Abstract
A large number of insect-specific viruses (ISVs) have recently been discovered, mostly from hematophagous insect vectors because of their medical importance, but little attention has been paid to important plant virus vectors such as the whitefly Bemisia tabaci, which exists as a complex of cryptic species. Public SRA datasets of B. tabaci and newly generated transcriptomes of three Chinese populations are here comprehensively investigated to characterize the whitefly viromes of different cryptic species. Twenty novel ISVs were confidently identified, mostly associated with a particular cryptic species while different cryptic species harbored one or more core ISVs. Microinjection experiments showed that some ISVs might cross-infect between the two invasive whitefly cryptic species, Middle East Asia Minor 1 (MEAM1) and Mediterranean (MED), but others appeared to have a more restricted host range, reflecting the possibility of distinct long-term coevolution of these ISVs and whitefly hosts. Moreover, analysis of the profiles of virus-derived small-interfering RNAs indicated that some of the ISVs can successfully replicate in whitefly and the antiviral RNAi pathway of B. tabaci is actively involved in response to ISV infections. Our study provides a comprehensive analysis of the RNA virome, the distinct relationships and cross-cryptic species infectivity of ISVs in an agriculturally important insect vector.
Subject terms: Microbiome, Metagenomics
Introduction
Viruses are the most abundant microbes on earth, and most of those previously discovered and studied viruses are pathogens causing diseases in their plant/animal hosts1. Over the past decade, the development of metagenomics next-generation sequencing (mNGS) has led to the discovery of a large number of novel RNA viruses, mostly from arthropod insects. These viruses, known as insect-specific viruses (ISVs), are confined exclusively to insects and are unable to replicate in vertebrates or vertebrates cells2. The majority of ISVs are believed to have close relationship within their host insect3. Most of the ISVs detected are members of particular virus taxa, including Baculoviridae, Parvoviridae, Flaviviridae, Ascoviridae, Togaviridae, Bunyavirales (formerly Bunyaviridae), Rhabdoviridae, and a novel group described as negeviruses4,5. Accumulating evidence has suggested that ISVs might be ancestors of arthropod-borne viruses (arboviruses) and that the presence of ISVs may influence the physiology of the host insect as well as its competence as a vector of arboviruses6–8. A number of ISVs have been discovered in hematophagous insects such as mosquitoes, ticks, and fleas due to their medical importance. Several ISVs have recently been reported in some important plant virus vectors: an iflavirus in a planthopper9, nege/kita-like viruses in aphids10, and a reovirus in a leafhopper11. However, considering their abundance in hematophagous insects, there has been comparatively little investigation into the diversity of ISVs in plant virus vectors12.
With the aid of this unbiased mNGS technology, a diverse assemblage of novel viruses were revealed in various hosts, including insects1,13. The discovered viruses are often shared among phylogenetically related host species, perhaps because of their similar ecology and food sources, as well as selective pressures from host immune response and microbial interactions14–16. A comparative analysis of the virome in mosquitoes indicated that the majority of the identified viruses were mosquito species specific, and that both Aedes aegypti and Culex quinquefasciatus were associated with a number of stable eukaryotic viruses, respectively17. In addition, a recent virome investigation in waterfowl and shorebird communities identified both multi-host generalist viruses as well as those that appear to be host-specific, demonstrating the importance of using multi-host, multi-virus systems in the study of virus ecology18.
The whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) causes substantial economic losses worldwide and poses a serious threat to global food security through direct feeding, excreting honeydew that promotes sooty mold, and more importantly, transmitting devastating pathogenic plant viruses19,20. In particular, ssDNA viruses belonging to the genus Begomovirus (Geminiviridae) are exclusively transmitted by B. tabaci in a persistent-circulative manner21. B. tabaci is a complex of morphologically indistinguishable cryptic species, and the threshold to define these cryptic species is 3.5% nucleotide divergence in their mitochondrial cytochrome oxidase subunit I (mtCOI) gene sequences22–24. Of these cryptic species, Middle East Asia Minor 1 (MEAM1; previously biotype B) and Mediterranean (MED; previously biotype Q) have the most important commercial impact due to their ability to spread globally, replace native whiteflies, and transmit economically important plant viruses20,25. Several studies suggest that many begomoviruses are transmitted by various B. tabaci cryptic species with different efficiencies, as for instance the differential efficiency of transmission of tomato yellow leaf curl virus and tomato yellow leaf curl Sardinia virus by MEAM1 and MED24,26.
Despite the increasing number of ISVs discovered in arthropod insects using mNGS, as far as we know, the diversity complex of ISVs in insect pests of agricultural importance has not yet been investigated. In this study, reassembly and extensive analysis of RNA viromes was performed with publicly available datasets (NCBI Sequence Read Archive, NCBI SRA) of B. tabaci distributed globally, as well as with transcriptomes generated as part of this work. Our results reveal the presence of 32 previously unreported RNA viruses in different cryptic species of B. tabaci. Analysis of sRNA profiles suggests that siRNA-based antiviral immunity is actively involved in the response of whiteflies to most of the ISVs. Comparative analysis and further experimental study confirmed that some ISVs might be specific to a particular cryptic species of whitefly whereas others may have a broader host range, highlighting the complex long-term coevolution between the different whitefly cryptic species and the ISVs that infect them.
Results
Transcriptome assembly and assignment of whitefly cryptic species
The 41 selected datasets of B. tabaci from the SRA repository were reassembled and the N50 of each library (assembled with Trinity) is listed in Table 1. These datasets were submitted by labs from seven countries in Asia, Europe, and America for various experimental purposes (Supplementary Fig. 1 and Table 1). The cryptic species of each assembled dataset was determined by homology search against the mtCOI database. The results indicated that the majority of the whitefly datasets belong to the invasive cryptic species MEAM1or MED, and the other whitefly cryptic species were identified include sub-Saharan Africa 1 (SSA1), New World 1 (NW1), AsiaII1, and AsiaII7 (Table 1). Meanwhile, the whitefly transcriptomes of NBU-B, NBU-Q (lab cultures) and FY-Q (field sample) were also sequenced, assembled, and assigned to the cryptic species as listed in Table 1.
