Abstract
Biomarkers that can guide cancer therapy based on patients’ individual cancer molecular signature can enable a more effective treatment with fewer adverse events. Data on actionable somatic mutations and germline genetic variants, studied by personalized medicine and pharmacogenomics, can be obtained from tumor tissue or blood samples. As tissue biopsy cannot reflect the heterogeneity of the tumor or its temporal changes, liquid biopsy is a promising alternative approach. In recent years, extracellular vesicles (EVs) have emerged as a potential source of biomarkers in liquid biopsy. EVs are a heterogeneous population of membrane bound particles, which are released from all cells and accumulate into body fluids. They contain various proteins, lipids, nucleic acids (miRNA, mRNA, and DNA) and metabolites. In cancer, EV biomolecular composition and concentration are changed. Tumor EVs can promote the remodeling of the tumor microenvironment and pre-metastatic niche formation, and contribute to transfer of oncogenic potential or drug resistance during chemotherapy. This makes them a promising source of minimally invasive biomarkers. A limited number of clinical studies investigated EVs to monitor cancer progression, tumor evolution or drug resistance and several putative EV-bound protein and RNA biomarkers were identified. This review is focused on EVs as novel biomarker source for personalized medicine and pharmacogenomics in oncology. As several pharmacogenes and genes associated with targeted therapy, chemotherapy or hormonal therapy were already detected in EVs, they might be used for fine-tuning personalized cancer treatment.
Keywords: extracellular vesicle, pharmacogenomics, liquid biopsy, cancer, biomarker, personalized medicine
Introduction
Advances in the field of genomics, proteomics, and other high-throughput methods for biomarker determination have enabled the era of personalized medicine. Biomarkers that can guide and customize therapy for individual patients can enable a more effective treatment with fewer adverse events (Zhang J. et al., 2018). One of the biggest breakthroughs in personalized medicine was in the field of oncology, as several actionable tumor somatic mutations that enabled targeted treatment and precision medicine were identified (Table 1) (Friedman et al., 2015; Syn et al., 2016; Panagiotara et al., 2017). In precision medicine, genomic approaches help the clinician to select the most appropriate treatment based on the individual cancer molecular signature (Panagiotara et al., 2017). For example, patient stratification based on actionable somatic mutations is already required before the initiation of some targeted treatments (Bonanno, 2013; Syn et al., 2016). Additionally, germline genetic variants are often associated with interindividual variability in treatment response and occurrence of adverse events (Sim et al., 2013). The field of pharmacogenomics focuses on genetic variability of genes involved in drug metabolism or transport, drug target genes and DNA repair genes that can affect response to various drugs, including chemotherapy and hormonal therapy and thus contribute to personalized medicine. Even though several drug prescribing recommendations are available, only a handful of pharmacogenetic tests are already required before the initiation of therapy in clinical practice (Lauschke et al., 2017). Novel biomarkers that would enable additional patient stratification, continuous disease monitoring or assessment of prognosis and thus improve treatment outcome are therefore still widely investigated.
TABLE 1.
List of pharmacogenes associated with targeted cancer therapy and the evidence connecting them with extracellular vesicles (EVs).
| Gene | Drug | Connection with extracellular vesicles: Examples | References a |
|---|---|---|---|
| ABL1 | Imatinib, bosutinib, dasatinib, nilotinib, ponatinib, vincristine | mRNA detected in various cancer cell EVs | Skog et al. (2008), Hong et al. (2009), Chacon-Heszele et al. (2014), Zhu et al. (2014), He et al. (2015), Fu et al. (2017), Milani et al. (2017), Kang et al. (2018) |
| Protein detected in urine EVs (healthy donors) | |||
| BCR-ABL1 mRNA and protein detected in EVs from CML cells and in serum EVs of patients with CML | |||
| ALK | Alectinib, atezolizumab, brigatinib, ceritinib, crizotinib, lorlatinib, pembrolizumab, ramucirumab | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors) | Gonzales et al. (2009), Hong et al. (2009), Hurwitz et al. (2016), Wu et al. (2018), Reclusa et al. (2019) |
| mRNA detected in colorectal cancer cell EVs | |||
| EML4-ALK translocation detected in plasma EVs in NSCLC | |||
| BCR | Imatinib, bosutinib, dasatinib, nilotinib, ponatinib, vincristine | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors) | Skog et al. (2008), Gonzales et al. (2009), Hong et al. (2009), Zhu et al. (2014), Hurwitz et al. (2016), Fu et al. (2017), Liem et al. (2017), Milani et al. (2017), Kang et al. (2018) |
| mRNA detected in colorectal cancer and glioblastoma cell EVs | |||
| BCR-ABL1 mRNA and protein detected in EVs from CML cells and in serum EVs of patients with CML | |||
| BRAF | Binimetinib, cobimetinib, dabrafenib, encorafenib, trametinib, vemurafenib | mRNA detected in colorectal cancer cell EVs, melanoma plasma and lymphatic drainage EVs and glioblastoma plasma EVs | Hong et al. (2009), Hao et al. (2017), Klump et al. (2018), García-Silva et al. (2019), Pasqualetti et al. (2019), Song et al. (2019) |
| DNA detected in melanoma serum EVs and lung adenocarcinoma pleural-effusion EVs | |||
| EGFR | Afatinib, atezolizumab, cetuximab, dacomitinib, erlotinib, gefitinib, osimertinib, panitumumab, pembrolizumab, ramucirumab | DNA detected in EVs from plasma, cerebrospinal fluid, bronchoalveolar lavage or pleural effusion of lung adenocarcinoma, NSCLC or glioblastoma patients mRNA detected in EVs from plasma or cerebrospinal fluid of NSCLC or glioblastoma patients | Skog et al. (2008), Hong et al. (2009), Welton et al. (2010), Adamczyk et al. (2011), Shao et al. (2012), Liang et al. (2013), Yamashita et al. (2013), Sinha et al. (2014), Hurwitz et al. (2016), Figueroa et al. (2017), Liem et al. (2017), Lee Y. T. et al. (2018), Castellanos-Rizaldos et al. (2018), Choi et al. (2018), Krug et al. (2018), Wan et al. (2018), Ortega et al. (2019), Qu et al. (2019), Song et al. (2019) |
| Protein detected in various cancer cell line EVs and plasma EVs of breast cancer patients | |||
| ERBB2 | Abemaciclib, alpelisib, everolimus, fulvestrant, lapatinib, neratinib, olaparib, palbociclib, pertuzumab, ribociclib, talazoparib, tipiracil/fluridine, trastuzumab | Protein detected in various cancer cell line EVs and plasma derived EVs in gastric and breast cancer patients | Skog et al. (2008), Hong et al. (2009), Baran et al. (2010), Ciravolo et al. (2012), Liang et al. (2013), Sinha et al. (2014), Gerratana et al. (2015), Hurwitz et al. (2016), Fang et al. (2017), Liem et al. (2017), Lee J. S. et al. (2018), Choi et al. (2018), Platko et al. (2019) |
| mRNA detected in colorectal cancer and glioblastoma cell EVs and plasma derived EVs in gastric and breast cancer patients | |||
| DNA amplification detected in urine EVs (urothelial bladder carcinoma) | |||
| KIT | Imatinib | Protein detected in various cancer cell line EVs | Atay et al. (2014), Hurwitz et al. (2016) |
| KRAS | Cetuximab, panitumumab, tipiracil/fluridine | Protein detected in various cancer cell line EVs, urine EVs (healthy donors) and EVs from pleural effusions of lung cancer patients | Gonzales et al. (2009), Hong et al. (2009), Demory Beckler et al. (2013), Fraser et al. (2013), Liang et al. (2013), Park et al. (2013), Kahlert et al. (2014), Lee et al. (2014), Sinha et al. (2014), Hurwitz et al. (2016), Kamerkar et al. (2017), Liem et al. (2017), Yang S.-J. et al. (2017), Choi et al. (2018), Möhrmann et al. (2018) |
| mRNA detected in colorectal cancer cell EVs | |||
| DNA detected in EVs from plasma or serum of pancreatic cancer patients, NSCLC, advanced cancers | |||
| NRAS | Cetuximab, panitumumab | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors) | Gonzales et al. (2009), Hong et al. (2009), Sinha et al. (2014), Hurwitz et al. (2016), Liem et al. (2017), Choi et al. (2018), Liu et al. (2020) |
| mRNA detected in colorectal cancer cell EVs |
References from individual functional/biomarker EV studies and EV-omics studies from VesiclePedia or Exocarta.
CML, chronic myeloid leukemia; NSCLC, non-small cell lung cancer.
Conventionally, cancer prognostic or predictive biomarkers are mostly determined in tumor tissue samples (Crowley et al., 2013). However, cancer is a very complex and heterogeneous disease, with a lot of factors contributing to its initiation, progression, development of metastasis or resistance to treatment (Zhang J. et al., 2018). Treatment resistance is not only associated with inherent genetic or non-genetic factors, but can also be acquired during or after treatment (Gong et al., 2012; Friedman et al., 2015). Tumor phenotype is therefore heterogeneous and constantly changing and can include different molecular mechanisms of carcinogenesis (Gong et al., 2012). With traditional tissue biopsy, only a specific section of the tumor is analyzed, therefore it does not reflect spatial tumor heterogeneity and patients may be stratified based on incomplete data (Crowley et al., 2013; Buder et al., 2016; Panagiotara et al., 2017). During the course of the treatment, new mutations can occur or accumulate, which can affect treatment selection or adjustment. As tissue biopsy is usually performed only at one time-point, these results are static and do not reflect the temporal changes of the tumor landscape (Crowley et al., 2013; Panagiotara et al., 2017). Tissue biopsies also have several other limitations: they are invasive and can even lead to complications. Enough material for all analyses cannot always be obtained, and some tumors are not accessible at all (Crowley et al., 2013; Buder et al., 2016; Panagiotara et al., 2017). In recent years, circulating tumor biomarkers are therefore extensively explored as an alternative minimally invasive approach called liquid biopsy (Mari et al., 2019).
A liquid biopsy is a test performed on biofluid samples to detect cancer cells or cancer-derived molecules (Vasconcelos et al., 2019). During the formation and growth of the tumor, various components may be released into the body fluids due to apoptosis, necrosis, or active release. These include circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating tumor RNA (ctRNA) and extracellular vesicles (EVs) (Alimirzaie et al., 2019; Vasconcelos et al., 2019). For example, ctRNA consists of mRNAs, miRNAs and long-noncoding RNAs and can be found either in ribonucleoprotein complexes, CTCs or EVs (Alimirzaie et al., 2019). All components from cancer tissue accumulate in body fluids and can be detected using liquid biopsies that reflect the genetic landscape of the whole cancer tissue. The possibility of minimally invasive serial sampling enables longitudinal monitoring of the disease at different time points. Liquid biopsy can therefore be used for screening or early diagnosis, assessment of prognosis, measurement of tumor burden and detection of minimal residual disease, early detection of disease recurrence, predicting or monitoring treatment response and detection of treatment resistance (Crowley et al., 2013; Buder et al., 2016; Mari et al., 2019; Vasconcelos et al., 2019). Most studies focus on blood-based biomarkers, however, urine, ascites, pleural effusion, and other biofluids can also be used (Lee J. S. et al., 2018; Mari et al., 2019).
Studies suggest that assessment of tumor-associated genetic changes in the blood could identify treatment resistance up to 10 months before radiological methods (Diaz et al., 2012; Misale et al., 2012). Liquid biopsy approach would therefore enable clinicians to modify or add treatment to achieve better response (Crowley et al., 2013). To date, most studies focused on identifying cancer-related genetic changes in CTCs or ctDNA. CTCs are intact tumor cells that actively or passively shed from primary or metastatic tumor into the bloodstream (Alimirzaie et al., 2019). Still, CTCs are rare events severely outnumbered by blood cells (1 per 1 ×106–1 × 109), resulting in limitations in CTC isolation and detection techniques and consequently low reproducibility of CTC-based tests (Geeurickx and Hendrix, 2020). Cell-free DNA (cfDNA) is fragmented DNA released from both healthy and tumor tissues from cells undergoing apoptosis or necrosis. ctDNA represents a subpopulation of cfDNA originating from cancer cells (Crowley et al., 2013; Alimirzaie et al., 2019). As ctDNA is highly fragmented [90–150 base pairs (bp)] and masked by a high background of total cfDNA, screening for clinically relevant mutations is challenging (Geeurickx and Hendrix, 2020). Currently, only five tests are Food and Drug Administration (FDA) approved, one CTC-based (CellSearch, Menarini Silicon Biosystems) and four cfDNA-based tests (cobas EGFR Mutation Test v2, Roche Molecular Systems; Epi proColon, Epigenomics; FoundationOne Liquid CDx, Foundation Medicine; Guardant360 CDx, Guardant Health). However, limited sensitivity or specificity as well as technological and regulatory challenges prevent a more widespread use in standard clinical practice (Vasconcelos et al., 2019).
In recent years, EVs emerged as a novel analyte used for liquid biopsies. In this review, we will focus on EVs as novel source of biomarkers for personalized medicine and pharmacogenomics in oncology (Figure 1). As several genes associated with targeted therapy, chemotherapy or hormonal therapy were already detected in EVs, they might be used for fine-tuning personalized cancer treatment.
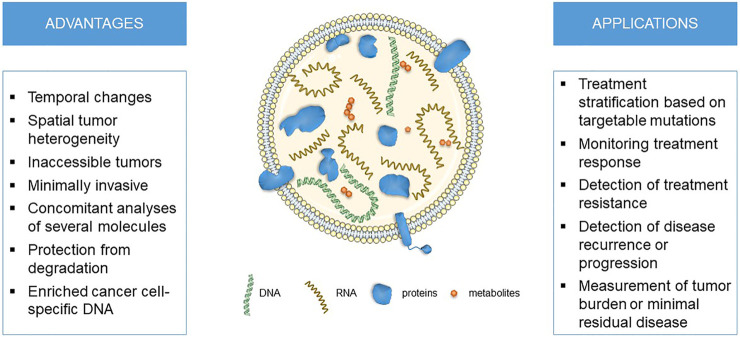
FIGURE 1.
The advantages and applications for liquid biopsies using extracellular vesicle DNA in cancer personalized medicine and pharmacogenomics.
