Abstract
Background:
Real-world evidence research plays an increasingly important role in diabetes care. However, a large fraction of real-world data are “locked” in narrative format. Natural language processing (NLP) technology offers a solution for analysis of narrative electronic data.
Methods:
We conducted a systematic review of studies of NLP technology focused on diabetes. Articles published prior to June 2020 were included.
Results:
We included 38 studies in the analysis. The majority (24; 63.2%) described only development of NLP tools; the remainder used NLP tools to conduct clinical research. A large fraction (17; 44.7%) of studies focused on identification of patients with diabetes; the rest covered a broad range of subjects that included hypoglycemia, lifestyle counseling, diabetic kidney disease, insulin therapy and others. The mean F1 score for all studies where it was available was 0.882. It tended to be lower (0.817) in studies of more linguistically complex concepts. Seven studies reported findings with potential implications for improving delivery of diabetes care.
Conclusion:
Research in NLP technology to study diabetes is growing quickly, although challenges (e.g. in analysis of more linguistically complex concepts) remain. Its potential to deliver evidence on treatment and improving quality of diabetes care is demonstrated by a number of studies. Further growth in this area would be aided by deeper collaboration between developers and end-users of natural language processing tools as well as by broader sharing of the tools themselves and related resources.
Keywords: diabetes, natural language processing, electronic health records
Introduction
As complexity and multidimensionality of patient care continue to grow, there is an increasing recognition that multiple sources of data are needed to provide a comprehensive picture of healthcare processes and outcomes. One important source of information that has been playing a progressively greater role in medical investigations is real-world evidence: the results of analysis of data that were generated in the course of routine patient care rather than specifically for research.1 The 21st Century Cures Act calls for increasing use of real-world evidence in development and evaluation of new medical treatments and technologies.2 The U.S. Food and Drug Administration (FDA) encourages using real-world evidence to support regulatory decision-making for drugs, biologics and medical devices.3 To utilize this novel source of data effectively, it is critical that we develop and validate methods that can leverage its strengths to reliably answer research questions.4
One important potential source of real-world evidence is so-called narrative electronic data. This category includes information in electronic health records that was generated as text rather than discrete data points (e.g. diagnoses on the problem list or laboratory values). It may comprise notes written by healthcare providers; reports describing results of imaging studies or conduct of surgical procedures; inpatient discharge summaries; etc. Narrative data in its native form cannot be analyzed using traditional analytical techniques and have to first be converted to structured (discrete) data. The suite of technologies that can accomplish this task is called natural language processing (NLP).5 In particular, over the last decade multiple applications of natural language processing in medicine have been developed and utilized in research, population management and clinical operations.6,7 Natural language processing applications have been used in research and clinical operations in a number of fields including radiology, psychiatry and oncology, among others.8-10
Diabetes mellitus is a good example of a disease that could benefit from generation of real-life evidence using natural language processing. Treatment of diabetes often involves extended discussions between patients and healthcare providers involving multiple aspects of the care process, including lifestyle changes, goals of care, adverse reactions to medications, barriers to care, etc.11 These discussions tend to be minimally represented in structured/discrete data and consequently are impossible to study or monitor on a population scale without a natural language processing solution. We therefore conducted a systematic review of studies of natural language processing systems focused on diabetes to describe the current state-of-the-art of the technology, its potential impact on diabetes care, and identify future directions for growth.
Methods
Study Design
We conducted a systematic review of original research studies focused on diabetes and natural language processing published prior to June 2020.
Data Sources and Searching Strategy
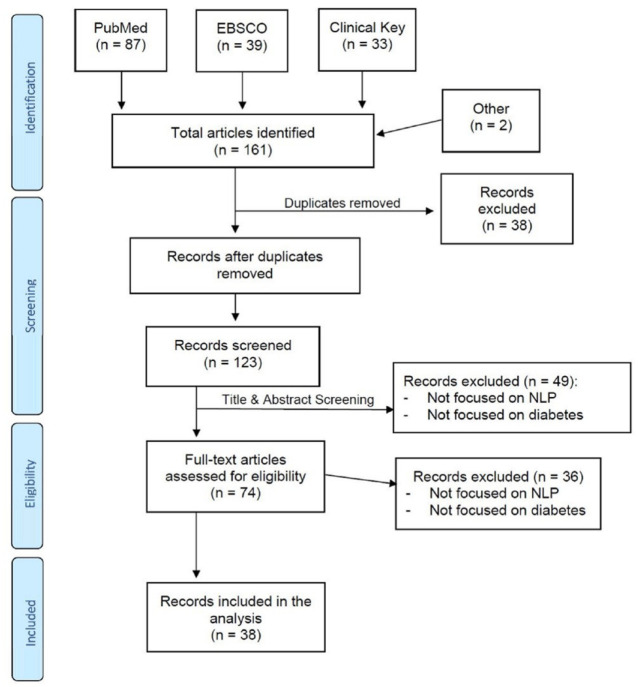
We searched for English language publications in June 2020 in PubMed, EBSCO and Clinical Key databases using Medical Subject Headings (MeSH) terms and keywords related to (a) diabetes and (b) natural language processing. Candidate articles were also identified based on the authors’ knowledge of the literature. The complete list of search terms is provided in the Appendix. The overall search strategy is illustrated in Figure 1.
