Abstract
Metabolic syndrome (MetS) is one of the most important risk factors for cardiovascular disease. The 11p23.3 chromosomal region plays a potential role in the pathogenesis of MetS. The present study aimed to assess the association between 18 single nucleotide polymorphisms (SNPs) located at the BUD13, ZPR1, and APOA5 genes with MetS in the Tehran Cardio-metabolic Genetics Study (TCGS). In 5421 MetS affected and non-affected participants, we analyzed the data using two models. The first model (MetS model) examined SNPs' association with MetS. The second model (HTg-MetS Model) examined the association of SNPs with MetS affection participants who had a high plasma triglyceride (TG). The four-gamete rules were used to make SNP sets from correlated nearby SNPs. The kernel machine regression models and single SNP regression evaluated the association between SNP sets and MetS. The kernel machine results showed two sets over three sets of correlated SNPs have a significant joint effect on both models (p < 0.0001). Also, single SNP regression results showed that the odds ratios (ORs) for both models are almost similar; however, the p-values had slightly higher significance levels in the HTg-MetS model. The strongest ORs in the HTg-MetS model belonged to the G allele in rs2266788 (MetS: OR = 1.3, p = 3.6 × 10–7; HTg-MetS: OR = 1.4, p = 2.3 × 10–11) and the T allele in rs651821 (MetS: OR = 1.3, p = 2.8 × 10–7; HTg-MetS: OR = 1.4, p = 3.6 × 10–11). In the present study, the kernel machine regression models could help assess the association between the BUD13, ZPR1, and APOA5 gene variants (11p23.3 region) with lipid-related traits in MetS and MetS affected with high TG.
Subject terms: Genetics, Molecular medicine
Introduction
Metabolic syndrome (MetS) is a combination of central obesity, hypertension, dyslipidemia, and insulin resistance which could increase the risk of type two diabetes (T2D) fivefold and is associated with a doubled risk of developing cardiovascular disease (CVD)1–4. It has been debated whether the combination of metabolic characteristics referred to as MetS represents a biologically meaningful entity in itself5,6 and generally accepted that the current criteria of MetS are integrated assessment strategies with only a binary outcome rather than precise levels of metabolic components7.
Many factors are involved in pathophysiological mechanisms of Mets, such as ectopic fat deposition, excess adipose tissue mass, the excessive flux of fatty acids, and inflammation.
As hyperlipidemia is the most frequent Mets component, some critical effects were reported on its pathophysiological mechanisms such as ectopic fat deposition, excess adipose tissue mass, the excessive flux of fatty acids, and inflammation3,8.
Apolipoproteins play a crucial role in blood, cerebrospinal fluid, and lymph lipid transportation.
In lipid transportation in the blood and cerebrospinal fluid and lymph, apolipoproteins play an essential role.
The previous genome-wide association analysis showed a genomic locus in correlation with MetS and its components in 11q23.39. This region encodes genes involved in lipid metabolism, and previous studies indicated that several single nucleotide polymorphisms (SNPs) located in ZPR1 Zinc Finger (Gene ID: 8882; OMIM: 603901), BUD13 homolog (Gene ID: 84811), and APOA5 (Gene ID: 116519; OMIM: 606368) have a significant association with quantitative phenotypes such as lipid profile components10–13.
ZPR1 is an essential regulatory protein that is responsible for normal cell proliferation and signal transduction and acts as a signaling molecule transporting proliferative growth signals from the cytoplasm to the nucleus. For normal nuclear function in cell proliferation and signal transduction, ZPR1 is an essential regulatory protein that acts as a signaling molecule that communicates proliferative growth signals from the cytoplasm to the nucleus14.
Peroxisome proliferator-activated receptor gamma (PPARG) and the hepatocyte nuclear factor 4 alpha (HNF4a, nuclear receptor 2A1) bind to ZPR1 promoter activation of various visceral systems genes in, involving in glucose, fatty acid, and cholesterol metabolism15,16. BUD13 was identified in yeast as a splicing factor that affects nuclear pre-mRNA retention; however, nowadays, this region is known as a member of the RES complex17,18 and its functionality is controversial. The increase of plasma triglyceride (TG) levels could be a major risk factor for coronary artery disease, and APOA5 is an essential determinant of its level. The APOA5 gene is the closest member of the APO gene cluster to the BUD13/ZPR1 gene cluster, which its variations are well known as the SNPs associated with an increased risk of coronary artery disease metabolic syndrome19–21.
The genetic variants of the BUD13, ZPR1, and APOA5 genes concerning cardio-metabolic phenotypes were reported in different populations10,11, but only a few studies have been conducted in the Middle Eastern populations. Therefore, we performed this study to estimate the association of the BUD13, ZPR1, and APOA5 genetic variations on the Tehran Cardio-metabolic Genetics Study (TCGS) population21. This study focuses on replicating the genetic markers associated with metabolic syndrome and determining their effects on triglyceride concentration among MetS affected subjects in an Iranian population22.
Results
In the first model of analysis (MetS versus non-Mets) 2996 individuals were affected with MetS, and 2425 individuals were non-affected. The MetS group was older with higher mean values for MetS components except for HDL concentration. About 0.7% and 35% were Lipid-lowering drug users, 3.5% and 35% were hypertension-lowering drug users, and 1.7% and 25% were diabetes drug users among MetS and non-MetS participants, respectively (Table 1).
Table 1.
Summary of demographic and clinical characteristics of study individuals.
