Abstract
The urgent need for a treatment of COVID-19 has left researchers with limited choice of either developing an effective vaccine or identifying approved/investigational drugs developed for other medical conditions for potential repurposing, thus bypassing long clinical trials. In this work, we compared the sequences of experimentally verified SARS-CoV-2 neutralizing antibodies and sequentially/structurally similar commercialized therapeutic monoclonal antibodies. We have identified three therapeutic antibodies, Tremelimumab, Ipilimumab and Afasevikumab. Interestingly, these antibodies target CTLA4 and IL17A, levels of which have been shown to be elevated during severe SARS-CoV-2 infection. The candidate antibodies were evaluated further for epitope restriction, interaction energy and interaction surface to gauge their repurposability to tackle SARS-CoV-2 infection. Our work provides candidate antibody scaffolds with dual activities of plausible viral neutralization and immunosuppression. Further, these candidate antibodies can also be explored in diagnostic test kits for SARS-CoV-2 infection. We opine that this in silico workflow to screen and analyze antibodies for repurposing would have widespread applications.
Subject terms: Computational biology and bioinformatics, Immunology
Introduction
COVID-19, caused by the novel coronavirus SARS-CoV-2, has emerged to be a global pandemic affecting over 46 million people worldwide so far (https://covid19.who.int/). The highly contagious virus SARS-CoV-2 belongs to the betacoronavirus genus of the Coronaviridae family1. It is an enveloped single-stranded positive-sense RNA virus with a genome size of ∼ 30,000 base pairs2,3. The viral genome encodes for 4 structural and 16 non-structural proteins. The spike structural protein (S), in particular, plays a vital role in fusion, entry, and transmission into the host cells. The S protein contains an N-terminal S1 subunit, responsible for the virus-receptor binding and a C-terminal S2 subunit, responsible for virus-cell membrane fusion4,5. The receptor-binding domain (RBD) in the S1 subunit of spike protein allows entry into the host cell via attachment to the angiotensin-converting enzyme 2 (ACE2) receptor6. Therefore, spike protein is currently a major therapeutic target for evolving interventions for COVID-197–10.
The combined efforts of the scientific community have substantially improved our understanding of the virus and the disease pathology in a short period. There have been several attempts to identify therapeutics for SARS-CoV-2 infection using experimental and computational approaches. Initial studies on COVID-19 suggested the importance of specific drugs such as ivermectin11, a combination of lopinavir, oseltamivir and ritonavir12; Remdesivir13 and hydroxychloroquine (HCQ)14 as potential ones against COVID-19. However, Remdesivir is the only drug currently approved by the FDA (https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19). A large-scale experimental study on 12,000 compounds for drug repurposing showed a set of 13 compounds to be effective against SARS-CoV-215. Other computational approaches included structure-based virtual screening16 and virus-host interactions network analysis to identify potential anti-SARS-CoV-2 repurposable drugs17. A recently developed online platform CoVex integrated virus-human protein interactions, human protein–protein interactions, and drug-target interactions to explore the host interactome and identification of drug(s) related to SARS-CoV-218. Few antibody therapies including Etesevimab, REGEN-COV (Casirivimab and Imdevimab) and Bamlanivimab are authorized for emergency use to treat mild-to-moderate COVID-19 cases (by March 2021; https://www.fda.gov/drugs/coronavirus-covid-19-drugs/coronavirus-treatment-acceleration-program-ctap)19.
As an alternate strategy for immediate relief in seriously ill patients, convalescent sera from recovered COVID-19 patients, supposedly rich in anti-SARS-CoV-2 antibodies, is in use. The antibodies present in the convalescent sera have been isolated and studied in detail for binding to spike protein, viral neutralization and cross-reactivity with spike proteins of SARS-CoV-220,21. S protein regions comprising RBD have shown to elicit multiple neutralizing antibodies that can neutralize SARS-CoV-2 by targeting different epitopes20,22. Raybould et al.23 recently developed a coronavirus antibody database, “CoV-AbDab”, with curated data on published antibodies and nanobodies related to different coronavirus strains. Taken together, in the development of clinical interventions against SARS-CoV-2, neutralizing antibodies play a significant role. On the other hand, pharmaceutical agencies supported this strategy and developed two neutralizing mAbs, VIR-7831 and VIR-7832, as a potential therapeutic intervention (https://www.nature.com/articles/d43747-020-01115-y). In addition, a cocktail of two neutralizing monoclonal antibodies (mAbs) are being tested in phase 2/3 trials for the treatment and prevention of SARS-CoV-2 infection.