Figure 1.
PRISMA flow diagram.
Screening Criteria
The articles identified using search criteria were first screened for duplicates, both within and across databases. After duplicates were removed, the title and abstract of each article were examined to determine whether both diabetes and natural language processing were a significant focus of the investigation. We subsequently examined the full text of articles that passed this initial screening to confirm their focus on diabetes and natural language processing. Articles were included if they focused on any aspect of diabetes care, including prevention, diagnosis, treatment and complications. All aspects of diabetes treatment were considered, including lifestyle changes, pharmacological and technology-based treatment approaches.
Study Measurements
Each article included in the analysis was rated on the following aspects:
Study Focus: we determined whether the study presented in the paper focused solely on development and validation of the natural language processing tool (NLP Tool) or utilized the natural language processing tool for analysis of any aspects of diabetes care (Research).
NLP Concepts: we identified concepts that were being ascertained by the natural language processing tool (for example, diagnosis of diabetes or hypoglycemia).
Concept Composition: we determined whether concepts that were being ascertained by the natural language processing tool were single- or multi-component. Single-component concepts are typically represented by either a single word (e.g. hypoglycemia) or a phrase/several words located next to each other (e.g. diabetes mellitus). Multi-component concepts are represented by sub-concepts (words/phrases) that can be located apart from each other in the document. An example of a multi-component concept could include side effects of medications, where the medication sub-concept and the side effect sub-concept may not be located next to each other in the sentence (e.g. She tried metformin several years ago but did not tolerate due to diarrhea.). Another example of a multi-component concept is a concept-value pair, which includes a concept and a corresponding numeric value (e.g. His HbA1c has been gradually increasing and is now 8.5%.).
Accuracy: F1 score12 (a harmonic mean of sensitivity and positive predictive value) was preferentially reported as a representation of the natural language processing tool accuracy if it was either directly available in the article or could be calculated from data presented. If accuracy data for several concepts were presented, micro-averaged F1 score was reported, if available; otherwise the highest accuracy described in the article was reported.
Competition: we reported whether the article described a natural language processing tool developed as part of a competition.
Analysis Category: for articles that described utilization of the natural language processing tool for analysis of diabetes care (i.e. articles whose Study Focus was rated as Research), we determined whether they presented an investigation that was Descriptive, Predictive Modeling or Hypothesis-Testing in nature.
Diabetes Care Impact: for articles that described utilization of the natural language processing tool for analysis of diabetes care, we determined whether any recommendations for changes in how diabetes care is delivered could potentially be derived from their findings. For example, if an article solely described predominance of hypoglycemia among patients with diabetes, it would be rated as No Diabetes Care Impact. On the other hand, if an article described higher prevalence of hypoglycemia among patients treated with insulin, it would be rated as Diabetes Care Impact because its findings implied that changing insulin to another diabetes medication could decrease the risk of hypoglycemia.
Results
We identified 38 articles describing original research studies focused on diabetes and natural language processing published prior to June 2020. The earliest article was published in 2005 and most were published in the last decade (Table 1). The majority of the studies (24; 63.2%) were focused solely on development and validation of natural language processing tools, but over a third used the tools to conduct clinical research. Most (23; 60.5%) studies identified single-component concepts. The most common concept identified by the natural language processing tools was the diagnosis of diabetes (17; 44.7%) and hypoglycemia was the second most common (5; 13.2%). Six (15.8%) studies described natural language processing tools developed for a competition held in 2014.13
Table 1.
