Abstract
The ctDNA plasma testing is one of the methods to examine biomarkers for lung adenocarcinoma in order to detect a mutation of epidermal growth factor receptor (EGFR) gene. The advantages of ctDNA testing over tissue biopsy and lung tumor cytology include less invasive, faster result, cheaper, and minimum risk of complication for the patient. We analyzed and compare the detection of EFGR mutation in peripheral blood plasma (liquid biopsy) with cytological specimens of patients with lung adenocarcinoma. We conducted ctDNA plasma testing in 124 lung adenocarcinoma patients who visited our hospital from January to December 2018. The ctDNA testing results were compared with the results of EGFR detection from the previous cytological specimen examination. Most of the patients were males, aged 55–59 years, nonsmokers, and had stage IVA lung adenocarcinoma, with most metastasis found in the pleura. We found a correlation between EGFR prevalence with nonsmoking status and patient’s age. The ctDNA plasma testing detected 27.4% common EGFR mutation and 72.6% wild-type EGFR. The figures of EGFR mutation detection from cytological specimens were 47.6% and 52.4%, respectively. Compared to cytological specimens, the EGFR mutation detection in ctDNA had a sensitivity of 48.3%, with a specificity of 90.9%, PPV of 82.35%, NPV of 66.7%, and 70.97% concordance rate. EGFR mutation with cytological specimen examination was more accurate than ctDNA.
Keywords: Concordance rate, ctDNA plasma, EGFR mutation, Lung adenocarcinoma
Introduction
Lung cancer is one of the leading causes of cancer death in the world. There are two main types of lung cancer, non-small cell lung cancer (NSCLC) consisted of 80% and small cell lung cancer (SCLC) consisted of 20% of all lung cancer. The NSCLC consisted mostly of adenocarcinoma, followed by squamous cell carcinoma, and large-cell carcinoma. Lung adenocarcinoma is the most common NSCLC type found in the world [1–3].
Lung cancer is the top 3 most common cancer found in Indonesia along with breast cancer and cervical cancer. Lung cancer is a cancer with the highest prevalence in men. Based on data from Dharmais National Cancer Hospital, Jakarta, Indonesia, the prevalence of lung cancer had been increasing from 2010 to 2013, where 117 cases took place in 2010 with 38 deaths, 163 cases in 2011 with 39 deaths, 165 cases in 2012 with 62 deaths, and 173 cases in 2013 with 65 deaths [4].
The incident of lung adenocarcinoma has been increasing as well in Asia and in the USA, particularly in women, young adults, and nonsmokers. The US data showed that lung adenocarcinoma was found in 31–54% of nonsmoker men, higher than in smoker men (25%–33%), and 49–74% of nonsmoker women, which also higher than in smoker women (33%–43%) [5].
Epidermal growth factor receptor (EGFR) mutation in NSCLC is particularly found in lung adenocarcinoma. It has an important role in the last 10 years as a target of cancer therapy. Advanced NSCLC patients with EGFR activating mutation, i.e., exon 18, exon 19, and exon 21, showed much better response when given EGFR tyrosine kinase inhibitor (TKI) as the treatment compared to standard chemotherapy [6, 7]. This advantage can only be achieved if the adequate materials or samples and tools are available. However, the success of EGFR mutation testing is often hindered by tissue availability, so that the patients often lose the opportunity to get the targeted therapy due to the insufficient tissue for histological or cytological examination.
Current mutation testing can alternatively be conducted with liquid biopsy using EGFR mutation detection from blood plasma. A study of EGFR mutation prevalence taken from tissue and blood conducted by Implementing GeNomics In pracTicE (IGNITE study) was done in Asia-Pacific and Russia and found that EGFR mutation in lung adenocarcinoma tissue was higher in Asia-Pacific than in Russia (49.3% vs. 18%). The study also found a concordance of EGFR mutation testing results between tissue and blood samples of 2581 patients (80.5% concordance rate, 46.5% sensitivity, and 95.6% specificity) [8]. The result is quite convincing, with 95.6% specificity, so that EGFR mutation testing from blood samples can be used as a screening method for NSCLC cases. Studies on this topic have not been conducted widely in Indonesia, particularly in Surabaya. Therefore, we conducted this research to determine the prevalence of EGFR mutation in NSCLC patients of adenocarcinoma histopathology at Dr. Soetomo General Hospital, Surabaya, Indonesia, and to compare the results of EGFR mutation detection in blood plasma samples to the results of cytological samples examination.
