Summary
In an elaborate form of inter-species exploitation, many insects hijack plant development to induce novel plant organs called galls that provide the insect with a source of nutrition and a temporary home. Galls result from dramatic reprogramming of plant cell biology driven by insect molecules, but the roles of specific insect molecules in gall development have not yet been determined. Here we study the aphid Hormaphis cornu, which makes distinctive “cone” galls on leaves of witch hazel Hamamelis virginiana. We found that derived genetic variants in the aphid gene determinant of gall color (dgc) are associated with strong downregulation of dgc transcription in aphid salivary glands, upregulation in galls of seven genes involved in anthocyanin synthesis, and deposition of two red anthocyanins in galls. We hypothesize that aphids inject DGC protein into galls, and that this results in differential expression of a small number of plant genes. Dgc is a member of a large, diverse family of novel predicted secreted proteins characterized by a pair of widely spaced cysteine-tyrosine-cysteine (CYC) residues, which we named BICYCLE proteins. Bicycle genes are most strongly expressed in the salivary glands specifically of galling aphid generations, suggesting that they may regulate many aspects of gall development. Bicycle genes have experienced unusually frequent diversifying selection, consistent with their potential role controlling gall development in a molecular arms race between aphids and their host plants.
eTOC Blurb
Korgaonkar et al. report on novel secreted aphid proteins encoded by bicycle genes. Variation in the bicycle gene determinant of gall color alters expression of targeted plant genes, suggesting that BICYCLE proteins modulate gall development.
Introduction
Organisms often exploit individuals of other species, for example through predation or parasitism. Parasites sometimes utilize molecular weapons against hosts, which themselves respond with molecular defenses, and the genes that encode or synthesize these molecular weapons may evolve rapidly in a continuous ‘arms race’1–3. Some of the most elaborate molecular defenses—such as adaptive immune systems, restriction modification systems, and CRISPR—have resulted from such host-parasite conflicts. In many less well-studied systems, parasites not only extract nutrients from their hosts but they also alter host behavior, physiology, or development to the parasite’s advantage4. Insect galls represent one of the most extreme forms of such inter-species manipulation.
Insect-induced galls are intricately patterned homes that provide insects with protection from environmental vicissitude and from some predators and parasites5–7. Galls are also resource sinks, drawing nutrients from distant plant organs, and providing insects with abundant food8. Insect galls are atypical plant growths that do not result simply from unpatterned cellular over-proliferation, as observed for microbial galls like the crown gall induced by Agrobacterium tumefaciens. Instead, each galling insect species appears to induce a distinctive gall, even when related insect species attack the same plant, implying that each species provides unique instructions to re-program latent plant developmental networks9–19.
At least some gall-inducing insects produce phytohormones20–25, although it is not yet clear whether insects introduce these hormones into plants to support gall development. However, injection of phytohormones alone probably cannot generate the large diversity of species-specific insect galls. In addition, galling insects induce plant transcriptional changes independently of phytohormone activity2,12,26–29. Thus, given the complex cellular changes required for gall development, insects probably introduce many molecules into plant tissue to induce galls.
In addition to the potential role of phytohormones in promoting gall growth, candidate gall effectors have been identified in several gall-forming insects30–32. However, none of these candidate effectors have yet been shown to contribute to gall development or physiology. In addition, while many herbivorous insects introduce effector molecules into plants to influence plant physiology33–36, there is no evidence that any previously described effectors contribute to gall development. Since there are currently no galling insect model systems that would facilitate a genetic approach to this problem, we turned to natural variation to identify insect genes that contribute to gall development.
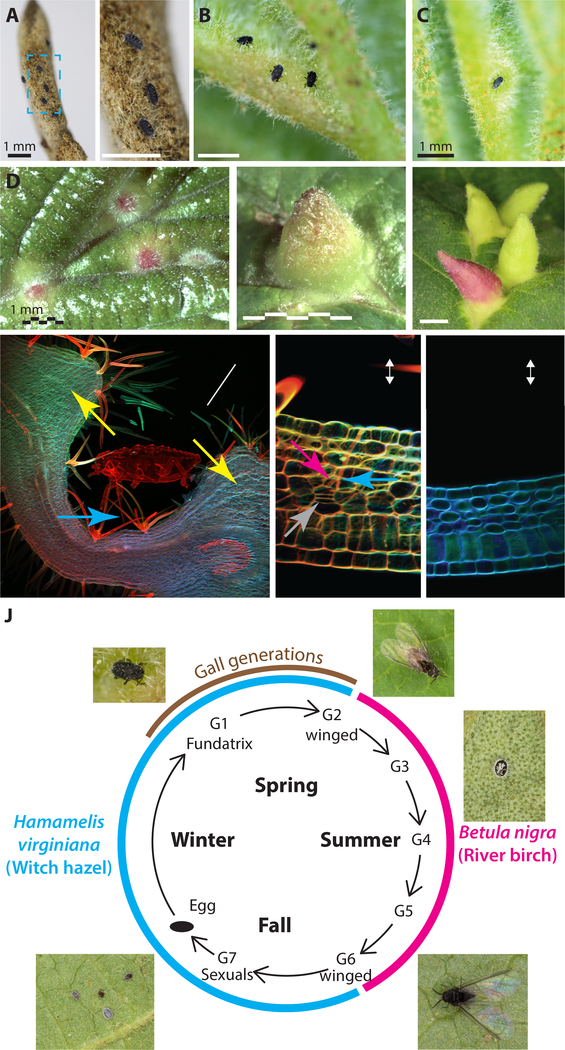
We studied the aphid, Hormaphis cornu, which induces galls on the leaves of witch hazel, Hamamelis virginiana, in the Eastern United States (Figure 1A–F, J). In early spring, each H. cornu gall foundress (fundatrix) probes an expanding leaf with her microscopic mouthparts (stylets) (Figure 1A, B, Video S1) and pierces individual mesophyll cells with her stylets (Figure 1G and H)37,38. We found that plant cells near injection sites, revealed by the persistent stylet sheaths, over-proliferate through periclinal cell divisions (Figure 1H). This pattern of cytokinesis is not otherwise observed in leaves at this stage of development (Figure 1I) and contributes to the thickening and expansion of leaf tissue that generates the gall (Figure 1D–G). The increased proliferation of cells near the tips of stylet sheaths suggests that secreted effector molecules produced in the salivary glands are deposited into the plant via the stylets.
Figure 1. Hormaphis cornu aphids drive patterned over-proliferation of plant cells to produce galls on leaves Hamamelis virginiana.
(A-C) Photographs of the abaxial surfaces of H. virginiana leaves being attacked by first-instar fundatrices of H. cornu. Nymphs gather on the unopened leaf buds (A) and soon after bud break the fundatrices gather near leaf veins (B) and inject material that begins to transform the leaf into a gall (C). Magnified region in blue rectangle of panel (A) shows three fundatrices waiting on unopened bud (A’).
(D-F) Photographs of the adaxial leaves of H. virginiana, showing galls at early (D), middle (E), and late (F) growth stages.
(G-I) Confocal micrographs of sections through a H. cornu gall (G, H) and un-galled H. virginiana leaf (I) stained with Congo red and calcofluor white. Extensive hypertropy is observed in the mesophyll (yellow arrows) at a considerable distance from the location of the aphid’s stylets (blue arrows) (G). The tips of aphid stylets (pink arrow) can be observed within cells of H. virginiana and plant tissue shows evidence of hyperproliferation and periclinal divisions (grey arrows) close to the tips of stylets and the termini of stylet sheaths (H). Periclinal divisions are observed in both spongy and palisade mesophyll cells during gall development, but never in ungalled leaf tissue (I). Plant tissue is presented in the aphid’s frame of reference, with abaxial leaf surface up.
(J) Diagram of life cycle of H. cornu. H. cornu migrates annually between H. virginiana (blue line) and Betula nigra (pink line) and the gall is produced only in the spring on H. virginiana (brown line). Each nymph of the first generation (G1, the fundatrix) hatches from an over-wintering egg and initiates development of a single gall. Her offspring (G2) feed and grow within the gall and develop with wings, which allows them to fly to B. nigra in late spring. For three subsequent generations (G3-G5) the aphids develop as small, coccidiform morphs on B. nigra. In the fall, aphids develop with wings (G6), fly to H. virginiana plants, and deposit male and female sexuals (G7), the only generation possessing meiotic cells. The sexuals feed and complete development on the senescing leaves of H. virginiana. As adults they mate and the females deposit eggs that overwinter and give rise to fundatrices the following spring.
See also Video S1.
After several days, the basal side of the gall encloses the fundatrix and the gall continues to grow apically and laterally, providing the fundatrix and her offspring with protection and abundant food. After several weeks, the basal side of the gall opens to allow aphids to remove excreta (honeydew) and molted nymphal skins from the gall and, eventually, to allow winged migrants to depart. Continued gall growth requires the constant presence of the fundatrix and gall tissue dies in her absence38,39, suggesting that the fundatrix must continuously inject salivary-gland produced effectors to overcome plant defenses.
Results
A natural gall color polymorphism is linked to regulatory variation in a novel aphid gene, determinant of gall color
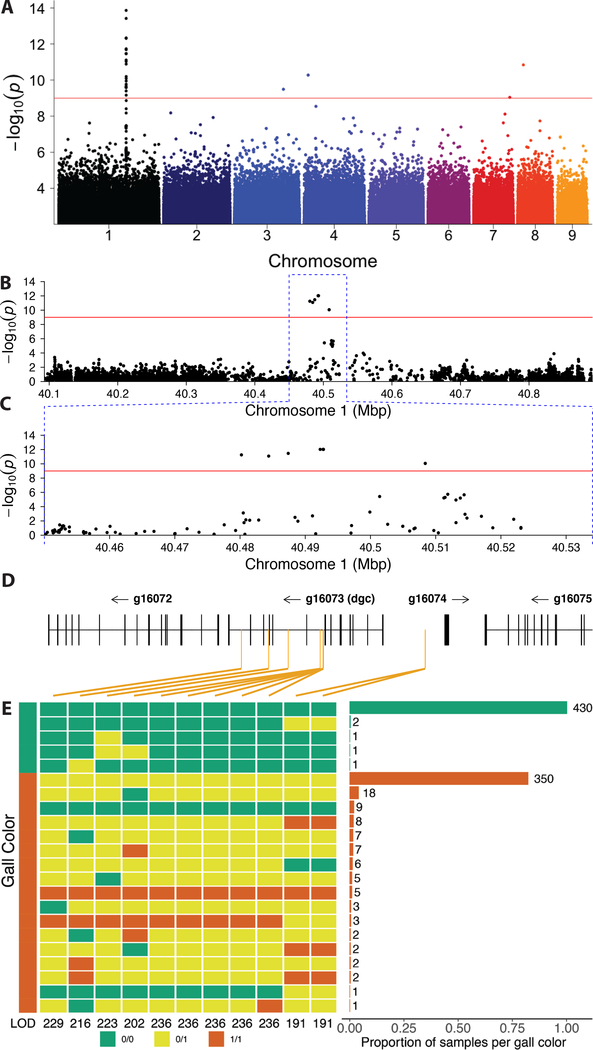
We found that populations of H. cornu include approximately 4% red galls and 96% green galls (Figure 1F). We inferred that this gall color polymorphism results from differences among aphids, rather than from differences associated with leaves or the location of galls on leaves, because red and green galls are located randomly on leaves and often adjacent to each other on a single leaf (Figure 1F). We sequenced and annotated the genome of H. cornu (Figure S1A–B; STAR Methods) and performed a genome-wide association study (GWAS) on fundatrices isolated from 43 green galls and 47 red galls by resequencing their genomes to approximately 3X coverage. There is no evidence for genome-wide differentiation of samples from red and green galls, suggesting that individuals making red and green galls were sampled from a single interbreeding population (Figure S1C–F). We identified SNPs near 40.5 Mbp on Chromosome 1 that were strongly associated with gall color (Figure 2A). We re-sequenced approximately 800 kbp flanking these SNPs to approximately 60X coverage and identified 11 single-nucleotide polymorphisms (SNPs) within the introns and upstream of gene g16073 that were strongly associated with gall color (Figure 2B–D). There is no evidence that large scale chromosomal aberrations are associated with gall color (Figure S1G–K; STAR Methods).
Figure 2. A genome-wide association study (GWAS) identifies variation within a novel aphid gene associated with gall color.
(A) A GWAS of fundatrices isolated from 43 green and 47 red galls identified a small region on chromosome 1 of the H. cornu genome that is strongly associated with gall color. Red line indicates FDR = 0.05. Colors of points on chromosomes are arbitrary.
(B-D) Resequencing 800 kbp of Chromosome 1 centered on the most significant SNPs from the original GWAS to approximately 60X coverage identified 11 spatially clustered SNPs significantly associated with gall color located within the introns and upstream of g16073, which was named dgc (D). (Some SNPs are closely adjacent and cannot be differentiated at this scale.) Significant SNPs are indicated with orange vertical lines in (D).
(E) Genotypes of all 11 SNPs associated with gall color from an independent sample of aphids from 435 green and 431 red galls. Color of gall for aphid samples shown on left and genotype at each SNP is shown adjacent in green (0/0, homozygous ancestral state), yellow (0/1, heterozygous), or red (1/1, homozygous derived state). LOD scores for association with gall color shown for each SNP at bottom of genotype plot. Histogram of frequencies of each multilocus genotype ordered by frequency and collected within gall color is shown on the right. All 11 SNPs are strongly associated with gall color (P < 10−192), and a cluster of 5 SNPs in a 61bp region are most strongly associated with gall color. Individuals homozygous for ancestral alleles at all or most loci and making red galls likely carry variants at other loci that influence gall color (STAR Methods).
See also Figures S1 and S2.
Since GWAS can sometimes produce spurious associations, we performed an independent replication study and found that all 11 SNPs were highly significantly associated with gall color in fundatrices isolated from 435 green and 431 red galls (LOD = 191 – 236; Figure 2E). All fundatrices from green galls were homozygous for the ancestral allele at 9 or more of these SNPs (Figure 2E). In contrast, 98% of fundatrices from red galls were heterozygous or homozygous for derived alleles at 9 or more SNPs (Figure 2E). This pattern suggests that alleles contributing to red gall color are genetically dominant to alleles that generate green galls. Two percent of fundatrices that induce red galls were homozygous for ancestral alleles at these SNPs and likely carry genetic variants elsewhere in the genome that confer red color to galls (Figure S1L; STAR Methods).
Based on these genetic associations and further evidence presented below, we assigned the name determinant of gall color (dgc) to g16073. Dgc encodes a predicted protein of 23 kDa with an N-terminal secretion signal sequence (Figure S1M). The putatively secreted portion of the protein shares no detectable sequence homology with any previously reported proteins.
Most SNPs associated with green or red galls were found in one of two predominant haplotypes (Figure 2E) and exhibited strong linkage disequilibrium (LD) (Figure S2A–D). LD can result from suppressed recombination. However, these 11 SNPs are in linkage equilibrium with many other intervening and flanking SNPs (Figures S2). Also, multiple observed genotypes are consistent with recombination between these 11 SNPs (Figure 2E) and we found no evidence for chromosomal aberrations that could suppress recombination (Figure S1G–K; STAR Methods). Thus, LD among the 11 dgc SNPs associated with gall color cannot be explained by suppressed recombination. It is more likely that the non-random association of the 11 dgcRed SNPs has been maintained by natural selection, suggesting that the combined action of all 11 SNPs may have a stronger effect on gene function than any single SNP alone.
