Summary
Whole blood cytokine release assays (CRA) assessing cellular immunity to gluten could simplify the diagnosis and monitoring of coeliac disease (CD). We aimed to determine the effectiveness of electrochemiluminescence CRA to detect responses to immunodominant gliadin peptides. HLA‐DQ2·5+ CD adults (cohort 1, n = 6; cohort 2, n = 12) and unaffected controls (cohort 3, n = 9) were enrolled. Cohort 1 had 3‐day gluten challenge (GC). Blood was collected at baseline, and for cohort 1 also at 3 h, 6 h and 6 days after commencing 3‐day GC. Gliadin peptide‐stimulated proliferation, interferon (IFN)‐γ enzyme‐linked immunospot (ELISPOT) and 14‐ and 3‐plex electrochemiluminescence CRA were performed. Poisson distribution analysis was used to estimate responding cell frequencies. In cohort 1, interleukin (IL)‐2 dominated the gliadin peptide‐stimulated cytokine release profile in whole blood. GC caused systemic IL‐2 release acutely and increased gliadin peptide‐stimulated IFN‐γ ELISPOT and whole blood CRA responses. Whole blood CRA after GC was dominated by IL‐2, but also included IFN‐γ, C‐X‐C motif chemokine ligand 10/IFN‐γ‐induced protein 10 (CXCL10/IP‐10), CXCL9/monokine induced by IFN‐γ (MIG), IL‐10, chemokine (C‐C motif) ligand 3/macrophage inflammatory protein 1‐alpha (CCL3/MIP‐1α), TNF‐α and IL‐8/CXCL8. In cohorts 2 and 3, gliadin peptide‐stimulated whole blood IL‐2 release was 100% specific and 92% sensitive for CD patients on a gluten‐free diet; the estimated frequency of cells in CD blood secreting IL‐2 to α‐gliadin peptide was 0·5 to 11 per ml. Whole blood IL‐2 release successfully mapped human leucocyte antigen (HLA)‐DQ2·5‐restricted epitopes in an α‐gliadin peptide library using CD blood before and after GC. Whole blood IL‐2 release assay using electrochemiluminescence is a sensitive test for rare gliadin‐specific T cells in CD, and could aid in monitoring and diagnosis. Larger studies and validation with tetramer‐based assays are warranted.
Keywords: coeliac disease, cytokine release assay, cytokines, diagnosis, IL‐2, T cells
Detection of rare circulating gluten‐specific T cells may simplify the diagnosis of coeliac disease but current approaches using tetramers are technically demanding. Here we show that a highly sensitive cytokine release assay assessing gliadin peptide‐stimulated interleukin‐2 release in whole blood was 100% specific and 92% sensitive for patients with coeliac disease adhering to a gluten free diet. Whole blood interleukin‐2 release assay using electrochemiluminescence is a sensitive test for rare gluten‐specific T cells in coeliac disease, and could aid in disease monitoring and diagnosis.

Introduction
Coeliac disease (CD) is associated with an acquired adaptive immune response directed against partially deamidated gluten peptides [1]. Staining peripheral blood mononuclear cells (PBMC) with human leucocyte antigen (HLA)‐DQ2·5‐gluten‐peptide tetramers indicates the frequency of effector memory CD4+ T cells specific for gluten is approximately 1 per 100 000 CD4+ T cells in CD and negligible in unaffected individuals [2, 3]. Tests for gluten‐specific CD4+ T cells could potentially support diagnosis of CD [4].
Diagnosis of tuberculosis can be based on detecting Mycobacterium tuberculosis‐specific T cells utilizing IFN‐γ release assays [5], but IFN‐γ enzyme‐linked immunosorbent spot (ELISPOT) assay responses to an immunodominant M. tuberculosis‐derived peptide in patients with tuberculosis are approximately 100 times greater than for an immunodominant gluten peptide in CD [6, 7]. HLA‐DQ2·5‐gluten‐peptide tetramers efficiently detect gluten epitope‐specific CD4+ T cells, but the technique is technically demanding [2, 3].
Alternatively, gluten challenge for 3 days boosts the frequency of gluten‐specific CD4+ T cells in blood and allows their detection in cytokine release assays (CRAs) by IFN‐γ ELISPOT or whole blood IFN‐γ, interleukin (IL)‐2 or C‐X‐C motif chemokine ligand (CXCL)10 release, but the need for gluten challenge is a barrier to translation into clinical practice [8, 9, 10].
Several new immunoassay platforms have improved performance characteristics for detection of low abundance cytokines such as IL‐2 in plasma and serum. Based on our recent experience using ultrasensitive multiplex assays in CD [10], we hypothesized that electrochemiluminescence assays might enhance the sensitivity of CRAs using freshly collected, unseparated ‘whole’ blood. The aim of this study was to determine the effectiveness of an optimized electrochemiluminescence whole blood CRA to detect responses to immunodominant gliadin peptides in CD patients with and without a prior gluten challenge.
Materials and methods
Study design
The study was approved by the Human Research Ethics Committee at the Walter and Eliza Hall Institute and Melbourne Health (identifiers 03/4 and 2003.009, respectively). In this exploratory study to establish an ex‐vivo CRA for gluten‐specific CD4+ T cells in treated CD, gliadin and control peptides were incubated with fresh blood or peripheral blood mononuclear cells (PBMCs) collected before and after gluten food challenge in order to (1) optimize cytokine selection and understand its relevance to in‐vivo cytokine release, (2) provide preliminary sensitivity and specificity data, (3) compare its performance to conventional IFN‐γ ELISPOT and dye‐dilution proliferation assays and (4) test whether a whole blood microwell CRA was capable of successfully mapping immunogenic regions of an α‐gliadin polypeptide.
Participants
All patients gave written, informed consent prior to undergoing any study procedures. Participants were required to be aged between 18 and 70 years. Volunteers were excluded if they had taken immunosuppressive medication within 3 months or had any medical condition that would impact the immune response or confound interpretation of study results, or pose an increased risk to the patient or interfere with study conduct. Enrolment in cohorts 1 or 2 required participants to have (1) a diagnosis of CD supported by previous small bowel biopsy showing villous atrophy and seropositive transglutaminase 2 (TG2) immunoglobulin (Ig)A, (2) at least 2 months since commencing a gluten‐free diet, (3) TG2 IgA or deamidated gliadin peptide (DGP) IgG‐negative at screening and (4) be positive for HLA‐DQ2·5. Enrolment in cohort 3 required participants to (1) regularly consume gluten‐containing food and (2) be seronegative for TG2 IgA and DGP IgG. Participants enrolled in cohort 1 were also required to have haemoglobin at screening in the normal range and to meet American Red Cross Blood Donation eligibility criteria (https://www.redcrossblood.org/donate‐blood/how‐to‐donate/eligibility‐requirements/eligibility‐criteria‐alphabetical.html).
Clinical procedures
Eligibility was assessed, medical details were collected and laboratory tests for HLA‐DQ, TG2 IgA and DGP IgG serology and haemoglobin were performed at visit 1. At visit 2 (baseline, day 1), 260 ml blood was collected from participants in cohort 1 or 70 ml for cohorts 2 and 3. Cohort 1 participants consumed 10 g vital wheat gluten in 100 ml water, as previously described [11], and were observed for 6 h. Blood for serum was collected at 3 and 6 h. Before breakfast on days 2 and 3, cohort 1 participants consumed 10 g vital wheat gluten in 100 ml water, the same as day 1. For cohort 1 at visit 3 on day 6, 170 ml blood was collected. Blood was collected via 21G ¾" Surflo winged infusion set (Terumo, Shibuya City, Japan) into 10 ml lithium–heparin vacutainers (Becton Dickinson, Franklin Lakes, NJ, USA) for functional assays or 8·5 ml vacutainer plus plastic serum separator tube for serum, as previously described [12].
Clinical laboratory assessments
QUANTA Lite® recombinant human tissue transglutaminase (R h‐tTG) IgA and gliadin IgG II (DGP) (Inova Diagnostics, San Diego, CA, USA) were performed by Dorevitch Pathology (Footscray, VIC, Australia). HLA‐DQA and HLA‐DQB alleles were determined using leucocyte‐derived DNA with a panel of sequence‐specific primers by the Australian Red Cross, Victorian Transplantation and Immunogenetics Service (Parkville, VIC, Australia).
Peptides and antigens
CS Bio (Menlo Park, CA, USA) manufactured immunodominant gliadin and hordein peptides, glia‐α (single‐letter amino acid code where Z is N‐pyroglutamate, ZLQPFPQPELPYPQPQ‐NH2), glia‐ω (ZQPFPQPEQPFPWQP‐NH2) and HorB (ZPEQPIPEQPQPYPQQ‐NH2) [13], and a pair of 8mer peptides that together were scrambled sequences of glia‐α and served as a negative control (ZPFPLPQP‐NH2 and ZPQYQPEQ‐NH2). Peptide purities were > 95% by high‐performance liquid chromatography (HPLC), and liquid chromatography–mass spectrometry (LC‐MS) confirmed their identities. JPT Peptide Technologies (Berlin, Germany) manufactured a 155‐member 14mer peptide library designed according to Beissbarth et al [14] (Table 1). Median purity was 94·5% (range = 70·9–98·8%) and LC‐MS confirmed their identities. The library comprised 69 peptides that spanned (with 10 amino acid overlaps) the wheat α‐gliadin polypeptide, CAB76964.1 and 84 partially deamidated, citrullinated or carbamylated variants of these native gliadin peptides. The library also included a positive control, immunodominant deamidated α‐gliadin peptide and a related scrambled‐sequence, negative control peptide. A deamidated protease‐digested vital wheat gluten used for gluten challenges was prepared as previously described [8]. Briefly, vital wheat gluten was dissolved in ammonium bicarbonate 0·1 M and urea 2 M, and incubated with 0·5% chymotrypsin (Merck, Darmstadt, Germany) at room temperature for 24 h; digestion was stopped by heating samples in a 98°C water bath for 10 min. Samples were centrifuged at 16 000 g for 10 min, and supernatants were collected and filtered through a 0·22 μm syringe membrane filter (VWR, Radnor, PA, USA). Protease‐digested gluten was diluted in phosphate‐buffered saline (PBS) containing 1 mM CaCl2 (Merck) to 640 µg/ml, according to the peptide concentration measured by a Pierce™ Modified Lowry Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and treated for 2 h at 37°C with guinea pig liver transglutaminase (Merck), 50 µg/ml. After filtering through a 0·22 μm syringe membrane filter, the final stock material was stored at −80°C.
