Summary
Follicular helper T cells (Tfh) cells have been identified in the circulation and in tertiary lymphoid structures in chronic inflammation. Gingival tissues with periodontitis reflect chronic inflammation, so genomic footprints of Tfh cells should occur in these tissues and may differ related to aging effects. Macaca mulatta were used in a ligature‐induced periodontitis model [adult group (aged 12–23 years); young group (aged 3–7 years)]. Gingival tissue and subgingival microbiome samples were obtained at matched healthy ligature‐induced disease and clinical resolution sites. Microarray analysis examined Tfh genes (n = 54) related to microbiome characteristics documented using 16S MiSeq. An increase in the major transcription factor of Tfh cells, BCL6, was found with disease in both adult and young animals, while master transcription markers of other T cell subsets were either decreased or showed minimal change. Multiple Tfh‐related genes, including surface receptors and transcription factors, were also significantly increased during disease. Specific microbiome patterns were significantly associated with profiles indicative of an increased presence/function of Tfh cells. Importantly, unique microbial complexes showed distinctive patterns of interaction with Tfh genes differing in health and disease and with the age of the animals. An increase in Tfh cell responsiveness occurred in the progression of periodontitis, affected by age and related to specific microbial complexes in the oral microbiome. The capacity of gingival Tfh cells to contribute to localized B cell activation and active antibody responses, including affinity maturation, may be critical for controlling periodontal lesions and contributing to limiting and/or resolving the lesions.
Keywords: follicular T cells, gingiva, non‐human primates, periodontitis
This study examined the characteristics of T follicular helper (Tfh) cells occurring in healthy and periodontitis affected gingival tissues using a nonhuman primate model of disease. Comparisons of the Tfh transcriptome were also examined across the lifespan to estimate age and disease effects on these cells. The study also identified oral microbiome bacterial consortia that strongly correlated with alterations in gene expression patterns related to Tfh functions.
Introduction
Follicular helper T cells (Tfh) are a subset of CD4+ helper cells that are involved in the regulation and development of antigen‐specific B cell immunity. The fundamental function of T helper cells is to provide help to B cells and to regulate their proliferation and immunoglobulin class‐switching with antibody affinity maturation. Historically, Tfh cells were considered to be generally localized to the B cell follicle of secondary lymphoid tissues, including tonsils, to interact with germinal center (GC) B cells [1, 2, 3]. Functional studies have demonstrated that Tfh cells provide signals that are essential for survival and proliferation of these GC B cells. Thus, they play a critical role in development of B cell protective immunity against pathogens via immunoglobulin‐switching and recombination to promote increased antibody affinity [2].
Tfh differentiation is a multi‐stage process and their functions depend upon the master regulatory transcription factor B cell lymphoma 6 (Bcl6) [4, 5, 6]. Bcl‐6 also suppresses the expression of factors that promote the differentiation of other CD4+ T helper cell subsets [7]. Canonical secreted Tfh molecules include C‐X‐C motif chemokine ligand (CXCL)13, interleukin (IL)‐21 and IL‐4 that are all important for B cell help. IL‐21 also regulates CD4+ T cell subsets, including the efficient development of Tfh, as well as B cell differentiation and activation. Tfh cells also display crucial cell surface markers, including CXCR5, programmed cell death 1 (PD‐1), inducible T cell co‐stimulator (ICOS) and CD40LG. The early differentiation of Tfh is driven by IL‐6, ICOS and the T cell receptor (TCR). The majority of differentiated Tfh cells are CXCR5hiPD‐1hiBcl6hiMafhiSAPhi and P‐selectin glycoprotein ligand‐1 (PSGL‐1)loCD200+B and T lymphocyte attenuator (BTLA)hiCCR7lo [8]. B cell help signals from Tfh include both cytokines and cell surface receptors [9]. Coupled with CD40LG, both PD‐1 and ICOS bind to ligands on the B cell surface to regulate the functions of both the B cells and Tfh cells, with direct co‐stimulation of B cells via CD40LG and production of IL‐21 driving B cell proliferation. The CXCR5 receptor enhances chemoattraction and migration of the Tfh cells into GC B cell zones [10, 11, 12].
Of particular interest to this study is that both circulating and tissue resident Tfh cells can be detected that produce cytokines to modulate antibody responses. Moreover, tissues including the periodontium that contain antigens in a milieu of chronic immune responses, are often infiltrated by T cells, macrophages, B cells and plasmacytes [13]. These cellular elements can organize themselves anatomically and functionally in a manner similar to secondary lymphoid organs [13, 14, 15]. This type of cellular infiltrate and functions have been identified for Tfh cells in the periodontium [16, 17], nasal [18] and gastrointestinal tract [19, 20] mucosa related to immune responses and disease outcomes. Furthermore, a number of reports have documented important aging effects on Tfh cell numbers and functions that can negatively impact adaptive immunity [21, 22, 23, 24]. As the gingival tissues with periodontitis reflect this type of aging microenvironment, we hypothesized that with development and progression of periodontitis, genomic footprints of Tfh cells would occur in the gingival tissues and may differ related to aging effects. It was also predicted that certain bacterial components of the complex oral microbiome would have the capacity to aid in triggering or dampening the various molecules required for functional Tfh cells in the gingival tissues. Examination of the characteristics of gene expression representing features of Tfh cells in the gingival tissues was implemented as reported for cellular transcriptomic studies in other systems [25, 26, 27, 28, 29, 30]. However, a limitation of the use of this approach to examine tissue level presentation of Tfh genes is that a number of these gene transcripts would also be produced by other cell types that reside in the gingival tissues.
Methods
Animals and diet
Rhesus monkeys (Macaca mulatta) (n = 36; 17 male, 19 female) housed at the Caribbean Primate Research Center at Sabana Seca, Puerto Rico were examined for periodontal health [31, 32, 33]. Adult animals (aged 12–23 years; n = 18) and young animals (aged 3–7 years; n = 18) were included in the study. The non‐human primates were fed a 20% protein, 5% fat and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified; Harlan Teklad, Madison, WI, USA). The diet was supplemented with fruit and vegetables, and water was provided ad libitum in an enclosed corral setting.
As we have reported previously, the protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico and a ligature disease model was utilized [34]. The clinical examination included probing pocket depth (PPD) and bleeding on probing (BOP; 0–5 scale) [35]. Periodontal health was defined by mean pocket depth (PD) ≤ 3·0 mm and mean bleeding on probing (BOP) ≤ 1 (0–5 scale) in a full mouth examination, excluding third molars and canines [34]. Ligature‐induced periodontal disease was initiated as we have previously reported [34] and gingival and subgingival plaque samples taken at 0·5, 1 and 3 months (initiation/progression) and 2 months after removal of ligatures and local factors (resolution). Determination of periodontal disease at the sampled site was documented by assessment of the presence of BOP and probing pocket depth of > 4 mm, as we have described previously [32].
Microbiome analysis
Subgingival bacterial samples were obtained from the 36 animals by a curette and analyzed using a MiSeq instrument [36, 37] for the total composition of the microbiome from each sample [38, 39]. Sequences were clustered into phylotypes based on their sequence similarity and these binned phylotypes were assigned to their respective taxonomic classification using the Human Oral Microbiome Database (HOMD V13) (http://www.homd.org/index.php?name=seqDownload &file&type = R), as we have described previously [40]. Raw data were deposited at the NIH NCBI (BioProject ID PRJNA516659). Statistical differences of bacterial operational taxonomic units (OTUs) were determined with a t‐test (P < 0·05). Correlations of OTUs within the oral microbiome were determined using a Pearson’s correlation coefficient analysis (P < 0·05). Correlations between the microbiome components and the gingival gene expression were determined only for matching samples derived from the same tooth in each of the animals. Matching samples with sufficient microbiome signals were compared for 58 samples in adults and 25 samples from the young group obtained at health and throughout the ligature model. As we have reported previously [40], of 396 OTUs identified in the non‐human primate oral samples the targeted OTU selection for this study was 58 for the adult samples that covered 88% of reads in all samples. Similarly, 49 OTUs were examined in the young samples that covered 91% of reads in all samples. The microbiome data have been deposited at BioProject ID PRJNA516659 through the NIH NCBI.
Gingival tissue sample collection and mRNA analysis
Gingival tissue samples of healthy and disease sites were surgically collected and total RNA extracted for microarray analysis via hybridization to the GeneChip® Rhesus Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, USA), similar to methods we have described previously [32, 41, 42, 43, 44].
Data analysis
A set of Tfh‐related genes (n = 54) (Table 1) were targeted in this study. The expression intensities across the samples were estimated using the robust multi‐array average (RMA) algorithm with probe‐level quintile normalization, as implemented in the Partek Genomics Suite software version 6.6 (Partek, St Louis, MO, USA). The age groups were initially compared using one‐way analysis of variance (anova). For genes that had significant mean differences, two‐sample t‐tests were used to investigate differences. A Pearson’s correlation analysis was used for the microbiome members with Tfh gene expression in matched samples. Z‐scores were calculated for each gene among all samples for each individual gene [Z = (x–µ)/σ)] with x gene expression value; µ means among the samples; and σ represents standard deviation among all samples. Statistical significance was considered by a P‐value < 0·05. The data have been uploaded into the ArrayExpress data base (www.ebi.ac.uk) under Accession number: E‐MTAB‐1977.
Table 1.
T follicular helper cell gene expression targets and lineage‐specific transcription factors for T helper cell subsets
| Gene ID | Fxn | Gene ID | Fxn |
|---|---|---|---|
| ASCL2 | Achaete–scute family BHLH transcription factor 2 | IL‐4 | Interleukin 4 |
| BATF | Basic leucine zipper ATF‐like transcription factor | IL‐6RA | IL‐6 cytokine receptor |
| BCL6 | BCL6 transcription repressor; master transcription factor Tfh | IL‐7R | IL‐7 cytokine receptor |
| BTLA | B and T lymphocyte associated (CD272) attenuator; inhibitory receptor on lymphocytes | IL‐10 | Interleukin 10 |
| CCR7 | C‐C motif chemokine receptor 7 | IL‐12 | Interleukin 12 |
| CD3D | TCR component; binds MHC‐II on APCs | IL‐17A | Interleukin 17A |
| CD3E | TCR component; binds MHC‐II on APCs | IL‐17F | Interleukin 17F |
| CD3G | TCR component; binds MHC‐II on APCs | IL‐21 | Interleukin 21; cytokine with immunoregulatory activity |
| CD4 | Essential role in immune responses; subset of T cells | IL‐21R | IL‐21 cytokine receptor |
| CD10/MME | Neprilysin; membrane metallopeptidase | IL‐23 | Interleukin 23; part of IL‐12 family of cytokines |
| CD40LG | T cell surface ligand; binds CD40 on B cells | IRF4 | Interferon regulatory factor 4 |
| CD57/B3GAT1 | Biosynthesis of L2/HNK‐1 carbohydrate epitope on glycoproteins | KCNK5 | Potassium two pore domain channel subfamily K member 5 |
| CD84/SLAMF5 | Signaling lymphocyte activation molecule (SLAM) family | c‐maf/MAF | Oncogene; acts as a transcriptional activator or repressor |
| CD150/SLAMF1 | Signaling lymphocyte activation molecule (SLAM) family | MYO7A | Myosin VIIA |
| CD200 | OX‐2; co‐stimulates T cell proliferation | PD‐1/PD‐CD1 | Inhibitory receptor on antigen activated T cells; binds to PD‐1L on B cells |
| CORO1B | Coronin 1B | PLCG2 | Phospholipase C gamma 2 |
| CXCL13 | C‐X‐C motif chemokine ligand 13 | RC3H1 | Ring finger and CCCH‐type domains 1 |
| CXCR4 | C‐X‐C motif chemokine receptor 4 | RC3H2 | Ring finger and CCCH‐type domains 2 |
| CXCR5 | C‐X‐C motif chemokine receptor 5 | RORA | RAR‐related orphan receptor A; master transcription factor Th17 |
| EBI2/GPR183 | G protein‐coupled receptor 183 | RORC | RAR‐related orphan receptor C; master transcription factor Th17 |
| FAM43A | Family with sequence similarity 43 member A | S1PR1 | Sphingosine‐1‐phosphate receptor 1 |
| FOXP1 | Forkhead box protein 1 | SELPLG/PSGL1 | Selectin P ligand |
| FOXP3 | Forkhead box protein 3; master transcription factor Treg | SH2D1A/SAP | SH2 domain containing 1A; adapter regulating receptors of the signaling lymphocytic activation molecule (SLAM) |
| FOXO1 | Forkhead box O1; master transcription factor Th9 | STAT‐3 | Signal transducer and activator of transcription 3; mediates cellular responses to interleukins |
| GATA3 | GATA binding protein 3; master transcription factor Th2 | TGF‐B1 | Transforming growth factor beta 1 |
| GNG4 | G protein subunit gamma 4 | T‐bet/TXB21 | T‐box transcription factor 21; master transcription factor Th1 |
| ICOS | Inducible T cell co‐stimulator; binds ICOSL on B cells | TNF‐RSF18 | TNF receptor superfamily member 18 |
| IFNG | Interferon gamma | TOX2 | TOX high‐mobility group box family member 2 |
| IL1R1 | Interleukin 1 receptor type 1 | TRIM8 | Tripartite motif containing 8 |
| IL2RA | IL‐2 cytokine receptor |
Genes in bold type represent the subset combined for Tfh footprint analysis in the tissues.
