Abstract
Background: Hepatitis B virus infection is associated with liver disease, including cancers. In this study, we assessed the power of sex-determining region Y (SRY)-related high-mobility group (HMG)-box 4(SOX4) gene to predict the clinical course of hepatocellular carcinoma (HCC).
Methods: To evaluate the differential expression of SOX4 and its diagnostic and prognostic potential in HCC, we analyzed the GSE14520 dataset. Stratified analysis and joint-effect analysis were done using SOX4 and clinical factor. We then designed a nomogram for predicting the clinical course of HCC. Differential SOX4 expression and its correlation with tumor stage as well as its diagnostic and prognostic value were analyzed on the oncomine and GEPIA websites. Gene set enrichment analysis was explored as well as candidate gene ontology and metabolic pathways modulated by in SOX4 HCC.
Results: Our analysis revealed that the level of SOX4 was significantly upregulated in tumor issue (P <0.001). This observation was validated through oncomine dataset and MERAV analysis (all P <0.05). Diagnostic receiver operating characteristic (ROC) analysis of SOX4 suggested it has diagnostic potential in HCC (GSE14520 dataset: P <0.001, area under curve (AUC) = 0.782; Oncomine: (Wurmbach dataset) P = 0.002, AUC = 0.831 and (Mas dataset) P <0.001, AUC = 0.947). In addition, SOX4 exhibited high correlation with overall survival of HBV-associated HCC (adjusted P = 0.004, hazard ratio (HR) (95% confidence interval (CI)) = 2.055 (1.261-3.349) and recurrence-free survival (adjusted P = 0.008, HR (95% CI) = 1.721 (1.151-2.574). These observations which were verified by GEPIA analysis for overall survival (P = 0.007) and recurrence-free survival (P= 0.096). Gene enrichment analysis revealed that affected processes included lymphocyte differentiation, pancreatic endocrine pathways, and insulin signaling pathway. SOX4 prognostic value was evaluated using nomogram analysis for HCC 1, 3, and 5-year, survival.
Conclusion: Differential SOX4 expression presents an avenue of diagnosing and predicting clinical course of HCC. In HCC, SOX4 may affect TP53 metabolic processes, lymphocyte differentiation and the insulin signaling pathway.
Keywords: SOX4, hepatocellular carcinoma, hepatitis B virus, mRNA expression
Introduction
Cancers affecting liver tissues have been on the rise, making liver cancer the fourth cause of deaths and sixth most prevalent cancer globally 1. Specifically, hepatocellular carcinoma is the most common type of primary liver cancers. Liver cancer is estimated to be the fourth commonest cancer among Chinese males 2. Majority of liver cancers have been associated hepatitis B virus (HBV) infection. While advances in diagnostic and treatment strategies have improved HCC clinical outcomes, its 5-year survival remains low (<15%) 3. Early detection and more effective management of liver cancer is therefore necessary. While some HCC prognostic biomarkers have been recommended, such as α-fetoprotein (AFP) 4 and PIVKA-II 5, HCC survival is still poor. Better understanding of the mechanisms of HCC development and progression, as well as the identification of novel prognostic biomarkers is needed.
Sex-determining region Y (SRY)-related high-mobility group (HMG)-box(SOX) genes are evolutionarily conserved and are thought to be regulate in cell fate determination during development 6. During embryogenesis, this family of genes participates in the development of neuronal tissue, nervous system and as well as skeletal tissue 6. SOX4 comprises three domains - a serine-rich region, a glycine-rich region and an HMG box 7. This gene participates in tumorigenesis and progression. It has also been verified that SOX4 regulates lymphocyte differentiation and development, and drive endocardial ridge development 8. It is emerging that SOX4 is markedly upregulated in various human cancers, including breast cancer 9, 10, colorectal cancer 11, gastric cancer 12 and HCC 13, 14. SOX4 expression has also been associated with prognosis of some cancer types 7, 15. Lack of SOX4 expression in normal adult liver does not affect normal liver function 16. Nevertheless, the exact role of SOX4 gene along the clinical course of HCC is yet to be fully uncovered. This study evaluated the prognostic and diagnostic value of SOX4 in HBV-associated HCC.
Materials and methods
Date source
The GSE14520 dataset for SOX4 expression, and corresponding clinical data on HBV-related HCC were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520, accessed December 15, 2017) 17, 18. Data on the expression of SOX4 in tumor vs. non-tumor tissue were downloaded from metabolic gene rapid visualizer (MERAV, http://merav.wi.mit.edu/, accessed December 15, 2017) 19.
Bioinformatic analysis and SOX4 diagnostic potential
In order to investigate the biological function and pathways associated with SOX4, we performed a gene ontology (GO) term analysis of SOX4 using the bionetwork gene ontology tool (BinGO) in Cytoscape_version3.4.0. GeneMANIA (http://www.genemania.org/, accessed December 17, 2017) 20, 21 and STRING (https://string-db.org/, accessed December 17, 2017) 22, 23 were used to investigate SOX4 gene-gene and protein-protein interactions, respectively. In order to explore the diagnostic value of the SOX4, we used T-test analysis to compare its expression in tumor vs non-tumor tissues in the GSE14520 dataset and then conducted a receiver operating characteristic (ROC) analysis. Diagnostic value was considered statistically significant when P<0.05 and area under curve (AUC) >0.7.
Survival analysis, joint-effect and stratified analysis
For survival analysis, patient data were divided into 2 categories on the basis of median SOX4 mRNA expression. RFS (recurrence-free survival) and OS (overall survival) were estimated using Cox proportional hazards regression and Kaplan-Meier models. Clinical factors found to be statistically significant were adjusted for survival analysis and joint effects survival analysis for SOX4. SOX4 gene was then combined with AFP for survival analysis. Furthermore, SOX4 gene expression was subclassified to effectively perform analyses for clinical factors. Next, factors found to be significant were included in multivariate analysis.
Gene set enrichment analysis (GSEA)
To investigate the prognostic value of SOX4 in HCC, the difference in biological functions and pathways between high and low SOX4 expression groups were analyzed using GSEA (http://software.broadinstitute.org/gsea/index.jsp,accessed December 19, 2017) 24, 25. This analysis evaluated the molecular signatures database (MSigDB) of c2 (c2.cp.kegg.v6.1.symbols) and c5 (c5.all.v6.1.symbols). Differences were considered statistically significant is the P value <0.05 and false discovery rate <0.25.
Nomogram construction of SOX4 and prognosis-related clinical factors
Nomogram analysis was used to 1, 3-, and 5-year OS and RFS. The nomogram was constructed using prognosis-related clinical factors and SOX4 expression. Different factors and genes had different expression scores.
Validation cohorts
To validate the diagnostic value of SOX4, we analyzed SOX4 expression in tumor tissue vs tumor-adjacent normal tissue using the Mas 26 and Wurmbach 27 liver datasets on Oncomine (http://www.oncomine.org/, accessed December 21, 2017). Differential SOX4 expression analysis was also performed using the aforementioned datasets. SOX4 expression scatter plots, staging verification and prognosis verification were conducted using gene expression profiling interactive analysis (GEPIA, http://gepia.cancer-pku.cn/index.html, accessed December 21, 2017) 28.
