chemokine expression, resulting in T-cell activation and recruitment toward pancreatic islets (Figure 1, 2).54 rs12720356 (A > C; I684S) and rs34536443 (G > C; P1104A) are missense mutations in the respective pseudokinase and kinase domains encoded by TYK2. The minor C alleles of both SNPs are speculated to confer T1D protection by reducing TYK2 activity (Table 1).55
A recent study presents the T1D-protective rs34536443 variant as significantly impairing TYK2 activity.55 PBMCs homozygous for the T1D-protective allele of rs34536443, but not rs12720356, showed decreased phosphorylation of TYK2, STAT1 and STAT3 in response to TI-IFN in vitro.55 Furthermore, decreased pSTAT4 and pSTAT3 induction were observed in response to IL-12 and IL-23, respectively, in CD4+ memory T cells in subjects homozygous for the rs34536443 T1D-protective allele, leading to reduced diabetogenic Th1 and Th17 populations.55 While the T1D-protective allele of rs34536443 clearly attenuates cytokine responses, the impact of rs12720356 on TYK2 function remains uncertain.
Interestingly, a natural loss-of-function Tyk2 mutation in the B10.Q/J mouse strain results in a decreased response to TI-IFN and pro-inflammatory cytokines, leaving mice more prone to Toxoplasma gondii infections,56 demonstrating the necessity for appropriate TYK2 expression in order to avoid infection while preventing autoimmunity. Since a cohort of individuals homozygous for the T1D-protective rs34536443 variant showed no sustained immune impairment, susceptibility to viral infections, or immunodeficiency aside from decreased cytokine signaling, the partial inhibition of TYK2 is considered a promising therapy for T1D.55 Notably, a clinical trial of subjects with recent-onset T1D treated with Imatinib, a tyrosine kinase inhibitor, showed some efficacy in sustaining stimulated C-peptide responses after 1 year of treatment (NCT01781975). Recently developed TYK2-specific inhibitors may avoid the undesired effects of nonspecific tyrosine kinase inhibitors.57
SH2B3
SH2B3, which encodes LNK (lymphocyte adaptor protein), is ubiquitously expressed with high levels observed in hematopoietic and endothelial cells (Figure 1).50 The T1D risk variant rs3184504 is a C > T (R262W) missense mutation (Table 1) within exon 3, which encodes the pleckstrin homology (PH) domain responsible for the membrane localization of this protein.58 In addition to interrupting membrane localization, rs3184504 is predicted to disrupt an exonic splicing enhancer (ESE) motif for a splicing regulator, SRp55, leading to speculation that the SNP may result in novel LNK isoforms with potentially disrupted functionality.56
Importantly, LNK regulates several signaling pathways involved in hematopoiesis, cytokine signaling, TCR signal transduction and cell migration.58 The presence of the T1D-risk variant (T) is associated with augmented IL-1β and IL-6 production as a consequence of activation of the nucleotide-binding oligomerization domain-containing protein 2 (NOD2) pathway (Figure 2).59 Consequently, the T allele is associated with protection from bacterial infections.60 In addition to the overall risk of T1D, the SH2B3 T allele has been associated with initial islet autoantibody development in genetically at-risk HLA-DR4/4 and -DR4/8 individuals (Table 2).60 Unfortunately, the NOD mouse model lacks an orthologous LNK risk variant,61 precluding the use of this model in studying the contributions of rs3184504 to T1D. This emphasizes the need for isogenic modeling of the risk variants with human samples, or the creation of orthologous Sh2b3 NOD models to investigate the mechanistic impacts of this risk variant.
FUT2
FUT2 encodes fucosyltransferase 2, an enzyme required for production of the H antigen (the base of the ABO blood group antigens) expressed specifically in the intestinal mucosa and secreted bodily fluids (Figures 1, 2).62 Disruption of FUT2 results in the absence of ABO secretion, as observed in individuals homozygous for the T1D-associated variant, rs601338 G > A (Table 1).63 The SNP converts a tryptophan (W) at position 154 to a stop codon, resulting in premature termination of FUT2 translation and loss of function.