TCR = T cell receptor; APC = antigen‐presenting cells; MHC‐II = major histocompatibility complex class II; ICOSL = inducible co‐stimulator‐ligand.
Results
Tfh gene expression profiles with age and periodontitis
We have previously documented significant differences in clinical findings of inflammation (BOP) and tissue destruction (PPD) with ligature‐induced periodontitis in younger and older non‐human primates [45]. Briefly, both groups of animals demonstrate a rapid increase in inflammation within the first 2 weeks (0·5 months) of ligature placement. Generally, BOP indices increase through 3 months of progressing periodontitis, and following removal of the ligatures within 60 days this inflammatory measure approaches baseline healthy tissues. In contrast, PPD measures were significantly different between the young and adult animals. The rate of PPD increase was slower in the young animals and the maximal PPD levels were significantly lower in the young group throughout the disease process. Thus, clinical differences suggest a potential for variations in the profiles of host responses that would be critical in contributing to and mitigating the disease process.
Figure 1 summarizes fold‐changes in gene expression for the 58 Tfh‐associated genes that were examined in gingival tissues from young (Fig. 1a) and adult (Fig. 1b) animals compared to baseline healthy tissue samples. The result demonstrated that a number of the Tfh‐related genes, including surface receptors, secreted products and transcription factors, were significantly increased during disease. In the young animals, striking increases were observed with BTLA, CXCL13 and interferon regulatory factor 4 (IRF4) expression levels, while IFNG, CD40LG and CD200 were all significantly decreased. There was considerable overlap in altered gene expression levels in the young and adult samples. BTLA, CXCL13 and IRF4 were also increased and CD200 decreased with disease in the adult animals. Also of note was the increase in B cell lymphoma 6 (BCL6) in both groups primarily at the 3‐month time‐point late in disease progression. The overall pattern of Tfh gene changes in both groups presented at 1 and 3 months during disease progression.
Fig. 1.
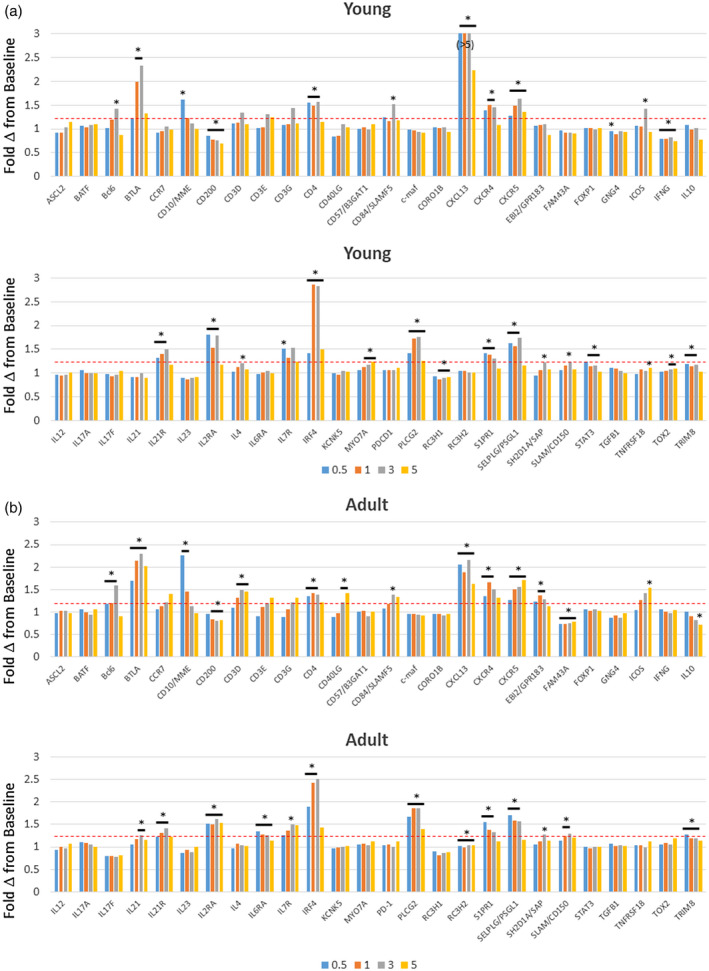
Expression levels of follicular T helper (Tfh) genes in gingival tissues from young (a) and adult (b) animals. The data are expressed as fold‐change in expression with disease (0.5, 1, 3 months) and resolution (5 months) compared to baseline healthy samples for each animal. The bars denote group means. The asterisk and horizontal bracket signifies significant difference from baseline at P < 0·01.
Figure 2 provides a summary of gene expression changes targeting the master regulatory transcription genes for each of the subsets of T helper cells. In both the adult and young gingival tissues samples the primary increase in transcription factor expression was with BCL6 for Tfh cells, particularly in the latter phases of disease progression, with levels returning to baseline in resolution samples. Also of note was the significant decrease in RAR‐related orphan receptors (RORA) in both age groups, and the decrease in retinoid‐related orphan receptor‐gamma (RORC) (RORγ) in the young animal tissues through disease initiation and progression. The T helper type 1 (Th1) transcription factor, T‐bet/TXB21, expression was found to be increased only in the young resolution samples. These results support some unique features of the characteristics of the T cell infiltrate, including Tfh cells, in gingival tissues with the initiation and progression of a periodontal lesion probably linked to both local regulatory features of immune responses, as well as setting the stage for enhanced B cell co‐operation and humoral immune responses occurring in progressing lesions [46].
Fig. 2.
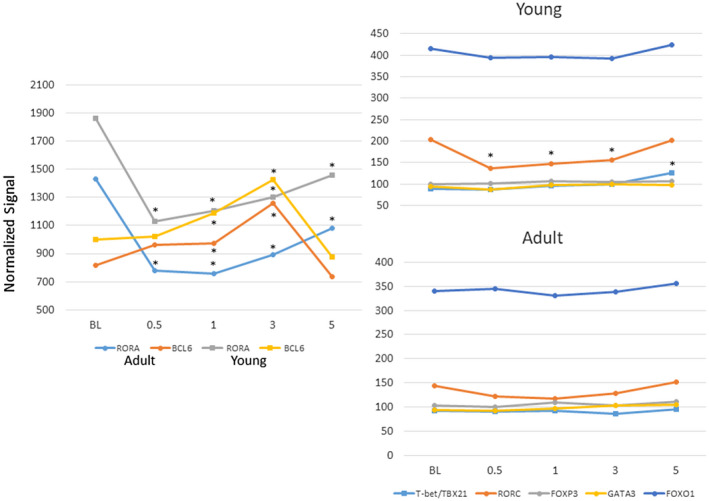
Expression of master transcription factor genes for various subsets of CD4+ T helper cells [T‐bet/TXB21–T helper type 1 (Th1), GATA3–Th2, RORα/γ–Th17, forkhead box (Fox)O1–Th9, FoxP3–regulatory T cells (Treg), BCL6–follicular T helper (Tfh) cells]. Points denote group means for young and adult animals at each time‐point expressed as fold change from baseline. Asterisk denotes significantly different from baseline at P < 0·01.
Gene expression comparison in adult and young samples
Figure 3a–e provides an analysis of the array of gene changes comparing responses in adult and young animals in health (BL), disease (0·5, 1, 3 months) and resolution (5 months). The data were transformed using a Z‐score to normalize across all adult and young samples and across all data points for each gene. In the adult animals a group of surface receptors associated with Tfh cells generally appeared to increase throughout disease progression and resolution (Fig. 3a), while changes that occurred in the young samples were primarily increases at 3 months of disease progression. However, in both young and adults, increased CD4 expression was observed at disease initiation. Examination of a second group of surface receptors (Fig. 3b) showed increased levels of CD10, sphingosine‐1‐phosphate receptor 1 (S1PR1), P‐selectin glycoprotein (SELPG), IL‐6RA by 0·5 months (initiation) and a subset that increased at 0·5 and continued to increase through 1 month of progression [BTLA, IL‐7R, IL‐21R, G‐protein coupled receptor 183 (GPR183), IL‐2RA] in adult animals. Similarly, CD10, S1PR1, SELPG and IL‐2RA increased in the young samples by 0·5 months, generally decreasing thereafter. BTLA showed increased levels at 3 months. Importantly, this tissue level approach to delineating characteristics of Tfh cells in the gingiva has limitations regarding the recognition that a number of these receptors can be expressed by other cell types although, within the gingival tissues in this experimental model, there is a lack of data demonstrating the breadth of cell types that display or alter expression of these receptors. Nevertheless, the somewhat different patterns of these receptors could reflect various cell types contributing to the transcript readout. We propose that, while all these receptors exist on the Tfh cells, the cellular interactions with the bacteria and other host factors within the gingival tissue milieu may actually drive kinetic differences in the magnitude of the gene expression for these receptors. Moreover, while there were somewhat different kinetics in the expression patterns, the large majority of the receptor genes increased in the tissues with disease in the adult and young samples, suggesting similar cellular responses to the microbial burden and disease process.
Fig. 3.