Statistical analysis
Data were analyzed using SPSS version 24.0 (IBM corporation, Armonk, NY, USA) and R 3.6.0. The log-rank P and median survival time (MST) were determined using Kaplan-Meier method. The hazard ratio (HR) and 95% confidence interval (CI) were estimated using univariate and multivariate Cox proportional hazards regression models. Differential SOX4 expression between tumor and non-tumor tissue was analyzed by T-test. P < 0.05 was considered significant.
Results
Differential expressions and diagnostic analysis
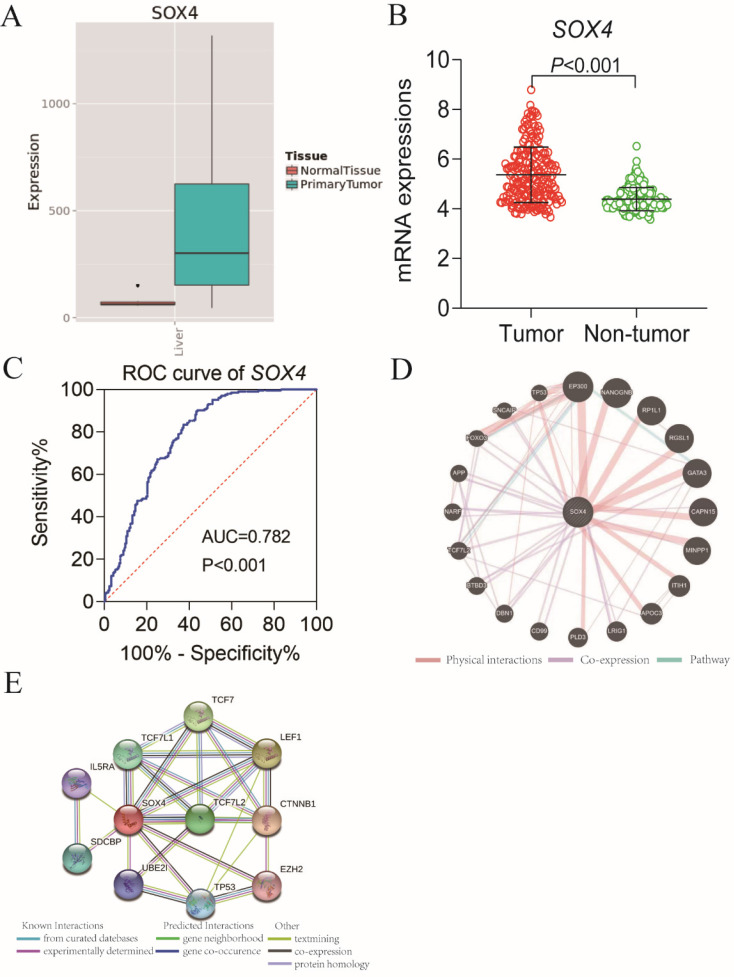
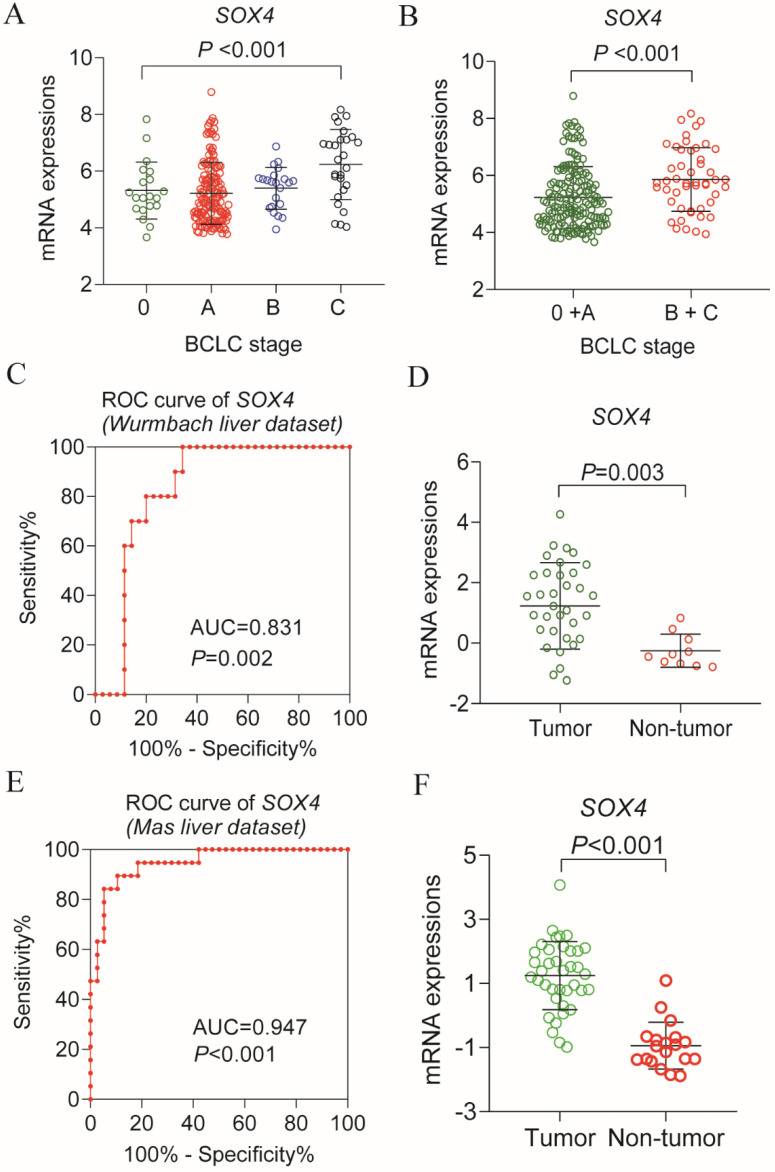
Our analysis of the GSE14520 and MERAV dataset revealed elevated SOX4 expression in HCC tissues relative to normal tissue (P <0.001, Figure 1A-B). The ROC analysis of SOX4 in the GSE14520 dataset, HBV-related HCC cohort indicated that SOX4 had a high accuracy of distinguishing tumor tissues from adjacent non-tumor liver tissues (P <0.001, AUC of the ROC curves = 0.782; Figure 1C).
Figure 1.
Scatter plots, ROC analysis, and interaction network of SOX4 gene. (A) Scatter plots of SOX4 expression by MERAV website. (B) Scatter plots of SOX4 expression in GSE14520 cohort. (C) Diagnostic receiver operating characteristic curve of SOX4 in GSE14520 cohort. (D) Gene-gene interaction network of SOX4 gene. (E) Protein-protein interaction network of SOX4 protein.