Extracellular Vesicles
EVs are a heterogeneous population of membrane bound particles, which are released from all cells and accumulate into various body fluids in vivo (Lee J. S. et al., 2018; Dhondt et al., 2018; Hur et al., 2018; Johnsen et al., 2019). According to their size and site of formation, EVs can be broadly divided into three main categories: exosomes, microvesicles and apoptotic bodies. Exosomes range from 30 to 150 nm in diameter and are formed as intraluminal vesicles in multivesicular bodies, which fuse with plasma membrane to release them extracellularly. Microvesicles are larger membrane structures, from 100 to 1,000 nm in diameter, that are formed directly at the plasma membrane by outward budding. If they are released from tumors and transport tumor molecular cargo (e.g., oncoproteins), they are referred to as oncosomes, which were reported to reach up to 10 µm in diameter (Vagner et al., 2018). In contrast to exosomes and microvesicles, apoptotic bodies (up to 5 um in diameter) are formed during late phase of programmed cell death by membrane blebbing. Cells can simultaneously use more than one pathway of EV formation, which can be rerouted during pathogenic processes (Colombo et al., 2014; van Niel et al., 2018). Due to this complexity, EV subcellular origin is difficult to establish, and it is therefore preferred to use the terms small EVs and larger EVs for EVs <200 and >200 nm in size, respectively, (Théry et al., 2018).
Increase in plasma EV levels and changes in EV size were observed in several cancers (Silva et al., 2012; Muller et al., 2015; Huang et al., 2018; Navarro et al., 2019; Badovinac et al., 2021). To at least some extent, this is due to present tumor mass (Lai et al., 2017; Osti et al., 2019; Rodríguez Zorrilla et al., 2019), but systemic response to disease like inflammation (Roxburgh and McMillan, 2014; Chan et al., 2019) and response to treatment (Stevic et al., 2020) could also contribute. Increase in plasma EV levels is also observed in other diseases such as cardiovascular and autoimmune (Dickhout and Koenen, 2018; Maione et al., 2020) and might be connected to common physiological factors like hypoxia, autophagy or stress that are also often altered in tumors (Vasconcelos et al., 2019). Importantly, the pathological state of cell of origin additionally affects molecular composition of released EVs (van Niel et al., 2018). In general, EVs consist of a lipid bilayer membrane that surrounds a small amount of cytosol, and they contain various typical proteins [proteins involved in membrane trafficking, tetraspanins, adhesion molecules, cytoskeletal proteins, endosomal proteins (Kowal et al., 2016; Zhang H. et al., 2018; Théry et al., 2018)], lipids [ceramide, cholesterol, phosphatidylserine (Skotland et al., 2020)], nucleic acids [miRNA, mRNA, and DNA (Elzanowska et al., 2020; O'Brien et al., 2020)] and metabolites (Puhka et al., 2017). In cancer patients, EVs in body fluids were shown to accumulate oncogenes, tumor suppressor genes and their products, signature proteins and RNAs, and mutated genomic DNA (Zocco et al., 2014; An et al., 2015; González and Falcón-Pérez, 2015; Rowland et al., 2019; Vasconcelos et al., 2019; Chennakrishnaiah et al., 2020).
DNA only recently gained attention as an important constituent of EVs or other extracellular particles and remains poorly understood. EV-DNA varies in its localization and structure (Elzanowska et al., 2020; Malkin and Bratman, 2020). It is attached to the surface or is present in the lumen of EVs and can range in size from 200 bp in small EVs to up to >2 million bp in large EVs. EV-DNA exists as single or double stranded molecule of genomic or mitochondrial origin, mostly protected from degradation by bound histones. Reported heterogeneity might reflect diversity in EV biogenesis, as DNA content was shown to differ between EV subtypes (Lázaro-Ibáñez et al., 2014; Vagner et al., 2018; Lázaro-Ibáñez et al., 2019). EV-DNA is actively secreted from living cells (in contrast to passive secretion via necrosis), but little is known of the exact mechanisms. Those might include sequestering of cytosolic DNA during outward plasma membrane budding or intraluminal vesicles formation, or shuttling of collapsed micronuclei (small buddings of nucleus) to multivesicular bodies (Elzanowska et al., 2020; Malkin and Bratman, 2020). The latter mechanism is supported by observations of increased micronuclei production and EV-DNA release after genotoxic drug exposure (Yokoi et al., 2019). The biological function of EV-DNA is poorly understood, but the few reported studies propose their role i) in cellular homeostasis, as excretion of damaged DNA might prevent induction of apoptosis (Takahashi et al., 2017), ii) in transfer of genetic material, as recipient cells might transcribe or integrate DNA into the genome (Cai et al., 2013; Sansone et al., 2017), and iii) in immune response as it stimulates pro-inflammatory signaling pathways or anti-tumor immunity (Kitai et al., 2017; Torralba et al., 2018). Importantly, EV-DNA is protected from degradation in biofluids (Jin et al., 2016) and was shown to represent the entire genome and mutational status of the cell of origin (Kahlert et al., 2014; Thakur et al., 2014; San Lucas et al., 2016; Lázaro-Ibáñez et al., 2019).
EVs are important agents of inter-cellular communication that were implicated in numerous physiological and pathological processes. They were shown to cross extracellular space and even biological barriers, to interact with and/or transfer molecular content of the target cells, thereby affecting their cellular processes (Alvarez-Erviti et al., 2011; Colombo et al., 2014; van Niel et al., 2018). In cancer, EVs can act locally to remodel the tumor microenvironment at primary sites [reviewed in (Vader et al., 2014; Clancy and D'Souza-Schorey, 2018; Nogués et al., 2018)]. EVs (oncosomes) are released from tumor cells and transport bioactive cargo to neighboring tumor cells, stromal cells and cells of the immune system, thereby promoting angiogenesis, immunosuppression, tumor growth and local invasion. Alternatively, EVs accumulate in diverse body fluids (e.g., blood, lymphatic fluid) and are transported to local (e.g., lymph node) or distant sites of metastasis (e.g., lung, brain, bone, liver), depending on EV-organotropism. There, they promote pre-metastatic niche formation by inducing angiogenesis, remodeling of stroma and extracellular matrix, and by modulating immune responses.
EVs traceability of their cellular origin and its (patho)physiological state, high concentrations in body fluids and their ability to protect molecular cargo from degradation makes them a promising source of minimally invasive biomarkers. This is especially true in cancer, as EVs dynamically reflect changes in tumor molecular profiles depending on disease progression and treatment effectiveness (An et al., 2015; Syn et al., 2016; Rofi et al., 2019; Vasconcelos et al., 2019; Chennakrishnaiah et al., 2020; LeBleu and Kalluri, 2020). Compared to other liquid biopsy analytes, EVs represent multicomponent biomarker platforms that enable concomitant analyses of several molecules carried in the same vesicles (LeBleu and Kalluri, 2020). EVs are more abundant in various biofluids and thus could better represent intra-tumor heterogeneity than CTCs and also reflect stromal response. They protect their molecular cargo from degradation, may enrich for cancer cell-specific DNA and provide better signal-to-noize ratio compared to ctDNA (Vasconcelos et al., 2019; LeBleu and Kalluri, 2020). EVs could also help monitor variability in host responses through immune, epithelial or endothelial-derived EVs (LeBleu and Kalluri, 2020). Altogether, diagnostics based on EVs could enable a more comprehensive assessment of cancer diagnosis, prognosis and progression, which could guide a more personalized and effective treatment of cancer patients.
Importantly, EVs also contribute to transfer of oncogenic potential or drug resistance during chemotherapy. For example, in breast cancer treated with trastuzumab, EV-specific proteins were upregulated in metastatic patients that benefited from therapy, but not in those where treatment failed (Drucker et al., 2020). Alternatively, neoadjuvant breast cancer therapies with taxanes and anthracyclines induced tumor-derived EVs with enhanced pro-metastatic capacity (Keklikoglou et al., 2019). On the other hand, tumor EVs can spread drug resistance to drug sensitive cells. Systemic temozolomide treatment in glioblastoma mouse models induced mRNA expression profile reflective of drug resistance, which was recapitulated in the transcriptome of released EVs (Garnier et al., 2018). In oxidative phosphorylation-dependent breast cancer, horizontal transfer of EV-bound mitochondrial DNA promoted exit from dormancy of therapy-induced cancer stem-like cells and lead to endocrine therapy resistance (Sansone et al., 2017). Additionally, EVs were implicated in chemotherapy drug (doxorubicin, cisplatin, cetuximab, pixantrone) efflux in vitro experiments (Shedden et al., 2003; Safaei et al., 2005; Koch et al., 2016; Fujiwara et al., 2018), a possible mechanism for reducing their effective concentration at target sites. Another study showed that malignant lymphoma EVs carried CD20, which bound therapeutic anti-CD20 antibodies and protected target cells from antibody attack (Aung et al., 2011). Comprehensive overviews of experimental data on EVs relevance in drug resistance are provided in reviews by Vasconcelos et al. (Vasconcelos et al., 2019) and Maacha et al. (Maacha et al., 2019).
A limited number of clinical studies investigated EVs to monitor cancer progression, tumor evolution or drug resistance and several putative protein and RNA biomarkers were identified (An et al., 2015; González and Falcón-Pérez, 2015; Dhondt et al., 2018; Vasconcelos et al., 2019). For example, GPC1-positive EVs were shown to successfully diagnose pancreatic ductal adenocarcinoma (PDAC) and correlated with overall survival (Melo et al., 2015), while EpCAM-positive EVs could predict progression free survival (Giampieri et al., 2019). In PDAC patients treated with gemcitabine, high expression of miRNA-155 in tumor tissue and plasma EVs correlated with poorer prognosis due to chemoresistance development (Mikamori et al., 2017). In breast cancer, levels of TRPC5-positive EVs negatively correlated with chemotherapy outcome and could predict progression free survival (Wang et al., 2017). Additionally, levels of EV-associated lncRNA HOTAIR were associated with tumor burden and aggressiveness of the disease (Tang et al., 2019). Specific EV-miRNAs were also shown to correlate with tumor burden and response to therapy in breast cancer (Rodríguez-Martínez et al., 2019). EV-associated UCH-L1 mRNA was associated with ER-/PR-aggressive breast cancers subtypes and poor prognosis (Miyoshi et al., 2006), and with poor response to adjuvant anthracycline/taxane-based chemotherapy (Ning et al., 2017). HER2-positive breast cancer patients that did not respond to treatment had higher levels of EV lncRNA SNHG14 (Dong et al., 2018) or TGF-β1 (Martinez et al., 2017). Specific EV-associated non-coding RNAs were also predictive of treatment response to gemcitabine (Wei et al., 2017), erlotinib (Zhang W. et al., 2018) or cisplatin (Yuwen et al., 2017) in non-small cell lung cancer (NSCLC). Interestingly, EVs and their cargo were also associated with immunosuppression and response to immunotherapy with immune checkpoint inhibitors (Chen et al., 2018). In melanoma, EVs carrying immune check-point ligand PD-L1 on their surface, captured the therapeutic anti-PD-1 antibodies and drove antibody away from target tumor (Chen et al., 2018). Temporal changes in EV-associated PD-L1 mRNA levels were associated with response to anti-PD-1 antibodies nivolumab and pembrolizumab in patients with melanoma and NSCLC (Del Re et al., 2018). This suggests EV liquid biopsy could be implemented also in other areas of cancer treatment. Key biomarkers implicated in cancer personalized medicine and pharmacogenomics are described in Extracellular Vesicles and Cancer Personalized Medicine and Extracellular Vesicles and Cancer Pharmacogenomics.
Extracellular Vesicles and Cancer Personalized Medicine
In recent years, targetable mutations and personalized medicine have significantly affected treatment options and outcomes for cancer patients, especially in solid tumors (Rofi et al., 2019). Targeted cancer treatment refers to the use of drugs that target specific molecules in tumor cells or tumor microenvironment involved in signaling, cell survival and proliferation to prevent the growth and spread of tumors (Lee Y. T. et al., 2018). Targeted drugs can be small molecules or monoclonal antibodies. Small molecules are usually tyrosine kinase inhibitors, proteasome inhibitors, cyclin-dependent kinase inhibitors or poly ADP-ribose polymerase inhibitors (Lee Y. T. et al., 2018). Monoclonal antibodies usually directly or indirectly interrupt key signaling pathways (Lee Y. T. et al., 2018). Numerous drugs are used for targeted treatment across several cancer types (Table 1). However, treatment toxicity and inherent or acquired resistance to therapy pose a challenge for monitoring patients and their disease and better biomarkers or tumor molecular profiling strategies are still needed (Panagiotara et al., 2017). Currently, individual genetic alterations are used to guide treatment selection (Crowley et al., 2013). As targeted treatment is often used in advanced tumors, determination of genetic profile using tissue biopsy samples obtained at diagnosis is not optimal as they do not reflect the state of the tumor at the time of the treatment. Liquid biopsies can be used to identify acquired mutations or resistance that would allow the selection of the most efficient treatment (Crowley et al., 2013).
For this review, cancer genome very important pharmacogenes (VIPs), required pharmacogenetic testing, and pharmacogenetic recommendations available until June 1st 2020 were extracted from PharmGKB (Whirl-Carrillo et al., 2012). Data on the association of the selected human genes with extracellular vesicles was compiled using ExoCarta (Keerthikumar et al., 2016) and Vesiclepedia (Kalra et al., 2012; Pathan et al., 2019) databases that represent manually curated web-based compendiums of EV proteins, RNAs and lipids. Additional literature was selected using PubMed based on the following searches: “extracellular vesicles and gene name” or “exosomes and gene name”. Data from animal studies was not included.
According to PharmGKB (Whirl-Carrillo et al., 2012), nine VIPs are associated with cancer genome and personalized medicine. Genetic testing based on these genes is required by United States FDA and/or European Medicines Agency (EMA) for 40 different target drugs (Table 1). All cancer VIPs were already identified in EVs, usually both on protein and mRNA level. Additionally, EV-DNA could be used in liquid biopsies predicting or monitoring response to their target drugs by monitoring the specific landscape of somatic mutations. A more detailed description of VIPs related to their functional and biomarker role as part of EVs is provided in chapters 3.1 through 3.7.