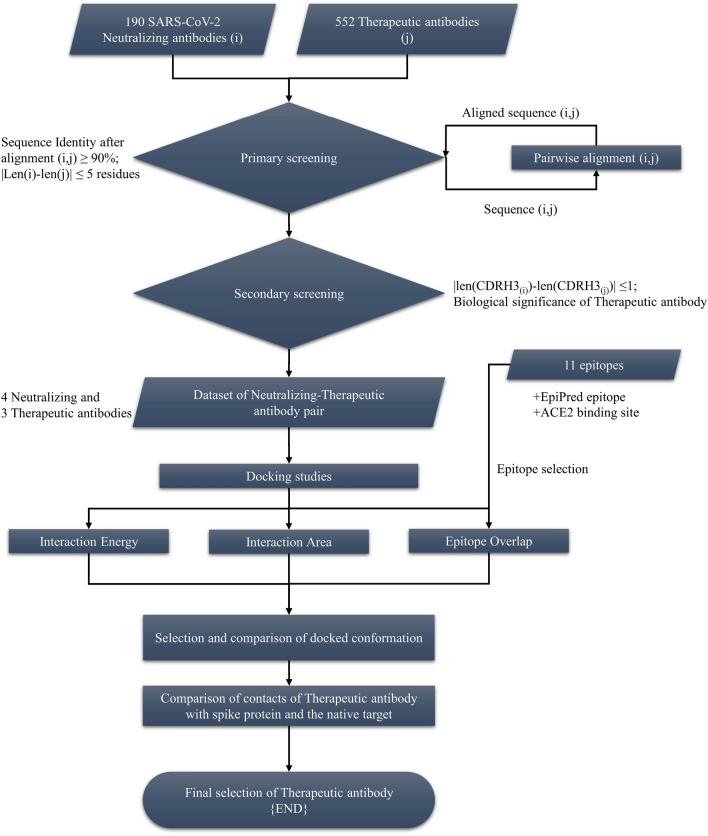
In this work, we have attempted antibody repurposing with a goal to identify therapeutic antibodies that are already approved or in clinical trials for potential cross-reactivity towards neutralization of the SARS-CoV-2. Sequences of 190 neutralizing antibodies specific to SARS-CoV-2 spike protein23 were compared with the dataset comprising 552 therapeutic antibodies24. Finally, four antibody pairs, including four neutralizing antibodies and three therapeutic antibodies, were selected and further scrutinized by a comprehensive analysis that includes (1) docking to the spike protein; (2) epitope overlaps; (3) interaction energy; (4) interaction area; (5) common contacts for therapeutic antibodies (with native target and spike protein), to effectively estimate the potential binding and a plausible replication of SARS-CoV-2 neutralization activity. Interestingly, the selected candidates, Tremelimumab, Ipilimumab and Afasevikumab are anti CTLA4 and anti IL17A antibodies. Severe SARS-CoV-2 infection is consistent with elevated levels of CTLA4 and IL17A. We posit that these candidate antibodies offer promise as ready to use and/or with the potential for better developability for COVID-19 management. The novel workflow presented here can be implemented for antibody repurposing for several other pathologies.
Materials and methods
Dataset preparation
We collected a set of 190 antibody sequences from CoV-abDab database23 using the following criteria: (1) Source of the antibody should be B cells from convalescent individual(s); (2) variable region sequence information of the heavy chain (VH) and light chain (VL) should be available; (3) neutralization activity against the SARS-CoV-2 should have been demonstrated and (iv) binding to the RBD region of the spike protein should have been confirmed. Similarly, a dataset of 552 therapeutic antibody sequences was collected from the Thera-SAbDab database24 for which VH and VL sequence information was available.
Regions considered as plausible epitopes in the RBD region of spike protein, regarded as “known epitope dataset”, were obtained from the crystal structures of neutralizing antibody-spike protein complexes deposited in Protein Data Bank, PDB25 and the information available in the literature22,26. The epitope residues in the spike protein were identified by setting a cutoff of 4 Å for distance with the antibody residues in a spike protein–antibody complex27.
Comparative analysis of antibody sequences and structures
The sequences of the SARS-CoV-2 neutralizing and therapeutic antibodies were compared by generating pairwise alignment for all 104,880 combinations (552*190) using Biopython28 as shown in Fig. 1. The maximum percent identity for each neutralizing-therapeutic antibody pair was calculated by dividing the total number of matches at the respective position in the aligned sequence with the length of the shorter antibody sequence (Eq. 1). The maximum identity was selected to accommodate as many antibodies as possible in the primary screening. The final cutoff of 90% was selected for screening and the antibody pairs with the length difference of more than five residues were removed.
| 1 |
Figure 1.
Workflow illustrating the steps followed to screen therapeutic antibodies for potential repurposing against SARS-CoV-2.
The light and heavy chain sequences were annotated using ANARCI tool29. The sequences and regions of third complementarity-determining regions for heavy chain (CDRH3) and light chain (CDRL3) were extracted based on the IMGT numbering scheme30. The antibody pairs were visualized in JalView31 after aligning with MAFFT32. Antibodies without structure information were modelled computationally using an automated antibody modelling pipeline called “ABodyBuilder”33. To identify the structural similarity between any two antibody structures, we superimposed them and estimated the root mean square deviation (RMSD) for the aligned regions.
Epitope prediction and comparison
The epitopes on the spike protein were predicted using a structure-based method called “EpiPred” for each neutralizing and therapeutic antibody34. The unique epitopes on the RBD region were selected from the “known epitope dataset” based on the following conditions: (1) least overlap (< 80% identity cutoff) with other regions considered to be epitopes to reduce redundancy, (2) epitope length is close (± 4 amino acid residues) to the average epitope length on the spike protein (~ 22 amino acid residues, from “known epitope dataset”) to avoid inclusion of too short and/or long epitopes and (3) overlap with the ACE2 binding site for potential virus neutralization ability. This procedure yielded three potential epitopes, CB6, S2H14 and S2H13 (Table S5, Figure S4).