| Unrelated individuals (n = 5421) | ||||||
|---|---|---|---|---|---|---|
| Non MetS (n = 2425) | MetS (n = 2996) | |||||
| TG < 150 (n = 2170)a | TG ≥ 150 (n = 255)b | All (n = 2425)c | TG < 150 (n = 453)d | TG ≥ 150 (n = 2543)e | All (n = 2996)f | |
| Age (years) | 41.2 ± 15.1 | 42.2 ± 12.4 | 41.3 ± 14.9 | 56.2 ± 14.7 | 55.7 ± 14.4 | 55.85 ± 14.4 |
| Male (%) | 857 (39.4) | 145 (56.8) | 1002 (41.3) | 215 (47.4) | 1225 (48.1) | 1440 (48) |
| Fast blood sugar (mg/dl) | 90.6 ± 14.6 | 89.37.5 | 90.5 ± 14.1 | 108.6 ± 29.3 | 112.6 ± 39 | 112 ± 37.7 |
| Waist circumference (cm) | 89.8 ± 11.5 | 89.9 ± 9.9 | 89.8 ± 11.3 | 102.3 ± 9.5 | 101.2 ± 10.2 | 101.3 ± 10.1 |
| Hip circumference (cm) | 99.9 ± 9.1 | 98.1 ± 8 | 99.7 ± 9 | 104.7 ± 10.2 | 103.2 ± 9.3 | 103.4 ± 9.5 |
| Body Mass Index | 26.4 ± 4.6 | 26.4 ± 3.8 | 26.4 ± 4.6 | 30.8 ± 4.8 | 30.2 ± 4.7 | 30.3 ± 4.7 |
| Diastolic blood pressure | 73.3 ± 9.1 | 73.6 ± 7.7 | 73.3 ± 8.9 | 81.9 ± 10.9 | 79.4 ± 10.1 | 79.8 ± 10.2 |
| Systolic blood pressure | 108.6 ± 14.6 | 108.5 ± 10.3 | 108.6 ± 14.2 | 129.1 ± 17.6 | 123.7 ± 17.1 | 124.5 ± 17.3 |
| Triglyceride (mg/dl) | 93.2 ± 28.3 | 194.3 ± 55 | 103.8 ± 44.7 | 111.3 ± 24.2 | 198.8 ± 116 | 185.5 ± 111.7 |
| HDL-C (mg/dl) | 51 ± 10.4 | 45.7 ± 9.9 | 50.5 ± 10.5 | 43.85 ± 9.4 | 42.2 ± 10.6 | 42.5 ± 10.4 |
| Cholesterol (mg/dl) | 178.4 ± 34.4 | 205 ± 36.6 | 181.2 ± 35.6 | 180.8 ± 36.7 | 188.7 ± 44 | 187.5 ± 43.1 |
| LDL-C (mg/dl) | 105.2 ± 27.7 | 123.9 ± 30.4 | 107.2 ± 28.6 | 112 ± 29.6 | 111.6 ± 35.5 | 111.7 ± 34.6 |
| non-HDL-C (mg/dl) | 127.3 ± 32.3 | 159.3 ± 35.4 | 130.7 ± 34.1 | 136.9 ± 33.9 | 146.3 ± 43.7 | 144.9 ± 42.5 |
| Lipid lowering drug user (%) | 0 | 16 (6.2) | 16 (0.7) | 0 | 1048 (41.2) | 1048 (35) |
| Hypertension lowering drug user (%) | 86 (4) | 0 | 86 (3.5) | 139 (30.6) | 898 (35.3) | 1037 (34.7) |
| Diabetes drug user (%) | 38 (1.8) | 4 (1.6) | 42 (1.7) | 82 (18.1) | 629 (24.7) | 711 (23.8) |
Values expressed as mean ± SD or number (%).
LDL-C low-density lipoprotein cholesterol, HDL-C high-density lipoprotein cholesterol.
p-values of t-student or chi-square test between the case and control groups in both models were significant (p < 0.001).
aHTg-MetS model control group.
bExcluded from HTg-MetS model.
cMetS model control group.
dExcluded from HTg-MetS model.
eHTg-MetS model control group.
fMetS model case group.
In the HTg-MetS model, comparing the 2543 cases and 2170 controls showed a significant difference within all metabolic syndrome risk factors. In this case, all considered risk factors for MetS had higher mean values in the case group. Between lipid drug users in the case and control groups, a significant difference is observed. None of the case group subjects used lipid-lowering drugs, but 41.2% of controls that consumed drugs indicated that the lipid-lowering drug consumption is directly related to the increased TG levels in HTg-MetS individuals. Table 1 summarizes the demographic and clinical characteristics of the studied individuals for both analytical models.
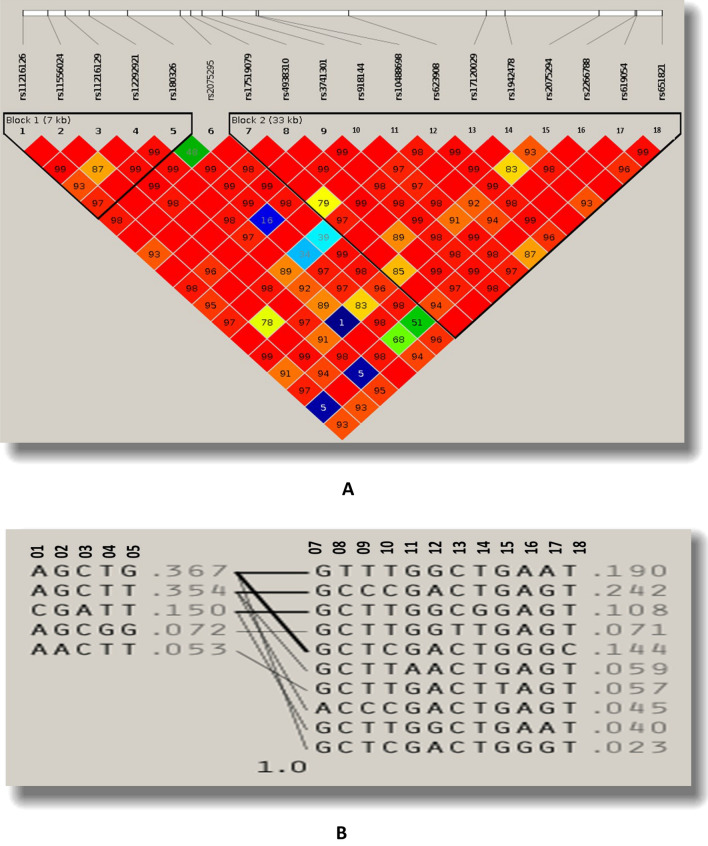
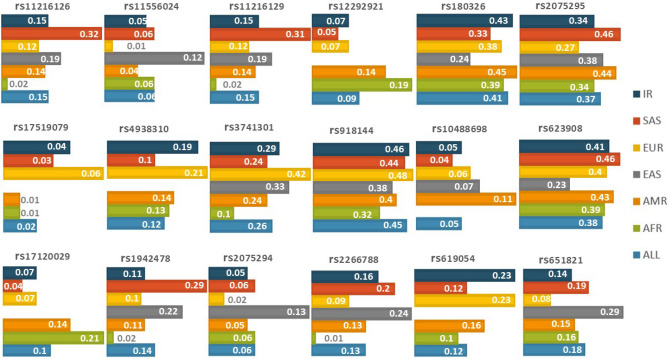
The selected SNPs were clustered into three variant sets according to the four gamete rules procedure with the sizes of 5, 1, and 12, shown in Table 2. Figure 1 presents the LD and haplotype plots related to the four gamete rules procedure. Moreover, Fig. 2 compares 18 selected SNPs' allele frequencies among populations from five continents with our population.