Regulatory variants at dgc dominantly silence dgc expression
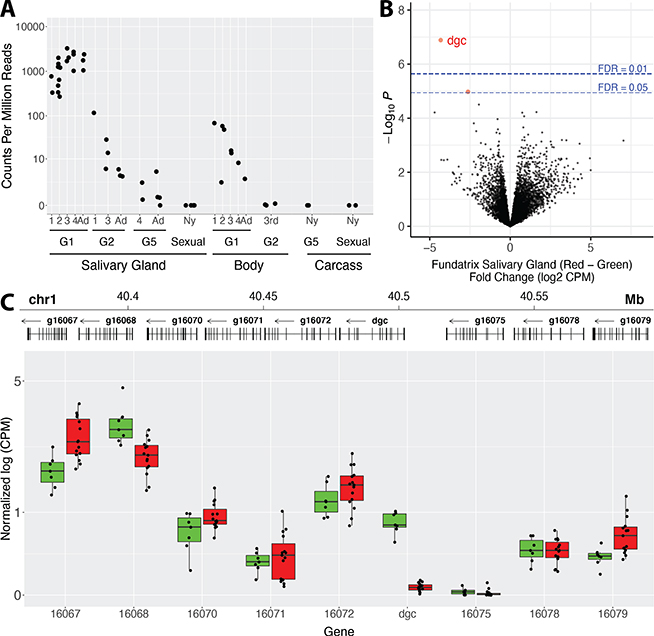
Since all 11 dgc polymorphisms associated with gall color occur outside of dgc exons (Figure 2D), we tested whether these polymorphisms influence expression of dgc or of any other genes in the genome. We first determined that dgc is expressed highly and specifically in fundatrix salivary glands and lowly or not at all in other tissues or other life cycle stages (Figure 3A). We then performed RNA-seq on salivary glands from fundatrices with dgcGreen /dgcGreen or dgcRed/dgcGreen genotypes. Dgc stands out as the most strongly differentially expressed gene between these genotypes (Figure 3B). Since dgcRed alleles appeared to be dominant to the dgcGreen alleles for gall color, we expected that dgc transcripts would be upregulated in animals with dgcRed alleles. In contrast, dgc transcripts were almost absent in fundatrices carrying dgcRed alleles (Figure 3C). That is, red galls are associated with strongly reduced dgc expression in fundatrix salivary glands.
Figure 3. Dgc is the most strongly differentially expressed gene in salivary glands of fundatrices collected from red versus green galls.
(A) Expression of dgc in salivary glands, whole bodies, or carcasses (body minus salivary glands) throughout the H. cornu life cycle. Salivary glands were dissected from multiple nymphal stages and adults of four generations representing major morphs of the life cycle, G1 (fundatrix), G2, G5, and sexuals. Dgc is expressed at highest levels in salivary glands of fundatrices. Dgc expression declines in salivary glands through the instars of G2 animals and later generations and was not detected in salivary glands of sexuals. Expression observed in full bodies of G1 animals (fundatrices) probably reflects expression in the salivary glands and expression was not observed in carcasses.
(B) Genome-wide differential expression analysis of H. cornu fundatrix salivary gland transcripts from individuals heterozygous for dgcRed/dgcGreen (Red; N = 15) versus homozygous for dgcGreen (Green; N = 7) illustrated as a volcano plot. Dgc is strongly downregulated in dgcRed/dgcGreen fundatrices.
(C) Salivary gland expression of genes in the paralog cluster that includes dgcRed/dgcGreen. Gene models of the paralogous genes are shown at the top and expression levels normalized by mean expression across all paralogs is shown below. Only dgc is strongly downregulated in this gene cluster between individuals carrying dgcRed/dgcGreen (Red) versus dgcGreen/dgcGreen (Green) genotypes.
See also Figures S1 and S3.
Dgc expression is reduced approximately 20-fold in fundatrix salivary glands with dgcRed/dgcGreen (27 ± 22.6 CPM, mean ± SD) versus dgcGreen/dgcGreen genotypes (536 ± 352.3 CPM, mean ± SD). This result suggested that dgcRed alleles downregulate both the dgcRed and dgcGreen alleles in heterozygotes. To confirm whether the dgcRed allele downregulates the dgcGreen allele in trans, we identified exonic SNPs that were specific to each allele and could be identified in the RNA-seq data. We found that both dgcRed and dgcGreen alleles were strongly downregulated in heterozygotes, confirming the trans activity of the dgcRed allele (Figure S3A). We observed no systematic transcriptional changes in neighboring genes (Figure 3C), most of which are dgc paralogs, indicating that dgcRed alleles exhibit a perhaps unique example of locus-specific repressive transvection40.
High levels of dgc transcription are associated with downregulation specifically of plant anthocyanin genes and two anthocyanins
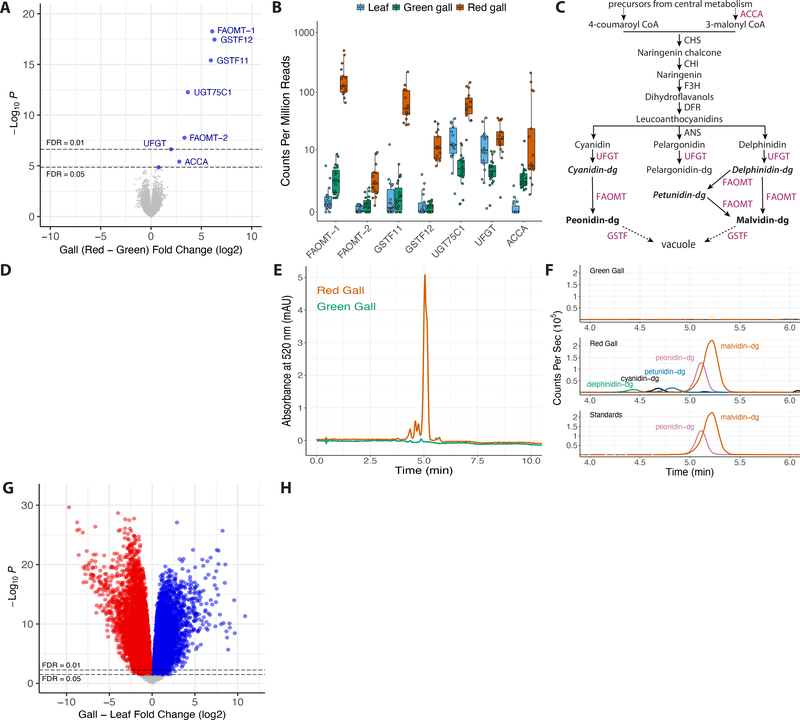
Since red galls are associated with strong differential expression of only dgc, we wondered how the plant responds to changes in this single putative effector. To examine this question, we sequenced and annotated the genome of the host plant Hamamelis virginiana and then performed whole-genome differential expression on plant mRNA isolated from galls induced by aphids with dgcRed/dgcGreen versus dgcGreen/dgcGreen genotypes (STAR Methods). We did not observe genome-wide differentiation between red and green galls (Figure S3B–C), and only eight plant genes were differentially expressed between red and green galls and all eight genes were downregulated in green galls (Figure 4A–B). That is, high levels of dgc are associated with downregulation of only eight plant genes in galls.
Figure 4. Genome-wide differential expression analysis of H. virginiana red versus green galls and galls versus leaves.
(A) Genome-wide differential expression analysis of H. virginiana transcripts isolated from galls made by fundatrices heterozygous for dgcRed/dgcGreen (Red; N = 17) versus homozygous for dgcGreen (Green; N = 23) illustrated as a volcano plot. Only eight genes are differentially expressed at FDR < 0.05, and all are overexpressed in red galls. The seven most strongly differentially expressed genes encode anthocyanin biosynthetic enzymes (FAOMT-1 = g23591; FAOMT-2 = g7147; GSTF11 = g134919; GSTF12 = g109682; UGT75C1 = g14194; UFGT = g22774; ACCA = g97071).
(B) Expression levels, in counts per million reads, of the seven anthocyanin biosynthetic genes overexpressed in red galls, in green (green) and red (red) galls and ungalled leaves (blue). Each data point within each gene is from a separate genome-wide RNA-seq sample.
(C) Simplified diagram of the anthocyanin biosynthetic pathway. Enzyme classes upregulated in red galls are shown in purple font. The two terminal anthocyanins that generate the red color in galls, peonidin-3,5-diglucoside and malvidin-3,5-diglucoside, are shown in bold font, and three precursor molecules found in red galls are shown in bold italic font. Anthocyanin names are abbreviated (dg = 3,5-diglucoside).
(D) Photos of cross-sections of green (top left) and red (top right) and the pigments extracted from green and red galls (below).
(E) UHPLC-DAD chromatograms at 520 nm of extract from red (red line) and green (green line) galls.
(F) Overlaid UHPLC-MS chromatograms of green (top) and red (middle) gall extracts and authentic standards (bottom). Each pigment is indicated with a different color: green = delphinidin-3,5-diglucoside (m/z = 627.1551); black = cyanidin-3,5-diglucoside (m/z = 611.1602); blue = petunidin-3,5-diglucoside (m/z = 641.1709); purple = peonidin-3,5-diglucoside (m/z = 625.1768); and red = malvidin-3,5-diglucoside (m/z = 655.1870). Anthocyanin names are abbreviated (dg = 3,5-diglucoside). Peonidin-3,5-diglucoside and malvidin-3,5-diglucoside together account for 87% of pigment detected in red galls.
(G) Genome-wide differential expression analysis of H. virginiana transcripts isolated from galls (N = 36) versus leaves (N = 17). Approximately 60% of expressed genes are differentially expressed between gall and leaf tissue at FDR < 0.05.
(H) Gene ontology analysis of GO terms down (left) and up-regulated (right) in galls, presented as volcano plots. Genes involved in cell division and morphogenesis were strongly upregulated in galls and genes involved in photosynthesis were strongly down-regulated in galls.
See also Figure S3.
Red pigmented galls could result from production of carotenoids41, anthocyanins42,43, or betacyanins. However, in red galls induced by H. cornu, the seven most strongly upregulated plant genes are all homologous to genes annotated as enzymes of the anthocyanin biosynthetic pathway (Figure 4C). One gene encodes an enzyme (ACCA) that irreversibly converts acetyl-CoA to malonyl CoA, a biosynthetic precursor of multiple anthocyanins. Two genes encode anthocyanidin 3–0-glucosyltransferases (UFGT and UGT75C1), which glycosylate unstable anthocyanidins to allow their accumulation44. Two genes encode flavonoid 3’−5’ methyltransferases (FAOMT-1, FAOMT-2), which methylate anthocyanin derivatives45. Finally, two genes encode phi class glutathione S-transferases (GSTF11, GSTF12), which conjugate glutathione to anthocyanins, facilitating anthocyanin transport and stable accumulation in vacuoles46.
Six of the enzymes upregulated in red galls are required for final steps of anthocyanin production and deposition (Figure 4C) and their upregulation in red galls may account for the accumulation of pigments in red galls. To test this hypothesis, we extracted and analyzed pigments from galls (Figure 4D) and identified high levels of two pigments only in red galls (Figure 4E), the anthocyanins malvidin-3,5-diglucoside and peonidin-3,5-diglucoside (Figures 4F, S3F–J). Thus, the pigments present in red galls are products of enzymes in the anthocyanin biosynthetic pathway, such as those encoded by genes that are upregulated in red galls. The two abundant anthocyanins are produced from distinct intermediate precursor molecules (Figure 4C), three of which were also detected in red galls (Figures 4F, S3F,G,I), and synthesis of these two anthocyanins likely requires activity of different methyltransferases and glucosyltransferases. The three pairs of glucosyltransferases, methyltransferases, and glutathione transferases upregulated in red galls may provide the specific activities required for production of these two anthocyanins.
Taken together, these observations suggest that dgc represses transcription of seven anthocyanin biosynthetic enzymes. It is not clear how dgc induces specific transcriptional changes in seven plant genes; it may act by altering activity of an upstream regulator of these plant genes.
Aphids induce widespread transcriptomic changes in galls
Gall color represents only one aspect of the gall phenotype, apparently mediated by changes in expression of seven plant genes, and the full complement of cell biological events during gall development presumably requires changes in many more plant genes. To estimate how many plant genes are differentially expressed during development of the H. cornu gall on H. virginiana, we performed differential expression analysis of plant genes in galls versus the surrounding leaf tissue. Approximately 31% of plant genes were upregulated and 34% were downregulated at FDR = 0.05 in galls versus leaf tissue (Figure 4G); 27% up and 29% down in gall at FDR = 0.01). Results of gene ontology analysis of up and down-regulated genes is consistent with the extensive growth of gall tissue and down regulation of chloroplasts seen in aphid galls (Figure 4H; STAR Methods), a pattern observed in other galling systems12.
Thus, approximately 15,000 plant genes are differentially expressed in galls, representing a system-wide re-programming of plant cell biology. If other aphid effector molecules act in ways similar to dgc, which is associated with differential expression of only eight plant genes, then gall development may require injection of hundreds or thousands of effector molecules.
Dgc is a member of a large class of novel bicycle genes expressed specifically in the salivary glands of gall-inducing aphids
To identify additional proteins that aphids may inject into plants to contribute to gall development, we exploited the fact that only some individuals in the complex life cycle of H. cornu induce galls (Figure 1J). Only the fundatrix generation induces galls and only her immediate offspring live alongside her in the developing gall. In contrast, individuals of generations that live on river birch (Betula nigra) through the summer and the sexual generation that feed on H. virginiana leaves in the autumn do not induce any leaf malformations. Thus, probably only the salivary glands of the generations that induce galls (the fundatrix (G1) and possibly also her immediate offspring (G2)) produce gall-effector molecules. We identified 3,048 genes upregulated in fundatrix salivary glands versus the fundatrix body (Figure S4A) and 3,427 genes upregulated in salivary glands of fundatrices, which induce galls, versus sexuals, which do not induce galls although they feed on the same host plant (Figure S4B). Intersection of these gene sets identified 1,482 genes specifically enriched in the salivary glands of fundatrices (Figure S4C).
Half of these genes (744) were homologous to previously identified genes, many of which had functional annotations (Figure 5A). Gene Ontology analysis of the “annotated” genes suggests that they contribute mostly to the demands for high levels of protein secretion in fundatrix salivary glands (Figure S4D). Most do not encode proteins with secretion signals (671; Figure S4E) and are thus unlikely to be injected into plants. We searched for homologs of genes that have been proposed as candidate gall-effector genes in other insects but found little evidence that these classes of genes contribute to aphid gall development (Figure S4F–H). We therefore focused on the remaining 738 unannotated genes, which included 459 genes encoding proteins with predicted secretion signals (Figure S4E).
