Abstract
The origin of genes has been a major topic of research for many years, albeit in some cases, it has been a difficult process to elucidate. Insightful is a recent publication that experimentally shows how one gene, linc-UR-UB was born. This gene is regulated in a complex manner in male germ cells during spermatogenesis and is believed to participate in the regulation of levels of the ubiquitin specific peptidase 18 (USP18) mRNA. The process of formation of linc-UR-UB appears relatively simple. It involves a transcription read through from an upstream gene to a downstream functional element, the USP18 3' UTR sequence. This small element also shares the same sequence as the 3' ends of the lincRNA FAM247 family genes. In addition to linc-UR-UB, it is possible that other genes formed in a similar fashion that involves a genomic sequence read through to a functional element.
Keywords: gene birth, lincRNA, USP18, evolution, gene structure, 3' UTR
Introduction
The recent paper by Sandra Pellegrini and co-workers in Frontiers in Genetics (Rubino et al., 2021) is multifaceted. The authors describe non-coding RNA components involved in the regulation of interferon and the JAK/STAT signaling pathway by the ubiquitin specific protease USP18, and they show binding of certain miRNAs to the USP18 mRNA 3' UTR, which is proposed to regulate USP18 expression. They also discovered a new long intergenic non-coding RNA (lincRNA) gene termed linc-UR-B1 that may form part of a network that regulates USP18 mRNA levels and act by a sponging process. Intriguing is the formation of linc-UR-B1, which the authors experimentally elucidated. The study by Rubino and co-workers on the linc-UR-B1 transcript also touches on the complexity and definition of eukaryotic genes (Carninci and Hayashizaki, 2007; Portin and Wilkins, 2017), and here, we discuss aspects of read through transcripts that may form new genes.
Structure and Formation of linc-Ur-B1
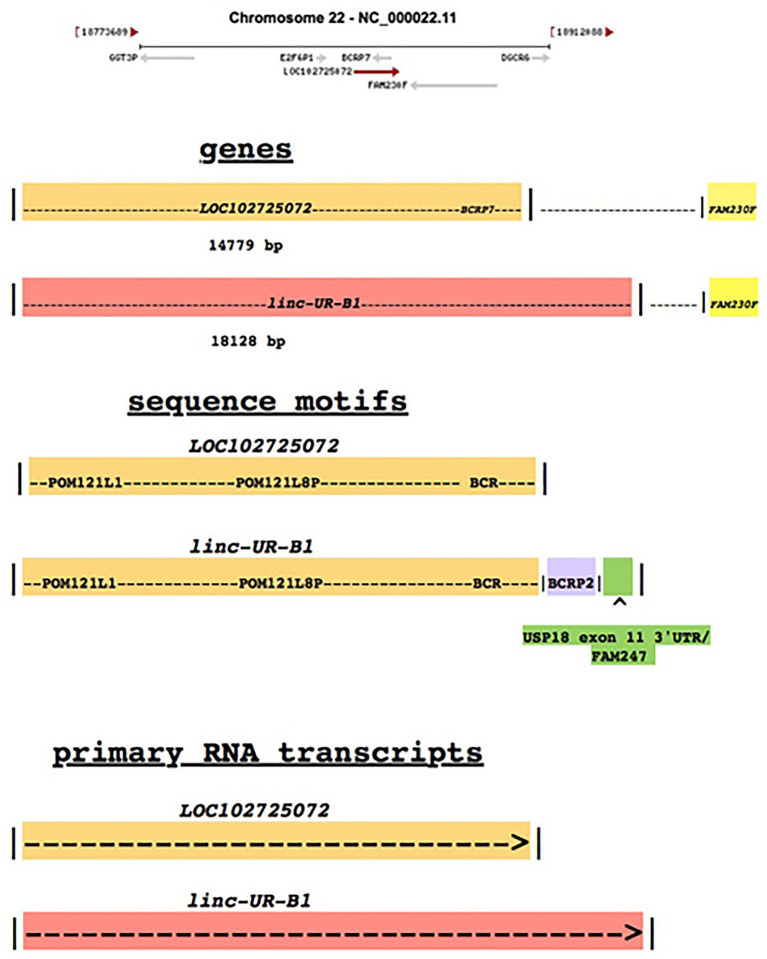
Linc-UR-B1 contains the upstream sequence of gene LOC102725072 and uses the transcriptional start of LOC102725072. In the downstream genomic region of LOC102725072, a sequence homologous to the USP18 exon 11 3' UTR/3' end of the lincRNA gene family FAM247A-D is present and this element is incorporated into the linc-UR-B1 sequence as a result of transcription read through (Figure 1). The terminal ends of FAM247A, C, and D carry the USP18 mRNA exon 11 and 3' UTR sequence. The 3' ends of USP18 and FAM247 family genes are nearly identical; there is 99.8% identity between the USP18 exon 11 3' UTR/3' end of FAM247A. Because of this high identity, the original source of the USP18 exon 11 3' UTR/3' end FAM247 sequence downstream of LOC102725072 is uncertain. A segment of the BCRP2 pseudogene sequence is also present in this downstream region (Figure 1, highlighted in light blue), but the function of this segment is unknown. Thus, linc-UR-B1 is formed by read through transcription of the LOC102725072 gene to include the BCRP2 segment and the USP18 exon 11 3' UTR/3' end of the FAM247 sequence, which may be the functional key player. Linc-UR-B1 encodes two transcript isoforms termed TCONS_00029753 and TCONS_00029754 (Rubino et al., 2021). LOC102725072 by itself encodes RNA transcripts NR_135922 and NR_170942.1. As linc-UR-B1 is regulated in a complex fashion in male germ cells, there are specific transcriptional regulatory elements that regulate linc-UR-B1 expression during spermatogenesis.