Methods
This study was an analytical observational research conducted in 124 consecutive lung adenocarcinoma patients who visited Dr. Soetomo General Hospital, Surabaya, Indonesia, from January to December 2018 (Fig. 1). The inclusion criteria were lung adenocarcinoma patients who had been previously diagnosed with cytological samples, had not received any cancer treatment, and had cytological sample slides with sufficient cell numbers that can be analyzed (minimum of 50 cells). Patients who had adenocarcinoma lesions in the lungs as metastases from other organs and having nonrepresentative cytological samples were excluded from this study. All eligible subjects were giving their consent. This study was approved by the Ethic Committee of Dr. Soetomo General Hospital, Surabaya (certificate No. 0209/KEPK/IV/2018).
Fig. 1.
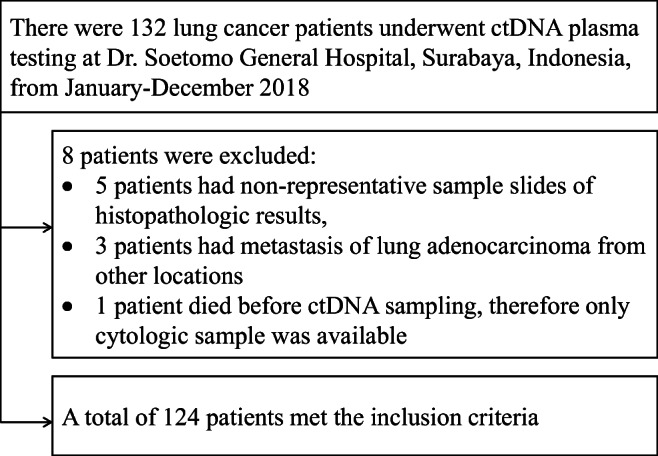
Flow diagram of subject recruitment
The examination of EGFR mutations status from cytological samples was done in the Pathology Anatomy Laboratory of Dr. Soetomo Hospital. The EGFR examination of cytological samples used the DNA extraction method (GeneAll Exgene™ Cell SV), real-time quantitative PCR (high-resolution melting analysis) analyzed with LightCycler 480 II real-time PCR (Roche, Basel, Switzerland), and AmoyDx® EGFR Adx-ARMS® (Amoy Diagnostics, Xiamen, China).
The EGFR mutation detection from patient’s blood plasma specimens was done in Prodia Laboratory, Surabaya, Indonesia. Approximately 10-mL blood sample was withdrawn from each patient, which was then centrifuged into blood plasma, stored in a special frozen container, and transferred to the laboratory. The blood plasma was then analyzed for EGFR mutation using real-time PCR Scorpion-ARMS with therascreen® EGFR plasma RGQ PCR kit (QIAGEN, Manchester, UK). The target detections of this examination were exon 19 deletions, T790 M mutation in exon 20, and L858R point mutation. The detection limit was in accordance with the provisions of the corporation. The work steps for DNA extraction and for EGFR mutation detection were done according to the manufacturer’s instruction. This method had been reported previously elsewhere [9]. The results of ctDNA were then compared with the results of EGFR mutation testing from cytological samples.
A diagnostic test was conducted to determine the correlation between EGFR ctDNA results and cytological specimens, in terms of sensitivity and specificity. The concordance rate test was performed to examine the conformity between ctDNA and cytological specimens. Cross-tabulation and logistic regression tests were done to examine the relationship between disease stage, sex, age, and smoking status with the results of EGFR mutations on ctDNA with 95% confidence intervals (CI) and p value < 0.05. Statistical analysis was conducted using IBM SPSS Statistics software version 23.0 (IBM Corp., Armonk, NY, USA).