Table 1.
Whitefly datasets used in this study derived from public database, lab cultures, and field investigation.
| Library | BioProject accession | Run accession number | Cryptic species ideification | University/Institute | Location | Brief description | Total base (Gb) | N50 |
|---|---|---|---|---|---|---|---|---|
| ZJU-B1 | PRJNA407873 | SRR6117406 | MEAM1 | Zhejiang University | Hangzhou, China | Healthy whitefly (gut) | 7.7 | 2690 |
| ZJU-B2 | SRR6117407 | MEAM1 | TYLCV infected whitefly (gut) | 7.7 | 2844 | |||
| ZJU-B3 | PRJNA282153 | SRR2001504 | MEAM1 | Whitefly nymph | 4.7 | 1076 | ||
| ZJU-B4 | PRJNA255986 | SRR1523522 | MEAM1 | Whitefly adult | 4.7 | 1462 | ||
| ZJU-Q1 | PRJNA79601 | SRR1104130 | MED | Whitefly adult | 10.4 | 2009 | ||
| ZJU-Q2 | PRJNA338731 | SRR4039449 | MED | Whitefly fed on cotton and tobacco | 18.6 | 1111 | ||
| CAAS-B1 | PRJNA298415 | SRR4293755 | MEAM1 | Chinese Academy of Agricultural Sciences | Beijing, China | Whitefly egg | 7.1 | 2942 |
| CAAS-B2 | SRR4293724 | MEAM1 | Whitefly male adult | 4.1 | 1557 | |||
| CAAS-B3 | SRR4293725 | MEAM1 | Whitefly female adult | 5.1 | 2142 | |||
| CAAS-Q1 | SRR4293748 | MED | Whitefly male adult | 5.6 | 1474 | |||
| CAAS-Q2 | SRR2619082 | MED | Unknown | 5.6 | 1475 | |||
| CAAS-BQ | SRR4293752 | MEAM1 and MED | Whitefly female adult | 5.5 | 1779 | |||
| CAAS-B4 | PRJEB17859 | ERR1726444 | MEAM1 | Imidacloprid treatment | 6.3 | 2508 | ||
| CAAS-Q3 | ERR1726458 | MED | Imidacloprid treatment | 5.5 | 3085 | |||
| CAAS-B5 | PRJNA344376 | SRR4426118 | MEAM1 | Whitefly fed on cotton | 6.2 | 648 | ||
| CAAS-Q4 | SRR4426099 | MED | Whitefly fed on cotton | 5.5 | 2308 | |||
| CAAS-B6 | PRJNA89143 | SRR453543 | MEAM1 | Different plant host and whitefly sex strains | 5.2 | 1114 | ||
| CAAS-Q5 | PRJNA276952 | SRR2895294 | MED | Sexual differences of whitefly | 4.1 | 1166 | ||
| CAAS-B7 | PRJNA391229 | SRR5723126 | MEAM1 | Whitefly antenna | 5.0 | 1186 | ||
| CAU-Q1 | PRJNA417353 | SRR6262725 | MED | China Agricultural University | Beijing, China | Wolbachia related study | 8.3 | 1128 |
| CAS-Q1 | PRJNA606896 | SRR11092386 | MED | Chinese Academy of Sciences | Beijing, China | Whitefly head | 6.2 | 2083 |
| QAU-BQ | PRJNA490883 | SRR7829909 | MEAM1 and MED | Qingdao Agricultural University | Qingdao, China | Whiteflies co-infected by TYLCV and ToCV | 5.2 | 2055 |
| QAU-Q1 | PRJNA279224 | SRR1930109 | MED | Interaction between whitefly and plant hosts | 5.0 | 2141 | ||
| NIAS-Q1 | PRJDB2008 | DRR018506 | MED | National Institute of Agrobiological Sciences | Tsukuba, Japan | Study of mitochondrial transporters | 3.4 | 2430 |
| NABI-A1 | PRJNA237273 | SRR1159208 | AsiaII7 | National Agri-Food Biotechnology Institute | Ajitgarh, India | Comprehensive transcriptome analysis | 8.5 | 2237 |
| UE-Q1 | PRJEB13160 | ERR1337902, ERR1337901, ERR1337900 | MED | University of Exeter | Penryn, UK | Study of insecticide resistance | 11.4 | 2851 |
| UC-Q1 | PRJNA293094 | SRR2174325 | MED | University of Crete | Heraklion, Greece | Study of insecticide resistance | 7.8 | 1877 |
| CU-B1 | PRJNA312467 | SRR3179979 | MEAM1 | Cornell University | New York, USA | Fed on tomato | 4.2 | 1339 |
| CU-S1 | PRJNA419386 | SRR6313815 | SSA1 | Whitefly adults | 7.4 | 597 | ||
| HU-Q1 | PRJNA427517 | SRR6432772 | MED—France | The Hebrew University | Jerusalem, Israel | Differential expression in whitefly species | 2.2 | 1036 |
| HU-Q2 | SRR6432775 | MED—France | 2.1 | 989 | ||||
| HU-Q3 | SRR6432776 | MED—France | 2.0 | 878 | ||||
| HU-S1 | SRR6432785 | SSA—Tanzania | 2.2 | 1560 | ||||
| HU-S2 | SRR6432829 | SSA1—Tanzania | 2.1 | 853 | ||||
| HU-S3 | SRR6432827 | SSA1—Tanzania | 2.1 | 846 | ||||
| HU-A1 | SRR6432767 | AsiaII1 | 2.1 | 667 | ||||
| HU-A2 | SRR6432762 | AsiaII1 | 2.1 | 1088 | ||||
| HU-A3 | SRR6432768 | AsiaII1 | 2.1 | 736 | ||||
| HU-N1 | SRR6432809 | NW1—Brazil | 2.3 | 1367 | ||||
| HU-N2 | SRR6432804 | NW1—Brazil | 2.0 | 1152 | ||||
| HU-N3 | SRR6432800 | NW1—Brzil | 1.9 | 1425 | ||||
| NBU-B | PRJNA677841 | SRR13050950 | MEAM1 | Ningbo University | Ningbo, China | Lab culture of this study | 14 | 2902 |
| NBU-Q | SRR13052369 | MED | 15 | 3385 | ||||
| FY-Q | SRR13039280 | MED | Fuyang, China | Field investigation | 13 | 2676 |
MEAM1 Middle East Asia Minor 1, MED Mediterranean, SSA1 sub-Saharan Africa 1, NW1 New World 1.
Diversity of RNA viromes discovered in B. tabaci
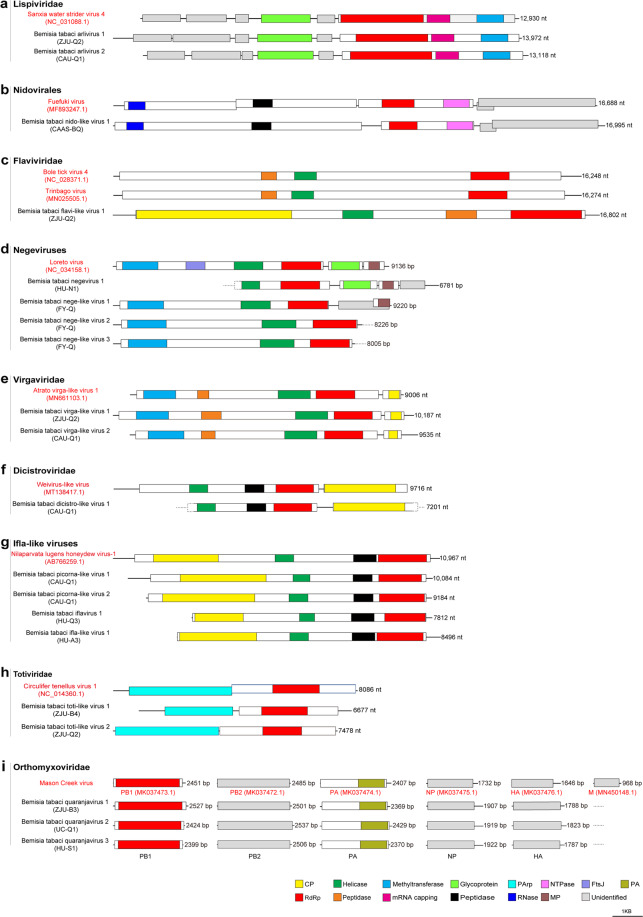
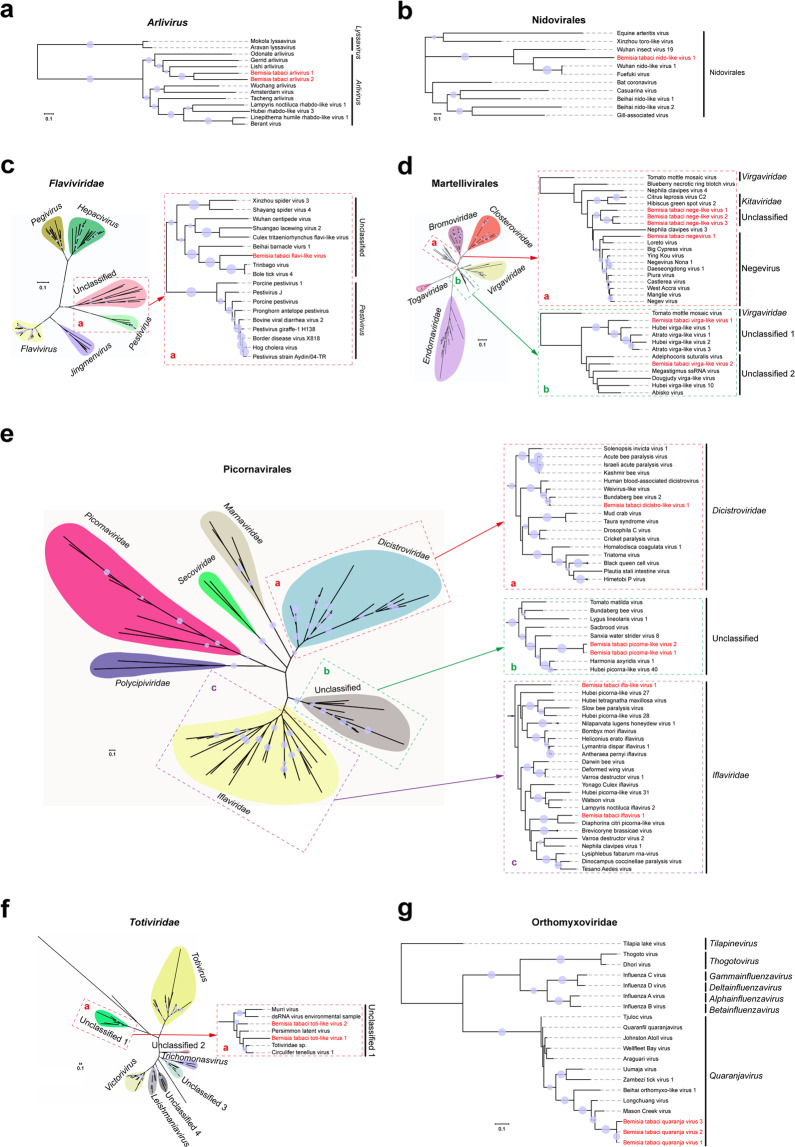
Across all of the assembled libraries and using the strict criteria described above, 32 novel RNA viruses (20 ISVs and 12 plant/fungal virus-like contigs) (Table 2), and a new isolate of Potato virus S (PVS) were identified in B. tabaci. Based on the taxonomy of the most closely related reference viruses, the 20 newly discovered RNA viruses were putative members of or related to the following orders/families: Lispiviridae (N = 2), Nidovirales (N = 1), Flaviviridae (N = 1), Negevirus (newly proposed taxon, N = 4), Virgaviridae (N = 2), Picornavirales (N = 5), Orthomyxoviridae (N = 3), and Totiviridae (N = 2). The genomic structures and phylogeny of these ISVs, together with related reference viruses, are shown in Figs.1 and 2. Two novel negative-sense single-stranded RNA (ssRNA) viral genomes were identified, and clearly cluster together with other viruses in the genus Arlivirus, which mainly infect invertebrates such as insects, spiders, and nematodes (Figs. 1a and 2a). Positive-sense ssRNA virus discovered in whitefly including a nido-like virus (Figs. 1b and 2b), a flavi-like virus (Figs. 1c and 2c), a negevirus and three nege-like viruses (Figs. 1d and 2d), two virga-like viruses (Figs. 1e and 2d), a dicistro-like virus (Figs. 1f and 2e), and four ifla-like viruses (Figs. 1g and 2e). Furthermore, we also detected two toti-like dsRNA viruses (Figs. 1h and 2f), and three closely related segmented negative ssRNA quaranjaviruses (Figs. 1i and 2g and Supplementary Fig. 2) in B. tabaci, suggesting the high diversity of ISVs in whiteflies. Besides the 20 novel ISVs described above, we also identified 12 diverse plant/fungal virus-like contigs in the whiteflies collected from field (sample FY-Q), and a new isolate of PVS in the whitefly dataset CU-B1 (Supplementary Fig. 3). Dataset FY-Q contained a total of six beny-like viruses and six bromo-like viruses with relatively high abundance (coverage) (Table 2 and Supplementary Fig. 4). Detailed descriptions for each of the identified novel virus are provided in Supplementary Result 1. In addition, tissue expression analysis of ISVs by quantitative reverse transcription PCR (qRT-PCR) showed that Bemisia tabaci quaranjavirus 1 (BtQuV1) was ubiquitously expressed in all tissues of NBU-B, whereas Bemisia tabaci virga-like virus 2 (BtViLV2) was mostly accumulated in the gut of NBU-Q (Supplementary Fig. 5).