ALK
ALK is a receptor tyrosine kinase important for the development of the nervous system (Cesi et al., 2018). Genetic rearrangements or mutations can lead to its constitutive activation and activation of downstream signaling pathways that affect proliferation and apoptosis in other differentiated tissues (Cesi et al., 2018; Wu et al., 2018). ALK therefore functions as an oncogene in several cancers, including NSCLC and neuroblastoma (Cesi et al., 2018). ALK fusion occurs in 3–7% of lung cancers, most commonly with EML4 (Chen et al., 2017). ALK inhibitors (e.g., crizotinib) improved treatment response and survival in patients with ALK rearrangements, but acquired treatment resistance is frequently a problem (Chen et al., 2017). Real-time monitoring using liquid biopsy could be used to detect the emergence of resistant mutations, however, there is limited knowledge about the connection of ALK with EVs (Table 1) (Chen et al., 2017). Irradiation of ALK-positive NSCLC cells lead to increased expression of ALK in EVs that were able to activate signaling pathways in recipient cells (Wu et al., 2018). In a pilot study, EML4-ALK translocation was detected in plasma-derived EVs of patients with NSCLC, suggesting that EVs may serve as an additional tool to guide treatment with ALK inhibitors (Reclusa et al., 2019).
BCR-ABL1
ABL1 is a proto-oncogene tyrosine kinase involved in cell differentiation, division, and proliferation. The role of BCR in not completely understood, but it has serine/threonine kinase activity and is a GTPase-activating protein. BCR-ABL1 fusion known as the Philadelphia chromosome that occurs due to a reciprocal translocation between chromosomes 9 and 22 is the driver fusion in chronic myeloid leukemia (CML) (Fu et al., 2017; Jurj et al., 2020). The fusion leads to dysregulation and over-activation of ABL1 tyrosine kinase domain. Several tyrosine kinase inhibitors such as imatinib target the fusion protein and have significantly improved survival of CML patients (Fu et al., 2017; Jurj et al., 2020). On the other hand, several mutations in ABL1 gene were associated with imatinib resistance. BCR, ABL1 and BCR-ABL1 fusion were frequently detected in EVs (Table 1). EV transfer of the oncogenic BCR-ABL1 transcript was able to affect signaling in mesenchymal stem cells and increase their proliferation (Zhu et al., 2014; Fu et al., 2017; Milani et al., 2017). BCR-ABL1 expression was already detected in serum-derived EVs of CML patients (Fu et al., 2017; Kang et al., 2018) and was associated also with response to tyrosine kinase inhibitors, imatinib resistance and disease remission (Kang et al., 2018; Liu et al., 2018; Jurj et al., 2020). EVs were also proposed as a novel biomarker for monitoring minimal residual disease through the number of BCR-ABL1 copies (Fu et al., 2017; Jurj et al., 2020).
BRAF
BRAF encodes a serine/threonine-specific mitogen-activated protein kinase (MAPK) kinase, which is part of the MAPK/ERK signaling cascade regulating cell proliferation. Presence of growth factors activates target membrane receptors, which in turn activate Ras GTPases that recruit BRAF to the membrane and induce homo or hetero-dimerization and activation of downstream ERK (Brummer and McInnes, 2020). BRAF alterations are important drivers of human cancer, particularly in melanoma (found in 40–50%), thyroid cancers (10–70%), colorectal cancers (10%) and NSCLC (3–5%) (Dankner et al., 2018). The most studied BRAF V600 allele functions as a hyperactive monomer, uncoupled from upstream regulation by Ras GTPase, thereby promoting constitutive activation of cell growth and proliferation (Dankner et al., 2018). BRAF inhibitors (e.g., dabrafenib and vemurafenib) alone or combined with MEK inhibitors provide a significant survival benefit, but over time tumors develop acquired resistance. Some BRAF V600 and non-V600 tumors also show intrinsic resistance, are insensitive to inhibitors or even cause paradoxical activation of ERK (Dankner et al., 2018). Further liquid biopsy biomarkers are needed to better guide targeted therapies for BRAF mutant cancers. EVs could serve as a novel source for genotyping BRAF or as biomarker of disease progression, since V600 mutation was detected in cancer cell line- and in body fluid-derived EVs in various cancers (Table 1), and corresponded to disease state, prognosis or survival (Hong et al., 2009; Thakur et al., 2014; Hao et al., 2017; Klump et al., 2018; García-Silva et al., 2019; Pasqualetti et al., 2019; Song et al., 2019).
EGFR
EGFR (also known as ERBB1) encodes a member of the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases, which regulate epithelial cell survival, proliferation, differentiation, and motility. Binding of one of its seven ligands (e.g., EGF and TGFα) promotes EGFR homo- or hetero-dimerization, which through structural rearrangements promotes trans-autophosphorylation of the cytosolic tyrosine kinase domains and activation of several signaling pathways including the Ras/MAPK, PI3K/AKT and PLC/PKC (Sigismund et al., 2018). EGFR mutations, frequently combined with overexpression, are important drivers of human cancers, particularly in glioblastoma (found in 50%), NSCLC (15–20%) and colorectal cancers (3%). Oncogenic EGFR (e.g., EGFRvIII, L858R) stimulate receptor homo- and heterodimerization and abnormal EGFR endocytic trafficking, which contributes to increased kinase activation and downstream signaling (Sebastian et al., 2006; Sigismund et al., 2018). Currently, monoclonal humanized antibodies (cetuximab, panitumumab) or tyrosine kinase inhibitors (e.g., erlotinib) targeting oncogenic EGFR show limited response and frequently evoke resistance in patients (Sebastian et al., 2006; Sigismund et al., 2018). Liquid biopsy, more specifically EVs, could contribute to improved cancer progression and therapy follow-up, as EVs containing EGFR DNA, RNA or protein were detected in blood, cerebrospinal fluid, bronchoalveolar lavage or pleural effusions in different cancers (Table 1), which in some studies correlated with prognosis (Skog et al., 2008; Shao et al., 2012; Yamashita et al., 2013; Figueroa et al., 2017; Lee J. S. et al., 2018; Castellanos-Rizaldos et al., 2018; Krug et al., 2018; Wan et al., 2018; Ortega et al., 2019; Qu et al., 2019; Song et al., 2019). For example, anticancer agents blocking oncogenic EGFR (e.g., dacomitinib, canertinib) stimulated the release of EVs carrying EGFRvIII and genomic DNA in glioblastoma animal models and patients (Skog et al., 2008; Montermini et al., 2015; Choi et al., 2019), and in turn, EV-EGFRvIII were shown to fuse with cancer cells lacking EGFRvIII and transfer the oncogenic activity (Al-Nedawi et al., 2008). EV-bound DNA was superior to cfDNA for EGFR mutation detection in early stage NSCLC (Wan et al., 2018). In lung adenocarcinoma, mutation status of EV-associated DNA from pleural effusion correlated with cfDNA (Song et al., 2019), while presence of mutated EGFR informed about the EGFR tyrosine kinase inhibitor treatment efficacy (Qu et al., 2019). EGFR EVs release additionally responded to cetuximab, TKIs or temozolomide treatment of cells in vitro (Shao et al., 2012; Montermini et al., 2015; van Dommelen et al., 2016; Fujiwara et al., 2018). In vitro, EGFR EVs were shown to be involved in several cancer hallmarks like inducing angiogenesis, sustaining proliferating signaling, evading growth suppression and immune destruction, promoting inflammation, resisting cell death, and activating invasion and metastasis (reviewed in (Zanetti-Domingues et al., 2020)). Further studies on EVs as therapy delivery system are also needed, as EVs containing let-7a reduced tumor size in mice EGFR-expressing breast cancer model (Ohno et al., 2013).
ERBB2
ERBB2 encodes a receptor tyrosine kinase also known as HER2 that belongs to EGFR family. Presence of growth factors leads to heterodimerization and activation of MAPK and PI3K signaling pathways (Graus-Porta et al., 1997). ERBB2 is a proto-oncogene that is overexpressed in approximately 25% of breast cancers, but also in gastric and other cancers, leading to malignant transformation of epithelial cells (Slamon et al., 2011; Fang et al., 2017; Galli de Amorim et al., 2019). Overexpression occurs mostly due to gene amplification, but other activating mutations were also described (Slamon et al., 2011; Galli de Amorim et al., 2019). ERBB2 overexpression is associated with a more aggressive phenotype with worse survival because of increased proliferation, invasiveness and metastasis (Slamon et al., 2011). On the other hand, ERBB2 represents a target for several different drugs, both monoclonal antibodies (e.g., trastuzumab) and tyrosine kinase inhibitors (e.g., lapatinib and neratinib). Introduction of targeted treatment significantly improved response and survival in HER2-positive breast cancer patients (Slamon et al., 2011). However, not all patients respond to treatment with trastuzumab and some patients develop resistance, therefore additional biomarkers are needed to further improve treatment outcome (Ciravolo et al., 2012; Galli de Amorim et al., 2019). ERBB2 was frequently identified in EVs (Table 1). ERBB2 expression was increased in EVs from HER2-positive cell lines or in blood-derived EVs of cancer patients (Baran et al., 2010; Ciravolo et al., 2012; Wang et al., 2018). Studies have shown that ERBB2 expression in blood or urine-derived EVs correlates with tissue expression and liquid biopsy may sometimes even be more sensitive to detect ERBB2 overexpression (Fang et al., 2017; Lee D. H. et al., 2018; Platko et al., 2019). Additionally, ERBB2 overexpression alters EV protein composition (Amorim et al., 2014). Studies also suggest activated ERBB2 in EVs can sequester trastuzumab, potentially affecting its efficacy (Ciravolo et al., 2012). EVs could therefore serve as an additional biomarker for personalized treatment of HER2-positive cancer patients.
KIT
KIT is a proto-oncogene receptor tyrosine kinase that influences cell growth, survival, proliferation, differentiation and migration through the activation of downstream signaling pathways (Wu et al., 2019). Activating KIT mutations are present in 75–85% of gastrointestinal stromal tumors (GISTs), representing the main oncogenic driver (von Mehren and Joensuu, 2018; Wu et al., 2019). Up to 90% of KIT mutations occur in exon 11, making cells sensitive to tyrosine kinase inhibitor imatinib (von Mehren and Joensuu, 2018; Wu et al., 2019). However, secondary mutations in other exons can lead to imatinib resistance (Wu et al., 2019). KIT was already detected in various cancer cell line EVs (Table 1). GIST cells were shown to secrete a lot of oncogenic EVs containing KIT. Uptake of these EVs by stromal cells lead to their conversion to tumor promoting cells, activation of signaling pathways, induction of MMP1 expression and increased tumor cell invasion (Atay et al., 2014). However, the data on monitoring KIT mutations in EVs is lacking. In one study that included only three patients, one KIT mutation associated with treatment resistance was only present in cfDNA (Klump et al., 2018). Further studies are needed to determine the usefulness of KIT mutations detection in EVs.
RAS (KRAS, NRAS)
KRAS and NRAS genes encode small membrane-localized GTPases, important for regulating cell growth, differentiation and survival. When growth factors bind target cellular receptors (e.g., receptor tyrosine kinases, G-protein coupled receptors, integrins), they activate RAS through recruitment of scaffolding proteins that mediate conversion of inactive GDP-bound to active GTP-bound form, which in turn activates diverse MAPK pathways. RAS activity can be further regulated by various proteins and post-translational modifications (e.g., farnesylation, palmitoylation, ubiquitination, acetylation) (Gimple and Wang, 2019). RAS alterations are drivers of 20–30% of all human cancers. KRAS mutations (mostly on the 12th codon) are exceedingly common in pancreatic adenocarcinomas (found in 94%), colorectal cancers (37%) and lung adenocarcinomas (20–30%), while NRAS mutations (mostly on 61st codon) are more common in melanomas (20–30%), thyroid cancers (10–40%, varies among different types), and leukemias (15%) (Cerami et al., 2012; Gao et al., 2013). They enhance RAS activity, effectively uncoupling downstream signaling from growth factor receptors, thereby promoting tumor cell proliferation, survival, metabolism, microenvironmental interactions and immune evasion (Gimple and Wang, 2019). RAS is believed to be an undruggable target due to its molecular structure, but RAS signaling can be indirectly targeted through its regulators (e.g., cetuximab) or downstream effectors (e.g., vemurafenib). Still, development of therapeutic resistance is common and indirect targeting can cause counterproductive effects (e.g., BRAF inhibitors) (Crowley et al., 2013; Gimple and Wang, 2019). Importantly, mutant KRAS was shown to affect composition of EVs released by cancer cells and its DNA and protein molecules were detected in EVs from blood and pleural effusion of different cancers (Table 1) (Demory Beckler et al., 2013; Park et al., 2013; Kahlert et al., 2014; Lee et al., 2014; Yang S. et al., 2017; Möhrmann et al., 2018). Level of mutant KRAS EVs in plasma even correlated with response to therapy and tumor burden with pancreatic cancer (Kahlert et al., 2014; Allenson et al., 2017; Yang S. et al., 2017; Bernard et al., 2019). Oncogenic NRAS was detected in EVs from various cell lines and similarly released by EVs from mouse brain tumor cells (Lee et al., 2014). When tested in vitro for function, mutant KRAS EVs stimulated cell proliferation, altered metabolic state and enhanced invasiveness of recipient cells (Demory Beckler et al., 2013; Lee et al., 2014; Zhang Q. et al., 2018). Thus mutant KRAS EVs could be used for rapid, non-invasive and continuous identification of cancer driver mutations, relevant for personalized treatment strategies. Further studies on EVs as therapy delivery system are also needed, as EVs containing siRNA/shRNA against oncogenic KRAS suppressed cancer and increased overall survival in multiple mouse models of pancreatic cancer (Kamerkar et al., 2017).
Extracellular Vesicles and Cancer Pharmacogenomics
Among key pharmacogenes are drug metabolizing enzymes such as cytochromes P450 (CYP) and UDP-glucuronosyltransreases (UGT) and ATP-binding cassette (ABC) transporters. CYPs and UGTs are involved in metabolic clearance of more than 90% of drugs (Rowland et al., 2019). Activity of CYPs and UGTs is influenced by their function and expression. Pharmacogenetics studies have found several genetic variants that modify enzyme activity (Rodriguez-Antona and Ingelman-Sundberg, 2006), however, genetic factors are often not sufficient to predict patient drug exposure (Rowland et al., 2019). Recent evidence suggests several pharmacogenes are also present in EVs (Kumar et al., 2017; Gerth et al., 2019; Rowland et al., 2019). For example, EVs isolated from human plasma contained functional proteins and mRNAs of different CYPs and UGTs (Kumar et al., 2017; Rowland et al., 2019). Due to relatively high abundance and confirmed enzymatic activity, circulating EV CYPs may also have a physiological role in extrahepatic drug metabolism (Kumar et al., 2017). For example, CYP2E1 is highly expressed in plasma EVs and was also associated with exacerbating alcohol and acetaminophen-induced toxicity in hepatic and extrahepatic cells (Kumar et al., 2017; Rahman et al., 2019).