ABO antigen expression on the mucosa impacts binding of environmentally acquired pathogens and commensal microbiota. The non-secretor genotype has shown associations with susceptibility to mucosally acquired infections such as measles and mumps.64 Although the association of these particular infectious diseases with T1D development has been rather tenuous, the fertile field hypothesis supports the notion that viral response(s) rather than a specific virus could trigger the development of T1D.65 Since the Azad et al.64 study implicating measles and mumps used self-reporting of infection,64 additional work should address the question of ABO secretor status and viral infection in a non-biased manner such as whole-virome analysis. Additionally, the fecal microbiota of non-secretor rs601338 AA subjects were found to carry a decreased abundance of probiotic Bifidobacteria, a genus capable of producing immunoregulatory short-chain fatty acids (SCFA) and promoting gut barrier integrity critical for preventing commensal-induced autoimmunity (Figure 2).66 FUT2 is an interesting example of genetics determining the ability for environmental factors to potentially contribute to T1D.
The rs601338 risk allele has been associated with a steeper deterioration of first-phase insulin response (FPIR) in children with multiple T1D-associated autoantibodies,67 which could possibly explain the age-at-diagnosis dependence on secretor status (Table 2).63 These observations suggest that therapeutics rescuing FUT2 function or downstream impacts would likely require intervention in genotype-selected patients at an early age. Generally regarded as safe (GRAS) agents such as human milk oligosaccharides or probiotics to augment Bifidobacteria abundance68 should be evaluated in clinical trials for the prevention of islet autoimmunity initiation, as well as the maintenance of glucose-stimulated insulin secretion in subjects with multiple autoantibodies.
IFIH1
Interferon induced with helicase C domain 1 (IFIH1), also known as melanoma differentiation-associated protein 5 (MDA5), is an innate immune receptor that senses intracellular RNA, initiating an antiviral response by inducing TI-IFN expression (Figures 1, 2). Three SNPs in IFIH1, rs35667974 (T > C; I923V), rs1990760 (T > C; T946A) and rs35337543 (C > G; splice donor variant) have been identified as protective in T1D GWAS (Table 1). rs35667974 and rs1990760 are missense mutations in the C-terminal regulatory domain (CTD), and rs35337543 causes an in-frame deletion of exon 8, resulting in the loss of part of the helicase domain.6,69 While both of these regions are thought to facilitate IFIH1 binding to dsRNA, electrophoretic mobility shift assays (EMSA) using 32P-labeled dsRNA analog, poly(I: C), showed that rs35667974 and rs1990760 did not impact RNA binding to IFIH1.70 rs35337543 has yet to be interrogated in similar EMSA assays. Importantly, in vitro cultures of Mda5−/− mouse embryonic fibroblasts (MEFs) transfected with human IFIH1 variants and a luciferase reporter with an IFN-β promoter showed that the T1D-protective allele of rs35667974 caused decreased luciferase activity in response to poly(I:C) compared with the T1D-protective variant at rs1990760 and both T1D-risk variant-transfected MEFs.70 While the mechanism by which these SNPs may impair IFIH1 function remains unclear, the CTD contains a zinc binding site that is unnecessary for dsRNA binding but required for IFIH1 signaling,71 the function of which should be studied in the context of the CTD-located T1D variants.
There is a growing body of evidence implicating dsRNA, either from enteroviral infection or as a byproduct of dysregulated mRNA processing, as an important factor in the etiology of T1D.72 In support of this notion, the risk allele of rs35667974 is more frequently observed in patients with T1D with detectable enteroviral RNA than in those without (Figure 2).73 Moreover, PBMCs from individuals homozygous for the rs35667974 T risk allele had a greater IFN-β secretion in response to poly(I:C) stimulation than those with at least one copy of the T1D-protective allele. However, enteroviruses more commonly observed in T1D, such as Coxsackie B virus (CVB), are capable of inhibiting IFIH1 to evade this antiviral response,74-76 raising important questions surrounding whether viral modulation of host TI-IFNs contributes or deflects from autoimmunity. Similar to rs35667974, the T risk allele of rs1990760 has been shown to elevate basal and ligand-induced TI-IFN response by human PBMCs, and knock-in of the risk allele in C57BL/6J mice enhanced streptozotocin-induced diabetes incidence.77 These studies suggest that the C alleles for rs35667974 and rs1990760 may confer resistance to T1D via efficient clearance of enterovirus, decreased TI-IFN persistence, and the avoidance of prolonged β-cell damage. Accordingly, there is significant interest in developing CVB vaccines to avoid persistent infection and exposure of the islets to TI-IFN-mediated stress.78 Additional strategies to reduce proinsulin processing defects observed in T1D79 may also reduce intracellular stress to prevent “sterile” innate immune responses that would be expected to increase β-cell vulnerability for autoimmune attack.