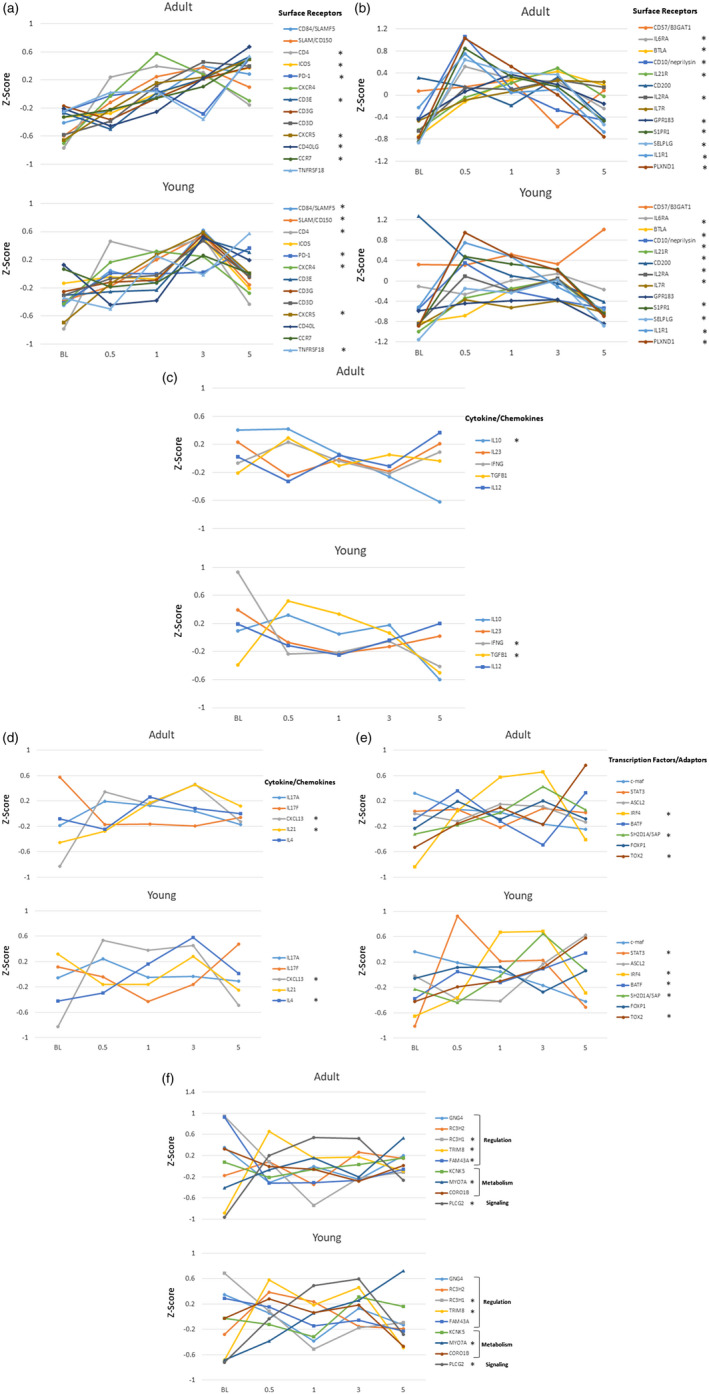
Z‐score values for follicular T helper (Tfh) surface receptors (a,b), cytokines/chemokines (c,d), transcription factors/adaptors (e) and regulatory/signaling/metabolism genes (f) in adult and young animal gingival samples obtained in health (BL), during disease (0·5, 1, 3 months) and clinical resolution (5 months). Points denote normalized group means comparing Z‐scores for expression of a specific gene across all animals and time‐points. The asterisk (*) in the legend denotes significant changes in gene expression from baseline at one or more of the disease time‐points and/or resolution.
Figure 3c,d presents a similar analysis for T cell‐associated cytokine/chemokine expression. In adults, only IL‐10 decreased significantly from baseline, while IL‐21 and CXCL13 each increased during the disease process. In comparison with the young samples, interferon gamma (IFNG) decreased with disease and transforming growth factor beta 1 (TGFB1), CXCL13 and IL‐4 were increased in disease samples. The expression of crucial additional transcription factors and adaptor molecules for Tfh functions are shown in Fig. 3e. Both IRF4 and SH2D1A/SAP showed substantial increases as disease progressed and basic leucine zipper ATF‐like transcription factor (BATF) increased with disease resolution in both adults and young samples. Signal transducer and activator of transcription 3 (STAT‐3) demonstrated a significant increase at disease initiation, with achaete–scute complex homolog 2 (ASCL2) significantly elevated at disease resolution in the young animal samples.
Finally, Fig. 3f summarizes changes in important regulatory, metabolism and signaling genes related to Tfh functions. Generally, the expression patterns of these genes was similar in adult and young samples. Striking changes were noted in tripartite motif containing 8 (TRIM8) increases and ring finger and CCCH‐type domains 1 (RC3H1) decreases with disease initiation. Additionally, phospholipase C gamma 2 (PLCG2) increased significantly with disease, while myosin VIIA (MYO7A) increased with disease and continued in resolution samples.
Examination of the existing literature provided a subset of 12 genes to provide a more directed description of changes in Tfh infiltrates or functional activities [47, 48, 49] in the gingival tissues, similar to approaches in complex tissues using molecular deconvolution approaches [25, 26, 27, 28, 29, 30]. Figure 4 depicts individual adult and young animal gingival expression of the composite sum of transcript levels for these 12 genes. The results showed similarities in Tfh gene expression profiles in healthy adult and young tissues. Significant increases occurred in both young and adult Tfh gene patterns with disease initiation and remained elevated with disease progression. This Tfh‐associated composite gene profile generally returned to baseline in both groups in the resolution samples. Supporting information, Table S1 summarizes the changes in responses of each of the Tfh‐related composite genes in adult and young animals. The results demonstrate a pattern for most of these genes with lower levels in healthy and resolution tissues, with increases during disease initiation and progression. Some differences in the temporal changes of elevated expression were noted for the individual genes.
Fig. 4.
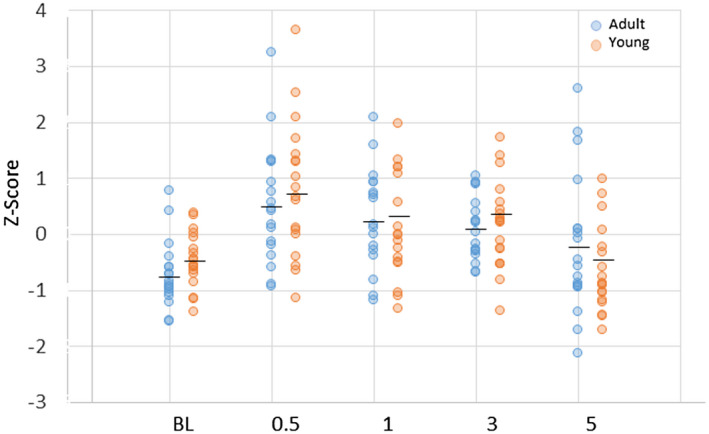
Z‐score summation of a composite of 12 follicular T helper (Tfh)‐associated genes to provide a footprint analysis in the gingival tissues. Each point denotes the value for an animal sample at each time‐point. The horizontal bar designates the group mean. No statistical differences were seen between levels in the adult versus young group at any time‐point. P‐value identifies comparison of gene expression profile at each time‐point compared to baseline levels in each age group.
Based upon analysis of this model of early lesion development and progression, significant differences were noted in the expression of an array of surface receptors in the gingival tissues of both young and adult animals. CD4 increased in both groups with disease initiation and progression consistent with an increase in T helper cells in the gingival tissues. However, generally two patterns of sets of Tfh receptors either showed increases throughout disease progression and even resolution, particularly in the adult group, and a second grouping of the receptors demonstrated increases in expression occurring early in the disease process, and then plateauing or decreasing in both young and adult samples. The reflection of Tfh activities via targeted cytokine/chemokine expression was more limited and differed somewhat between the adult and young samples. Finally, an array of important transcription factors beyond BCL6 were altered, particularly in young samples, although a similar pattern for IRF4 and SH2S1A/SAP was observed in both age groups. These data suggested a probable increase in the numbers of infiltrating Tfh cells with disease development and progression and a role for specific transcription factors in affecting Tfh functions, although a somewhat limited change in gene expression for Tfh‐associated cytokines and chemokines within this gingival milieu.
Profiles of microbiome and host Tfh gene expression
Previously, we had identified that 58 OTUs comprised 79·7% and 81·5% of the signals in the adult and young animals, respectively, of the 396 OTUs that were detected across the non‐human primate microbiome specimens. Thus, we focused upon a comparison of Tfh gene expression related to this panel of dominant OTUs within the oral microbiome. Figure 5a–c provides a heat‐map of the overall bacterial–gene correlations, and highlight specific bacterial complexes associated with the Tfh identifying subset of gene expression profiles in adults. The complexes that showed significant correlations with the composite profile of the 12 Tfh genes were specifically identified. In healthy adults, two microbial complexes were significantly positively or negatively correlated with the Tfh footprints (Table 2). Similarly, two bacterial clusters were significantly associated with the Tfh footprints with disease and resolution in the adult samples. Of note, within the framework of the 58 OTUs examined, only 19–24% of the OTUs appeared to be correlated with the Tfh composite gene expression.
Fig. 5.
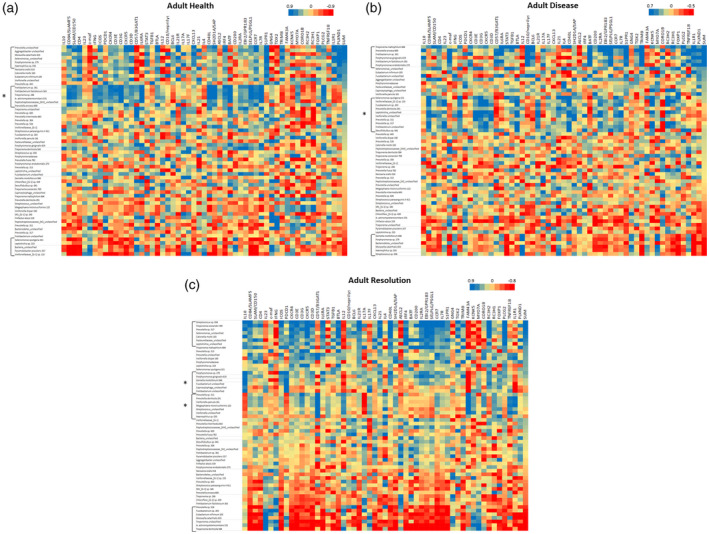
Heat‐map of correlations between bacterial operational taxonomic units (OTUs) relative abundance and individual gene expression adult healthy (a), diseased (b) and resolution (c) samples. Data are sorted based upon SUM that represents a summation of transcript levels for the composite of 12 follicular T helper (Tfh) genes. Brackets identify bacterial clusters correlated positively or negatively with the composite Tfh gene footprint. Brackets with asterisk (*) identify bacterial clusters broadly correlated with an array of the Tfh genes but not directly correlated with the composite profile.
Table 2.