Results
Subject Characteristics
Most of the subjects were males (58.1%), in the age group of 55–59 years (22.6%). The majority of the study subjects were nonsmokers (51.6%), while 21.0% of them were active smokers (Table 1). Most of smoking subjects were in the age group of 40–49 (15.3%), and the highest number of cigarettes smoked was around 10–19 per day (24.2%).
Table 1.
Subject characteristics
| Characteristics | n (%) |
|---|---|
| Age | |
| 30–34 years | 2 (1.6%) |
| 35–39 years | 6 (4.8%) |
| 40–44 years | 5 (4.0%) |
| 45–49 years | 6 (4.8%) |
| 50–54 years | 25 (20.2%) |
| 55–59 years | 28 (22.6%) |
| 60–64 years | 22 (17.7%) |
| 65–69 years | 10 (8.1%) |
| 70–74 years | 13 (10.5%) |
| 75–79 years | 6 (4.8%) |
| 80–84 years | 1 (0.8%) |
| Sex | |
| Male | 72 (58.1%) |
| Female | 52 (41.9%) |
| Smoking status | |
| Active Smoker | 26 (21.0%) |
| Nonsmoker | 64 (51.6%) |
| Ex-smoker | 34 (27.4%) |
| Tissue origin | |
| Open biopsy | 5 (4.0%) |
| Core biopsy | 18 (14.5%) |
| Bronchoalveolar lavage (BAL) | 7 (5.6%) |
| Bronchial brushing | 2 (1.6%) |
| FNAB lung | 74 (59.7%) |
| FNAB cervical lymph node | 4 (3.2%) |
| Pleural fluid cytology | 14 (11.3%) |
| Diagnosis | |
| Lung adenocarcinoma (right lung) | 87 (70.2%) |
| Lung adenocarcinoma (left lung) | 37 (29.8%) |
| Stage | |
| IIA | 2 (1.6%) |
| IIB | 3 (2.4%) |
| IIIA | 11 (8.9%) |
| IIIB | 5 (4.0%) |
| IIIC | 6 (4.8%) |
| IVA | 83 (66.9%) |
| IVB | 14 (11.3%) |
FNAB fine-needle aspiration biopsy
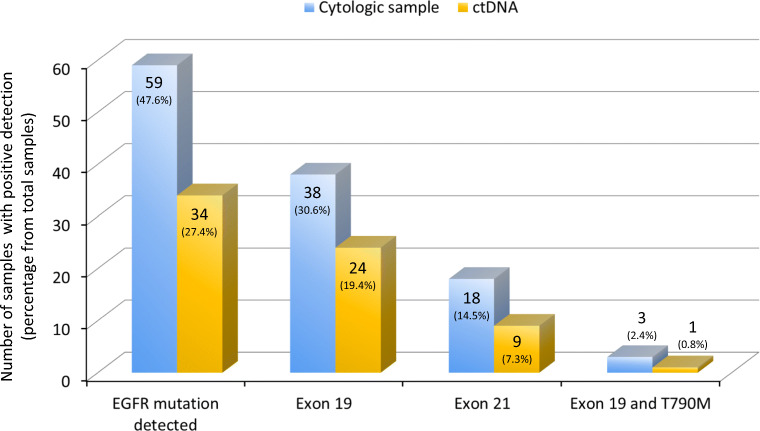
Most of the study subjects had adenocarcinoma of the right lung (70.2%). The majority of cytological specimens’ examination for the diagnosis of lung adenocarcinoma was taken from lung fine-needle aspiration biopsy (FNAB) which was 59.7%. Most of the patients (66.9%) had stages IVA disease (Table 1). Tumor metastases were found to be diverse; most metastases were in the pleura (52.4%), followed by bone metastases (including vertebral bones, ribs, and other large bones) in 9.7% of the patients. The EGFR mutation was detected in 47.6% of patients from cytological specimens and in 27.4% of patients from ctDNA. The type of EGFR mutation was dominated by the common mutations, i.e., in exon 19 (30.6% in cytological specimens and 19.4% in ctDNA) and exon 21 L858R (14.5% in cytological specimens and 7.3% in ctDNA). The results of EGFR mutation detection by the two methods can be seen in Fig. 2.
Fig. 2.