Table 2.
Characteristics of the Markers in selected chromosomal region.
| Gene | Consequence | SNP | Set name | Position | Major:minor | MAFa | GMAFb | Risk allel | RAFC |
|---|---|---|---|---|---|---|---|---|---|
| BUD13 | Downstream gene variant | rs11216126 | S1 | 11:116746524 | A:C | 0.15 | 0.15 | C | 0.15 |
| 3 Prime UTR variant | rs11556024 | 11:116748335 | G:A | 0.05 | 0.06 | A | 0.05 | ||
| Intronic variant | rs11216129 | 11:116749540 | C:A | 0.15 | 0.15 | A | 0.15 | ||
| Intronic variant | rs12292921 | 11:116751247 | T:G | 0.07 | 0.09 | G | 0.07 | ||
| Intronic variant | rs180326 | 11:116753987 | T:G | 0.43 | 0.41 | G | 0.43 | ||
| Intronic variant | rs2075295 | S2 | 11:116757685 | T:C | 0.34 | 0.37 | C | 0.34 | |
| Intronic variant | rs17519079 | S3 | 11:116758437 | G:A | 0.04 | 0.02 | A | 0.04 | |
| Intronic variant | rs4938310 | 11:116759233 | C:T | 0.19 | 0.12 | C | 0.81 | ||
| Intronic variant | rs3741301 | 11:116760675 | T:C | 0.29 | 0.001 | T | 0.71 | ||
| Synonymous variant | rs918144 | 11:116763109 | T:C | 0.46 | 0.45 | C | 0.46 | ||
| Missense variant | rs10488698 | 11:116763231 | G:A | 0.05 | 0.05 | A | 0.05 | ||
| Downstream gene variant | rs623908 | 11:116769652 | A:G | 0.41 | 0.4 | A | 0.59 | ||
| ZPR1 | Intronic variant | rs17120029 | 11:116779402 | C:T | 0.07 | 0.1 | T | 0.07 | |
| Intronic variant | rs1942478 | 11:116780747 | T:G | 0.11 | 0.1 | T | 0.89 | ||
| Intronic variant | rs2075294 | 11:116787406 | G:T | 0.05 | 0.06 | T | 0.05 | ||
| ZPR1/APOA5 |
ZPR1: 2 KB Upstream Variant APOA5 : 3 Prime UTR Variant |
rs2266788 | 11:116789970 | A:G | 0.16 | 0.13 | G | 0.16 | |
|
ZPR1: 2 KB Upstream Variant APOA5 : 3 Prime UTR Variant |
rs619054 | 11:116790097 | G:A | 0.23 | 0.12 | G | 0.77 | ||
| APOA5 | 5 prime UTR variant | rs651821 | 11:116791863 | T:C | 0.14 | 0.18 | C | 0.14 |
aMinor allele frequency in Iranian population.
bGlobal minor allele frequency.
cRisk allele frequency.
Figure 1.
Linkage Disequilibrium blocks of 18 tag SNPs of BUD13/ZPR1/APOA5 in the chromosomal region (11q23.3) (3.3 Kb) according to four gamete rules, LD plot of Haploview software 2(A) and Haplotypes 2(B).
Figure 2.
Compares MAF (Minor allele frequency) for 18 SNPs for different populations, Iranian population (IR), and Global minor allele frequency (ALL).
The kernel machine regression between SNP sets and case–control groups in the MetS model showed that the first and the last sets had a significant association with MetS (p-value < 0.0001). This finding were confirmed in the HTg-MetS model with the same significance level. In both models, most SNPs showed the association, especially two variations of the APOA5 gene: rs2266788, MetS model: [OR = 1.3, p = 3.66E − 07], HTg-MetS model: [OR = 1.4, p = 2.32E − 11] and rs651821, MetS model: [OR = 1.3, p = 2.86E − 07], HTg-MetS model: [OR: 1.4, p = 3.63E − 11] (Tables 3 and 4).
Table 3.
Association result for MetS model.
| Set name | SNP | LKMRa p-value |
Beta | SE | CIc (lower, upper) | OR b | Max (T)d p-value |
|
|---|---|---|---|---|---|---|---|---|
| S1 | rs11216126 | < 0.0001 | − 0.062 | 0.05284 | − 0.1659 | 0.04124 | 0.9 | 0.2382 |
| rs11556024 | − 0.067 | 0.0846 | − 0.2329 | 0.09873 | 0.9 | 0.4278 | ||
| rs11216129 | − 0.057 | 0.05328 | − 0.1616 | 0.04726 | 0.9 | 0.2833 | ||
| rs12292921 | 0.2862 | 0.07402 | 0.1411 | 0.4313 | 1.3 | 0.0001 | ||
| rs180326 | 0.1293 | 0.0387 | 0.05347 | 0.2052 | 1.1 | 0.0008 | ||
| S2 | rs2075295 | 0.938 | − 0.003 | 0.0399 | − 0.08132 | 0.07511 | 0.9 | 0.93 |
| S3 | rs17519079 | < 0.0001 | − 0.057 | 0.0894 | − 0.2319 | 0.1185 | 0.9 | 0.52 |
| rs4938310 | − 0.133 | 0.04784 | − 0.2263 | − 0.03876 | 0.8 | 0.005 | ||
| rs3741301 | − 0.056 | 0.04173 | − 0.1374 | 0.0262 | 0.9 | 0.18 | ||
| rs918144 | 0.0974 | 0.0381 | 0.0227 | 0.1721 | 1.1 | 0.01 | ||
| rs10488698 | − 0.119 | 0.07948 | − 0.2751 | 0.0365 | 0.8 | 0.13 | ||
| rs623908 | − 0.042 | 0.03834 | − 0.1176 | 0.03275 | 0.9 | 0.26 | ||
| rs17120029 | 0.245 | 0.07447 | 0.09909 | 0.391 | 1.2 | 0.0009 | ||
| rs1942478 | − 0.105 | 0.05997 | − 0.2225 | 0.01254 | 0.9 | 0.07 | ||
| rs2075294 | − 0.098 | 0.08 | − 0.255 | 0.05858 | 0.9 | 0.21 | ||
| rs2266788 | 0.2609 | 0.05131 | 0.1604 | 0.3615 | 1.3 | 3.663E − 07 | ||
| rs619054 | − 0.097 | 0.04493 | − 0.1854 | − 0.009276 | 0.9 | 0.03 | ||
| rs651821 | 0.2773 | 0.05403 | 0.1714 | 0.3832 | 1.3 | 2.867E − 07 | ||
aLogistic Kernel machine regression.
bOdd ratio for minor allele.
c95% Confidence interval for BETA.
dMAX(T) p-value with 1,000,000 replications.