Selected Studies.
| Author | Year | Study focus | NLP concepts | Concept composition | Accuracy | Competition |
|---|---|---|---|---|---|---|
| Chang et al14 | 2015 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.941 | Yesa |
| Wei et al15 | 2010 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.956 | No |
| Akay et al16 | 2013 | Research | Patient opinions about a diabetes medication | Multi-component | Not reported | No |
| Kotfila and Uzuner17 | 2015 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.964 | No |
| Khalifa and Meystre18 | 2015 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.970 | Yesa |
| Cormack et al19 | 2015 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.947 | Yesa |
| Chen et al20 | 2015 | NLP tool | Diabetes diagnosis | Single component | F1 = 0.929 | Yesa |
| Zhou et al21 | 2019 | Research | Classification of discussion board user questions about diabetes | Multi-component | Not reported | No |
| Makino et al22 | 2019 | Research | Multipleb | Single-component | Not reported | No |
| Nunes et al23 | 2016 | Research | Hypoglycemia | Single-component | F1 = 0.717 | No |
| Upadhyaya et al24 | 2017 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.990 | No |
| Pakhomov et al25 | 2010 | NLP tool | Aspirin use and contraindications to it | Multi-component | F1 = 0.802c | No |
| Jin et al26 | 2019 | NLP tool | Hypoglycemia | Single-component | F1 = 0.91 | No |
| Hazlehurst et al27 | 2014 | NLP tool | Lifestyle counseling | Multi-component | F1 = 0.944 | No |
| Malmasi et al28 | 2019 | NLP tool | Rejection of insulin by patients | Multi-component | F1 = 0.974 | No |
| Shivade et al29 | 2015 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.937 | Yesa |
| Jonnagaddala et al30 | 2015 | Research | Diabetes diagnosis | Single-component | F1 = 0.941 | No |
| Hosomura et al31 | 2017 | Research | Rejection of insulin by patients | Multi-component | F1 = 0.974 | No |
| Salmasian et al32 | 2013 | NLP tool | Diabetes diagnosis | Single-component | Not reported | No |
| Chen et al33 | 2019 | NLP tool | Hypoglycemia | Single-component | F1 = 0.590 | No |
| Turchin et al34 | 2005 | NLP tool | Diabetes diagnosis | Single-component | Sensitivity 0.962 | No |
| Specificity 0.980 | ||||||
| Topaz et al35 | 2019 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.94 | No |
| Eskildsen et al36 | 2020 | NLP tool | Medication error | Multi-component | F1 = 0.064 | No |
| Zhang et al37 | 2019 | Research | Lifestyle counseling | Multi-component | Sensitivity 0.914 | No |
| Specificity 0.943 | ||||||
| Morrison et al38 | 2012 | Research | Lifestyle counseling | Multi-component | Sensitivity 0.914 | No |
| Specificity 0.943 | ||||||
| Smith et al39 | 2008 | Research | Visual acuity | Multi-component | Not reported | No |
| Liao et al40 | 2015 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.885 | No |
| Urbain41 | 2015 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.955 | Yesa |
| Dura et al42 | 2014 | NLP tool | Diabetes-related effects of chemical compounds | Multi-component | PPV = 0.98 | No |
| Misra-Hebert et al43 | 2020 | Research | Mild hypoglycemia | Multi-component | PPV = 0.93 | No |
| Czerniecki et al44 | 2019 | Research | Lower extremity amputation | Multi-component | Not reported | No |
| Li et al45 | 2019 | Research | Hypoglycemia | Single-component | Not reported | No |
| Turchin et al46 | 2020 | Research | Rejection of insulin by patients | Multi-component | F1 = 0.974 | No |
| Mishra et al47 | 2012 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.947 | No |
| Wright et al48 | 2013 | NLP tool | Diabetes diagnosis | Single-component | F1 = 0.934 | No |
| Xu et al49 | 2014 | Research | Diabetes medications | Single-component | PPV = 0.98 | No |
| Hao et al50 | 2016 | NLP tool | HbA1c and glucose lab tests | Multi-component | F1 = 0.988d | No |
| Zheng et al51 | 2016 | NLP tool | Diabetes diagnosis | Single-component | PPV = 0.90 | No |
Participant in the 2014 i2b2/UTHealth shared task Track 2 competition.13
In this study the goal was not to identify a specific concept with any degree of accuracy but rather to extract a broad range of information from text to assist with predictive modeling of an adverse outcome.
F1 score for contraindications to aspirin.
F1 score for HbA1c lab test result.