CTSH
Cathepsin H (CTSH) is a lysosomal proteinase that plays a role in protein recycling, prohormone processing and MHC class II antigen presentation (Figure 2). While CTSH is ubiquitously expressed, it is most highly observed on type II lung alveolar cells for surfactant protein maturation80 and in APCs such as B cells, monocytes and DCs (Figure 1). Accordingly, the balance between cathepsins and endogenous inhibitors, such as cystatins, is thought to determine the peptide repertoire presented on class II MHC (Figure 2). The role of cathepsins in disease development has been demonstrated via knockout of cathepsin L in NOD mice, which induced diabetes protection concomitant with increased Treg frequency, presumably via altered thymocyte selection.81 Knockout of Ctsh in C57BL/6 mice did not induce any overt effects on the overall physiology, including lung performance,80 but additional studies are required in mice with T1D permissive MHC to determine whether Ctsh KO impacts the immune system in a manner similar to that observed in the Ctsl KO strain.
For rs2289702 in exon 1 of CTSH, the minor allele (C > T; G11R) was found to be protective in T1D (Table 1). This missense mutation, located in the signal peptide sequence, may affect CTSH cleavage to its active form and trafficking to lysosomes82 or decrease mRNA expression, potentially due to signal sequence coding region effects on mRNA trafficking from the nucleus.83 The T allele demonstrates protection from the early onset of T1D, particularly in patients younger than 7 years of age.83 One potential explanation for this finding is that reduced CTSH expression may decrease N-terminal cleavage of toll-like receptor 3 (TLR3), impairing TLR3 functionality and decreasing TI-IFN expression in response to viral infections in early childhood.83 The relationship between CTSH expression and T1D is further complicated by a report of CTSH overexpression inducing β-cell-intrinsic protection from cytokine-mediated damage and stimulation of insulin production.84 These studies support the continued investigation of CTSH modulation as a potential means of T1D prevention while paying careful attention to off-target effects due to the widespread expression of this protein.
CONCLUSIONS
Although the field has progressed considerably since initial T1D GWAS, our current understanding of genotype:phenotype interactions in T1D leaves many outstanding questions. Hence, the development of novel murine in vivo, human ex vivo and isogenic in vitro model systems and clinical studies will be required to further elucidate the impacts of T1D-associated risk variants, with investigation of the 13 SNPs altering amino acid sequence representing high value targets. To circumvent confounding variability in background genetics, continued improvement of induced pluripotent stem cell (iPSC) differentiation methods and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene-editing protocols may enable isogenic modeling, as reviewed by our group.50 Briefly, isogenic in vitro models of thymic selection, leukocyte extravasation through endothelium and β-cell:immune cell interactions should be improved to more accurately mimic in vivo processes.50 Newly developed “islet-on-a-chip” engineering methods may further enhance the modeling of the islet microenvironment and vasculature using microfluidics,85 and humanized mouse models may prove important for demonstrating complex in vivo interactions.86
With comprehensive understanding of T1D genetics comes the possibility of expression quantitative trait loci (eQTL) and nsSNP haplotype analysis to understand how not only protein quality, but also quantity, are modified in T1D. Combinatorial SNP analysis will allow for improved disease endotype definitions87 as well as the enhanced prediction of T1D through the use of genetic risk scores.88 Application of genotype to therapeutic strategies will likely require a precision medicine-based approach with combinatorial therapies targeting dysregulated pathways. Accordingly, subjects with pre-T1D or those recently diagnosed with TYK2 and IFIH1 risk variants may preferentially benefit from therapies inhibiting the TI-IFN pathway, while those with PTPN22, CD226, CTLA4, UBASH3A and/or SIRPG risk variants may show enhanced responses to drugs modulating T-cell costimulation. Antigen-specific tolerance efforts may also prove useful in HLA-DR3 subjects with CTLA4 risk or HLA-DR4 with PTPN22 risk prone to initial seroconversion to GADA or IAA, respectively.12,26,87 Ultimately, the incorporation of genotype into clinical trial design and responder analyses provides promise in future attempts to prevent or suspend heterogeneous T1D progression.