Table 4.
Association result for HTg-MetS model.
| Set name | SNP | LKMRa p-value |
Beta | SE | CIc (lower, upper) | ORb | Max (T)d p-value |
|
|---|---|---|---|---|---|---|---|---|
| S1 | rs11216126 | < 0.0001 | − 0.1095 | 0.05718 | − 0.2216 | 0.002587 | 0.8 | 0.05553 |
| rs11556024 | − 0.05451 | 0.0909 | − 0.2327 | 0.1236 | 0.9 | 0.5487 | ||
| rs11216129 | − 0.1049 | 0.05772 | − 0.2181 | 0.008186 | 0.9 | 0.06904 | ||
| rs12292921 | 0.331 | 0.07953 | 0.1751 | 0.4869 | 1.3 | 0.00003 | ||
| rs180326 | 0.1724 | 0.04191 | 0.09029 | 0.2546 | 1.1 | 0.00003 | ||
| S2 | rs2075295 | 0.944 | − 0.003 | 0.04306 | − 0.08741 | 0.0814 | 0.9 | 0.94 |
| S3 | rs17519079 | < 0.0001 | − 0.1562 | 0.09806 | − 0.3484 | 0.03601 | 0.8 | 0.11 |
| rs4938310 | − 0.1845 | 0.05172 | − 0.2858 | − 0.08309 | 0.8 | 0.0003 | ||
| rs3741301 | − 0.09505 | 0.04517 | − 0.1836 | − 0.00651 | 0.9 | 0.03 | ||
| rs918144 | 0.13 | 0.04119 | 0.04925 | 0.2107 | 1.1 | 0.001 | ||
| rs10488698 | − 0.07839 | 0.0851 | − 0.2452 | 0.08839 | 0.9 | 0.35 | ||
| rs623908 | − 0.08625 | 0.04155 | − 0.1677 | − 0.00481 | 0.9 | 0.03 | ||
| rs17120029 | 0.299 | 0.08015 | 0.1419 | 0.4561 | 1.3 | 0.0001 | ||
| rs1942478 | − 0.1816 | 0.06504 | − 0.3091 | − 0.05415 | 0.9 | 0.005 | ||
| rs2075294 | − 0.08536 | 0.08589 | − 0.2537 | 0.08298 | 0.9 | 0.32 | ||
| rs2266788 | 0.3747 | 0.05605 | 0.2648 | 0.4845 | 1.4 | 2.32E − 11 | ||
| rs619054 | − 0.1348 | 0.04835 | − 0.2296 | − 0.04005 | 0.8 | 0.005 | ||
| rs651821 | 0.3901 | 0.05894 | 0.2746 | 0.5056 | 1.4 | 3.63E − 11 | ||
aLogistic Kernel machine regression.
bOdd ratio for minor allele.
c95% confidence interval for BETA.
dMAX(T) p-value with 1,000,000 replications.
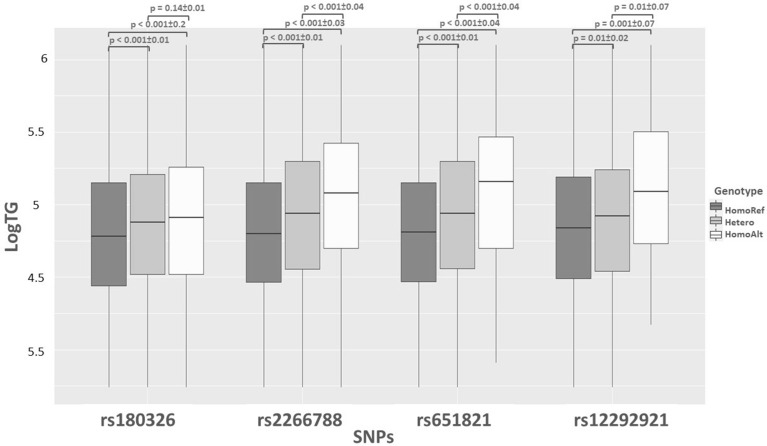
Four SNPs (rs2266788, rs651821, rs12292921, and rs180326) with the highest significance levels were selected to indicate the relation between MetS components level variation and the number of risk alleles. The result of association analysis showed the significant difference between studied markers and the triglyceride and HDL-C level (p < 0.001), and three of them were in association with non-HDL-C levels (p < 0.001) (Table 5).
Table 5.
Single SNPs association analyses for metabolic components (mean ± SD).