CYPs play an important role in the activation or inactivation of several cancer drugs (Rodriguez-Antona and Ingelman-Sundberg, 2006). For example, one of the most important enzymes, CYP3A4, is involved in metabolism of docetaxel, etoposide, cyclophosphamide, vincristine, and paclitaxel (Rodriguez-Antona and Ingelman-Sundberg, 2006). Rare CYP3A4*22 and *1B alleles alter CYP3A4 activity, but genotyping cannot explain all interindividual differences in CYP3A4 expression or function and no drug prescribing recommendations are currently in use (Rowland et al., 2019). CYP3A4 activity was confirmed also in plasma EVs (Kumar et al., 2017; Rowland et al., 2019). Additionally, treatment with CYP3A4 inductor rifampicin significantly increased CYP3A4 expression in EVs and CYP3A4 expression in plasma EVs correlated with midazolam clearance (Rowland et al., 2019). These results suggest that measuring CYP3A4 expression in EVs could help predict response to drugs metabolized by this enzyme. However, the patients in published studies were selected for wild-type CYP3A4 genotype and further studies evaluating EV expression of CYP3A4 are needed to evaluate the role of CYP3A4 from EVs. Combination of CYP3A4 genotyping and liquid biopsy could potentially serve as a novel biomarker for identifying variability in drug exposure.
Several other pharmacogenes can influence response to cancer therapy and for some of them, drug prescribing recommendations (e.g., dose adjustment, alternative drugs) are already available (Table 2). According to PharmGKB (Whirl-Carrillo et al., 2012), four cancer pharmacogenes have pharmacogenetic drug prescribing recommendations from Clinical Pharmacogenetics Implementation Consortium (CPIC) or Dutch Pharmacogenetics Working Group (DPWG) (PharmGKB level 1A and 1B clinical annotations). Additional six genes are also important for predicting response or adverse events to chemotherapy (PharmGKB level 2A clinical annotations). Among these ten genes, all except SLC O 1B1 were previously identified in EVs. Most genes were detected in EVs from different cancer cell lines (protein and/or mRNA), several were also detected in urine. A more detailed evaluation of the connection with EVs is described for UGT1A1, ABCB1, and GSTP1.
TABLE 2.
List of pharmacogenes associated with cancer therapy and the evidence connecting them with extracellular vesicles (EVs).
| Gene | Drug | Therapy type | Connection with extracellular vesicles: Examples | References a |
|---|---|---|---|---|
| Genes with pharmacogenetic drug prescribing recommendations b | ||||
| CYP2D6 | Tamoxifen | Hormone therapy | Protein detected in urine EVs (healthy donors, patients with nephropathy) | Moon et al. (2011) |
| DPYD | Capecitabine, fluorouracil, tegafur | Chemotherapy | Protein detected in glioblastoma cells EVs and in urine EVs (healthy donors) | Fraser et al. (2013), Pavlyukov et al. (2018) |
| TPMT | Mercaptopurine, thioguanine | Chemotherapy | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors) | Hong et al. (2009), Wang et al. (2012), Bijnsdorp et al. (2013), Liang et al. (2013), Sinha et al. (2014), Liu et al. (2015), Hurwitz et al. (2016), Liem et al. (2017) |
| mRNA detected in colorectal cancer cell EVs | ||||
| UGT1A1 | Irinotecan | Chemotherapy | Protein detected in platelet EVs | Dean et al. (2009), Liu et al. (2020) |
| mRNA detected in plasma derived EVs in lung adenocarcinoma | ||||
| Genes with PharmGKB level 2A clinical annotations | ||||
| ABCB1 | Methotrexate | Chemotherapy | Protein detected in various cancer cell line EVs, platelet EVs, serum EVs in prostate cancer and urine EVs (healthy donors) | Dean et al. (2009), Gonzales et al. (2009), Hong et al. (2009), Kato et al. (2015), Kharaziha et al. (2015), Hurwitz et al. (2016), Wang et al. (2019) |
| mRNA detected in colorectal cancer cell EVs | ||||
| NQ O 1 | Alkylating agents, anthracyclines and related substances, fluorouracil, platinum compounds | Chemotherapy | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors) | Hong et al. (2009), Welton et al. (2010), Wang et al. (2012), Ji et al. (2013), Hurwitz et al. (2016), Liem et al. (2017), Choi et al. (2018) |
| mRNA detected in colorectal cancer cell EVs | ||||
| MTHFR | Carboplatin, cisplatin, methotrexate | Chemotherapy | Protein detected in various cancer cell line EVs | Hong et al. (2009), Hurwitz et al. (2016), Liem et al. (2017) |
| mRNA detected in colorectal cancer cell EVs | ||||
| GSTP1 | Fluorouracil, oxaliplatin, cyclophosphamide, epirubicin, platinum compounds | Chemotherapy | Protein detected in various cancer cell line EVs and in urine EVs (healthy donors, prostate cancer) | Skog et al. (2008), Gonzales et al. (2009), Hong et al. (2009), Welton et al. (2010), Fraser et al. (2013), Ji et al. (2013), Paggetti et al. (2015), Øverbye et al. (2015), Hurwitz et al. (2016), Yang S. et al. (2017), Lázaro-Ibáñez et al. (2017), Liem et al. (2017), Choi et al. (2018), Zhang et al. (2019) |
| mRNA detected in colorectal cancer and glioblastoma cell EVs and serum derived EVs in breast cancer patients | ||||
| SLCO1B1 | Methotrexate | Chemotherapy | Not reported | — |
| TYMS | Capecitabine, fluorouracil | Chemotherapy | Protein detected in various cancer cell line EVs | Skog et al. (2008), Hong et al. (2009), Paggetti et al. (2015), Hurwitz et al. (2016), Liem et al. (2017), Choi et al. (2018) |
| mRNA detected in colorectal cancer and glioblastoma cell EVs | ||||
References from individual functional/biomarker EV studies and EV-omics studies from VesiclePedia or Exocarta.
Pharmacogenetic recommendations from CPIC or DPWG reported in PharmGKB, level 1A and 1B clinical annotations.
UGT1A1
Among pharmacogenes with drug prescribing recommendations, only uridine diphosphate glucuronosyltransferase 1A (UGT1A1) was already assessed as a potential biomarker in EVs (Table 2) (Liu et al., 2020). UGT1A1 catalyzes the glucuronidation of bilirubin and various xenobiotics, which enables their elimination (de Man et al., 2018). UGT1A1 is also involved in inactivation of SN-38, the active metabolite of irinotecan, a chemotherapeutic drug used for treatment of lung, colon, gastric, pancreatic, and gynecological cancers (Campbell et al., 2017; de Man et al., 2018). UGT1A1 is a very polymorphic enzyme, with over 100 genetic variants described (de Man et al., 2018). Most studied genetic variants are UGT1A1*6 and *28. Both are associated with increased irinotecan toxicity, especially neutropenia and diarrhea, due to increased systemic exposure to irinotecan and its metabolite (Campbell et al., 2017; de Man et al., 2018). For carriers of two polymorphic alleles, lower irinotecan starting dose is recommended (de Man et al., 2018). When comparing EVs from plasma of patients with lung adenocarcinoma and benign lung diseases using RNA-Seq, mRNA expression of UGT1A1 could differentiate between both groups and was suggested as an additional biomarker in NSCLC patients (Liu et al., 2020).
ABCB1
ABC transporter ABCB1, also known as multidrug resistance protein 1 (MDR1) or P-glycoprotein, is one of the key genes, involved in the development of chemoresistance to anticancer drugs (Kharaziha et al., 2015). ABCB1 is a cell surface glycoprotein that mediates ATP-dependent efflux of various molecules, including different xenobiotics (Gong et al., 2012; Kharaziha et al., 2015). Several ABCB1 genetic variants have been described (e.g., rs1128503, rs2032582, and rs1045642), but due to inconsistent results, no drug prescribing recommendations are currently available (Whirl-Carrillo et al., 2012). However, rs1045642 was associated with methotrexate toxicity in lymphoma and leukemia (Suthandiram et al., 2014; Zgheib et al., 2014; Gregers et al., 2015). On the other hand, ABCB1 was consistently associated with intrinsic or acquired multidrug resistance and treatment failure in several cancer types (Gong et al., 2012). EVs were proposed as one of the mechanisms associated with drug resistance, as they can help efflux chemotherapeutic (e.g., cisplatin, doxorubicin) from tumor cells (Samuel et al., 2018; Gluszko et al., 2019). Additionally, EVs were involved in intercellular transfer of drug resistance from drug-resistant to sensitive cells via ABCB1 (Table 2) (Bebawy et al., 2009; Gong et al., 2012; Kato et al., 2015; Kharaziha et al., 2015; Maacha et al., 2019; Vasconcelos et al., 2019; Wang et al., 2019). ABCB1 was enriched in EVs from various drug-resistant cancer cells after chemotherapy (Wang et al., 2019) and was associated with acquired resistance to docetaxel (Kato et al., 2015; Kharaziha et al., 2015). ABCB1 expression was also increased in serum derived EVs from prostate cancer patients, resistant to docetaxel (Kato et al., 2015). Detection of ABCB1 expression in EVs with liquid biopsy could therefore serve as an important biomarker of cancer treatment resistance.
GSTP1
Glutathione S-transferase pi 1 (GSTP1) is a phase II metabolic enzyme involved in the detoxification of various anti-cancer drugs and other carcinogens. GSTP1 catalyzes their conjugation with glutathione (Yang S.-J. et al., 2017). Two common non-synonymous polymorphisms, rs1695 and rs1138272, affect the electrophile-binding active site of the enzyme and alter enzyme activity (Ali-Osman et al., 1997). GSTP1 genetic variability was associated with toxicity and efficacy of numerous chemotherapeutics, especially platinum compounds (Stoehlmacher et al., 2004; Lamas et al., 2011; Goricar et al., 2015). However, no drug prescribing recommendations are currently available due to inconsistent results among cancer types (Whirl-Carrillo et al., 2012; Campbell et al., 2016). GSTP1 was frequently identified in EVs (Table 2). GSTP1 expression was increased in EVs from 5-fluorouracil or adriamycin resistant cancer cell lines (Yang S.-J. et al., 2017; Zhang et al., 2019). Additionally, EV-mediated transfer of GSTP1 was associated with intercellular transfer of adriamycin resistance, which is also an important clinical problem (Yang S.-J. et al., 2017). In breast cancer patients treated with anthracycline/taxane chemotherapy, GSTP1 mRNA expression was increased in serum EVs from patients with worse treatment response (Yang S.-J. et al., 2017). Detection of GSTP1 expression in EVs with liquid biopsy could therefore serve as an additional biomarker, potentially improving the predictive value of pharmacogenetics variants.
Conclusion and Future Perspectives
EVs have a proven role in transfer of oncogenic potential or drug resistance during chemotherapy and were proposed as biomarkers of cancer progression, tumor evolution or drug resistance in several cancer types. Several pharmacogenes and genes associated with treatment response were already detected in EVs in vitro and in vivo, thus they might be used for fine-tuning personalized cancer treatment. Before implementation in clinical practice, technical challenges regarding standardization and repeatability of EV analysis and detection of rare genetic variants should be addressed. Additionally, sufficient sensitivity and specificity of the novel biomarker assays should be obtained. As genetic factors cannot account for all observed variability, composite biomarkers incorporating genetic factors and other EV cargo are needed for some drugs. This could further improve personalized treatment for various drugs used in targeted treatment and chemotherapy, especially drugs targeting inducible enzymes of enzymes with poor genotype-phenotype associations (Gerth et al., 2019; Rodrigues and Rowland, 2019). All biomarkers and methods for their determination also have to be validated in independent studies and approved by appropriate regulatory agencies (Novelli et al., 2010). Several genomic biomarkers have been recognized, but there are considerable differences between EMA and FDA recommendations in summary of product characteristics labels (Shekhani et al., 2020). Consensus among regulators would further contribute to clinical implementation of key biomarkers.
In conclusion, EVs are emerging as liquid biopsy analytes that have several advantages and could be used in various applications in oncology, from treatment stratification to detection of treatment response or resistance, enabling new possibilities in personalized medicine or pharmacogenomics.
Acknowledgments
The authors would like to thank Marija Holcar, for her help with figure preparation.