Table 1.
Wheat α‐gliadin peptide library
| No. | Sequence | No. | Sequence | No. | Sequence | No. | Sequence |
|---|---|---|---|---|---|---|---|
| Native | 41 | TYQLVQQLCCQQLW | 81 | PFPSEQPYLQLQPF | Citrullinated | ||
| 1 | MVRVPVPQLQPQNP | 42 | VQQLCCQQLWQIPE | 82 | EQPYLQLQPFPQPQ | 121 | MV‐Cit‐VPVPQLQPQNP |
| 2 | PVPQLQPQNPSQQQ | 43 | CCQQLWQIPEQSRC | 83 | LQLQPFPQPELPYP | 122 | QPQLPYPQPQPF‐Cit‐P |
| 3 | LQPQNPSQQQPQEQ | 44 | LWQIPEQSRCQAIH | 84 | PFPQPELPYPQPQL | 123 | PYPQPQPF‐Cit‐PQQPY |
| 4 | NPSQQQPQEQVPLV | 45 | PEQSRCQAIHNVVH | 85 | PELPYPQPELPYPQ | 124 | PQPF‐Cit‐PQQPYPQSQ |
| 5 | QQPQEQVPLVQQQQ | 46 | RCQAIHNVVHAIIL | 86 | YPQPELPYPQPQLP | 125 | Cit‐PQQPYPQSQPQYS |
| 6 | EQVPLVQQQQFPGQ | 47 | IHNVVHAIILHQQQ | 87 | QPELPYPQPQPFRP | 126 | QQILQQQLIPC‐Cit‐DV |
| 7 | LVQQQQFPGQQQPF | 48 | VHAIILHQQQQQQQ | 88 | PYPQPQPFRPEQPY | 127 | QQQLIPC‐Cit‐DVVLQQ |
| 8 | QQFPGQQQPFPPQQ | 49 | ILHQQQQQQQQQQQ | 89 | PQPFRPEQPYPQSQ | 128 | IPC‐Cit‐DVVLQQHSIA |
| 9 | GQQQPFPPQQPYPQ | 50 | QQQQQQQQQQQPLS | 90 | RPEQPYPQSQPQYS | 129 | CCQQLWQIPEQS‐Cit‐C |
| 10 | PFPPQQPYPQPQPF | 51 | QQQQQQQPLSQVSF | 91 | SQPQYSQPEQPISQ | 130 | LWQIPEQS‐Cit‐CQAIH |
| 11 | QQPYPQPQPFPSQQ | 52 | QQQPLSQVSFQQPQ | 92 | YSQPEQPISQQQQQ | 131 | PEQS‐Cit‐CQAIHNVVH |
| 12 | PQPQPFPSQQPYLQ | 53 | LSQVSFQQPQQQYP | 93 | EQPISQQQQQQQQQ | 132 | Cit‐CQAIHNVVHAIIL |
| 13 | PFPSQQPYLQLQPF | 54 | SFQQPQQQYPSGQG | 94 | QKQQQQQEEQILQQ | 133 | VQPQQLPQFEEI‐Cit‐N |
| 14 | QQPYLQLQPFPQPQ | 55 | PQQQYPSGQGSFQP | 95 | QQQEEQILEQILQQ | 134 | QLPQFEEI‐Cit‐NLALE |
| 15 | LQLQPFPQPQLPYP | 56 | YPSGQGSFQPSQQN | 96 | EQILEQILEEQLIP | 135 | FEEI‐Cit‐NLALETLPA |
| 16 | PFPQPQLPYPQPQL | 57 | QGSFQPSQQNPQAQ | 97 | EQILEEQLIPCRDV | 136 | Cit‐NLALETLPAMCNV |
| 17 | PQLPYPQPQLPYPQ | 58 | QPSQQNPQAQGSVQ | 98 | EEQLIPCRDVVLQQ | 137 | CTIAPVGIFGTNY‐Cit |
| 18 | YPQPQLPYPQPQLP | 59 | QNPQAQGSVQPQQL | 99 | IPCRDVVLQEHSIA | Carbamylated | |
| 19 | QPQLPYPQPQPFRP | 60 | AQGSVQPQQLPQFE | 100 | DVVLQEHSIAYGSS | 138 | SQQQQQQQQQQQQ‐hCit |
| 20 | PYPQPQPFRPQQPY | 61 | VQPQQLPQFEEIRN | 101 | QEHSIAYGSSQVLQ | 139 | QQQQQQQQQ‐hCit‐QQQQ |
| 21 | PQPFRPQQPYPQSQ | 62 | QLPQFEEIRNLALE | 102 | IAYGSSQVLQESTY | 140 | QQQQQ‐hCit‐QQQQQQQQ |
| 22 | RPQQPYPQSQPQYS | 63 | FEEIRNLALETLPA | 103 | SSQVLQESTYQLVQ | 141 | Q‐hCit‐QQQQQQQQILQQ |
| 23 | PYPQSQPQYSQPQQ | 64 | RNLALETLPAMCNV | 104 | LQESTYQLVQQLCC | Combinations | |
| 24 | SQPQYSQPQQPISQ | 65 | LETLPAMCNVYIPP | 105 | TYQLVQQLCCEQLW | 142 | QPELPYPQPQPF‐Cit‐P |
| 25 | YSQPQQPISQQQQQ | 66 | PAMCNVYIPPYCTI | 106 | VQQLCCEQLWEIPE | 143 | PYPQPQPF‐Cit‐PEQPY |
| 26 | QQPISQQQQQQQQQ | 67 | NVYIPPYCTIAPVG | 107 | CCEQLWEIPEQSRC | 144 | PQPF‐Cit‐PEQPYPQSQ |
| 27 | SQQQQQQQQQQQQK | 68 | PPYCTIAPVGIFGT | 108 | LWEIPEQSRCQAIH | 145 | Cit‐PEQPYPQSQPQYS |
| 28 | QQQQQQQQQKQQQQ | 69 | CTIAPVGIFGTNYR | 109 | QQQQQQQQQEQPLS | 146 | Q‐hCit‐QQQQQEEQILQQ |
| 29 | QQQQQKQQQQQQQQ | Deamidated | 110 | QQQQQEQPLSEVSF | 147 | EQILEEQLIPC‐Cit‐DV | |
| 30 | QKQQQQQQQQILQQ | 70 | PVPQLQPENPSQQQ | 111 | QEQPLSEVSFEQPQ | 148 | EEQLIPC‐Cit‐DVVLQQ |
| 31 | QQQQQQILQQILQQ | 71 | LQPENPSQEQPQEQ | 112 | LSEVSFEQPEQQYP | 149 | IPC‐Cit‐DVVLQEHSIA |
| 32 | QQILQQILQQQLIP | 72 | NPSQEQPEEEVPLV | 113 | SFEQPEQEYPSGQG | 150 | CCEQLWEIPEQS‐Cit‐C |
| 33 | QQILQQQLIPCRDV | 73 | EQPEEEVPLVQQQQ | 114 | PEQEYPSGQGSFQP | 151 | LWEIPEQS‐Cit‐CQAIH |
| 34 | QQQLIPCRDVVLQQ | 74 | EEVPLVQEQEFPGQ | 115 | QGSFQPSQENPQAQ | 152 | VQPQELPQFEEI‐Cit‐N |
| 35 | IPCRDVVLQQHSIA | 75 | LVQEQEFPGQEQPF | 116 | QPSQENPQAQGSVQ | 153 | ELPQFEEI‐Cit‐NLALE |
| 36 | DVVLQQHSIAYGSS | 76 | QEFPGQEQPFPPQQ | 117 | ENPQAQGSVQPQQL | Positive control | |
| 37 | QQHSIAYGSSQVLQ | 77 | GQEQPFPPEQPYPQ | 118 | AQGSVQPQELPQFE | 154 | LQPFPQPELPYPQP |
| 38 | IAYGSSQVLQQSTY | 78 | PFPPEQPYPQPQPF | 119 | VQPQELPQFEEIRN | Negative control | |
| 39 | SSQVLQQSTYQLVQ | 79 | EQPYPQPQPFPSQQ | 120 | ELPQFEEIRNLALE | 155 | EPQPYPLPQPFPQL |
| 40 | LQQSTYQLVQQLCC | 80 | PQPQPFPSEQPYLQ |
Cit = citrulline; hCit = homocitrulline. All peptides have free amino‐ and carboxyl terminals.
Underlined sequences are related to or are exact matches for human leucocyte antigen (HLA)‐DQ2·5‐ or HLA‐DQ2·2‐restricted epitopes: DQ2·5‐glia‐α1a (PFPQPQLPY), DQ2·5‐glia‐α1b (PYPQPQLPY), DQ2·5‐glia‐α2 (PQPQLPYPQ), DQ2·5‐glia‐α3 (FRPQQPYPQ), DQ2·2‐glia‐α1 (QGSVQPQQL), DQ2·2‐glia‐α2 (QYSQPQQPI) and DQ2·2‐glia‐α1 (QGSFQPSQQ) where Q is preferentially deamidated by TG2 and represented by E.
Peptides and gluten for cellular assays were dissolved in dimethyl sulphoxide (DMSO) and diluted in PBS to ×10 or ×2 (for ELISPOT assays) final concentration in PBS 1% DMSO. Peptide solutions were transferred, 25 μl per well, to sterile 96‐well U‐bottomed microwell plates (Thermo Fisher Scientific). Plates were sealed with sterile adhesive covers (Thermo Fisher Scientific), stored at −80°C, and shipped on dry ice to the study site.
Whole blood stimulation in microplates
Immediately before use, to avoid condensation and leakage between wells, microplates containing test solutions were thawed for 15 min at room temperature while being centrifuged at 300 g. Fresh heparinized blood was dispensed (225 μl) into individual wells with glia‐α (final concentration 15 μM) or negative control (15 μM) either in 16 replicate wells for later pooling or into 24 replicate wells for individual assessment. Blood was incubated in duplicate wells with individual peptides of the α‐gliadin library for later pooling. Incubation plates were immediately placed in a humidified incubator at 37°C in 5% CO2. After 24 h, plates were centrifuged at 500 g for 10 min at room temperature. Plasma, 90–120 μl per well, was carefully collected to avoid erythrocyte contamination and transferred to a corresponding well in a ‘mirror image’ sterile 96‐well plate. Plates were sealed and frozen at −80°C, and shipped back to the central laboratory for processing and cytokine assays.