Table 2.
Novel viruses identified in whiteflies from publicly available databases and field samples.
| Tentative virus names | NCBI accession | Library | Length (nt) | Coverage | E-value | Homologous virus (genome size, nt) | Protein identities | Virus family | Virus genus | Homologous virus reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Bemisia tabaci arlivirus 1 (BtArV1) | MW256666 | ZJU-Q2 | 13,972 | 965× | 0.0 | Lishi Spider Virus 2 (9924) | 30% | Lispiviridae | Arlivirus | 60 |
| Bemisia tabaci arlivirus 2 (BtArV2) | MW256667 | CAU-Q1 | 13,118 | 250× | 0.0 | Hubei odonate virus 10 (14,440) | 28% | Lispiviridae | Arlivirus | 61 |
| Bemisia tabaci nido-like virus 1 (BtNiLV1) | MW256673 | CAAS-BQ | 16,995 | 414× | 4e−157 | Wuhan insect virus 19 (15,441) | 33% | Unassigned | Unassigned | 61 |
| Bemisia tabaci flavi-like virus 1 (BtFlLV1) | MW256672 | ZJU-Q2 | 16,802 | 22× | 2e−161 | Bole tick virus 4 (16,248) | 34% | Flaviviridae | Unassigned | 62 |
| Bemisia tabaci negevirus 1 (BtNeV1) | MW256675 | HU-N1 | 6781 | 72× | 0.0 | Loreto virus (9136) | 41% | Unassigned | Unassigned | 63 |
| Bemisia tabaci nege-like virus 1 (BtNeLV1) | MW256676 | FY-Q | 9220 | 163× | 3e−174 | Wuhan house centipede virus 1 (10,310) | 36% | Unassigned | Unassigned | 64 |
| Bemisia tabaci nege-like virus 2 (BtNeLV2) | MW256677 | FY-Q | 8226 | 46× | 4e−170 | Big Cypress virus (9506) | 36% | Unassigned | Unassigned | 65 |
| Bemisia tabaci nege-like virus 3 (BtNeLV3) | MW256678 | FY-Q | 8005 | 58× | 4e−171 | Big Cypress virus (9506) | 35% | Unassigned | Unassigned | 65 |
| Bemisia tabaci virga-like virus 1 (BtViLV1) | MW256664 | ZJU-Q2 | 10,187 | 137× | 0.0 | Hubei virga-like virus 1 (9141) | 40% | Virgaviridae | Unassigned | 61 |
| Bemisia tabaci virga-like virus 2 (BtViLV2) | MW256665 | CAU-Q1 | 9535 | 3297× | 3e−172 | Megastigmus ssRNA virus (10,187) | 35% | Virgaviridae | Unassigned | 66 |
| Bemisia tabaci dicistro-like virus 1 (BtDiLV1) | MW256674 | CAU-Q1 | 7201 | 125× | 0.0 | Bundaberg bee virus 2 (7955) | 84% | Dicistroviridae | Unassigned | 67 |
| Bemisia tabaci picorna-like virus 1 (BtPiLV1) | MW256668 | CAU-Q1 | 10,084 | 86× | 1e−123 | Hubei tetragnatha maxillosa virus 2 (9763) | 27% | Unassigned | Unassigned | 61 |
| Bemisia tabaci picorna-like virus 2 (BtPiLV2) | MW256669 | CAU-Q1 | 9184 | 50× | 3e−137 | Sanxia water strider virus 8 (9166) | 38% | Unassigned | Unassigned | 61 |
| Bemisia tabaci iflavirus 1 (BtIfV1) | MW256671 | HU-Q3 | 7812 | 78× | 1e−165 | Nephila clavipes virus 1 (10,198) | 29% | Iflaviridae | Iflavirus | 68 |
| Bemisia tabaci ifla-like virus 1 (BtIfLV1) | MW256670 | HU-A3 | 8496 | 44× | 1e−142 | Sanxia water strider virus 8 (9166) | 36% | Iflaviridae | Unassigned | 61 |
| Bemisia tabaci toti-like virus 1 (BtToLV1) | MW227222 | ZJU-B4 | 6677 | 26× | 1.9e−118 | Circulifer tenellus virus 1 (8086) | 34% | Totiviridae | Unassigned | 69 |
| Bemisia tabaci toti-like virus 2 (BtToLV2) | MW227223 | ZJU-Q2 | 7478 | 47× | 2.1e−155 | Persimmon late virus (7475) | 55% | Totiviridae | Unassigned | 70 |
| Bemisia tabaci Quaranjavirus 1 (BtQuV1) | MW256682 | ZJU-B3 | 2501 | 111× | 1e−86 | Mason Creek virus (Seg. 1, PB2, 2485) | 28% | Orthomyxoviridae | Quaranjavirus | 71 |
| MW256679 | 2369 | 138× | 3e−163 | Mason Creek virus (Seg. 2, PA, 2407) | 37% | |||||
| MW256680 | 2541 | 177× | 7e−141 | Beihai orthomyxo-like virus 1 (Seg. 3, PB1, 2432) | 45% | |||||
| MW256681 | 1907 | 519× | 7e−96 | Mason Creek virus (Seg. 4, NP, 1732) | 34% | |||||
| MW256683 | 1788 | 274× | 1e−75 | Mason Creek virus (Seg. 5, HA, 1646) | 29% | |||||
| Bemisia tabaci Quaranjavirus 2 (BtQuV2) | MW256687 | UC-Q1 | 2537 | 69× | 1e−84 | Mason Creek virus (Seg. 1, PB2, 2485) | 28% | Orthomyxoviridae | Quaranjavirus | 71 |
| MW256685 | 2429 | 107× | 1e−162 | Mason Creek virus (Seg. 2, PA, 2407) | 37% | |||||
| MW256686 | 2424 | 223× | 0.0 | Mason Creek virus (Seg. 3, PB1, 2451) | 56% | |||||
| MW256688 | 1919 | 196× | 1e−93 | Mason Creek virus (Seg. 4, NP, 1732) | 34% | |||||
| MW256684 | 1823 | 290× | 1e−73 | Mason Creek virus (Seg. 5, HA, 1646) | 29% | |||||
| Bemisia tabaci Quaranjavirus 3 (BtQuV3) | MW256692 | HU-S1 | 2506 | 33× | 3e−92 | Mason Creek virus (Seg. 1, PB2, 2485) | 29% | Orthomyxoviridae | Quaranjavirus | 71 |
| MW256690 | 2370 | 63× | 2e−159 | Mason Creek virus (Seg. 2, PA, 2407) | 37% | |||||
| MW256691 | 2399 | 33× | 0.0 | Mason Creek virus (Seg. 3, PB1, 2451) | 56% | |||||
| MW256693 | 1922 | 124× | 2e−79 | Mason Creek virus (Seg. 4, NP, 1732) | 34% | |||||
| MW256689 | 1787 | 60× | 3e−73 | Mason Creek virus (Seg. 5, HA, 1646) | 29% | |||||
| Bemisia tabaci beny-like virus 1 (BtBeLV1) | MW256694 | FY-Q | 6479 | 42× | 0.0 | Agaricus bisporus virus 8 (8280, partial) | 32% | Benyviridae | Benyvirus | N/A |
| Bemisia tabaci beny-like virus 2 (BtBeLV2) | MW256695 | FY-Q | 6369 | 23× | 0.0 | Agaricus bisporus virus 8 (8280, partial) | 32% | Benyviridae | Benyvirus | N/A |
| Bemisia tabaci beny-like virus 3 (BtBeLV3) | MW256696 | FY-Q | 6209 | 44× | 0.0 | Agaricus bisporus virus 8 (8280, partial) | 32% | Benyviridae | Benyvirus | N/A |
| Bemisia tabaci beny-like virus 4 (BtBeLV4) | MW256697 | FY-Q | 6343 | 219× | 0.0 | Agaricus bisporus virus 8 (8280, partial) | 31% | Benyviridae | Benyvirus | N/A |
| Bemisia tabaci beny-like virus 5 (BtBeLV5) | MW256698 | FY-Q | 6229 | 31× | 0.0 | Agaricus bisporus virus 8 (8280, partial) | 31% | Benyviridae | Benyvirus | N/A |
| Bemisia tabaci beny-like virus 6 (BtBeLV6) | MW256699 | FY-Q | 5127 | 96× | 0.0 | Hubei Beny-like virus 1 (4365) | 76% | Benyviridae | Benyvirus | 61 |
| Bemisia tabaci bromo-like virus 1 (BtBromoLV1) | MW256700 | FY-Q | 6158 | 181× | 1e−82 | Beihai charybdis crab virus 1 (6969) | 38% | Bromoviridae | Unassigned | 61 |
| Bemisia tabaci bromo-like virus 2 (BtBromoLV2) | MW256701 | FY-Q | 6215 | 523× | 2e−79 | Beihai charybdis crab virus 1 (6969) | 36% | Bromoviridae | Unassigned | 61 |
| Bemisia tabaci bromo-like virus 3 (BtBromoLV3) | MW256702 | FY-Q | 6005 | 494× | 7e−82 | Beihai charybdis crab virus 1 (6969) | 38% | Bromoviridae | Unassigned | 61 |
| Bemisia tabaci bromo-like virus 4 (BtBromoLV4) | MW256703 | FY-Q | 6631 | 36× | 5e−66 | Beihai charybdis crab virus 1 (6969) | 35% | Bromoviridae | Unassigned | 61 |
| Bemisia tabaci bromo-like virus 5 (BtBromoLV5) | MW256704 | FY-Q | 6529 | 126× | 3e−51 | Beihai charybdis crab virus 1 (6969) | 35% | Bromoviridae | Unassigned | 61 |
| Bemisia tabaci bromo-like virus 6 (BtBromoLV6) | MW256705 | FY-Q | 5972 | 45× | 2e−37 | Beihai charybdis crab virus 1 (6969) | 30% | Bromoviridae | Unassigned | 61 |
Fig. 1. Genomic structures of novel insect-specific viruses identified in whitefly B. tabaci.