Author Contributions
KG performed database search and KG and ML performed the literature search. KG, VD, and ML participated in writing and editing of manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Slovenian Research Agency (ARRS Grant Nos. P1-0170, J3-9255, and J3-1753).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ABC, ATP-binding cassette; bp, base pair; cfDNA, cell-free DNA; CML, chronic myeloid leukemia; CPOC, clinical pharmacogenetics implementation consortium; CTC, circulating tumor cell; ctDNA, circulating tumor DNA; ctRNA, circulating tumor RNA; CYP, cytochromes; DPWG, dutch pharmacogenetics working group; EGFR, epidermal growth factor receptor; EMA, European Medicines Agency; EV, extracellular vesicle; FDA, Food and Drug Administration; GIST, gastrointestinal stromal tumor; MAPK, mitogen-activated protein kinase; NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; UGT, UDP-glucuronosyltransrease; VIP, very important pharmacogene
References
- Adamczyk K. A., Klein-Scory S., Tehrani M. M., Warnken U., Schmiegel W., Schnölzer M., et al. (2011). Characterization of Soluble and Exosomal Forms of the EGFR Released from Pancreatic Cancer Cells. Life Sci. 89, 304–312. 10.1016/j.lfs.2011.06.020 [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., et al. (2008). Intercellular Transfer of the Oncogenic Receptor EGFRvIII by Microvesicles Derived from Tumour Cells. Nat. Cel Biol. 10, 619–624. 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- Ali-Osman F., Akande O., Antoun G., Mao J.-X., Buolamwini J. (1997). Molecular Cloning, Characterization, and Expression in Escherichia coli of Full-Length cDNAs of Three Human Glutathione S-Transferase Pi Gene Variants. J. Biol. Chem. 272, 10004–10012. 10.1074/jbc.272.15.10004 [DOI] [PubMed] [Google Scholar]
- Alimirzaie S., Bagherzadeh M., Akbari M. R. (2019). Liquid Biopsy in Breast Cancer: A Comprehensive Review. Clin. Genet. 95, 643–660. 10.1111/cge.13514 [DOI] [PubMed] [Google Scholar]
- Allenson K., Castillo J., San Lucas F. A., Scelo G., Kim D. U., Bernard V., et al. (2017). High Prevalence of mutantKRAS in Circulating Exosome-Derived DNA from Early-Stage Pancreatic Cancer Patients. Ann. Oncol. 28, 741–747. 10.1093/annonc/mdx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M. J. A. (2011). Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 29, 341–345. 10.1038/nbt.1807 [DOI] [PubMed] [Google Scholar]
- Amorim M., Fernandes G., Oliveira P., Martins-de-Souza D., Dias-Neto E., Nunes D. (2014). The Overexpression of a Single Oncogene (ERBB2/HER2) Alters the Proteomic Landscape of Extracellular Vesicles. Proteomics 14, 1472–1479. 10.1002/pmic.201300485 [DOI] [PubMed] [Google Scholar]
- An T., Qin S., Xu Y., Tang Y., Huang Y., Situ B., et al. (2015). Exosomes Serve as Tumour Markers for Personalized Diagnostics Owing to Their Important Role in Cancer Metastasis. J. Extracellular Vesicles 4, 27522. 10.3402/jev.v4.27522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atay S., Banskota S., Crow J., Sethi G., Rink L., Godwin A. K. (2014). Oncogenic KIT-Containing Exosomes Increase Gastrointestinal Stromal Tumor Cell Invasion. Proc. Natl. Acad. Sci. 111, 711–716. 10.1073/pnas.1310501111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung T., Chapuy B., Vogel D., Wenzel D., Oppermann M., Lahmann M., et al. (2011). Exosomal Evasion of Humoral Immunotherapy in Aggressive B-Cell Lymphoma Modulated by ATP-Binding Cassette Transporter A3. Proc. Natl. Acad. Sci. 108, 15336–15341. 10.1073/pnas.1102855108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac D., Goričar K., Zavrtanik H., Petrič M., Lavrin T., Mavec N., et al. (2021). Plasma Extracellular Vesicle Characteristics Correlate with Tumor Differentiation and Predict Overall Survival in Patients with Pancreatic Ductal Adenocarcinoma Undergoing Surgery with Curative Intent. J Pers Med 11, 77. 10.3390/jpm11020077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran J., Baj-Krzyworzeka M., Weglarczyk K., Szatanek R., Zembala M., Barbasz J., et al. (2010). Circulating Tumour-Derived Microvesicles in Plasma of Gastric Cancer Patients. Cancer Immunol. Immunother. 59, 841–850. 10.1007/s00262-009-0808-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebawy M., Combes V., Lee E., Jaiswal R., Gong J., Bonhoure A., et al. (2009). Membrane Microparticles Mediate Transfer of P-Glycoprotein to Drug Sensitive Cancer Cells. Leukemia 23, 1643–1649. 10.1038/leu.2009.76 [DOI] [PubMed] [Google Scholar]
- Bernard V., Kim D. U., San Lucas F. A., Castillo J., Allenson K., Mulu F. C., et al. (2019). Circulating Nucleic Acids Are Associated with Outcomes of Patients with Pancreatic Cancer . Gastroenterology 156, 108–118.e4. 10.1053/j.gastro.2018.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijnsdorp I. V., Geldof A. A., Lavaei M., Piersma S. R., van Moorselaar R. J. A., Jimenez C. R. (2013). Exosomal ITGA3 Interferes with Non-cancerous Prostate Cell Functions and Is Increased in Urine Exosomes of Metastatic Prostate Cancer Patients. J. Extracellular Vesicles 2, 22097. 10.3402/jev.v2i0.22097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno L. (2013). Predictive Models for Customizing Chemotherapy in Advanced Non-small Cell Lung Cancer (NSCLC). Transl Lung Cancer Res. 2, 160–171. 10.3978/j.issn.2218-6751.2013.03.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer T., McInnes C. (2020). RAF Kinase Dimerization: Implications for Drug Discovery and Clinical Outcomes. Oncogene 39, 4155–4169. 10.1038/s41388-020-1263-y [DOI] [PubMed] [Google Scholar]
- Buder A., Tomuta C., Filipits M. (2016). The Potential of Liquid Biopsies. Curr. Opin. Oncol. 28, 130–134. 10.1097/cco.0000000000000267 [DOI] [PubMed] [Google Scholar]
- Cai J., Han Y., Ren H., Chen C., He D., Zhou L., et al. (2013). Extracellular Vesicle-Mediated Transfer of Donor Genomic DNA to Recipient Cells Is a Novel Mechanism for Genetic Influence between Cells. J. Mol. Cel. Biol. 5, 227–238. 10.1093/jmcb/mjt011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. M., Bateman E., Peters M. D., Bowen J. M., Keefe D. M., Stephenson M. D. (2016). Fluoropyrimidine and Platinum Toxicity Pharmacogenetics: an Umbrella Review of Systematic Reviews and Meta-Analyses. Pharmacogenomics 17, 435–451. 10.2217/pgs.15.180 [DOI] [PubMed] [Google Scholar]
- Campbell J. M., Stephenson M. D., Bateman E., Peters M. D. J., Keefe D. M., Bowen J. M. (2017). Irinotecan-induced Toxicity Pharmacogenetics: an Umbrella Review of Systematic Reviews and Meta-Analyses. Pharmacogenomics J. 17, 21–28. 10.1038/tpj.2016.58 [DOI] [PubMed] [Google Scholar]
- Castellanos-Rizaldos E., Grimm D. G., Tadigotla V., Hurley J., Healy J., Neal P. L., et al. (2018). Exosome-Based Detection of EGFR T790M in Plasma from Non-small Cell Lung Cancer Patients. Clin. Cancer Res. 24, 2944–2950. 10.1158/1078-0432.Ccr-17-3369 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2, 401–404. 10.1158/2159-8290.Cd-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesi G., Philippidou D., Kozar I., Kim Y. J., Bernardin F., Van Niel G., et al. (2018). A New ALK Isoform Transported by Extracellular Vesicles Confers Drug Resistance to Melanoma Cells. Mol. Cancer 17, 145. 10.1186/s12943-018-0886-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Heszele M. F., Choi S. Y., Zuo X., Baek J.-I., Ward C., Lipschutz J. H. (2014). The Exocyst and Regulatory GTPases in Urinary Exosomes. Physiol. Rep. 2, e12116. 10.14814/phy2.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B. D., Wong W. Y., Lee M. M. L., Cho W. C. S., Yee B. K., Kwan Y. W., et al. (2019). Exosomes in Inflammation and Inflammatory Disease. Proteomics 19, e1800149. 10.1002/pmic.201800149 [DOI] [PubMed] [Google Scholar]
- Chen G., Huang A. C., Zhang W., Zhang G., Wu M., Xu W., et al. (2018). Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 560, 382–386. 10.1038/s41586-018-0392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo W., Fan J., Chen Y., Zhang X., Chen X., et al. (2017). The Applications of Liquid Biopsy in Resistance Surveillance of Anaplastic Lymphoma Kinase Inhibitor. Cancer Manag. Res. 9, 801–811. 10.2147/CMAR.S151235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennakrishnaiah S., Tsering T., Aprikian S., Rak J. (2020). Leukobiopsy - A Possible New Liquid Biopsy Platform for Detecting Oncogenic Mutations. Front. Pharmacol. 10, 1608. 10.3389/fphar.2019.01608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Montermini L., Jeong H., Sharma S., Meehan B., Rak J. (2019). Mapping Subpopulations of Cancer Cell-Derived Extracellular Vesicles and Particles by Nano-Flow Cytometry. ACS nano 13, 10499–10511. 10.1021/acsnano.9b04480 [DOI] [PubMed] [Google Scholar]
- Choi D., Montermini L., Kim D.-K., Meehan B., Roth F. P., Rak J. (2018). The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell Proteomics 17, 1948–1964. 10.1074/mcp.RA118.000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciravolo V., Huber V., Ghedini G. C., Venturelli E., Bianchi F., Campiglio M., et al. (2012). Potential Role of HER2-Overexpressing Exosomes in Countering Trastuzumab-Based Therapy. J. Cel. Physiol. 227, 658–667. 10.1002/jcp.22773 [DOI] [PubMed] [Google Scholar]
- Clancy J., D’Souza-Schorey C. (2018). Extracellular Vesicles in Cancer . Cancer J. 24, 65–69. 10.1097/ppo.0000000000000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Théry C. (2014). Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cel Dev. Biol. 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. (2013). Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol. 10, 472–484. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- Dankner M., Rose A. A. N., Rajkumar S., Siegel P. M., Watson I. R. (2018). Classifying BRAF Alterations in Cancer: New Rational Therapeutic Strategies for Actionable Mutations. Oncogene 37, 3183–3199. 10.1038/s41388-018-0171-x [DOI] [PubMed] [Google Scholar]
- de Man F. M., Goey A. K. L., van Schaik R. H. N., Mathijssen R. H. J., Bins S. (2018). Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics. Clin. Pharmacokinet. 57, 1229–1254. 10.1007/s40262-018-0644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W. L., Lee M. J., Cummins T. D., Schultz D. J., Powell D. W. (2009). Proteomic and Functional Characterisation of Platelet Microparticle Size Classes. Thromb. Haemost. 102, 711–718. 10.1160/TH09-04-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re M., Marconcini R., Pasquini G., Rofi E., Vivaldi C., Bloise F., et al. (2018). PD-L1 mRNA Expression in Plasma-Derived Exosomes Is Associated with Response to Anti-PD-1 Antibodies in Melanoma and NSCLC. Br. J. Cancer 118, 820–824. 10.1038/bjc.2018.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M., Higginbotham J. N., Franklin J. L., Ham A.-J., Halvey P. J., Imasuen I. E., et al. (2013). Proteomic Analysis of Exosomes from Mutant KRAS Colon Cancer Cells Identifies Intercellular Transfer of Mutant KRAS. Mol. Cell Proteomics 12, 343–355. 10.1074/mcp.M112.022806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt B., Van Deun J., Vermaerke S., de Marco A., Lumen N., De Wever O., et al. (2018). Urinary Extracellular Vesicle Biomarkers in Urological Cancers: From Discovery towards Clinical Implementation. Int. J. Biochem. Cel Biol. 99, 236–256. 10.1016/j.biocel.2018.04.009 [DOI] [PubMed] [Google Scholar]
- Diaz Jr L. A., Jr., Williams R. T., Wu J., Kinde I., Hecht J. R., Berlin J., et al. (2012). The Molecular Evolution of Acquired Resistance to Targeted EGFR Blockade in Colorectal Cancers. Nature 486, 537–540. 10.1038/nature11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickhout A., Koenen R. R. (2018). Extracellular Vesicles as Biomarkers in Cardiovascular Disease; Chances and Risks. Front. Cardiovasc. Med. 5, 113. 10.3389/fcvm.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Wang W., Chen R., Zhang Y., Zou K., Ye M., et al. (2018). Exosome-mediated Transfer of lncRNASNHG14 Promotes Trastuzumab Chemoresistance in Breast Cancer. Int. J. Oncol. 53, 1013–1026. 10.3892/ijo.2018.4467 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Drucker A., Yoo B. H., Khan I. A., Choi D., Montermini L., Liu X., et al. (2020). Trastuzumab-induced Upregulation of a Protein Set in Extracellular Vesicles Emitted by ErbB2-Positive Breast Cancer Cells Correlates with Their Trastuzumab Sensitivity. Breast Cancer Res. 22, 105. 10.1186/s13058-020-01342-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzanowska J., Semira C., Costa‐Silva B. (2020). DNA in Extracellular Vesicles: Biological and Clinical Aspects. Mol. Oncol. 10.1002/1878-0261.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Tian H., Li X., Jin D., Li X., Kong J., et al. (2017). Clinical Application of a Microfluidic Chip for Immunocapture and Quantification of Circulating Exosomes to Assist Breast Cancer Diagnosis and Molecular Classification. PloS one 12, e0175050. 10.1371/journal.pone.0175050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa J. M., Skog J., Akers J., Li H., Komotar R., Jensen R., et al. (2017). Detection of Wild-type EGFR Amplification and EGFRvIII Mutation in CSF-Derived Extracellular Vesicles of Glioblastoma Patients. Neuro-oncology 19, 1494–1502. 10.1093/neuonc/nox085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser K. B., Moehle M. S., Daher J. P. L., Webber P. J., Williams J. Y., Stewart C. A., et al. (2013). LRRK2 Secretion in Exosomes Is Regulated by 14-3-3. Hum. Mol. Genet. 22, 4988–5000. 10.1093/hmg/ddt346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. A., Letai A., Fisher D. E., Flaherty K. T. (2015). Precision Medicine for Cancer with Next-Generation Functional Diagnostics. Nat. Rev. Cancer 15, 747–756. 10.1038/nrc4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F.-f., Zhu X.-j., Wang H.-x., Zhang L.-m., Yuan G.-l., Chen Z.-c., et al. (2017). BCR-ABL1-positive Microvesicles Malignantly Transform Human Bone Marrow Mesenchymal Stem Cells In Vitro. Acta Pharmacol. Sin. 38, 1475–1485. 10.1038/aps.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Eguchi T., Sogawa C., Ono K., Murakami J., Ibaragi S., et al. (2018). Anti-EGFR Antibody Cetuximab Is Secreted by Oral Squamous Cell Carcinoma and Alters EGF-Driven Mesenchymal Transition. Biochem. Biophysical Res. Commun. 503, 1267–1272. 10.1016/j.bbrc.2018.07.035 [DOI] [PubMed] [Google Scholar]