Electrochemiluminescence immunoassays
Electrochemiluminescence assay kits from Meso Scale Diagnostics LLC (Rockville, MD, USA) were used to measure cytokines according to the manufacturer’s instructions. Plasmas pooled from 16‐replicate wells were assessed for IFN‐γ, IL‐10, IL‐2, IL‐6, IL‐8/CXCL8, TNF‐α, CXCL10/IFN‐γ‐induced protein 10 (IP‐10), CCL3/macrophage inflammatory protein (MIP)‐1α, CCL4/MIP‐1β, CCL2/monocyte chemoattractant protein‐1 (MCP1), IL‐17A, IL‐22, MIP‐3α and CXCL9. IFN‐γ, IL‐10 and IL‐2 were assessed in plasma from other whole blood stimulations and also sera. Samples were diluted 1 : 1 with assay diluent for IFN‐γ, IL‐10, IL‐2, IL‐6, IL‐8/CXCL8 and TNF‐α, or 1 : 3 for other cytokines. Duplicate diluted samples, 50 µl, were assessed in adjacent wells. Mean signal strength and cytokine concentration for duplicate well measurements were generated using an MSD MESO™ Sector S600 plate reader and Discovery Workbench 4.0. The lower limit of detection (LLOD) was calculated for each cytokine on each assay plate. Cytokine concentrations for signal values below the LLOD were reported as equal to the LLOD.
Peptide‐stimulated proliferation of CD4+ T cells
PBMC were prepared from heparinized blood by Ficoll‐Paque (GE Healthcare, Chicago, IL, USA) density gradient centrifugation using SepMate™ tubes (StemCell Technologies, Vancouver, BC, Canada), according to the manufacturer’s instructions. PBMC (1 million/ml in PBS) were labelled with 2·5 μM CellTrace™ Violet dye (CTV; Thermo Fisher Scientific) for 20 min at 37°C, washed, then resuspended in RPMI‐1640/10% human antibody serum. Proliferation assays consisted of 24 replicate wells per condition containing ~0·3 × 106 in 225 μl per well of CTV‐labelled PBMC and 25 μl containing glia‐α or glia‐ω peptides, negative control 8mer peptide mix (final peptide concentrations all 15 μM) or 1 μg/ml anti‐CD3 antibody. Outer wells contained PBS to minimize evaporation. After 8 days cells were stained with CD3‐fluorescein isothiocyanate (FITC) (UCHT1), CD4‐antigen‐presenting cells (APC) (SK3) and 7‐amino‐actinomycin D (7‐AAD) to discriminate dead cells (all from BD Biosciences, San Jose, CA, USA). Cells were analysed on a BD FACSVerse cytometer and flow cytometry data were analysed by FlowJo software (version 10; TreeStar, Inc., Ashland, OR, USA).
Lymphocyte subset and monocyte frequencies in fresh blood and PBMC
Whole blood or PBMC were stained with antibody mix comprising anti‐human CD3‐Bv421, CD4‐phycoerythrin (PE), CD8‐APC, CD14‐APCH7, CD19‐Bv480, CD20‐PE cyanin 7(Cy7) and CD45‐FITC (clones UCHT1, SK3, SK1, MphiP9, SJ25C1, L27, 2D1, respectively) and 7‐AAD (all from BD Biosciences). For whole blood, BD Trucount™ tubes were used according to the manufacturer’s instructions. Briefly, antibody mix was added followed by 50 μl whole blood by reverse pipetting. Tubes were vortexed and incubated for 15 min at room temperature in the dark. Erythrocytes were lysed by incubating with 450 μl 1×Pharm Lyse™ (BD Biosciences) for 15 min before analysing samples on a BD FACS Verse.
IFN‐γ ELISPOT assay
IFN‐γ ELISPOT (Mabtech, Nacka Strand, Sweden) assays were performed and analysed as previously described [15], with minor modifications to peptide and cell concentrations. Briefly, fresh PBMC were resuspended to a concentration of 6–8 million/ml in complete RPMI‐1640 10% pooled human serum media and were passed through a 70 μm cell strainer. Peptides or the control antibody was added in a final volume of 50 μl per well to which 50 μl PBMC/well were added and incubated overnight. Conditions and replicates tested were similar to the proliferation assays. Spot‐forming units (SFUs) were counted using an automated ELISPOT reader (Autoimmun Diagnostika, Straßberg, Germany).
Statistics
The sample size was empirical for this exploratory study. Non‐parametric statistical tests were used to assess paired (Wilcoxon’s signed‐rank test) or unpaired data (Mann–Whitney U‐test) without correction for multiple comparisons. Spearman’s correlation coefficients were calculated. The ratio of signal strength for test peptide to control peptide was analysed to empirically determine cut‐offs for whole blood CRA performance. Poisson distribution analysis of signal strength data from 24 replicate assays were used to infer the frequency of responding cells per well and in blood using the number of CD4+CD3+ cells per well [16].
Results
Participant characteristics
Participants were recruited and studied in July 2019 at one site. Cohorts 1 and 2 were enrolled sequentially. Table 2 summarizes participant characteristics. All 18 CD volunteers were eligible. Eleven unaffected adults were screened for cohort 3, but one was excluded with elevated CD serology and one was discontinued before blood was collected. CD participants had a median age of 48·5 years (range = 37–69), 15 were female, their median period on gluten‐free diet was 7 years (range = 11 months to 26 years) and all but one were seronegative for both TG2 IgA and DGP‐IgG. Unaffected participants in cohort 3 had a median age of 46 years (range = 35–66); all nine were female, all were seronegative and seven were positive for HLA‐DQ2·5.
Table 2.
Participant characteristics
| Cohort | 1 CD on GFD | 2 CD on GFD | 3 Healthy controls |
|---|---|---|---|
| Number | 6 | 12 | 10 a |
| Females, n (%) | 5 (83%) | 10 (83%) | 10 (100%) |
| Median age, years (range) | 44 (23–66) | 50 (26–69) | 46 (35–65) |
| Medically diagnosed CD, n (%) | 6 (100%) | 12 (100%) | 0 |
| GFD duration in months, median (range) | 79 (23–203) | 87 (11–316) | 0 |
| GFD > 1 year, n (%) | 6 (100%) | 11 (92%) | 0 |
| Elevated baseline TG2‐IgA, n (%) | 0 | 0 | 1 (10%) |
| Elevated baseline DGP‐IgG, n (%) | 0 | 1 (8%) | 1 (10%) |
| HLA‐DQ2·5‐positive, n (%) | 6 (100%) | 12 (100%) | 7 (70%) |
| HLA‐DQ8‐positive and DQ2‐negative, n (%) | 0 | 0 | 2 (20%) |
| HLA‐DQ2·5‐, 2·2‐, 8‐ and 7‐negative, n (%) | 0 | 0 | 1 (10%) |
CD = coeliac disease; HLA = human leucocyte antigen; GFD = gluten‐free diet.
One healthy control participant was excluded due to positive tissue transglutaminase immunoglobulin (TGA‐Ig)A and deamidated gliadin peptide (DGP)‐IgG at screening. An additional healthy control discontinued before any blood was collected (data not shown).
IFN‐γ ELISPOT assay and proliferation assays
IFN‐γ ELISPOT assay and dye‐dilution CD4+ T cell proliferation assays are well‐established functional assays for antigen‐specific T cells using freshly isolated PBMC [17, 18]. These assays were performed in 24 replicate wells to allow robust statistical comparisons of responses to glia‐α and glia‐ω peptides and the negative control. An example of images of IFN‐γ ELISPOT wells and proliferation assays are shown in Supporting information, Figs S1 and S2, respectively. For the six participants in cohort 1, the median frequency of live CD4+ CD3+ T cells in each well was 0·15 million (range = 0·12–0·24) for IFN‐γ ELISPOT assays and 0·13 million (range = 0·10–0·15) for proliferation assays. Table 3 shows that at baseline only one participant (103) showed significantly increased responses compared to control, and this was for both glia‐α and glia‐ω peptides in both the ELISPOT and proliferation assays.
Table 3.
Baseline gluten peptide‐specific T cell responses and point frequencies by Poisson distribution analysis in 24 replicate wells for the six treated CD patients in cohort 1
| Control | Glia‐α peptide | Glia‐ω peptide | |||||
|---|---|---|---|---|---|---|---|
| SFU/106 PBMC mean (s.e.m.) | SFU/106 PBMC Mean (s.e.m.) | % wells positive (n = 24) | Point frequency /106 CD4+CD3+ (95% CI) | SFU/106 PBMC mean (s.e.m.) | % wells positive (n = 24) | Point frequency /106 CD4+CD3+ (95% CI) | |
| IFN‐γ ELISPOT | |||||||
| 101 | 10 (1·3) | 11 (1·3) | 0 | 0 (0–1·1) | 12 (0·8) | 0 | 0 (0–1·1) |
| 102 | 3 (0·6) | 3 (0·5) | 0 | 0 (0–0·8) | 4 (0·8) | 0 | 0 (0–0·8) |
| 103 | 3 (0·6) | 6 (1·0)*** | 17 | 1·5 (0·4–3·8) | 6 (1·0)** | 17 | 1·5 (0·4–3·8) |
| 104 | 1 (0·4) | 1 (0·4) | 0 | 0 (0–0·6) | 1 (0·3) | 0 | 0 (0–0·6) |
| 105 | 4 (0·6) | 4 (0·8) | 4 | 0·2 (0·0–1·2) | 4 (0·8) | 0 | 0 (0–0·7) |
| 106 | 4 (0·9) | 7 (0·9) | 0 | 0 (0–1·1) | 7 (1·0) | 0 | 0 (0–1·1) |
| % dividing CD4+CD3+ cells | % dividing CD4+CD3+ cells | % dividing CD4+CD3+ cells | |||||
| Proliferation | |||||||
| 101 | 1·6 (0·4) | 1·0 (0·2) | 0 | 0 (0–1·1) | 1·1 (0·2) | 0 | 0 (0–1·0) |
| 102 | 1·2 (0·2) | 0·6 (0·1) | 0 | 0 (0–1·2) | 1·1 (0·1) | 0 | 0 (0–1·2) |
| 103 | 0·2 (0·1) | 4·1 (0·7)† | 92 | 23 (21–41) | 0·2 (0·4)† | 38 | 4·3 (1·9–8·2) |
| 104 | 0·1 (0·01) | 0·02 (0·01) | 0 | 0 (0–1·0) | 0·03 (0·01) | 0 | 0 (0–1·0) |
| 105 | 0·4 (0·1) | 0·4 (0·1) | 4 | 0·4 (0·1–2·4) | 0·3 (0·04) | 0 | 0·4 (0·1–2·4) |
| 106 | 0·2 (0·1) | 0·10 (0·02) | 0 | 0 (0–1·0) | 0·2 (0·01) | 0 | 0 (0–1·0) |
CD = coeliac disease; s.e.m. = standard error of the mean; CI = confidence interval; ELISPOT = enzyme‐linked immunospot.