Survival analysis of SOX4 in OS and RFS
In order to avoid the batch effect of microarray data, only the dataset of Affymetrix HT Human Genome U133A Array of GSE14520 was included in the current study. Because most of the patients in GSE14520 were HBV-related HCC, we excluded those patients without HBV infection reports and survival information. As a result, there were 212 HBV-related HCC patients were included in the current study, and all of the 212 HBV-related HCC patients and had prognosis information. Our analysis of suggested that the cirrhosis and Barcelona Clinic Liver Cancer (BCLC) stage significantly correlate with OS and RFS (OS: P = 0.041, <0.001; RFS: 0.036, <0.001 respectively; Supplementary Table 1) 29. Tumor size and APF significantly associate with OS (P = 0.002, 0.049, respectively; Supplementary Table 1). This analysis further revealed that gender significantly correlates with RFS (P = 0.021; Supplementary Table 1).
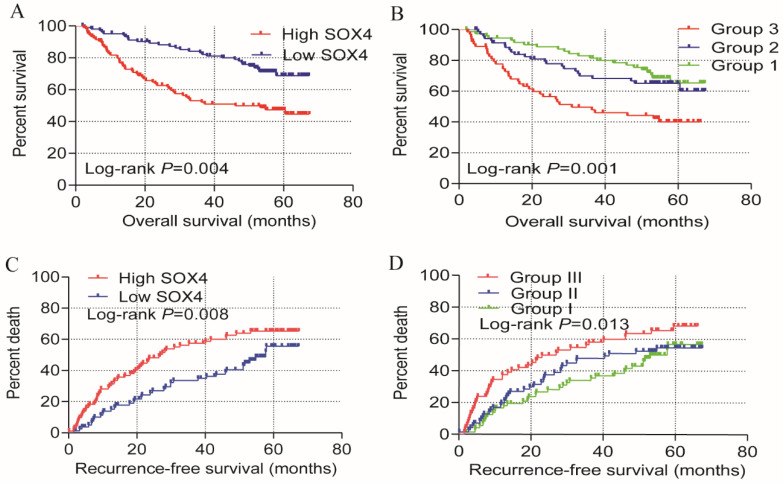
Univariate OS analysis, revealed that SOX4 expression significantly correlates with survival (crude P<0.001, HR = 2.397, 95% CI = 1.522-3.775; Table 1, Figure 2). Similar results were obtained from multivariate OS analysis (adjusted P = 0.004, HR = 2.055, 95% CI = 1.261-3.349; Table 1, Figure 2). Univariate analysis of RFS revealed that SOX4 expression significantly correlates with survival (crude P = 0.001, HR = 1.896, 95% CI = 1.307-2.750; Table 1, Figure 2) and similar results were obtained by multivariate RFS analysis (adjusted P = 0.008, HR = 1.721, 95% CI=1.151-2.574; Table 1, Figure 2).
Table 1.
Prognostic analysis of SOX-4 gene in HBV-related HCC of GSE14250 cohort
| Type | Gene expression | Patients (n=212) | OS | |||||
|---|---|---|---|---|---|---|---|---|
| NO. of event | MRT (months) | Crude HR | Crude P | Adjusted HR (95% CI) | Adjusted P# | |||
| OS | Low | 106 | 29 | NA | Ref. | Ref. | ||
| High | 106 | 53 | 46.1 | 2.397 (1.522-3.775) | 0.000 | 2.055 (1.261-3.349) | 0.004 | |
| RFS | Low | 106 | 47 | 57.9 | Ref. | Ref. | ||
| High | 106 | 69 | 26.4 | 1.896 (1.307-2.750) | 0.001 | 1.721 (1.151-2.574) | 0.008 | |
Note: #: P values were adjusted for age, gender, tumor size, multinodular, cirrhosis, AFP and BCLC stage; Bold indicates significant P values.
Figure 2.
Univariate prognosis analysis and joint-effect analysis of SOX4. (A) OS analysis plot of SOX4. (B) Joint-effect OS analysis plot of SOX4 and AFP. (C) RFS analysis plot of SOX4. (D) Joint-effect RFS analysis plot of SOX4 and AFP. OS = overall survival, RFS = recurrence-free survival.
Stratified analysis and joint-effect analysis
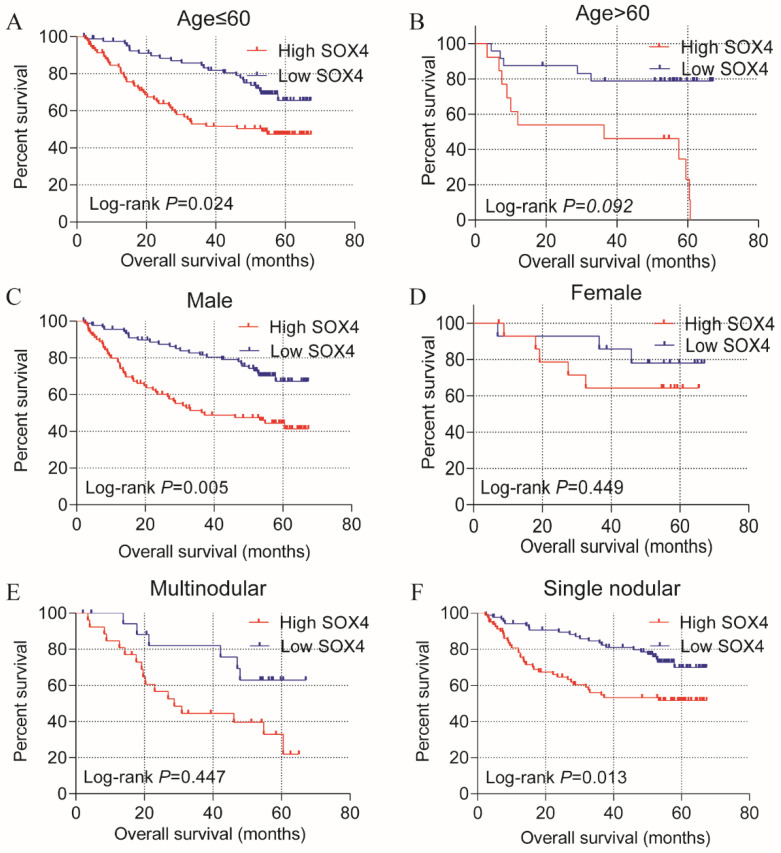
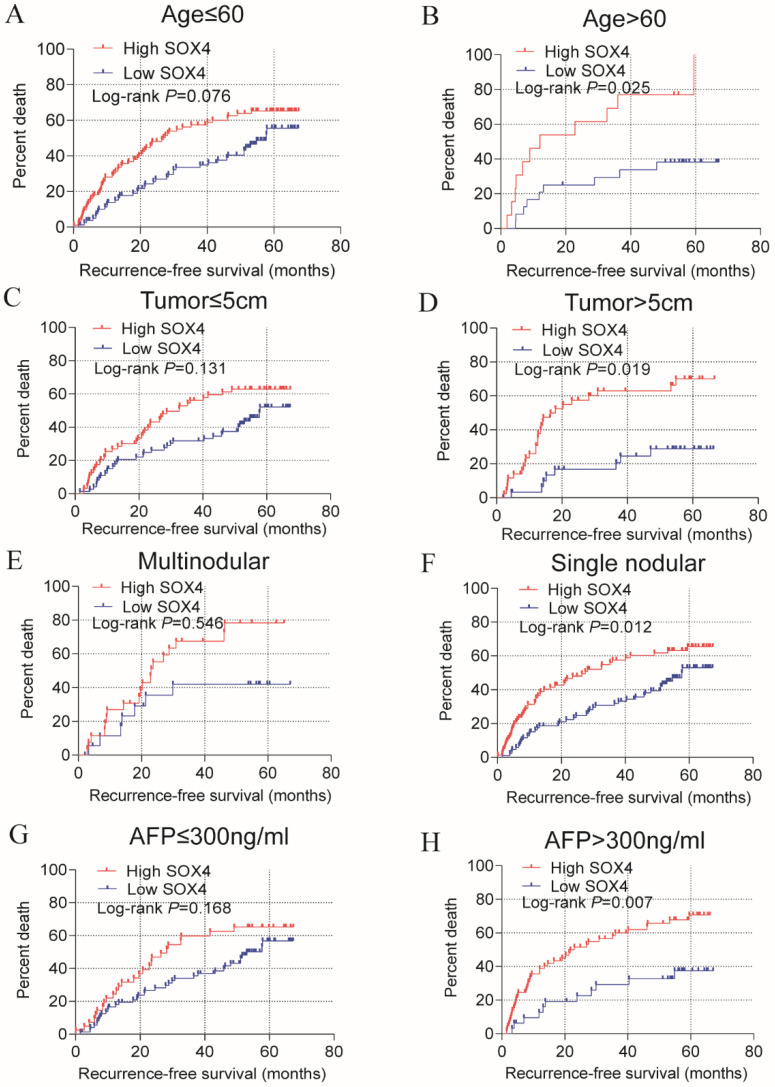
Stratified analysis of how SOX4 influences OS and RFS indicated age (≤60), being male and single nodular significantly correlate with HCC OS (P = 0.024, 0.005 and 0.013, respectively; Figure 3; Table 2). An age of >60 years, tumor size >5 cm, single nodular and AFP >300 ng/mL were associated with a longer RFS relative to others (P = 0.025, 0.019, 0.012 and 0.007 respectively; Figure 4; Table 2).