Bacterial operational taxonomic units (OTUs) correlated with composite follicular T helper (Tfh) profile genes (SUM; Figs 5 and 6) in adult and young healthy, diseased, and resolution samples. The asterisk (*) identifies bacterial clusters strongly correlated with an array of the Tfh genes but not directly correlated with the composite profile. Mixed denotes significant positive and negative correlations with Tfh genes
| Sample | Health | Disease | Resolution |
|---|---|---|---|
| Adult | |||
| Positive | Prevotella unclassified | Treponema maltophilum 664 | Streptococcus sp. 058 |
| Aggregatibacter unclassified | Prevotella enoeca 600 | Treponema socranskii 769 | |
| Moraxella catarrhalis 833 | Fretibacterium sp. 361 | Prevotella sp. 317 | |
| Selenomonas_unclassified | Porphyromonas gingivalis 619 | Selenomonas_unclassified | |
| Porphyromonas sp. 279 | Fretibacterium fastidiosum 363 | Catonella morbi 165 | |
| Haemophilus sp. 035 | Porphyromonas endodontalis 273 | Pasteurellaceae_unclassified | |
| Leptotrichia_unclassified | |||
| Positive* | Fretibacterium sp. 361 | Selenomonas sputigena 151 | |
| Fretibacterium fastidiosum 363 | Veillonellaceae_[G‐1] sp. 155 | ||
| Treponema sp. 246 | Fusobacterium sp. 203 | ||
| A. actinomycetemcomitans 531 | Prevotella denticola 291 | ||
| Peptostreptococcaceae_[XIII]_ | Leptotrichia_unclassified | ||
| unclassified | Veillonella unclassified | ||
| Prevotella enoeca 600 | Prevotella sp. 311 | ||
| Prevotella sp. 317 | |||
| Fretibacterium unclassified | |||
| Desulfobulbus sp. 041 | |||
| Mixed* | Porphyromonas sp. 279 | ||
| Porphyromonas gingivalis 619 | |||
| Gemella morbillorum 046 | |||
| Fusobacterium unclassified | |||
| Capnocytophaga_unclassified | |||
| Fretibacterium unclassified | |||
| Mixed* | Prevotella sp. 311 | ||
| Prevotella denticola 291 | |||
| Veillonella parvula 161 | |||
| Megasphaera micronuciformis 122 | |||
| Streptococcus_unclassified | |||
| Veillonella unclassified | |||
| Haemophilus sp. 035 | |||
| Negative | Selenomonas sputigena 151 | Gemella morbillorum 046 | Prevotella sp. 526 |
| Leptotrichia sp. 223 | Porphyromonas sp. 279 | Fusobacterium sp. 203 | |
| Bacteria_unclassified | Bacteroidetes_unclassified | Eubacterium infirmum 105 | |
| Pyramidobacter piscolens 357 | Moraxella catarrhalis 833 | Moraxella catarrhalis 833 | |
| Veillonellaceae_[G‐1] sp. 155 | Haemophilus sp. 035 | Treponema unclassified | |
| Streptococcus sp. 058 | A. actinomycetemcomitans 531 | ||
| Treponema denticola 584 | |||
| Young | |||
| Positive | Haemophilus sp. 035 | Prevotella sp. 317 | Prevotella enoeca 600 |
| Capnocytophaga_unclassified | A. actinomycetemcomitans 531 | Selenomonas_unclassified | |
| Moraxella catarrhalis 833 | Veillonella parvula 161 | Catonella morbi 165 | |
| Prevotella_unclassified | Moraxella catarrhalis 833 | SR1_[G‐1]sp. 345 | |
| Eubacterium_[XI][G‐1]infirmum 105 | Prevotella sp. 317 | ||
| Filifactor alocis 539 | Moraxella catarrhalis 833 | ||
| Positive* | Treponema sp. 246 | ||
| Fretibacterium_unclassified | |||
| Leptotrichia_unclassified | |||
| Porphyromonas endodontalis 273 | |||
| Positive* | Peptostreptococcaceae_[XIII]_ | ||
| unclassified | |||
| Bacteroidetes_unclassified | |||
| Treponema_unclassified | |||
| Pyramidobacter piscolens 357 | |||
| Mixed* | Veillonellaceae_[G‐1]_unclassified | ||
| Prevotella sp. 820 | |||
| Fretibacterium sp. 361 | |||
| Prevotella sp. 526 | |||
| Porphyromonas gingivalis 619 | |||
| Eubacterium_[XI][G‐1]infirmum 105 | |||
| Leptotrichia sp. 223 | |||
| Chloroflexi_[G‐1]sp. 439 | |||
| Fretibacterium_unclassified | |||
| Fretibacterium fastidiosum 363 | |||
| Fusobacterium_unclassified | |||
| Filifactor alocis 539 | |||
| Prevotella sp. 304 | |||
| Prevotella_unclassified | |||
| Negative* | Prevotella sp. 526 | Eubacterium_[XI][G‐1]infirmum 105 | Peptostreptococcaceae_[XIII]_ |
| Bacteria_unclassified | Prevotella sp. 313 | unclassified | |
| Chloroflexi_[G‐1]sp. 439 | Prevotella sp. 820 | Treponema denticola 584 | |
| Selenomonas_unclassified | Veillonellaceae_[G‐1]_unclassified | Treponema maltophilum 664 | |
| Veillonellaceae_[G‐1]sp. 155 | Prevotella intermedia 643 | Treponema_unclassified | |
| Prevotella sp. 820 | A. actinomycetemcomitans 531 | ||
| Prevotella sp. 317 | |||
| Veillonella parvula 161 | |||
| Pyramidobacter piscolens 357 | |||
| Prevotella sp. 304 | |||
| Negative | Bacteroidetes_unclassified | Catonella morbi 165 | Streptococcus parasanguinis_II 411 |
| Porphyromonas endodontalis 273 | Bacteria_unclassified | Prevotella fusca 782 | |
| Leptotrichia_unclassified | Treponema maltophilum 664 | Pasteurellaceae_unclassified | |
| Fusobacterium sp. 203 | Fretibacterium sp. 361 | Gemella morbillorum 046 | |
| Selenomonas sputigena 151 | Streptococcus parasanguinis_II 411 | ||
| Fretibacterium fastidiosum 363 | |||
| Selenomonas_unclassified | |||
| Fusobacterium_unclassified | |||
| Filifactor alocis 539 |
Figure 6a–c summarizes the bacterial clusters that correlate with the composite Tfh gene footprint and individual Tfh‐related genes in samples from young animals. As noted with the adult specimens, specific bacterial clusters significantly correlated with the Tfh gene composite (Table 2). While there were specific microbial clusters that correlated with the Tfh footprint genes, we also observed in the healthy young group that a major microbial complex was significantly negatively correlated with a wide array of individual Tfh‐related genes, although not specifically related to the Tfh composite portfolio (Table 2). Additionally, in resolution samples, there was a large complex of bacteria that correlated with both the Tfh composite footprint, as well as showing a broad reflection for an array of individual Tfh‐related genes. Finally, in these resolution samples a cluster of bacterial species showed a broad negative correlation with a large group of Tfh‐related genes in the absence of correlation with the Tfh composite footprint. In contrast to the somewhat limited microbial representation in the clusters in adult samples, 39 of 58 of these dominant OTUs were represented in these Tfh‐correlated clusters in the young animals.
Fig. 6.
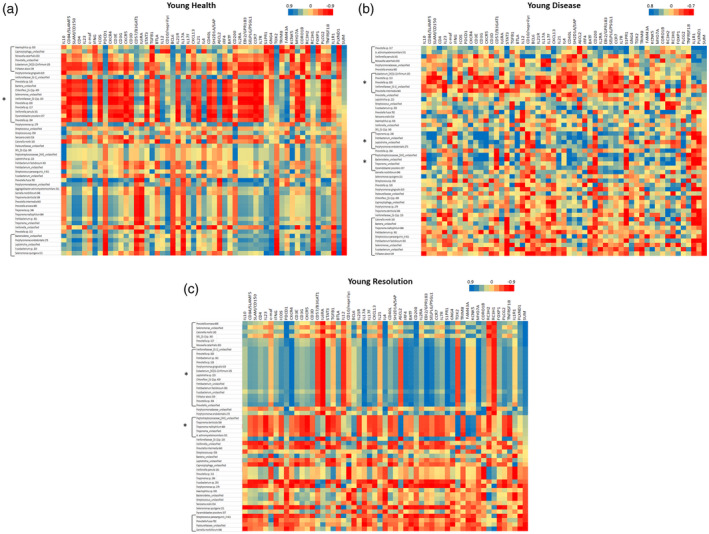
Heat‐map of correlations between bacterial operational taxonomic units (OTUs) relative abundance and individual gene expression young healthy (a), diseased (b) and resolution (c) samples. Data are sorted based upon SUM that represents a summation of transcript levels for the composite of 12 follicular T helper (Tfh) genes. Brackets identify bacterial clusters correlated positively or negatively with the composite Tfh gene footprint. Brackets with asterisk (*) identify bacterial clusters broadly correlated with an array of the Tfh genes but not directly correlated with the composite profile.
Focusing upon the individual bacterial OTUs, Fig. 7a,b depicts the dominant OTUs that were correlated with the 58 Tfh genes in both age groups and through health, disease and resolution. In healthy adult samples, only seven of the dominant OTUs showed 10 or greater correlations, with some predominantly positive (e.g. Prevotella_unclassified, Aggregatibacter_unclassified) and some skewed towards negative correlations [e.g. Veillonella (G‐1), P. piscolens 357]. With disease, this number increased to 13, with disease generally representing a different group of bacteria. In samples from resolution sites, a similar number of bacteria showed this magnitude of correlations although, in contrast to the predominantly positive correlations in disease, a substantial number of negative correlations were seen, particularly related to Treponema_unclassifed, Prevotella sp. 526, A. actinomycetemcomitans 531, T. denticola 584, Fusobacterium sp. 203 and E. infirmum 105.
Fig. 7.
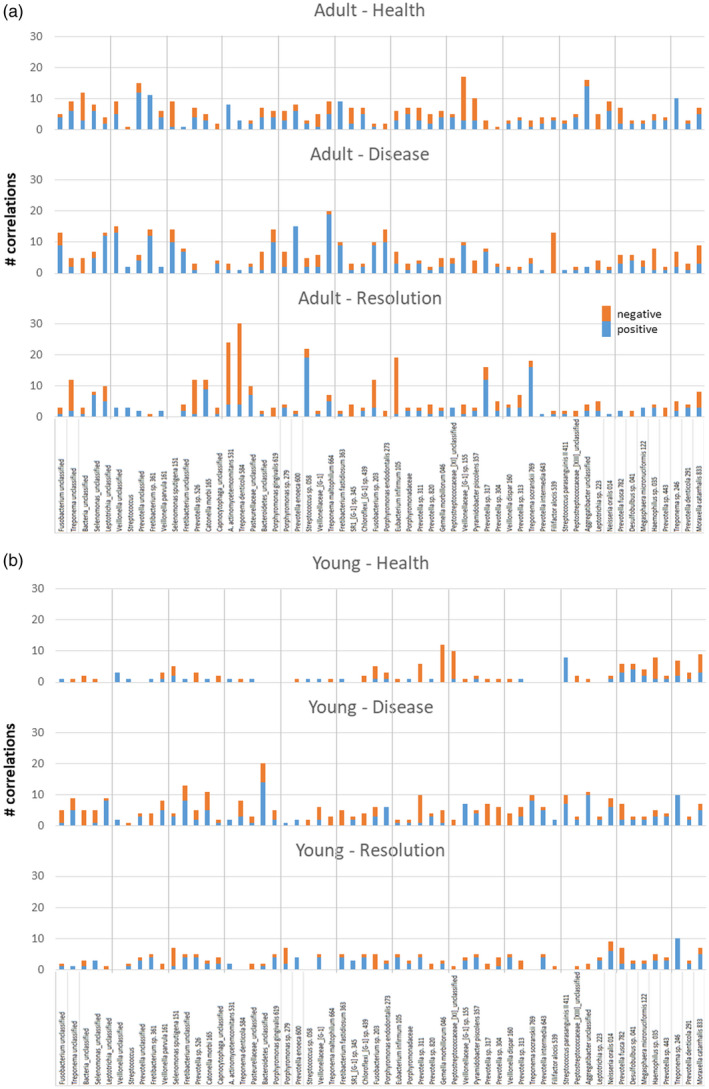
Summary of the number of follicular T helper (Tfh)‐related genes significantly correlated with the individual operational taxonomic units (OTUs) in the microbiomes of adult (a) or young (b) samples. Correlations stratified based upon healthy, diseased or resolution samples and identifying the number of both positive and negative correlations.
A similar analysis with the young samples (Fig. 7b) showed a very different pattern. In health and resolution samples only two and one, respectively, of the OTUs exhibited an elevated number of correlations. However, with disease samples, eight OTUs, including Fretibacterium_unclassifed, Catonella morbi 165, Bacteroidetes_unclassified, Prevotella sp. 311, T. socranskii 789, S. parasanguinis 411, Aggregatibacter_unclassified and Treponema sp. 246 showed an elevated frequency of significant correlations with Tfh‐related genes.
This approach provided a summary picture regarding the potential host–bacterial interactions that occur in the adult and young samples related to Tfh cell responses. It demonstrated unique features of these relationships in health versus disease, as well as distinctive age effects on the specifics of the oral microbial pattern related to changes in Tfh numbers or functions in the juxtaposed gingival tissues.