All studies that reported on development of NLP tools described the patient population used in the analysis in sufficient detail. Majority of studies (24; 63.2%) reported F1 scores as one of their measures of accuracy of natural language processing tools. The mean F1 score was 0.882. Studies that focused on identification of diagnosis of diabetes reported uniformly high accuracy, with 13 out 14 studies reporting F1 score >0.92; their overall mean F1 score was 0.945. By comparison, the remaining 10 studies that reported F1 scores had a broad range of F1 between 0.064 and 0.988 with the mean of 0.793. None of the studies that focused on identification of diagnosis of diabetes involved identification of clinically important diabetes characteristics, such as diabetes type (e.g. type 1 vs. type 2) or duration of diabetes. Within the second most common subject of study, hypoglycemia, F1 scores ranged from 0.59 to 0.91 with a mean of 0.739.
Out of 14 studies that used natural language processing tools for clinical research, five included only descriptive statistics, five tested a hypothesis and four developed a predictive model (Table 2). Of the four predictive modeling studies, only one reported any measure of accuracy of their natural language processing tool. All five hypothesis-testing studies and two out of four predictive modeling studies, but none of the five descriptive studies, had findings with potential implications for delivery of diabetes care.
Table 2.
Research Studies.
| Author | Year | Analysis category | Potential for diabetes care impact |
|---|---|---|---|
| Akay et al16 | 2013 | Descriptive | No |
| Zhou et al21 | 2019 | Descriptive | No |
| Makino et al22 | 2019 | Modeling | No |
| Nunes et al23 | 2016 | Descriptive | No |
| Jonnagaddala et al30 | 2015 | Descriptive | No |
| Hosomura et al31 | 2017 | Descriptive | No |
| Zhang et al37 | 2019 | Hypothesis | Yes |
| Morrison et al38 | 2012 | Hypothesis | Yes |
| Smith et al39 | 2008 | Hypothesis | Yes |
| Misra-Hebert et al43 | 2020 | Modeling | Yes |
| Czerniecki et al44 | 2019 | Modeling | No |
| Li et al45 | 2019 | Modeling | Yes |
| Turchin et al46 | 2020 | Hypothesis | Yes |
| Xu et al49 | 2014 | Hypothesis | Yes |
Descriptive, the study presents descriptive statistics; hypothesis, the study described a hypothesis-testing analysis; modeling, the study developed a predictive model.
Discussion
Applications of natural language processing in medicine have a long history, having started their development in 1990s.52-55 In this systematic review we found that natural language processing tools focused on diabetes are a relative newcomer to the field, with the first study appearing in 2005.34 Nevertheless, the field has grown quickly and is now represented by several dozen studies, reviewed in this analysis.
On the other hand, while the number of applications of natural language processing in diabetes is growing quickly, their diversity is lagging behind. Nearly half of the applications we found focused on the same task: identification of documented diagnosis of diabetes. This is particularly surprising because patients with diabetes can be identified with a reasonable degree of accuracy using structured data, such as diagnoses, medications and laboratory test results.47,56 Furthermore, most studies in this area showed a similarly high level of accuracy, with almost all F1 scores above 0.9. It is therefore likely that return on further investment in this particular area will be low and researchers’ efforts would be more productively redirected elsewhere. While a number of clinically important characteristics of diabetes, such as diabetes type or duration, remain difficult to determine from structured electronic medical record data, these were not addressed by the studies we identified.
Hypoglycemia, on the other hand, is a good example of a clinically important event that is poorly documented in structured electronic data.57 A high-fidelity natural language processing tool that could identify documentation of hypoglycemia in provider notes could do much to advance our understanding of its prevalence, risk factors and consequences. Unfortunately, the efforts to develop this tool appear to remain disparate and uncoordinated. Most natural language processing tools described in the studies have not been validated on external (to the institution/locale where they were originally developed) dataset or made available to the public (either through open-source or licensing). Lack of coordination and sharing of tools and resources needed for their development (e.g. annotated de-identified datasets) remains one of the major impediments to advancement of medical natural language processing as a field58,59—in contrast with non-medical natural processing, where such sharing is common.60-62
Majority of studies focused on relatively simple, single-component concepts (of which diagnosis of diabetes was the most common example). There were fewer studies that attempted identification of the more linguistically complex multi-component concepts, and their accuracy tended to be lower (mean F1 of 0.817 vs. 0.909 for single-component concept studies). Analyzing more complex multi-component concepts remains a challenge in the natural language processing field in general because neither of the two predominant approaches—machine-learning (statistical) models or grammar (rule-based) models—include a complete representation of the syntactic and semantic relationships that govern language. Technologies that do involve parsing sentence structure in an attempt to identify these relationships exist, but remain too computationally intensive for practical implementation on a large scale. New technological developments are therefore likely needed to achieve a qualitative improvement in natural language processing accuracy. It should also be noted that many texts (e.g. a punctuation-less sentence I have two hours to kill someone come see me), particularly so in medicine where many documents are not carefully proofread, may be ambiguous. As a result, even highly trained human annotators of medical texts do not usually reach a complete concordance, likely representing the upper bound of the possible accuracy of information extraction.63,64
Most articles described only development of diabetes natural language processing tools that did not appear to have been utilized for any practical purpose—whether direct patient care, population management or research. This was true even for the majority (16 out of 19/84.2%) of natural language processing tools that achieved high accuracy ratings with F1 ≥ 0.9 and were thus apparently ready for prime time. This could have several possible explanations. One is that the natural language processing tools that were developed were not made available to potential users. Another is that the natural language processing tools that were developed were not actually the ones that the users needed. Either way, greater collaboration and cooperation between developers and users of natural language processing tools is needed to ensure that resources being devoted to design and evaluation of this sophisticated technology are utilized most effectively for the benefit of patients and the public-at-large.