Figure 3.
Statistical OS analysis plot of SOX4 by clinical factors. (A-B): Statistical OS analysis plot of SOX4 by, (A) age ≤60, (B) >60, (C) male, (D) female, (E) multinodular, and (F) single nodular.
Table 2.
Stratified analysis of SOX-4 for overall survival and recurrence-free survival
| Variables | Overall survival | Recurrence-free survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Low | High | Adjusted HR (95%CI) | Adjusted P value | Low | High | Adjusted HR (95%CI) | Adjusted P value | |
| Age (years) | ||||||||
| ≤60 | 80 | 92 | 1.830 (1.084-3.090) | 0.024 | 80 | 92 | 1.469 (0.960-2.246) | 0.076 |
| >60 | 23 | 13 | 2.947 (0.839-10.352) | 0.092 | 24 | 13 | 2.890 (1.114-7.297) | 0.025 |
| Gender | ||||||||
| Male | 90 | 90 | 2.069 (1.251-3.420) | 0.005 | 92 | 91 | 1.617 (1.085-2.409) | 0.018 |
| Female | 13 | 15 | 2.068 (0.315-13.578) | 0.449 | 14 | 15 | 1.744 (0.352-8.643) | 0.496 |
| Tumor size (cm) | ||||||||
| ≤5 | 73 | 63 | 1.322 (0.684-2.591) | 0.399 | 74 | 63 | 1.450 (0.895-2.349) | 0.131 |
| >5 | 30 | 42 | 3.413 (1.523-7.646) | 0.003 | 31 | 43 | 2.237 (1.143-4.377) | 0.019 |
| Cirrhosis | ||||||||
| Yes | 93 | 98 | 1.164 (0.695-1.949) | 0.563 | 96 | 99 | 1.636 (1.103-2.425) | 0.014 |
| No | 10 | 7 | 2.121 (0.127-35.360) | 0.600 | 10 | 7 | 0.878 (0.126-6.136) | 0.896 |
| Multinodular | ||||||||
| Yes | 19 | 26 | 1.489 (0.534-4.153) | 0.447 | 19 | 26 | 1.322 (0.535-3.268) | 0.546 |
| No | 84 | 79 | 2.044 (1.163-3.592) | 0.013 | 87 | 80 | 1.725 (1.130-2.636) | 0.012 |
| AFP (ng/ml) | ||||||||
| ≤300 | 74 | 41 | 1.539 (0.791-2.992) | 0.204 | 74 | 41 | 1.448 (0.856-2.451) | 0.168 |
| >300 | 29 | 64 | 3.553 (1.542-18.186) | 0.003 | 30 | 64 | 2.557 (1.297-5.039) | 0.007 |
| BCLC stage | ||||||||
| 0 | 11 | 9 | 1.871 (0.117-29.956) | 0.658 | 11 | 9 | 4.624 (0.686-31.164) | 0.116 |
| A | 78 | 61 | 1.773 (0.969-3.245) | 0.063 | 81 | 62 | 1.399 (0.884-2.216) | 0.152 |
| B | 8 | 14 | 1.206 (0.247-5.898) | 1.206 | 8 | 14 | 1.026 (0.310-3.393) | 0.996 |
| C | 6 | 21 | 3.801 (0.838-17.234) | 3.801 | 6 | 21 | 2.170 (0.630-7.471) | 0.220 |
Figure 4.
Statistical RFS analysis plot of SOX4 by clinical factors. (A-B) Statistical OS analysis plot of SOX4 by (A) age ≤60, (B) >60, (C) tumor ≤5 cm, (D) tumor >5 cm, (E) multinodular, (F) single nodular, (G) AFP ≤300 ng/ml and (I) AFP >300 ng/ml.
Analysis of survival on the GSE14520 cohort revealed that SOX4 expression is significantly associated with HCC OS and RFS. Previous studies have reported that AFP is associated with the HCC diagnosis and prognosis. We therefore investigated the combined role of SOX4 and AFP expression on HCC OS and RFS. Analysis of the GSE14520 cohort indicated that the risk of death and recurrence was significantly higher in patients exhibiting high AFP and SOX4 expression when compared to those with low (Figure 2; Table 3).
Table 3.
Joint-effect analysis of SOX4 and AFP for overall survival and recurrence-free survival
| Type | Group | SOX4 expression | APF expression | No. of event | MST (month) | Crude HR (95%CI) | Crude P value | Adjusted HR (95%CI) | Adjusted P value# |
|---|---|---|---|---|---|---|---|---|---|
| OS | 1 | Low | Low | 22/74 | 55.6 | Ref. | 0.001 | Ref. | 0.009 |
| 2 | Low | High | 24/71 | 51.0 | 1.238 (0.694-2.208) | 0.470 | 1.380 (0.718-2.651) | 0.333 | |
| High | Low | ||||||||
| 3 | High | High | 36/64 | 37.6 | 2.617 (1.537-4.455) | <0.001 | 4.667 (1.631-13.358) | 0.004 | |
| RFS | 1 | Low | Low | 36/74 | 45.5 | Ref. | 0.013 | Ref. | 0.163 |
| 2 | Low | High | 37/71 | 41.3 | 1.136 (0.718-1.797) | 0.587 | 0.960 (0.598-1.542) | 0.867 | |
| High | Low | ||||||||
| 3 | High | High | 43/64 | 31.4 | 1.861 (1.194-2.901) | 0.006 | 1.537 (0.964-2.452) | 0.071 |
Note: Group 1 stands for a combination of AFP low expression and SOX4 low expression; Group 2 stands for combinations of AFP low expression and SOX4 high expression, as well as AFP high expression and SOX4 low expression; Group 3 stand for a combination of AFP high expression and SOX4 high expression.