Docking studies and antibody-SARS-CoV-2 spike protein complex analysis
We have carried out molecular docking using ClusPro 2.035 and generated the structures (30 conformations each) of SARS-CoV-2 spike protein complexed with both SARS-CoV-2 specific neutralizing antibodies and therapeutic antibodies. The optimal docked conformation was selected for each SARS-CoV-2 spike protein-antibody complex based on (1) binding to unique epitopes in “known epitope dataset” (maximum overlap), (2) interaction energy and (3) interaction area. The interaction energies of the docked structures at chosen conformations were calculated using FoldX36. Accessible surface areas of the RBD region in the spike protein (SE), antibody as well as the entire complex () were calculated by rolling a water molecule of radius 1.4 Å on the protein/complex surface. Further, the area of the interface () was calculated using Eq. (2).
| 2 |
Comparative assessment of contacts of therapeutic antibodies with their native targets and SARS-CoV-2 spike protein
The contact pattern between the amino acid residues of these therapeutic antibodies and their respective native targets was compared with contacts of the therapeutic antibody—SARS-CoV-2 spike protein complex obtained from the docking studies. The epitope and paratope residues are compared accordingly for each therapeutic antibody to assess the similarity in their mode of binding and common interactions, which in turn will help in understanding the basis for therapeutic antibody binding to RBD region of SARS-CoV-2 spike protein.
Results
Screening of the therapeutic antibodies
The first step in identifying potential antibodies for repurposing is screening therapeutic antibodies similar to known SARS-CoV-2 neutralizing antibodies. The screening was done in two stages:
In the primary screening, variable region of heavy chain and light chain (VH and VL) sequences of the antibodies were combined into a single sequence in order to pick up full-length antibody pairs, which were used further to compute the sequence identity for all combinations of neutralizing-therapeutic antibody pairs (552*190 combinations). The maximum percent identities of the antibody pairs, obtained from the pairwise sequence alignment, were grouped based on the descending identity cut offs, ranging from 95 to 85%, as shown in Figure S1. The number of antibody pairs increased exponentially with a decrease in cutoff. We selected the optimal number of antibodies at 90% max identity cut off (containing 88 antibody pairs with 37 unique neutralizing antibodies and 41 unique therapeutic antibodies) to allow multiple levels of screening downstream without exhausting all the antibodies and to facilitate manual screening. We also removed antibody pairs with a high difference in sequence length (at least five residues) in this primary screening. Antibodies selected after primary screening are given in Table S1.
The secondary screening of the antibodies consisted of two levels: (1) difference in the length of the third complementarity-determining regions for heavy chain (CDRH3) and (2) feasibility of the selected therapeutic antibody acting as a neutralizing antibody for SARS-CoV-2. Antibodies generally have canonical structures for the 5 CDRs (except CDRH3). CDRH3 regions show high diversity in sequence length, composition and loop structure37. These regions are also the major contributors to antibody specificity38. We have attempted to limit the diversity of the CDRH3 loop by selecting only the CDRH3 sequences of similar length (length difference of ± 1 amino acid residue; Table S2). The specificity of the CDRH3 is assessed using docking studies and interaction energy calculations. A similar analysis was also done for the CDRL3, where most of the CDRL3 sequences were of equal length. The final dataset after all these screening procedures contained 11 neutralizing-therapeutic antibody pairs (7 unique neutralizing antibodies and 10 unique therapeutic antibodies). Nurulimab is one of the selected therapeutic antibodies, similar to Ipilimumab, with a sequence identity of > 99% (Table S3a). It was removed from the dataset to reduce redundancy and the selected candidate therapeutic antibodies were further assessed for their biological significance (Table 1).
Table 1.
List of shortlisted therapeutic antibodies and their specifics.
| Therapeutic mAbs | Length (aa) (VH + VL) | Clinical trial status (Jan-2020) | Target | Clinical condition(s) (active/approved) | Year proposed |
|---|---|---|---|---|---|
| Afasevikumab | 231 | Phase-I | IL17A | Inflammation | 2015 |
| Daratumumab | 229 | Approved | CD38 | AL amyloidosis; cancer | 2009 |
| Denosumab | 230 | Approved | TNFSF11 | Cancer | 2005 |
| Enapotamab | 224 | Phase-II | AXL | Solid tumors | 2017 |
| Ipilimumab | Approved | CTLA4 | Cancer | 2005 | |
| Marstacimab | Phase-III | ERBB2 | Haemophilia | 2013 | |
| Ofatumumab | 229 | Approved | MS4A1 | Cancer | 2005 |
| Simlukafusp | Phase-II | FAP | Cancer | 2019 | |
| Tremelimumab | 232 | Phase-III | CTLA4 | Cancer | 2005 |
Highlighted antibodies were chosen as promising candidates. Further comprehensive analysis was carried out on these candidate antibodies to gauge their potential use in COVID-19 management.