Microbiome and host Tfh gene expression in adult and young animal samples
Supporting information, Table S2 provides a summary of the microbiome relationships to gingival tissue transcriptome in samples from the two groups of animals. The data present complexes of bacterial OTUs that were significantly correlated with individual genes related to Tfh phenotype and functions. An initial comparison of the Tfh transcriptome genes and the microbiome among all adult and young samples demonstrated few significant correlations within the potential 2320 comparisons, with only 2% showing a significant correlation (data not shown). However, the complexes and relationships were substantially different in health, disease and resolution in adult or young animal samples. Additionally, the same complexes were significantly related to multiple Tfh genes with both positive and negative correlations (see examples Tfh‐H3A, Tfh‐R4A), while during disease the complexes were only positively correlated with gene expression levels. Similarities on the component members of the complexes showed some overlap in adults (see Tfh‐H1A/Tfh‐D2A; Tfh‐H4A/Tfh‐D3A), although these similarities did not appear in the microbiome complexes in the young animals. Moreover, minimal overlap was noted in the complexes between adult and young animals. In adult animals, the complexes were associated with multiple genes in health, disease and resolution; however, in the young samples, only with the disease‐associated microbiome were multiple gene correlations generally observed. Finally, IL‐6RA, CD84 and CD10 were the Tfh‐related genes showing the most frequent correlations with the microbial complexes. IL‐6RA and CD84 were predominantly associated with the microbiome in adults, while CD10 related to microbiome complexes in both adult and young samples.
In addition to exploring the relationship between bacterial complexes and profiles of host Tfh genes with related abundance or expression levels, we identified the individual bacterial OTUs that predominated among these correlations (Table 3). The results showed that multiple bacteria historically associated with changes in the disease microbiota, including Fretibacterium species, Treponema species, Porphyromonas species and Prevotella species demonstrated a high frequency of correlations with Tfh genes. Also of note, Haemophilus, Leptotrichia and G. morbillorum, generally considered as commensals, also demonstrated an elevated prevalence of correlations with the Tfh gene patterns. A number of the bacteria also appeared to primarily correlate with Tfh gene expression in adult samples compared to young samples. Finally, the was a predominance of individual bacterial–host correlations in resolution samples compared to health or disease in the young animals.
Table 3.
Significant correlations of individual bacterial operational taxonomic units (OTUs) with follicular T helper (Tfh) genes in samples from both age groups and in health, disease and resolution site
| Bacteria | Adult | Young | Total | ||||
|---|---|---|---|---|---|---|---|
| Disease | Health | Resolution | Disease | Health | Resolution | ||
| Fretibacterium sp. 361 | + | + | + | + | + | + | 6 |
| Fretibacterium fastidiosum 363 | + | + | + | + | + | + | 6 |
| Haemophilus sp. 035 | + | + | − | + | + | + | 5 |
| Treponema unclassified | + | + | + | − | + | + | 5 |
| Fretibacterium unclassified | + | − | + | + | + | + | 5 |
| Prevotella sp. 526 | − | + | + | + | + | + | 5 |
| Porphyromonas gingivalis 619 | + | + | + | + | − | + | 5 |
| Chloroflexi_[G‐1] sp. 439 | − | + | + | + | + | + | 5 |
| Fusobacterium unclassified | + | − | + | + | + | − | 4 |
| Leptotrichia_unclassified | + | + | + | − | − | + | 4 |
| Treponema denticola 584 | + | − | + | − | + | + | 4 |
| Porphyromonas sp. 279 | + | + | + | − | − | + | 4 |
| Prevotella enoeca 600 | + | + | − | + | + | − | 4 |
| Gemella morbillorum 046 | + | + | + | − | − | + | 4 |
| Treponema sp. 246 | + | + | + | − | − | + | 4 |
| Bacteria_unclassified | − | + | − | − | + | + | 3 |
| Veillonella unclassified | + | + | + | − | − | − | 3 |
| Prevotella unclassified | − | + | + | + | − | − | 3 |
| A. actinomycetemcomitans 531 | − | + | + | − | + | − | 3 |
| Streptococcus sp. 058 | + | − | + | − | − | + | 3 |
| Treponema maltophilum 664 | + | + | − | − | + | − | 3 |
| Porphyromonas endodontalis 273 | + | − | + | − | − | + | 3 |
| Eubacterium infirmum 105 | − | + | + | + | − | − | 3 |
| Prevotella sp. 820 | − | + | − | + | + | − | 3 |
| Prevotella sp. 304 | − | − | + | + | − | + | 3 |
| Prevotella intermedia 643 | − | + | + | − | + | − | 3 |
| Leptotrichia sp. 223 | + | − | + | + | − | − | 3 |
| Prevotella sp. 317 | + | − | − | + | − | + | 3 |
| Filifactor alocis 539 | − | + | − | + | − | + | 3 |
| Peptostreptococcaceae_[XIII]_unclassified | + | + | − | − | − | + | 3 |
| Moraxella catarrhalis 833 | + | + | + | − | − | − | 3 |
| Selenomonas_unclassified | − | + | + | − | − | − | 2 |
| Streptococcus_unclassified | − | + | + | − | − | − | 2 |
| Veillonella parvula 161 | − | − | + | − | + | − | 2 |
| Selenomonas sputigena 151 | + | + | − | − | − | − | 2 |
| Catonella morbi 165 | − | − | + | − | − | + | 2 |
| Capnocytophaga_unclassified | − | − | + | − | − | + | 2 |
| Veillonellaceae_[G‐1] | − | − | − | + | − | + | 2 |
| Fusobacterium sp. _203 | + | − | + | − | − | − | 2 |
| Veillonellaceae_[G‐1] sp. 155 | + | − | − | − | − | + | 2 |
| Prevotella fusca 782 | − | − | + | − | − | + | 2 |
| Pyramidobacter piscolens 357 | − | + | − | − | − | − | 1 |
| Prevotella sp. 313 | − | − | − | − | + | − | 1 |
| Streptococcus parasanguinis II 411 | − | − | + | − | − | − | 1 |
| Aggregatibacter unclassified | − | + | − | n.a. | n.a. | n.a. | 1 |
| Pasteurellaceae_unclassified | − | − | − | − | − | + | 1 |
| Bacteroidetes_unclassified | − | − | − | − | − | + | 1 |
| SR1_[G‐1] sp. 345 | − | − | − | − | − | + | 1 |
| Prevotella sp. 311 | − | − | + | n.a. | n.a. | n.a. | 1 |
| Neisseria oralis 014 | − | + | − | − | − | − | 1 |
| Desulfobulbus sp. 041 | + | − | − | n.a. | n.a. | n.a. | 1 |
| Megasphaera micronuciformis 122 | − | − | − | n.a. | n.a. | n.a. | 0 |
| Porphyromonadaceae | − | − | − | − | − | − | 0 |
| Peptostreptococcaceae_[XI]_unclassified | − | − | − | n.a. | n.a. | n.a. | 0 |
| Veillonella dispar 160 | − | − | − | n.a. | n.a. | n.a. | 0 |
| Treponema socranskii 769 | − | − | − | n.a. | n.a. | n.a. | 0 |
| Prevotella sp. 443 | − | − | − | n.a. | n.a. | n.a. | 0 |
| Prevotella denticola 291 | − | − | − | n.a. | n.a. | n.a. | 0 |
| Total | 25 | 29 | 32 | 17 | 17 | 28 | 148 |
+ = Significant correlation between the OTU and gene expression levels, − = no significant correlations; n.a. = the OTU was not represented in the microbiome members who were evaluated.
Figure 8 provides a schematic of a protein interaction network related to the Tfh genes examined that significantly correlated with bacterial complexes. This includes interactions of transcription factors, cell surface receptors and soluble products produced by Tfh cells. We highlight the gene expression profiles that were altered in adults and young animal samples stratified into health, disease and resolution samples. These findings showed that cell receptors CD40LG, CD10/MME, IL‐6RA, CD84 and the composite of CD3D/E/G were particularly enriched for these correlations. Additionally, of the cytokines/chemokines IL‐12 and IL‐17A showed enrichment for microbiome correlations. Also of interest was that these Tfh gene–bacterial relationships were represented in both adult and young samples.
Fig. 8.
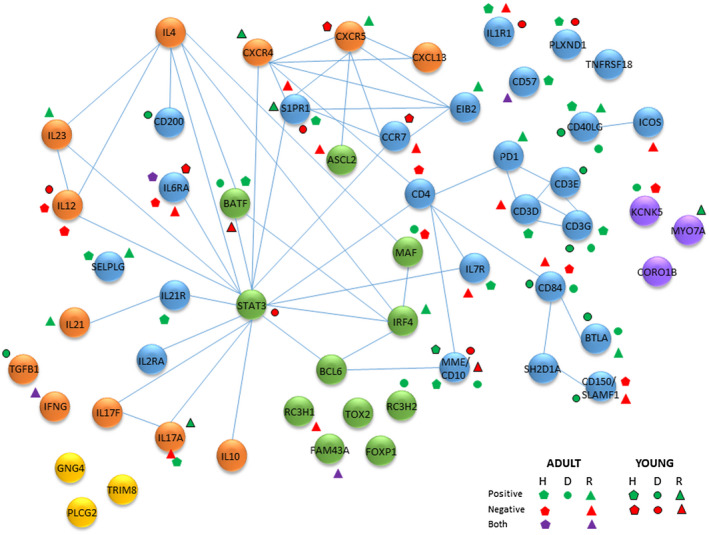
Schematic of potential protein interactome among follicular T helper (Tfh)‐associated genes. Blue nodes are surface receptors, green nodes are transcription/regulatory factors, orange nodes are secreted products, yellow nodes are signaling molecules and purple nodes are structural/metabolic components. The symbols identify significant positive or negative correlations of the gene with bacterial complexes in healthy (H), diseased (D), and resolution (R) samples from adult and young animal gingival tissues. Both denotes that individual bacteria within the complex demonstrated both significant positive and negative correlations the particular gene.
Discussion
The CD4+ T helper cell population in humans is composed of multiple subsets with clear phenotypical and functional differences, including Th1, Th2, Th17, Th9, regulatory T cells (Treg) and Tfh. The development of each of the Th cell subsets is driven by lineage‐specific transcription factors: T‐bet/TXB21 for Th1, GATA binding factor 3 (GATA3) for Th2, RORα/γ for Th17, forkhead box protein O1 (FoxO1) for Th9 and forkhead box protein 3 (FoxP3) for Treg cells. The critical transcription factor for Tfh cells is Bcl6 [1, 4, 6, 50]. These various cell types respond to different stimuli, including antigenic challenge from pathogens and support development and maturation of both B and T cell effector functions [51]. Cell subsets with similar features have been identified in most mammals, particularly related to rodent and non‐human primate disease models. Using a non‐human primate model of ligature‐induced periodontitis, we have shown aging effects on the genomic footprints of various T cell subsets in gingival tissues with health, progressing disease lesions and clinical resolution [34]. Additionally, we have identified major gingival responses in the model, particularly later in disease progression, that reflected adaptive immune system triggering with substantial expression of rearranged immunoglobulin (Ig) genes [41, 42, 46]. Of particular note for this study is that chronically inflamed tissues commonly accumulate lymphoid aggregates that would facilitate localized T–B cell interactions. These lymphocytes that migrate into chronically inflamed tissues can form ectopic lymphoid structures with functional germinal centers and have been termed tertiary lymphoid structures (TLS) [14], that demonstrate prototypical Tfh cells with elevated expression of Bcl6, CXCR5, PD‐1 and ICOS [1]. Understanding the range of T cell populations that can provide help to B cells within these chronically inflamed tissues is essential to a mechanistic clarification of the biology of these inflammatory conditions. More specifically within the gingival tissues of periodontitis lesions, Tfh cells may be important for effective responses to pathogens, as well as commensal bacteria, as they are required for antibody response quality. Also noted in these data were a general lack of increases in primary transcription factors for Th17 and Treg cells. These cell types have been shown to increase in the developed lesions in both rodent models and human samples [52, 53, 54]. However, a fundamental difference exists in the animal and human models that have been explored to determine the immune and inflammatory characteristics of periodontal lesions. First, the murine model of disease that is routinely used includes either ligation (similar to this study) or oral infection with human bacteria as, importantly, these studies are routinely conducted in very young mice, and the ligature disease takes place in approximately 7 days. Thus, the underlying microbial–host environment that leads to a disease process in the mice is probably quite different than might occur in humans and these non‐human primates over months of disease progression in response to members of the autochthonous microbiome. The dynamics of these localized interactions in the non‐human primates evolve over many years prior to the activation of a disease process. As such, the distribution of local signals that induce various T cell subsets might be expected to be different. Secondly, with regard to human disease, all the human studies have compared healthy periodontal sites to those that demonstrate clinical measures of the existence of a past or present disease process. Thus, whether that disease process is occurring at present or has occurred weeks or months prior and is now stable cannot be determined from these human studies. Therefore, potentially later in lesion development or in stable lesions, Th17 or Treg cells may be a more critical component. Our data show a rapid decrease in the critical transcription factor for Th17 cells following initiation of the periodontal lesion that increases in tissues with periodontitis progression and clinical resolution in this model. This may fundamentally reflect differences in early lesion development compared to more established lesions later in a disease process in humans, when clinical changes can be determined.