Prognostic nomogram for survival prediction
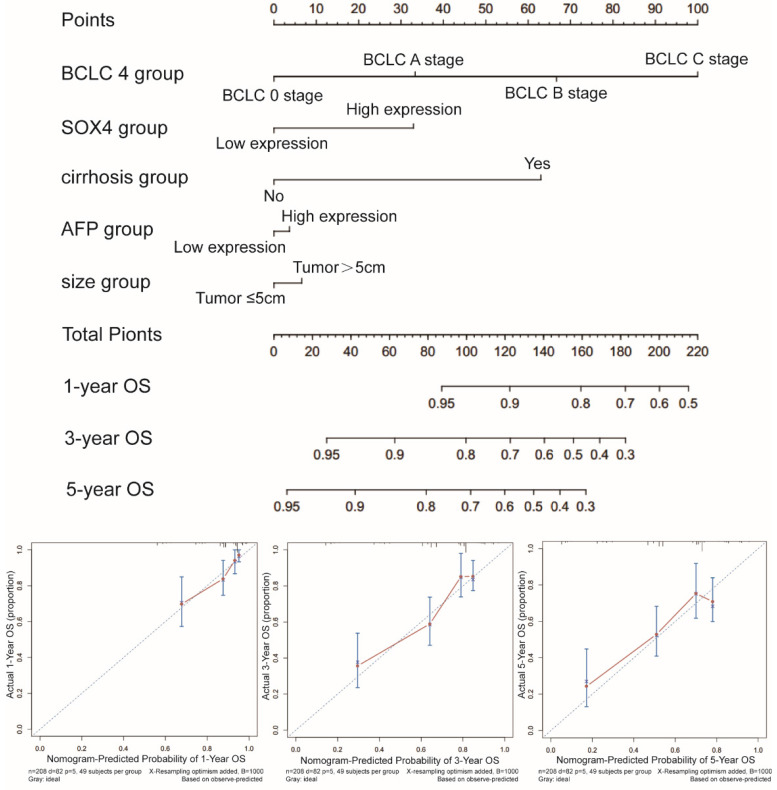
Next, we constructed a nomogram for OS based on the following clinical features: BCLC stage, cirrhosis, serum AFP level, tumor size and SOX4 expression. The following clinical features were used to construct a nomogram for RFS: BCLC stage, cirrhosis, gender and SOX4 expression. The nomograms may enable individualized prognosis prediction. Nomogram analysis was performed for the probabilities of 1-, 3-and 5-year OS (Figure 7) and RFS (Figure 8). These analyses revealed that SOX4 expression levels were correlated with the patients' clinical prognosis.
Figure 7.
Nomograms constructed using OS-related clinical factors and SOX4.
Figure 8.
Nomograms constructed using RFS-related clinical factors and SOX4.
Bioinformatics analysis of SOX4 gene
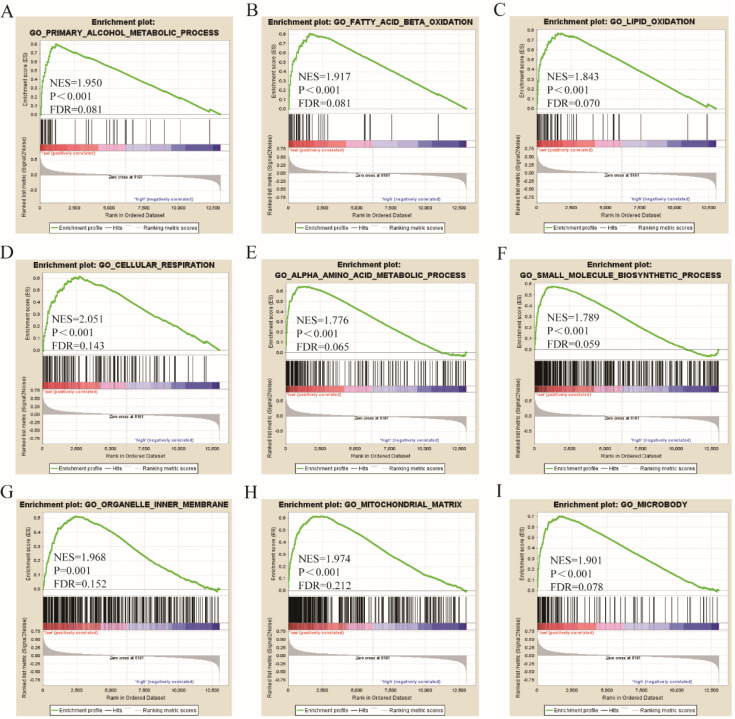
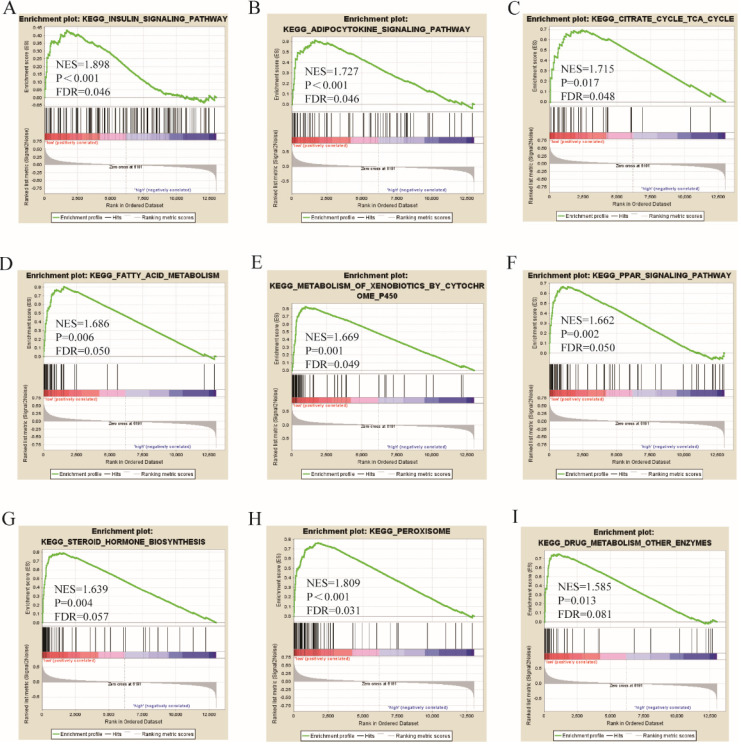
Go term analysis indicated that SOX4 gene is involved in the modulation primary alcohol metabolic processes, fatty acid beta oxidation, lipid oxidation, cellular respiration, alpha amino acid metabolic process, small molecule biosynthetic process, organelle inner membrane, mitochondrial matrix and microbody (Figure 5). KEGG functional analysis indicated that the SOX4 gene is involved in various signaling pathways, including insulin and adipocytokine signaling etc. (Figure 6). Detailed representations of the GSEA results are shown in Figure 5 and 6. The visualized interactions of GO terms were constructed using BinGO (Figure 11). This analysis revealed that SOX4 may be involved WNT signaling, lymphocyte differentiation and pancreatic endocrine development. Analysis of gene-gene interaction found that SOX4 is associated with TP53 etc. (Figure 1D) while analysis of protein-protein interaction found that SOX4 is associated with CTNNB1 and TP53 etc. (Figure 1E).