The viruses were taxonomically classified into nine groups as shown in panels a–i. Each panel contains a genome representing a phylogenetically close reference virus on the top (with red font) and the insect-specific viruses discovered from whiteflies in this study. GenBank accession numbers are provided in parentheses after the name of the reference viruses. The name of the whitefly dataset corresponding to the identified novel viruses is also indicated in parentheses below the virus name (details in Table 2). Conserved functional domains are color-coded and the names of the domains are indicated at the bottom of the figure. Abbreviation of the conserved domain names: CP coat protein, FtsJ RNA ribosomal methyltransferase, MP membrane protein, PA polymerase, PArp proline–alanine-rich protein, RdRp RNA-dependent RNA polymerase.
Fig. 2. Phylogeny of novel insect-specific viruses (ISVs) identified in whitefly B. tabaci with other related viruses.
Trees for Arlivirus (a), Nidovirales (b), Flaviviridae (c), Martellivirales (d), Picornavirales (e), Totiviridae (f), and Orthomyxoviridae (g) are based on the maximum likelihood method and inferred from conserved viral RdRp domains. Novel ISVs identified in this study are shown in red font. Nodes with bootstrap values >50% are marked with solid blue circles, and the larger circles indicate higher bootstrap values. In panels c–f, a taxonomic overview of viruses at order or family level are shown on the left, and a close-up view of the viruses of interest in this study are shown in the dotted frames on the right. The viral sequences used in this study were extracted from GenBank: the accession numbers and other related details are listed in Supplementary Table 2.
Abundance and association of whitefly ISVs with host cryptic species
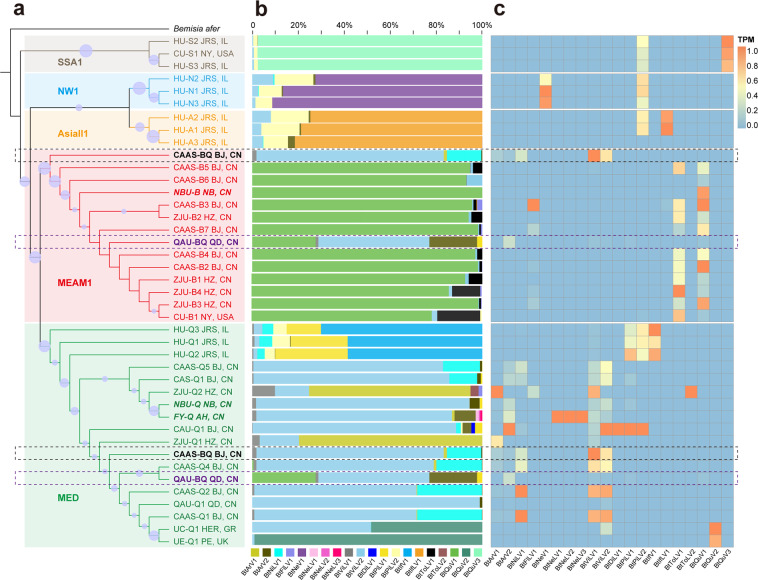
To illustrate the abundance, prevalence, and association of whitefly ISVs with host cryptic species, a phylogenetic tree was reconstructed based on the whitefly mtCOI sequence and its relationship with the RNA virome of each whitefly dataset was investigated. All of the whitefly datasets separate distinctly and form as five groups, corresponding to the five cryptic species of whitefly MEAM1 (N = 14), MED (N = 18), AsiaII1 (N = 3), NW1 (N = 3), and SSA1 (N = 3). Two of the whitefly datasets, CAAS-BQ (SRR4293752) and QAU-BQ (SRR7829909), contain a mixture of whiteflies cryptic species from both the MEAM1 and MED clades as indicated in Fig. 3a.
Fig. 3. Correlation between whitefly cryptic species and insect-specific viruses.
a Phylogeny of whitefly cryptic species based on mtCOI sequences using the maximum likelihood method. The mtCOI of Bemisia afer (MK360160.1) was used to root the tree. Two datasets (CAAS-BQ BJ,CN and QAU-BQ QD,CN) containing mixed cryptic species (MEAM1 and MED) are highlighted by dotted frames colored with black and purple, respectively. Nodes with bootstrap values >50% are marked with solid blue circles. b Composition of insect-specific viruses (ISVs) in each whitefly cryptic species dataset. ISVs are color-coded and the names of viruses are indicated at the bottom of the figure. c Relative abundance of ISVs across the different whitefly cryptic species datasets. The transcripts per million (TPM) of each ISV are displayed by the heat map. Abbreviations of the cities and countries: JRS, IL: Jerusalem, Israel; NY: New York, USA; AJ, IN: Ajitgarh, India; BJ, CN: Beijing, China; NB, CN: Ningbo, China; HZ, CN: Hangzhou, China; QD, CN: Qingdao, China; AH, CN: Anhui, China; HER, GR: Heraklion, Greece; PE: Penryn. Abbreviation and details of the whitefly datasets and newly discovered ISVs are listed in Tables 1 and 2, respectively. Abbreviations of the whitefly cryptic species: MEAM1 Middle East Asia Minor 1, MED Mediterranean, NW1 New World 1, SSA1 sub-Saharan Africa 1.
Analysis of ISVs composition (percentage) indicated that most of the whitefly populations (datasets) harbor stable dominant (core) ISVs which are obviously associated with a particular cryptic species of B. tabaci. More than 95% of the viral reads are derived from Bemisia tabaci quaranjavirus 3 (BtQuV3) for the cryptic species of SSA1, while the dominant virus for the cryptic species NW1 and AsiaII1 are, respectively, Bemisia tabaci negevirus 1 (BtNeV1) and Bemisia tabaci ifla-like virus 1 (BtIfLV1) (Fig. 3b). Although most of the datasets of these three whitefly cryptic species were submitted by the Hebrew University (Jerusalem, Israel) with limited numbers of datasets, the high similarity of ISVs composition derived from another SSA1 dataset (CU-S1) submitted by Cornell University (New York, USA) with HU-S2 and HU-S3 strongly supports the hypothesis that there is a stable and distinct group of core viruses in each of the three whitefly cryptic species (Fig. 3b). For the whitefly clade MEAM1, the dominant ISV is apparently BtQuV1 in all of the datasets (excluding CAAS-BQ and QAU-BQ) irrespective of the whitefly origin, and Bemisia tabaci toti-like virus 1 (BtToLV1) is also prevalent in most of the MEAM1 whitefly datasets (Fig. 3b). On the other hand, the RNA virome and core viruses in MED are more diverse than in the other cryptic species and the ISVs of MED are related to the geographical location of the whitefly populations as well as the clusters in the phylogenetic tree (Fig. 3b). BtViLV2 is the most prevalent and dominant virus in most of the MED datasets from China. But the two datasets from Zhejiang University (Hangzhou, China) (ZJU-Q1 and ZJU-Q2), both have Bemisia tabaci arlivirus 1 (BtArV1) as the dominant virus. The other two MED datasets from Europe, UC-Q1 (Heraklion, Greece) and UE-Q1 (Penryn, UK), group together and harbor another specific core virus Bemisia tabaci quaranjavirus 2 (BtQuV2), whereas the three MED datasets from Hebrew University (Jerusalem, Israel) also contain distinctly different viral composition including Bemisia tabaci iflavirus 1 (BtIfV1), Bemisia tabaci picorna-like virus 1 (BtPiLV1), and Bemisia tabaci picorna-like virus 2 (BtPiLV2) (Fig. 3b). Principal component analysis (PCA) also confirms the relationship between the core viruses and the various cryptic species (Supplementary Fig. 6). It is worth mentioning that in the datasets with mixed cryptic species, CAAS-BQ has a similar RNA virome composition to the other MED datasets, whereas QAU-BQ clearly has the combined core viruses of both MEM1 (BtQuV1) and MED (BtViLV2) (Fig. 3b).