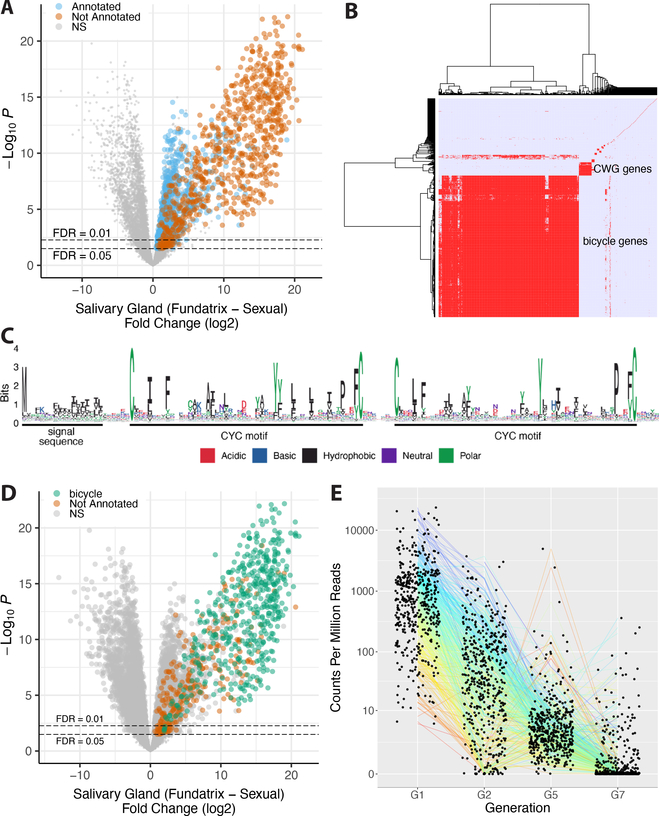
Figure 5. bicycle genes are salivary gland enriched transcripts of gall-associated H. cornu generations.
(A) Differential expression of fundatrix versus sexual salivary glands with only genes significantly upregulated in fundatrix salivary glands marked with colors, shown as a volcano plot. Genes with and without homologs in public databases are labeled as “Annotated” (blue) and “Not Annotated” (brown), respectively.
(B) Hierarchical clustering of unannotated salivary-gland specific genes reveals one large cluster of bicycle genes, one small cluster of CWG genes, and many largely unique, unclustered genes.
(C) Amino-acid logo for predicted BICYCLE proteins. Sequence alignment used for logo was filtered to highlight conserved positions. Most genes encode proteins with an N-terminal signal sequence and a pair of conserved cysteine-tyrosine-cysteine motifs (CYC).
(D) Bicycle (green) and remaining unannotated (brown) genes labelled on a differential expression volcano plot illustrate that, on average, bicycle genes are the most strongly differentially expressed genes expressed specifically in fundatrix salivary glands.
(E) Bicycle gene expression levels in salivary glands of aphids from four generations. Bicycle genes are expressed at highest levels in the fundatrix and mostly decline in expression during subsequent generations. (Sample sizes: G1 (N = 20); G2 (N = 4); G5 (N = 6); and G7 (N = 6).
See also Figures S1, S4, S5 and Table S2.
Hierarchical clustering of the unannotated genes by sequence similarity identified one large (476 genes) and one small (43 genes) cluster of related genes, and 222 genes sharing few or no homologs amongst the unannotated genes (Figure 5B). Genes in both the large and small clusters encode proteins with N-terminal secretion signals, as expected for effector proteins that might be injected into plants. The small cluster encodes a divergent set of proteins containing several conserved cysteines (C) and a well conserved tryptophan (W) and glycine (G), and we named these CWG genes (Figure S5A–C).
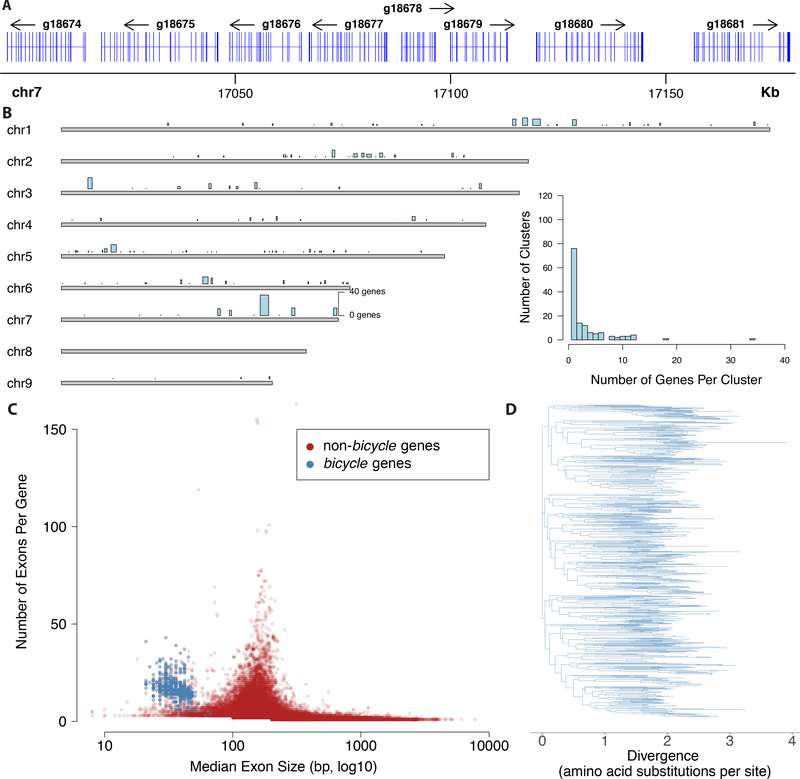
Proteins encoded by the large cluster display conservation mainly of a pair of widely spaced cysteine-tyrosine-cysteine (CYC) motifs and spacing between the C, Y, and C residues of each motif is not well conserved (Figure 5C). This pair of CYC motifs led us to name these bicycle (bi-CYC-like) genes. The bicycle genes were the most strongly upregulated class of genes in fundatrix salivary glands (Figures 5D, S5D) and were expressed specifically in the salivary glands of the two generations associated with galls (G1 and G2) (Figure 5E). Many bicycle genes are found in paralog clusters throughout the H. cornu genome (Figure 6A, B) and each bicycle gene contains approximately 5–25 microexons (Figure 6A, C)—a large excess relative to the genomic background (Figure 6C)—interrupted by long introns (Figure 6A).
Figure 6. H. cornu bicycle genes are found in paralog clusters, contain many microexons, and are highly diverse.
(A) Example of part of a paralog cluster of bicycle genes from chromosome 7 of the H. cornu genome, illustrating abundance of small exons in each gene.
(B) Distribution of singleton bicycle genes and paralog clusters in the H. cornu genome. Number of genes per cluster and genomic range is indicating by height and width, respectively, of blue bars above chromosomes. Histogram of number of bicycle genes per paralog cluster is shown in inset.
(C) Number of exons per gene versus median exon size for H. cornu bicycle (blue) and non-bicycle (red) genes. Bicycle genes possess an unusually large number of unusually small exons.
(D) Maximum likelihood phylogenetic tree of H. cornu bicycle gene amino acid sequences reveals extensive sequence divergence of bicycle genes.
We found that dgc shares many features with bicycle genes—it is strongly expressed specifically in fundatrix salivary glands, and it exhibits many microexons and a pair of CYC motifs (Figures 2D, S1M)—and that it is evolutionarily related to other bicycle genes. Thus, dgc is a member of a diverse family of genes encoding secreted proteins expressed specifically in the salivary glands of gall forming generations. Bicycle genes are therefore good candidates to encode many of the molecules required to generate the extensive transcriptional changes observed in galls.
bicycle genes experienced intense diversifying selection, consistent with a potential arms race between aphids and plants
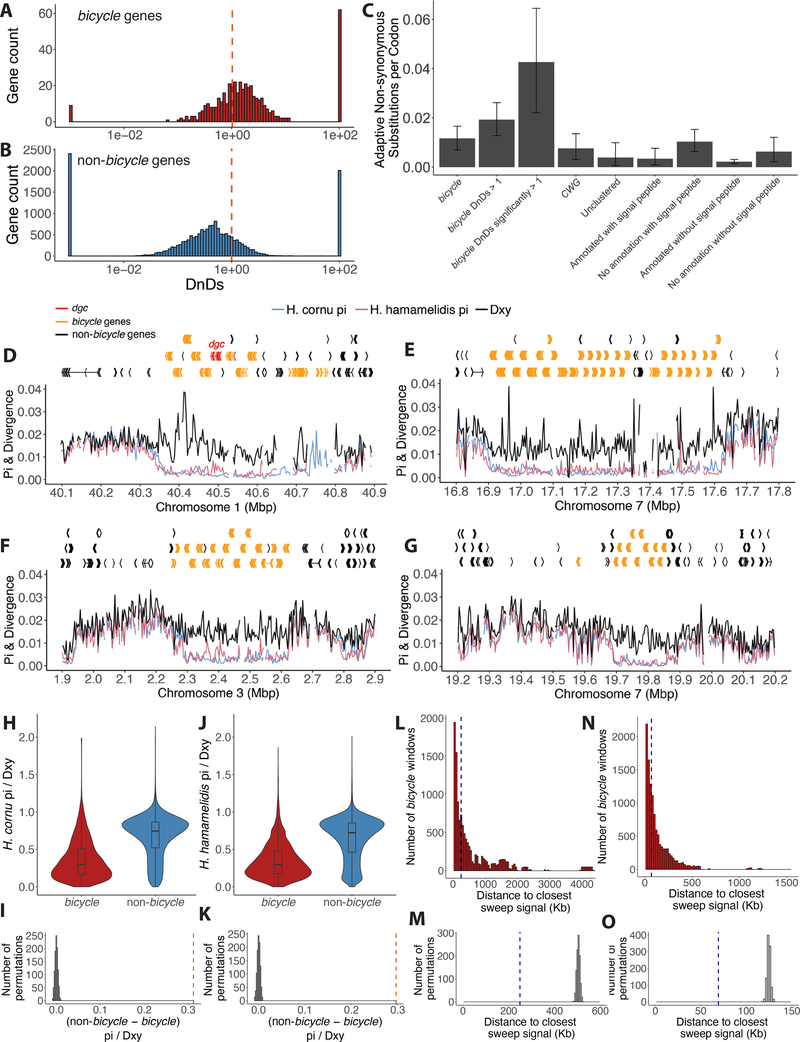
Bicycle genes are extremely diverse at the amino acid sequence level, as has been observed for other candidate insect gall effector genes47. Each BICYCLE protein has accumulated approximately two substitutions per amino acid site since divergence from paralogs (Figure 6D). To explore whether this diversity resulted from natural selection rather than genetic drift, we compared rates of non-synonymous (dN) versus synonymous (dS) substitutions between the sister species H. cornu and H. hamamelidis (which also induces galls) in bicycle versus non-bicycle genes, because dN/dS values greater than one provide evidence for positive selection48. To calculate polymorphism of orthologous genes in each species, we mapped sequencing reads from individuals of each species to the H. cornu genome. For divergence estimates, we estimated the H. hamamelidis genome by mapping H. hamamelidis sequencing reads to the H. cornu genome (STAR Methods) and compared this genome with the original H. cornu genome. A large excess of bicycle genes displayed dN/dS significantly greater than 1 relative to the genomic background (Figure 7A–B, Table S1; P < 2.2e-16), revealing recurrent adaptive amino acid substitution at many bicycle genes since these species diverged.
Figure 7. Genome-wide signals of selective sweeps are enriched near bicycle genes.
(A) The majority of bicycle genes display dN/dS values great than 1, with few showing strong sequence conservation (dN/dS <<1). Dashed vertical red line indicates dN/dS = 1.
(B) Non-bicycle genes are more conserved, on average, than bicycle genes (Mann-Whitney U test p=2.6e-76. Dashed vertical red line indicates dN/dS = 1.
(C) Mean number of adaptive non-synonymous substitutions scaled by protein length for different categories of genes over-expressed in fundatrix salivary glands. As a proportion of protein length, bicycle genes display the fastest rate of adaptive evolution of any category of these genes. Error bars represent 95% confidence intervals. Note that the four categories on the right include all genes shown on the left, but categorized by whether genes were annotated and included a signal peptide. Thus, for example, the category “No annotation with signal peptide” is composed mostly of bicycle and CWG genes.
(D-G) Gene models and population genomic statistics for the 800 kb dgc bicycle gene cluster (D) and for three additional genomic regions containing bicycle gene clusters (E-G). Divergence between (black line) and polymorphism within H. cornu (blue line) and H. hamamelidis (pink line) in 3000bp windows shown below gene models.
(H and J) Ratio of Pi to Dxy for bicycle and non-bicycle gene regions in H. cornu (H) and H. hamamelidis (J).
(I and K) The observed difference in Pi/Dxy between non-bicycle and bicycle genes (dashed red line) is much larger than the expectation generated by permuting the locations of Pi/Dxy values relative to gene locations for both H. cornu (I) and H. hamamelidis (K).
(L and N) Distance from each bicycle gene to the closest significant selective sweep signal is shown as red histogram and dashed blue line indicates the median of this distribution for H. cornu (L) and H. hamamelidis (N).
(M and O) The median distance from each bicycle gene to the closest significant selective sweep signal from (L) for H. cornu (M) and from (N) for H. hamamelidis (O) is shown with dashed blue line and the values after 1000 permutations of sweep signals relative to gene locations are shown as grey histograms. The observed sweep signals are closer to bicycle genes than expected by chance.
See also Figures S6 and S7 and Tables S1 and S2.
We then quantified the frequency of adaptive amino acid substitutions at bicycle genes and other categories of genes overexpressed in fundatrix salivary glands by calculating, the proportion of non-synonymous substitutions fixed by positive selection49,50. We estimate that for all bicycle genes α = 0.33 (95%CI 0.24 – 0.41) and that for the subset of bicycle genes displaying dN/dS significantly greater than 1, α = 0.62 (95%CI 0.45–0.73, Table S2). Other categories of genes overexpressed in fundatrix salivary glands display values of in the range of 0.27–0.38, however, bicycle genes display a considerably higher ratio of non-synonymous to synonymous substitutions than other categories of genes (Table S2). Most strikingly, as a fraction of protein length, bicycle genes display a considerably greater fraction of adaptive amino acid substitutions than other categories of genes (Figure 7C). Since speciation between H. cornu and H. hamamelidis, positive selection has resulted, on average, in fixation of approximately 2–3 substitutions in each bicycle gene, and approximately 10 substitutions in the most rapidly evolving bicycle genes. This represents a considerable fraction of the average length of these proteins (~200 residues) and reveals intense selection on bicycle genes, presumably for novel functions that require multiple amino acid substitutions.
The previous tests cannot detect selection on non-coding regions and do not discriminate between selection acting in the deep past versus more recently. To search for recent selection in bicycle gene regions, we examined patterns of polymorphism and divergence within and between H. cornu and H. hamamelidis. Polymorphism was strongly reduced relative to divergence in bicycle gene regions compared with the genomic background (Figures 7D–K; S6) and patterns of reduced polymorphism were strikingly similar in both species. This pattern is suggestive of recent selective sweeps in bicycle gene regions in both species, which we tested by performing genome-wide scans for positive selection51. Genome-wide signals of selective sweeps were enriched near bicycle genes and multiple signals fell within bicycle gene clusters in both species (Figures 7L–O; S7). Thus, in addition to long-term adaptive protein evolution of BICYCLE proteins, it appears that strong positive selection has acted recently and presumably frequently near many bicycle genes throughout the genome.
In summary, we find evidence for widespread, strong, recent, and frequent positive selection on bicycle genes. Since bicycle genes are likely secreted from salivary glands specifically in gall-forming aphids, these observations are consistent with the hypothesis that bicycle genes encode proteins that are intimately involved in reciprocal molecular evolution between the aphid and their host plant.