- Galli de Amorim M., Branco G., Valieris R., Tarcitano E., Tojal da Silva I., Ferreira de Araújo L., et al. (2019). The Impact of HER2 Overexpression on the miRNA and circRNA Transcriptomes in Two Breast Cell Lines and Their Vesicles. Pharmacogenomics 20, 493–502. 10.2217/pgs-2018-0182 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signaling 6, pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Silva S., Benito-Martín A., Sánchez-Redondo S., Hernández-Barranco A., Ximénez-Embún P., Nogués L., et al. (2019). Use of Extracellular Vesicles from Lymphatic Drainage as Surrogate Markers of Melanoma Progression and BRAFV600E Mutation. J. Exp. Med. 216, 1061–1070. 10.1084/jem.20181522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier D., Meehan B., Kislinger T., Daniel P., Sinha A., Abdulkarim B., et al. (2018). Divergent Evolution of Temozolomide Resistance in Glioblastoma Stem Cells Is Reflected in Extracellular Vesicles and Coupled with Radiosensitization. Neuro-oncology 20, 236–248. 10.1093/neuonc/nox142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeurickx E., Hendrix A. (2020). Targets, Pitfalls and Reference Materials for Liquid Biopsy Tests in Cancer Diagnostics. Mol. Aspects Med. 72, 100828. 10.1016/j.mam.2019.10.005 [DOI] [PubMed] [Google Scholar]
- Gerratana L., Toffoletto B., Bulfoni M., Cesselli D., Beltrami A. P., Di Loreto C., et al. (2015). Metastatic Breast Cancer and Circulating Exosomes. Hints from an Exploratory Analysis. Ann. Oncol. 26, vi14. 10.1093/annonc/mdv336.37 [DOI] [Google Scholar]
- Gerth K., Kodidela S., Mahon M., Haque S., Verma N., Kumar S. (2019). Circulating Extracellular Vesicles Containing Xenobiotic Metabolizing CYP Enzymes and Their Potential Roles in Extrahepatic Cells via Cell-Cell Interactions. Int. J. Mol. Sci. 20, 6178. 10.3390/ijms20246178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri R., Piva F., Occhipinti G., Bittoni A., Righetti A., Pagliaretta S., et al. (2019). Clinical Impact of Different Exosomes' Protein Expression in Pancreatic Ductal Carcinoma Patients Treated with Standard First Line Palliative Chemotherapy. PloS one 14, e0215990. 10.1371/journal.pone.0215990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimple R. C., Wang X. (2019). RAS: Striking at the Core of the Oncogenic Circuitry. Front. Oncol. 9, 965. 10.3389/fonc.2019.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluszko A., Szczepanski M. J., Ludwig N., Mirza S. M., Olejarz W. (2019). Exosomes in Cancer: Circulating Immune-Related Biomarkers. Biomed. Res. Int. 2019, 1628029. 10.1155/2019/1628029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Jaiswal R., Mathys J.-M., Combes V., Grau G. E. R., Bebawy M. (2012). Microparticles and Their Emerging Role in Cancer Multidrug Resistance. Cancer Treat. Rev. 38, 226–234. 10.1016/j.ctrv.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Gonzales P. A., Pisitkun T., Hoffert J. D., Tchapyjnikov D., Star R. A., Kleta R., et al. (2009). Large-scale Proteomics and Phosphoproteomics of Urinary Exosomes. Jasn 20, 363–379. 10.1681/asn.2008040406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Falcón-Pérez J. M. (2015). Cell-derived Extracellular Vesicles as a Platform to Identify Low-Invasive Disease Biomarkers. Expert Rev. Mol. Diagn. 15, 907–923. 10.1586/14737159.2015.1043272 [DOI] [PubMed] [Google Scholar]
- Goricar K., Kovac V., Jazbec J., Zakotnik B., Lamovec J., Dolzan V. (2015). Genetic Variability of DNA Repair Mechanisms and Glutathione-S-Transferase Genes Influences Treatment Outcome in Osteosarcoma. Cancer Epidemiol. 39, 182. 10.1016/j.canep.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. (1997). ErbB-2, the Preferred Heterodimerization Partner of All ErbB Receptors, Is a Mediator of Lateral Signaling. EMBO J. 16, 1647–1655. 10.1093/emboj/16.7.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregers J., Gréen H., Christensen I. J., Dalhoff K., Schroeder H., Carlsen N., et al. (2015). Polymorphisms in the ABCB1 Gene and Effect on Outcome and Toxicity in Childhood Acute Lymphoblastic Leukemia. Pharmacogenomics J. 15, 372–379. 10.1038/tpj.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y.-X., Li Y.-M., Ye M., Guo Y.-Y., Li Q.-W., Peng X.-M., et al. (2017). KRAS and BRAF Mutations in Serum Exosomes from Patients with Colorectal Cancer in a Chinese Population. Oncol. Lett. 13, 3608–3616. 10.3892/ol.2017.5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Qin H., Poon T. C. W., Sze S.-C., Ding X., Co N. N., et al. (2015). Hepatocellular Carcinoma-Derived Exosomes Promote Motility of Immortalized Hepatocyte through Transfer of Oncogenic Proteins and RNAs. Carcin 36, 1008–1018. 10.1093/carcin/bgv081 [DOI] [PubMed] [Google Scholar]
- Hong B., Cho J.-H., Kim H., Choi E.-J., Rho S., Kim J., et al. (2009). Colorectal Cancer Cell-Derived Microvesicles Are Enriched in Cell Cycle-Related mRNAs that Promote Proliferation of Endothelial Cells. BMC Genomics 10, 556. 10.1186/1471-2164-10-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Fang C., Yi K., Liu X., Qi H., Tan Y., et al. (2018). The Role of PTRF/Cavin1 as a Biomarker in Both Glioma and Serum Exosomes. Theranostics 8, 1540–1557. 10.7150/thno.22952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J. Y., Kim H. J., Lee J. S., Choi C.-M., Lee J. C., Jung M. K., et al. (2018). Extracellular Vesicle-Derived DNA for Performing EGFR Genotyping of NSCLC Patients. Mol. Cancer 17, 15. 10.1186/s12943-018-0772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz S. N., Rider M. A., Bundy J. L., Liu X., Singh R. K., Meckes D. G., Jr. (2016). Proteomic Profiling of NCI-60 Extracellular Vesicles Uncovers Common Protein Cargo and Cancer Type-specific Biomarkers. Oncotarget 7, 86999–87015. 10.18632/oncotarget.13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Greening D. W., Barnes T. W., Lim J. W., Tauro B. J., Rai A., et al. (2013). Proteome Profiling of Exosomes Derived from Human Primary and Metastatic Colorectal Cancer Cells Reveal Differential Expression of Key Metastatic Factors and Signal Transduction Components. Proteomics 13, 1672–1686. 10.1002/pmic.201200562 [DOI] [PubMed] [Google Scholar]
- Jin Y., Chen K., Wang Z., Wang Y., Liu J., Lin L., et al. (2016). DNA in Serum Extracellular Vesicles Is Stable under Different Storage Conditions. BMC Cancer 16, 753. 10.1186/s12885-016-2783-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen K. B., Gudbergsson J. M., Andresen T. L., Simonsen J. B. (2019). What Is the Blood Concentration of Extracellular Vesicles? Implications for the Use of Extracellular Vesicles as Blood-Borne Biomarkers of Cancer. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1871, 109–116. 10.1016/j.bbcan.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Jurj A., Pasca S., Teodorescu P., Tomuleasa C., Berindan-Neagoe I. (2020). Basic Knowledge on BCR-ABL1-Positive Extracellular Vesicles. Biomarkers Med. 14, 451–458. 10.2217/bmm-2019-0510 [DOI] [PubMed] [Google Scholar]
- Kahlert C., Melo S. A., Protopopov A., Tang J., Seth S., Koch M., et al. (2014). Identification of Double-Stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and P53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 289, 3869–3875. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H., Simpson R. J., Ji H., Aikawa E., Altevogt P., Askenase P., et al. (2012). Vesiclepedia: a Compendium for Extracellular Vesicles with Continuous Community Annotation. Plos Biol. 10, e1001450. 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S., LeBleu V. S., Sugimoto H., Yang S., Ruivo C. F., Melo S. A., et al. (2017). Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature 546, 498–503. 10.1038/nature22341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K.-W., Jung J.-H., Hur W., Park J., Shin H., Choi B., et al. (2018). The Potential of Exosomes Derived from Chronic Myelogenous Leukaemia Cells as a Biomarker. Anticancer Res. 38, 3935–3942. 10.21873/anticanres.12679 [DOI] [PubMed] [Google Scholar]
- Kato T., Mizutani K., Kameyama K., Kawakami K., Fujita Y., Nakane K., et al. (2015). Serum Exosomal P-Glycoprotein Is a Potential Marker to Diagnose Docetaxel Resistance and Select a Taxoid for Patients with Prostate Cancer. Urol. Oncol. Semin. Original Invest. 33, 385.e15-20. 10.1016/j.urolonc.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., et al. (2016). ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 428, 688–692. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keklikoglou I., Cianciaruso C., Güç E., Squadrito M. L., Spring L. M., Tazzyman S., et al. (2019). Chemotherapy Elicits Pro-metastatic Extracellular Vesicles in Breast Cancer Models. Nat. Cel Biol. 21, 190–202. 10.1038/s41556-018-0256-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharaziha P., Chioureas D., Rutishauser D., Baltatzis G., Lennartsson L., Fonseca P., et al. (2015). Molecular Profiling of Prostate Cancer Derived Exosomes May Reveal a Predictive Signature for Response to Docetaxel. Oncotarget 6, 21740–21754. 10.18632/oncotarget.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai Y., Kawasaki T., Sueyoshi T., Kobiyama K., Ishii K. J., Zou J., et al. (2017). DNA-containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-dependent Pathway and Reinforce Antitumor Immunity. J Immunol 198, 1649–1659. 10.4049/jimmunol.1601694 [DOI] [PubMed] [Google Scholar]
- Klump J., Phillipp U., Follo M., Eremin A., Lehmann H., Nestel S., et al. (2018). Extracellular Vesicles or Free Circulating DNA: where to Search for BRAF and cKIT Mutations? Nanomedicine: Nanotechnology, Biol. Med. 14, 875–882. 10.1016/j.nano.2017.12.009 [DOI] [PubMed] [Google Scholar]
- Koch R., Aung T., Vogel D., Chapuy B., Wenzel D., Becker S., et al. (2016). Nuclear Trapping through Inhibition of Exosomal Export by Indomethacin Increases Cytostatic Efficacy of Doxorubicin and Pixantrone. Clin. Cancer Res. 22, 395–404. 10.1158/1078-0432.Ccr-15-0577 [DOI] [PubMed] [Google Scholar]
- Kowal J., Arras G., Colombo M., Jouve M., Morath J. P., Primdal-Bengtson B., et al. (2016). Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 113, E968–E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A. K., Enderle D., Karlovich C., Priewasser T., Bentink S., Spiel A., et al. (2018). Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann. Oncol. 29, 700–706. 10.1093/annonc/mdx765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sinha N., Gerth K. A., Rahman M. A., Yallapu M. M., Midde N. M. (2017). Specific Packaging and Circulation of Cytochromes P450, Especially 2E1 Isozyme, in Human Plasma Exosomes and Their Implications in Cellular Communications. Biochem. Biophysical Res. Commun. 491, 675–680. 10.1016/j.bbrc.2017.07.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X., Wang M., McElyea S. D., Sherman S., House M., Korc M. (2017). A microRNA Signature in Circulating Exosomes Is Superior to Exosomal Glypican-1 Levels for Diagnosing Pancreatic Cancer. Cancer Lett. 393, 86–93. 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas M. J., Duran G., Balboa E., Bernardez B., Touris M., Vidal Y., et al. (2011). Use of a Comprehensive Panel of Biomarkers to Predict Response to a Fluorouracil-Oxaliplatin Regimen in Patients with Metastatic Colorectal Cancer. Pharmacogenomics 12, 433–442. 10.2217/pgs.10.196 [DOI] [PubMed] [Google Scholar]
- Lauschke V. M., Milani L., Ingelman-Sundberg M. (2017). Pharmacogenomic Biomarkers for Improved Drug Therapy-Recent Progress and Future Developments. AAPS J. 20, 4. 10.1208/s12248-017-0161-x [DOI] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Lässer C., Shelke G. V., Crescitelli R., Jang S. C., Cvjetkovic A., et al. (2019). DNA Analysis of Low- and High-Density Fractions Defines Heterogeneous Subpopulations of Small Extracellular Vesicles Based on Their DNA Cargo and Topology. J. Extracellular Vesicles 8, 1656993. 10.1080/20013078.2019.1656993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Lunavat T. R., Jang S. C., Escobedo-Lucea C., Oliver-De La Cruz J., Siljander P., et al. (2017). Distinct Prostate Cancer-Related mRNA Cargo in Extracellular Vesicle Subsets from Prostate Cell Lines. BMC Cancer 17, 92. 10.1186/s12885-017-3087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Sanz-Garcia A., Visakorpi T., Escobedo-Lucea C., Siljander P., Ayuso-Sacido A., et al. (2014). Different gDNA Content in the Subpopulations of Prostate Cancer Extracellular Vesicles: Apoptotic Bodies, Microvesicles, and Exosomes. Prostate 74, 1379–1390. 10.1002/pros.22853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu V. S., Kalluri R. (2020). Exosomes as a Multicomponent Biomarker Platform in Cancer . Trends Cancer 6, 767–774. 10.1016/j.trecan.2020.03.007 [DOI] [PubMed] [Google Scholar]