The heat map shown in Fig. 3c consolidates the above observation that the RNA virome diversity is more complex in MED than in the other four cryptic species. Most of the whitefly ISVs (N = 13) appeared to be MED-specific, while some of the other ISVs are present specifically in MEAM1 (BtToLV1 and BtQuV1), NW1 (BtNeV1), AsiaII1 (BtIfLV1), or SSA1 (BtQuV3). Bemisia tabaci flavi-like virus 1 (BtFlLV1) is present in the datasets of both MEAM1 (N = 5) and MED (N = 1), whereas BtPiLV2 is present in most datasets of the four whitefly cryptic species from Hebrew University (Jerusalem, Israel) (Fig. 3c), implying that BtPiLV2 may have a broader host range amongst the different whitefly cryptic species. It is also interesting that Bemisia tabaci toti-like virus 2 (BtToLV2) and Bemisia tabaci dicistro-like virus 1 (BtDiLV1) are only present in datasets ZJU-Q2 and CAU-Q1, respectively, and the three nege-like viruses of B. tabaci (BtNeLV1-3) were exclusively discovered in the field sample FY-Q (Fig. 3c). In addition, the three phylogenetically related quaranjaviruses (BtQuV1-3) (Fig. 2g) were found in three cryptic species of whitefly MEAM1, MED, and SSA1, respectively. BtQuV1 and BtQuV3 were identified in each dataset of their respective cryptic species, whereas BtQuV2 was only found in two MED whitefly datasets originating from Europe (Fig. 3c).
Experimental evaluation of the ability of whitefly ISVs to cross-infect the cryptic species MEAM1 and MED
Since most of whitefly ISVs were identified from only one specific cryptic species, we next tested whether the whitefly ISVs of the cryptic species MEAM1 and MED were able to cross-infect. The whitefly population NBU-B (representing of cryptic species MEAM1) and NBU-Q (representing of cryptic species MED) in our lab were used for this study through microinjection as described in “Methods” and Fig. 4a. NBU-B contains only one virus BtQuV1 that is the dominant virus in MEAM1, while the four ISVs, BtPiLV1, Bemisia tabaci arlivirus 2 (BtArV2), BtViLV2, and Bemisia tabaci virga-like virus 1 (BtViLV1) present in NBU-Q appear to be “MED-specific” (Fig. 3). Results of RT-PCR and qRT-PCR detection indicated that BtQuV1 could barely be detected in NBU-Q microinjected with whitefly homogenate (NBU-B) 0, 3, 6, and 12 DPI. In addition, nearly no viral RNA of BtQuV1 was detected in the F1 of injected NBU-Q, confirming that the core virus of MEAM1 (BtQuV1) might not successfully replicate and be transmitted transovarially in a MED whitefly population (Fig. 4b and Supplementary Fig. 7e). When the four “MED-specific” ISVs were injected into NBU-B, BtPiLV1 replicated well with increasing accumulation of detected viral RNAs, and the virus could also be easily identified in the F1 of NBU-B whiteflies, providing the possibility of its ability to replicate and of transovarial transmission in MEAM1 whiteflies (Fig. 4b and Supplementary Fig. 7a). BtArV2 and BtViLV2 were also present at 3, 6, and 12 DPI in NBU-B, providing evidence of replication in MEAM1 but they could hardly be detected in the F1 of NBU-B whiteflies, suggesting that they might not be transmitted in a transovarial manner (Fig. 4b and Supplementary Fig. 7b, c). For BtViLV1, only several weak bands were detected (likely the injected virus) in some of the NBU-B whiteflies and no virus could be detected in the F1 generation, indicating that BtViLV1 may be MED-specific and unable to replicate in MEAM1 whiteflies (Fig. 4b and Supplementary Fig. 7d).
Fig. 4. Evaluation of the ability of insect-specific viruses to cross-infect the cryptic whitefly species Middle East Asia Minor 1 (MEAM1) and Mediterranean (MED).
a Diagram illustrating the experimental workflow. In the upper panel whitefly individuals of NBU-Q (representing cryptic species MED) were microinjected with homogenate of NBU-B (representing cryptic species MEAM1). The reciprocal injection of NBU-B with homogenate of NBU-Q whiteflies is shown in the lower panel. b Detection of insect-specific viruses (ISVs) 0, 3, 6, and 12 days post injection (DPI), and in the next generation (F1). The presence of ISVs in each whitefly sample was determined by RT-PCR. Untreated NBU-B and NBU-Q whiteflies were used as controls. A pool of 20–30 whiteflies were collected for MEAM1 and MED whiteflies at each time point, and three independent biological replicates were performed. Abbreviation of virus names: BtArV2, Bemisia tabaci arlivirus 2, BtPiLV1 Bemisia tabaci picorna-like virus 1, BtQuV1 Bemisia tabaci quaranjavirus 1, BtViLV1 Bemisia tabaci virga-like virus 1, BtViLV2 Bemisia tabaci virga-like virus 2.
Analysis of virus-derived siRNAs in B. tabaci
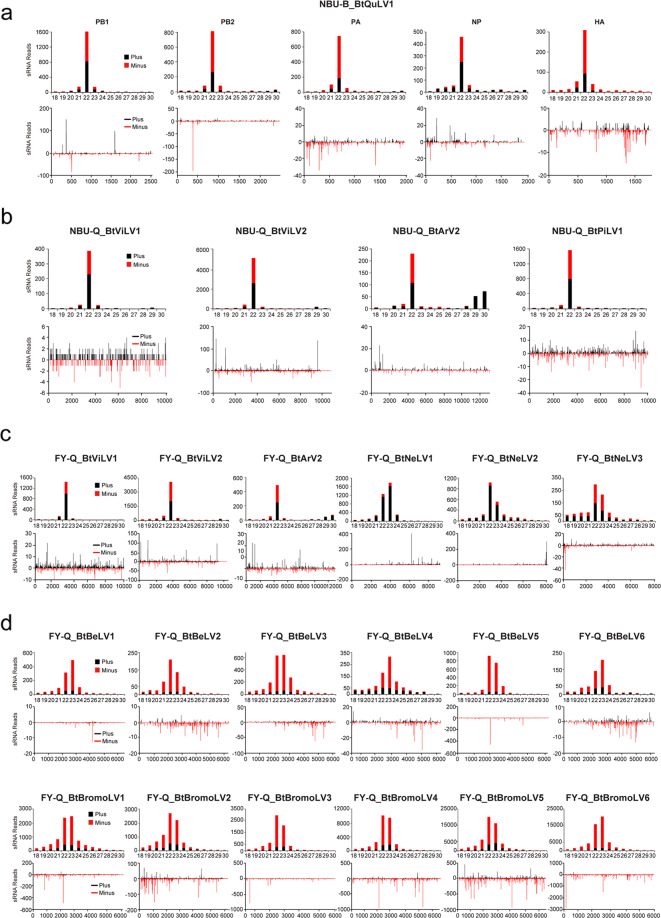
To further explore siRNA-based antiviral immunity in B. tabaci, virus-derived siRNAs (vsiRNAs) from the whitefly datasets of NBU-B (BtQuLV1), NBU-Q (BtViLV1, BtViLV2, BtArV2, and BtPiLV1), and FY-Q (BtViLV1, BtViLV2, BtArV2, BtNeLV1-3, BtBeLV1-6, and BtBromoLV1-6) were comprehensively characterized. For BtQuLV1 in the whitefly dataset NBU-B, siRNAs derived from the five segments are mostly 22 nt long. vsiRNAs are almost equally derived from the sense and antisense strands of the viral genomic RNA in segments PB1 and NP, whereas more sense vsiRNAs are detected for PB2, PA, and HA (Fig. 5a). The other three ISVs (BtViLV1, BtViLV2, BtArV2) in both datasets of NBU-Q and FY-Q, and BtPiLV1 in NBU-Q, are also mostly 22 nt long and are equally derived from both vsiRNA strands (Fig. 5b, c). The typical size distribution and polarity of vsiRNAs strongly suggested that these ISVs can successfully replicate in whitefly, and the antiviral RNAi pathway of B. tabaci is actively involved in response to ISV infections. In contrast, for BtNeLV1-3, BtBeLV1-6, and BtBromoLV1-6 there are many more 23nt vsiRNAs (Fig. 5c, d) and the vast majority of siRNAs of BtNeLV1 and BtNeLV2 are from the positive strand of the genome (Fig. 5c), while there is an obvious preference of vsiRNAs derived from antisense strands of the viral genomes of BtBeLV1-6 and BtBromoLV1-6. The discrepant and non-canonical characteristics of vsiRNAs derived from BtNeLV1, BtNeLV2, BtBeLV1-6, and BtBromoLV1-6 raises the possibility that these vsiRNAs (or perhaps some of them) may be produced by the microorganism/parasitism of the insect host, rather than directly from cleavage by the whitefly siRNA immune pathway. Moreover, our analysis showed a strong U bias in the 5′-terminal nucleotide of vsiRNAs for BtNeLV1-3, BtBeLV1-6, and BtBromoLV1-6 (Supplementary Fig. 8c) in whitefly FY-Q, whereas classical A/U bias of vsiRNAs was observed for the other viruses (BtQuLV1, BtViLV1, BtViLV2, BtArV2, and BtPiLV1) discovered in the three whitefly datasets (Supplementary Fig. 8). Previous study indicated that the distinct patterns of vsiRNAs produced by various hosts can be used for virus detection27. An unusual profile of vsiRNAs was also reported for a Twyford virus discovered in D. melanogaster28, and more recently, a study showed that non-canonical characteristics of these vsiRNAs (21–23 peak, negative strand bias, and a strong 5′ U bias) were more likely to be processed by a Dicer pathway in a fungi rather than insect host29, indicating that BtBeLV1-6 and BtBromoLV1-6 might be originated from fungi of whitefly (Fig. 5 and Supplementary Fig. 8). In addition, analysis of the distribution of vsiRNAs shows them to be widely distributed in the corresponding genomes (segments) but with notable asymmetric hotspots on both strands, which may indicate that these regions are preferentially targeted for cleavage by the host immune system (Fig. 5).