Discussion
We presented multiple lines of evidence that suggest that bicycle gene products provide instructive molecules required for aphid gall development. The strongest evidence is that eleven derived regulatory polymorphisms at dgc are associated with red galls (Figure 2), with almost complete silencing of dgc in aphid salivary glands (Figure 3), and with upregulation of seven anthocyanin biosynthetic genes and two red-purple anthocyanins in galls (Figure 4). Gall color is one small, but convenient, aspect of the panoply of cell biological changes required for gall development. We hypothesize that the product of each bicycle gene has its own unique set of targets in the plant and that the combined action of all bicycle gene products regulates many aspects of gall development. Testing this hypothesis will require the development of new methods to explore and manipulate this aphid-plant system.
BICYCLE protein functions
Dgc likely encodes a protein that is deposited by aphids into gall tissue, and current evidence suggests that this protein specifically and dramatically results in the downregulation of seven anthocyanin biosynthetic genes. The mechanisms by which this novel aphid protein could alter plant transcription remains to be determined. The primary sequences of DGC and other BICYCLE proteins provide few clues to their molecular mode of action. Outside of the N-terminal secretion signal, BICYCLE proteins possess no similarity with previously reported proteins and display no conserved domains that might guide functional studies. The relatively well-conserved C-Y-C motif appears to define a pair of ~50–80 aa domains in each protein and the paired cysteines may form disulfide bonds, which is commonly observed for secreted proteins. Secondary structure prediction methods provide little evidence for structural conservation across BICYCLE proteins and the extensive variation in the spacing between conserved cysteines further suggests that BICYCLE proteins may display structural heterogeneity. Identification of their molecular mode of action will require identification of BICYCLE protein binding targets.
Evolution of bicycle genes
We were unable to detect any sequence homology between bicycle genes and previously reported genes, which is one reason that it is difficult to infer the molecular function of bicycle genes from sequence alone. It is possible that bicycle genes evolved de novo in an ancestor of gall-forming aphids, perhaps through capture of a 5’ exon encoding an N-terminal signal sequence. However, if bicycle genes experienced strong diversifying selection since their origin, perhaps in an ancestor of gall forming aphids about 280 MYA, then the rate of amino-acid substitution that we detected between two closely-related species would likely be sufficient to have eliminated sequence similarity that could be detected by homology-detection algorithms. Identifying the evolutionary antecedents of bicycle genes will likely require tracing their evolutionary history across genomes of related species. It may also be more fruitful to use the unusual gene structure of bicycle genes to search for the antecedents of bicycle genes.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David L. Stern (sternd@hhmi.org)
Material Availability
This study did not generate new unique reagents.
Data and Code Availability
All sequencing data generated during this study are available at the NCBI Short Read Archive and the accession numbers are provided for each sample in Methods S1.
The genomes are available at Genbank and the genomes and gene annotations (GFF files) are available at FigShare (Methods S1J).
All of the analysis scripts we produced for this study are freely available at FigShare (Methods S1J).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Aphids and plants used in this study are listed in Methods S1.
METHOD DETAILS
Imaging of leaves and fundatrices inside developing galls
Young Hamamelis virginiana (witch hazel) leaves or leaves with early stage galls of Hormaphis cornu were fixed in Phosphate Buffered Saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) containing 0.1% Triton X-100, 2% paraformaldehyde and 0.5% glutaraldehyde (paraformaldehyde and glutaraldehyde were EM grade from Electron Microscopy Services) at room temperature for two hours without agitation to prevent the disruption of the aphid stylet inserted into leaf tissue. Fixed leaves or galls were washed in PBS containing 0.1% Triton X-100, hand cut into small sections (~10 mm2), and embedded in 7% agarose for subsequent sectioning into 0.3 mm thick sections using a Leica Vibratome (VT1000s). Sectioned plant tissue was stained with 0.1 mg/mL Calcofluor White (Sigma-Aldrich, F3543) and 0.25 mg/mL Congo Red (Sigma-Aldrich, C6767) in PBS containing 0.1% Triton X-100 with 0.5% DMSO and 0.05 mg/mL Escin (Sigma-Aldrich, E1378) at room temperature with gentle agitation for 2 days. Stained sections were washed with PBS containing 0.1% Triton X-100. Soft tissues were digested and cleared to reduce light scattering during subsequent imaging using a mixture of 0.25 mg/mL collagenase/dispase (Roche #10269638001) and 0.25 mg/mL hyaluronidase (Sigma Aldrich #H3884) in PBS containing 0.1% Triton X-100 for 5 hours at 37°C. To avoid artifacts and warping caused by osmotic shrinkage of soft tissue and agarose, samples were gradually dehydrated in glycerol (2% to 80%) and then ethanol (20% to 100 %)52 and mounted in methyl salicylate (Sigma-Aldrich, M6752) for imaging. Serial optical sections were obtained at 2 μm intervals on a Zeiss 880 confocal microscope with a Plan-Apochromat 10x/0.45 NA objective, at 1 μm with a LD-LCI 25x/0.8 NA objective or at 0.5 μm with a Plan-Apochromat 40x/0.8 NA objective. Maximum projections of confocal stacks or rotation of images were carried out using FIJI53.
Hormaphis cornu genome sequencing, assembly, and annotation
We collected H. cornu aphids from a single gall for genome sequencing (Methods S1A). All aphids within the gall were presumed to be clonal offspring of a single fundatrix, because all H. cornu galls we have ever examined have contained only a single fundatrix and the ostiole of the galls was closed at the time we collected this gall, so there is little chance of inter-gall migration. High molecular weight (HMW) DNA was prepared by gently grinding aphids with a plastic pestle against the inside wall of a 2 mL Eppendorf tube in 1 mL of 0.5% SDS, 200 mM Tris-HCl pH 8, 25 mM EDTA, 250 mM NaCl with 10 uL of 1 mg/mL RNAse A. Sample was incubated at 37°C for 1 hour and then 30uL of 10 mg/mL Proteinase K was added and the sample was incubated for an additional 1 hr at 50°C with gentle agitation at 300 RPM. One mL of Phenol:Chloroform:Isoamyl alcohol (25:24:1) was added and the sample was centrifuged at 16,000 RCF for 10 min. The supernatant was removed to a new 2mL Eppendorf tube and the Phenol:Chloroform:Isoamyl alcohol extraction was repeated. The supernatant was removed to a new 2 mL tube and 2.5 X volumes of absolute ethanol were added. The sample was centrifuged at 16,000 RCF for 15 min and then washed with fresh 70% ethanol. All ethanol was removed with a pipette and the sample was air dried for approximately 15 minutes and DNA was resuspended in 50 uL TE. This sample was sent to HudsonAlpha Institute for Biotechnology for genome sequencing.
DNA quality control, library preparation, and Chromium 10X linked read sequencing were performed by HudsonAlpha Institute for Biotechnology. Most of the mass of the HMW DNA appeared greater than approximately 50 kb on a pulsed field gel and paired end sequencing on an Illumina HiSeq X10 yielded 816M reads. The genome was assembled using Supernova54 using 175M reads, which generated the best genome N50 of a range of values tested. This 10X genome consisted of 21,072 scaffolds of total length 319.352 MB. The genome scaffold N50 was 839.101 KB and the maximum scaffold length was 3.495 MB.
We then contracted with Dovetail Genomics to apply Chicago (in vitro proximity ligation) and HiC (in vivo proximity ligation) to generate larger scaffolds (https://dovetailgenomics.com/ga_tech_overview/). We submitted HMW gDNA from the same sample used for 10X genome sequencing for Chicago and a separate sample of frozen aphids for HiC (Methods S1A). The Dovetail genome consisted of 11,244 scaffolds of total length 320.34 MB with a scaffold N50 of 36.084 Mb. This genome, named hormaphis_cornu_26Sep2017_PoQx8, contains 9 main scaffolds, each longer than 17.934 Mb, which appear to represent the expected 9 chromosomes of H. cornu55. This assembly also includes the circular genome of the bacterial endosymbiont Buchnera aphidicola of 643,259 bp. Assembly and BUSCO analysis statistics56 using the gVolante web interface57 with the Arthropod gene set are shown below.
| Assembly statistics | hormaphis_cornu_26Sep2017_PoQx8 |
|---|---|
| # of scaffolds: | 11242 |
| Total length: | 320,336,030 |
| Longest sequence: | 60,222,264 |
| N50 sequence length: | 36,083,769 |
| Sum length of sequences > 1M | 297,831,669 (93.0% of total length) |
| Sum length of sequences > 10M | 294,347,208 (91.9% of total length) |
| BUSCO Analysis | hormaphis_cornu_26Sep2017_PoQx8 |
|---|---|
| Total # of core genes queried | 1066 |
| # of complete core genes detected | 1026 (96.25%) |
| # of complete and partial core genes detected | 1038 (97.37%) |
| # of missing core genes: | 28 (2.63%) |
| Average # of orthologs per core genes: | 1.02 |
| % of detected core genes that have more than 1 ortholog: | 2.34 |
As further checks on the quality of this genome assembly, we examined the K-mer spectra (Figure S1A) and the HiC contact map (Figure S1B).
We annotated this genome for protein-coding genes using RNA-seq data collected from salivary glands and carcasses of many stages of the H. cornu life cycle (Methods S1B) using BRAKER58–65. To increase the efficiency of mapping RNA-seq reads for differential expression analysis, we predicted 3’ UTRs using UTRme66. We found that UTRme sometimes predicted UTRs within introns. We therefore applied a custom R script to remove UTRs located within introns. Later, after discovering the bicycle genes, we manually annotated all predicted bicycle genes, including 5’ and 3’ UTRs, in APOLLO67 by examining evidence from RNA-seq reads aligned to the genome with the Integrative Genomics Viewer68,69. We found that the start sites of many bicycle genes were incorrectly annotated by BRAKER at a downstream methionine, inadvertently excluding predicted putative signal peptides from these genes. RNA-seq evidence often supported transcription start sites that preceded an upstream methionine and these exons were corrected in APOLLO. The combined collection of 18,895 automated and 687 manually curated gene models (19,582 total) were used for all subsequent analyses of H. cornu genomic data. The genome assembly (JABAOA000000000) and sequence reads (PRJNA614456) are available from NCBI. The genome assembly and our annotations are also available on FigShare (Methods S1J).
Genome-wide association study of aphids inducing red and green galls
Galls produced by H. cornu were collected in the early summer (Methods S1C) and dissected by making a single vertical cut down the side of each gall with a razor blade to expose the aphids inside. DNA was extracted using the Zymo ZR-96 Quick gDNA kit from the foundress of each gall. We performed tagmentation of genomic DNA derived from 47 individuals from red galls and 43 from green galls using barcoded adaptors compatible with the Illumina sequencing platform70. Tagmented samples were pooled without normalization, PCR amplified for 14 cycles, and sequenced on an Illumina NextSeq 500 to generated paired end 150 bp reads to an average depth of 2.9X genomic coverage. The average sequencing depth before filtering was calculated by multiplying the number of read pairs generated by SAMtools flagstat version 1.371 by the read length of 150bp, then dividing by the total genome size (323,839,449bp).
We performed principle component analysis on the genome-wide polymorphism data to detect any potential population structure that might confound a GWAS. Reads were mapped using bwa mem version 0.7.17-r118863 and joint genotyped using SAMtools mpileup version 1.3, with the flag -ugf, followed by BCFtools call version 1.972, with the flag -m. Genotype calls were then filtered for quality and missingness using BCFtools filter and view version 1.9, where only SNPs with MAF > 0.05, QUAL > 20, and genotyped in at least 80% of the individuals were kept. To limit the number of SNPs for computational efficiency, the SNPs were additionally thinned using VCFtools –thin version 0.1.1573 to exclude any SNPs within 1000 bp of each other. PCA was performed using the snpgdsPCA function from the R package SNPRelate version 1.20.1 in R version 3.6.174.
We performed a GWAS with these low coverage data by mapping reads with bwa mem version 0.7.17-r1188 and calculating the likelihood of association with gall color with SAMtools mpileup version 0.1.19 and BCFtools view -vcs version 0.1.19 using BAM files as the input. Association for each SNP was measured by the likelihood-ratio test (LRT) value in the INFO field of the output VCF file, which is a one-degree of freedom association test P value. This method calculates association likelihoods using genotype likelihoods rather than hard genotype calls, ameliorating the issue of low-confidence genotype calls resulting from low-coverage data72. The false discovery rate was set as the Bonferroni corrected value for 0.05, which was calculated as 0.05 / 50,957,130 (the total number of SNPs in the genome-wide association mapping).
Enrichment and sequencing of the genomic region containing highly significant GWAS hits
The low coverage GWAS identified multiple linked SNPS on chromosome 1 that were strongly associated with gall color (Figure 2A). To identify all candidate SNPs in this genomic region and to generate higher-confidence GWAS calls, we enriched this genomic region from a library of pooled tagmented samples of fundatrix DNA from red and green galls using custom designed Arbor Bioscience MyBaits for a 800,290 bp region on chromosome 1 spanning the highest scoring GWAS SNP (40,092,625 – 40,892,915 bp). This enriched library was sequenced on an Illumina NextSeq 550 generating paired-end 150 bp reads and resulted in usable resequencing data for 48 red gall-producing individuals and 42 green gall-producing individuals, with average pre-filtered sequencing depth of 58.2X.
We mapped reads with bwa mem version 0.7.17-r1188 and sorted bam files with SAMtools sort version 1.7, marked duplicate reads with Picard MarkDuplicates version 2.18.0 (http://broadinstitute.github.io/picard/), re-aligned indels using GATK IndelRealigner version 3.475, and called variants using SAMtools mpileup version 1.7 and BCFtools call version 1.7 (https://github.com/SAMtools/bcftools). This genotyping pipeline is available at https://github.com/YourePrettyGood/PseudoreferencePipeline (thereafter referred to as PseudoreferencePipeline). SNPs were quality filtered from the VCF file using BCFtools view version 1.7 at DP > 10 and MQ > 40 and merged using BCFtools merge.
For PCA analysis, the joint genotype calls were filtered for quality and missingness using BCFtools filter and view version 1.9, where only SNPs with MAF > 0.05 and genotyped in at least 80% of the individuals were retained. PCA was performed using the snpgdsPCA function from the R package SNPRelate version 1.20.1 in Rstudio version 3.6.1.
Association testing was performed using PLINK version 1.9076 with minor allele frequency filtered at MAF > 0.2. We did not apply a Hardy-Weinberg equilibrium filter because the samples were not randomly collected from nature. Red galls are rare in our population and we oversampled fundatrices from red galls to roughly match the number of fundatrices sampled from green galls. Results of the GWAS were plotted using the plotManhattan function of Sushi version 1.24.077.
To calculate LD for the 45kbp region surrounding the 11 most strongly associated SNPs, we extracted positions 40,475,000 – 40,520,000 of chromosome 1 from the merged VCF and retained SNPs that both exhibited MAF > 0.05 and were genotyped in at least 80% of the samples using BCFtools view and filter version 1.9.
To plot LD for the entire target enrichment region, we filtered the VCF for only the SNPs with MAF>0.2 and that were genotyped in at least 80% of the samples using bcftools view and filter version 1.9, and thinned the resulting SNP set using VCFtools–thin version 0.1.15 to exclude any SNPs within 500bp of each other. We further merged back the 11 significant GWAS SNPs using bcftools concat, since the thinning process could have removed one or more of these SNPs. We also removed SNPs in regions where the H. cornu reference genome did not align with the genome of the sister species H. hamamelidis using BEDTools intersect version 2.29.278.