EGFR mutation detection in cytologic samples and ctDNA
Association of Age, Sex, Smoking Status, and Disease Stage on EGFR Mutation Results
The age and the smoking behavior of the study subjects were significantly correlated with EGFR mutations detected in ctDNA (β = 0.220; p = 0.042 and β = 1.740; p < 0.001, respectively). There was no significant correlation between sex and the EGFR mutation (β = − 0.030; p = 0.961) as can be seen in Table 2.
Table 2.
Association between subject characteristics and EGFR mutation status of ctDNA sample
| Characteristics | β | p | OR | CI for OR |
|---|---|---|---|---|
| Age ≤ 65 vs > 65 | 0.220 | 0.042 | 1.246 | 0.415–2.316 |
| Female vs male | − 0.030 | 0.961 | 0.970 | 0.407–4.571 |
| Nonsmoker vs smoker | 1.740 | < 0.001 | 5.698 | 1.283–5.590 |
Dependent, EGFR ctDNA
The ctDNA detection of EGFR mutation is more likely positive in more advanced stage lung cancer. In other words, subjects with advanced disease stages had the highest probability of EGFR mutations detected in ctDNA. Most of our subjects were in stage IVA lung adenocarcinoma. Positive EGFR mutations were detected in 30.12% (25/83) of stage IVA patients but only in 16.6% (1/6) of stage IIIC patients. No ctDNA detection of EGFR mutations was found in patients with stage IIA to IIIB diseases (Table 3).
Table 3.
Disease stage and the positivity of EGFR mutation detection in ctDNA sample
| CA | Positive EGFR mutation (%) n = 34 | Wild type (%) n = 90 | Total (%) n = 124 |
|---|---|---|---|
| IIA | 0 (0.00) | 2 (100.00) | 2 (1.61) |
| IIB | 0 (0.00) | 3 (100.00) | 3 (2.42) |
| IIIA | 0 (0.00) | 11 (100.00) | 11 (8.87) |
| IIIB | 0 (0.00) | 5 (100.00) | 5 (4.03) |
| IIIC | 1 (16.60) | 5 (83.33) | 6 (4.84) |
| IVA | 25 (30.12) | 58 (69.88) | 83 (66.93) |
| IVB | 8 (57.14) | 6 (42.86) | 14 (11.29) |
Concordance of ctDNA EGFR Mutation Results and Cytological Specimens
Analysis of concordance of EGFR mutation detection results between liquid biopsy (ctDNA) and cytological specimens was done to obtain the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and concordance rate. Compared to detection in cytological specimens, the EGFR mutation detection in ctDNA had a sensitivity of 48.3%, with a specificity of 90.9%, PPV of 82.35%, NPV of 66.7%, and 70.97% concordance rate (Table 4).
Table 4.
Concordance of EGFR mutation status between cytological specimens and ctDNA (plasma)
| Concordance rate | Sensitivity | Specificity | PPV | NPV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | CI 95% | n (%) | CI 95% | n (%) | CI 95% | n (%) | CI 95% | n (%) | CI 95% | |
| EGFR ctDNA | 88/124 (70.97) | 62–79 | 28/58 (48.3) | 35–62 | 60/66 (90.9) | 81–97 | 28/34 (82.35) | 68–91 | 60/90 (66.67) | 61–72 |
Discussion
Our study showed a good correlation and concordance between EGFR mutation status detected in plasma samples (liquid biopsy) and cytological specimens. Our finding was also in accord with the results of the IGNITE study which also included patients from our hospital [8]. This is of particular interest, because now we can use ctDNA or plasma biopsy to detect EGFR mutations in adenocarcinoma NSCLC patients in whom an adequate tissue sample cannot be obtained.
Most subjects in this study were in the age group of 55–59 years. This finding is consistent with some previous studies which stated that the majority of lung adenocarcinoma patients receiving EGFR-TKI treatment were more than 45 years old with an average of 61.8-year old [10]. Another study also found that adenocarcinoma patients mostly fell in the age group of 40–49 years [11]. The largest epidemiological study of adenocarcinoma patients in Asia, the PIONEER study, reported that the average age of adenocarcinoma patients in Asia was 60 years old, ranged from 17 to 94 years old [12].