| rs12292921 | rs180326 | rs2266788 | rs651821 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | GT | GG | TT | GT | GG | AA | GA | GG | TT | CT | CC | |
| Fast blood sugar (mg/dl) | 102 ± 31.5 | 102.7 ± 29.8 | 97.8 ± 14.9 | 101.7 ± 30.4 | 102.3 ± 32 | 101.8 ± 30.4 | 102.2 ± 31 | 101.5 ± 30.3 | 104.5 ± 40.7 | 102.1 ± 31 | 101.6 ± 31.4 | 104.8 ± 35.5 |
| Waist circumference (cm) | 95.5 ± 12.6 | 95.8 ± 12.5 | 99.6 ± 13.7 | 95.6 ± 12.3 | 95.6 ± 12.9 | 95.4 ± 12.5 | 95.7 ± 12.5 | 95.1 ± 12.9 | 95.4 ± 12.3 | 95.7 ± 12.6 | 95 ± 12.8 | 96.7 ± 12.9 |
| Diastolic blood pressure | 76.6 ± 10.3 | 77.3 ± 10.9 | 76.9 ± 10.4 | 76.4 ± 10.2 | 76.7 ± 10.6 | 76.8 ± 10.2 | 76.8 ± 10.4 | 76.2 ± 10.4 | 76.7 ± 10 | 76.8 ± 10.4 | 76.2 ± 10.6 | 77.3 ± 9.7 |
| Systolic blood pressure | 116.9 ± 18 | 117.9 ± 17.8 | 114.5 ± 15.6 | 116.9 ± 17.7 | 117.3 ± 18.2 | 116.5 ± 17.5 | 117.1 ± 17.9 | 116.7 ± 18.1 | 116.3 ± 16.8 | 117 ± 17.9 | 116.8 ± 18.1 | 117.6 ± 17.4 |
| Cholesterol (mg/dl) | 184.5 ± 39.9 | 183.2 ± 40.5 | 186 ± 41.2 | 184.5 ± 40.4 | 184.2 ± 39.4 | 184.6 ± 40 | 183.5 ± 39.8 | 186.1 ± 39.7 | 186.5 ± 44.6 | 183.6 ± 39.8 | 186.2 ± 40.3 | 186.2 ± 38.5 |
| Triglyceride (mg/dl) | 146.3 ± 98.2 | 153.6 ± 82.2 | 197.8 ± 124.3 | 136 ± 69.1 | 150 ± 90.2 | 160.6 ± 139.9 | 138.9 ± 76 | 162.1 ± 99.2 | 215 ± 277.3 | 140.1 ± 77.8 | 163.8 ± 128.8 | 205.5 ± 158.4 |
| HDL-C (mg/dl) | 46.4 ± 11.2 | 45.2 ± 11.1 | 42.1 ± 14.3 | 47.3 ± 10.9 | 46 ± 11.3 | 44.9 ± 11.4 | 46.8 ± 11.2 | 45.1 ± 11.2 | 42 ± 10.8 | 46.8 ± 11.2 | 45 ± 11.3 | 41.2 ± 10.6 |
| LDL-C (mg/dl) | 109.6 ± 32 | 108.6 ± 32.5 | 107 ± 35.4 | 109.8 ± 32.8 | 109.2 ± 31.7 | 109.3 ± 31.9 | 109 ± 32.3 | 110.6 ± 31.9 | 108.2 ± 30.2 | 109 ± 32.3 | 110.5 ± 32 | 110 ± 27.6 |
| Non-HDL-C (mg/dl) | 138 ± 39.6 | 137.9 ± 39.5 | 143.8 ± 39.8 | 137.2 ± 39.8 | 138.1 ± 39.2 | 139.3 ± 39.5 | 136.7 ± 39.1 | 140.9 ± 40.3 | 142.8 ± 39.2 | 136.8 ± 39.3 | 140.9 ± 40 | 145 ± 39.3 |
Bold values: p-values of between genotype groups analysis were significant (p < 0.001).
Figure 3 tries to present that the TG has the highest level in the HomoAlt groups for all four SNPs, and there is an association between raised TG levels in MetS-affected and the presence of the risk allele. The SNPs were tested for linkage disequilibrium (LD) to indicate the same signal between rs2266788 and rs651821 in the S3 and showed a strong LD (D′ > 0.9; r2 > 0.8).
Figure 3.
Log of TG levels (LogTG) against the four notable significant SNPs. This figure also shows TG levels by increasing risk alleles in each genotype type in subjects. The p-values and standard errors for between genotypes can be found (p-value ± S.E).
Discussion
This is the first kernel machine study to evaluate association genetic variations within the BUD13, ZPR1, and APOA5 genes (11q23.3 region) with MetS and HTg-MetS. This kernel machine is a logistic sequence kernel association test that is denoted as SKAT method. SKAT is a supervised, flexible, computationally efficient regression method that handles tests for evaluating the association between common and rare genetic variants within a region and a continuous or dichotomous trait while easily adjusting for covariates. This method is also useful for assessing the association between joint effects of all SNPs within a set and the considered phenotype. Moreover, SKAT can quickly calculate calibrated p-values for rare variant association analysis in case–control studies. This regression-based method overcomes the limited power of classical single-marker association analysis for rare variants poses a central challenge in association studies23,24.
The results of both the kernel machine and MAX (T) tests in our study show that rs2266788, which belongs to both ZPR1 and APOA5 genes, is associated with MetS and MetS affected subjects that had a high TG. Furthermore, MetS affected subjects with homozygote G allele in rs2266788 have 1.4 times high risk for Tg increasing. These findings are consistent with similar studies on Korean and Kuwaiti populations25,26.
For the studied markers, most of the Iranian allele frequencies were common in the population, and the association result confirms the result of previous studies7,13,25,26. According to the association models, the S1 and S3 sets demonstrated a significant correlation with MetS and the high TG model. The two lowest p-values belong to rs2266788 (ZPR1/APOA5 genes) and rs651821 (APOA5 gene), to increase the risk of High TG 1.4 times in the case group of the HTg-MetS model.
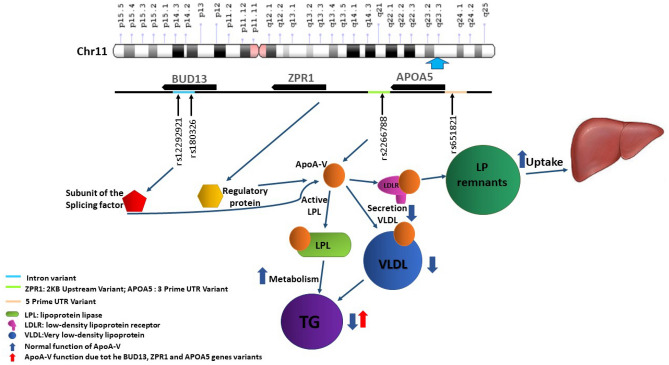
The BUD13, ZPR1, and APOA5 genes encode three proteins in the APOA5 protein pathway in the 11q23.3 chromosomal region. BUD13 is a subunit of the splicing factor that plays a role in preserving nuclear pre-mRNA, and ZPR1 is a regulatory protein related to signal transduction and cell proliferation27. APOA5 protein plays an important role in several mechanisms, including insulin sensitivity, glucose, fatty acid, and cholesterol metabolism15,16. The APOA5 protein is an LPL activator, which inhibits hepatic VLDL production, and accelerates the hepatic uptake of lipoprotein remnants and insulin secretion. Furthermore, it is related to free fatty acid decrease, triglyceride synthesis, and stimulates adipose tissue25,28. It is claimed that interaction between BUD13/ZPR1 variants and APOA5 may change its function and increase the TG level7,29,30 (Fig. 4).
Figure 4.
Schematic picture for the BUD13, ZPR1, and APOA5 genes on the 11p23.3 region, location of four notable significant SNPs, and shows the mechanism of ApoA-V action on triglyceride level.