The LD heatmaps were generated using the R packages vcfR version 1.10.079, snpStats version 1.36.080, and LDheatmap version 0.99.881 in Rstudio version 3.6.1. The R code used to generate the LD heatmap figure was adapted from code provided at sfustatgen.github.io/LDheatmap/articles/vcfOnLDheatmap.html. The gene models were plotted using the plotGenes function from the R package Sushi version 1.24.0.
Lack of evidence for chromosomal aberrations
To identify possible chromosomal rearrangements or transposable elements that might be linked to the GWAS SNPs, we first trimmed adapters from the H. cornu target enrichment data using Trim Galore! version 0.6.5 and cutadapt version 2.782. The trimming pipeline is available at github.com/YourePrettyGood/ParsingPipeline. We then mapped the reads to the H. cornu reference genome with bwa mem version 0.7.17, sorted BAM files with SAMtools sort version 1.9, and marked duplicate reads with Picard MarkDuplicates version 2.22.7 (http://broadinstitute.github.io/picard/), all done with the MAP function of the PseudoreferencePipeline. The analysis includes 43 high coverage red individuals and 42 high coverage green individuals. The five individuals isolated from red galls that did not carry the associated GWAS SNPs in dgc were excluded since the genetic basis for their gall coloration is unknown.
We selected the subset of the BAM file for each individual that contained only the target enrichment region on chromosome 1 from 40,092,625 to 40,892,915 bp using SAMtools view version 1.9 and generated a merged BAM file for each color using SAMtools merge. The discordant reads were then extracted from each BAM file using SAMtools view with flag -F 1286 and the percentage of discordant reads was calculated as the ratio of the number of discordant reads over the total number of mapped reads for each 5000 bp window.
To further explore the possibility that chromosomal aberrations near the GWAS signal might differ between red- and green-gall producing individuals, we plotted the mapping locations of discordant reads in the 100 kbp region near the 11 GWAS hits (40,450,000 – 40,550,000 bp) for red individuals, since the H. cornu reference was made from a green individual. We obtained the read ID for all the discordant reads within the 100 kbp region and extracted all occurrences of these reads from the whole genome BAM file, regardless of their mapping location. We then extracted the paired-end reads from the discordant reads BAM file using bedtools bamtofastq version 2.29.2 and used bwa mem to map these reads as single-end reads for read 1 and read 2 separately to a merged reference containing the H. cornu genome and the 343 Acyrthosiphon pisum transposable elements annotated in RepBase83. We then removed duplicates and sorted the BAM file using SAMtools rmdup and sort and determined the mapping location of all discordant reads using SAMtools view. We masked the windows on chromosome 1 from 40,400,000–40,599,999 bp in the genome-wide scatter plot of discordant reads mapping because the majority of the discordant reads are expected to map to these regions and displaying their counts would obscure potential signals in the rest of the genome.
Large scale survey of 11 dgc SNPs associated with gall color
Aphids were collected from red and green galls as described above for the GWAS study directly into Zymo DNA extraction buffer and ground with a plastic pestle. DNA was prepared using the ZR-96 Quick gDNA kit. We developed qPCR assays and amplicon-seq assays to genotype all individuals at all 11 SNPs (Methods S1L–M). PCR amplicon products were barcoded and samples were pooled for sequencing on an Illumina platform.
Adaptors were trimmed from amplicon reads using Trim Galore! version 0.6.5 and cutadapt version 2.7. The wrapper pipeline is available at github.com/YourePrettyGood/ParsingPipeline. We mapped reads to a 34 kbp region of chromosome 1 of the H. cornu genome that includes the amplicon SNPs (40,477,000 – 40,511,000 bp) with bwa mem version 0.7.17, sorted BAM files with SAMtools sort version 1.9, and re-aligned indels using GATK IndelRealigner version 3.4. No marking of duplicates was done given the nature of amplicon sequencing data. To maximize genotyping efficiency and improve accuracy, we performed variant calling with two distinct pipelines: SAMtools mpileup version 1.7 plus BCFtools call version 1.7, and GATK HaplotypeCaller version 3.4. The mapping and indel re-alignment pipelines are available as part of the MAP (with flag only_bwa) and IR functions of the PseudoreferencePipeline. Using the same PseudoreferencePipeline, variant calling was performed using the MPILEUP function of BCFtools and HC function of GATK. FASTA sequences for each individual, where the genotyped SNPs were updated in the reference space, were then generated for both BCFtools and GATK variant calls using the above PseudoreferencePipeline’s PSEUDOFASTA function, with flags “MPILEUP, no_markdup” and “HC, no_markdup” respectively. The BCFtools SNP updating pipeline used bcftools filter, query, and consensus version 1.9, and we masked all sites where MQ <= 20 or QUAL <= 26 or DP <= 5. The HC SNP updating pipeline used GATK SelectVariants and FastaAlternateReferenceMaker version 3.4, and we masked all sites where MQ < 50, DP <= 5, GQ < 90 or RGQ < 90.
We then merged the variant calls from BCFtools and GATK, as well as the qPCR genotyping results, and manually identified all missing or discrepant genotypes. We manually curated these missing or discrepant genotype calls from the indel realigned BAM files using the following criteria: for heterozygous calls, the site had at least two reads supporting each allele, and for homozygous calls, the site had at least ten reads supporting the allele and no reads supporting alternative alleles.
RNA-seq of salivary glands from aphids inducing red and green galls
We dissected salivary glands from fundatrices isolated from green and red galls in PBS, gently pipetted the salivary glands from the dissection tray in < 0.5uL volume of PBS, and deposited glands into 3 uL of Smart-seq2 lysis buffer (0.2% Triton-X 100, 0.1 U/uL RNasin® Ribonuclease Inhibitor). RNA-seq libraries were prepared with a single-cell RNA-seq method developed by the Janelia Quantitative Genomics core facility and described previously84. RNA-seq libraries were prepared as described above for red and green gall samples except that the entire 3uL sample of salivary glands in lysis buffer was provided as input. Barcoded samples were pooled and sequenced on an Illumina NextSeq 550. We detected 9.0 million reads per sample on average. We replaced the original oligonucleotides with modified oligonucleotides to generate unstranded reads from the entire transcript (Methods S1K). Samples were PCR amplified for 18 cycles and the library was prepared using ¼ of the standard Nextera XT sample size and 150 pg of cDNA.
Differential expression analyses of fundatrix salivary glands from red and green galls
All differential expression analyses for plant and aphid samples were performed in R version 3.6.185 and all R Notebooks are provided on FigShare (Methods S1J). Adapters were trimmed using cutadapt version 2.7 and read counts per transcript were calculated by mapping reads to the genome with hisat2 version 2.1.086 and counting reads per gene with htseq-count version 0.12.487. In R, technical replicates were examined and pooled, since all replicates were very similar to each other. We performed exploratory data analysis using interactive multidimensional scaling plots, using the command glMDSPlot from the package Glimma88, and outlier samples were excluded from subsequent analyses. Differentially expressed genes were identified using the glmQLFTest and associated functions of the package edgeR89. Volcano plots were generated using the EnhancedVolcano command from the package EnhancedVolcano version 1.4.090.
Hamamelis virginiana genome sequencing, assembly, and annotation
Leaves from a single tree of Hamamelis virginiana were sampled from the Janelia Research Campus forest as follows. Branches containing leaves that were less than 50% expanded were wrapped with aluminum foil and harvested after 40 hours. Leaves were cleared of obvious contamination, including aphids and other insects, and then plunged into liquid N2. Samples were stored at −80°C and sent to the Arizona Genomics Institute, University of Arizona on dry ice, which prepared HMW DNA from nuclei isolated from the frozen leaves. The Janelia Quantitative Genomics core facility generated a 10X Chromium linked-read library from this DNA and sequenced the library on an Illumina NextSeq 550 to generate 608M linked reads.
The H. virginiana genome was assembled with the supernova commands run and mkoutput version 2.1.1, with options minsize=1000 and style=pseudohap54. We used 332M reads in the assembly to achieve raw coverage of 56X as recommended by the supernova instruction manual. BUSCO completeness analysis56 was performed using the gVolante Web interface57 using the plants database. Genome assembly and BUSCO statistics are reported below.
| Assembly statistics | Hvir_nuclei_sn_run2_2_pseudohap |
|---|---|
| # of scaffolds: | 84,975 |
| Total length: | 907,642,797 |
| Longest sequence: | 7,097,227 |
| N50 sequence length: | 167,515 |
| Sum length of sequences > 1M | 142 |
| Sum length of sequences > 1M (nt) | 303,391,067 (33.4% of total length) |
| BUSCO Analysis | Hvir_nuclei_sn_run2_2_pseudohap |
|---|---|
| Total # of core genes queried | 1440 |
| # of complete core genes detected | 1309 (90.90%) |
| # of complete and partial core genes detected | 1365 (94.79%) |
| # of missing core genes: | 75 (5.21%) |
| Average # of orthologs per core genes: | 1.05 |
| % of detected core genes that have more than 1 ortholog: | 3.90 |
The assembled genome reference was repeat masked with soft masking using RepeatMasker version 4.0.991. Twenty-five RNA-seq libraries from galls and leaves were used for genome annotation. RNA-seq reads were adapter trimmed using cutadapt version 2.7 and mapped to the genome using HISAT2 version 2.1.0. Genome annotation was performed with BRAKER version 2.1.4 using the RNA-seq data to provide intron hints58–62,64,65,71,92,93 and 3’ UTRs were predicted using UTRme66. The genome assembly (JAESVK000000000) and sequence reads (PRJNA614456) are available from NCBI. The genome assembly and our annotations are also available on FigShare (Methods S1J).
H. virginiana RNA extraction and RNA-seq library preparation
RNA was extracted from frozen H. virginiana leaf or gall tissue as follows. Plant tissue frozen at −80°C was placed into ZR BashingBead Lysis Tubes (pre-chilled in liquid N2) and pulverized to a fine powder in a Talboys High Throughput homogenizer (Troemer) with minimal thawing. Powdered plant tissue was suspended in extraction buffer (100 mM Tris-HCl, pH 7.5, 25 mM EDTA, 1.5 M NaCl, 2% (w/v) Hexadecyltrimethylammonium bromide, 10% Polyvinylpyrrolidone (w/v) and 0.3% (v/v) β-mercaptoethanol) and heated to 55°C for 8 min followed by centrifugation at 13,000 × g for 5 minutes at room temperature to remove insoluble debris94. Total RNA was extracted from the supernatant using the Quick-RNA Plant Miniprep Kit (Zymo Research) with the inclusion of in-column DNAse I treatment. RNA-seq libraries were prepared with the Universal Plus mRNA-Seq kit (Nugen).
Differential expression analysis of red versus green galls
We performed differential expression analysis on red and green galls by collecting paired red and green gall samples from the same leaves. In total, we collected 17 red galls and 23 green galls from 17 leaves. RNA was prepared as described above for plant material and RNA-seq libraries were prepared with the single-cell RNA-seq method described above.
These RNA-seq libraries contained on average 4.4 million mapped reads per sample. Reads were quality trimmed and mapped to the transcriptome as described above. Only genes with greater than 1 count per million in at least 15 samples were included in subsequent analyses. Red and green samples clustered together in a principal components analysis (Figure S3B and C) and no samples were identified as outliers. The expression analysis model included the effect of leaf blocking.
Gall pigment extraction and analysis
Frozen gall tissue was ground to a powder under liquid nitrogen. We first tested for the presence of carotenoids by dehydrating gall tissue with methanol and extracting with hexane/acetone95. However, the lipophilic extract was colorless and all color remained in the polar phase and pellet, indicating that carotenoids do not contribute to red gall color.
We therefore next tested for presence of anthocyanins. Approximately 20 mg of ground gall tissue was suspended in 100 μL methanol (Optima grade, Fisher Scientific) and 400 μl of 5% aqueous formic acid (Optima grade, Fisher Scientific), vortexed for 30 seconds and centrifuged at 8000 × g for 2 min at 10° C. The supernatant containing pigment was filtered using a 0.2 μm, 13 mm diameter PTFE syringe filter to remove debris. Colorless pellet was discarded. Authentic anthocyanin pigment standards for malvidin 3,5-diglucoside chloride (Sigma Aldrich, St. Louis, MO, USA) and peonidin-3,5-diglucoside chloride (Carbosynth LLC,San Diego, CA, USA) were prepared at 1mg/mL in 5% aqueous formic acid.
Pigment separation and identification alongside standards was performed on a reverse phase C18 column (Acquity Plus BEH, 50 mm × 2.1 mm, 1.7 μm particle size, Waters, Milford, MA) using an Agilent 1290 UHPLC coupled to an Agilent 6545 quadrupole time-of-flight mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) using an ESI probe in positive ion mode. Five μl of filtered pigment extract or a 1:100 dilution of anthocyanin standard was injected. Solvent (A) consisted of 5% aqueous formic acid and Solvent (B) 1:99 water/acetonitrile acidified with 5% aqueous formic acid (v/v). The gradient conditions were as follows: 1 min hold at 0% B, 4 min linear increase to 20% B, 5 min linear increase to 40% B, ramp up to 95% B in 0.1 min, hold at 95% B for 5 min, return to 0% B and hold for 0.9 min. The column flow rate was 0.3 mL/min, and the column temperature 30° C. The MS source parameters for initial anthocyanin detection were as follows: capillary = 4000 V, nozzle = 2000 V, gas temperature = 350° C, gas flow = 13 L/min, nebulizer = 30 psi, sheath gas temperature = 400° C, sheath gas flow = 12 L/min. DAD detection at 300 nm and 520 nm, and MS scanning from 50–1700 m/z at a rate of 2 spectra per second. Iterative fragmentation, followed by targeted MS/MS experiments, were performed using a collision energy = 35. Authentic standards confirmed the presence of peonidin-3,5-diglucoside and malvidin 3,5-diglucoside. The remaining anthocyanin species were identified using UV-Vis spectra, retention time relative to the other species in the sample, [M]+ precursor ions, and aglycone fragment ions matching the respective entries in the RIKEN database96.
Differential expression analysis of galls versus leaves
We performed RNA seq on 36 gall samples and 17 adjacent leaf samples. These gall samples did not overlap with the gall samples used in red versus green gall comparison described earlier. For larger galls, RNA was isolated separately from basal, medial, and apical gall regions (Methods S1H). Libraries were sequenced on an Illumina NextSeq 550 to generate 150 bp paired-end reads with an average of 8.1 million mapped reads per sample. Only genes expressed at greater than 1 count per million in at least 18 samples were included in subsequent analysis. Gall and leaf samples clustered separately in a principal components analysis (Figure S3D and E) and no samples were excluded as outliers. We included only samples for which paired gall and leaf samples were available from the same leaf and potential leaf effects were modeled in the different expression analysis.
To facilitate Gene Ontology (GO) analysis, the UniProt IDs of the differentially expressed genes were obtained by mapping the coding sequences of the H. virginana genome to the UniProt/Swiss-Prot database97 using Protein-Protein BLAST 2.7.158,60 and by extracting the differentially expressed genes (Figure 4G). We used the WebGestalt 2019 webtool98 to perform GO analysis on the differentially expressed genes.