Biological significance of the candidate therapeutic antibodies
We have further assessed the 9 selected therapeutic antibodies (Table 1) for their biological significance and feasible use in treating SARS-CoV-2 infection. Most of the strategies currently available for drug repurposing for COVID-19 target human proteins involved in the host–virus interaction network due to limited options available for targeting the viral proteins17,18, which might lead to adverse effects. Therefore, a careful selection of therapeutic antibodies is important. We found that Afasevikumab, Ipilimumab and Tremelimumab are of special interest for SARS-CoV-2 infection (highlighted in Table 1). Among the three therapeutic mAbs, Afasevikumab targets the IL17A, a proinflammatory cytokine majorly secreted by Th17-cells39. Similarly, sequentially distant antibodies, Tremelimumab and Ipilimumab target CTLA4, an immune checkpoint receptor found majorly on T cells40. The three-dimensional structures of the Afasevikumab, Ipilimumab and Tremelimumab therapeutic antibodies with their native targets (IL17A and CTLA4 respectively) (PDB codes: 6PPG, 5TRU and 5GGV, respectively) are also available in Protein Data Bank, PDB25.
Comprehensive analysis of the selected antibody pairs
Among the three therapeutic antibodies mentioned in the previous section, Tremelimumab paired with two SARS-CoV-2 specific neutralizing antibodies, C002 and COVA2-29 (Table 2). These neutralizing antibodies had sequence identity of 93.5% with each other (Table S3b). The sequence alignment for the selected antibody pairs is given in Figure S2. The structural similarity between these selected antibody pairs was assessed by superimposing the modelled/crystal structures of respective antibody pairs (neutralizing and therapeutic antibodies) and calculating the root mean square deviation (RMSD) for the aligned regions (Figure S3). Overall, Tremelimumab had the least RMSD (0.44 Å with C002 and 0.35 Å with COVA2-29) over the aligned segments. Afasevikumab and Ipilimumab showed a RMSD of 0.59 Å and 0.92 Å with their neutralizing antibody counterpart, COV2-2015 and HbnC3t1p1_G4, respectively.
Table 2.
Final list of screened SARS-CoV-2 specific neutralizing and therapeutic antibodies.
| Neutralizing antibody | Therapeutic antibody | Sequence identity (%) | ||
|---|---|---|---|---|
| Name | Length (aa) | Name | Length (aa) | |
| C002 | 231 | Tremelimumab | 232 | 90 |
| COV2-2015 | 230 | Afasevikumab | 231 | 92 |
| COVA2-29 | 232 | Tremelimumab | 232 | 90 |
| HbnC3t1p1_G4 | 224 | Ipilimumab | 226 | 90 |
Identification of epitopes on SARS-CoV-2 spike protein for docking with antibodies
A “known epitope dataset” was constructed using the information deduced from crystal structures of eight SARS-CoV-2 neutralizing antibodies and epitope information available in the literature (Table S4). These data were used to identify the most optimal antibody-spike protein binding conformation obtained from the docking studies. We have also included epitopes predicted by EpiPred34 and the ACE2 binding site on spike protein to examine the overlap of binding site of the selected conformation with in silico predicted epitopes and potential competition with ACE2 binding site. The structure-based epitope prediction method EpiPred predicted the same region on the spike protein as an epitope for all neutralizing and therapeutic antibodies (Table S4).
Overlapping epitope regions, interaction energy and interface area of neutralizing and therapeutic antibodies
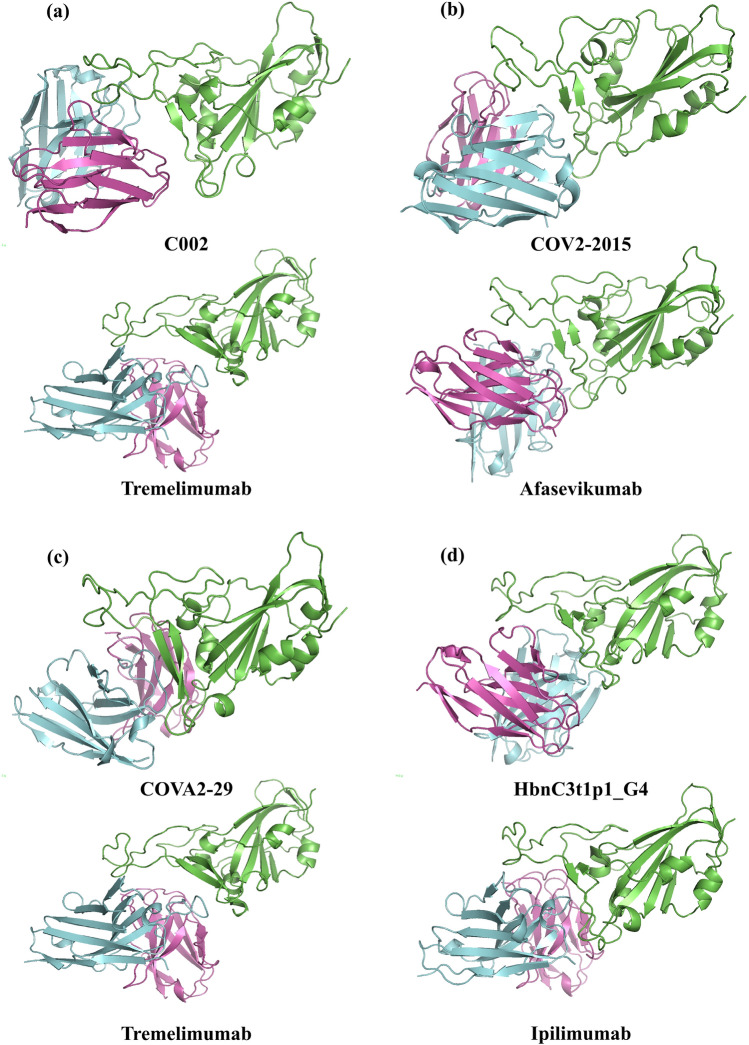
The docked conformations of each antibody with spike protein were assessed based on interaction energy and overlap with the epitopes in “known epitope dataset” (Table 3). The interaction area and length of the epitope were considered in addition, where the above criteria were inadequate to select the optimum conformation. The selected complex structures of neutralizing/therapeutic antibodies with spike protein are shown in Fig. 2 and the details on overlapping residues with known epitopes, interaction energy and interaction area are given in Table 3.