ACKNOWLEDGMENTS
We regret that the full extent of the relevant literature could not be cited due to editorial limitations; we have judiciously selected references to recent publications that we viewed as most pertinent.
FUNDING
Efforts related to the content reviewed herein are supported by grants from the National Institutes of Health (P01 AI042288, R01 DK106191 and HIRN UC4 DK104194 to TMB; T32 DK108736 to LP) as well as The Leona M. and Harry B. Helmsley Charitable Trust.
Footnotes
CONFLICT OF INTEREST
The authors declare that no relevant conflicts of interest exist.
REFERENCES
- 1.Wasserfall C, Nead K, Mathews C, Atkinson MA. The threshold hypothesis: solving the equation of nurture vs nature in type 1 diabetes. Diabetologia 2011; 54: 2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyaga DM, Vickers MH, Jefferies C, Perry JK, O’Sullivan JM. The genetic architecture of type 1 diabetes mellitus. Mol Cell Endocrinol 2018; 477: 70–80. [DOI] [PubMed] [Google Scholar]
- 3.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 1996; 59: 1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 4.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 1984; 33: 176–183. [DOI] [PubMed] [Google Scholar]
- 5.Onengut-Gumuscu S, Chen WM, Burren O, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015; 47: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson CC, Inshaw JRJ, Onengut-Gumuscu S, et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. bioRxiv 2020: 2020.2006.2019.158071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant SFA, Wells AD, Rich SS. Next steps in the identification of gene targets for type 1 diabetes. Diabetologia 2020; 63: 2260–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015; 38: 1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp RC, Abdulrahim M, Naser ES, Naser SA. Genetic Variations of PTPN2 and PTPN22: Role in the Pathogenesis of Type 1 Diabetes and Crohn’s Disease. Front Cell Infect Microbiol 2015; 5: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol 2006; 18: 207–213. [DOI] [PubMed] [Google Scholar]
- 11.Sharp RC, Beg SA, Naser SA. Polymorphisms in Protein Tyrosine Phosphatase Non-receptor Type 2 and 22 (PTPN2/22) Are Linked to Hyper-Proliferative T-Cells and Susceptibility to Mycobacteria in Rheumatoid Arthritis. Front Cell Infect Microbiol 2018; 8: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krischer JP, Liu X, Vehik K, et al. Predicting Islet Cell Autoimmunity and Type 1 Diabetes: An 8-Year TEDDY Study Progress Report. Diabetes Care 2019; 42: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvani G, Fousteri G. PTPN22 and islet-specific autoimmunity: What have the mouse models taught us? World J Diabetes 2017; 8: 330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorillo E, Orrú V, Stanford SM, et al. Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J Biol Chem 2010; 285: 26506–26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arechiga AF, Habib T, He Y, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol 2009; 182: 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Shaked I, Stanford SM, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 2013; 39: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gianchecchi E, Crinò A, Giorda E, et al. Altered B cell homeostasis and toll-like receptor 9-driven response in type 1 diabetes carriers of the C1858T PTPN22 allelic variant: implications in the disease pathogenesis. PLoS One 2014; 9: e110755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Ewart D, Crabtree JN, et al. PTPN22 Variant R620W Is Associated With Reduced Toll-like Receptor 7-Induced Type I Interferon in Systemic Lupus Erythematosus. Arthritis Rheumatol 2015; 67: 2403–2414. [DOI] [PubMed] [Google Scholar]
- 19.Spalinger MR, Kasper S, Gottier C, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest 2016; 126: 4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeren S, Miles K, Chu JY, Salter D, Zamoyska R, Gray M. PTPN22 Is a Critical Regulator of Fcγ Receptor-Mediated Neutrophil Activation. J Immunol 2016; 197: 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayley R, Kite KA, McGettrick HM, et al. The autoimmune-associated genetic variant PTPN22 R620W enhances neutrophil activation and function in patients with rheumatoid arthritis and healthy individuals. Ann Rheum Dis 2015; 74: 1588–1595. [DOI] [PubMed] [Google Scholar]