Differential expression analysis of H. cornu organs and life stages
RNA-seq libraries were generated for fundatrix salivary glands (N = 20) and whole bodies (N = 8), G2 salivary glands (N = 6) and carcasses (N = 3), G5 salivary glands (N = 6) and carcasses (N = 2), and G7 salivary glands (N = 5). Libraries were generated as described above for salivary glands except that RNA samples of carcasses and whole bodies were prepared using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems). Only genes expressed with at least 1 count per million in at least 29 samples were included in subsequent analyses.
Bioinformatic identification of bicycle genes in H. cornu
Genes that were upregulated specifically in the salivary glands of the fundatrix generation were prime candidates for inducing galls. We therefore identified genes that were upregulated both in salivary glands of fundatrices versus sexuals and in fundatrix salivary glands versus fundatrix body. These differentially expressed genes were then separated into genes with and without homologs containing some functional annotation. Homologs with previous functional annotations were identified using three methods: we performed (1) translated query-protein (blastx) and (2) protein-protein (blastp) based homology searches using BLAST 2.7.158,60 against the UniProt/Swiss-Prot database97, and (3) Hidden-Markov based searches with the predicted proteins using hmmscan in HMMER version 3.1b299 against the pfam database100. For all predicted proteins, we also searched for secretion signal peptides using SignalP-5.0101 and for transmembrane domains using tmhmm version 2.0102. Gene Ontology analysis of genes with annotations that were enriched in fundatrix salivary glands was performed by searching for Drosophila melanogaster homologs of differentially expressed genes and using these D. melanogaster homologs as input into gene ontology analysis.
To determine whether any of the differentially expressed genes without detectable homologs in existing protein databases were homologous to each another, we performed sensitive homology searches of all-against-all of these genes using jackhmmer in HMMER version 3.1b2. We performed hierarchical clustering on the quantitative results of the jackhmmer analysis by first calculating distances amongst genes with the dist function using method canberra and clustering using the hclust function with method ward.D2, both from the library stats in R85. We aligned sequences of the clustered homologs using MAFFT version 7.419 with default parameters103,104, trimmed aligned sequences using trimAI105 with parameters -gt 0.50, and generated sequence logos by importing alignments using the functions read.alignment and ggseqlogo in the R packages seqinr106 and ggseqlogo107. After identification of the bicycle genes, we searched for additional bicycle genes in the entire H. cornu genome, which might not have been enriched in fundatrix salivary glands, using jackhmmer followed by hierarchical clustering to identify additional putative homologs. As described above, we manually annotated all of these candidate bicycle genes.
RNA-seq analysis of a single H. cornu fundatrix with a dgcGreen genotype inducing a red gall
Approximately 2.1% of fundatrices inducing red galls were homozygous for ancestral “green” alleles at all of the 11 dgc SNPs (Figure 2E). Five such individuals were found in our original GWAS study and we did not observe any variants in the dgc gene region that were associated specifically with these individuals, suggesting that they carried variants elsewhere in the genome that caused them to generate red galls. Since isolating salivary glands from fundatrices is challenging and time consuming, we were unable to systematically examine transcriptome changes in the salivary glands of the rare individuals homozygous for dgcGreen that induced red galls. However, we fortuitously isolated salivary glands from one fundatrix from a red gall that was homozygous for dgcGreen. We performed whole-transcriptome sequencing of the salivary glands from this one individual and compared expression levels of all genes in the bicycle gene paralog group to which dgc belongs. We also examined the dgc transcripts produced by this individual and found no exonic SNPs in the dgc transcripts produced by this individual, indicating that this individual probably expressed a functional dgc transcript.
H. cornu and H. hamamelidis polymorphism and divergence measurements in GWAS region
To summarize the polymorphism and divergence patterns in H. cornu and H. hamamelidis in the target enrichment region, we generated SNP updated FASTA sequences for each individual by mapping the H. cornu reads to the H. cornu genome reference and the H. hamamelidis reads to a H. hamamelidis SNP-updated reference genome in H. cornu genome coordinate space. We used the same genotyping pipeline (PseudoreferencePipeline) as described above for the enrichment region GWAS and sites with DP <= 10, MQ <= 20.0, or QUAL <= 29.5 were masked. High coverage individuals of H. cornu (n = 90) and H. hamamelidis (N = 92) were included in this analysis, including both of the color phenotypes. Polymorphism within each species and divergence (Dxy) between species were calculated using a custom script calculateDiversity (available at https://github.com/YourePrettyGood/DyakInversions/tree/master/tools). Windowed measurements of each of these statistics were generated with the custom script nonOverlappingWindows.cpp, using window size 3000bp (script available at github.com/YourePrettyGood/RandomScripts). Sites with missing genotypes due to masking in 50% or more of the samples were not included in the windowed average and windows where 50% or more of the sites were missing were excluded from the plots in Figure 7D–G.
Whole-genome polymorphism and divergence measurements in H. cornu and H. hamamelidis
The sister species H. cornu and H. hamamelidis produce similar looking galls in adjacent geographic areas and were long confused as populations of a single species 108. However, H. cornu exhibits a life cycle where aphids alternate between Hamamelis virginiana and Betula nigra (Figure 1J), whereas H. hamamelidis does not host alternate to B. nigra and displays a truncated life cycle, where the offspring of the fundatrix develop as sexuparae (the equivalent of G6 in Figure 1J) and deposit sexuals on H. virginiana in the autumn. Populations of H. cornu tend to live in the lowlands with prolonged summers and H. hamamelidis tend to live in the highlands and more northern latitudes, which experience shorter summers. In some locations, both species can be found making galls on the same trees.
Previous mtDNA sequencing supported the hypothesis that these are distinct species109. We estimate genome-wide divergence (Dxy) between these species to be 0.0189 and polymorphism to be 0.0132 and 0.0125 for H. cornu and H. hamamelidis, respectively, providing further support that these are reproductively separate species. Assuming a mutation rate of μ=2.8e-9110, we estimated effective population sizes using θ = 4Neμ for H. cornu and H. hamamelidis as 1,174,297 and 1,115,822, respectively.
To examine genome-wide patterns of polymorphism and divergence, we used the same whole-genome sequencing data used in the genome-wide association study for gall color. These data included samples from fundatrices isolated from 43 green galls and 47 red galls for H. cornu and 48 galls for H. hamamelidis. Reads were mapped using bwa mem version 0.7.17-r1188. The H. cornu reads were mapped to the H. cornu reference genome and the H. hamamelidis reads were mapped to the H. hamamelidis SNPs updated reference genome. We joint genotyped each species separately using bcftools version 1.9 mpileup and call -m to generate raw, multi-sample VCF for each species. Sites filtering for QUAL >= 20 was then done using bcftools filter, and insertions and deletions sites were removed using VCFtools version 0.1.16 –remove-indels73. To mask sites lacking genotype calls for each sample individually, we used bcftools query to extract the positions of all sites, bcftools view with flag - g ^miss to extract the positions of genotyped sites for each sample, and bedtools version 2.29.2 complement to generate the positions of non-genotyped sites, which were then masked. Consensus sequences were generated for each sample using bcftools consensus, with the flag – iupac-codes.
Polymorphism within each species and divergence (Dxy) between species was calculated using a custom script calculateDiversity (available at https://github.com/YourePrettyGood/DyakInversions/tree/master/tools). Windowed measurements of each of these statistics were generated with the custom script nonOverlappingWindows.cpp, using window size 1000bp (script available at github.com/YourePrettyGood/RandomScripts). Windows were designated as “bicycle” if they overlapped in coordinates with any bicycle genes; otherwise they were designated as “non-bicycle”. Sites masked in 80% or more of the samples were excluded in the windowed average and windows with 80% or more of the sites missing were excluded from genome-wide summary statistic calculations. This genotyping rate filter is more lenient than the enrichment region genotyping to accommodate the shallower sequencing depth of the whole-genome data, which resulted in a higher rate of missing data in the whole-genome data.
Bioinformatic analysis of bicycle gene structure and evolution
The H. cornu median exon size and number of exons per bicycle gene were calculated from the GFF annotation file Augustus.updated_w_annots.21Aug20.gff3. The bicycle genes alignment for the divergence tree was generated using FastTree version 2.1.11111 and plotted using ggtree version 2.2.2112 in R.
To calculate DnDs between H. cornu and H. hamamelidis, we first generated a masked H. hamamelidis reference genome by updating the H. cornu reference genome with H. hamamelidis SNPs. We selected a random subset of H. hamamelidis reads from the 150 bp PE 10X linked reads sequencing data to approximately 65X coverage using seqtk version 1.3 sample -s100 (https://github.com/lh3/seqtk) and trimmed adapters using Long Ranger version 2.2.2 basic (10X Genomics). We then mapped the H. hamamelidis reads to the H. cornu reference and updated the SNPs using the PseudoreferencePipeline. Sites where MQ <= 20, QUAL <= 26 or DP <= 5 were masked, resulting in 21.0% of the genome being masked. This H. hamamelidis reference genome was used in the DnDs calculation.
DnDs between all orthologous genes in H. cornu and H. hamamelidis was calculated using the codeml function from the PAML package, version 4.9j113. The CDS sequences were extracted from both reference genomes for each gene using constructCDSesFromGFF3.pl and we split all degenerate bases into the two alleles to generate two pseudo-haplotypes using fakeHaplotype.pl -s 42 (both scripts available at github.com/YourePrettyGood/RandomScripts). Haplotype 1 was used as input into codeml for both species. The settings used in the codeml control file were: runmode=−2, seqtype=1, CodonFreq=0, clock=0, model=0, NSsites=0, fix_kappa=0, kappa=2, fix_omega=0, omega=1, Small_Diff=0.5e-6, method=0, fix_blength=0. Then, to evaluate whether DnDs is significantly different from 1 for each gene, we additionally ran a null model with the same control file, except with fix_omega=1. We then calculated the likelihood scores between the two models as 2×(lnL1-lnL2) and compared it to the 95th percentile of the Chi-square distribution with 1 degree of freedom. We performed a Mann-Whitney U test comparing DnDs of bicycle and non-bicycle genes using the wilcox.test function in R version 3.6.1.
To perform McDonald-Kreitman tests for bicycle genes, we generated a population sample of the bicycle gene coding regions for H. cornu by mapping RNA-seq data from 21 fundatrix salivary gland samples, which provided high coverage. We performed genotyping using the STAR, IRRNA, and HC functions in the PseudoreferencePipeline. This pipeline used STAR version 2.7.3a for mapping and IndelRealigner and HaplotypeCaller in GATK version 3.4 for indel realignment and variant calling. SNP-updated FASTA sequences were generated for each individual from the genotype VCF using the PSEUDOFASTA function of the PseudoreferencePipeline without any additional masking. Then, for each individual, we split all degenerate bases into the two alleles to generate two pseudo-haplotypes using fakeHaplotype.pl -s 42. We calculated the number of synonymous and nonsynonymous polymorphisms (PS and PN, respectively) and divergent substitutions (DS and DN, respectively) using Polymorphorama version 6 (https://ib.berkeley.edu/labs/bachtrog/data/polyMORPHOrama/polyMORPHOrama.html)114 for multiple categories of genes significantly over-expressed in the fundatrix salivary gland. The fraction of non-synonymous divergent substitutions that were fixed by positive selection50,115 (α) was estimated using the Cochran-Mantel-Haenszel test framework116. Using the mantelhaen.test function in R version 3.6.1, we estimated a common odds ratio across a series of McDonald-Kreitman tables117, and estimated . A MAF filter of 0.03 was applied to the polymorphism counts to reduce the downward bias in the estimator due to segregating weakly deleterious amino acid polymorphisms115.
Genome-wide tests of selection
To scan for genome-wide signatures of positive selection in H. cornu and H. hamamelidis, we used the joint-genotyped VCF for each species as described above. We filtered for QUAL >= 20 using bcftools filter, and 80% maximum missing genotype and biallelic sites using VCFtools version 0.1.16 -max-missing 0.2, -max-alleles 2, and -min-alleles 2. We then performed the selection scan using SweeD-P version 3.151, with one grid point per kilobase and -folded flag.
To determine the significance cutoff for the composite likelihood ratio (CLR) output by SweeD, we simulated 10 Mbp regions under neutrality for 100 haplotypes for each species using MaCS version 0.4f118. The effective population size was set to 1,174,297 and 1,115,822 for H. cornu and H. hamamelidis, respectively. The mutation and recombination rates were set to 2.8e-9 and 1.1e-8, respectively, per bp per generation. The simulation output was converted to multi-sample VCF format using a custom script and run through SweeD-P using the same parameters as for the observed data. We ran ten iterations of the simulation for each species and combined the results of each iteration within each species to account for stochasticity of the simulations. The CLR cutoffs for the observed data were chosen as the 99th quantiles of the CLR values from the neutral simulations of their respective species.
Identification of H. cornu homologs of genes previously proposed as gall effector genes in other species
The SSGP-71s effector gene family was described from the Hessian fly genome119 and it was suggested that these encode E3-ligase-mimicking effector proteins. We identified four homologs of the SSGP-71s effector gene family in H. cornu by searching H. cornu protein sequences using a Hidden Markov Model search with a protein profile generated using the 426 SSGP-71 genes from Document S2, SSGP-71s tab, from Zhao et al. 201532 (hmmsearch with HMMER v3.2.1). Two of these genes were not expressed at sufficient levels to be detected in our differential expression studies and neither of the remaining two were among the 1,482 genes specifically enriched in the salivary glands of fundatrices (Figure S4F). Thus, it is unlikely that SSGP homologs contribute to gall development in H. cornu.
We also identified eight ubiquitin E3-ligase genes119 in H. cornu based on protein similarity to proteins in the PFAM database100 that include “E3” in the description. Seven of these genes had sufficient expression to be detected in the differential expression studies, and only two were among the 1,482 genes enriched in fundatrix salivary glands (Figure S4G). In addition, none of these seven genes contain N-terminal secretion signals, making it unlikely that they could be injected into plants. Thus, secreted ubiquitin E3-ligase genes are unlikely to contribute to H. cornu gall development.
Ring domain proteins usually mediate E2 ubiquitin ligase activity, which can modify protein function or target proteins for proteosomal degradation120. The genome of a galling phylloxeran, a species from the sister family to aphids, has been shown to encode secreted RING domain proteins121, although it is not yet clear how the phylloxeran secreted RING domain proteins act and whether they are involved in gall-specific processes. We identified two H. cornu secreted RING domain proteins based on their similarity to proteins in the PFAM database100 that include “Ring finger domain” in the description, and only one was among the 1,482 genes specifically upregulated in fundatrix salivary glands (Figure S4H). Thus, secreted RING domain proteins are unlikely to contribute to H. cornu gall development.
QUANTIFICATION AND STATISTICAL ANALYSIS
All quantitative methods and statistical analyses were explained in the Method Details section
Supplementary Material
Methods S1: List of samples, genome and annotation files, analysis scripts, and primers used in this study. Related to STAR Methods.
(A-J) Details of aphid and plant samples and sequence files.
(K) Oligonucleotides used to generate RNA-seq libraries.
(L-M) Primers and probes used for qPCR and amplicon-seq assays.