Table 3.
Summary of epitope regions, interaction energy and interface area for selected conformation of neutralizing and therapeutic antibodies bound with SARS-CoV-2 spike protein.
| Antibody | Epitope region | Length | Overlapping residues with known epitopes | Interaction energy (kcal/mol) | Interaction area (Å2) | ||||
|---|---|---|---|---|---|---|---|---|---|
| EPIPRED (29 residues) | 7C01 (26 residues) | S2H14 (23 residues) | S2H13 (20 residues) | ACE2 (17 residues) | |||||
| C002 | 417,449,453,455,456,473,475,484,485,486,487,488,489,490,493,494,496,498,501,505 | 20 | 11 | 11 | 14 | 10 | 14 | − 17.2 | 855 |
| Tremelimumab | 403,406,417,449,453,455,456,484,485,486,487,488,489,493,494,495,496,498,500,501,502,505 | 22 | 8 | 12 | 17 | 9 | 15 | − 15.9 | 848 |
| COV2-2015 | 346,444,445,446,447,448,449,450,452,453,484,485,486,490,493,494,496,498,505 | 19 | 4 | 3 | 12 | 12 | 8 | − 12.7 | 827 |
| Afasevikumab | 403,444,445,446,447,449,450,453,455,483,484,485,486,490,493,494,496,498,505 | 19 | 6 | 5 | 14 | 13 | 9 | − 14.9 | 923 |
| COVA2-29 | 403,405,406,408,409,416,417,446,447,449,453,455,493,494,496,498,500,501,502,503,504,505 | 22 | 2 | 12 | 14 | 6 | 12 | − 15.6 | 934 |
| Tremelimumab | 403,406,417,449,453,455,456,484,485,486,487,488,489,493,494,495,496,498,500,501,502,505 | 22 | 8 | 12 | 17 | 9 | 15 | − 15.9 | 848 |
| HbnC3t1p1_G4 | 403,405,408,409,416,417,420,421,449,453,455,456,475,484,486,487,489,490,493,494,496,498,500,501,502,504,505 | 27 | 10 | 18 | 16 | 8 | 16 | − 19.9 | 955 |
| Ipilimumab | 403,406,417,421,444,446,447,449,453,455,456,484,485,486,487,489,493,496,498,500,501,502,505 | 23 | 9 | 13 | 18 | 10 | 16 | − 13.7 | 1039 |
Epitopes for neutralizing and therapeutic antibodies, which show the highest overlap with known epitope regions are highlighted.
Figure 2.
Docked conformation of the SARS-CoV-2 neutralizing antibody (top) and therapeutic antibody (bottom) (light chain: violet; heavy chain: cyan) with spike protein (green) for the pairs (a) C002 and Tremelimumab (b) COV2-2015 and Afasevikumab (c) COVA2-29 and Tremelimumab, and (d) HbnC3t1p1_G4 and Ipilimumab. The figures are generated using PyMOL 2.4 (https://pymol.org/2/).
Very interestingly, our results revealed that SARS-CoV-2 neutralizing antibodies and their corresponding therapeutic antibodies exibhit the highest binding overlap with the same epitope regions on SARS-CoV-2 spike protein (Table 3). Tremelimumab and Afasevikumab showed similar interaction energies and overlap with the known epitopes. Afasevikumab and corresponding neutralizing antibody COV2-2015 showed almost equal overlap with S2H14 and S2H13 epitopes. However, Tremelimumab and corresponding neutralizing antibodies C002 and COVA2-29 exclusively bind to S2H14 epitope. The interaction energy of the Tremelimumab was 1.3 kcal/mol weaker than the C002 and 0.3 kcal/mol stronger than the COVA2-29. Ipilimumab showed poor binding to the spike protein than its neutralizing antibody counterpart with an interaction energy difference of 6.2 kcal/mol. On the other hand, Afasevikumab showed better binding than the corresponding neutralizing antibody with an increase of 2.2 kcal/mol in interaction energy. Tremelimumab exhibited marginally stronger binding affinity with spike protein (− 15.9 kcal/mol) than COVA2-29 (− 15.6 kcal/mol) but weaker than C002 (− 17.2 kcal/mol). We observed that the interaction energies of the therapeutic antibodies, tremelimumab, Ipilimumab and afasevikumab with their cognate receptors are − 24.70 kcal/mol, − 18.13 kcal/mol and − 14.23 kcal/mol, respectively. Further, afasevikumab is the only therapeutic antibody with better binding to spike protein (− 14.9 kcal/mol) than its cognate receptor (− 14.23 kcal/mol).