P < 0·01, *** P < 0·001, † P < 0·0001 versus control.
Based on previous studies using HLA‐DQ2·5 gluten peptide tetramers assessing the frequency of effector memory CD4+ T cells specific for gluten epitopes, we expected one CD4+ T cell specific for glia‐α or glia‐ω peptides on average in each well [2, 3, 19]. Therefore, a Poisson distribution analysis tool was applied to establish the number of active and inactive wells, and model the average frequency of responding cell per well and per million CD4+CD3+ cells [16]. For the only responsive participant (103), the frequency of cells responding to glia‐α or glia‐ω by ELISPOT was 1·5 per million live CD4+CD3+ cells and by dye‐dilution proliferation assay was 23 and 4.3 per million to glia‐α and glia‐ω, respectively.
Immune activation after gluten challenge
It was unclear whether patients in cohort 1 who had no detectable responses to glia‐α and glia‐ω had specific T cells that were below the limits of detection for IFN‐γ ELISPOT and proliferation assays or whether they truly were deficient in IFN‐γ‐secreting T cells with these specificities. A 3‐day gluten food challenge was undertaken to expand the circulating gluten‐specific CD4+ T cell population [8]. In keeping with previous studies using this gluten challenge composition [11], five CD patients in cohort 1 showed increased serum concentrations of IL‐2 (Fig. 1a), IFN‐γ (Fig. 1b) and IL‐10 (Fig. 1c). These changes in serum cytokine concentrations reflected a median 84‐fold (range = 1.8–364; P = 0·03, Wilcoxon’s signed‐rank test) increase in serum levels of IL‐2 at 3 h after consuming gluten on day 1 (Fig. 1d) and smaller median fold increases at 6 h for serum IFN‐γ 1·5 (0·9–16) and IL‐10 1·6 (1·3–4·1) (Fig. 1e,f). In addition, IFN‐γ ELISPOT responses on day 6 were increased compared to negative control for glia‐ω (P = 0·03) and for deamidated chymotrypsin‐digested gluten (P = 0·03), and there was a trend to increase for glia‐α (P = 0·06) (Fig. 1g). A dose–response study with PBMC from day 6 confirmed that the concentrations of glia‐α and glia‐ω used in the IFN‐γ ELISPOT assays on day 1 elicited near‐maximal responses (Fig. 1h). Together, these findings indicated that five of six patients in cohort 1 had a recall response to gluten, and that in addition to subject 103, four others possessed T cells specific for glia‐α and glia‐ω detectable by IFN‐γ ELISPOT after, but not before, gluten challenge.
Fig. 1.
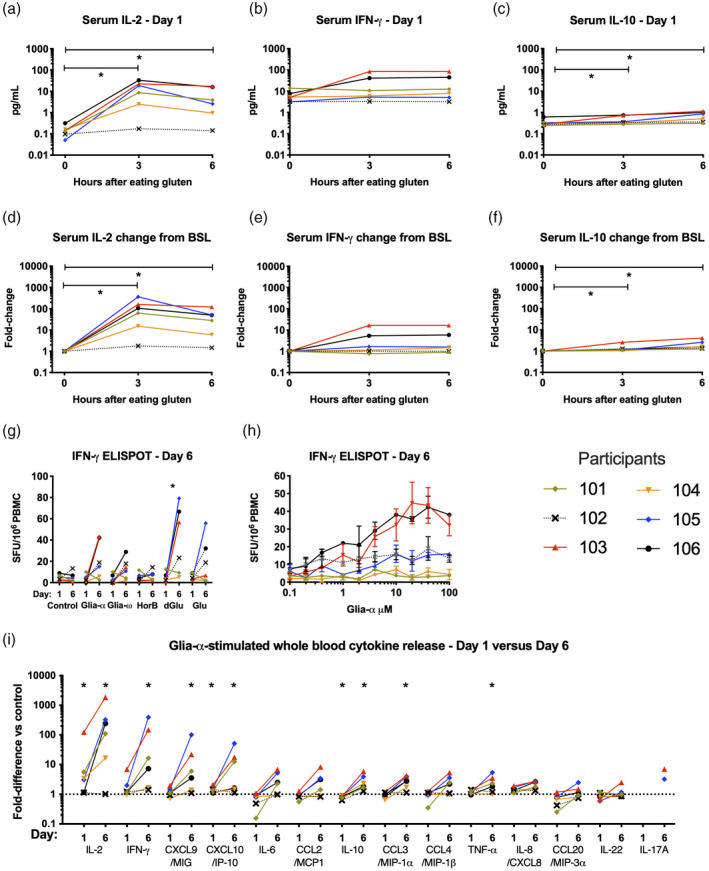
Changes in cytokine levels in vivo and in ex‐vivo whole blood cytokine release when coeliac disease (CD) patients in cohort 1 consumed 10 g vital wheat gluten as a bolus on days 1, 2 and 3. Interleukin (IL)‐2 (a), interferon (IFN)‐γ (b) and IL‐10 (c) concentrations in serum measured by electrochemiluminescence immunoassay are shown for the 6 h after consuming 10 g vital wheat gluten on day 1. The same data for serum concentrations of IL‐2 (d), IFN‐γ (e) and IL‐10 (f) are shown as fold change from baseline. Changes in fresh peripheral blood mononuclear cells (PBMC) IFN‐γ enzyme‐linked immunospot (ELISPOT) responses between baseline on day 1 and on day 6 are shown for individual patients (g). Dose–response relationship for glia‐α peptide‐stimulated IFN‐γ ELISPOT spot‐forming units per million PBMC is shown for individual patients (h). Changes between baseline on day 1 and on day 6 are shown for whole blood cytokine release stimulated by glia‐α relative to control peptide (i). *P = 0·03, Wilcoxon’s signed‐rank test (n = 6) comparing baseline with later time‐points (a–h) or comparing glia‐α and control peptide‐stimulated response at baseline or day 6 (i).
Whole blood cytokine release assay
CRAs using whole blood stimulated with antigen are effective for detecting gluten‐specific CD4+ T cell responses after gluten challenge when IL‐2, CXCL10 or IFN‐γ are measured by multiplex bead assays or enzyme‐linked immunosorbent assay (ELISA) [9, 10]. Recently, we showed that electrochemiluminescence immunoassays are more sensitive than multiplex bead assays for detection of low abundance cytokines such as IL‐2 in plasma or serum [10, 20]. As a first step to selecting which cytokines may be best to increase the sensitivity of whole blood CRAs, plasma from fresh blood incubated with glia‐α (15 μM) or negative control was assessed for 14 cytokines implicated in gluten challenge‐induced cytokine release in patients [12]. Cytokine release was significantly higher for glia‐α than control peptide at baseline for IL‐2, CXCL10 and IL‐8/CXCL8, and on day 6 after gluten challenge for IL‐2, IFN‐γ, CXCL10/IP‐10, CXCL9/MIG, IL‐10, CCL3/MIP‐1α, TNF‐α and IL‐8/CXCL8 (for each cytokine: P = 0·03, Wilcoxon’s signed‐rank test) (Fig. 1i). IL‐2 release stimulated by glia‐α was highest, with a median of 3·2 (range = 1·1–124) times more than for control peptide, and this increased by a median of 17·2 (range = 0·9–209) times after gluten challenge. Whole blood cytokine secretion stimulated by glia‐α compared to control also increased substantially for IFN‐γ (median = 10·3, range = 1·1–196), CXCL10/IP‐10 (5·0, 0·8–33·6), CXCL9/MIG (4·4, 1·2–69), IL‐10 (2·4, 2·0–6·9), CCL3/MIP‐1α (3·1, 1·0–4·4), TNF‐α (1·9, 0·8–4·8) and IL‐8/CXCL8 (1·5, 1·0–2·0). IL‐2, IFN‐γ and IL‐10 peak serum concentrations after gluten ingestion on day 1 showed a trend towards or were significantly correlated with glia‐α peptide‐stimulated whole blood cytokine release on day 6, but not at baseline (Supporting information, Fig. S3). Collectively, these findings indicated that IL‐2 assessed by electrochemiluminescence was likely to provide a more sensitive whole blood CRA than IFN‐γ or IL‐10.
The performance of a microplate whole blood cytokine release test measuring IL‐2, IFN‐γ and IL‐10 was assessed in 24 replicate wells at baseline and after gluten challenge in cohort 1 (Table 4). The median number of CD4+ CD3+ T cells in each well containing 225 μl fresh blood was 0·18 (0·13–0·31) million. Supporting information, Fig. S4 shows an indicative flow cytometry plot used to enumerate cell subsets in whole blood. The lower and upper limits of detection of IL‐2, IFN‐γ and IL‐10 in the 3‐plex electrochemiluminescence assay were 0·067–2940, 0·453–2860 and 0·029–680 pg/ml. Glia‐α stimulated significantly higher IL‐2, IFN‐γ and IL‐10 release than control peptide at baseline in four, two and none of the six patients, respectively. After gluten challenge, glia‐α stimulated significantly higher IL‐2, IFN‐γ and IL‐10 release than control peptide in five, six and four patients, respectively. Poisson distribution analysis was consistent with the frequency of IL‐2‐secreting T cells responsive to glia‐α being between 2·4 and greater than 15 cells per million circulating CD4+ T cells at baseline for the four responding patients in cohort 1 (Table 4). After gluten challenge, all 24 replicate wells with glia‐α were active for five patients, which is consistent with the frequency of glia‐α‐responsive cells being more than 11 per million CD4+ T cells in blood. Similar cell frequencies were deduced by Poisson distribution analysis for glia‐α‐responsive cells releasing IFN‐γ or IL‐10 in blood after gluten challenge (Table 4). Collectively, these findings indicated that a whole blood CRA with electrochemiluminescence immunoassay measuring IL‐2, and sometimes IFN‐γ, could often detect responses in small volumes of blood with low numbers of responding T cells.