Video S1: Description of the phenomenon of insect gall development, including time lapse video of H. cornu fundatrices directing gall formation on budding leaves of H. virginiana. Related to Figure 1.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Hormaphis cornu | This study | N/A, Methods S1 |
| Hormaphis hamamelidis | This study | N/A, Methods S1 |
| Hamamelis virginiana | This study | N/A, Methods S1 |
| Chemicals | ||
| Calcofluor White | Sigma-Aldrich | F3543 |
| Congo Red | Sigma-Aldrich | C6767 |
| Escin | Sigma-Aldrich | E1378 |
| Collagenase/Dispase | Roche | 10269638001 |
| Hyaluronidase | Sigma-Aldrich | H3884 |
| Methyl salicylate | Sigma-Aldrich | M6752 |
| Phenol:Chloroform:Isoamyl alcohol | Thermo Fisher | AM9730 |
| Proteinase K | Sigma-Aldrich | 03115887001 |
| RNasin® Ribonuclease Inhibitor | Lucigen | 30281 |
| Hexadecyltrimethylammonium bromide | Sigma-Aldrich | 52365 |
| Polyvinylpyrrolidone | Sigma-Aldrich | PVP40 |
| β-mercaptoethanol | Sigma-Aldrich | M6250 |
| EDTA | Sigma-Aldrich | 324506 |
| Malvidin 3,5-diglucoside chloride | Sigma-Aldrich | PHL89727 |
| Peonidin-3,5-diglucoside chloride | Carbosynth | FP65437 |
| Critical Commercial Assays | ||
| Zymo ZR-96 Quick gDNA kit | Zymo Research | D3012 |
| Nextera XT DNA Library Preparation Kit | Illumina | FC-131–1096 |
| Quick-RNA Plant Miniprep Kit | Zymo Research | R2024 |
| PicoPure RNA Isolation Kit | Thermo Fisher | KIT0204 |
| MyBaits | Arbor Bioscience | |
| Deposited Data | ||
| Raw and analyzed data | This paper | Methods S1A–J |
| Oligonucleotides | ||
| WGB library sequencing primers WGB_RTr1:TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN WGB_PCRr1:TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG WGB_PCRr2:GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG WGB_TSOr2:/5Biosg/GTCTCGTGGGCTCGGAGATGTGTATAAGAGACArGrGrG |
84 | IDT, Methods S1K |
|
H. cornu genotyping qPCR probes gwas-1 VIC: CTTTTATCGAGGGTCAATT FAM: TCTTTTATCGAGAGTCAATT context_sequence: GCTTCTAGGTTAATCTTTTATCGAG[G/A]GTCAATTCTAAGTATTTAACAGTGC FWD: TGGATCTTATGAGCTGCTTCTAGGT VIC: CCTATACCACCATCTGCACTGTTAA gwas-7 VIC: TTCCTATCTGATGGACCTAA FAM: TCCTATCTGATGAACCTAA context_sequence: TTCGCTTAGTCATTCCTATCTGATG[G/A]ACCTAATGGTATATCGGTTTGCTGT FWD:CCATCTCCTTGATTCGCTTAGTCA REV: ATGGTGACAATTATACAAACAGCAAACC gwas-8 VIC: ACGCTTTATTTTAAAATATT FAM: CGCTTTATTTTACAATATT context_sequence: TTTGTAAAGTATACGCTTTATTTTA[A/C]AATATTTAATTTTTCATTCAAAATA FWD:TGTATACTATAAAAGACTAAAAGATAAAAATGTTTGGTTT REV: TGGAAAACGTTATGTTCTAGTTTATTTTGAATGAAA gwas-9 VIC: AACATGTACCAAATTTAA FAM: AACATGTACCTAATTTAA context sequence: TATGACCAATATCCGAACATGTACC[A/T]AATTTAAACAACTAAACATAAATAT FWD:CCTAAAAATTTTGAAAGGTATGACCAATATCCG REV: AAAATTTCGGAAATCCGTTGTCACAA gwas-other VIC: ACACATTCTCTTTTTTTC FAM: ACACATTCTTATTTTTTC context_sequence: TTAGCTATGGCTTGGTGGAAAAAA[AG/TA]AGAATGTGTGTATNCGAGGGAAAAG FWD: ACAGGAGTAAGAAGTCATCCATTAGCT REV: CGACGTCATTTAAACAGGAGATGGT |
This paper | Thermo Fisher, Methods S1L |
| H. cornu amplicon sequencing primers | This paper | IDT, Methods S1M |
| Software and Algorithms | ||
| FIJI | 53 | https://imagej.net/Fiji |
| Chicago and HiC genome scaffolding | Dovetail Genomics | https://dovetailgenomics.com/ga_tech_overview/ |
| Supernova version 2.1.1 | 54 | https://support.10xgenomics.com/de-novo-assembly/software/pipelines/latest/using/running |
| RepeatMasker version 4.0.9 | 91 | http://www.repeatmasker.org |
| BUSCO | 56 | https://busco.ezlab.org |
| gVolante web interface | 57 | https://gvolante.riken.jp |
| BRAKER version 2.1.4 | 61,62 | https://github.com/Gaius-Augustus/BRAKER |
| UTRme | 66 | https://github.com/sradiouy/UTRme |
| APOLLO | 67 | https://genomearchitect.readthedocs.io/en/latest/ |
| Integrative Genomics Reviewer | 68,69 | http://software.broadinstitute.org/software/igv/ |
| BWA version 0.7.17-r1188 | 63 | https://github.com/lh3/bwa |
| SAMtools version 0.1.19 / 1.3 / 1.7 | 71 | http://www.htslib.org/download/ |
| BCFtools version 0.1.19 / 1.7 / 1.9 | 72 | http://www.htslib.org/download/ |
| VCFtools version 0.1.15 / 0.1.16 | 73 | https://vcftools.github.io |
| R version 3.6.1 | 85 | https://www.r-project.org |
| SNPRelate version 1.20.1 (R package) | 74 | https://bioconductor.org/packages/release/bioc/html/SNPRelate.html |
| Picard version 2.18.0 | Broad Institute | http://broadinstitute.github.io/picard/ |
| GATK version 3.4 | 75 | https://gatk.broadinstitute.org/hc/en-us |
| PLINK version 1.90 | 76 | http://zzz.bwh.harvard.edu/plink/ |
| Sushi version 1.24.0 (R package) | 77 | https://github.com/dphansti/Sushi |
| BEDtools version 2.29.2 | 78 | https://bedtools.readthedocs.io/en/latest/ |
| vcfR version 1.10.0 (R package) | 79 | https://github.com/knausb/vcfR |
| snpStats version 1.36.0 (R package) | 80 | http://bioconductor.org/packages/release/bioc/html/snpStats.html |
| LDheatmap version 0.99.8 (R package) | 81 | https://sfustatgen.github.io/LDheatmap/index.html |
| Trim Galore! version 0.6.5 | 82 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| cutadapt version 2.7 | 82 | https://cutadapt.readthedocs.io/en/stable/ |
| RepBase | 83 | https://www.girinst.org/repbase/ |
| PseudoreferencePipeline | Reilly, P. F. (unpublished) | https://github.com/YourePrettyGood/PseudoreferencePipeline |
| HISAT version 2.1.0 | 86 | http://daehwankimlab.github.io/hisat2/ |
| HTSeq version 0.12.4 | 87 | https://htseq.readthedocs.io/en/master/ |
| Glimma version 1.14.0 (R package) | 88 | https://bioconductor.org/packages/release/bioc/html/Glimma.html |
| edgeR version 3.28.1 (R package) | 89 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| EnhancedVolcano version 1.4.0 (R package) | 90 | https://github.com/kevinblighe/EnhancedVolcano |
| BLAST version 2.7.1 | 58,60 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| HMMER version 3.1b2 | 99 | http://hmmer.org |
| ReSpect database | 96 | https://rdrr.io/github/WMBEdmands/compMS2Miner/man/ReSpect.html |
| UniProt/Swiss-Prot database | 97 | https://www.uniprot.org |
| WebGestalt 2019 | 98 | http://www.webgestalt.org |
| PFAM database | 100 | http://pfam.xfam.org |
| SignalP-5.0 | 101 | http://www.cbs.dtu.dk/services/SignalP/abstract.php |
| tmhmm version 2.0 | 102 | http://www.cbs.dtu.dk/services/TMHMM/ |
| MAFFT version 7.419 | 103, 104 | https://mafft.cbrc.jp/alignment/software/ |
| stats (R package) | 85 | https://www.rdocumentation.org/packages/stats/versions/3.6.2 |
| trimAI version 1.4 | 105 | http://trimal.cgenomics.org/downloads |
| Seqinr version 3.6–1 (R package) | 106 | http://seqinr.r-forge.r-project.org |
| Ggseqlogo version 0.1 (R package) | 107 | https://github.com/omarwagih/ggseqlogo |
| seqtk version 1.3 | Li, H. (unpublished) | https://github.com/lh3/seqtk |
| FastTree version 2.1.11 | 111 | http://www.microbesonline.org/fasttree/ |
| ggtree version 2.2.2 (R package) | 112 | https://bioconductor.org/packages/release/bioc/html/ggtree.html |
| PAML version 4.9j | 113 | http://abacus.gene.ucl.ac.uk/software/paml.html |
| Polymorphorama version 6 | 114 | https://ib.berkeley.edu/labs/bachtrog/data/polyMORPHOrama/polyMORPHOrama.html |
| SweeD-P version 3.1 | 51 | https://cme.h-its.org/exelixis/web/software/sweed/ |
| MaCS version 0.4f | 118 | https://github.com/gchen98/macs |
| Other | ||
| H. cornu RNA sequencing | This paper | Methods S1B |
| H. cornu red-green GWAS | This paper | Methods S1C |
| Red-green targeted resequencing | This paper | Methods S1D |
| H. hamamelidis genome resequencing | This paper | Methods S1E |
| H. hamamelidis targeted resequencing | This paper | Methods S1F |
| H. cornu salivary gland red-green RNA seq | This paper | Methods S1G |
| H. virginiana gall-leaf RNA seq | This paper | Methods S1H |
| H. virginiana red-green galls RNA seq | This paper | Methods S1I |
| FigShare resources: genomes, annotations, protein sequences and analysis scripts | This paper | Methods S1J |
Highlights.
Novel aphid bicycle genes contribute to plant gall development
Variation in a bicycle gene alters plant gene expression and a gall phenotype
Bicycle genes encode a large family of diverse, secreted, cysteine-rich proteins
Many bicycle genes have experienced repeated diversifying selection
Acknowledgements
We thank Jim Truman for performing the initial aphid salivary gland dissections and for teaching us the tricks, which enabled the entire project, Erika Gajda and Henry Horn for help collecting galls, Patrick Reilly for his genotyping pipeline and helpful conversations, Goran Ceric for getting all software packages to run on the Janelia compute cluster, Tom Dolafi for maintaining the Apollo genome annotation service, Mountain Lakes Biological Station for permission to collect specimens, and Jim Truman, Nicolas Frankel, Richard Mann, Brett Mensh, and Vanessa Ruta for helpful comments on the manuscript. Gall pigment analyses were partially supported by the Ohio Agricultural Research and Development Center at OSU (Hatch Accession #1007234), NIH Award Number Grant P30 CA016058, OSU, and OSUCCC.