Many studies in different populations, including European and Asian populations, confirmed that BUD13, ZNF259, and APOA5 variants correlate with changes in plasma TG levels11,31,32. Studies on SNPs in noncoding, exonic, and intergenic regions showed transcriptional binding sites of adjacent genes lead to transcriptional processes affected by the SNPs without being linked explicitly to protein regulation27.
The GWAS study conducted on the Chinese population indicated that rs651821 on APOA5 and rs180326 on BUD13 are associated with MetS and serum TG levels7. Another GWAS study showed the homozygotes carriers of rs2266788-G APOA5 has pleiotropic effects on (increasing) TG and (lowering) HDL-C levels13. Also, as a functional point mutation, rs2266788 is highly correlated with rs6518217.
Our results are consistent with Korean and Kuwaiti population studies25,26, which reported rs2266788 as related variant hypertriglyceridemia (TG > 150 mg/dL) with similar OR. Our results are also consistent with a study performed by Karja et al. to confirm the rs2266788 risk allele concerning increased TG13. The results of a linkage disequilibrium replicated by Zhu et al. indicated a strong LD between rs2266788 and rs651821. Furthermore, the association of rs2266788, rs651821, rs180326, and rs12292921 with MetS and HTg-MetS were reported in our outcome. Although the first three SNPs' association in our study and the global studies were deep-rooted, rs12292921 is not well known7,33–36.
Finally, the present study was done in well-characterized and extended TCGS participants who were monitored for more than twenty years. The kernel machine SNP set analysis by two-step models was used for the first time to evaluate this genetic relationship; this would be the most important strength of these findings. The limitation of this research includes the failure to check the APO cluster associated with the lipid profile.
In conclusion, the kernel machine and Single SNP Regression models found the association of genetic variations on the BUD13, ZPR1, and APOA5 genes (11q23.3 region) with MetS and High TG levels in MetS affected individuals in the TCGS population. We suggested using these methods in the same studies. Moreover, rs2266788 and rs651821 in, APOA5 gene was in strong LD and showed a significant association with MetS and HTg-MetS and can be used for prediction. These two SNPs also showed the most significant association with HTg-MetS, which indicates that this region is related to changing lipid profiles in metabolic syndrome. Therefore, we recommend a more in-depth study in the APO genes cluster to confirm these findings.
Materials and methods
Subjects and population
Subjects selected from Tehran Cardio-metabolic Genetic Study (TCGS)37. This study is an ongoing genetic cohort study designed to determine the risk factors of major non-communicable disorders in the Tehran Lipid and Glucose Study (TLGS)38, whose subjects were followed-up cardio-metabolic risk factors over twenty years. The attendance enrolled in the study from 1999 until now, and over twenty thousand participants were monitored for seven phases (1999–until now; periodic follow-up interval: 3 years). An expert obtained informed consent from each subject and then interviewed for collecting demographic data and referred each one to trained physicians and laboratories for clinical examinations and blood sampling at each visit; details are published elsewhere37,39,40. All procedures performed in this study approved by the ethics committee on human subject research at Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences (code of “IR.SBMU.ENDOCRINE.REC.1395.366”), which were following the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
In the present study, 5421 unrelated individuals more than 18 years old were recruited. According to the Iranian modified metabolic syndrome joint interim statement (JIS) definition, the metabolic syndrome is defined for all participants41,42 who attended the sixth phase of TCGS (2015–2017).
We defined two models to assign individuals to the case and control groups. In the first model (Mets versus non-MetS), the cases were selected as MetS affected if they had at least three of the following criteria41: (1) abdominal obesity (increased WC ≥ 91 cm in females and males) based on national cut-offs42, (2) TG ≥ 150 mg/dl or receiving treatment for hypertriglyceridemia, (3) HDL < 50/40 mg/dl in F/M, (4) SBP ≥ 130 mmHg or DBP ≥ 85 mmHg or receiving treatment for hypertension, (5) FPG ≥ 100 mg/dl or previously diagnosed with type 2 diabetes and diabetic drug users. The rest of the population were selected as controls42.
For the second model to measure precise triglyceride effect on association analysis (hTG-MetS), subjects were divided into case and control if they affected by MetS at the same time have TG higher than 150 mg/dl and Non-MetS with TG less than 150 mg/dl, respectively, and other individuals were excluded (Fig. 5).
Figure 5.
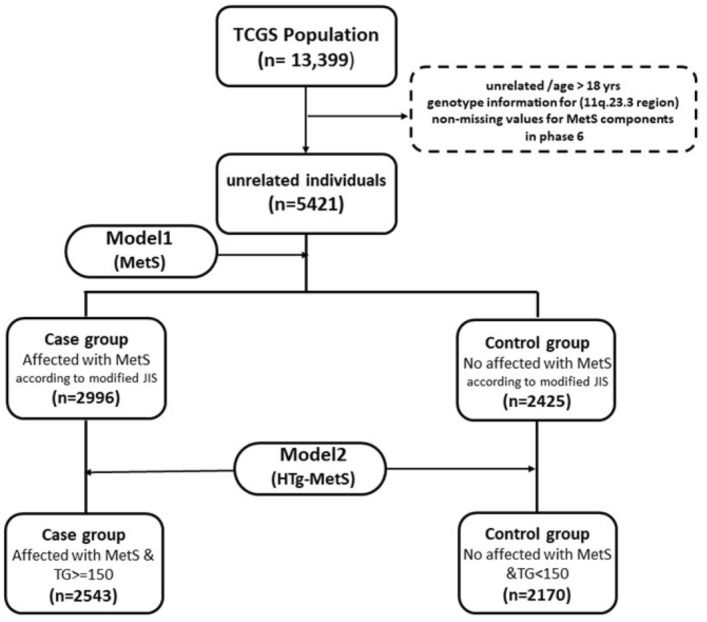
Flow diagram of the study. Model 1 and Model 2 shows by MetS and HTg-MetS symbols, respectively.
Genotyping and SNP selection
DNA samples were extracted from white blood cells by a standard Proteinase K, salting-out method. The accuracy and quality control43 of extracted samples were evaluated with electrophoresis and spectrophotometry. DNA samples of TCGS participants were genotyped with HumanOmniExpress-24-v1-0 bead chips at the deCODE genetics company (Iceland) according to the manufacturer's specifications (Illumina Inc., San Diego, CA, USA)37. After quality control for markers and individuals in our desired region, 20 SNPs located on the BUD13, ZPR1, and APOA5 genes were selected. According to our defined inclusion criteria and genotyping After omitting 2 SNPs with departure from Hardy–Weinberg equilibrium, 18 SNP remained in our analysis44. (Exact test p < 0.01) (Table 2). The plink software was used to create a list of minor allele frequencies (MAF) for SNPs45, and the 1000 Genomes database was used to find global minor allele frequencies for comparison46.