Among studies that utilized natural language processing tools for research, predictive modeling was a popular area of interest. Notably, many predictive modeling studies did not assess accuracy of their natural language processing tools. This could be because many predictive models that use data derived from natural language processing are not looking for a pre-specified set of variables. Instead, they use what could be described as a “generalized” natural language processing data collection, whereupon the model includes numerous—hundreds or even thousands of variables—that are derived from the data empirically rather than based on pre-existing evidence or expert opinion. These variables could be as simple as frequencies of unique words (often adjusted for how common they are in a particular document vs. the entire dataset). In that approach the accuracy of any given variable is less critical because mistakes in one variable can be compensated for by many others. Predictive modeling has the potential to significantly impact measurement and interventions to improve quality of diabetes care by helping define at-risk populations that would reap the greatest benefit from interventions. It is a very active area of research and natural language processing has the potential to significantly enhance its accuracy by allowing the models to incorporate information not found in other data sources.65-67
Finally, several studies of natural language processing in diabetes used the technology for “traditional” hypothesis-testing analyses. Several of these investigations were able to link information that was often only found in narrative data (e.g. visual acuity of counseling on lifestyle changes) to outcomes of significant importance to patients (quality of life, risk of malignancy and cardiovascular events). While observational in their nature and thus not able to definitively demonstrate a causal relationship, studies like these can serve to both generate hypotheses that can subsequently be tested in interventional clinical trials and help accumulate body of evidence in areas where clinical trials are not feasible or not likely to be conducted. Once sufficient evidence to support the relationship between the metric identified by natural language processing and diabetes care outcomes is accrued, the technology can then be used to measure quality of treatment of diabetes as delivered in real-world settings and target appropriate interventions, coming full circle to support improvement of outcomes for patients with diabetes.
The findings of the present study should be interpreted in the context of several limitations. Heterogeneity of the studies reviewed did not allow direct analytical comparison of methods or results. We were also not able to find a study that quantified the contribution of narrative vs. structured data in modern electronic health records, and specifically in the care of patients with diabetes.
Conclusion
In summary, natural language processing of data related to treatment and quality of diabetes is a blossoming field of research. It has already been used to power real-world evidence studies addressing a broad range of subjects in care and outcomes of diabetes. Further growth in this area would be aided by deeper collaboration between developers and end-users of natural language processing tools as well as by broader sharing of the tools themselves and related resources.
Supplemental Material
Supplemental material, sj-docx-1-dst-10.1177_19322968211000831 for Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review by Alexander Turchin and Luisa F. Florez Builes in Journal of Diabetes Science and Technology
Supplemental material, sj-docx-2-dst-10.1177_19322968211000831 for Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review by Alexander Turchin and Luisa F. Florez Builes in Journal of Diabetes Science and Technology
Footnotes
Abbreviation: NLP, natural language processing.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Alexander Turchin: equity in Brio Systems; consulting fees from Proteomics International; research funding from Astra Zeneca, Edwards, Eli Lilly, Novo Nordisk, Pfizer and Sanofi.
Luisa F. Florez Builes: none.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Patient-Centered Outcomes Research Institute (ME-2019C1-15328, Turchin).