Table 4.
Whole blood cytokine release using electrochemiluminescence immunoassay to assess gluten peptide‐specific T cell responses and point frequencies by Poisson distribution analysis in 24 replicate wells in six treated CD patients
| Baseline | Day 6 after commencing gluten challenge | |||||||
|---|---|---|---|---|---|---|---|---|
| Control | Glia‐α peptide | Control | Glia‐α peptide | |||||
| Cytokine pg/ml mean (s.e.m.) | Cytokine pg/ml mean (s.e.m.) | % wells pos. (n = 24) | Point frequency/106 CD4+ CD3+T cells (95% CI) | Cytokine pg/ml mean (s.e.m.) | Cytokine pg/ml mean (s.e.m.) | % wells pos. (n = 24) | Point frequency/106 CD4+ CD3+T cells (95% CI) | |
| IL‐2 whole blood release assay | ||||||||
| 101 | 0·5 (0·3) | 2·3 (0·6)† | 58 | 4·1 (2·1–7·0) | 0·04 (0·00) | 44·0 (4·5)† | 100 | > 11 |
| 102 | 0·11 (0·02) | 0·2 (0·1) | 17 | 1·3 (0·3–3·2) | 0·08 (0·01) | 0·09 (0·03) | 4 | 0·3 (0·01–1·5) |
| 103 | 0·11 (0·05) | 26 (0·4)† | 100 | > 15 | 0·17 (0·07) | 302 (9·5)† | 100 | > 12 |
| 104 | 0·39 (0·19) | 1·2 (0·6) | 21 | 0·7 (0·2–1·7) | 0·21 (0·06) | 5·5 (0·8)† | 100 | > 11 |
| 105 | 0·06 (0·01) | 1·3 (0·4)** | 50 | 2·4 (1·2–4·3) | 0·11 (0·03) | 200 (5)† | 100 | > 11 |
| 106 | 0·17 (0·03) | 0·6 (0·1)*** | 29 | 2·6 (1·0–5·4) | 0·24 (0·05) | 103 (5)† | 100 | > 13 |
| IFN‐γ whole blood release assay | ||||||||
| 101 | 8·2 (0·1) | 8·8 (0·7) | 13 | 0·6 (0·1–1·8) | 12 (0·1) | 200 (17)† | 100 | > 11 |
| 102 | 20·0 (0·7) | 22 (1) | 0 | 0 (0–0·9) | 65 (3) | 81 (5)† | 17 | 0·9 (0·2–2·5) |
| 103 | 4·6 (0·2) | 28 (5)† | 100 | > 15 | 12 (0·2) | 1495 (62)† | 100 | > 11 |
| 104 | 3·1 (0·1) | 4·1 (0·6) | 8 | 0·3 (0·03–1·0) | 51 (1) | 83 (5)† | 96 | 19 (9·1–40) |
| 105 | 2·6 (0·1) | 3·4 (0·5) | 17 | 0·6 (0·2–1·6) | 3·3 (0·1) | 883 (24)† | 100 | > 11 |
| 106 | 45·6 (2·3) | 64 (4)*** | 4 | 0·32 (0·01–1·8) | 69 (2) | 496 (19)† | 100 | > 13 |
| IL‐10 whole blood release assay | ||||||||
| 101 | 0·50 (0·13) | 0·31 (0·02) | 4 | 0·2 (0·01–1·1) | 0·21 (0·01) | 0·28 (0·03) | 17 | 1·0 (0·3–2·6) |
| 102 | 0·20 (0·05) | 0·13 (0·01) | 0 | 0 (0–0·9) | 0·14 (0·02) | 0·07 (0·01) | 0 | <0·9 |
| 103 | 0·15 (0·01) | 0·12 (0·01) | 0 | 0 (0–1·0) | 0·17 (0·01) | 1·2 (0·1)† | 100 | > 11 |
| 104 | 0·09 (0·01) | 0·11 (0·01) | 0 | 0 (0–0·5) | 0·18 (0·01) | 0·36 (0·02)† | 42 | 3·2 (1·5–5·9) |
| 105 | 0·13 (0·01) | 0·12 (0·01) | 0 | 0 (0–0·5) | 0·19 (0·01) | 0·83 (0·06)† | 100 | > 11 |
| 106 | 0·21 (0·01) | 0·20 (0·01) | 8 | 6·5 (0·01–2·3) | 0·23 (0·01) | 0·54 (0·04)† | 75 | 9·3 (5·1–16) |
CD = coeliac disease; IL = interleukin; IFN = interferon; s.e.m. = standard error of the mean; CI = confidence interval.
P < 0·05, ** P < 0·01, *** P < 0·001, † P < 0·0001 versus control.
Performance characteristics of the whole blood IL‐2 release test were assessed with fresh blood from a further group of 12 HLA‐DQ2·5+ adult patients with CD (cohort 2) and nine unaffected adults (cohort 3). Fresh blood incubated with glia‐α or control peptide was assessed in 24 replicate wells (Fig. 2, Supporting information, Table S1). The IL‐2 signal strength for glia‐α was compared to control peptide and the mean for both expressed as a ratio for each participant. The sensitivity was 92% and specificity 100% for CD patients using an empirical upper cut‐off of 1·5. Poisson distribution analysis was consistent with the 11 responsive CD patients having between 0·5 and 11 responding cells per ml fresh blood.
Fig. 2.
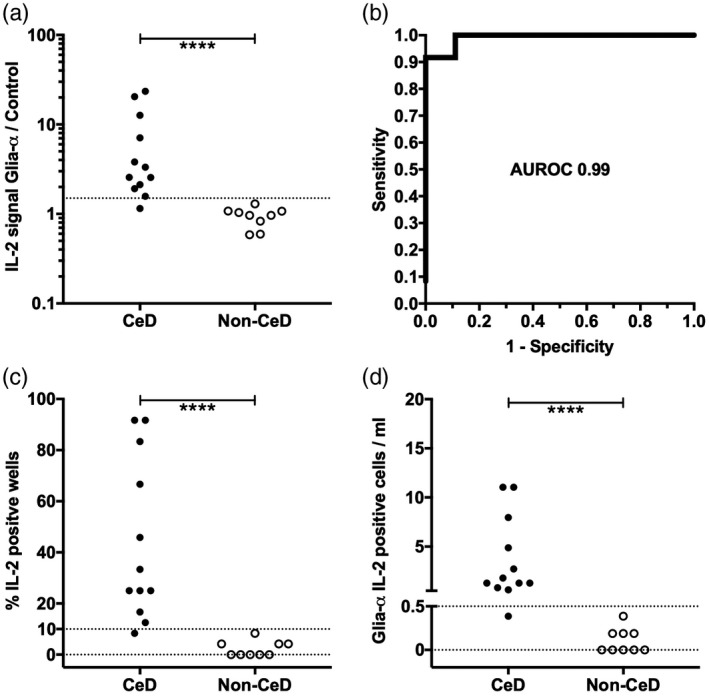
Whole blood interleukin (IL)‐2 release stimulated by glia‐α and control peptide in 24 replicate wells for 12 coeliac disease (CD) patients in cohort 2 and nine unaffected volunteers in cohort 3: mean IL‐2 signal stimulated by glia‐α in 24 replicate wells relative to mean IL‐2 signal stimulated by control in 24 replicate wells (a); receiver operating characteristic curve (ROC) for the whole blood IL‐2 cytokine release assay (CRA) with calculated area under the ROC (AUROC) (b); percentage of glia‐α wells with IL‐2 signal above the threshold set using control wells in individual participants (c); and frequency of cells in fresh blood responding to glia‐α according to Poisson distribution analysis in individual participants (d). ****P < 0·0001, Mann–Whitney U‐test.
Screening immunogenic α‐gliadin peptides using pooled plasma and whole blood CRA
Finally, to assess the utility of a microplate whole blood IL‐2 release assay for T cell epitope discovery in CD, a library of 153 wheat‐derived α‐gliadin peptides (Table 1) was assessed with blood collected at baseline and after gluten challenge for cohort 1. The library consisted of 69 native sequence peptides, their predicted deamidation products resulting from the action of TG2 [21], and variants where citrulline replaced arginine or homocitrulline replaced lysine. Some peptides in the library included one or two copies of each of the four well‐characterized HLA‐DQ2·5‐restricted α‐gliadin epitopes [22]. To efficiently assess IL‐2, IFN‐γ and IL‐10 release, the initial screen was with plasma pooled from all six patients at baseline or on day 6 for each individual peptide. At baseline, IL‐2 release was elevated exclusively for peptides that comprised native or modified sequences related to each of the four known α‐gliadin epitopes: DQ2·5‐glia‐α 1a (PFPQPELPY), DQ2·5‐glia‐α 1b (PYPQPELPY), DQ2·5‐glia‐α 2 (PQPELPYPQ) or DQ2·5‐glia‐α 3 (FRPEQPYPQ) (Fig. 3). Pooled plasma IL‐2 and IFN‐γ responses to these same peptides were strongly enhanced after gluten challenge, but peptide library responses at baseline were absent for IFN‐γ, and for IL‐10 they were rare and not consistent between days 1 and 6. Further validating the use of plasma pooled from all six donors, IL‐2, IFN‐γ and IL‐10 measured in pooled plasma correlated closely with the average of plasma cytokine levels in each individual assessed separately (Fig. 4).
Fig. 3.