Figure 1.
Comparison of LOC102725072 and linc-UR-B1. Top diagram: Annotated genes that neighbor LOC102725072 in human chr22 with chromosomal coordinates of the genomic segment shown. The schematic is from the NLM/NCBI website: https://www.ncbi.nlm.nih.gov/gene/?term=LOC102725072 (O’Leary et al., 2016). Bottom highlighted diagrams: The BCRP7 gene is a counter transcript within the LOC102725072 gene. LOC102725072 consists of the sequence of the paralogs gene POM121L8P and the POM121L1 (LOC102724151) sequence at its 5' end that is carried by POM121L8P; and in addition, a segment of the BCRP2 gene that carries a portion of the 3' end of the BCR gene (Supplementary Figure S1). The BCRP2 sequence extends from BCRP7 to the 3' end of LOC102725072 (and beyond) and is contiguous with the BCRP7 sequence. The lower diagrams represent the linc-UR-B1 gene and the signature gene/sequence motifs that LOC102725072 and linc-UR-B1 contain. Also shown are the bp lengths of the genes. The BCRP2 gene sequence (of which 2,124 bp of the sequence resides beyond the 3' end of LOC102725072) forms part of linc-UR-B. In addition, 1,225 bp of a homologous sequence to FAM247 is also in linc-UR-B1, which contains the USP18 exon 11 3' UTR sequence but also contains additional sequences of FAM247. POM121L1 is the putative POM121 transmembrane nucleoporin like 1 protein gene LOC10272415 but has not yet been characterized outside of computational methods. The diagrams are not drawn to scale.
Although linc-UR-B1 and LOC102725072 use the same transcriptional start site, we consider linc-UR-B1 to be a separate gene as it is a read through transcript, carries additional sequences not included in either LOC102725072 or its transcripts, and one added sequence provides a functional element, the USP18 3'UTR. Of interest, Ensembl/GENCODE provides a discussion on how they annotate read through transcripts as genes and the difficulties in this process.1
From BLAST searches performed by this author, ~83% of the LOC102725072 gene sequence is found present in chimpanzee chr 22. Additionally, the BCRP2 sequence is also present in the downstream region; importantly, however, neighbor sequences, the FAM247 sequence bearing the USP18 terminal exon 11 3' UTR and the FAM230F lincRNA gene are not present (Supplementary Figure S2). Thus, most of the sequence that forms LOC102725072 and the entire associated BCRP2 downstream segment are in place in the primate ancestor, but the 3' end of USP18/FAM247 is not; thus, the functional USP exon11 3' UTR motif was added in the human genome, presumably to enable the formation and the function of linc-UR-B1.