Analysis of common contacts for therapeutic antibodies
The contacting amino acid residues of the therapeutic antibodies were compared to the contacts on the spike protein and their cognate native target (Table 4). The amino acid residues on antibody (paratope) interacting with both epitopes of spike protein and cognate target were 53.8% (14 out of 26 residues) for Afasevikumab, 59.1% (13 out of 22 residues) for Ipilimumab and 68.2% (15 out of 22 residues) for Tremelimumab. The identical contacts on the spike protein and cognate receptor were further identified for each paratope residue. Our results showed that Afasevikumab harbored the least number of paratope residues with common contacts (2 out of 14 contacts; 14.3%) in the respective epitope, followed by Ipilimumab (5 out of 13 contacts; 38.5%). Tremelimumab had the highest number of paratope residues with common contacts in respective epitopes (9 out of 15; 60%). Overall, Tremelimumab had the highest number of common paratope and epitope residues for spike protein and the cognate receptor CTLA4. The contacts on epitopes, forming multiple contacts with paratope residues of Tremelimumab, include E498Q in spike and Y41Q/Y45Q in CTLA4 in contact with paratope residues H105L, H106Y and H107Y; and E489Y in spike and Y104Y in CTLA4 forming contacts with paratope residues H59Y, L93S and L94T. Such residues, forming multiple contacts with the paratope of antibody, are potentially important for the recognition and effective binding of the antibody to the target.
Table 4.
Comparison of the contacting amino acid residues of therapeutic antibodies with SARS-CoV-2 spike protein and their respective native target.
First letter indicates the polypeptide chain followed by amino acid residue number and notation. Highlighted amino acid residues show the common contacts in spike protein and respective native target.
Discussion
In this work, we have assessed the antibodies, either approved and/or in clinical trials, for their potential to mimic SARS-CoV-2 neutralizing antibodies. The therapeutic antibodies, sequentially similar to neutralizing antibodies, were screened on the basis of length of CDRH3 region and biological significance of their ability to bind to the known epitopes on the RBD region of spike protein. The candidate antibodies Afasevikumab, Ipilimumab, and Tremelimumab, were thus shortlisted and further scrutinized. Interestingly, sequentially distant candidate therapeutic antibodies Ipilimumab and Tremelimumab have the same native target, CTLA4. Moreover, these selected candidate therapeutic antibodies paired with different neutralizing antibodies (Table 2), yet showed maximum binding overlap with the same S2H14 epitope (Table 3). CTLA4 is an inhibitory co-receptor that has been scored as one of the exhaustion markers on T cells and whose levels have been assessed in COVID-19 patients41. Severe COVID-19 has been shown to be associated with sustained multifaceted cellular immunosuppression. CTLA4 and PD-1 levels have been shown to be enhanced, with CD4 T cells and Treg cells transiently overexpressing CTLA4 in first few days of post-infection42–44. Also, SARS-CoV-2–specific CD4 + T cells from intensive care unit (ICU) patients had significantly higher expression of CTLA-4 than CD4 T cells from convalescent individuals45. Further, CTLA-4, has been proposed as a candidate molecule with potential for controlling inflammation in severe COVID-19 patients in the context of Treg based disease management in COVID-1946.
On the other hand, cytokine storm is a hallmark of acute respiratory distress syndrome (ARDS) of SARS-CoV-2 infection. IL17, a native target of Afasevikumab, being a pro-inflammatory cytokine, has been considered as a target for drug development for COVID-1947,48. Also, given that IL17 is upstream of IL6, an established marker of severity in COVID-1949,50, it is noteworthy that an anti-IL17 antibody shares resemblance with a SARS-CoV-2 neutralizing antibody.
As for the binding, antibodies are unique and differ in several ways compared to chemical compounds. Their interaction with antigen is not majorly dependent on the structural complementarity alone. In fact, their interacting surfaces are generally flat and the interactiveness relies mainly on the atomic interactions51. They also bind to specific regions on the antigens called epitopes. Therefore, to effectively identify biologically relevant antibodies for the present context, we sampled all possible docked conformations of the shortlisted therapeutic/neutralizing antibodies on the spike protein and selected the best conformation based on the overlap with regions considered as epitopes on the RBD of the spike protein as well as the interaction energy of the whole complex. Among the selected candidate antibodies, Tremelimumab exhibited a good binding to the SARS-CoV-2 spike protein (− 15.9 kcal/mol) comparable to the neutralizing antibodies C002 (− 17.2 kcal/mol) and COVA2-29 (− 15.6 kcal/mol). Tremelimumab also showed similar contacting paratope-epitope residue pairs for spike protein and native cognate receptor CTLA4. Further, Afasevikumab showed better binding energy than its corresponding neutralizing antibody (COV2-2015). However, it exhibited unique paratope-epitope contacts in spike protein and its native receptor.