ORCID iD: Alexander Turchin  https://orcid.org/0000-0002-8609-564X
https://orcid.org/0000-0002-8609-564X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sherman RE, Anderson SA, Dal Pan GJ, et al. Real-world evidence: what is it and what can it tell us? N Engl J Med. 2016;375(23):2293-2297. [DOI] [PubMed] [Google Scholar]
- 2. Krause JH, Saver RS. Real-world evidence in the real world: beyond the FDA. Am J Law Med. 2018;44(2-3):161-179. [DOI] [PubMed] [Google Scholar]
- 3. Klonoff DC. The new FDA real-world evidence program to support development of drugs and biologics. J Diabetes Sci Technol. 2020;14(2):345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Califf RM, Robb MA, Bindman AB, et al. Transforming evidence generation to support health and health care decisions. N Engl J Med. 2016;375(24):2395-2400. [DOI] [PubMed] [Google Scholar]
- 5. Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform. 2017;73:14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7(2):e12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pons E, Braun LM, Hunink MG, Kors JA. Natural language processing in radiology: a systematic review. Radiology. 2016;279(2):329-343. [DOI] [PubMed] [Google Scholar]
- 9. Abbe A, Grouin C, Zweigenbaum P, Falissard B. Text mining applications in psychiatry: a systematic literature review. Int J Methods Psychiatr Res. 2016;25(2):86-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spasic I, Livsey J, Keane JA, Nenadic G. Text mining of cancer-related information: review of current status and future directions. Int J Med Inform. 2014;83(9):605-623. [DOI] [PubMed] [Google Scholar]
- 11. Inzucchi SE, Bergenstal RM, Buse JB, et al. Management of hyperglycemia in type 2 diabetes, 2015: a patient-centered approach: update to a position statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2015;38(1):140-149. [DOI] [PubMed] [Google Scholar]
- 12. Derczynski L, ed. Complementarity, F-score, and NLP evaluation. In: Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC’16); May 2016; 23-28. Portorož, Slovenia. [Google Scholar]
- 13. Stubbs A, Kotfila C, Xu H, Uzuner O. Identifying risk factors for heart disease over time: overview of 2014 i2b2/UTHealth shared task Track 2. J Biomed Inform. 2015;58(suppl):S67-S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang NW, Dai HJ, Jonnagaddala J, Chen CW, Tsai RT, Hsu WL. A context-aware approach for progression tracking of medical concepts in electronic medical records. J Biomed Inform. 2015;58(suppl):S150-S157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei WQ, Tao C, Jiang G, Chute CG. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA Annu Symp Proc. 2010;2010:857-861. [PMC free article] [PubMed] [Google Scholar]
- 16. Akay A, Dragomir A, Erlandsson BE. A novel data-mining platform leveraging social media to monitor outcomes of Januvia. Paper presented at: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 3-7 July 2013;7484-7487. Osaka, Japan. [DOI] [PubMed] [Google Scholar]
- 17. Kotfila C, Uzuner O. A systematic comparison of feature space effects on disease classifier performance for phenotype identification of five diseases. J Biomed Inform. 2015;58(suppl):S92-S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khalifa A, Meystre S. Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes. J Biomed Inform. 2015;58(suppl):S128-S132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cormack J, Nath C, Milward D, Raja K, Jonnalagadda SR. Agile text mining for the 2014 i2b2/UTHealth Cardiac risk factors challenge. J Biomed Inform. 2015;58(suppl):S120-S127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Q, Li H, Tang B, et al. An automatic system to identify heart disease risk factors in clinical texts over time. J Biomed Inform. 2015;58(suppl):S158-S163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou X, Ni Y, Xie G, et al. Analysis of the health information needs of diabetics in China. Stud Health Technol Inform. 2019;264:487-491. [DOI] [PubMed] [Google Scholar]
- 22. Makino M, Yoshimoto R, Ono M, et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci Rep. 2019;9(1):11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nunes AP, Yang J, Radican L, et al. Assessing occurrence of hypoglycemia and its severity from electronic health records of patients with type 2 diabetes mellitus. Diabetes Res Clin Pract. 2016;121:192-203. [DOI] [PubMed] [Google Scholar]
- 24. Upadhyaya SG, Murphree DH, Jr, Ngufor CG, et al. Automated diabetes case identification using electronic health record data at a tertiary care facility. Mayo Clin Proc Innov Qual Outcomes. 2017;1(1):100-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pakhomov SV, Shah ND, Hanson P, Balasubramaniam SC, Smith SA. Automated processing of electronic medical records is a reliable method of determining aspirin use in populations at risk for cardiovascular events. Inform Prim Care. 2010;18(2):125-133. [DOI] [PubMed] [Google Scholar]