In this study, most subjects were male patients, consistent with the results of a study conducted by The International Agency for Research on Cancer (IARC). IARC reported a high incidence of lung cancer cases in male, particularly in Asia, North America, Middle East, and South Europe, with the incident rate ranging from 48.5 to 56.5 per 100,000 cases [13]. Another study also found that lung adenocarcinoma cancer was mostly (63%) found in males [11]. The ratio of lung adenocarcinoma cancer cases between males and females was 7.4:1 [14].
Smoking is one obvious risk factor for lung cancer. Some research reported that secondhand smoke decreases the function of ciliary follicles. The lung’s physiological ability to expel foreign material would decrease, and it leads to adenocarcinoma in long-term period [15, 16]. In this study, most of our subjects were nonsmokers (51.6%). Our result was in accord with a study which also reported that the majority of female lung adenocarcinoma patients (86.6%) were nonsmokers [17].
In our study, most of the samples for histopathological diagnosis of adenocarcinoma were taken from lung mass (86.8%). Most of them (59.7%) were obtained via fine-needle aspiration biopsy (FNAB) technique. As had been reported in IGNITE study, FNAB was used as sampling technique as much as 51% in Indonesia compared to 40.4% in Thailand [8, 10]. Most subjects in our study were in advanced stage of disease (stage IVA 66.9% and stage IVB 11.3%). Elhidsi et al. [10] also reported that most of their patients were diagnosed with stage IV lung adenocarcinoma when they were first brought to health services (44.6% stage IVA and 43.1% stage IVB). Our findings were also consistent with a previous study conducted in India, stating that lung adenocarcinoma patients presented mostly (82%) in stage IV disease at diagnosis [14].
The positive plasma EGFR mutation detected in ctDNA in our study was 27.4%. Other study, the PIONEER study, reported a proportion of positive EGFR mutation of lung adenocarcinoma in seven Asian countries ranging from 22 to 64% [12]. Similar results were also found in a study conducted by Oktaviyanti in Indonesia, which reported that 34% of adenocarcinoma patients had positive EGFR mutation [11]. The type of EGFR mutations in our study was dominated by common EGFR mutation (97%), consisted of 72.7% mutations in exon 19 and 27.2% mutations in exon 21. Our finding was also consistent with results of study conducted by Pirker et al. [18] and the IGNITE study [8]. Pirker et al. found that EGFR common mutation was around 85–90%, consisted of exon 19 deletion mutations (45%) and L858R exon 21 mutations (40–45%) [18]. Likewise, the IGNITE study on populations in the Asia-Pacific region found EGFR common mutations of 91.2%, consisted of exon 19 deletion mutations (48.7%) and L858R exon 21 mutations (42.5%) [8]. Most of the metastasis in our study was in the pleura (52.4%) which is also consistent with the result of Elhidsi et al., where pleural metastases was the highest frequency of metastatic lesion in lung adenocarcinoma patients at 66.1% [10].
The ctDNA detection of EGFR mutation is more likely positive in more advanced stage lung cancer. In other words, subjects with advanced disease stages had the highest probability of EGFR mutations detected in ctDNA and would be lowest in the early stages. In our study, positive EGFR mutations were detected strongly in stage IVB (57.1%) and stage IVA (30.1%) and were not detected at all in stage I–IIIB. These findings were similar to those reported by Elhidsi et al., who found that most of EGFR mutations in lung adenocarcinoma were detected in advanced stages, i.e., stage IIIB–IV [10]. Plasma samples of patients with lung cancer contained very high DNA when compared to non-cancer patients and would increase especially in the advanced cancer stage. Most circulatory DNA release is believed to originate from cancer cells that die in the primary place or tumor metastasis [19]. Therefore, plasma DNA has proven to be a noninvasive source of genotypic information that can be used to replace tumor tissue in detecting tumor-specific molecular markers. It can also be used to access the therapeutic response and patients’ prognosis [20–22].