- 22.Anjos S, Nguyen A, Ounissi-Benkalha H, Tessier MC, Polychronakos C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J Biol Chem 2002; 277: 46478–46486. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Chen S, Gu Y, et al. CTLA-4 +49 G/A, a functional T1D risk SNP, affects CTLA-4 level in Treg subsets and IA-2A positivity, but not β-cell function. Sci Rep 2018; 8: 10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balic I, Angel B, Codner E, Carrasco E, Perez-Bravo F. Association of CTLA-4 polymorphisms and clinical-immunologic characteristics at onset of type 1 diabetes mellitus in children. Hum Immunol 2009; 70: 116–120. [DOI] [PubMed] [Google Scholar]
- 25.Walldén J, Ilonen J, Roivainen M, Ludvigsson J, Vaarala O, Group AS. Effect of HLA genotype or CTLA-4 polymorphism on cytokine response in healthy children. Scand J Immunol 2008; 68: 345–350. [DOI] [PubMed] [Google Scholar]
- 26.Krischer JP, Lynch KF, Schatz DA, et al. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 2015; 58: 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orban T, Bundy B, Becker DJ, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet 2011; 378: 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas AJ, Dawson IR-S, Novakovsky GE, et al. Functional effects of chimeric antigen receptor co-receptor signaling domains in human Tregs. BioRxiv 2019; 749721. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya A, Campbell D, Hannum C, et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996; 4: 573–581. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho-Silva D, Pierleoni A, Pignatelli M, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res 2019; 47: D1056–D1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirakawa J, Shibuya K, Shibuya A. Requirement of the serine at residue 329 for lipid raft recruitment of DNAM-1 (CD226). Int Immunol 2005; 17: 217–223. [DOI] [PubMed] [Google Scholar]
- 32.Gaud G, Roncagalli R, Chaoui K, et al. The costimulatory molecule CD226 signals through VAV1 to amplify TCR signals and promote IL-17 production by CD4+ T cells. Sci Signal 2018; 11: eaar3083. [DOI] [PubMed] [Google Scholar]
- 33.Dean JW, Peters LD, Fuhrman CA, et al. Innate inflammation drives NK cell activation to impair Treg activity. J Autoimmun 2020; 108: 102417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro MR, Yeh W-I, Longfield JR, et al. CD226 deletion reduces Type 1 diabetes in the NOD mouse by impairing thymocyte development and peripheral T cell activation. Front Immunol 2020; 11: e2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuhrman CA, Yeh WI, Seay HR, et al. Divergent phenotypes of human regulatory T cells expressing the receptors TIGIT and CD226. J Immunol 2015; 195: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redlberger-Fritz M, Vietzen H, Puchhammer-Stöckl E. Association of severe influenza virus infections with CD226 (DNAM-1) variants. J Infect Dis 2019; 220: 1162–1165. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz PLD, Tapia G, Bakken IJ, et al. Pandemic influenza and subsequent risk of type 1 diabetes: A nationwide cohort study. Diabetologia 2018; 61: 1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattana TC, Santos AS, Fukui RT, et al. CD226 rs763361 is associated with the susceptibility to type 1 diabetes and greater frequency of GAD65 autoantibody in a Brazilian cohort. Mediators Inflamm 2014; 2014: 694948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Luis B, Sondgeroth B, Nassar N, Carpino N. Sts-2 is a phosphatase that negatively regulates zeta-associated protein (ZAP)-70 and T cell receptor signaling pathways. J Biol Chem 2011; 286: 15943–15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge Y, Paisie TK, Newman JRB, McIntyre LM, Concannon P. UBASH3A mediates risk for Type 1 diabetes through inhibition of T-cell receptor-induced NF-κB signaling. Diabetes 2017; 66: 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge Y, Concannon P. Molecular-genetic characterization of common, noncoding UBASH3A variants associated with type 1 diabetes. Eur J Hum Genet 2018; 26: 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steck AK, Dong F, Wong R, et al. Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers. Pediatr Diabetes 2014; 15: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YG, Ciecko AE, Khaja S, Grzybowski M, Geurts AM, Lieberman SM. UBASH3A deficiency accelerates type 1 diabetes development and enhances salivary gland inflammation in NOD mice. Sci Rep 2020; 10: 12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naseem S, Frank D, Konopka JB, Carpino N. Protection from systemic Candida albicans infection by inactivation of the Sts phosphatases. Infect Immun 2015; 83: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge Y, Paisie TK, Chen S, Concannon P. UBASH3A Regulates the Synthesis and Dynamics of TCR-CD3 Complexes. J Immunol 2019; 203: 2827–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertelsen V, Breen K, Sandvig K, Stang E, Madshus IH. The Cbl-interacting protein TULA inhibits dynamin-dependent endocytosis. Exp Cell Res 2007; 313: 1696–1709. [DOI] [PubMed] [Google Scholar]