Antibodies based therapeutics would be required to tackle COVID-19 even after the availability of the vaccines, mainly for treating elderly and people with compromised immune system. Based on our results, we put forward the following contexts of how these antibodies can be applied in COVID-19 management: (1) for lone activity of plausible virus neutralization or immunomodulation; (2) for dual activity exhibiting both neutralization and immunomodulation. This will be better applicable in conditions where both of these activities are preferred, for example, in case of SARS-CoV-2 sepsis52,53, where both viral persistence and exacerbated inflammation are present. We strongly opine this will be a major ground for application of these candidate antibodies. (3) For cocktails of anti-immune checkpoint antibodies and anti-cytokine antibodies; (4) for possible inclusion in diagnostic kits for targeted diagnostics in specific suspect groups after careful standardization.
It is of course discernible that caution must be exercised before exploring these candidate antibodies in clinic. Ipilimumab and Tremelimumab have known adverse side effects such as skin reaction, diarrhoea, nausea, fatigue etc.54,55 when used for cancer conditions and we believe though such adverse effects cannot be directly anticipated/extrapolated for the present situation, further scrutiny is required before the on field applications. Afasevikumab has only completed phase one clinical trials and its side effects are not known56. Experimental evidences are also required for proving their neutralization capacity. Also, before using them for specific purposes owing to their dual activity (as anti-viral as well as immune modulators), crucial factors such as time point post-infection (as the initial stages of infection require anti-viral activity vis-a-vis, later stages require immuno-modulatory activity), dose, temporal windows for administration, turnover rate of the antibodies in vivo and need for repeat injections require to be carefully gauged out.
The concept of in silico antibody design has been around for quite some time now57,58. However, computational resources developed recently for such analyses have led to very few studies mostly related to optimization of already available antibodies59,60. In the present work, although experimental validation is not reported, results from the computational analyses stem from employing experimentally derived datasets: (1) neutralizing antibodies are experimentally verified to neutralize the SARS-CoV-2 and most of these sequences are derived from patients; (2) therapeutic antibodies and their cognate receptors are experimentally evaluated. The interacting residues of the therapeutic antibody-cognate receptor complex are taken from the experimentally determined PDB structures; (3) The epitope (on the RBD region of Spike protein of SARS-CoV-2) dataset used in the study is taken from the experimentally determined PDB structures of spike protein-neutralizing antibody complexes and from literature involving peptide microarray experiments to map linear epitopes on spike protein of SARS-Cov-2 using sera from convalescent COVID-19 patients and (4) the immunosuppression activity is experimentally established for the therapeutic antibodies. All of these in turn add a level of credibility to the observations. Taken together, we have presented a novel computational pipeline which will have significantly high potential in the near future when a large pool of therapeutic antibodies will be available for potential repurposing of therapeutic antibodies and hence might find broader applications in the intervention of other similar clinical conditions.
Supplementary Information
Acknowledgements
We thank Department of Biotechnology and Indian Institute of Technology Madras for computational facilities and Ministry of human resource and development (MHRD) for HTRA scholarship to PR, DS and AS. This work is partially supported by the Robert Bosch Center for Data Science and Artificial Intelligence (RBCDSAI), Indian Institute of Technology Madras to MMG and VJ (Project no: CR1718CSE001RBEIBRAV).
Abbreviations
- RBD
Receptor-binding domain of spike protein
- ACE2
Angiotensin-converting enzyme 2
- mAbs
Monoclonal antibody
- VH and VL
Variable region of heavy and light chains of antibody
- CDRH3 andCDRL3
Complementarity-determining regions of heavy and light chain of antibody
- RMSD
Root mean square deviation
- aa
Amino acid
Author contributions
Conceptualization: M.M.G., V.J., P.R.; methodology: M.M.G., P.R.; software/code: P.R., D.S., A.S.; investigation: V.J., M.M.G.; writing original draft: P.R.; review and editing M.M.G., V.J.; supervision: M.M.G., V.J. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vani Janakiraman, Email: vani@iitm.ac.in.