The FAM247A-D lincRNA gene family is believed to have formed in humans by the process of gene duplication via chromosomal segmental duplications or low copy repeats in chromosome 22 (Delihas, 2020). These genes show ubiquitous RNA expression in somatic tissues but major expression in fat, brain, and testes (O’Leary et al., 2016).2 The functions of these genes are unknown. The FAM247A sequence has been used as a standard for sequence and evolutionary comparisons and termed FAM247 for practical purposes. Different sections of the FAM247 sequence are found to be components of diverse genes, which include two ancient protein genes, one of which is USP18.
Linc-UR-B1 is an experimentally determined RNA gene that is formed by the simple addition of a functional element to the downstream region of an existing gene and a transcriptional read through to the functional element, the 3' UTR USP18/3' end FAM247 (Figure 1). It should be noted that there is an analogy between linc-UR-B1 and a human neuronal transcript, PTENJ2 that encodes an altered PTEN protein (Lerch et al., 2012). This transcript is described by the authors as a diverse or non-conventional isoform of PTEN that has novel 5' and 3' UTRs. PTEN is a phosphatase and tensin homolog.
Sequence Motifs in linc-Ur-B1 Are Related to Those in Genomic Non-Coding Regions With Fam247 as the Consistent Element
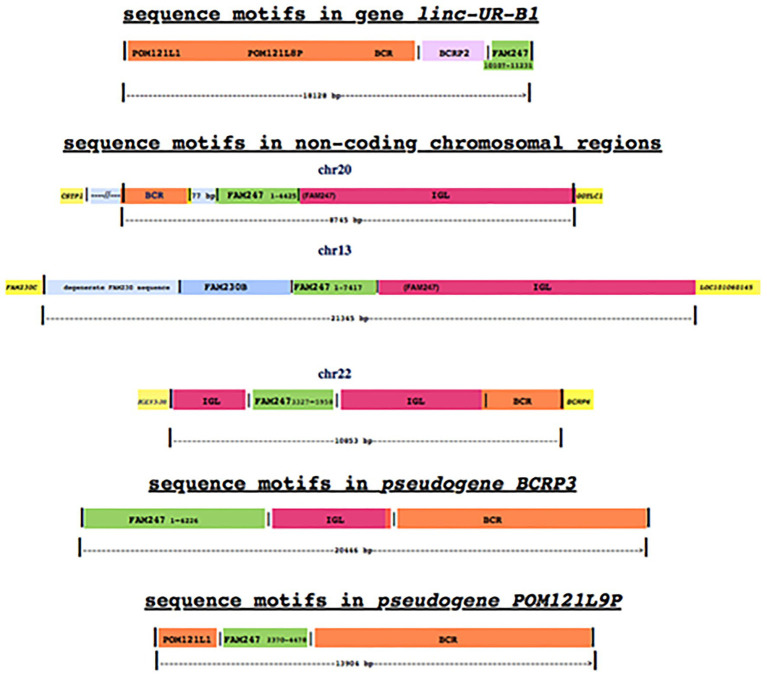
Three non-coding chromosomal loci have been detected by a BLAT sequence/gene search of the human genome (Madeira et al., 2019) by using a 2.87 kb sequence query from the ancestral primate Philippine tarsier, which consists of the chromosomal sequence between genes GGT1 and GGT5. This sequence contains an ancestral homolog to the 5' end of FAM247. The region (between GGT1 and GGT5) is of major evolutionary significance as it shows a very large genomic expansion in the Rhesus monkey, to 216.20 kb (Delihas, 2020). The three non-coding loci are in human chromosomes 20, 13, and 22 (Figure 2); in chr22, the non-coding region is part of the large immunoglobulin lambda (IGL) locus. These regions show no gene expression by RNA-seq analyses in somatic tissues according to RNA expression analyses3 but display sequence motifs that represent segments of genes, some of which are similar to those shown for linc-UR-B1. What is striking is the similarity in the “cast of characters” consisting of segments of various genes/sequences linked to different segments of FAM247, where FAM247 (highlighted in green) is the motif present in all examples (Figure 2; Supplementary Figures S3A–C). However, the individual segments of FAM247 differ, including the FAM247 sequence present in the two pseudogenes, BCRP3 and POM121L9P (Figure 2, bottom); linc-UR-B1 is the only example that carries the USP18 exon 11 3' UTR/3' end of FAM247). Although there are similarities in sequence motifs between the linc-UR-B1 gene, the three non-coding regions and the two pseudogenes, especially between linc-UR-B1 and POM121L9P (Figure 2, bottom diagram), these non-coding chromosomal regions are not well-understood, e.g., whether or not the FAM247 sequence formed the basis for the addition of gene motifs to the non-coding loci. The regions do date back to the chimpanzee genome, and also to the Rhesus monkey where partial sequences and signatures are also present. Aside from FAM247, functions of other gene motifs in the linc-UR-B1 gene are not known.