- 47.Toomer KH, Lui JB, Altman NH, Ban Y, Chen X, Malek TR. Essential and non-overlapping IL-2Rα-dependent processes for thymic development and peripheral homeostasis of regulatory T cells. Nat Commun 2019; 10: 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinha S, Borcherding N, Renavikar PS, et al. An autoimmune disease risk SNP, rs2281808, in SIRPG is associated with reduced expression of SIRPγ and heightened effector state in human CD8 T-cells. Sci Rep 2018; 8: 15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westra HJ, Martínez-Bonet M, Onengut-Gumuscu S, et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat Genet 2018; 50: 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallet MA, Santostefano KE, Terada N, Brusko TM. Isogenic Cellular Systems Model the Impact of Genetic Risk Variants in the Pathogenesis of Type 1 Diabetes. Front Endocrinol (Lausanne) 2017; 8: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatherley D, Graham SC, Turner J, Harlos K, Stuart DI, Barclay AN. Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47. Mol Cell 2008; 31: 266–277. [DOI] [PubMed] [Google Scholar]
- 52.Brooke G, Holbrook JD, Brown MH, Barclay AN. Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J Immunol 2004; 173: 2562–2570. [DOI] [PubMed] [Google Scholar]
- 53.Legrand N, Huntington ND, Nagasawa M, et al. Functional CD47/signal regulatory protein α (SIRPα) interaction is required for optimal human T- and natural killer- (NK) cell homeostasis in vivo. Proc Natl Acad Sci USA 2011; 108: 13224–13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marroqui L, Dos Santos RS, Floyel T, et al. TYK2, a Candidate Gene for Type 1 Diabetes, Modulates Apoptosis and the Innate Immune Response in Human Pancreatic β-Cells. Diabetes 2015; 64: 3808–3817. [DOI] [PubMed] [Google Scholar]
- 55.Dendrou CA, Cortes A, Shipman L, et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med 2016; 8: (Abstract 363ra149). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santin I, Eizirik DL. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and β-cell apoptosis. Diabetes Obes Metab 2013; 15: 71–81. [DOI] [PubMed] [Google Scholar]
- 57.Coomans de Brachène A, Castela A, Op de Beeck A, et al. Preclinical evaluation of tyrosine kinase 2 inhibitors for human β-cell protection in type 1 diabetes. Diabetes Obes Metab 2020; 22: 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devalliere J, Charreau B. The adaptor Lnk (SH2B3): An emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem Pharmacol 2011; 82: 1391–1402. [DOI] [PubMed] [Google Scholar]
- 59.Maslah N, Cassinat B, Verger E, Kiladjian JJ, Velazquez L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia 2017; 31: 1661–1670. [DOI] [PubMed] [Google Scholar]
- 60.Torn C, Hadley D, Lee HS, et al. Role of Type 1 Diabetes-Associated SNPs on Risk of Autoantibody Positivity in the TEDDY Study. Diabetes 2015; 64: 1818–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li YJ, Li XY, Guo XR, et al. Absence of SH2B3 mutation in nonobese diabetic mice. Genet Mol Res 2012; 11: 1266–1271. [DOI] [PubMed] [Google Scholar]
- 62.Cooling L. Blood Groups in Infection and Host Susceptibility. Clin Microbiol Rev 2015; 28: 801–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smyth DJ, Cooper JD, Howson JM, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes 2011; 60: 3081–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azad MB, Wade KH, Timpson NJ. secretor genotype and susceptibility to infections and chronic conditions in the ALSPAC cohort. Wellcome Open Res 2018; 3: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Herrath MG, Fujinami RS, Whitton JL. Microorganisms and autoimmunity: making the barren field fertile? Nat Rev Microbiol 2003; 1: 151–157. [DOI] [PubMed] [Google Scholar]
- 66.Wacklin P, Tuimala J, Nikkilä J, et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One 2014; 9: e94863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koskinen MK, Mikk ML, Laine AP, et al. Longitudinal Pattern of First-Phase Insulin Response Is Associated With Genetic Variants Outside the Class II HLA Region in Children With Multiple Autoantibodies. Diabetes 2020; 69: 12–19. [DOI] [PubMed] [Google Scholar]