This study targeted gene expression of an array of Tfh genes including various transcription factors (BCL6, STAT‐3, ASCL2, BATF, c‐MAF, IRF4) [4, 55, 56, 57, 58, 59]. BCL6 was increased in gingival tissues from both age groups, particularly later in disease progression, and the pattern differed from other T helper cell regulatory transcription factors. A major role for high Bcl6 levels is directing the positioning of Tfh cells to B cells through up‐regulation of CXCR5 and down‐regulation of CCR7, PSGL1 and EBI2 [13]. While BCL6 has been described as a transcriptional repressor in B cells [60, 61], this has focused only on a role in germinal center reactions, and appears to only occur immediately after antigen exposure. It appears clear from a wide array of studies that plasmacytes are significantly increased in gingival tissues of periodontal disease lesions. Moreover, the type of tissue architecture in lymph node germinal centers needed for reactions between T and B cells, and thus the needed immune regulation, is not quite like the more diffuse lymphocytic aggregates in the gingival tissues. Generally, low levels of these three genes were also noted with disease in tissues from both age groups. IRF4 and BATF are both essential transcription factors for early Tfh differentiation [10, 57, 58]. BATF showed minimal change with disease; however, IRF4 increased significantly in both age groups, particularly at the disease progression sampling points. STAT‐3 is also crucial for Tfh cells role for inducing GC B cells. Both STAT‐3 and IRF4 are also critical transcription factors for production of IL‐21 that impacts Tfh cell development [56, 62, 63]. STAT‐3 was increased at disease initiation and progression, and decreased to baseline levels with resolution.
IL‐21, IL‐12, IL‐23 and TGF‐β are all required for development of Tfh cells, and IL‐21 plays a critical role in regulating Ig production and GC formation. It also acts in an autocrine fashion for Tfh cell development [64, 65, 66]. While IL‐21 is induced by IL‐6 through the STAT‐3 transcription factor, the mRNA for this cytokine showed minimal change with disease in young animals, but was significantly increased later in disease in the adult animals. IL‐12 induces generation of Tfh cells from naive CD4+ T cells [67, 68], while IL‐23 has been suggested to contribute to the induction of autoantibody formation in various inflammatory diseases [69, 70, 71]. Neither of these factors were altered at the mRNA level in this chronic inflammatory disease model.
Tfh cells show elevated expression of various cytokine (IL‐6R, IL‐21R) and chemokine (CXCR4, CXCR5) receptors [7, 10]. Tfh cells co‐localize with B cells by Bcl6 up‐regulating CXCR5 and down‐regulating CCR7, with CXCR5 enhancing migration of Tfh to B cell regions via interaction with its ligand CXCL13 [1, 9, 15, 51]. We identified significant increases in CXCR5 in both adult and young tissues that increased later in disease and actually remained elevated in the resolution samples, while no change in expression of CCR7 was seen. CXCR4 was also elevated, but appeared earlier in the disease process, and then dropped to baseline in resolution samples. ICOS is expressed on activated T cells and its ligand, ICOS‐L (B7h) is broadly expressed on antigen‐presenting cells (APCs) non‐lymphoid tissues and is constitutively expressed on B cells [72, 73]. The interaction of ICOS/ICOS‐L is critical for increases in antigen‐specific CXCR5+PD‐1+ Tfh cells and GC‐B cells and has been shown to be dependent upon IL‐21, IL‐6 and STAT‐3 [12]. ICOS is also important for IL‐21 and IL‐4 production under the control of signaling lymphocyte activation (SLAM) molecules [63]. Importantly, ICOS engagement induces c‐Maf expression that supports Tfh formation and maintenance of these cells through production of IL‐21 [59]. ICOS mRNA is increased later in disease in both adult and young animals, although levels of c‐Maf mRNA were generally unaffected.
CD40LG expressed on Tfh cells binds to the CD40 receptor on B cells as a major co‐stimulatory molecule [74, 75]. This interaction induces expression of CD25 on the B cells that helps to control B cell receptor expression, and represents a phenotype of memory B cell population with improved proliferative and antigen‐presenting capacity. We noted that increases in message for CD40LG was a response change with disease primarily in adult animals. IL‐6 production is generally felt to arise from follicular dendritic cells for Tfh efficient cell responses [5, 76], although dendritic cell numbers and functions have also been reported within periodontitis lesions [77, 78, 79]. Furthermore, T cell‐specific deletion of the IL‐6 receptor α chain (IL‐6RA) results in impaired T cell responses in vivo, including a defect in Tfh functions [80]. The periodontitis model showed increases in IL‐6RA only in adult disease, thus representing one of the varied differences in gene expression related to Tfh functions based upon age. PD‐1 on Tfh cells binds to programmed cell death ligand 1 (PD‐L1) on B cells and engages APCs via TCR signaling that provides transient help to B cells, but not Tfh proliferation [81]. It has been found that elevated PD‐1 actually limits Tfh proliferation through dampening TCR signaling. However, no changes in the mRNA for this important Tfh receptor were observed in this disease model.
CD84/SLAM5 and CD150/SLAM molecules have been identified on GC Tfh cells (Calpe 2008). Through signaling activity, lymphocyte activation molecule (SLAM)‐associated protein (SAP/SH2D1A) adaptor molecule enables these receptors to have a critical role in B cell help and antibody class‐switching related to Tfh development [82, 83, 84]. While the Tfh interactions with dendritic cells (DCs) appear primarily via integrin and TCR receptors, the sustained phase of Tfh interaction with B cells requires SAP. The SLAM receptors on Tfh cells are engaged in prolonged T–B cell contact and optimal Tfh functions [85, 86]. In samples from both adult and young animals, both CD84 and CD150 genes were up‐regulated later in the disease process, consistent with an increased role of Tfh cells and adaptive immune responses at this stage of the periodontal lesions.
Sphingosine‐1‐phosphate (S1P) is a biologically active metabolite of plasma‐membrane sphingolipids that is essential for immune‐cell trafficking. Its receptor, S1PR1, signaling is a crucial factor in regulating naive Tfh‐cells egress from draining lymph nodes [87]. The gene expression for this receptor was significantly increased in gingival tissues of both adults and young animals primarily at early time‐points in the disease process, and then returned to baseline levels in resolution samples. This is consistent with a role in localization of Tfh cell help within the developing periodontal lesion that could contribute to enhancing control over the host–bacterial interactions during disease. Additionally, CD10/MME/neprilysin is a marker of immature T and B cells and GC B cells and is virtually absent on circulating mature T cells [88]. However, it is not only expressed on Tfh cells, but was substantially increased early in the disease process, again potentially related to the localization and communication of Tfh cells in this inflammatory environment.
Finally, a number of biomolecules associated with Tfh cells specifically down‐regulate functions associated with T–B cell interactions. CD200 is a glycoprotein surface molecule that is a member of the immunoglobulin superfamily and is expressed on B cells and a subset of T cells, including Tfh, particularly in reactive lymphoid tissues [89, 90]. It binds to CD200R1, which is an inhibitory receptor for inflammatory mediator production from lymphoid cells. Of interest is the significant decrease in this molecule across all disease time‐points, thus potentially contributing to less control over the developing chronic inflammatory lesion. Similarly, BTLA (e.g. CD272) is a co‐inhibitory receptor also belonging to the immunoglobulin superfamily. It is induced during T cell activation except on Th2 cells [91]. BTLA interacts with the B7H4 co‐stimulatory molecule and inhibits T cell effector functions. In the non‐human primate gingival tissues BTLA mRNA levels were significantly increased during disease, which may contribute to overall regulation of the immune responses occurring through Tfh help in the gingival tissues. SELPLG encodes for PSGL‐1, which is an adhesion and regulatory molecule involved in immune cell trafficking. However, PSGL‐1 has emerged as an important checkpoint in negatively regulating Tfh cell function during adaptive immune responses [5, 9]. As with BTLA, this gene was significantly up‐regulated only during disease that might be predicted to be an important portion of the T cell regulatory environment in the gingival tissues. It has been reported that IL‐2 signaling is another potent inhibitor of Tfh cell differentiation [92]. While IL‐2RA mRNA levels increased significantly with disease in both adults and young samples, IL‐2 gene levels were generally low in the gingival tissues. STAT‐5 is the downstream target of IL‐2 engagement of IL‐2RA. Thus, although STAT‐5 would also repress Tfh differentiation, there was no change in STAT‐5 mRNA with disease in either age group. As noted in the previous discussion, we employed a strategy of utilizing the existing literature to identify an array of 54 genes associated with Tfh phenotype and functions. While a gene set enrichment analysis is often used to identify important biological functions and pathways in complex tissues, this approach could not be effectively used with this macaque microarray data. More specifically, we determined that > 12 000 of the probes were only annotated as LOC genes lacking published symbols or orthologs. Additionally, only approximately half the significantly altered affymetrix probes were successfully mapped by david. As such, our strategy did identify a portfolio of genes related to Tfh functions as reported in this study.