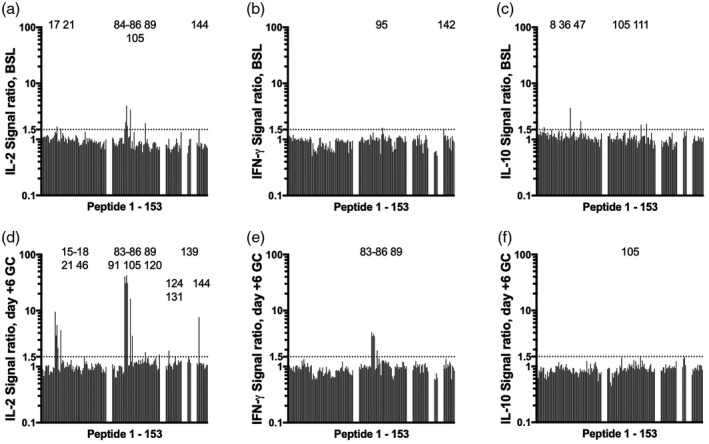
Whole blood cytokine release responses to individual peptides relative to control peptide for the α‐gliadin library at baseline on day 1 (BSL): interleukin (IL)‐2 (a), interferon (IFN)‐γ (b) and IL‐10 (c) and day 6 after commencing gluten challenge (GC): IL‐2 (d), IFN‐γ (e) and IL‐10 (f). Data shown are for plasma pooled from all six patients in cohort 1 for each individual peptide. Peptides 1–69 were 14mers overlapping by 10 amino acids that spanned the complete sequence of a wheat α‐gliadin (CAB76964.1). Modified versions of peptides 1–69 corresponded to partially deamidated (glutamine selectively replaced by glutamate according to Vader et al. [21]) (peptides 70–120), citrullinated (arginine replaced by citrulline) (peptides 121–137), carbamylated (lysine replaced by homocitrulline) (peptides 138–141) or partially deamidated and citrullinated or carbamylated variants (peptides 142–153). Numbers at the top of each graph indicate peptides eliciting responses above the cut‐off.
Fig. 4.
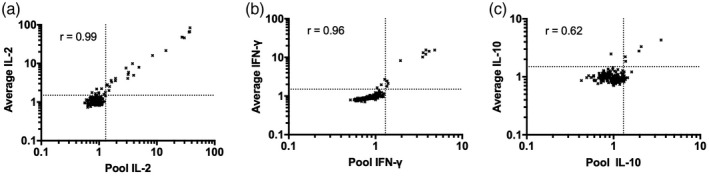
Interleukin (IL)‐2 (a), interferon (IFN)‐γ (b) and IL‐10 (c) signal in pooled plasma from six patients in cohort 1 compared to the average of plasma cytokine levels for these individuals measured separately. Data are responses to individual peptides 1–92 and positive control relative to negative control peptide for the α‐gliadin library at baseline on days 1 and 6 after commencing gluten challenge measured by electrochemiluminescence immunoassay. Spearman’s correlation coefficients are shown.
Personalized mapping of immunogenic α‐gliadin peptides using whole blood CRA
Table 5 shows individual patient’s responses to peptides that elicited elevated IL‐2, IFN‐γ or IL‐10 with pooled plasma at baseline or after gluten challenge. For this assessment, only peptides 1–92 were evaluated. As expected, responses to peptides with known immunodominant α‐gliadin epitopes were reproducible from baseline to day 6, and after gluten challenge they were stronger and more consistent, and for the most active peptides clearly showed preference for deamidated variants. Gluten challenge also resulted in a more diverse range of cytokine responses elicited by individual peptides especially for those that stimulated only IL‐2 at baseline. Collectively, these findings reinforced the value of gluten challenge to map immunogenic gluten peptides, but also indicated that gluten challenge is not essential for identification of the most active gluten peptides using the whole blood IL‐2 release assay.
Table 5.
Consistency of individual patient’s IL‐2, IFN‐γ, and/or IL‐10 responses to active peptides using pooled plasma
| Peptide | Sequence * | Peptide response ‐ ratio of peptide signal to negative control signal (>1.5 indicated in red) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL‐2 Participant | IFN‐γ Participant | IL‐10 Participant | |||||||||||||||||
| 101 | 102 | 103 | 104 | 105 | 106 | 101 | 102 | 103 | 104 | 105 | 106 | 101 | 102 | 103 | 104 | 105 | 106 | ||
| Blood collected at baseline | |||||||||||||||||||
| 8 | QQFPGQQQPFPPQQ | 1.5 | 0.4 | 0.6 | 1.1 | 0.8 | 1.1 | 1.1 | 1.2 | 0.9 | 1.3 | 0.9 | 0.9 | 2.9 | 0.3 | 0.9 | 1.0 | 1.3 | 1.0 |
| 15 | LQLQPFPQPQLPYP | 1.6 | 0.4 | 5.9 | 1.0 | 1.0 | 0.9 | 1.1 | 0.9 | 2.4 | 1.1 | 0.8 | 0.8 | 1.1 | 0.8 | 0.8 | 1.0 | 0.9 | 0.9 |
| 16 | PFPQPQLPYPQPQL | 1.4 | 0.8 | 4.3 | 1.0 | 1.0 | 1.0 | 1.1 | 0.9 | 1.3 | 1.0 | 1.0 | 0.9 | 1.5 | 0.5 | 1.0 | 1.0 | 0.9 | 0.9 |
| 17 | PQLPYPQPQLPYPQ | 2.4 | 0.5 | 15.9 | 0.9 | 1.0 | 0.9 | 1.3 | 1.1 | 1.3 | 1.0 | 1.0 | 0.8 | 1.2 | 0.7 | 1.1 | 1.0 | 1.2 | 0.8 |
| 18 | YPQPQLPYPQPQLP | 1.7 | 0.7 | 2.8 | 1.0 | 1.0 | 0.7 | 1.0 | 1.0 | 1.1 | 1.0 | 1.1 | 0.8 | 1.1 | 0.6 | 1.1 | 1.0 | 1.0 | 0.8 |
| 21 | PQPFRPQQPYPQSQ | 1.3 | 0.6 | 5.7 | 1.0 | 2.6 | 0.7 | 1.0 | 0.9 | 1.1 | 1.1 | 0.8 | 1.0 | 0.9 | 0.6 | 1.0 | 1.0 | 0.7 | 0.7 |
| 36 | DVVLQQHSIAYGSS | 1.3 | 0.5 | 0.9 | 0.9 | 0.7 | 0.7 | 1.0 | 0.5 | 1.0 | 1.0 | 0.8 | 0.4 | 2.0 | 19.5 | 0.8 | 1.0 | 1.1 | 1.3 |
| 46 | RCQAIHNVVHAIIL | 1.5 | 1.1 | 1.0 | 0.8 | 0.8 | 0.8 | 1.1 | 0.6 | 0.8 | 1.2 | 0.9 | 0.3 | 1.0 | 0.7 | 0.9 | 1.0 | 0.9 | 0.9 |
| 47 | IHNVVHAIILHQQQ | 1.8 | 1.1 | 1.0 | 1.1 | 1.0 | 2.6 | 1.3 | 0.2 | 1.1 | 1.3 | 0.8 | 0.6 | 7.7 | 7.8 | 1.2 | 0.9 | 1.2 | 1.1 |
| 83 | LQLQPFPQPELPYP | 1.4 | 1.0 | 11.9 | 0.7 | 1.2 | 0.8 | 1.0 | 0.8 | 1.5 | 1.0 | 1.0 | 1.1 | 1.1 | 0.7 | 1.1 | 0.8 | 1.0 | 0.8 |
| 84 | PFPQPELPYPQPQL | 1.3 | 0.7 | 17.4 | 0.7 | 3.6 | 0.9 | 1.0 | 0.9 | 1.3 | 1.1 | 1.2 | 1.0 | 0.9 | 0.5 | 1.0 | 0.9 | 0.9 | 0.7 |
| 85 | PELPYPQPELPYPQ | 1.2 | 0.8 | 52.7 | 1.0 | 1.7 | 2.0 | 1.0 | 1.0 | 5.3 | 1.1 | 1.1 | 1.2 | 0.9 | 0.8 | 0.9 | 1.1 | 1.0 | 1.0 |
| 86 | YPQPELPYPQPQLP | 1.8 | 0.8 | 9.7 | 1.1 | 5.5 | 0.9 | 1.1 | 0.9 | 2.1 | 1.2 | 1.1 | 0.9 | 1.1 | 0.8 | 0.8 | 1.2 | 1.1 | 0.9 |
| 89 | PQPFRPEQPYPQSQ | 1.5 | 1.1 | 19.7 | 0.8 | 11.5 | 0.8 | 1.0 | 1.4 | 1.5 | 1.0 | 2.6 | 0.7 | 1.3 | 0.8 | 0.9 | 0.9 | 1.0 | 0.8 |
| 91 | SQPQYSQPEQPISQ | 1.5 | 0.9 | 3.7 | 0.8 | 0.9 | 0.8 | 1.1 | 1.1 | 0.8 | 1.1 | 0.8 | 1.0 | 1.1 | 0.8 | 0.7 | 1.1 | 0.9 | 0.8 |
| Blood collected on Day 6 after commencing gluten challenge | |||||||||||||||||||
| 8 | QQFPGQQQPFPPQQ | 0.7 | 2.1 | 0.9 | 1.1 | 0.7 | 0.8 | 1.1 | 1.3 | 0.9 | 1.0 | 1.2 | 0.9 | 1.1 | 1.1 | 1.0 | 1.0 | 1.0 | 1.1 |
| 15 | LQLQPFPQPQLPYP | 11.5 | 2.6 | 50.3 | 1.0 | 26.8 | 4.1 | 1.4 | 0.9 | 6.8 | 0.9 | 5.3 | 0.9 | 1.1 | 1.1 | 1.5 | 0.8 | 1.1 | 1.1 |
| 16 | PFPQPQLPYPQPQL | 4.8 | 2.1 | 14.1 | 0.9 | 5.8 | 3.4 | 1.4 | 1.0 | 3.3 | 1.0 | 2.4 | 0.9 | 1.4 | 0.8 | 1.3 | 0.9 | 0.9 | 1.2 |
| 17 | PQLPYPQPQLPYPQ | 1.4 | 4.1 | 20.6 | 0.9 | 14.8 | 6.0 | 1.0 | 1.1 | 4.7 | 1.0 | 5.9 | 1.1 | 1.0 | 1.5 | 2.2 | 1.4 | 0.5 | 0.8 |
| 18 | YPQPQLPYPQPQLP | 1.8 | 3.9 | 6.4 | 1.8 | 6.0 | 1.5 | 0.9 | 1.1 | 1.8 | 1.0 | 1.6 | 1.2 | 0.9 | 1.5 | 1.3 | 0.7 | 0.5 | 1.0 |
| 21 | PQPFRPQQPYPQSQ | 1.0 | 2.4 | 2.8 | 0.8 | 21.8 | 0.9 | 1.0 | 1.1 | 1.3 | 1.0 | 6.5 | 0.9 | 1.4 | 0.7 | 1.0 | 0.8 | 1.0 | 0.8 |
| 36 | DVVLQQHSIAYGSS | 0.9 | 3.0 | 0.9 | 0.9 | 1.3 | 0.6 | 1.0 | 0.6 | 0.9 | 1.0 | 1.0 | 0.7 | 1.3 | 1.3 | 1.2 | 1.0 | 0.6 | 0.8 |
| 46 | RCQAIHNVVHAIIL | 9.3 | 2.6 | 2.1 | 0.9 | 0.6 | 0.7 | 1.4 | 0.6 | 1.1 | 1.0 | 0.9 | 0.4 | 1.3 | 1.0 | 1.3 | 0.9 | 0.6 | 1.0 |
| 47 | IHNVVHAIILHQQQ | 3.2 | 2.6 | 1.4 | 0.9 | 0.7 | 1.6 | 1.1 | 0.0 | 0.9 | 1.1 | 0.9 | 0.7 | 3.3 | 0.6 | 1.4 | 0.9 | 0.8 | 1.1 |
| 83 | LQLQPFPQPELPYP | 41.3 | 2.8 | 213.3 | 10.8 | 97.0 | 23.2 | 4.3 | 1.0 | 34.2 | 0.9 | 43.7 | 2.2 | 1.0 | 1.0 | 2.8 | 0.8 | 1.5 | 1.0 |
| 84 | PFPQPELPYPQPQL | 16.7 | 3.5 | 155.4 | 1.3 | 91.3 | 17.6 | 3.2 | 1.0 | 31.9 | 1.0 | 40.9 | 1.5 | 0.9 | 1.0 | 2.1 | 1.0 | 1.5 | 1.0 |
| 85 | PELPYPQPELPYPQ | 45.0 | 2.6 | 276.5 | 2.1 | 157.4 | 20.5 | 4.2 | 1.0 | 43.4 | 1.0 | 41.4 | 2.0 | 1.4 | 0.6 | 5.8 | 1.4 | 2.8 | 1.2 |
| 86 | YPQPELPYPQPQLP | 25.4 | 2.7 | 156.1 | 2.5 | 69.1 | 33.3 | 3.0 | 0.8 | 24.9 | 1.0 | 28.9 | 2.7 | 0.8 | 0.9 | 2.4 | 0.9 | 1.2 | 0.9 |
| 89 | PQPFRPEQPYPQSQ | 4.6 | 0.9 | 16.0 | 1.3 | 105.7 | 0.9 | 1.7 | 1.4 | 2.2 | 1.1 | 42.0 | 1.0 | 1.2 | 1.1 | 1.1 | 1.1 | 1.7 | 1.0 |
| 91 | SQPQYSQPEQPISQ | 0.9 | 1.4 | 0.7 | 0.9 | 19.4 | 1.4 | 1.0 | 1.1 | 0.9 | 1.0 | 7.4 | 1.2 | 0.8 | 1.0 | 1.2 | 1.1 | 0.8 | 1.1 |