Fig. 5. Profiles of virus-derived small interfering RNAs (vsiRNAs).
vsiRNAs derived from whitefly datasets NBU-B (a), NBU-Q (b), and FY-Q (c, d). The upper panel shows the size distribution of vsiRNAs, while the lower panel shows the distribution of vsiRNAs along the corresponding viral genome. Color coding shows vsiRNAs derived from the sense (black, plus) and antisense (red, minus) genomic strands. All reads in this analysis are redundant. The abbreviation of the virus names is listed in Table 2.
Discussion
Over the past decade, a large number of insect-associated viruses (mostly ISVs) have been discovered by taking advantage of mNGS and advanced bioinformatics tools, contributing new insights on insect viromes and viral evolution5. In this study, the RNA virome of a notorious insect pest, the whitefly B. tabaci, and the complex association of ISVs with its cryptic species were investigated by screening the publicly available whitefly datasets, as well as the whitefly populations derived from lab and field populations in China. Twenty novel ISVs, together with an isolate of a plant RNA virus and 12 novel plant/fungal virus-like contigs were identified among these whitefly samples. All the ISVs discovered in whitefly are novel and distinct from previously described ones (Table 2), suggesting the rich diversity of ISVs in whiteflies and that different ISVs may be associated with specific hosts. In addition, our results add a number of new virus members to unclassified groups which will facilitate the official establishment of new viral taxa in the future. These include unclassified clades in Flaviviridae (Fig. 2c), Martellivirales (Fig. 2d), Picornavirales (Fig. 2e), and Totiviridae (Fig. 2f). Previous studies have shown that it is common for a host insect to harbor several closely related ISVs, such as partitiviruses in Drosophila28, negeviruses in a dungfly30, and totiviruses, anpheviruses, and quaranja-like viruses in mosquitoes17. Our results also discovered that a number of intimately related viruses were present in different datasets of whitefly, including two arliviruses (Figs. 1a and 2a), three nege-like viruses (Figs. 1d and 2d), two picorna-like viruses (Figs. 1g and 2e), and three quaranjaviruses (Figs. 1i and 2g). The three quaranjaviruses discovered in whitefly are of particular interest because our results indicate that the diversity of quaranjaviruses in the family Orthomyxoviridae might be greatly underestimated in arthropods other than the previously described hematophagous insects.
The majority of economically important plant viruses are transmitted by insect vectors, and the recently developed “vector-enabled metagenomics” method facilitates the discovery of new plant viruses or new insect vectors of known plant viruses31,32. Our detection of a new isolate of PVS in the dataset CU-B1 was unexpected. Several different species of aphids, including Myzus persicae and Aphis nasturtii, are well-known to be efficient vectors for the transmission of PVS33,34 and our results provide the evidence that whiteflies might also be vectors of PVS, although further field investigations and lab experiments are needed to confirm it. In view of the relatively low coverage (20×) of PVS, it is also possible for the accident acquisition of PVS by the whitely. Nevertheless, understanding the complex diversity of plant viruses in insect vectors will contribute to the early surveillance of emerging plant viruses and the management of viral diseases.
Previous studies have shown that the viromes of the two important mosquitoes, A. aegypti and C. quinquefasciatus, have their own relatively stable core virome, which might have important implications for the competence to host the relevant arboviruses17. Moreover, further investigation revealed that the core virome was very stable across all developmental stages of both lab-derived and field-collected A. albopictus35. Our results suggested that different cryptic species of B. tabaci clearly harbor specific core virus/viruses (Fig. 3b), indicating a long-term coevolution between these ISVs and cryptic species of whitefly. Because the majority of the whitefly datasets used in this study are from public databases and are mostly derived from lab cultures, it is expected that the cryptic-species-specific core viruses in these whitefly cultures might constitute a vertically transmitted core virome, whereas the field whitefly dataset (FY-Q) likely harbors distinct environmentally-derived viruses (BtNeLV1-3, BtBeLV1-6, and BtBromoLV1-6) as described previously35. Nevertheless, more investigation is needed to confirm this hypothesis due to limited whitefly datasets from the field used in this study. It should also be noted that the dominant ISVs of MED whitefly exhibit more diversity and are associated directly with the original location of the whitefly as well as its taxonomical status (Fig. 3b), implying that other dominant/core viruses might be obtained from the environment and establish stable infections in the local whitefly populations.
Cross-species transmission (or interspecies transmission) is the ability for a foreign virus to infect a new host species individually and spread in the new host population36. Interspecies transmission has been well demonstrated in several important emerging zoonotic viruses, including Severe Acute Respiratory Syndrome (SARS-CoV), Ebola, swine flu, rabies, avian influenza, as well as SARS-CoV-2, the causative agent of the current COVID-19 outbreak37,38. Our study showed that most of the ISVs discovered in this study exhibit close association with a specific cryptic species, but some have the ability to infect different cryptic species (Fig. 3c). Microinjection experiments confirmed that the “MEAM1-specific” virus BtQuV1 cannot establish infection or be transmitted in a transovarial manner in MED whitefly, whereas some “MED-specific” ISVs (BtPiLV1, BtArV2, BtViLV2, and BtViLV1) could replicate and sometimes be transmitted transovarially in MEAM1 whitefly (Fig. 4). The distinct interspecies transmission abilities of ISVs in MED and MEAM1 whitefly might reflect the different origin and discrete long-term coevolution of these ISVs with their whitefly hosts. Further investigations are necessary to answer the intriguing issue about how some, but not all, of these ISVs can cross the cryptic species barrier of whitefly. In addition, the high diversity and efficient cross-species ability of some ISVs in various cryptic whitefly species may provide an excellent model system for future studies on the molecular and evolutionary mechanisms of interspecies transmission in zoonotic viruses. However, more investigations are needed to confirm the complex relationships between the ISVs and whitefly cryptic species, and to comprehensively evaluate and understand the ability of ISVs to cross the cryptic species boundaries.
One of the most important reasons that ISVs have gained increasing attention recently is because they can affect vector competence and could therefore have potential as biocontrol agents. Previous studies in mosquitos indicated that ISVs can negatively regulate several arbovirus infections both in vitro and in vivo5,39. For example, two ISVs identified in an A. albopictus C6/36 cell line, Menghai rhabdovirus and Shinobi tetravirus, suppressed Zika virus replication in vitro40. It is proposed that the infection of ISVs might indirectly upregulate the innate immune system of mosquitoes and further interfere with the replication of mosquito-borne pathogenic viruses by decreasing vector competence5,39,41,42, which is similar to the molecular mechanism by which Wolbachia controls arboviruses in mosquitos43. The activation of the siRNA-based antiviral response by ISVs in whiteflies (Fig. 5) indicates that the immune system is also induced and raises the possibility that this might interfere with the transmission of devastating plant viruses, including begomoviruses, which are vectored by whiteflies. Our results also highlight the need to recognize that ISVs and other viruses in an insect vector could have important effects on laboratory experiments, especially for studies related to the immune response of vector insects challenged by various pathogens. It would be important, therefore, to investigate and understand the virome background of any insect line maintained in the lab, particularly where it is being used in vector studies.
Methods
RNA sequencing (RNA-seq) libraries of B. tabaci from the public database
Information about approximately 400 RNA-seq datasets of B. tabaci was retrieved from the NCBI SRA repository. Filtering of the datasets was based on the following criteria: Firstly, the dataset should be >4 Gb since RNA-seq depth is essential for virus discovery; secondly, where there were several biological replicates, the dataset with largest total number of bases was selected (since a similar virome should be present within the replicates); thirdly, the datasets were not used unless some novel virus was identified. Exceptions to these rules included a dataset of 3.4 Gb (PRJDB2008), representing a unique submitter (National Institute of Agrobiological Sciences, Tsukuba, Japan) and 12 datasets with the project number PRJNA427517 (each with total bases <3.0 Gb) submitted by The Hebrew University, Jerusalem, Israel, because the project included several different cryptic species of B. tabaci that were of interest for this study. In total, 41 high-quality SRA datasets representing the different whitefly cryptic species and various geographical locations worldwide were chosen for further bioinformatics analysis. Abbreviations and detail information of these whitefly datasets are provided in Table 1.
RNA-seq libraries of B. tabaci generated from lab and field samples
The B. tabaci culture of cryptic species MEAM1 (NBU-B) was kindly provided by Xiao-Wei Wang and Shu-Sheng Liu (Institute of Insect Sciences, Zhejiang University) in June 2019, and the MED population of B. tabaci (NBU-Q) maintained in our lab at Ningbo University (NBU) was originally collected from soybean plants in Suzhou (An’hui province, China) in June 2019. The NBU-B and NBU-Q were maintained in the laboratory in Ningbo University thereafter. The two whitefly cultures were reared separately in insect-proof cages on cotton plants (Gossypium hirsutum L. cv. Zhemian 1793) at 25 ± 1 °C, 50–70% relative humidity, and 14 h light/10 h darkness. The field sample of B. tabaci was collected from cucumber plants in Fuyang (An’hui province, China) on November 2019 and the cryptic species was determined to be MED (FY-Q) by mtCOI sequences23. Total RNAs were extracted using approximately 100 adult whiteflies from each of the two lab cultures (NBU-B, NBU-Q), as well as the field sample (FY-Q). Each RNA sample was subdivided for Illumina high throughput sequencing (transcriptome and sRNA). Specifically, for transcriptome, paired-end (150 bp) sequencing of the RNA library was performed on the Illumina HiSeq 4000 platform (Illumina, CA, USA) by Novogene (Tianjin, China). For sRNA, the cDNA libraries were prepared using the Illumina TruSeq Small RNA Sample Preparation Kit (Illumina, CA, USA), and sRNA sequencing was performed on an Illumina HiSeq 2500 by Novogene (Tianjin, China). The transcriptome raw reads of NBU-B, NBU-Q, and FY-Q were deposited in SRA under accession numbers SRR13050950, SRR13052369, and SRR13039280, respectively. Meanwhile, the sRNA raw reads of NBU-B, NBU-Q, and FY-Q were deposited in SRA under accession numbers SRR13050947, SRR13050948, and SRR13082984, respectively.