- 26. Jin Y, Li F, Vimalananda VG, Yu H. Automatic detection of hypoglycemic events from the electronic health record notes of diabetes patients: empirical study. JMIR Med Inform. 2019;7(4):e14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hazlehurst BL, Lawrence JM, Donahoo WT, et al. Automating assessment of lifestyle counseling in electronic health records. Am J Prev Med. 2014;46(5):457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malmasi S, Ge W, Hosomura N, Turchin A. Comparing information extraction techniques for low-prevalence concepts: the case of insulin rejection by patients. J Biomed Inform. 2019;99:103306. [DOI] [PubMed] [Google Scholar]
- 29. Shivade C, Malewadkar P, Fosler-Lussier E, Lai AM. Comparison of UMLS terminologies to identify risk of heart disease using clinical notes. J Biomed Inform. 2015;58(suppl):S103-S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jonnagaddala J, Liaw ST, Ray P, Kumar M, Chang NW, Dai HJ. Coronary artery disease risk assessment from unstructured electronic health records using text mining. J Biomed Inform. 2015;58(suppl):S203-S210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosomura N, Malmasi S, Timerman D, et al. Decline of insulin therapy and delays in insulin initiation in people with uncontrolled diabetes mellitus. Diabet Med. 2017;34(11):1599-1602. [DOI] [PubMed] [Google Scholar]
- 32. Salmasian H, Freedberg DE, Friedman C. Deriving comorbidities from medical records using natural language processing. J Am Med Inform Assoc. 2013;20(e2):e239-e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Lalor J, Liu W, et al. Detecting hypoglycemia incidents reported in patients’ secure messages: using cost-sensitive learning and oversampling to reduce data imbalance. J Med Internet Res. 2019;21(3):e11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turchin A, Pendergrass ML, Kohane IS. DITTO: a tool for identification of patient cohorts from the text of physician notes in the electronic medical record. AMIA Annu Symp Proc. 2005;2005:744-748. [PMC free article] [PubMed] [Google Scholar]
- 35. Topaz M, Murga L, Grossman C, et al. Identifying diabetes in clinical notes in Hebrew: a novel text classification approach based on word embedding. Stud Health Technol Inform. 2019;264:393-397. [DOI] [PubMed] [Google Scholar]
- 36. Eskildsen NK, Eriksson R, Christensen SB, et al. Implementation and comparison of two text mining methods with a standard pharmacovigilance method for signal detection of medication errors. BMC Med Inform Decis Mak. 2020;20(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H, Goldberg SI, Hosomura N, et al. Lifestyle counseling and long-term clinical outcomes in patients with diabetes. Diabetes Care. 2019;42(9):1833-1836. [DOI] [PubMed] [Google Scholar]
- 38. Morrison F, Shubina M, Turchin A. Lifestyle counseling in routine care and long-term glucose, blood pressure, and cholesterol control in patients with diabetes. Diabetes Care. 2012;35(2):334-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith DH, Johnson ES, Russell A, et al. Lower visual acuity predicts worse utility values among patients with type 2 diabetes. Qual Life Res. 2008;17(10):1277-1284. [DOI] [PubMed] [Google Scholar]
- 40. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One. 2015;10(8):e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Urbain J. Mining heart disease risk factors in clinical text with named entity recognition and distributional semantic models. J Biomed Inform. 2015;58(suppl):S143-S149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dura E, Muresan S, Engkvist O, Blomberg N, Chen H. Mining molecular pharmacological effects from biomedical text: a case study for eliciting anti-obesity/diabetes effects of chemical compounds. Mol Inform. 2014;33(5):332-342. [DOI] [PubMed] [Google Scholar]
- 43. Misra-Hebert AD, Milinovich A, Zajichek A, et al. Natural language processing improves detection of nonsevere hypoglycemia in medical records versus coding alone in patients with type 2 diabetes but does not improve prediction of severe hypoglycemia events: an analysis using the electronic medical record in a large health system. Diabetes Care. 2020;43(8):1937-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Czerniecki JM, Thompson ML, Littman AJ, et al. Predicting reamputation risk in patients undergoing lower extremity amputation due to the complications of peripheral artery disease and/or diabetes. Br J Surg. 2019;106(8):1026-1034. [DOI] [PubMed] [Google Scholar]