- Lee D. H., Yoon H., Park S., Kim J. S., Ahn Y.-H., Kwon K., et al. (2018). Urinary Exosomal and Cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci. Rep. 8, 14707. 10.1038/s41598-018-32900-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Hur J. Y., Kim I. A., Kim H. J., Choi C. M., Lee J. C., et al. (2018). Liquid Biopsy Using the Supernatant of a Pleural Effusion for EGFR Genotyping in Pulmonary Adenocarcinoma Patients: a Comparison between Cell-free DNA and Extracellular Vesicle-Derived DNA. BMC Cancer 18, 1236. 10.1186/s12885-018-5138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Chennakrishnaiah S., Audemard E., Montermini L., Meehan B., Rak J. (2014). Oncogenic Ras-Driven Cancer Cell Vesiculation Leads to Emission of Double-Stranded DNA Capable of Interacting with Target Cells. Biochem. Biophysical Res. Commun. 451, 295–301. 10.1016/j.bbrc.2014.07.109 [DOI] [PubMed] [Google Scholar]
- Lee Y. T., Tan Y. J., Oon C. E. (2018). Molecular Targeted Therapy: Treating Cancer with Specificity. Eur. J. Pharmacol. 834, 188–196. 10.1016/j.ejphar.2018.07.034 [DOI] [PubMed] [Google Scholar]
- Liang B., Peng P., Chen S., Li L., Zhang M., Cao D., et al. (2013). Characterization and Proteomic Analysis of Ovarian Cancer-Derived Exosomes. J. Proteomics 80, 171–182. 10.1016/j.jprot.2012.12.029 [DOI] [PubMed] [Google Scholar]
- Liem M., Ang C.-S., Mathivanan S. (2017). Insulin Mediated Activation of PI3K/Akt Signalling Pathway Modifies the Proteomic Cargo of Extracellular Vesicles. Proteomics 17, 1600371. 10.1002/pmic.201600371 [DOI] [PubMed] [Google Scholar]
- Liu X., Chinello C., Musante L., Cazzaniga M., Tataruch D., Calzaferri G., et al. (2015). Intraluminal Proteome and Peptidome of Human Urinary Extracellular Vesicles. Prot. Clin. Appl. 9, 568–573. 10.1002/prca.201400085 [DOI] [PubMed] [Google Scholar]
- Liu X. Q., Tufman A., Kiefl R., Li G. F., Ma Q. L., Huber R. M. (2020). Identification of Lung Adenocarcinoma-specific Exosome RNAs in Peripheral Blood by RNA-Seq Analysis. Eur. Rev. Med. Pharmacol. Sci. 24, 1877–1886. 10.26355/eurrev_202002_20366 [DOI] [PubMed] [Google Scholar]
- Liu Y., Song B., Wei Y., Chen F., Chi Y., Fan H., et al. (2018). Exosomes from Mesenchymal Stromal Cells Enhance Imatinib-Induced Apoptosis in Human Leukemia Cells via Activation of Caspase Signaling Pathway. Cytotherapy 20, 181–188. 10.1016/j.jcyt.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Maacha S., Bhat A. A., Jimenez L., Raza A., Haris M., Uddin S., et al. (2019). Extracellular Vesicles-Mediated Intercellular Communication: Roles in the Tumor Microenvironment and Anti-cancer Drug Resistance. Mol. Cancer 18, 55. 10.1186/s12943-019-0965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione F., Cappellano G., Bellan M., Raineri D., Chiocchetti A. (2020). Chicken‐or‐egg Question: Which Came First, Extracellular Vesicles or Autoimmune Diseases? J. Leukoc. Biol. 108, 601–616. 10.1002/jlb.3mr0120-232r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin E. Z., Bratman S. V. (2020). Bioactive DNA from Extracellular Vesicles and Particles. Cell Death Dis 11, 584. 10.1038/s41419-020-02803-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari R., Mamessier E., Lambaudie E., Provansal M., Birnbaum D., Bertucci F., et al. (2019). Liquid Biopsies for Ovarian Carcinoma: How Blood Tests May Improve the Clinical Management of a Deadly Disease. Cancers 11, 774. 10.3390/cancers11060774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez V. G., O'Neill S., Salimu J., Breslin S., Clayton A., Crown J., et al. (2017). Resistance to HER2-Targeted Anti-cancer Drugs Is Associated with Immune Evasion in Cancer Cells and Their Derived Extracellular Vesicles. Oncoimmunology 6, e1362530. 10.1080/2162402x.2017.1362530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A., Luecke L. B., Kahlert C., Fernandez A. F., Gammon S. T., Kaye J., et al. (2015). Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 523, 177–182. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikamori M., Yamada D., Eguchi H., Hasegawa S., Kishimoto T., Tomimaru Y., et al. (2017). MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 7, 42339. 10.1038/srep42339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani G., Lana T., Bresolin S., Aveic S., Pastò A., Frasson C., et al. (2017). Expression Profiling of Circulating Microvesicles Reveals Intercellular Transmission of Oncogenic Pathways. Mol. Cancer Res. 15, 683–695. 10.1158/1541-7786.Mcr-16-0307 [DOI] [PubMed] [Google Scholar]
- Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D., et al. (2012). Emergence of KRAS Mutations and Acquired Resistance to Anti-EGFR Therapy in Colorectal Cancer. Nature 486, 532–536. 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y., Nakayama S., Torikoshi Y., Tanaka S., Ishihara H., Taguchi T., et al. (2006). High Expression of Ubiquitin Carboxy-Terminal Hydrolase-L1 and -L3 mRNA Predicts Early Recurrence in Patients with Invasive Breast Cancer. Cancer Sci. 97, 523–529. 10.1111/j.1349-7006.2006.00202.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhrmann L., Huang H. J., Hong D. S., Tsimberidou A. M., Fu S., Piha-Paul S. A., et al. (2018). Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res. 24, 181–188. 10.1158/1078-0432.Ccr-17-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montermini L., Meehan B., Garnier D., Lee W. J., Lee T. H., Guha A., et al. (2015). Inhibition of Oncogenic Epidermal Growth Factor Receptor Kinase Triggers Release of Exosome-like Extracellular Vesicles and Impacts Their Phosphoprotein and DNA Content. J. Biol. Chem. 290, 24534–24546. 10.1074/jbc.M115.679217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon P.-G., Lee J.-E., You S., Kim T.-K., Cho J.-H., Kim I.-S., et al. (2011). Proteomic Analysis of Urinary Exosomes from Patients of Early IgA Nephropathy and Thin Basement Membrane Nephropathy. Proteomics 11, 2459–2475. 10.1002/pmic.201000443 [DOI] [PubMed] [Google Scholar]
- Muller L., Muller-Haegele S., Mitsuhashi M., Gooding W., Okada H., Whiteside T. L. (2015). Exosomes Isolated from Plasma of Glioma Patients Enrolled in a Vaccination Trial Reflect Antitumor Immune Activity and Might Predict Survival. Oncoimmunology 4, e1008347. 10.1080/2162402x.2015.1008347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Molins L., Marrades R., Moises J., Viñolas N., Morales S., et al. (2019). Exosome Analysis in Tumor-Draining Pulmonary Vein Identifies NSCLC Patients with Higher Risk of Relapse after Curative Surgery. Cancers 11, 249. 10.3390/cancers11020249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K., Wang T., Sun X., Zhang P., Chen Y., Jin J., et al. (2017). UCH-L1-containing Exosomes Mediate Chemotherapeutic Resistance Transfer in Breast Cancer. J. Surg. Oncol. 115, 932–940. 10.1002/jso.24614 [DOI] [PubMed] [Google Scholar]
- Nogués L., Benito-Martin A., Hergueta-Redondo M., Peinado H. (2018). The Influence of Tumour-Derived Extracellular Vesicles on Local and Distal Metastatic Dissemination. Mol. Aspects Med. 60, 15–26. 10.1016/j.mam.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G., Borgiani P., Ciccacci C., Di Daniele N., Sirugo G., Papaluca Amati M. (2010). Pharmacogenomics: Role in Medicines Approval and Clinical Use. Public health genomics 13, 284–291. 10.1159/000245271 [DOI] [PubMed] [Google Scholar]
- O'Brien K., Breyne K., Ughetto S., Laurent L. C., Breakefield X. O. (2020). RNA Delivery by Extracellular Vesicles in Mammalian Cells and its Applications. Nat. Rev. Mol. Cel Biol. 21, 585–606. 10.1038/s41580-020-0251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S.-i., Takanashi M., Sudo K., Ueda S., Ishikawa A., Matsuyama N., et al. (2013). Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor microRNA to Breast Cancer Cells. Mol. Ther. 21, 185–191. 10.1038/mt.2012.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega F. G., Piguillem S. V., Messina G. A., Tortella G. R., Rubilar O., Jiménez Castillo M. I., et al. (2019). EGFR Detection in Extracellular Vesicles of Breast Cancer Patients through Immunosensor Based on Silica-Chitosan Nanoplatform. Talanta 194, 243–252. 10.1016/j.talanta.2018.10.016 [DOI] [PubMed] [Google Scholar]
- Osti D., Del Bene M., Rappa G., Santos M., Matafora V., Richichi C., et al. (2019). Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 25, 266–276. 10.1158/1078-0432.Ccr-18-1941 [DOI] [PubMed] [Google Scholar]
- Øverbye A., Skotland T., Koehler C. J., Thiede B., Seierstad T., Berge V., et al. (2015). Identification of Prostate Cancer Biomarkers in Urinary Exosomes. Oncotarget 6, 30357–30376. 10.18632/oncotarget.4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paggetti J., Haderk F., Seiffert M., Janji B., Distler U., Ammerlaan W., et al. (2015). Exosomes Released by Chronic Lymphocytic Leukemia Cells Induce the Transition of Stromal Cells into Cancer-Associated Fibroblasts. Blood 126, 1106–1117. 10.1182/blood-2014-12-618025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotara A., Markou A., Lianidou E. S., Patrinos G. P., Katsila T. (2017). Exosomes: A Cancer Theranostics Road Map. Public health genomics 20, 116–125. 10.1159/000478253 [DOI] [PubMed] [Google Scholar]
- Park J. O., Choi D.-Y., Choi D.-S., Kim H. J., Kang J. W., Jung J. H., et al. (2013). Identification and Characterization of Proteins Isolated from Microvesicles Derived from Human Lung Cancer Pleural Effusions. Proteomics 13, 2125–2134. 10.1002/pmic.201200323 [DOI] [PubMed] [Google Scholar]
- Pasqualetti F., Restante G., Gonnelli A., Rofi E., Molinari A., Crucitta S., et al. (2019). Dabrafenib Treatment in a Patient with BRAF V600E Ganglioglioma: Circulating Exosome-Derived Cancer RNA Supports Treatment Choice and Clinical Monitoring. Neuro-oncology 21, 1610–1611. 10.1093/neuonc/noz157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M., Fonseka P., Chitti S. V., Kang T., Sanwlani R., Van Deun J., et al. (2019). Vesiclepedia 2019: a Compendium of RNA, Proteins, Lipids and Metabolites in Extracellular Vesicles. Nucleic Acids Res. 47, D516–d519. 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlyukov M. S., Yu H., Bastola S., Minata M., Shender V. O., Lee Y., et al. (2018). Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma via Intercellular Transfer of Splicing Factors. Cancer Cell 34, 119–135 e110. 10.1016/j.ccell.2018.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platko K., Haas‐Neill S., Aziz T., Al‐Nedawi K. (2019). The Role of Circulating Extracellular Vesicles in Breast Cancer Classification and Molecular Subtyping. Breast J. 25, 691–695. 10.1111/tbj.13309 [DOI] [PubMed] [Google Scholar]
- Puhka M., Takatalo M., Nordberg M.-E., Valkonen S., Nandania J., Aatonen M., et al. (2017). Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics 7, 3824–3841. 10.7150/thno.19890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Li Q., Yang J., Zhao H., Wang F., Zhang F., et al. (2019). Double-Stranded DNA in Exosomes of Malignant Pleural Effusions as a Novel DNA Source for EGFR Mutation Detection in Lung Adenocarcinoma. Front. Oncol. 9, 931. 10.3389/fonc.2019.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. A., Kodidela S., Sinha N., Haque S., Shukla P. K., Rao R., et al. (2019). Plasma Exosomes Exacerbate Alcohol- and Acetaminophen-Induced Toxicity via CYP2E1 Pathway. Sci. Rep. 9, 6571. 10.1038/s41598-019-43064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reclusa P., Laes J.-F., Malapelle U., Valentino A., Rocco D., Gil-Bazo I., et al. (2018). EML4-ALK Translocation Identification in RNA Exosomal Cargo (ExoALK) in NSCLC Patients: a Novel Role for Liquid Biopsy. Transl. Cancer Res. 8, S76–S78. 10.21037/tcr.2018.11.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D., Rowland A. (2019). From Endogenous Compounds as Biomarkers to Plasma‐Derived Nanovesicles as Liquid Biopsy; Has the Golden Age of Translational Pharmacokinetics‐Absorption, Distribution, Metabolism, Excretion‐Drug-Drug Interaction Science Finally Arrived? Clin. Pharmacol. Ther. 105, 1407–1420. 10.1002/cpt.1328 [DOI] [PubMed] [Google Scholar]
- Rodríguez Zorrilla S., Pérez-Sayans M., Fais S., Logozzi M., Gallas Torreira M., García García A. (2019). A Pilot Clinical Study on the Prognostic Relevance of Plasmatic Exosomes Levels in Oral Squamous Cell Carcinoma Patients. Cancers 11, 429. 10.3390/cancers11030429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Antona C., Ingelman-Sundberg M. (2006). Cytochrome P450 Pharmacogenetics and Cancer. Oncogene 25, 1679–1691. 10.1038/sj.onc.1209377 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez A., de Miguel-Pérez D., Ortega F. G., García-Puche J. L., Robles-Fernández I., Exposito J., et al. (2019). Exosomal miRNA Profile as Complementary Tool in the Diagnostic and Prediction of Treatment Response in Localized Breast Cancer under Neoadjuvant Chemotherapy. Breast Cancer Res. 21, 21. 10.1186/s13058-019-1109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rofi E., Vivaldi C., Del Re M., Arrigoni E., Crucitta S., Funel N., et al. (2019). The Emerging Role of Liquid Biopsy in Diagnosis, Prognosis and Treatment Monitoring of Pancreatic Cancer. Pharmacogenomics 20, 49–68. 10.2217/pgs-2018-0149 [DOI] [PubMed] [Google Scholar]