The association between tumor stage and the success rate of ctDNA detection has evolved through various recent studies. Those studies reported that ctDNA could be detected in 82–100% of stage IV patients, while only 47% of ctDNA was detected in patients with stage I lung tumors [23]. In our study, only age and smoking status variables had a significant correlation with positive EGFR detections from ctDNA testing. The significance value of nonsmokers was p < 0.001, odd ratio = 5.698, and R square = 15.2%. These results indicated a relationship of nonsmoking status with lung adenocarcinoma EGFR mutation case, but the effect was only 15.2%, with the assumption that the EGFR mutation positivity will increased by 5.698 times greater in nonsmoker patients compared to smoker patients. These findings were consistent with a study conducted by Shigematsu et al., who found that NSCLC patients had more gene mutations in nonsmoker patients (51%), compared to smokers (10%) [24]. Thus, there is no correlation between the occurrence of EGFR mutation and the act of smoking. Patients with positive EGFR mutation will easily develop lung cancer, including the nonsmoker group. This proves that genetic inheritance is more influential than the environment, as also reported by Tsao et al. [25].
The significance value of age was p = 0.042, odd ratio = 1.246, and R square was 15.2%. These findings indicated that the age of 65 years could increase EGFR mutation by 1.246 times greater compared to age group less than 65 years, but the effect was only 15.2%. The tendency of the data showed that the older the age, the higher the risk for cancer. A study conducted in 2014 reported that the tendency of smoking patterns according to age also influenced the occurrence of lung cancer. The age group of 50–64 years had the highest gene inactivation in the CDH1 and GSTP1 genes, while the age group of > 70 years had the highest tendency to inactivate the GTSP1 and RASSF1A genes among other age groups. This indicates that the age group above 65 years has a higher risk of developing lung cancer compared to the population less than 65 years of age [26]. A study on cancer incidence in Korea also showed a tendency that lung cancer occurred in men and women over 65 years [27].
In our study, compared to detection in cytological specimens, the EGFR mutation detection in ctDNA had a sensitivity of 48.3%, with a specificity of 90.9%, PPV of 82.35%, NPV of 66.7%, and 70.97% concordance rate. This finding was also consistent with the IGNITE study that showed a concordance rate of 77.7% [8]. Some previous clinical trials argued that ctDNA detection is an appropriate alternative method for the determination of EGFR mutation status [28–30]. In real-life condition, the ASSESS study reported the use of plasma ctDNA derivatives, and their concordance values were quite good according to the results of tissue/cytology samples which is of 89% (with sensitivity of 46%, specificity of 97%, PPV of 78%, and NPV of 90%) [31]. Some other recent studies, including two meta-analyses of the concordance of EGFR mutation between plasma and tissue, showed a high concordance rate even though with somewhat low sensitivity value of 62–67% [32, 33].
Conclusion
Lung adenocarcinoma patients who underwent EGFR examination at Dr. Soetomo General Hospital, Surabaya, Indonesia, were mostly male, in the age range of 55–59 years, and nonsmokers. FNAB was the most common sampling technique for cytological specimens testing. Most of the specimens were taken from lung mass. Most of the patients had stage IVA disease with pleural metastases. The EGFR mutation was detected in 47.6% of patients from cytological specimens and in 27.4% of patients from ctDNA. The majority of EGFR mutations were in exon 19 and exon 21. EGFR mutation was increasingly detected as the disease stage increased, with the strongest detection in stage IVB (57.1%). The EGFR mutation detection in ctDNA had a high concordance rate with cytological specimens’ examination and thus had the potential to be used as an alternative method to determine the EGFR mutation in adenocarcinoma NSCLC patients in whom an adequate tissue sample cannot be obtained.
Acknowledgements
We would like to thank Astra Zeneca Indonesia Ltd. for the support of ctDNA testing.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laksmi Wulandari, Email: laksmi.wulandari@fk.unair.ac.id.
Gatot Soegiarto, Email: gatot_soegiarto@fk.unair.ac.id.
Anna Febriani, Email: febrianianna@gmail.com.
Farah Fatmawati, Email: farahummufeno@yahoo.co.id.
Sahrun, Email: cahrun.carter@gmail.com.