- 45. Li X, Yu S, Zhang Z, et al. Predictive modeling of hypoglycemia for clinical decision support in evaluating outpatients with diabetes mellitus. Curr Med Res Opin. 2019;35(11):1885-1891. [DOI] [PubMed] [Google Scholar]
- 46. Turchin A, Hosomura N, Zhang H, Malmasi S, Shubina M. Predictors and consequences of declining insulin therapy by individuals with type 2 diabetes. Diabet Med. 2020;37(5):814-821. [DOI] [PubMed] [Google Scholar]
- 47. Mishra NK, Son RY, Arnzen JJ. Towards automatic diabetes case detection and ABCS protocol compliance assessment. Clin Med Res. 2012;10(3):106-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright A, McCoy AB, Henkin S, Kale A, Sittig DF. Use of a support vector machine for categorizing free-text notes: assessment of accuracy across two institutions. J Am Med Inform Assoc. 2013;20(5):887-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu H, Aldrich MC, Chen Q, et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc. 2015;22(1):179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hao T, Liu H, Weng C. Valx: a system for extracting and structuring numeric lab test comparison statements from text. Methods Inf Med. 2016;55(3):266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng L, Wang Y, Hao S, et al. Web-based real-time case finding for the population health management of patients with diabetes mellitus: a prospective validation of the natural language processing-based algorithm with statewide electronic medical records. JMIR Med Inform. 2016;4(4):e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996;35(4-5):285-301. [PubMed] [Google Scholar]
- 53. Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Work in progress. Radiology. 1990;174(2):543-548. [DOI] [PubMed] [Google Scholar]
- 55. Sager N, Lyman M, Nhan NT, Tick LJ. Medical language processing: applications to patient data representation and automatic encoding. Methods Inf Med. 1995;34(1-2):140-146. [PubMed] [Google Scholar]
- 56. Pacheco JA, Thompson W, Kho A. Automatically detecting problem list omissions of type 2 diabetes cases using electronic medical records. AMIA Annu Symp Proc. 2011;2011:1062-1069. [PMC free article] [PubMed] [Google Scholar]
- 57. Unger J. Uncovering undetected hypoglycemic events. Diabetes Metab Syndr Obes. 2012;5:57-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Friedman C, Rindflesch TC, Corn M. Natural language processing: state of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform. 2013;46(5):765-773. [DOI] [PubMed] [Google Scholar]
- 59. Luo Y, Thompson WK, Herr TM, et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf. 2017;40(11):1075-1089. [DOI] [PubMed] [Google Scholar]
- 60. Dabre R, Kurohashi S. Mmcr4nlp: multilingual multiway corpora repository for natural language processing. arXiv Preprint. Posted online 2017. arXiv 171001025. [Google Scholar]
- 61. Serban IV, Lowe R, Henderson P, Charlin L, Pineau J. A survey of available corpora for building data-driven dialogue systems. arXiv Preprint. Posted online 2015. arXiv 151205742. [Google Scholar]
- 62. Tiedemann J. Recycling Translations: Extraction of Lexical Data from Parallel Corpora and Their Application in Natural Language Processing. Dissertation. Acta Universitatis Upsaliensis; 2003. [Google Scholar]
- 63. Kim H, Bian J, Mostafa J, Jonnalagadda S, Del Fiol G. Feasibility of extracting key elements from ClinicalTrials.gov to support clinicians’ patient care decisions. AMIA Annu Symp Proc. 2016;2016:705-714. [PMC free article] [PubMed] [Google Scholar]
- 64. Deleger L, Li Q, Lingren T, et al. Building gold standard corpora for medical natural language processing tasks. AMIA Annu Symp Proc. 2012;2012:144-153. [PMC free article] [PubMed] [Google Scholar]
- 65. Rumshisky A, Ghassemi M, Naumann T, et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Transl Psychiatry. 2016;6(10):e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang X, Kim J, Patzer RE, Pitts SR, Patzer A, Schrager JD. Prediction of emergency department hospital admission based on natural language processing and neural networks. Methods Inf Med. 2017;56(5):377-389. [DOI] [PubMed] [Google Scholar]
- 67. Demner-Fushman D, Elhadad N. Aspiring to unintended consequences of natural language processing: a review of recent developments in clinical and consumer-generated text processing. Yearb Med Inform. 2016;(25):224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-dst-10.1177_19322968211000831 for Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review by Alexander Turchin and Luisa F. Florez Builes in Journal of Diabetes Science and Technology
Supplemental material, sj-docx-2-dst-10.1177_19322968211000831 for Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review by Alexander Turchin and Luisa F. Florez Builes in Journal of Diabetes Science and Technology