Figure 5.
Gene ontology analysis results of SOX4 gene.
Figure 6.
KEGG pathway analysis results of SOX4 gene.
Figure 11.
BiNGO analysis results for SOX4 gene.
Analysis of correlation between SOX4 expression and tumor stage
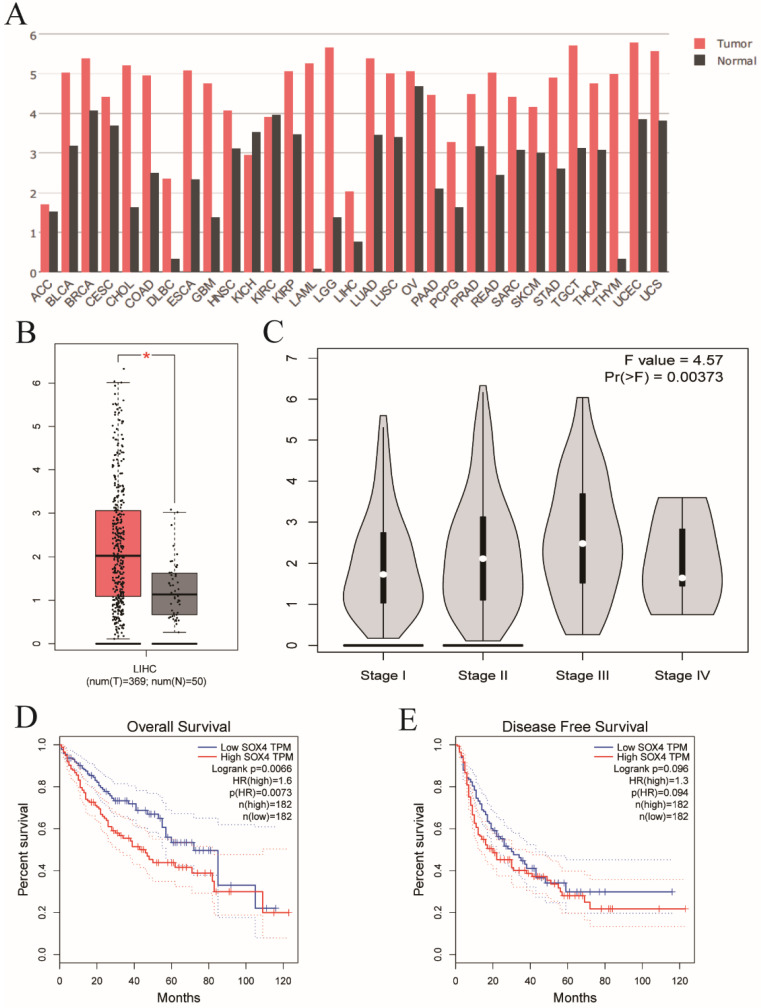
Analysis of the GSE14520 dataset for SOX4 expression at various BCLC stages revealed significantly elevated expression in each BCLC stage P <0.001, Figure 9A), but least expressed in the BCLC stage C. Next, we combined BCLC stage 0 and A to constitute the early-stage cancer category and BCLC stage B and C to constitute the advanced-stage cancer category. Interestingly, there was significance lower in former one. Similar results were obtained by GEPIA analysis (P = 0.00373; Figure 10C).
Figure 9.
Scatter plot of SOX4 gene in BCLC stage of GSE14520 cohort and validation using Oncomine database. (A) Scatter plot of SOX4 gene by BCLC stage (0, A, B, C) of GSE14520 cohort. (B) Scatter plot of SOX4 gene by BCLC stage (0+A (early stage), B+ C (advance stage) of GSE14520 cohort. (C) ROC curve of SOX4 in Wurmbach liver dataset. (D) Relative mRNA expressions of SOX4 in tumor tissue and non-tumor tissue in Wurmbach liver dataset. (E) ROC curve of SOX4 in Mas liver dataset. (F) Relative mRNA expressions of SOX4 in tumor tissue and non-tumor tissue in Mas liver dataset.
Figure 10.
Validation of SOX4 gene in GEPIA website. (A) The SOX4 expression profile across all tumor and normal tissues. (B) Differential expressions of SOX4 in GEPIA website. (C) Violin plot of SOX4 by tumor stage in GEPIA website. (D) OS analysis plot of SOX4 in GEPIA website. (E) RFS analysis plot of SOX4 in GEPIA website.
Differential expression, diagnostic and prognostic validation analysis
Next, we analyzed SOX4 expression in the Wurmbach and Mas liver datasets and found markedly elevated SOX4 mRNA levels in tumor tissue in relative to normal tissue (P = 0.003, <0.001, respectively; Figure 9D, F). Moreover, the potential diagnostic value of SOX4 expression was revealed by ROC analysis of these two databases (AUC = 0.831, 0.947 respectively; P = 0.002, <0.001, respectively; Figure 9C, E). SOX4 expression was also found to be significantly upregulated in tumor tissue following GEPIA analysis (Figure 10A and B). Analysis of the possible impact of SOX4 expression on survival indicated that patients with low SOX4 expression levels in the GEPIA analysis, exhibit longer OS relative to those with high expression (P = 0.007, Figure 10D). Similar results were obtained for RFS (P = 0.096, Figure 10E), although this was not statistically significant. In addition, differences in SOX4 gene at various stages of HCC were statistically significant (P = 0.004; Figure 10C).
Discussion
Here, we assessed the relationship between SOX4 levels and various parameters of HBV-related HCC. Results reveal that SOX4 gene possesses significant value for HCC diagnosis, a finding that is in agreement with previous reports (Wurmbach E et al. and Mas VR et al.) 26, 27. In addition, we find that low SOX4 expression correlates with better HCC prognosis. Next, we carried out joint-effect and stratified analyses of the value of SOX4 as a prognostic indicator in HCC. GSEA analysis indicated that SOX4 positively modulates primary alcohol metabolic process, fatty acid beta oxidation, lipid oxidation, cellular respiration, small molecule biosynthetic process, alpha amino acid metabolic process, organelle inner membrane, mitochondrial matrix and microbody.