As we noted, these substantive changes in the footprint of genes related to Tfh functions, it must be recognized that these changes result from alterations in the quality and quantity of the microbiome at disease sites and the resulting clinical changes reflecting the disease process. The current paradigm in periodontitis is the transition from a healthy symbiotic microbiome to a dysbiotic microbiome at disease sites [93, 94, 95, 96, 97]. This dysbiosis reflects altered biology of both pathogens and commensals in the disease ecology, the summation of which drives host response changes, dysregulates these responses and triggers tissue changes of periodontitis. However, a more detailed profile of the bacteria and/or bacterial complexes related to specific changes in host responses in situ remains to be elucidated. Studies of human gingival transcriptomes and associated targeted bacteria have identified relationships of changes in the expression of specific genes with altered levels of some microbes as indicative of disease versus health; however, these are ‘point‐in‐time’ samples with minimal knowledge regarding the true dynamics of the lesion development [98, 99, 100, 101, 102]. Thus, using this non‐human primate model of progressing disease, we examined the inter‐relationship of the microbiome with the pattern of gene expression through significant associations. A goal was to discern the potential that certain bacteria or complexes of bacteria might be more contributory to the Tfh gene expression profiles, and thus be more involved in triggering adaptive immune responses. Multiple outcomes of this analysis were detected. First, there were a finite number of complexes of bacteria that strongly correlated with the Tfh gene expression. The complexes differed in health, disease and resolution sites, as well as showing unique differences in the adult and young microbiomes. Moreover, these complexes were not simply composed of only bacteria historically considered as periodontopathogens versus oral commensal bacteria, but were generally mixtures of these types of microorganisms. Secondly, certain of these complexes were significantly correlated with a composite of Tfh genes representing a more holistic assessment of relative levels of Tfh cells or functions occurring in the gingival tissues. The complexes showed differences depending upon the clinical health, disease or resolution phase of the lesions. Additionally, while there were some similarities in OTU representation within the complexes correlating with the Tfh composite gene expression, generally the OTUs were somewhat distinctive in the adult and young samples. This is consistent with our previous report regarding some fundamental differences in the oral microbiomes in adult and young animals, as well as major differences in these complexes related to other biological pathways in the gingival tissues [40, 103]. Extending these observations, we noted multiple Tfh‐related genes that generally were significantly correlated with the panel of Tfh genes (e.g. Tfh‐H1A, Tfh‐H5A, Tfh‐R2A, Tfh‐R4A, Tfh‐D1Y; Supporting information, Table S2). Finally, while we could estimate the statistical significance of specific correlations of complexes of bacteria with specific Tfh genes, a systems approach to the relationship of bacterial abundance to gene expression levels provided a visualization of broad correlations between specific bacterial complexes and panels of the Tfh genes. Of note, the same bacterial complex demonstrated positive and negative correlation patterns with different groups of genes within the Tfh footprint, supporting an important interaction between the microbiome components and regulation of host responses in the gingival tissues. Thus, Tfh cell functions are mediated through the production of cytokines that also promote B cell survival and antibody production and through the engagement of T‐APC and T–B cell co‐stimulatory molecules (e.g. TCR, ICOS, CD40LG, PD‐1) and other receptors (e.g. SLAM, CXCR5, etc.), which favor these strong interactions leading to successful and mature B cell immunoglobulin responses. Coupled with existing data in humans [104, 105, 106, 107, 108, 109, 110] and non‐human primates [35, 111, 112], detailing local and systemic antibody production that can be enhanced by immunization supports that localized Tfh functions in gingival tissues may provide an important adaptive immune regulatory mechanism for controlling the progression and magnitude of disease.
Future studies using this non‐human primate model of chronic mucosal inflammation to interrogate the kinetics and dynamics of various aspects of the innate immune, inflammatory and adaptive immune responses in periodontal lesions should be enhanced with new technologies, including scRNASeq. These types of approaches would allow a fuller documentation of the nature of the Tfh role in the gingival tissues, as well as estimation of differences in the localization of these cells within the complex tissue microenvironment.
Disclosures
The authors acknowledge no conflicts of interest with the content of this report.
Author contributions
J. L. E. and O. A. G. were responsible for the design, conduct of the experiment, clinical data and sample collection, interpretation of the data and preparation of the manuscript. S. S. K. was responsible for preparation of the gingival tissue mRNA and bacterial DNA for analysis. L. O. contributed to the clinical measures and J. G. M. contributed to the animal husbandry and conduct of the experiment.
Supporting information
Table S1. Characteristics of expression of Tfh composite gene profile.
Table S2. Significant correlations of microbiome OTU complexes with genes related to Tfh phenotype and functions. Blue denotes significant positive correlation and red denotes a significant negative correlation with the specific Tfh‐related gene.
Acknowledgements
This work was supported by National Institute of Health grant P20GM103538. We express our gratitude to the Caribbean Primate Research Center (CPRC) supported by grant P40RR03640 and the Center for Oral Health Research in the College of Dentistry at the University of Kentucky. The authors thank the Microarray Core of University Kentucky for their invaluable technical assistance and Dr A. Stromberg for initial normalization of the microarray data.
Data Availability Statement
The microbiome data has been deposited at BioProject ID PRJNA516659 through the NIH NCBI. The data have been uploaded into the ArrayExpress data base (www.ebi.ac.uk) under accession number: E‐MTAB‐1977.
References
- 1. Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity 2019; 50:1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014; 41:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29:621–663. [DOI] [PubMed] [Google Scholar]
- 4. Hatzi K, Nance JP, Kroenke MA et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med 2015; 212:539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poholek AC, Hansen K, Hernandez SG et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol 2010; 185:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nurieva RI, Chung Y, Martinez GJ et al. Bcl6 mediates the development of T follicular helper cells. Science 2009; 325:1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu D, Rao S, Tsai LM et al. The transcriptional repressor Bcl‐6 directs T follicular helper cell lineage commitment. Immunity 2009; 31:457–468. [DOI] [PubMed] [Google Scholar]
- 8. Qin L, Waseem TC, Sahoo A et al. Insights into the molecular mechanisms of T follicular helper‐mediated immunity and pathology. Front Immunol 2018; 9:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol 2016; 34:335–368. [DOI] [PubMed] [Google Scholar]
- 10. Schmitt N, Liu Y, Bentebibel SE, Ueno H. Molecular mechanisms regulating T helper 1 versus T follicular helper cell differentiation in humans. Cell Rep 2016; 16:1082–1095. [DOI] [PubMed] [Google Scholar]
- 11. Morita R, Schmitt N, Bentebibel SE et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011; 34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akiba H, Takeda K, Kojima Y et al. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo . J Immunol 2005; 175:2340–2348. [DOI] [PubMed] [Google Scholar]
- 13. Rao DA. T cells that help B cells in chronically inflamed tissues. Front Immunol 2018; 9:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cicalese MP, Salek‐Ardakani S, Fousteri G. Editorial: Follicular helper T cells in immunity and autoimmunity. Front Immunol 2020; 11:1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng J, Wei Y, Fonseca VR, Graca L, Yu D. T follicular helper cells and T follicular regulatory cells in rheumatic diseases. Nat Rev Rheumatol 2019; 15:475–490. [DOI] [PubMed] [Google Scholar]
- 16. Li W, Zhang Z, Wang ZM. Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC Oral Health 2020; 20:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng D, Sun Q, Su Z et al. Enhancing specific‐antibody production to the ragB vaccine with GITRL that expand Tfh, IFN‐gamma(+) T cells and attenuates Porphyromonas gingivalis infection in mice. PLoS One 2013; 8:e59604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang YN, Song J, Wang H et al. Nasal IL‐4(+)CXCR5(+)CD4(+) T follicular helper cell counts correlate with local IgE production in eosinophilic nasal polyps. J Allergy Clin Immunol 2016; 137:462–473. [DOI] [PubMed] [Google Scholar]
- 19. Bai X, Chi X, Qiao Q et al. T follicular helper cells regulate humoral response for host protection against intestinal Citrobacter rodentium infection. J Immunol 2020; 204:2754–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu J, He S, Liu P et al. Interleukin21 promotes the development of ulcerative colitis and regulates the proliferation and secretion of follicular T helper cells in the colitides microenvironment. Mol Med Rep 2015; 11:1049–1056. [DOI] [PubMed] [Google Scholar]
- 21. Sage PT, Tan CL, Freeman GJ, Haigis M, Sharpe AH. Defective TFH cell function and increased TFR cells contribute to defective antibody production in aging. Cell Rep 2015; 12:163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varricchi G, Bencivenga L, Poto R, Pecoraro A, Shamji MH, Rengo G. The emerging role of T follicular helper (TFH) cells in aging: Influence on the immune frailty. Ageing Res Rev 2020; 61:101071. [DOI] [PubMed] [Google Scholar]
- 23. Pallikkuth S, de Armas L, Rinaldi S, Pahwa S. T follicular helper cells and B cell dysfunction in aging and HIV‐1 infection. Front Immunol 2017; 8:1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou M, Zou R, Gan H et al. The effect of aging on the frequency, phenotype and cytokine production of human blood CD4+ CXCR5+ T follicular helper cells: comparison of aged and young subjects. Immunity Ageing 2014; 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang C, Zheng JH, Lin ZH et al. Profiles of immune cell infiltration and immune‐related genes in the tumor microenvironment of osteosarcoma. Aging 2020; 12:3486–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Danziger SA, Gibbs DL, Shmulevich I et al. ADAPTS: automated deconvolution augmentation of profiles for tissue specific cells. PLoS One 2019; 14:e0224693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ayana R, Singh S, Pati S. Deconvolution of human brain cell type transcriptomes unraveled microglia‐specific potential biomarkers. Front Neurol 2018; 9:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi‐subject single‐cell expression reference. Nat Commun 2019; 10:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ge P, Wang W, Li L et al. Profiles of immune cell infiltration and immune‐related genes in the tumor microenvironment of colorectal cancer. Biomed Pharmacother 2019; 118:109228. [DOI] [PubMed] [Google Scholar]
- 30. Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell‐type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genom 2006; 7:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gonzalez OA, Novak MJ, Kirakodu S et al. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis 2013; 18:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez‐Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res 2011; 90:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebersole JL, Steffen MJ, Reynolds MA et al. Differential gender effects of a reduced‐calorie diet on systemic inflammatory and immune parameters in nonhuman primates. J Periodontal Res 2008; 43:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ebersole JL, Kirakodu S, Novak MJ et al. Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J Clin Periodontol 2014; 41:853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebersole JL, Steffen MJ, Gonzalez‐Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol 2008; 15:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kirakodu S, Chen J, Gonzalez Martinez J, Gonzalez OA, Ebersole J. Microbiome profiles of ligature‐induced periodontitis in nonhuman primates across the lifespan. Infect Immun 2019; 87:e00067‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schloss PD, Westcott SL, Ryabin T et al. Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kirakodu S, Chen J, Gonzalez Martinez J, Gonzalez OA, Ebersole J. Microbiome profiles of ligature‐induced periodontitis in nonhuman primates across the life span. Infect Immun 2019; 87:e00067‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gonzalez OA, Novak MJ, Kirakodu S et al. Differential gene expression profiles reflecting macrophage polarization in aging and periodontitis gingival tissues. Immunol Invest 2015; 44:643–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gonzalez OA, Novak MJ, Kirakodu S et al. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol 2014; 41:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gonzalez OA, John Novak M, Kirakodu S et al. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis 2013; 18:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meka A, Bakthavatchalu V, Sathishkumar S et al. Porphyromonas gingivalis infection‐induced tissue and bone transcriptional profiles. Mol Oral Microbiol 2010; 25:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ebersole JL, Kirakodu SS, Novak MJ et al. Gingival tissue autophagy pathway gene expression profiles in peirodontitis and aging. J Periodont Res 2020; 55:34–45. [DOI] [PubMed] [Google Scholar]