References
- 1.Fatmawati F. Profil Pasien Kanker Paru Jenis Karsinoma Bukan Sel Kecil yang Mendapatkan Inhibitor Tirosin Kinase Sebagai Terapi Lini Pertama di RSUD Dr. Soetomo: Universitas Airlangga; 2016. [Google Scholar]
- 2.Hudoyo A, Wibawanto A, Lutfi A, Rima A, Putra AC, Ratnawati A, Febriani A, Jusuf A, Harsal A, Westi A, Icksan AG, Heru B, Tobing DL, Soeis DS, Paramita D, Syahruddin E, tenda ED, Diana ES, Suzanna E, Dewi FL, Djuita F, Prajogi GB, Iskandar H, Hidayat H, Indriani PI, Zaini J, Juniarti KJ, Kardinah RKW, KBH P, Wulandari L, Lisnawati M, Supriana N, Lubis N, Soeroso NN, Soetandyo N, Nuhonni SA, Siregar N, Dilangga P, Prasenohadi AP, Pahlesia R, Sembiring RE, Ermayanti S, Anggoro SC, Tarigan SP, Andarini SL, Gondhowiardjo S, Munir SM, Pratiwi SD, Seto RS, Mety SH, Endardjo S, Aniwidyaningsih W, Soeharno W, Amin Z. Pedoman Nasional Pelayanan Kedokteran Kanker Paru. Jakarta: Kementerian Kesehatan Republik Indonesia; 2017. [Google Scholar]
- 3.Jusuf A, Harryanto A, Syahruddin E, Endardjo S, Mudjiantoro S, Sutandio N (2005) Kanker paru jenis karsinoma bukan sel kecil. Pedoman nasional untuk diagnosis dan penatalaksanaan di Indonesia
- 4.VandenBussche CJ, Illei PB, Lin M-T, Ettinger DS, Maleki Z. Molecular alterations in non–small cell lung carcinomas of the young. Hum Pathol. 2014;45(12):2379–2387. doi: 10.1016/j.humpath.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1(7):497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le X, Freed JA, VanderLaan PA, Huberman MS, Rangachari D, Jorge SE, Lucena-Araujo AR, Kobayashi SS, Balasubramanian S, He J, Chudnovsky Y, Miller VA, Ali SM, Costa DB. Detection of crizotinib-sensitive lung adenocarcinomas with MET, ALK, and ROS1 genomic alterations via comprehensive genomic profiling. Clin Lung Cancer. 2015;16(5):e105–e109. doi: 10.1016/j.cllc.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh JE, An CH, Yoo NJ, Lee SH. Detection of low-level EGFR T790M mutation in lung cancer tissues. APMIS. 2011;119(7):403–411. doi: 10.1111/j.1600-0463.2011.02738.x. [DOI] [PubMed] [Google Scholar]
- 8.Han B, Tjulandin S, Hagiwara K, Normanno N, Wulandari L, Laktionov K, Hudoyo A, He Y, Zhang Y-P, Wang M-Z, Liu CY, Ratcliffe M, McCormack R, Reck M. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: the IGNITE study. Lung Cancer. 2017;113:37–44. doi: 10.1016/j.lungcan.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, McCormack R, Gu Y, Liu X. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung Cancer. J Mol Diagn. 2015;17(3):265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Elhidsi M, Andarini SL, Hudoyo A. Profil Mutasi epidermal growth factor receptor Pasien Adenokarsinoma Paru Usia Muda. Jurnal Respirologi Indonesia. 2016;4(36):244–247. [Google Scholar]
- 11.Oktaviyanti IK. Mutasi Egfr Pada Pemeriksaan Sitologi Adenokarsinoma Paru. Berkala Kedokteran Unlam. 2015;11(2):213–219. [Google Scholar]
- 12.Shi Y, Au JS-K, Thongprasert S, Srinivasan S, Tsai C-M, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang P-C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 14.Mohan A, Latifi A, Guleria R (2016) Increasing incidence of adenocarcinoma lung in India: following the global trend? Indian J Cancer 53(1):92–92, 95 [DOI] [PubMed]
- 15.Chen J, Qi Y, Wampfler JA, Jatoi A, Garces YI, Busta AJ, Mandrekar SJ, Yang P. Effect of cigarette smoking on quality of life in small cell lung cancer patients. Eur J Cancer (Oxford, England : 1990) 2012;48(11):1593–1601. doi: 10.1016/j.ejca.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezaei S, Karami Matin B, Kazemi Karyani A, Woldemichael A, Khosravi F, Khosravipour M, Rezaeian S. Impact of smoking on health-related quality of life: a general population survey in West Iran. Asian Pac J Cancer Prev. 2017;18(11):3179–3185. doi: 10.22034/APJCP.2017.18.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prudkin L, Tang X, Wistuba II. Germ-line and somatic presentations of the EGFR T790M mutation in lung cancer. J Thorac Oncol. 