Footnotes
Declaration of Interests
HHMI has filed a provisional patent, number 63/092,942, for the inventors AK and DLS covering unique aphid polypeptides for use in modifying plants.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Obbard DJ, Jiggins FM, Halligan DL, and Little TJ (2006). Natural selection drives extremely rapid evolution in antiviral RNAi genes. Current Biology 16, 580–585. [DOI] [PubMed] [Google Scholar]
- 2.Papkou A, Guzella T, Yang W, Koepper S, Pees B, Schalkowski R, Barg MC, Rosenstiel PC, Teotónio H, and Schulenburg H (2019). The genomic basis of red queen dynamics during rapid reciprocal host–pathogen coevolution. Proceedings of the National Academy of Sciences of the United States of America 116, 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, Quail M, Smith F, Walker D, Libberton B, et al. (2010). Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heil M (2016). Host Manipulation by Parasites: Cases, Patterns, and Remaining Doubts. Front. Ecol. Evol 4. [Google Scholar]
- 5.Bailey R, Schönrogge K, Cook JM, Melika G, Csóka G, Thuróczy C, and Stone GN (2009). Host Niches and Defensive Extended Phenotypes Structure Parasitoid Wasp Communities. PLOS Biology 7, e1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani MS (1964). Ecology of Plant Galls Weisbach WW and Oye PV, eds. (Springer-Science+Business Media, B.V.). [Google Scholar]
- 7.Stone GN, and Schönrogge K (2003). The adaptive significance of insect gall morphology. Trends in Ecology and Evolution 18, 512–522. [Google Scholar]
- 8.Larson KC, and Whitham TG (1991). Manipulation of food resources by a gall-forming aphid: the physiology of sink-source interactions. Oecologia 88, 15–21. [DOI] [PubMed] [Google Scholar]
- 9.Cook LG, and Gullan PJ (2008). Insect, not plant, determines gall morphology in the Apiomorpha pharetrata species-group (Hemiptera: Coccoidea). Australian Journal of Entomology 47, 51–57. [Google Scholar]
- 10.Crespi B, and Worobey M (1998). Comparative analysis of gall morphology in Australian gall thrips: The evolution of extended phenotypes. Evolution 52, 1686–1696. [DOI] [PubMed] [Google Scholar]
- 11.Dodson GN (1991). Control of gall morphology: tephritid gallformers (Aciurina spp.) on rabbitbrush (Chrysothamnus). Ecol. Entomol 16, 177–181. [Google Scholar]
- 12.Hearn J, Blaxter M, Schönrogge K, Nieves-Aldrey J-L, Pujade-Villar J, Huguet E, Drezen J-M, Shorthouse JD, and Stone GN (2019). Genomic dissection of an extended phenotype: Oak galling by a cynipid gall wasp. PLOS Genetics 15, e1008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leatherdale D (1955). Plant hyperplasia induced with a cell-free insect extract. Nature 175, 553–554. [Google Scholar]
- 14.Martin JP (1938). Stem galls of sugar cane induced with an insect extract. Hawaiian planters’ record 42, 129–134. [Google Scholar]
- 15.Martinson E, Werren J, and Egan S (2020). Tissue-specific gene expression shows cynipid wasps repurpose host gene networks to create complex and novel parasite-specific organs on oaks (Preprints). [DOI] [PubMed] [Google Scholar]
- 16.Parr T (1940). Asterolecanium variolosum Ratzeburg, a gall-forming coccid, and its effect upon the host trees. Yale University School of Forestry Bulletin 46, 1–49. [Google Scholar]
- 17.Plumb GH (1953). The formation and development of the Norway Spruce gall caused by Adelges abietis L. Bulletin of the Connecticut Experiment Station 566, 1–77. [Google Scholar]
- 18.Stern DL (1995). Phylogenetic evidence that aphids rather than plants, determine gall morphology. Proceedings of Royal Society London B 260, 85–89. [Google Scholar]
- 19.Stone GN, and Cook JM (1998). The structure of cynipid oak galls: patterns in the evolution of an extended phenotype. Proceedings of the Royal Society, London Series B 265, 979–988. [Google Scholar]
- 20.Dorchin N, Hoffmann JH, Stirk WA, NovÁk O, Strnad M, and Van Staden J (2009). Sexually dimorphic gall structures correspond to differential phytohormone contents in male and female wasp larvae. Physiological Entomology 34, 359–369. [Google Scholar]
- 21.McCalla DR, Genthe MK, and Hovanitz W (1962). Chemical Nature of an Insect Gall Growth-Factor. Plant Physiol 37, 98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H, Yokokura J, Ito T, Arai R, Yokoyama C, Toshima H, Nagata S, Asami T, and Suzuki Y (2014). Biosynthetic pathway of the phytohormone auxin in insects and screening of its inhibitors. Insect Biochemistry and Molecular Biology 53, 66–72. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka Y, Okada K, Asami T, and Suzuki Y (2013). Phytohormones in Japanese Mugwort Gall Induction by a Gall-Inducing Gall Midge. Bioscience, Biotechnology and Biochemistry 77, 1942–1948. [DOI] [PubMed] [Google Scholar]
- 24.Tooker JF, and Helms AM (2014). Phytohormone Dynamics Associated with Gall Insects, and their Potential Role in the Evolution of the Gall-Inducing Habit. Journal of Chemical Ecology 40, 742–753. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi H, Tanaka H, Hasegawa M, Tokuda M, Asami T, and Suzuki Y (2012). Phytohormones and willow gall induction by a gall-inducing sawfly. New Phytologist 196, 586–595. [DOI] [PubMed] [Google Scholar]
- 26.Bailey S, Percy DM, Hefer CA, and Cronk QCB (2015). The transcriptional landscape of insect galls: psyllid (Hemiptera) gall formation in Hawaiian Metrosideros polymorpha (Myrtaceae). BMC Genomics 16, 943–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabity PD, Haus MJ, Berenbaum MR, and DeLucia EH (2013). Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proceedings of the National Academy of Sciences of the United States of America 110, 16663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih TH, Lin SH, Huang MY, Sun CW, and Yang CM (2018). Transcriptome profile of cup-shaped galls in Litsea acuminata leaves. PLoS ONE 13, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda S, Yoza M, Amano T, Ohshima I, Hirano T, Sato MH, Sakamoto T, and Kimura S (2019). Comparative transcriptome analysis of galls from four different host plants suggests the molecular mechanism of gall development. PloS one 14, e0223686–e0223686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cambier S, Ginis O, Moreau SJM, Gayral P, Hearn J, Stone GN, Giron D, Huguet E, and Drezen J-M (2019). Gall Wasp Transcriptomes Unravel Potential Effectors Involved in Molecular Dialogues With Oak and Rose. Front. Physiol 10, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eitle MW, Carolan JC, Griesser M, and Forneck A (2019). The salivary gland proteome of root-galling grape phylloxera (Daktulosphaira vitifoliae Fitch) feeding on Vitis spp. PLoS ONE 14, e0225881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Escalante LN, Chen H, Benatti TR, Qu J, Chellapilla S, Waterhouse RM, Wheeler D, Andersson MN, Bao R, et al. (2015). A Massive Expansion of Effector Genes Underlies Gall-Formation in the Wheat Pest Mayetiola destructor. Current Biology 25, 613–620. [DOI] [PubMed] [Google Scholar]
- 33.Elzinga DA, and Jander G (2013). The role of protein effectors in plant-aphid interactions. Current Opinion in Plant Biology 16, 451–456. [DOI] [PubMed] [Google Scholar]
- 34.Hogenhout SA, and Bos JIB (2011). Effector proteins that modulate plant-insect interactions. Current Opinion in Plant Biology 14, 422–428. [DOI] [PubMed] [Google Scholar]
- 35.Kaloshian I, and Walling LL (2016). Hemipteran and dipteran pests: Effectors and plant host immune regulators. Journal of Integrative Plant Biology 58, 350–361. [DOI] [PubMed] [Google Scholar]
- 36.Stuart J (2015). Insect effectors and gene-for-gene interactions with host plants. Current Opinion in Insect Science 9, 56–61. [DOI] [PubMed] [Google Scholar]
- 37.Lewis IF, and Walton L (1947). Initiation of the cone gall of witch hazel. Science 106, 419–420. [DOI] [PubMed] [Google Scholar]
- 38.Lewis IF, and Walton L (1958). Gall-formation on Hamamelis virginiana resulting from material injected by the aphid Hormaphis hamamelidis. Trans. Am. Microscop. Soc 77, 146–200. [Google Scholar]
- 39.Rehill BJ, and Schultz JC (2001). Hormaphis hamamelidis and gall size: a test of the plant vigor hypothesis. Oikos 95, 94–104. [Google Scholar]
- 40.Duncan IW (2002). Transvection effects in Drosophila. Annual review of genetics 36, 521–56. [DOI] [PubMed] [Google Scholar]
- 41.Smits BL, and Peterson WJ (1942). Carotenoids of Telial Galls of Gymnosporangium Juniperi-Virginianae Lk. Science 96, 210–211. [DOI] [PubMed] [Google Scholar]
- 42.Blunden G, and Challen SB (1965). Red pigment in leaf galls of Salix fragilis L. Nature 208, 388–389. [Google Scholar]
- 43.Bomfim PMS, Cardoso JCF, Rezende UC, Martini VC, and Oliveira DC (2019). Red galls: the different stories of two gall types on the same host. Plant Biology 21, 284–291. [DOI] [PubMed] [Google Scholar]
- 44.Springob K, Nakajima J-I, Yamazaki M, and Saito K (2003). Recent advances in the biosynthesis and accumulation of anthocyanins. [DOI] [PubMed] [Google Scholar]
- 45.Hugueney P, Provenzano S, Verriès C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, and Ageorges A (2009). A novel cation-dependent o-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiology 150, 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marrs KA, Alfenito MR, Lloyd AM, and Walbot V (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- 47.Chen M-S, Liu X, Yang Z, Zhao H, Shukle RH, Stuart JJ, and Hulbert S (2010). Unusual conservation among genes encoding small secreted salivary gland proteins from a gall midge. BMC evolutionary biology 10, 296–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, and Bielawski JP (2000). Statistical methods for detecting molecular adaptation. Trends in Ecology & Evolution 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fay JC, Wyckoff GJ, and Wu C-I (2001). Positive and Negative Selection on the Human Genome. Genetics 158, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith NG, and Eyre-Walker A (2002). Adaptive protein evolution in Drosophila. Nature 415, 1022–1024. [DOI] [PubMed] [Google Scholar]
- 51.Pavlidis P, Živkovic D, Stamatakis A, and Alachiotis N (2013). SweeD: Likelihood-Based Detection of Selective Sweeps in Thousands of Genomes. Molecular Biology & Evolution 30, 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott SR (2008). Confocal microscopy in large insect brains: Zinc-formaldehyde fixation improves synapsin immunostaining and preservation of morphology in whole-mounts. Journal of Neuroscience Methods 172, 220–230. [DOI] [PubMed] [Google Scholar]
- 53.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisenfeld NI, Kumar V, Shah P, Church DM, and Jaffe DB (2017). Direct determination of diploid genome sequences. Genome research 27, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blackman RL, and Eastop VF (1994). Aphids on the world’s trees: An identification and information guide (CAB International; ). [Google Scholar]
- 56.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, and Zdobnov EM (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura O, Hara Y, and Kuraku S (2017). GVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 33, 3635–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 59.Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, and Marth GT (2011). Bamtools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, and Stanke M (2016). BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoff KJ, Lomsadze A, Borodovsky M, and Stanke M (2019). Whole-genome annotation with BRAKER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997. [Google Scholar]
- 64.Lomsadze A, Burns PD, and Borodovsky M (2014). Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Research 42, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanke M, Schöffmann O, Morgenstern B, and Waack S (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Radío S, Fort RS, Garat B, Sotelo-Silveira J, and Smircich P (2018). UTRme: A Scoring-Based Tool to Annotate Untranslated Regions in Trypanosomatid Genomes. Frontiers in genetics 9, 671–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al. (2002). Apollo: a sequence annotation editor. Genome biology 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nature biotechnology 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hennig BP, Velten L, Racke I, Tu CS, Thoms M, Rybin V, Besir H, Remans K, and Steinmetz LM (2018). Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol. G3: Genes, Genomes, Genetics 8, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, and Weir BS (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ, et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phanstiel DH, Boyle AP, Araya CL, and Snyder MP (2014). Sushi.R: Flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knaus BJ, and Grünwald NJ (2017). VCFR : a package to manipulate and visualize variant call format data in R. Mol Ecol Resour 17, 44–53. [DOI] [PubMed] [Google Scholar]
- 80.Clayton D (2019). snpStats: SnpMatrix and XSnpMatrix classes and methods. [Google Scholar]
- 81.Shin J-H, Blay S, Graham J, and McNeney B (2006). LDheatmap : An R Function for Graphical Display of Pairwise Linkage Disequilibria Between Single Nucleotide Polymorphisms. J. Stat. Soft 16. [Google Scholar]
- 82.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–10. [Google Scholar]
- 83.Jurka J (1998). Repeats in genomic DNA: mining and meaning. Current Opinion in Structural Biology 8, 333–337. [DOI] [PubMed] [Google Scholar]
- 84.Cembrowski MS, Phillips MG, DiLisio SF, Shields BC, Winnubst J, Chandrashekar J, Bas E, and Spruston N (2018). Dissociable Structural and Functional Hippocampal Outputs via Distinct Subiculum Cell Classes. Cell 173, 1280–1292.e18. [DOI] [PubMed] [Google Scholar]
- 85.R Core Team (2018). R: A Language and Environment for Statistical Computing. [Google Scholar]
- 86.Kim D, Langmead B, and Salzberg SL (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Su S, Law CW, Ah-Cann C, Asselin-Labat M-L, Blewitt ME, and Ritchie ME (2017). Glimma: interactive graphics for gene expression analysis. Bioinformatics 33, 2050–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson MD, McCarthy DJ, and Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blighe K, Rana S, and Lewis M (2018). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. Bioconductor, 1–8. [Google Scholar]
- 91.Smit AFA, Hubley R,. Green P RepeatMasker Open-4.0. [Google Scholar]
- 92.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stanke M, Diekhans M, Baertsch R, and Haussler D (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. [DOI] [PubMed] [Google Scholar]
- 94.Jordon-Thaden IE, Chanderbali AS, Gitzendanner MA, and Soltis DE (2015). Modified CTAB and TRIzol Protocols Improve RNA Extraction from Chemically Complex Embryophyta. Applications in Plant Sciences 3, 1400105–1400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhong S, Vendrell-Pacheco M, Heskitt B, Chitchumroonchokchai C, Failla M, Sastry SK, Francis DM, Martin-Belloso O, Elez-Martínez P, and Kopec RE (2019). Novel Processing Technologies as Compared to Thermal Treatment on the Bioaccessibility and Caco-2 Cell Uptake of Carotenoids from Tomato and Kale-Based Juices. J. Agric. Food Chem 67, 10185–10194. [DOI] [PubMed] [Google Scholar]
- 96.Sawada Y, Nakabayashi R, Yamada Y, Suzuki M, Sato M, Sakata A, Akiyama K, Sakurai T, Matsuda F, Aoki T, et al. (2012). RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 82, 38–45. [DOI] [PubMed] [Google Scholar]
- 97.Bateman A (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liao Y, Wang J, Jaehnig EJ, Shi Z, and Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic acids research 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eddy SR (2011). Accelerated profile HMM searches. PLoS Computational Biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014). Pfam: The protein families database. Nucleic Acids Research 42, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, and Nielsen H (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology 37, 420–423. [DOI] [PubMed] [Google Scholar]
- 102.Krogh A, Larsson B, Von Heijne G, and Sonnhammer ELL (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 103.Katoh K (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Capella-Gutiérrez S, Silla-Martínez JM, and Gabaldón T (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Charif D, and Lobry JR (2007). Seqin{R} 1.0–2: a contributed package to the {R} project for statistical computing devoted to biological sequences retrieval and analysis. In Structural approaches to sequence evolution: Molecules, networks, populations Biological and Medical Physics, Biomedical Engineering., Bastolla U, Porto M, Roman HE, and Vendruscolo M, eds. (Springer Verlag; ), pp. 207–232. [Google Scholar]
- 107.Wagih O (2017). Ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647. [DOI] [PubMed] [Google Scholar]
- 108.von Dohlen CD, and Stoetzel MB (1991). Separation and redescription of Hormaphis hamamelidis (Fitch 1851) and Hormaphis cornu (Shimer 1867) (Homoptera: Aphididae) on witch-hazel in the Eastern United States. Proc. Entomol. Soc. Wash 93, 533–548. [Google Scholar]
- 109.von Dohlen CD, Kurosu U, and Aoki S (2002). Phylogenetics and evolution of the eastern Asian-eastern North American disjunct aphid tribe, Hormaphidini (Hemiptera: Aphididae). Molecular phylogenetics and evolution 23, 257–67. [DOI] [PubMed] [Google Scholar]
- 110.Keightley PD, Ness RW, Halligan DL, and Haddrill PR (2014). Estimation of the Spontaneous Mutation Rate per Nucleotide Site in a Drosophila melanogaster Full-Sib Family. Genetics 196, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Price MN, Dehal PS, and Arkin AP (2010). FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu G (2020). Using ggtree to Visualize Data on Tree-Like Structures. Current Protocols in Bioinformatics 69, e96. [DOI] [PubMed] [Google Scholar]
- 113.Yang Z (2007). PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- 114.Andolfatto P (2007). Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome research 17, 1755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fay JC, Wyckoff GJ, and Wu C-I (2002). Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415, 1024–1026. [DOI] [PubMed] [Google Scholar]
- 116.Shapiro JA, Huang W, Zhang C, Hubisz MJ, Lu J, Turissini DA, Fang S, Wang H-Y, Hudson RR, Nielsen R, et al. (2007). Adaptive genic evolution in the Drosophila genomes. PNAS 104, 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McDonald JH, and Kreitman M (1991). Adaptive Protein Evolution at the Adh Locus in Drosophila. Nature 351, 652–654. [DOI] [PubMed] [Google Scholar]
- 118.Chen GK, Marjoram P, and Wall JD (2008). Fast and flexible simulation of DNA sequence data. Genome Research 19, 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao C, Escalante LN, Chen H, Benatti TR, Qu J, Chellapilla S, Waterhouse RM, Wheeler D, Andersson MN, Bao R, et al. (2015). A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Current Biology 25, 613–620. [DOI] [PubMed] [Google Scholar]
- 120.Metzger MB, Pruneda JN, Klevit RE, and Weissman AM (2014). RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochimica et Biophysica Acta - Molecular Cell Research 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao C, Rispe C, and Nabity PD (2019). Secretory RING finger proteins function as effectors in a grapevine galling insect. BMC Genomics 20, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S1: List of samples, genome and annotation files, analysis scripts, and primers used in this study. Related to STAR Methods.
(A-J) Details of aphid and plant samples and sequence files.
(K) Oligonucleotides used to generate RNA-seq libraries.
(L-M) Primers and probes used for qPCR and amplicon-seq assays.
Video S1: Description of the phenomenon of insect gall development, including time lapse video of H. cornu fundatrices directing gall formation on budding leaves of H. virginiana. Related to Figure 1.
Data Availability Statement
All sequencing data generated during this study are available at the NCBI Short Read Archive and the accession numbers are provided for each sample in Methods S1.
The genomes are available at Genbank and the genomes and gene annotations (GFF files) are available at FigShare (Methods S1J).
All of the analysis scripts we produced for this study are freely available at FigShare (Methods S1J).