Dataset reassembly and assignment of whitefly cryptic species
Raw reads of the 41 selected datasets from the SRA repository, as well as the transcriptomes of the lab populations (NBU-B, NBU-Q) and field sample (FY-Q), were quality trimmed. The remaining reads were reassembled/assembled de novo using the two assembler software packages Trinity and MetaviralSPAdes with default parameters44,45. The assembled contigs were searched against the mtCOI reference database of the B. tabaci species complex to assign the correct cryptic species of whitefly for each dataset46. Furthermore, to facilitate the discovery of the whitefly RNA virome, the host-derived reads were removed by mapping against the two representative genomes of whitefly (GCA_001854935.1 and GCA_003994315.1) using BWA software47.
RNA virome discovery
All the assembled contigs were compared with the NCBI viral RefSeq database using diamond Blastx48. Since most of the datasets were retrieved from public databases, strict criteria were used for the identification of putative novel viruses in each dataset. Firstly, E-value cutoff of 1 × 10−20 was rigorously set for the diamond Blastx. Secondly, the minimum coverage and length threshold for the viral homology contigs was no less than 20× and 500 bp, respectively, and the viral homology contigs had to contain almost complete open reading frames (ORF) of predicted viral proteins. Thirdly, the viral homology contigs needed to be confirmed by both of the assemblers (Trinity and MetaviralSPAdes). Finally, the regions of the candidate viral-like contigs matched to the reference virus were extracted and further compared with the entire NCBI nucleotide and non-redundant protein databases to eliminate false positives. In addition, RT-PCR followed by Sanger sequencing was performed to verify the presence of the newly discovered viruses in the whitefly populations NBU-B, NBU-Q, and FY-Q. The primers used for RT-PCR are listed in Supplementary Table 1. Sequences of all identified novel viruses from this study have been deposited in GenBank (MW256664–MW256706 and MW227222–MW227223).
Viral genome annotation and phylogenetic analysis
The newly identified viral contigs were annotated with InterPro49. Conserved RNA-dependent RNA polymerase (RdRp) regions of the discovered viruses, together with RdRp protein sequences of reference viruses, were used for phylogenetic analysis. The RdRp sequences were aligned with MAFFT50, and ambiguously aligned regions were trimmed by Gblock51. The best-fit model of amino acid substitution was evaluated by ModelTest-NG52. Maximum likelihood (ML) trees were constructed using RAxML-NG with 1000 bootstrap replications53. Details of all the reference sequences used in phylogenetic analysis are listed in Supplementary Table 2.
Correlation between whitefly cryptic species and ISVs
Phylogeny of whitefly cryptic species was constructed as described above. For the identification of ISV-derived viral reads, raw reads of each whitefly dataset were aligned to corresponding ISV contigs using bowtie2 software54. To better understand the relative abundance of the newly identified ISVs across the different whitefly datasets, the unassembled transcriptome reads of each datasets were mapped back to the corresponding viral contigs. Specifically, relative abundance for the ISV of each whitefly dataset was calculated and normalized based on the transcripts per million (TPM) values calculated as . In this equation, represents the TPM of viral contig j, represents the number of uniquely mapped fragments in a dataset, represents the length of viral contig j, and n is the total number of viral contigs55,56. In addition, relative abundance of the ISV in each whitefly dataset was further subjected to PCA using R 3.5.
Ability of ISVs to cross-infect the whitefly cryptic species MEAM1 and MED
The whitefly lab cultures of NBU-B (MEAM1) and NBU-Q (MED) were used to investigate whether the ISVs were specific to one cryptic species of whitefly or able to infect both. Before the experiment, the presence of viruses in each cryptic species was verified by RT-PCR. To explore the potential ability of ISVs in NBU-B to infect NBU-Q whiteflies, a pool of 10 NBU-B whiteflies were homogenized in 150 μl phosphate-buffered saline solutions (137 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, and 1.47 mM KH2PO4 at pH 7.4). After centrifugation at 12,000 r.p.m. for three times, the supernatant was collected and microinjected into individual NBU-Q whiteflies (approximately 0.02 μl/per insect) as described previously57. The injected whiteflies were then transferred and maintained on cotton plants. The same method was also used in a reciprocal manner to evaluate the potential ability of ISVs in NBU-Q to infect NBU-B. The microinjected whiteflies were collected at 0, 3, 6, and 12 days post injection (DPI) and the viruses were detected by RT-PCR and qRT-PCR. Additionally, the microinjected whiteflies were allowed to oviposit, and the presence of ISVs in the next generation (F1) were also determined using RT-PCR and qRT-PCR. For qRT-PCR analysis, primers were designed using Primer Premier v6.0, and the B. tabaci 18sRNA was used as an internal control. The reaction was run on a Roche Light Cycler® 480 Real-Time PCR System using the SYBR Green Supermix Kit (Yeasen, Shanghai, China) under the following programs: denaturation for 5 min at 95 °C, followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. A relative quantitative method (2−ΔΔCt) was used to evaluate quantitative variation. A pool of 20–30 whiteflies were collected for MEAM1 and MED at each time point, and three independent biological replicates were performed. The primers for RT-PCR and qRT-PCR are listed in Supplementary Table 1.
Tissue expression of ISVs in whitefly NBU-Q and NBU-B
To investigate the relative spatial expression of ISVs in B. tabaci, tissue samples from salivary glands, guts, fat bodies, ovaries, and carcasses were dissected from the whitefly lab cultures of NBU-Q and NBU-B in a phosphate-buffered saline (PBS) solution (137 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, and 1.47 mM KH2PO4 at pH 7.4) under a stereomicroscope (Olympus SZX7, Tokyo, Japan) using sharp forceps (Ideal-Tek, Switzerland). The collected samples were immediately transferred to TRIzol Reagent (Invitrogen Corp., CA, USA) using Eppendorf tips. After RNA extraction, the relative abundance of ISVs in each tissue was determined by qRT-PCR as described above.
Small RNA analysis
The sRNA raw reads of the three libraries NBU-B, NBU-Q, and FY-Q were first treated to remove the adapter, low quality, and junk sequences as described previously58. The clean sRNA reads 18- to 30-nt long were extracted using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). The extracted sRNA were then mapped to the identified viral contigs using Bowtie software with perfect match (i.e. allowing zero mismatch)59. Downstream analyses were performed using custom perl scripts and Linux shell bash scripts.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Mike J. Adams (Minehead, UK) and Xiao-Wei Wang (Zhejiang University, China) for their valuable and constructive suggestions for improving the manuscript. This work was funded by National Natural Science Foundation of China (U20A2036), Project of State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (ZS20190102), and Ningbo Science and Technology Innovation 2025 Major Project (2019B10004). This work was sponsored by K.C.Wong Magna Fund in Ningbo University.
Author contributions
J.-P.C., C.X.Z., and J.-M.L. conceived the idea. H.J.H., Z.X.Y., X.W., X.-T.Y., J.-C.Z., G.L., and J.-B.L. performed the bioinformatics analysis. J.-M.L. and H.-J.H. designed experiments. H.-J.H., Y.Z., Y.-J.H., Y.-H.Q., and X.-D.Z. performed experiments. H.-J.H., J.-M.L., and Z.-X.Y. wrote the original draft. Q.-Z.M., Z.-T.S., F.Y., C.-X.Z., and J.-P.C. reviewed and edited the manuscript. H.-J.H. and Z.-X.Y. contribute equally to this work. All authors reviewed and approved the final version of the manuscript.
Data availability
The raw reads of RNA-seq generated in this study were deposited in NCBI SRA with accession numbers SRR13050950 (NBU-B), SRR13052369 (NBU-Q), SRR13039280 (FY-Q) for transcriptome, and SRR13050947 (NBU-B), SRR13050948 (NBU-Q), SRR13082984 (FY-Q) for sRNA, respectively. Sequences of all identified novel viruses from this study have been deposited in NCBI GenBank with accession numbers MW256664–MW256706 and MW227222–MW227223.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hai-Jian Huang, Zhuang-Xin Ye.
Contributor Information
Jian-Ping Chen, Email: jpchen2001@126.com.
Chuan-Xi Zhang, Email: chxzhang@zju.edu.cn.
Jun-Min Li, Email: lijunmin@nbu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-021-00216-5.