Statistical analysis
Descriptive
All continuous variables were expressed as mean and standard deviation, whereas categorical variables are summarized as frequencies and percentages. Differences between the two groups were calculated using the chi-square test or t-student test. The significance of deviations of observed genotype frequencies from those predicted by the Hardy–Weinberg equation evaluates the χ2 test with statistical significance at the level of 0.01. Rare variants, SNPs with minor allele frequency (MAF) less than 0.01, were not considered for further analysis.
Haploview
In Haploview software version 4.2, four gamete reules procedure was used to make SNP sets from correlated adjacent SNPs in our region47. For each marker pair, the four possible two-marker haplotypes’ population frequencies are computed; if all four were observed with at least a frequency of 0.01, it is deemed to recombination have occurred. According to this analysis, SNPs were clustered into three sets. According to linkage disequilibrium (LD), the LD plot and haplotypes were drawn under the four gamete rule, with a connection between genotyped SNPs and the tested regions of the BUD13, ZPR1, and APOA5 genes in the 11q23.3 chromosomal region47,48.
Kernel machine regression
To increase the association test's power and accommodate the epistasis effect between adjacent correlated SNPs in a set, we used a logistic kernel machine regression method to assess SNP sets' association binary phenotype23,28,49,50. In the additive model, genotypes AA, Aa, and aa recoded to 0, 1, and 2, respectively28,50.
Single SNP analysis
We considered an additive logistic regression model to assess each SNP's association in the significance sets with binary phenotype. We used the PLINK2 program and P values based on MAX (T) procedure and permutation test on the additive effect (with 1 million iterations)45,51. The risk allele for each associated marker was labeled, and the presence of the risk allele was classified into three genotype groups (Homozygote for reference allele (HomoRef = 0), Heterozygote = 1, Homozygote for the alternate allele (HomoAlt = 2)) in all significant SNPs. For the four most significant SNPs genotypes, MetS components level were compared by one-way ANOVA and Tukey test (Table 5). The triglyceride level was normalized, and the logarithm (log) of TG for each individual was plotted against genotypes. Then, LogTG was compared in different genotypes by ANOVA and Tukey test to find significant genotype groups (Fig. 3). Finally, We used the PLINK2 program to calculate Pairwise r2 and D′ values between rs2266788 and rs651821.
Ethics approval
All procedures followed the ethics committee's ethical standards on human subject research at Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences.
Consent to participate
Informed consent was obtained from all the participants.
Consent for publication
All co-authors and responsible authorities have approved this study's publication at the institute.
Acknowledgements
The authors would like to express their gratitude to the patients participating in the Tehran lipid and glucose and Tehran Genetic Cardio metabolic studies. Also, special thanks to the DeCODE genetic company for doing the genetic screening.
Author contributions
S.M., B.S. Designed, analyzed data, and co-wrote the paper; N.J. performed the statistical analysis, L.N. performed the genetic analysis; F.A. and M.D. supervised the research.
Funding
In this study, we used a data set funded by the Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences (Tehran, Iran).
Data availability
In this study, the information of participants was from TLGS and TCGS studies.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Daneshpour M, Sedaghatikhayat B, Hedayati M, Azizi F. From genome to gene: A review of genes and genetic variations to be associated with metabolic syndrome. Iran. J. Diabetes Metab. 2015;14:225–235. [Google Scholar]
- 2.Alberti KG, Zimmet P, Shaw J. Metabolic syndrome—A new world-wide definition. A consensus statement from the International Diabetes Federation. Diabet. Med. 2006;23:469–480. doi: 10.1111/j.1464-5491.2006.01858.x. [DOI] [PubMed] [Google Scholar]
- 3.Cornier MA, et al. The metabolic syndrome. Endocr. Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang HH, et al. Novel Insights into the Pathogenesis and Management of the Metabolic Syndrome. Pediatr. Gastroenterol. Hepatol. Nutr. 2020;23:189. doi: 10.5223/pghn.2020.23.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Despres JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 6.Ding EL, Smit LA, Hu FB. The metabolic syndrome as a cluster of risk factors: is the whole greater than the sum of its parts? Comment on "The metabolic syndrome, its component risk factors, and progression of coronary atherosclerosis". Arch. Intern. Med. 2010;170:484–485. doi: 10.1001/archinternmed.2009.552. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: A multi-stage genome-wide association study. J. Cell Mol. Med. 2017;21:1106–1116. doi: 10.1111/jcmm.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet (London, England) 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 9.Daneshpour MS, et al. 8q24.3 and 11q25 chromosomal loci association with low HDL-C in metabolic syndrome. Eur. J. Clin. Investigat. 2011;41:1105–1112. doi: 10.1111/j.1365-2362.2011.02516.x. [DOI] [PubMed] [Google Scholar]
- 10.Major JM, et al. Genome-wide association study identifies common variants associated with circulating vitamin E levels. Hum. Mol. Genet. 2011;20:3876–3883. doi: 10.1093/hmg/ddr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun TR, et al. A replication study of GWAS-derived lipid genes in Asian Indians: The chromosomal region 11q23.3 harbors loci contributing to triglycerides. PLoS ONE. 2012 doi: 10.1371/journal.pone.0037056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W, Gyenesei A, Semple CA, Haley CS. Properties of local interactions and their potential value in complementing genome-wide association studies. PLoS ONE. 2013 doi: 10.1371/journal.pone.0071203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraja AT, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galcheva-Gargova Z, et al. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell. 1998;9:2963–2971. doi: 10.1091/mbc.9.10.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aung LH, et al. Association of the variants in the BUD13-ZNF259 genes and the risk of hyperlipidaemia. J. Cell Mol. Med. 2014;18:1417–1428. doi: 10.1111/jcmm.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corton JC, Anderson SP, Stauber A. Central role of peroxisome proliferator-activated receptors in the actions of peroxisome proliferators. Annu. Rev. Pharmacol. Toxicol. 2000;40:491–518. doi: 10.1146/annurev.pharmtox.40.1.491. [DOI] [PubMed] [Google Scholar]