M. Michael Gromiha, Email: gromiha@iitm.ac.in.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89621-6.
References
- 1.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yesudhas D, Srivastava A, Gromiha MM. COVID-19 outbreak: History, mechanism, transmission, structural studies and therapeutics. Infection. 2020 doi: 10.1007/s15010-020-01516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanmugam A, Muralidharan N, Velmurugan D, Gromiha MM. Therapeutic targets and computational approaches on drug development for COVID-19. Curr. Top. Med. Chem. 2020;20:2210–2220. doi: 10.2174/1568026620666200710105507. [DOI] [PubMed] [Google Scholar]
- 6.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey A, et al. Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sci. 2020;256:117883. doi: 10.1016/j.lfs.2020.117883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvatori G, et al. SARS-CoV-2 SPIKE PROTEIN: An optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang S, Zhang X, Du L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin. Ther. 2020 doi: 10.1080/14728222.2020.1820482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 13.Madsen LW. Remdesivir for the treatment of Covid-19-final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, et al. A pilot study of hydroxychloroquine in treatment of patients with moderate COVID-19. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020;49:215–219. doi: 10.3785/j.issn.1008-9292.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riva L, et al. Discovery of SARS-CoV-2 anti-viral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary S, Malik YS, Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front. Immunol. 2020;11:1664. doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadegh S, et al. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 2020;11:3518. doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd EC, Gandhi TN, Petty LA. Monoclonal antibodies for COVID-19. JAMA. 2021;325:1015. doi: 10.1001/jama.2021.1225. [DOI] [PubMed] [Google Scholar]
- 20.Brouwer P, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wec AZ, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piccoli L, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raybould MI, Kovaltsuk A, Marks C, Deane CM. CoV-AbDab: The coronavirus antibody database. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raybould MI, et al. Thera-SAbDab: The therapeutic structural antibody database. Nucleic Acids Res. 2020;48:D383–D388. doi: 10.1093/nar/gkz827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burley SK, et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020;17:1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 28.Cock PJ, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbar J, Deane CM. ANARCI: Antigen receptor numbering and receptor classification. Bioinformatics. 2016;32:298–300. doi: 10.1093/bioinformatics/btv552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefranc MP, et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/S0145-305X(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leem J, Dunbar J, Georges G, Shi J, Deane CM. ABodyBuilder: Automated antibody structure prediction with data–driven accuracy estimation. MAbs. 2016;8:1259–1268. doi: 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krawczyk K, Liu X, Baker T, Shi J, Deane CM. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics. 2014;30:2288–2294. doi: 10.1093/bioinformatics/btu190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vajda S, et al. New additions to the Clus Pro server motivated by CAPRI. Proteins. 2017;85:435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado J, Radusky LG, Cianferoni D, Serrano L. FoldX 5.0: Working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35:4168–4169. doi: 10.1093/bioinformatics/btz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16:1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 38.Xu JL, Davis MM. Diversity in the CDR3 region of VH is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/S1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 39.Shin HCK, Benbernou N, Esnault S, Guenounou M. Expression of IL-17 in human memory CD45RO+ T lymphocytes and its regulation by protein kinase A pathway. Cytokine. 1999;11:257–266. doi: 10.1006/cyto.1998.0433. [DOI] [PubMed] [Google Scholar]
- 40.Ribas A. Clinical development of the anti-CTLA-4 antibody tremelimumab. Semin. Oncol. 2010;37:450–454. doi: 10.1053/j.seminoncol.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Zheng HY, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeannet R, Daix T, Formento R, Feuillard J, François B. Severe COVID-19 is associated with deep and sustained multifaceted cellular immunosuppression. Intensive Care Med. 2020;46:1769–1771. doi: 10.1007/s00134-020-06127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Wherry EJ. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schub D, et al. High levels of SARS-CoV-2–specific T cells with restricted functionality in severe courses of COVID-19. JCI insight. 2020;5:e142167. doi: 10.1172/jci.insight.142167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephen-Victor E, et al. Potential of regulatory T-cell-based therapies in the management of severe COVID-19. Eur. Respir. J. 2020;56:2002182. doi: 10.1183/13993003.02182-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacha O, Sallman MA, Evans SE. COVID-19: A case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Megna M, Napolitano M, Fabbrocini G. May IL-17 have a role in COVID-19 infection? Med. Hypotheses. 2020;140:109749. doi: 10.1016/j.mehy.2020.109749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo P, et al. Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herold T, et al. Elevated levels of interleukin-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020;146:128–136. doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kringelum JV, Nielsen M, Padkjær SB, Lund O. Structural analysis of B-cell epitopes in antibody: Protein complexes. Mol. Immunol. 2013;53:24–34. doi: 10.1016/j.molimm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Florindo HF, et al. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fecher LA, Agarwala SS, Hodi FS, Weber JS. Ipilimumab and its toxicities: A multidisciplinary approach. Oncologist. 2013;18:733. doi: 10.1634/theoncologist.2012-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gan EH, Mitchell AL, Plummer R, Pearce S, Perros P. Tremelimumab-induced graves hyperthyroidism. Eur. Thyroid J. 2017;6:167–170. doi: 10.1159/000464285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng K, et al. Measurement of IL-17AA and IL-17FF as pharmacodynamic biomarkers to demonstrate target engagement in the phase I study of MCAF5352A. AAPS J. 2019;21:1–9. doi: 10.1208/s12248-018-0280-z. [DOI] [PubMed] [Google Scholar]
- 57.Roy A, Nair S, Sen N, Soni N, Madhusudhan MS. In silico methods for design of biological therapeutics. Methods. 2017;131:33–65. doi: 10.1016/j.ymeth.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Norman RA, et al. Computational approaches to therapeutic antibody design: Established methods and emerging trends. Brief. Bioinform. 2020;21:1549–1567. doi: 10.1093/bib/bbz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luan B, Huynh T. In silico antibody mutagenesis for optimizing its binding to spike protein of severe acute respiratory syndrome coronavirus 2. J. Phys. Chem. Lett. 2020;11:9781–9787. doi: 10.1021/acs.jpclett.0c02706. [DOI] [PubMed] [Google Scholar]
- 60.Desautels T, Zemla A, Lau E, Franco M, Faissol D. Rapid in silico design of antibodies targeting SARS-CoV-2 using machine learning and supercomputing. BioRxiv. 2020 doi: 10.1101/2020.04.03.024885. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.