References
- 1.Geoghegan JL, Holmes EC. Predicting virus emergence amid evolutionary noise. Open Biol. 2017;7:170189. doi: 10.1098/rsob.170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasilakis N, Tesh RB. Insect-specific viruses and their potential impact on arbovirus transmission. Curr. Opin. Virol. 2015;15:69–74. doi: 10.1016/j.coviro.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roossinck MJ, Bazan ER. Symbiosis: viruses as intimate partners. Annu. Rev. Virol. 2017;4:123–139. doi: 10.1146/annurev-virology-110615-042323. [DOI] [PubMed] [Google Scholar]
- 4.Vasilakis N, et al. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013;87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nouri S, Matsumura EE, Kuo YW, Falk BW. Insect-specific viruses: from discovery to potential translational applications. Curr. Opin. Virol. 2018;33:33–41. doi: 10.1016/j.coviro.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology. 2012;427:90–97. doi: 10.1016/j.virol.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marklewitz M, Zirkel F, Kurth A, Drosten C, Junglen S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl Acad. Sci. USA. 2015;112:7536–7541. doi: 10.1073/pnas.1502036112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenney JL, Solberg OD, Langevin SA, Brault AC. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J. Gen. Virol. 2014;95:2796–2808. doi: 10.1099/vir.0.068031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu N, et al. Complete genome sequence and characterization of a new iflavirus from the small brown planthopper (Laodelphax striatellus) Virus Res. 2019;272:197651. doi: 10.1016/j.virusres.2019.197651. [DOI] [PubMed] [Google Scholar]
- 10.Kondo H, et al. Virome analysis of aphid populations that infest the barley field: the discovery of two novel groups of Nege/Kita-like viruses and other novel RNA viruses. Front. Microbiol. 2020;11:509. doi: 10.3389/fmicb.2020.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, et al. Discovery and characterization of a novel insect-specific reovirus isolated from Psammotettix alienus. J. Gen. Virol. 2020;101:884–892. doi: 10.1099/jgv.0.001442. [DOI] [PubMed] [Google Scholar]
- 12.Du J, et al. Characterization of viromes within mosquito species in China. Sci. China Life Sci. 2020;63:1089–1092. doi: 10.1007/s11427-019-1583-9. [DOI] [PubMed] [Google Scholar]
- 13.Belden LK, Harris RN. Infectious diseases in wildlife: the community ecology context. Front. Ecol. Env. 2007;5:533–539. doi: 10.1890/060122. [DOI] [Google Scholar]
- 14.Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. B Biol. Sci. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennison NJ, Jupatanakul N, Dimopoulos G. The mosquito microbiota influences vector competence for human pathogens. Curr. Opin. Insect Sci. 2014;3:6–13. doi: 10.1016/j.cois.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez JL, et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 2012;6:e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi C, et al. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome. 2019;7:121. doi: 10.1186/s40168-019-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wille M, Shi M, Klaassen M, Hurt AC, Holmes EC. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 2019;13:2603–2616. doi: 10.1038/s41396-019-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira MRV, Henneberry TJ, Anderson P. History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot. 2001;20:709–723. doi: 10.1016/S0261-2194(01)00108-9. [DOI] [Google Scholar]
- 20.Navas-Castillo J, Fiallo-Olive E, Sanchez-Campos S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011;49:219–248. doi: 10.1146/annurev-phyto-072910-095235. [DOI] [PubMed] [Google Scholar]
- 21.Rosen R, et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 2015;15:1–8. doi: 10.1016/j.coviro.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 22.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- 23.Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 2010;103:196–208. doi: 10.1603/AN09061. [DOI] [Google Scholar]
- 24.Fiallo-Olive E, Pan LL, Liu SS, Navas-Castillo J. Transmission of Begomoviruses and other whitefly-borne viruses: dependence on the vector species. Phytopathology. 2020;110:10–17. doi: 10.1094/PHYTO-07-19-0273-FI. [DOI] [PubMed] [Google Scholar]
- 25.Gilbertson RL, Batuman O, Webster CG, Adkins S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015;2:67–93. doi: 10.1146/annurev-virology-031413-085410. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Campos S, et al. Displacement of tomato yellow leaf curl virus (TYLCV)-Sr by TYLCV-Is in tomato epidemics in spain. Phytopathology. 1999;89:1038–1043. doi: 10.1094/PHYTO.1999.89.11.1038. [DOI] [PubMed] [Google Scholar]
- 27.Aguiar ER, et al. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res. 2015;43:6191–6206. doi: 10.1093/nar/gkv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster CL, et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyle, M. C., Elya, C. N., Bronski, M. & Eisen, M. B. Entomophthovirus: an insect-derived iflavirus that infects a behavior manipulating fungal pathogen of dipterans. Preprint at biorxiv10.1101/371526 (2018).
- 30.Lu G, et al. Discovery of two novel negeviruses in a dungfly collected from the Arctic. Viruses. 2020;12:692. doi: 10.3390/v12070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng TF, et al. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE. 2011;6:e19050. doi: 10.1371/journal.pone.0019050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosario K, Capobianco H, Ng TF, Breitbart M, Polston JE. RNA viral metagenome of whiteflies leads to the discovery and characterization of a whitefly-transmitted carlavirus in North America. PLoS ONE. 2014;9:e86748. doi: 10.1371/journal.pone.0086748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wardrop EA, Gray AB, Singh R, Peterson J. Aphid transmission of potato virus S. Am. J. Potato Res. 1989;66:449. doi: 10.1007/BF02855437. [DOI] [Google Scholar]
- 34.Santillan FW, et al. The biology and phylogenetics of potato virus S isolates from the Andean region of South America. Plant Dis. 2018;102:869–885. doi: 10.1094/PDIS-09-17-1414-RE. [DOI] [PubMed] [Google Scholar]
- 35.Shi C, et al. Stability of the virome in lab- and field-collected Aedes albopictus mosquitoes across different developmental stages and possible core viruses in the publicly available virome data of Aedes mosquitoes. mSystems. 2020;5:e00640–20. doi: 10.1128/mSystems.00640-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. R. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faria NR, Suchard MA, Rambaut A, Streicker DG, Lemey P. Simultaneously reconstructing viral cross-species transmission history and identifying the underlying constraints. Philos. Trans. R. Soc. B. 2013;368:20120196. doi: 10.1098/rstb.2012.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latinne A, et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Patterson EI, Villinger J, Muthoni JN, Dobel-Ober L, Hughes GL. Exploiting insect-specific viruses as a novel strategy to control vector-borne disease. Curr. Opin. Insect Sci. 2020;39:50–56. doi: 10.1016/j.cois.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita R, et al. Persistent viruses in mosquito cultured cell line suppress multiplication of flaviviruses. Heliyon. 2018;4:e00736. doi: 10.1016/j.heliyon.2018.e00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolling BG, Weaver SC, Tesh RB, Vasilakis N. Insect-specific virus discovery: significance for the arbovirus community. Viruses. 2015;7:4911–4928. doi: 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlund P, Lunden H, Blomstrom AL. Insect-specific virus evolution and potential effects on vector competence. Virus Genes. 2019;55:127–137. doi: 10.1007/s11262-018-01629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreira LA, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 44.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antipov D, Raiko M, Lapidus A, Pevzner PA. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics. 2020;36:4126–4129. doi: 10.1093/bioinformatics/btaa490. [DOI] [PubMed] [Google Scholar]
- 46.Boykin LM, Savill A, De Barro P. Updated mtCOI reference dataset for the Bemisia tabaci species complex. F1000Res. 2017;6:1835. doi: 10.12688/f1000research.12858.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell AL, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 52.Darriba D, et al. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quan J, et al. Metagenomic characterization of intestinal regions in pigs with contrasting feed efficiency. Front. Microbiol. 2020;11:32. doi: 10.3389/fmicb.2020.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theor. Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 57.Xu HJ, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519:464–467. doi: 10.1038/nature14286. [DOI] [PubMed] [Google Scholar]
- 58.Li J, et al. Characterization of rice black-streaked dwarf virus- and rice stripe virus-derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus. PLoS ONE. 2013;8:e66007. doi: 10.1371/journal.pone.0066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li CX, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. elife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi M, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 62.Shi M, et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J. Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vasilakis N, et al. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013;87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kondo H, et al. Virome analysis of aphid populations that infest the barley field: the discovery of two novel groups of Nege/Kita-like viruses and other novel RNA viruses. Front. Microbiol. 2020;11:509. doi: 10.3389/fmicb.2020.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nunes MRT, et al. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specific viruses in the taxon Negevirus. Virology. 2017;504:152–167. doi: 10.1016/j.virol.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Miranda JR, et al. Characterization of a novel RNA virus discovered in the autumnal moth Epirrita autumnata in Sweden. Viruses. 2017;9:214. doi: 10.3390/v9080214. [DOI] [Google Scholar]
- 67.Phan TG, et al. Sera of Peruvians with fever of unknown origins include viral nucleic acids from non-vertebrate hosts. Virus Genes. 2018;54:33–40. doi: 10.1007/s11262-017-1514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debat HJAn. RNA virome associated to the golden orb-weaver spider Nephila clavipes. Front. Microbiol. 2017;8:2097. doi: 10.3389/fmicb.2017.02097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spear A, Sisterson MS, Yokomi R, Stenger DC. Plant-feeding insects harbor double-stranded RNA viruses encoding a novel proline-alanine rich protein and a polymerase distantly related to that of fungal viruses. Virology. 2010;404:304–311. doi: 10.1016/j.virol.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 70.Ito T, Suzaki K, Nakano M. Genetic characterization of novel putative rhabdovirus and dsRNA virus from Japanese persimmon. J. Gen. Virol. 2013;94:1917–1921. doi: 10.1099/vir.0.054445-0. [DOI] [PubMed] [Google Scholar]
- 71.Walker PJ, et al. Characterization of three novel viruses from the families Nyamiviridae, Orthomyxoviridae, and Peribunyaviridae, isolated from dead birds collected during West Nile Virus Surveillance in Harris County, Texas. Viruses. 2019;11:927. doi: 10.3390/v11100927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads of RNA-seq generated in this study were deposited in NCBI SRA with accession numbers SRR13050950 (NBU-B), SRR13052369 (NBU-Q), SRR13039280 (FY-Q) for transcriptome, and SRR13050947 (NBU-B), SRR13050948 (NBU-Q), SRR13082984 (FY-Q) for sRNA, respectively. Sequences of all identified novel viruses from this study have been deposited in NCBI GenBank with accession numbers MW256664–MW256706 and MW227222–MW227223.