- 17.Tripsianes K, et al. A novel protein–protein interaction in the RES (REtention and Splicing) complex. J. Biol. Chem. 2014;289:28640–28650. doi: 10.1074/jbc.M114.592311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysoczanski P, Zweckstetter M. Retention and splicing complex (RES)—The importance of cooperativity. RNA Biol. 2016;13:128–133. doi: 10.1080/15476286.2015.1096484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson SK, et al. Endocytosis of apolipoprotein AV by members of the low density lipoprotein receptor and the VPS10p domain receptor families. J. Biol. Chem. 2008;283:25920–25927. doi: 10.1074/jbc.M802721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, C. et al. Effects of APOA5-1131T> C (rs662799) on fasting plasma lipids and risk of metabolic syndrome: Evidence from a case–control study in China and a meta-analysis.PLoS One 8, e56216. 10.1371/journal.pone.0056216 (2013). [DOI] [PMC free article] [PubMed]
- 21.Fallah M-S, et al. The relation between metabolic syndrome risk factors and genetic variations of apolipoprotein V in relation with serum triglyceride and HDL-C level. Arch. Iran. Med. 2016;19:46–50. [PubMed] [Google Scholar]
- 22.Zarkesh M, et al. Heritability of the metabolic syndrome and its components in the Tehran Lipid and Glucose Study (TLGS) Genet Res (Camb). 2012;94:331–337. doi: 10.1017/S001667231200050X. [DOI] [PubMed] [Google Scholar]
- 23.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin XJ. Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 2013;92:841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh S-W, et al. Genome-wide association study of metabolic syndrome in Korean populations. PLoS ONE. 2020;15:e0227357. doi: 10.1371/journal.pone.0227357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jasim AA, Al-Bustan SA, Al-Kandari W, Al-Serri A, AlAskar HJ. Sequence analysis of APOA5 among the Kuwaiti population identifies association of rs2072560, rs2266788, and rs662799 With TG and VLDL levels. Front. Genet. 2018;9:1–12. doi: 10.3389/fgene.2018.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H-K, Anwar MA, Choi SJ. Association of BUD13-ZNF259-APOA5-APOA1-SIK3 cluster polymorphism in 11q233 and structure of APOA5 with increased plasma triglyceride levels in a Korean population. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-44699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javanrouh N, Daneshpour MS, Soltanian AR, Tapak LJ. Kernel machine SNP set analysis provides new insight into the association between obesity and polymorphisms located on the chromosomal 16q.12.2 region: Tehran lipid and glucose study. Gene. 2018;658:146–151. doi: 10.1016/j.gene.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, et al. Zinc finger protein 259 (ZNF259) polymorphisms are associated with the risk of metabolic syndrome in a Han Chinese Population. Clin. Lab. 2015;61:615–621. doi: 10.7754/clin.lab.2014.141138. [DOI] [PubMed] [Google Scholar]
- 30.Guardiola M, Ribalta JJ. Update on APOA5 genetics: Toward a better understanding of its physiological impact. Curr. Atheroscl. Rep. 2017;19:30. doi: 10.1007/s11883-017-0665-y. [DOI] [PubMed] [Google Scholar]
- 31.Galcheva-Gargova Z, et al. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science. 1996;272:1797–1802. doi: 10.1126/science.272.5269.1797. [DOI] [PubMed] [Google Scholar]
- 32.Aung, L.-H.-H. et al. Association between the MLX interacting protein-like, BUD13 homolog and zinc finger protein 259 gene polymorphisms and serum lipid levels. Sci. Rep. 4, 5565 (2014). [DOI] [PMC free article] [PubMed]
- 33.Gombojav B, et al. Multiple susceptibility loci at chromosome 11q23.3 are associated with plasma triglyceride in East Asians. J. Lipid Res. 2016;57:318–324. doi: 10.1194/jlr.P063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kefi R, et al. Association of apolipoprotein A5 gene variants with metabolic syndrome in Tunisian population. Ann. d'endocrinologie. 2017 doi: 10.1016/j.ando.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Su M-W, et al. Blood multiomics reveal insights into population clusters with low prevalence of diabetes, dyslipidemia and hypertension. PLoS ONE. 2020 doi: 10.1371/journal.pone.0229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pranavchand R, Kumar AS, Reddy BM. Genetic determinants of clinical heterogeneity of the coronary artery disease in the population of Hyderabad. India. Hum. Genom. 2017 doi: 10.1186/s40246-017-0099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daneshpour MS, et al. Rationale and design of a genetic study on cardiometabolic risk factors: Protocol for the Tehran Cardiometabolic Genetic Study (TCGS) JMIR Res. Protocols. 2017 doi: 10.2196/resprot.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azizi F, et al. Prevention of non-communicable disease in a population in nutrition transition: Tehran lipid and glucose study phase II. Trials. 2009;10:5. doi: 10.1186/1745-6215-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azizi F, Rahmani M, Emami H, Madjid M. Tehran lipid and glucose study: Rationale and design. CVD Prev. 2000;3:242–247. [Google Scholar]
- 40.Azizi F, et al. Cardiovascular risk factors in an Iranian urban population: Tehran lipid and glucose study (phase 1) Soz Praventivmed. 2002;47:408–426. doi: 10.1007/s000380200008. [DOI] [PubMed] [Google Scholar]
- 41.Alberti KG, et al. Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 42.Azizi F, et al. Appropriate waist circumference cut-off points among Iranian adults: The first report of the Iranian National Committee of Obesity. Arch. Iran Med. 2010;13:243–244. [PubMed] [Google Scholar]
- 43.Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Givi NJ, et al. The joint effect of PPARG upstream genetic variation in association with long-term persistent obesity: Tehran cardio-metabolic genetic study (TCGS) Eat Weight Disord. . 2021 doi: 10.1007/s40519-020-01063-7. [DOI] [PubMed] [Google Scholar]
- 45.Purcell, S. & Chang C. (2015) PLINK 1.9. https://www.cog-genomics.org/plink2. Accessed 10 Oct (2009).
- 46.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 48.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: The interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S, Wu MC, Lin XJB. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javanrouh N, et al. A novel association of rs13334070 in the RPGRIP1L gene with adiposity factors discovered by joint linkage and linkage disequilibrium analysis in Iranian pedigrees: Tehran Cardiometabolic Genetic Study (TCGS) Genet. Epidemiol. 2019;43:342–351. doi: 10.1002/gepi.22179. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
In this study, the information of participants was from TLGS and TCGS studies.