The SOX4 gene belongs to group C SOX transcription factors 30. The products of these genes consist of three domains: a serine-rich region (SRR, aa 333-397), which encodes a protein of 474 amino acids (aa), a glycine-rich region (aa 152-227) and an HMG box (aa 57-135) 30, 31. The HMG box possessed DNA binding, which has been take part in various developmental processes through its transcriptional activity, while SRR domain acts as a deactivation domain. Glycine-rich central region (CD), located between the SRR region and the HMG box is a recently identified functional region that promotes apoptosis 15, 31.
The SOX4 gene modulates tumor development and growth, epithelial-mesenchymal transition and metastasis 14, 32-34. Furthermore, SOX4 drives several components of the RNAi machinery, transcriptional regulators, and cellular proteins 35-37. Thus, SOX4 is a momentous transcription factor that regulates various cellular functions.
Multiple studies have reported the action of SOX4 as an oncogene in solid tumors 7, 38, 39. It has been reported that SOX4 is upregulated in various malignancies, including HCC, pancreatic cancer, bladder carcinoma, prostate cancer, breast cancer, colorectal cancer, gastric cancer and melanoma 11, 12, 14, 32, 40-44, raising the potential of this gene as a diagnostic marker. The Mas and Wurmbch liver cancer datasets have reported that SOX4 is highly expressed in hepatitis C virus-associated HCC 26, 27, which is consistent with our results. Moreover, various reports suggest that SOX4 can aid in predicting marker in some cancer types. High SOX4 expression has been associated with poor prognosis in prostate cancer, gastric cancer, colorectal cancer, breast cancer and HCC 11, 12, 14, 32, 45. On the contrary, low expression has been associated with better prognosis in bladder carcinoma and melanoma 41, 43.
Majority of HCCs are attributable to HBV infection 46, 47. Shang et al. reported that HBV increases expression of SOX4 gene by upregulating transcription factor YY1 via the mitogen-activated protein kinase pathway, epigenetically suppressing miR-203, miR-335, and miR-129-2 by protecting SOX4 from HBsAg mediated degradation 48. On the other hand, SOX4 has been shown to promote HBV replication by stimulating viral DNA replication and protein expression in liver cancer cells 49. As a consequence, SOX4 interacts with HBV and synergistically promotes the occurrence and development of HCC.
It was initially found that SOX4 acted as a transcription factor that drive B and T lymphocyte differentiation 8, 50. Wilson et al. have reported that SOX4 is involved in pancreatic endocrine development 49. It has been reported that WNT pathway promotes SOX4 expression in colorectal cancer (Van der Flier LG et al. and Reichling T et al.) 51, 52. SOX4 in turn enhances WNT pathway by stabilizing beta-catenin and directly promoting transcription factor 4 expression 35, 52-55. Multiple studies have demonstrated that SOX4 interacts with the tumor suppressor p53 during DNA-damage and apoptosis in HCC 15, 56, 57. A study by Yang Jiao et al. reported that tribbles homolog3 is a SOX4 target 58. Tribbles homolog3 is a pseudo-kinase that disrupts the insulin signaling pathway in the liver by binding to Protein Kinase B and blocking its activation 59, 60. However, none has verified the link between TP53, insulin signaling and SOX4. Based on our findings, we hypothesis that SOX4 may modulate TP53 activity and insulin signaling pathway. However, the mechanism still needs further investigated. Consistent with the aforementioned studies, our data show that SOX4 might influence WNT signaling, lymphocyte differentiation and TP53 activity.
Herein, we report that SOX4 is elevated in HCC with BCLC stage B+C than with BCLC stage 0+A. The OS and RFS nomograms indicated that SOX4 is associated with HCC prognosis. Previous studies have shown that SOX4 expression is upregulated in breast cancer 9 and promotes HCC metastases 14, suggesting it might lead to poor metastasis-free survival. It has been reported that SOX4 contributes to hepatocarcinogenesis and its expression can reflect the clinical course of HCC after surgical resection 15.
This study is limited by the small sample size, consisting of 212 HBV-associated liver cancer cases. Future studies should utilize a larger sample size. Our analysis was limited to HBV-associated HCC. It is necessary to explore the diagnostic and prognostic value of SOX4 all the HCCs, irrespective of HBV status. Since the data of the two cohorts in this study are from public databases, there is no additional validation cohort. This study still needs to be independently verified in an additional cohort. Relative to past studies, this study only assessed the relationship between SOX4 RNA levels and HCC clinical course. Thus, further investigation is advocated to provide better understanding.
Conclusions
This study found that SOX4 expression is significantly upregulated in HCC tumor tissues. Our data indicate that this gene has potential value in HCC diagnosis. Further survival analysis of SOX4 gene in two cohorts suggests that it significantly correlates with HCC OS and RFS. Bioinformatics analysis suggested that SOX4 may affect HCC prognosis by modulating TP53 activity, lymphocyte differentiation, pancreatic endocrine development and insulin signaling.
Supplementary Material
Supplementary table S1.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (No.: 81560535), Natural Science Foundation of Guangxi Province of China (Grant No. 2017JJB140189y, 2018GXNSFAA050119), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Natural Sciences Foundation (No.: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018). The present study is also partly supported by Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318), the Guangxi Key R & D Program (GKEAB18221019), Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (GKE2018-01, GKE2019-11), Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (No. GXCDCKL201902). As well as, the present study is also partly supported by Research Institute of Innovative Think-tank in Guangxi Medical University (The gene-environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention). We would also acknowledge the supported by the Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education.