Plasma was pooled from whole blood stimulations from all six patients in Cohort 1. Note that only peptides 1 to 92 were assessed for individual patients; peptides that were deemed active for IL‐2, IFN‐γ, or IL‐10, but were not reassessed with individual plasmas include: 95 (QQQEEQILEQILQQ), 105 (TYQLVQQLCCEQLW), 111 (QEQPLSEVSFEQPQ), 120 (ELPQFEEIRNLALE), 124 (PQPF‐Cit‐PQQPYPQSQ), 131 (PEQS‐Cit‐CQAIHNVVH), 139 (QQQQQQQQQ‐hCit‐QQQQ), 142 (QPELPYPQPQPF‐Cit‐P), 144 (PQPF‐Cit‐PEQPYPQSQ).
Amino acid sequences related to known HLA‐DQ2.5‐restricted epitopes are underlined: DQ2.5‐glia‐α1a (PFPQPQLPY), DQ2.5‐glia‐α1b (PYPQPQLPY), DQ2.5‐glia‐α2 (PQPQLPYPQ), and DQ2.5‐glia‐α3 (FRPQQPYPQ), DQ2.2‐glia‐α2 (QYSQPQQPI), where Q is preferentially deamidated by TG2 and represented by E.
Discussion
We set out to adapt an established whole blood gluten peptide‐stimulated IFN‐γ release assay [9] to allow detection of gluten‐specific CD4+ T cells without CD patients having to undertake a gluten challenge. We again confirmed that, without gluten challenge, conventional IFN‐γ release and dye‐dilution proliferation assays rarely detect CD patients with circulating gluten‐specific CD4+ T cells. Improving the sensitivity of the cytokine immunoassay and selecting the cytokine most responsive to gluten peptide stimulation (IL‐2) allowed the whole blood CRA to detect responses in blood from CD patients without having a gluten challenge. When compared to unaffected volunteers, fresh blood IL‐2 release stimulated by gluten peptide was highly specific and sensitive for CD patients on a gluten‐free diet. This enhanced whole blood gluten peptide‐stimulated IL‐2 release assay was estimated to detect as few as one cell secreting IL‐2 in a microwell containing 0·225 ml fresh blood. We also demonstrated that freshly collected blood from CD patients on a gluten‐free diet can be used in conjunction with unseparated, whole blood IL‐2 release to efficiently map the most immunoreactive gluten peptides from a large candidate library, which was previously only possible with fresh peripheral blood if patients underwent gluten challenge.
Gluten‐reactive CD4+ T cells relevant to CD were originally thought to be limited to the gut [23, 24]. Employing a gluten food challenge to expand the pool of circulating gluten‐reactive CD4+ T cells allowed their detection by standard IFN‐γ ELISPOT using fresh PBMC [8]. Subsequently, application of HLA‐DQ2·5 gluten peptide tetramers confirmed that CD patients have gluten‐specific CD4+ T cells in peripheral blood that are rare [25], but share the same phenotype and specificity as they do in the gut [26]. These advances have allowed the cellular immunology of CD to be studied using blood instead of relying on cells extracted from gut biopsies. Currently, however, blood‐based T cell diagnostics have not entered clinical practice for CD, even though they may ultimately allow CD to be defined and diagnosed on the basis of a memory T cell response to gluten.
Consistent detection of rare gluten‐specific CD4+ T cells by HLA‐DQ2·5 gluten peptide tetramers and flow cytometry is possible using PBMC separated from relatively large volumes of blood [19]. However, the tetramer assay format is time‐consuming, technically demanding, requires proprietary flow cytometry and magnetic bead enrichment reagents and does not lend itself to high throughput. Recently, polymerase chain reaction (PCR) detection of genes encoding T cell receptors (TCRs) has emerged as another potential approach for diagnosis of CD, as variable regions of TCRs specific for gluten epitopes are conserved between CD patients who share the same HLA‐DQ genotype [27]. However, this diagnostic approach is yet to be assessed in patients. Whole blood IL‐2 release assay using an electrochemiluminescence assay is straightforward to set up and analyse, utilizes a standardized assay kit, and harvesting plasma from whole blood following peptide stimulation in a standard CO2 humidified incubator requires only a simple centrifugation step before plasma is frozen. Subsequently, samples can be shipped to a central laboratory for analysis. Because only 25 μl of plasma is required for duplicate assessments of IL‐2 in either a single‐ or multiplex format, a single plasma sample can be assayed several times in parallel or in series. We took advantage of this opportunity when we compared pooled and individual plasma samples while screening an α‐gliadin peptide library.
The utility of IL‐2 in whole blood CRAs may not be limited to CD. A recent study has demonstrated that diagnostic whole blood cytokine release tests for tuberculosis may be improved by assessing IL‐2 [28]. To our knowledge, this is the first study to assess an electrochemiluminescence immunoassay to measure IL‐2 in a whole blood CRA in a disease associated with low frequencies of disease‐relevant T cells in blood. We are also unaware of other studies that have applied Poisson distribution analysis to estimate the frequency of disease‐relevant responding T cells in fresh blood.
This study has several weaknesses. Only a relatively small number of volunteers from a single centre were included in this discovery study. Strategically, we relied upon CD patients positive for HLA‐DQ2·5 having a memory CD4+ T cell response to two HLA‐DQ2·5‐restricted gluten epitopes in a single immunodominant α‐gliadin peptide. CD patients negative for HLA‐DQ2·5 may respond differently, and would certainly require different gluten peptides. Our findings in HLA‐DQ2·5+ CD patients are, overall, supportive of the updated listing of HLA‐DQ2·5‐restricted α‐gliadin epitopes reported in Sollid et al. [22]. Whether additional α‐gliadin epitopes restricted by HLA‐DQ8 or DQ2·2 might be identified is an open question, and would require CD patients with these genetic backgrounds to be studied in the future. IL‐2 is, however, likely to be the preferred cytokine for whole blood release assays in these patients, as IL‐2 has recently been invoked in systemic cytokine release that follows gluten challenge in CD patients negative for HLA‐DQ2·5 [29]. Including a cross‐validation using HLA‐DQ2·5‐gluten‐peptide tetramers and flow cytometry would have been valuable to corroborate estimates of responding CD4+ T cells made by Poisson distribution analysis, but due to the availability of suitable samples was not possible. Finally, correlating whole blood IL‐2 release directly with assessment of PBMC by HLA‐DQ2·5‐gluten‐peptide tetramers and flow cytometry will be an important validation of this improved CRA.
Selecting IL‐2 as the biomarker and using an electrochemiluminescence immunoassay to measure IL‐2 improves the sensitivity of a whole blood CRA for CD. The improved test detects responses in treated CD patients who are known to have gluten‐specific CD4+ T cells at frequencies of approximately 1 in 100 000 CD4+ T cells in blood. Larger studies in patients who avoid gluten or are regularly exposed to dietary gluten will be required to understand the clinical utility of this whole blood IL‐2 release assay in CD. Correlating whole blood IL‐2 release directly with assessment of PBMC by HLA‐DQ2·5‐gluten‐peptide tetramers and flow cytometry will be an important validation of this improved CRA.
Disclosures
R. P. A., G. G., S. W., E. S., R. Z., K. E. G., K. M. N., K. E. T., L. J. W. and J. L. D. were employees of ImmusanT, Inc. J. A. T.‐D. has served as an advisor to ImmusanT, Inc. R. P. A. is inventor of Patents, owned or licensed by ImmusanT, Inc, relating to the diagnostic application of gluten challenge.