- 46. Ebersole JL, Kirakodu SS, Novak MJ et al. Transcriptome analysis of B cell immune functions in periodontitis: mucosal tissue responses to the oral microbiome in aging. Front Immunol 2016; 7:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mesquita D Jr, Cruvinel WM, Resende LS et al. Follicular helper T cell in immunity and autoimmunity. Braz J Med Biol Res 2016; 49:e5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Georgiev H, Ravens I, Papadogianni G et al. Shared and unique features distinguishing follicular T helper and regulatory cells of peripheral lymph node and Peyer’s patches. Front Immunol 2018; 9:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weinstein JS, Lezon‐Geyda K, Maksimova Y et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood 2014; 124:3719–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choi J, Diao H, Faliti CE et al. Bcl‐6 is the nexus transcription factor of T follicular helper cells via repressor‐of‐repressor circuits. Nat Immunol 2020; 21:777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nurieva RI, Chung Y. Understanding the development and function of T follicular helper cells. Cell Mol Immunol 2010; 7:190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dutzan N, Kajikawa T, Abusleme L et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med 2018; 10:eaat0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alvarez C, Rojas C, Rojas L, Cafferata EA, Monasterio G, Vernal R. Regulatory T lymphocytes in periodontitis: a translational view. Mediators Inflamm 2018; 2018:7806912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garlet GP. Destructive and protective roles of cytokines in periodontitis: a re‐appraisal from host defense and tissue destruction viewpoints. J Dent Res 2010; 89:1349–1363. [DOI] [PubMed] [Google Scholar]
- 55. Liu X, Chen X, Zhong B et al. Transcription factor achaete‐scute homologue 2 initiates follicular T‐helper‐cell development. Nature 2014; 507:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma CS, Avery DT, Chan A et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood 2012; 119:3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Betz BC, Jordan‐Williams KL, Wang C et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med 2010; 207:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bollig N, Brustle A, Kellner K et al. Transcription factor IRF4 determines germinal center formation through follicular T‐helper cell differentiation. Proc Natl Acad Sci USA 2012; 109:8664–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bauquet AT, Jin H, Paterson AM et al. The costimulatory molecule ICOS regulates the expression of c‐Maf and IL‐21 in the development of follicular T helper cells and TH‐17 cells. Nat Immunol 2009; 10:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robinson MJ, Ding Z, Pitt C et al. The amount of BCL6 in B cells shortly after antigen engagement determines their representation in subsequent germinal centers. Cell Rep 2020; 30:1530–41 e4. [DOI] [PubMed] [Google Scholar]
- 61. Basso K, Dalla‐Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev 2012; 247:172–183. [DOI] [PubMed] [Google Scholar]
- 62. Schmitt N, Liu Y, Bentebibel SE et al. The cytokine TGF‐beta co‐opts signaling via STAT3–STAT4 to promote the differentiation of human TFH cells. Nat Immunol 2014; 15:856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nurieva RI, Chung Y, Hwang D et al. Generation of T follicular helper cells is mediated by interleukin‐21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 2008; 29:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gong F, Zheng T, Zhou P. T follicular helper cell subsets and the associated cytokine IL‐21 in the pathogenesis and therapy of asthma. Front Immunol 2019; 10:2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tangye SG. Advances in IL‐21 biology – enhancing our understanding of human disease. Curr Opin Immunol 2015; 34:107–115. [DOI] [PubMed] [Google Scholar]
- 66. Spolski R, Leonard WJ. Interleukin‐21: a double‐edged sword with therapeutic potential. Nat Rev Drug Discov 2014; 13:379–395. [DOI] [PubMed] [Google Scholar]
- 67. Schmitt N, Bustamante J, Bourdery L et al. IL‐12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood 2013; 121:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ma CS, Suryani S, Avery DT et al. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL‐12. Immunol Cell Biol 2009; 87:590–600. [DOI] [PubMed] [Google Scholar]
- 69. Menegat JS, Lira‐Junior R, Siqueira MA et al. Cytokine expression in gingival and intestinal tissues of patients with periodontitis and inflammatory bowel disease: an exploratory study. Arch Oral Biol 2016; 66:141–146. [DOI] [PubMed] [Google Scholar]
- 70. Jain R, Chen Y, Kanno Y et al. Interleukin‐23‐induced transcription factor blimp‐1 promotes pathogenicity of T helper 17 cells. Immunity 2016; 44:131–142. [DOI] [PubMed] [Google Scholar]
- 71. Wendling D, Abbas W, Godfrin‐Valnet M et al. Dysregulated serum IL‐23 and SIRT1 activity in peripheral blood mononuclear cells of patients with rheumatoid arthritis. PLoS One 2015; 10:e0119981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Uwadiae FI, Pyle CJ, Walker SA, Lloyd CM, Harker JA. Targeting the ICOS/ICOS‐L pathway in a mouse model of established allergic asthma disrupts T follicular helper cell responses and ameliorates disease. Allergy 2019; 74:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Choi YS, Kageyama R, Eto D et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 2011; 34:932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grewal IS, Flavell RA. The CD40 ligand. At the center of the immune universe? Immunol Res 1997; 16:59–70. [DOI] [PubMed] [Google Scholar]
- 75. Liu YJ, Grouard G, de Bouteiller O, Banchereau J. Follicular dendritic cells and germinal centers. Int Rev Cytol 1996; 166:139–179. [DOI] [PubMed] [Google Scholar]
- 76. Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL‐6‐mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol 2013; 190:3049–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meyle J, Chapple I. Molecular aspects of the pathogenesis of periodontitis. Periodontol 2000 2015; 69:7–17. [DOI] [PubMed] [Google Scholar]
- 78. El‐Awady AR, Arce RM, Cutler CW. Dendritic cells: microbial clearance via autophagy and potential immunobiological consequences for periodontal disease. Periodontol 2000 2015; 69:160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Souto GR, Queiroz CM Jr, Costa FO, Mesquita RA. Relationship between chemokines and dendritic cells in human chronic periodontitis. J Periodontol 2014; 85:1416–1423. [DOI] [PubMed] [Google Scholar]
- 80. Nish SA, Schenten D, Wunderlich FT et al. T cell‐intrinsic role of IL‐6 signaling in primary and memory responses. eLife 2014; 3:e01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jogdand GM, Mohanty S, Devadas S. Regulators of Tfh cell differentiation. Front Immunol 2016; 7:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hu J, Havenar‐Daughton C, Crotty S. Modulation of SAP dependent T:B cell interactions as a strategy to improve vaccination. Curr Opin Virol 2013; 3:363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. He J, Tsai LM, Leong YA et al. Circulating precursor CCR7(lo)PD‐1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity 2013; 39:770–781. [DOI] [PubMed] [Google Scholar]
- 84. Cannons JL, Qi H, Lu KT et al. Optimal germinal center responses require a multistage T cell: B cell adhesion process involving integrins, SLAM‐associated protein, and CD84. Immunity 2010; 32:253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Biram A, Davidzohn N, Shulman Z. T cell interactions with B cells during germinal center formation, a three‐step model. Immunol Rev 2019; 288:37–48. [DOI] [PubMed] [Google Scholar]
- 86. Qi H. From SAP‐less T cells to helpless B cells and back: dynamic T‐B cell interactions underlie germinal center development and function. Immunol Rev 2012; 247:24–35. [DOI] [PubMed] [Google Scholar]
- 87. Moriyama S, Takahashi N, Green JA et al. Sphingosine‐1‐phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. J Exp Med 2014; 211:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ame‐Thomas P, Hoeller S, Artchounin C et al. CD10 delineates a subset of human IL‐4 producing follicular helper T cells involved in the survival of follicular lymphoma B cells. Blood 2015; 125:2381–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chakera A, Bennett SC, Morteau O, Bowness P, Luqmani RA, Cornall RJ. The phenotype of circulating follicular‐helper T cells in patients with rheumatoid arthritis defines CD200 as a potential therapeutic target. Clin Dev Immunol 2012; 2012:948218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chtanova T, Tangye SG, Newton R et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non‐Th1/Th2 effector cells that provide help for B cells. J Immunol 2004; 173:68–78. [DOI] [PubMed] [Google Scholar]
- 91. M’Hidi H, Thibult ML, Chetaille B et al. High expression of the inhibitory receptor BTLA in T‐follicular helper cells and in B‐cell small lymphocytic lymphoma/chronic lymphocytic leukemia. Am J Clin Pathol 2009; 132:589–596. [DOI] [PubMed] [Google Scholar]
- 92. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol 2015; 15:185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Curtis MA, Diaz PI, Van Dyke TE. The role of the microbiota in periodontal disease. Periodontol 2000 2020; 83:14–25. [DOI] [PubMed] [Google Scholar]
- 94. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 2018; 16:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mira A, Simon‐Soro A, Curtis MA. Role of microbial communities in the pathogenesis of periodontal diseases and caries. J Clin Periodontol 2017;44(Suppl 18):S23–S38. [DOI] [PubMed] [Google Scholar]
- 96. Kilian M, Chapple IL, Hannig M et al. The oral microbiome – an update for oral healthcare professionals. Br Dent J 2016; 221:657–666. [DOI] [PubMed] [Google Scholar]
- 97. Papapanou PN, Park H, Cheng B et al. Subgingival microbiome and clinical periodontal status in an elderly cohort: the WHICAP ancillary study of oral health. J Periodontol 2020;91(Suppl 1):S56–S67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sawle AD, Kebschull M, Demmer RT, Papapanou PN. Identification of master regulator genes in human periodontitis. J Dent Res 2016; 95:1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kebschull M, Demmer RT, Grun B, Guarnieri P, Pavlidis P, Papapanou PN. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J Dent Res 2014; 93:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kebschull M, Guarnieri P, Demmer RT, Boulesteix AL, Pavlidis P, Papapanou PN. Molecular differences between chronic and aggressive periodontitis. J Dent Res 2013; 92:1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Stoecklin‐Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. J Dent Res 2012; 91:934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kebschull M, Papapanou PN. The use of gene arrays in deciphering the pathobiology of periodontal diseases. Methods Mol Biol 2010; 666:385–393. [DOI] [PubMed] [Google Scholar]
- 103. Ebersole J, Kirakodu S, Chen J, Nagarajan R, Gonzalez OA. Oral microbiome and gingival transcriptome profiles of ligature‐induced periodontitis. J Dent Res 2020; 99:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schenkein HA, Berry CR, Burmeister JA, Brooks CN, Best AM, Tew JG. Locally produced anti‐phosphorylcholine and anti‐oxidized low‐density lipoprotein antibodies in gingival crevicular fluid from aggressive periodontitis patients. J Periodontol 2004; 75:146–153. [DOI] [PubMed] [Google Scholar]
- 105. Ebersole JL. Humoral immune responses in gingival crevice fluid: local and systemic implications. Periodontol 2000 2003; 31:135–166. [DOI] [PubMed] [Google Scholar]
- 106. Darby IB, Mooney J, Kinane DF. Changes in subgingival microflora and humoral immune response following periodontal therapy. J Clin Periodontol 2001; 28:796–805. [DOI] [PubMed] [Google Scholar]
- 107. Ebersole JL, Cappelli D, Steffen MJ. Antigenic specificity of gingival crevicular fluid antibody to Actinobacillus actinomycetemcomitans . J Dent Res 2000; 79:1362–1370. [DOI] [PubMed] [Google Scholar]
- 108. Mooney J, Kinane DF. Levels of specific immunoglobulin G to Porphyromonas gingivalis in gingival crevicular fluid are related to site disease status. Oral Microbiol Immunol 1997; 12:112–116. [DOI] [PubMed] [Google Scholar]
- 109. Ebersole JL, Cappelli D. Gingival crevicular fluid antibody to Actinobacillus actinomycetemcomitans in periodontal disease. Oral Microbiol Immunol 1994; 9:335–344. [DOI] [PubMed] [Google Scholar]
- 110. Kinane DF, Mooney J, MacFarlane TW, McDonald M. Local and systemic antibody response to putative periodontopathogens in patients with chronic periodontitis: correlation with clinical indices. Oral Microbiol Immunol 1993; 8:65–68. [DOI] [PubMed] [Google Scholar]
- 111. Ebersole JL, Holt SC, Delaney JE. Acquisition of oral microbes and associated systemic responses of newborn nonhuman primates. Clin Vaccine Immunol 2014; 21:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ebersole JL, Cappelli D, Holt SC, Singer RE, Filloon T. Gingival crevicular fluid inflammatory mediators and bacteriology of gingivitis in nonhuman primates related to susceptibility to periodontitis. Oral Microbiol Immunol 2000; 15:19–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Characteristics of expression of Tfh composite gene profile.
Table S2. Significant correlations of microbiome OTU complexes with genes related to Tfh phenotype and functions. Blue denotes significant positive correlation and red denotes a significant negative correlation with the specific Tfh‐related gene.
Data Availability Statement
The microbiome data has been deposited at BioProject ID PRJNA516659 through the NIH NCBI. The data have been uploaded into the ArrayExpress data base (www.ebi.ac.uk) under accession number: E‐MTAB‐1977.