Author contribution
Jian-Lv Huang and Tao Peng designed this manuscript; Xiang-Kun Wang, Xi-Wen Liao, Chuang-Ye Han, Ting-Dong Yu, Ke-Tuan Huang, Cheng-Kun Yang, Xiao-Guang Liu, Long Yu, Guang-Zhi Zhu, Hao Su, Wei Qin, Quan-Fa Han, Zheng-Qian Liu, Xin Zhou, Jun-Qi Liu, Xin-Ping Ye and Tao Peng conducted the study and analyzed the data. Jian-Lv Huang wrote the manuscript, and Tao Peng guided the writing.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer communications. 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai Z, Liu Q. Understanding the Global Cancer Statistics 2018: implications for cancer control. Science China Life sciences. 2019. 10.1007/s11427-019-9816-1. [DOI] [PubMed]
- 3.DiStefano JK, Davis B. Diagnostic and Prognostic Potential of AKR1B10 in Human Hepatocellular Carcinoma. Cancers. 2019;11:486. doi: 10.3390/cancers11040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Molecular and clinical oncology. 2013;1:593–8. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi F, Zhou A, Yan L, Yuan X, Wang D, Chang R, The diagnostic value of PIVKA-II, AFP, AFP-L3, CEA, and their combinations in primary and metastatic hepatocellular carcinoma. Journal of clinical laboratory analysis. 2019: e23158. [DOI] [PMC free article] [PubMed]
- 6.Pevny LH, Lovell-Badge R. Sox genes find their feet. Current opinion in genetics & development. 1997;7:338–44. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 7.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 8.Schilham MW, Clevers H. HMG box containing transcription factors in lymphocyte differentiation. Seminars in immunology. 1998;10:127–32. doi: 10.1006/smim.1998.0114. [DOI] [PubMed] [Google Scholar]
- 9.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song GD, Sun Y, Shen H, Li W. SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:4167–73. doi: 10.1007/s13277-015-3051-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang B, Li Y, Tan F, Xiao Z. Increased expression of SOX4 is associated with colorectal cancer progression. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:9131–7. doi: 10.1007/s13277-015-4756-5. [DOI] [PubMed] [Google Scholar]
- 12.Pang L, Li B, Zheng B, Niu L, Ge L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncology reports. 2017;38:1295–302. doi: 10.3892/or.2017.5745. [DOI] [PubMed] [Google Scholar]
- 13.Zheng JH, Jian ZX, Jin HS, Chen SC, Wang GY. [Expression of SOX4 gene and early recurrence of hepatocellular carcinoma: their relationship and the clinical significance] Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2010;30:818–9. [PubMed] [Google Scholar]
- 14.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL. et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–89. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 15.Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW. et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298–307. doi: 10.1093/carcin/bgq072. [DOI] [PubMed] [Google Scholar]
- 16.Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biology of reproduction. 1999;61:476–81. doi: 10.1095/biolreprod61.2.476. [DOI] [PubMed] [Google Scholar]
- 17.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–66. doi: 10.1053/j.gastro.2011.12.039. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Parpart S, Takai A, Roessler S, Budhu A, Yu Z. et al. Integrative genomics identifies YY1AP1 as an oncogenic driver in EpCAM(+) AFP(+) hepatocellular carcinoma. Oncogene. 2015;34:5095–104. doi: 10.1038/onc.2014.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaul YD, Yuan B, Thiru P, Nutter-Upham A, McCallum S, Lanzkron C. et al. MERAV: a tool for comparing gene expression across human tissues and cell types. Nucleic acids research. 2016;44:D560–6. doi: 10.1093/nar/gkv1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research. 2010;38:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome biology. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research. 2017;45:D362–D8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L. et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Molecular medicine. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–47. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 28.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Liu X, Yang C, Wang X, Yu T, Han C. et al. Distinct Diagnostic and Prognostic Values of Minichromosome Maintenance Gene Expression in Patients with Hepatocellular Carcinoma. Journal of Cancer. 2018;9:2357–73. doi: 10.7150/jca.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Developmental biology. 2000;227:239–55. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 31.Farr CJ, Easty DJ, Ragoussis J, Collignon J, Lovell-Badge R, Goodfellow PN. Characterization and mapping of the human SOX4 gene. Mammalian genome: official journal of the International Mammalian Genome Society. 1993;4:577–84. doi: 10.1007/BF00361388. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X. et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer research. 2012;72:4597–608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 33.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9309–14. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lourenco AR, Coffer PJ. SOX4: Joining the Master Regulators of Epithelial-to-Mesenchymal Transition? Trends in cancer. 2017;3:571–82. doi: 10.1016/j.trecan.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC. et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Molecular and cellular biology. 2007;27:7802–15. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–39. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 37.Beekman JM, Vervoort SJ, Dekkers F, van Vessem ME, Vendelbosch S, Brugulat-Panes A. et al. Syntenin-mediated regulation of Sox4 proteasomal degradation modulates transcriptional output. Oncogene. 2012;31:2668–79. doi: 10.1038/onc.2011.445. [DOI] [PubMed] [Google Scholar]
- 38.Grimm D, Bauer J, Wise P, Kruger M, Simonsen U, Wehland M. et al. The role of SOX family members in solid tumours and metastasis. Seminars in cancer biology. 2020;67:122–53. doi: 10.1016/j.semcancer.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Zhang L, Zhang T, Hao M, Zhang X, Zhang J. et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver international: official journal of the International Association for the Study of the Liver. 2013;33:476–86. doi: 10.1111/liv.12097. [DOI] [PubMed] [Google Scholar]
- 40.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H. et al. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Developmental biology. 2005;285:211–23. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Gunes S, Yegin Z, Sullu Y, Buyukalpelli R, Bagci H. SOX4 expression levels in urothelial bladder carcinoma. Pathology, research and practice. 2011;207:423–7. doi: 10.1016/j.prp.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer research. 2009;69:709–17. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jafarnejad SM, Wani AA, Martinka M, Li G. Prognostic significance of Sox4 expression in human cutaneous melanoma and its role in cell migration and invasion. The American journal of pathology. 2010;177:2741–52. doi: 10.2353/ajpath.2010.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Lu D, Xiang T, Wu X, Ge S, Wang Y. et al. MicroRNA-132-3p regulates cell proliferation, apoptosis, migration and invasion of liver cancer by targeting Sox4. Oncology letters. 2020;19:3173–80. doi: 10.3892/ol.2020.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB. et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer research. 2006;66:4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 46.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World journal of gastroenterology. 2015;21:10584–97. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 48.Shang J, Zheng Y, Guo X, Mo J, Xie X, Xiong Y. et al. Hepatitis B virus replication and sex-determining region Y box 4 production are tightly controlled by a novel positive feedback mechanism. Scientific reports. 2015;5:10066. doi: 10.1038/srep10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC. et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54:3402–9. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 50.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M. et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–4. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 51.Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M. et al. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–32. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 52.Reichling T, Goss KH, Carson DJ, Holdcraft RW, Ley-Ebert C, Witte D. et al. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer research. 2005;65:166–76. [PubMed] [Google Scholar]
- 53.Lee AK, Ahn SG, Yoon JH, Kim SA. Sox4 stimulates ss-catenin activity through induction of CK2. Oncology reports. 2011;25:559–65. doi: 10.3892/or.2010.1091. [DOI] [PubMed] [Google Scholar]
- 54.Song DH, Sussman DJ, Seldin DC. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. The Journal of biological chemistry. 2000;275:23790–7. doi: 10.1074/jbc.M909107199. [DOI] [PubMed] [Google Scholar]
- 55.Saegusa M, Hashimura M, Kuwata T. Sox4 functions as a positive regulator of beta-catenin signaling through upregulation of TCF4 during morular differentiation of endometrial carcinomas. Laboratory investigation; a journal of technical methods and pathology. 2012;92:511–21. doi: 10.1038/labinvest.2011.196. [DOI] [PubMed] [Google Scholar]
- 56.Nikolova D, Chalovska-Ivanova V, Genadieva-Dimitrova M, Eftimov A, Jovanovik R, Janevska V. TP53 Mutation in Correlation to Immunohistochemical Expression of P53 Protein in Patients with Hepatocellular Carcinoma. Open access Macedonian journal of medical sciences. 2018;6:1181–6. doi: 10.3889/oamjms.2018.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T. et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3788–93. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao Y, Zhao J, Zhang Z, Li M, Yu X, Yang Y. et al. SRY-Box Containing Gene 4 Promotes Liver Steatosis by Upregulation of SREBP-1c. Diabetes. 2018;67:2227–38. doi: 10.2337/db18-0184. [DOI] [PubMed] [Google Scholar]
- 59.Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R. et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nature medicine. 2004;10:530–4. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 60.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–7. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.