Author contributions
R. P. A., G. G., L. J. W., J. L. D. and A. J. T.‐D. designed the studies; J. A. T.‐D., K. M. N. and K. E. T. conducted clinical studies; M. Y. H., A. K. R., S. W., R. Z., E. S. and J. L. D. performed and analyzed immune assays; R. P. A. and G. G. provided data integration and analysis; R. P. A. wrote the manuscript and prepared the tables and figures. All authors reviewed and approved the manuscript, tables and figures. The authors made the decision to submit the manuscript for publication and vouch for the accuracy of the data and analyses and for the fidelity of this report to the trial protocol. R. P. A. had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supporting information
Table S1. Whole blood cytokine release using electrochemiluminescence immunoassay to assess gluten peptide‐specific T‐cell responses and point frequencies by Poisson Distribution Analysis in 24‐replicate wells in treated CD patients (Cohort 2) and unaffected controls (Cohort 3).
Fig. S1. IFN‐γ ELISpot images from Subject #103 pre‐ and post‐ 3‐day gluten challenge taken with the AID ELISpot counter.
Fig. S2. Representative proliferation data from Subject #103. Cells were gated on the live (7‐AAD negative) CD3+CD4+ T cells as shown. Replicate well proliferation is shown for control and peptide conditions as a percentage of the CD3+CD4+ T cells (12 out of 24 replicates shown).
Fig. S3. Peak serum cytokine concentration at 3‐hours (IL‐2) or 6‐hours (IFN‐γ and IL‐10) after Cohort 1 CD patients ingested 10‐grams vital wheat gluten ingestion on Day 1 compared to Glia‐α peptide‐stimulated whole blood cytokine release at baseline on Day 1, and on Day 6. Spearman correlation coefficients and associated p values are shown.
Fig. S4. Whole blood cell subset enumeration by flow cytometry using BD Trucount™ tubes for Subject #103 at baseline. Whole blood was stained with an antibody mix comprising anti‐human CD3‐Bv421, CD4‐PE, CD8‐APC, CD14‐APCH7, CD19‐Bv480, CD20‐PECy7, and CD45‐FITC and 7‐AAD. Cell counts per ul whole blood for each gated subset were determined based on the proportion of the known number of beads recorded for each TruCount tube.
Acknowledgements
The authors are grateful to Cathy Pizzey and Lee Henneken, who were the Research Nurses in this study, and to the volunteers who participated in this research. ImmusanT, Inc., Cambridge, MA, USA provided funding for the study.
# Joint senior authors.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, R. P. A., upon reasonable request.
References
- 1. Jabri B, Sollid LM. T cells in celiac disease. J Immunol 2017; 198:3005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sarna VK, Lundin KEA, Morkrid L, Qiao SW, Sollid LM, Christophersen A. HLA‐DQ‐Gluten tetramer blood test accurately identifies patients with and without celiac disease in absence of gluten consumption. Gastroenterology 2018; 154:886–96 e6. [DOI] [PubMed] [Google Scholar]
- 3. Zuhlke S, Risnes LF, Dahal‐Koirala S, Christophersen A, Sollid LM, Lundin KE. CD38 expression on gluten‐specific T cells is a robust marker of gluten re‐exposure in coeliac disease. United Euro Gastroenterol J 2019; 7:1337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hardy MY, Tye‐Din JA. T cells in coeliac disease: a rational target for diagnosis and therapy. Nat Rev Gastroenterol Hepatol 2018; 15:583–584. [DOI] [PubMed] [Google Scholar]
- 5. Pai M, Zwerling A, Menzies D. Systematic review: T‐cell‐based assays for the diagnosis of latent tuberculosis infection: an update. Ann Intern Med 2008; 149:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pathan AA, Wilkinson KA, Klenerman P et al. Direct ex vivo analysis of antigen‐specific IFN‐gamma‐secreting CD4 T cells in Mycobacterium tuberculosis‐infected individuals: associations with clinical disease state and effect of treatment. J Immunol 2001; 167:5217–25. [DOI] [PubMed] [Google Scholar]
- 7. Anderson RP, van Heel DA, Tye‐Din JA et al. T cells in peripheral blood after gluten challenge in coeliac disease. Gut 2005; 54:1217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AVS. In vivo antigen challenge in celiac disease identifies a single transglutaminase‐modified peptide as the dominant A‐gliadin T‐cell epitope. Nat Med 2000; 6:337–42. [DOI] [PubMed] [Google Scholar]
- 9. Ontiveros N, Tye‐Din JA, Hardy MY, Anderson RP. Ex‐vivo whole blood secretion of interferon (IFN)‐gamma and IFN‐gamma‐inducible protein‐10 measured by enzyme‐linked immunosorbent assay are as sensitive as IFN‐gamma enzyme‐linked immunospot for the detection of gluten‐reactive T cells in human leucocyte antigen (HLA)‐DQ2.5(+) ‐associated coeliac disease. Clin Exp Immunol 2014; 175:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goel G, Tye‐Din JA, Qiao SW et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci Adv 2019; 5:eaaw7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daveson AJM, Tye‐Din JA, Goel G et al., Group RCS . Masked bolus gluten challenge low in FODMAPs implicates nausea and vomiting as key symptoms associated with immune activation in treated coeliac disease. Aliment Pharmacol Ther 2020; 51:244–52. [DOI] [PubMed] [Google Scholar]
- 12. Goel G, Daveson AJM, Hooi CE et al. Serum cytokines elevated during gluten‐mediated cytokine release in coeliac disease. Clin Exp Immunol 2020; 199:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tye‐Din JA, Stewart JA, Dromey JA et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010; 2:1–14. [DOI] [PubMed] [Google Scholar]
- 14. Beissbarth T, Tye‐Din JA, Smyth GK, Speed TP, Anderson RP. A systematic approach for comprehensive T‐cell epitope discovery using peptide libraries. Bioinformatics 2005; 21:i29–37. [DOI] [PubMed] [Google Scholar]
- 15. Hardy MY, Tye‐Din JA, Stewart JA et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross‐reactive T cells activated by avenin peptides and immuno‐dominant hordein peptides. J Autoimmun 2015; 56:56–65. [DOI] [PubMed] [Google Scholar]
- 16. Di Lullo G, Ieva F, Longhi R, Paganoni AM, Protti MP. Estimating point and interval frequency of antigen‐specific CD4+ T cells based on short in vitro expansion and improved poisson distribution analysis. PLOS ONE 2012; 7:e42340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox JH, Ferrari G, Janetzki S. Measurement of cytokine release at the single cell level using the ELISPOT assay. Methods 2006; 38:274–82. [DOI] [PubMed] [Google Scholar]
- 18. Quah BJ, Parish CR. New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE‐like fluorescent dyes. J Immunol Methods 2012; 379:1–14. [DOI] [PubMed] [Google Scholar]
- 19. Christophersen A, Raki M, Bergseng E et al. Tetramer‐visualized gluten‐specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. United Euro Gastroenterol J 2014; 2:268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tye‐Din JA, Skodje GI, Sarna VK et al. Cytokine release after gluten ingestion differentiates coeliac disease from self‐reported gluten sensitivity. United Euro Gastroenterol J 2020; 8:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vader LW, de Ru A, van der Wal Y et al. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med 2002; 195:643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sollid LM, Tye‐Din JA, Qiao SW, Anderson RP, Gianfrani C, Koning F. Update 2020: nomenclature and listing of celiac disease‐relevant gluten epitopes recognized by CD4(+) T cells. Immunogenetics 2020; 72:85–8. [DOI] [PubMed] [Google Scholar]
- 23. Gjertsen HA, Sollid LM, Ek J, Thorsby E, Lundin KE. T cells from the peripheral blood of coeliac disease patients recognize gluten antigens when presented by HLA‐DR, ‐DQ, or ‐DP molecules. Scand J Immunol 1994; 39:567–74. [DOI] [PubMed] [Google Scholar]
- 24. Molberg O, Lundin KE, Nilsen EM et al. HLA restriction patterns of gliadin‐ and astrovirus‐specific CD4+ T cells isolated in parallel from the small intestine of celiac disease patients. Tissue Antigens 1998; 52:407–15. [DOI] [PubMed] [Google Scholar]
- 25. Raki M, Fallang LE, Brottveit M et al. Tetramer visualization of gut‐homing gluten‐specific T cells in the peripheral blood of celiac disease patients. Proc Natl Acad Sci USA 2007; 104:2831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christophersen A, Lund EG, Snir O et al. Distinct phenotype of CD4(+) T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med 2019; 25:734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christophersen A, Risnes LF, Dahal‐Koirala S, Sollid LM. Therapeutic and diagnostic implications of t cell scarring in celiac disease and beyond. Trends Mol Med 2019; 25:836–52. [DOI] [PubMed] [Google Scholar]
- 28. Clifford V, Tebruegge M, Zufferey C et al. Cytokine biomarkers for the diagnosis of tuberculosis infection and disease in adults in a low prevalence setting. Tuberculosis 2019; 114:91–102. [DOI] [PubMed] [Google Scholar]
- 29. Tye‐Din JA, Daveson AJM, Goldstein KE et al., Group RCS . Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med 2020; 18:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Whole blood cytokine release using electrochemiluminescence immunoassay to assess gluten peptide‐specific T‐cell responses and point frequencies by Poisson Distribution Analysis in 24‐replicate wells in treated CD patients (Cohort 2) and unaffected controls (Cohort 3).
Fig. S1. IFN‐γ ELISpot images from Subject #103 pre‐ and post‐ 3‐day gluten challenge taken with the AID ELISpot counter.
Fig. S2. Representative proliferation data from Subject #103. Cells were gated on the live (7‐AAD negative) CD3+CD4+ T cells as shown. Replicate well proliferation is shown for control and peptide conditions as a percentage of the CD3+CD4+ T cells (12 out of 24 replicates shown).
Fig. S3. Peak serum cytokine concentration at 3‐hours (IL‐2) or 6‐hours (IFN‐γ and IL‐10) after Cohort 1 CD patients ingested 10‐grams vital wheat gluten ingestion on Day 1 compared to Glia‐α peptide‐stimulated whole blood cytokine release at baseline on Day 1, and on Day 6. Spearman correlation coefficients and associated p values are shown.
Fig. S4. Whole blood cell subset enumeration by flow cytometry using BD Trucount™ tubes for Subject #103 at baseline. Whole blood was stained with an antibody mix comprising anti‐human CD3‐Bv421, CD4‐PE, CD8‐APC, CD14‐APCH7, CD19‐Bv480, CD20‐PECy7, and CD45‐FITC and 7‐AAD. Cell counts per ul whole blood for each gated subset were determined based on the proportion of the known number of beads recorded for each TruCount tube.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, R. P. A., upon reasonable request.