- 68.Lawson MAE, O’Neill IJ, Kujawska M, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J 2020; 14: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asgari S, Schlapbach LJ, Anchisi S, et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc Natl Acad Sci USA 2017; 114: 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem 2009; 284: 13348–13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui S, Eisenächer K, Kirchhofer A, et al. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol Cell 2008; 29: 169–179. [DOI] [PubMed] [Google Scholar]
- 72.Dias Junior AG, Sampaio NG, Rehwinkel J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol 2019; 27: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chistiakov DA, Voronova NV, Savost’Anov KV, Turakulov RI. Loss-of-function mutations E6 27X and I923V of IFIH1 are associated with lower poly(I:C)-induced interferon-β production in peripheral blood mononuclear cells of type 1 diabetes patients. Hum Immunol 2010; 71: 1128–1134. [DOI] [PubMed] [Google Scholar]
- 74.Mukherjee A, Morosky SA, Delorme-Axford E, et al. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 2011; 7: e1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Q, Langereis MA, Lork M, et al. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol 2014; 88: 3369–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lind K, Svedin E, Domsgen E, et al. Coxsackievirus counters the host innate immune response by blocking type III interferon expression. J Gen Virol 2016; 97: 1368–1380. [DOI] [PubMed] [Google Scholar]
- 77.Gorman JA, Hundhausen C, Errett JS, et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol 2017; 18: 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hyöty H, Leon F, Knip M. Developing a vaccine for type 1 diabetes by targeting coxsackievirus B. Expert Rev Vaccines 2018; 17: 1071–1083. [DOI] [PubMed] [Google Scholar]
- 79.Wasserfall C, Nick HS, Campbell-Thompson M, et al. Persistence of Pancreatic Insulin mRNA Expression and Proinsulin Protein in Type 1 Diabetes Pancreata. Cell Metab 2017; 26(568–575): e563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bühling F, Kouadio M, Chwieralski CE, et al. Gene targeting of the cysteine peptidase cathepsin H impairs lung surfactant in mice. PLoS One 2011; 6: e26247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hsing LC, Kirk EA, McMillen TS, et al. Roles for cathepsins S, L, and B in insulitis and diabetes in the NOD mouse. J Autoimmun 2010; 34: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Faraco J, Lin L, Kornum BR, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet 2013; 9: e1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inshaw JRJ, Cutler AJ, Crouch DJM, Wicker LS, Todd JA. Genetic Variants Predisposing Most Strongly to Type 1 Diabetes Diagnosed Under Age 7 Years Lie Near Candidate Genes That Function in the Immune System and in Pancreatic β-Cells. Diabetes Care 2020; 43: 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fløyel T, Brorsson C, Nielsen LB, et al. CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc Natl Acad Sci USA 2014; 111: 10305–10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glieberman AL, Pope BD, Zimmerman JF, et al. Synchronized stimulation and continuous insulin sensing in a microfluidic human Islet on a Chip designed for scalable manufacturing. Lab Chip 2019; 19: 2993–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greiner DL, Brehm MA, Hosur V, Harlan DM, Powers AC, Shultz LD. Humanized mice for the study of type 1 and type 2 diabetes. Ann NY Acad Sci 2011; 1245: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Battaglia M, Ahmed S, Anderson MS, et al. Introducing the endotype concept to address the challenge of disease heterogeneity in Type 1 diabetes. Diabetes Care 2020; 43: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perry DJ, Wasserfall CH, Oram RA, et al. Application of a genetic risk score to racially diverse Type 1 diabetes populations demonstrates the need for diversity in risk-modeling. Sci Rep 2018; 8: 4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bradfield JP, Qu HQ, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet 2011; 7: e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015; 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 91.Regev A, Teichmann SA, Lander ES, et al. The Human Cell Atlas. Elife 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]