Figure 2.
A diagrammatic comparison of non-coding chromosomal loci with linc-UR-B1 and genes that display similar gene/sequence motifs. The numbers next to FAM247 are the bp positions of the FAM247 sequences that are present in each non-coding region and each gene; the segments of the FAM247 sequence that is present in the genes/non-coding regions differ in each example shown. In chr 20, 4,425 bp of FAM247 includes 3,305 bp present in and contiguous with the immunoglobulin lambda (IGL) sequence; in chr 13, the 7,442 bp shown includes 3,298 bp of FAM247 that are also in the IGL sequence. The genes highlighted in yellow flank the non-coding regions and represent guideposts. Supplementary Figures S3A–C provides an analysis of these regions. The POM121L9P and BCRP3 gene structures are from Delihas (2020).
We do not know why gene/sequence signatures in non-coding regions have been maintained during primate evolution; however, one possibility is that they have been reserved for the birth of future genes. Human pseudogenes BCRP3 and POM121L9P (Figure 2, bottom diagrams) as well as LOC102725072 (Figure 1) may be candidates for genes that were formed in humans from sequences in lower primate non-coding chromosomal regions. More needs to be learned about how fragments of specific genes came together in non-coding regions, their possible functions, and how these phylogenetically conserved sequences might transform into viable genes.
Conclusion
Although the evolutionary history of the formation of linc-UR-B1 is incomplete, the work of Rubino et al. (2021) significantly advances our knowledge of gene birth by showing how linc-UR-B1 was created in humans. And there is beauty in the simplicity of this process-insertion of a small functional unit close to the end of an existing gene, utilizing the existing transcriptional apparatus and setting in place specific regulatory mechanisms for expression of this new gene in specific cells. This basic process of gene formation may also have prevailed in the creation of other lincRNA genes, although, this remains to be determined. At least one other known transcript, PTENJ2 shows some analogous properties. The Rubino et al. (2021) study also adds to the multifaceted properties of 3'UTRs, properties that other investigators have previously described. For example, Mercer et al. (2011) and Kocabas et al. (2015) showed that many 3'UTR sequences are independently expressed in the absence of protein coding sequences and that their expression is regulated during development.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
ND initiated the concept of the paper and wrote the paper.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.661425/full#supplementary-material
References
- Carninci P., Hayashizaki Y. (2007). Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 17, 139–144. 10.1016/j.gde.2007.02.008, PMID: [DOI] [PubMed] [Google Scholar]
- Delihas N. (2020). Genesis of non-coding RNA genes in human chromosome 22-A sequence connection with protein genes separated by evolutionary time. Noncoding RNA 6:36. 10.3390/ncrna6030036, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas A., Duarte T., Kumar S., Hynes M. A. (2015). Widespread differential expression of coding region and 3'UTR sequences in neurons and other tissues. Neuron 88, 1149–1156. 10.1016/j.neuron.2015.10.048, PMID: [DOI] [PubMed] [Google Scholar]
- Lerch J. K., Kuo F., Motti D., Morris R., Bixby J. L., Lemmon V. P. (2012). Isoform diversity and regulation in peripheral and central neurons revealed through RNA-Seq. PLoS One 7:e30417. 10.1371/journal.pone.0030417, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. 10.1093/nar/gkz268, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Wilhelm D., Dinger M. E., Soldà G., Korbie D. J., Glazov E. A., et al. (2011). Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res. 39, 2393–2403. 10.1093/nar/gkq1158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. 10.1093/nar/gkv1189, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portin P., Wilkins A. (2017). The evolving definition of the term “Gene.” Genetics 205, 1353–1364. 10.1534/genetics.116.196956, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino E., Cruciani M., Tchitchek N., Le Tortorec A., Rolland A. D., Veli O., et al. (2021). Human USP18 is regulated by miRNAs via the 3'UTR, a sequence duplicated in lincRNA genes residing in chr22q11.21. Front. Genet. 11:627007. 10.3389/fgene.2020.627007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.