2009;4(1):139–141. doi: 10.1097/JTO.0b013e3181915f92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirker R, Herth FJF, Kerr KM, Filipits M, Taron M, Gandara D, Hirsch FR, Grunenwald D, Popper H, Smit E, Dietel M, Marchetti A, Manegold C, Schirmacher P, Thomas M, Rosell R, Cappuzzo F, Stahel R. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010;5(10):1706–1713. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 19.Sozzi G, Conte D, Leon M, Cirincione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung Cancer. J Clin Oncol. 2003;21(21):3902–3908. doi: 10.1200/jco.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, Wang X, Duan CJ, Wu NM, Guo ZQ, Liu YX, Liu HN, Wang YY, Wang J. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non–small-cell lung Cancer. J Clin Oncol. 2009;27(16):2653–2659. doi: 10.1200/jco.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 21.Yung TKF, Chan KCA, Mok TSK, Tong J, To K-F. Lo YMD. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non–small cell lung Cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. doi: 10.1158/1078-0432.ccr-08-2622. [DOI] [PubMed] [Google Scholar]
- 22.Jian G, Songwen Z, Ling Z, Qinfang D, Jie Z, Liang T, Caicun Z. Prediction of epidermal growth factor receptor mutations in the plasma/pleural effusion to efficacy of gefitinib treatment in advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136(9):1341–1347. doi: 10.1007/s00432-010-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong S-M, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih L-M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang T-L, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224):224ra224–224ra224. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed]
- 24.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 25.Tsao AS, Tang XM, Sabloff B, Xiao L, Shigematsu H, Roth J, Spitz M, Hong WK, Gazdar A, Wistuba I. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung Cancer. J Thorac Oncol. 2006;1(3):231–239. doi: 10.1016/S1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 26.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutation Research/Reviews in Mutation Research. 2008;659(1):40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Jung K-W, Won Y-J, Kong H-J, Oh C-M, Lee DH, Lee JS. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2011. Cancer Res Treat. 2014;46(2):109–123. doi: 10.4143/crt.2014.46.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douillard J-Y, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, Walker J, Dearden S, Webster A, Milenkova T, McCormack R. Gefitinib treatment in EGFR mutated Caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mok T, Wu Y-L, Lee JS, Yu C-J, Sriuranpong V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao M, Zhu Y, Zhou C, Fuerte F, Margono B, Wen W, Tsai J, Truman M, Klughammer B, Shames DS, Wu L. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.ccr-14-2594. [DOI] [PubMed] [Google Scholar]
- 30.Karachaliou N, Mayo-de las Casas C, Queralt C, de Aguirre I, Melloni B, Cardenal F, Garcia-Gomez R, Massuti B, Sánchez JM, Porta R. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 2015;1(2):149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 31.Reck M, Hagiwara K, Han B, Tjulandin S, Grohé C, Yokoi T, Morabito A, Novello S, Arriola E, Molinier O, McCormack R, Ratcliffe M, Normanno N. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol. 2016;11(10):1682–1689. doi: 10.1016/j.jtho.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang F, Xu L, Yin R. Circulating tumor DNA is effective for the detection of EGFR mutation in non–small cell lung cancer: a meta-analysis. Cancer epidemiology biomarkers &. Prevention. 2015;24(1):206–212. doi: 10.1158/1055-9965.epi-14-0895. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269–6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]