- Rowland A., Ruanglertboon W., Dyk M., Wijayakumara D., Wood L. S., Meech R., et al. (2019). Plasma Extracellular Nanovesicle (Exosome)‐derived Biomarkers for Drug Metabolism Pathways: a Novel Approach to Characterize Variability in Drug Exposure. Br. J. Clin. Pharmacol. 85, 216–226. 10.1111/bcp.13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh C. S. D., McMillan D. C. (2014). Cancer and Systemic Inflammation: Treat the Tumour and Treat the Host. Br. J. Cancer 110, 1409–1412. 10.1038/bjc.2014.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaei R., Larson B. J., Cheng T. C., Gibson M. A., Otani S., Naerdemann W., et al. (2005). Abnormal Lysosomal Trafficking and Enhanced Exosomal Export of Cisplatin in Drug-Resistant Human Ovarian Carcinoma Cells. Mol. Cancer Ther. 4, 1595–1604. 10.1158/1535-7163.Mct-05-0102 [DOI] [PubMed] [Google Scholar]
- Samuel P., Mulcahy L. A., Furlong F., McCarthy H. O., Brooks S. A., Fabbri M., et al. (2018). Cisplatin Induces the Release of Extracellular Vesicles from Ovarian Cancer Cells that Can Induce Invasiveness and Drug Resistance in Bystander Cells. Phil. Trans. R. Soc. B 373, 20170065. 10.1098/rstb.2017.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Lucas F. A., Allenson K., Bernard V., Castillo J., Kim D. U., Ellis K., et al. (2016). Minimally Invasive Genomic and Transcriptomic Profiling of Visceral Cancers by Next-Generation Sequencing of Circulating Exosomes. Ann. Oncol. 27, 635–641. 10.1093/annonc/mdv604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P., Savini C., Kurelac I., Chang Q., Amato L. B., Strillacci A., et al. (2017). Packaging and Transfer of Mitochondrial DNA via Exosomes Regulate Escape from Dormancy in Hormonal Therapy-Resistant Breast Cancer. Proc. Natl. Acad. Sci. USA 114, E9066–E9075. 10.1073/pnas.1704862114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S., Settleman J., Reshkin S. J., Azzariti A., Bellizzi A., Paradiso A. (2006). The Complexity of Targeting EGFR Signalling in Cancer: from Expression to Turnover. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1766, 120–139. 10.1016/j.bbcan.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Shao H., Chung J., Balaj L., Charest A., Bigner D. D., Carter B. S., et al. (2012). Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 18, 1835–1840. 10.1038/nm.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedden K., Xie X. T., Chandaroy P., Chang Y. T., Rosania G. R. (2003). Expulsion of Small Molecules in Vesicles Shed by Cancer Cells: Association with Gene Expression and Chemosensitivity Profiles. Cancer Res. 63, 4331–4337. [PubMed] [Google Scholar]
- Shekhani R., Steinacher L., Swen J. J., Ingelman‐Sundberg M. (2020). Evaluation of Current Regulation and Guidelines of Pharmacogenomic Drug Labels: Opportunities for Improvements. Clin. Pharmacol. Ther. 107, 1240–1255. 10.1002/cpt.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S., Avanzato D., Lanzetti L. (2018). Emerging Functions of the EGFR in Cancer. Mol. Oncol. 12, 3–20. 10.1002/1878-0261.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Garcia V., Rodriguez M., Compte M., Cisneros E., Veguillas P., et al. (2012). Analysis of Exosome Release and its Prognostic Value in Human Colorectal Cancer. Genes Chromosom. Cancer 51, 409–418. 10.1002/gcc.21926 [DOI] [PubMed] [Google Scholar]
- Sim S. C., Kacevska M., Ingelman-Sundberg M. (2013). Pharmacogenomics of Drug-Metabolizing Enzymes: a Recent Update on Clinical Implications and Endogenous Effects. Pharmacogenomics J. 13, 1–11. 10.1038/tpj.2012.45 [DOI] [PubMed] [Google Scholar]
- Sinha A., Ignatchenko V., Ignatchenko A., Mejia-Guerrero S., Kislinger T. (2014). In-depth Proteomic Analyses of Ovarian Cancer Cell Line Exosomes Reveals Differential Enrichment of Functional Categories Compared to the NCI 60 Proteome. Biochem. Biophysical Res. Commun. 445, 694–701. 10.1016/j.bbrc.2013.12.070 [DOI] [PubMed] [Google Scholar]
- Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Curry W. T., et al. (2008). Glioblastoma Microvesicles Transport RNA and Proteins that Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cel Biol. 10, 1470–1476. 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T., Sagini K., Sandvig K., Llorente A. (2020). An Emerging Focus on Lipids in Extracellular Vesicles. Adv. Drug Deliv. Rev. 159, 308–321. 10.1016/j.addr.2020.03.002 [DOI] [PubMed] [Google Scholar]
- Slamon D., Eiermann W., Robert N., Pienkowski T., Martin M., Press M., et al. (2011). Adjuvant Trastuzumab in HER2-Positive Breast Cancer. N. Engl. J. Med. 365, 1273–1283. 10.1056/NEJMoa0910383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Cai Z., Yan J., Shao Y. W., Zhang Y. (2019). Liquid Biopsies Using Pleural Effusion-Derived Exosomal DNA in Advanced Lung Adenocarcinoma. Transl. Lung Cancer Res. 8, 392–400. 10.21037/tlcr.2019.08.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevic I., Buescher G., Ricklefs F. L. (2020). Monitoring Therapy Efficiency in Cancer through Extracellular Vesicles. Cells 9, 130. 10.3390/cells9010130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehlmacher J., Park D. J., Zhang W., Yang D., Groshen S., Zahedy S., et al. (2004). A Multivariate Analysis of Genomic Polymorphisms: Prediction of Clinical Outcome to 5-FU/oxaliplatin Combination Chemotherapy in Refractory Colorectal Cancer. Br. J. Cancer 91, 344–354. 10.1038/sj.bjc.6601975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthandiram S., Gan G.-G., Zain S. M., Bee P.-C., Lian L.-H., Chang K.-M., et al. (2014). Effect of Polymorphisms within Methotrexate Pathway Genes on Methotrexate Toxicity and Plasma Levels in Adults with Hematological Malignancies. Pharmacogenomics 15, 1479–1494. 10.2217/pgs.14.97 [DOI] [PubMed] [Google Scholar]
- Syn N. L.-X., Yong W.-P., Goh B.-C., Lee S.-C. (2016). Evolving Landscape of Tumor Molecular Profiling for Personalized Cancer Therapy: a Comprehensive Review. Expert Opin. Drug Metab. Toxicol. 12, 911–922. 10.1080/17425255.2016.1196187 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Okada R., Nagao K., Kawamata Y., Hanyu A., Yoshimoto S., et al. (2017). Exosomes Maintain Cellular Homeostasis by Excreting Harmful DNA from Cells. Nat. Commun. 8, 15287. 10.1038/ncomms15287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Zheng K., Tang Y., Li Z., Zou T., Liu D. (2019). Overexpression of Serum Exosomal HOTAIR Is Correlated with Poor Survival and Poor Response to Chemotherapy in Breast Cancer Patients. J. Biosci. 44, 37. 10.1007/s12038-019-9861-y [DOI] [PubMed] [Google Scholar]
- Thakur B. K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., et al. (2014). Double-stranded DNA in Exosomes: a Novel Biomarker in Cancer Detection. Cell Res 24, 766–769. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., et al. (2018). Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 7, 1535750. 10.1080/20013078.2018.153575010.1080/20013078.2018.1461450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torralba D., Baixauli F., Villarroya-Beltri C., Fernández-Delgado I., Latorre-Pellicer A., Acín-Pérez R., et al. (2018). Priming of Dendritic Cells by DNA-Containing Extracellular Vesicles from Activated T Cells through Antigen-Driven Contacts. Nat. Commun. 9, 2658. 10.1038/s41467-018-05077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader P., Breakefield X. O., Wood M. J. A. (2014). Extracellular Vesicles: Emerging Targets for Cancer Therapy. Trends Mol. Med. 20, 385–393. 10.1016/j.molmed.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T., Spinelli C., Minciacchi V. R., Balaj L., Zandian M., Conley A., et al. (2018). Large Extracellular Vesicles Carry Most of the Tumour DNA Circulating in Prostate Cancer Patient Plasma. J. Extracellular Vesicles 7, 1505403. 10.1080/20013078.2018.1505403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dommelen S. M., van der Meel R., van Solinge W. W., Coimbra M., Vader P., Schiffelers R. M. (2016). Cetuximab Treatment Alters the Content of Extracellular Vesicles Released from Tumor Cells. Nanomedicine 11, 881–890. 10.2217/nnm-2015-0009 [DOI] [PubMed] [Google Scholar]
- van Niel G., D'Angelo G., Raposo G. (2018). Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cel Biol. 19, 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Vasconcelos M. H., Caires H. R., Ābols A., Xavier C. P. R., Linē A. (2019). Extracellular Vesicles as a Novel Source of Biomarkers in Liquid Biopsies for Monitoring Cancer Progression and Drug Resistance. Drug Resist. Updates 47, 100647. 10.1016/j.drup.2019.100647 [DOI] [PubMed] [Google Scholar]
- von Mehren M., Joensuu H. (2018). Gastrointestinal Stromal Tumors. J Clin Oncol 36, 136–143. 10.1200/jco.2017.74.9705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Liu B., Lei H., Zhang B., Wang Y., Huang H., et al. (2018). Nanoscale Extracellular Vesicle-Derived DNA Is Superior to Circulating Cell-free DNA for Mutation Detection in Early-Stage Non-small-cell Lung Cancer. Ann. Oncol. 29, 2379–2383. 10.1093/annonc/mdy458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ji S., Shao G., Zhang J., Zhao K., Wang Z., et al. (2018). Effect of Exosome Biomarkers for Diagnosis and Prognosis of Breast Cancer Patients. Clin. Transl. Oncol. 20, 906–911. 10.1007/s12094-017-1805-0 [DOI] [PubMed] [Google Scholar]
- Wang T., Ning K., Lu T.-x., Sun X., Jin L., Qi X., et al. (2017). Increasing Circulating Exosomes-Carrying TRPC5 Predicts Chemoresistance in Metastatic Breast Cancer Patients. Cancer Sci. 108, 448–454. 10.1111/cas.13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Qiao D., Chen L., Xu M., Chen S., Huang L., et al. (2019). Chemotherapeutic Drugs Stimulate the Release and Recycling of Extracellular Vesicles to Assist Cancer Cells in Developing an Urgent Chemoresistance. Mol. Cancer 18, 182. 10.1186/s12943-019-1114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hill S., Luther J. M., Hachey D. L., Schey K. L. (2012). Proteomic Analysis of Urine Exosomes by Multidimensional Protein Identification Technology (MudPIT). Proteomics 12, 329–338. 10.1002/pmic.201100477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F., Ma C., Zhou T., Dong X., Luo Q., Geng L., et al. (2017). Exosomes Derived from Gemcitabine-Resistant Cells Transfer Malignant Phenotypic Traits via Delivery of miRNA-222-3p. Mol. Cancer 16, 132. 10.1186/s12943-017-0694-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton J. L., Khanna S., Giles P. J., Brennan P., Brewis I. A., Staffurth J., et al. (2010). Proteomics Analysis of Bladder Cancer Exosomes. Mol. Cell Proteomics 9, 1324–1338. 10.1074/mcp.M000063-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M., McDonagh E. M., Hebert J. M., Gong L., Sangkuhl K., Thorn C. F., et al. (2012). Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 92, 414–417. 10.1038/clpt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-E., Tzen C.-Y., Wang S.-Y., Yeh C.-N. (2019). Clinical Diagnosis of Gastrointestinal Stromal Tumor (GIST): From the Molecular Genetic Point of View. Cancers 11, 679. 10.3390/cancers11050679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zeng C., Ye Y., Liu J., Mu Z., Xie Y., et al. (2018). Exosomes from Irradiated Nonsmall Cell Lung Cancer Cells Reduced Sensitivity of Recipient Cells to Anaplastic Lymphoma Kinase Inhibitors. Mol. Pharmaceutics 15, 1892–1900. 10.1021/acs.molpharmaceut.8b00059 [DOI] [PubMed] [Google Scholar]
- Yamashita T., Kamada H., Kanasaki S., Maeda Y., Nagano K., Abe Y., et al. (2013). Epidermal Growth Factor Receptor Localized to Exosome Membranes as a Possible Biomarker for Lung Cancer Diagnosis. Pharmazie 68, 969–973. 10.1691/ph.2013.3599 [DOI] [PubMed] [Google Scholar]
- Yang S.-J., Wang D.-D., Li J., Xu H.-Z., Shen H.-Y., Chen X., et al. (2017). Predictive Role of GSTP1-Containing Exosomes in Chemotherapy-Resistant Breast Cancer. Gene 623, 5–14. 10.1016/j.gene.2017.04.031 [DOI] [PubMed] [Google Scholar]
- Yang S., Che S. P. Y., Kurywchak P., Tavormina J. L., Gansmo L. B., Correa de Sampaio P., et al. (2017). Detection of Mutant KRAS and TP53 DNA in Circulating Exosomes from Healthy Individuals and Patients with Pancreatic Cancer. Cancer Biol. Ther. 18, 158–165. 10.1080/15384047.2017.1281499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A., Villar-Prados A., Oliphint P. A., Zhang J., Song X., De Hoff P., et al. (2019). Mechanisms of Nuclear Content Loading to Exosomes. Sci. Adv. 5, eaax8849. 10.1126/sciadv.aax8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuwen D. L., Sheng B. B., Liu J., Wenyu W., Shu Y. Q. (2017). MiR-146a-5p Level in Serum Exosomes Predicts Therapeutic Effect of Cisplatin in Non-small Cell Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 2650–2658. [PubMed] [Google Scholar]
- Zanetti-Domingues L. C., Bonner S. E., Martin-Fernandez M. L., Huber V. (2020). Mechanisms of Action of EGFR Tyrosine Kinase Receptor Incorporated in Extracellular Vesicles. Cells 9, 2505. 10.3390/cells9112505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgheib N. K., Akra-Ismail M., Aridi C., Mahfouz R., Abboud M. R., Solh H., et al. (2014). Genetic Polymorphisms in Candidate Genes Predict Increased Toxicity with Methotrexate Therapy in Lebanese Children with Acute Lymphoblastic Leukemia. Pharmacogenet. Genomics 24, 387–396. 10.1097/fpc.0000000000000069 [DOI] [PubMed] [Google Scholar]
- Zhang H., Freitas D., Kim H. S., Fabijanic K., Li Z., Chen H., et al. (2018). Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cel Biol. 20, 332–343. 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Späth S. S., Marjani S. L., Zhang W., Pan X. (2018). Characterization of Cancer Genomic Heterogeneity by Next-Generation Sequencing Advances Precision Medicine in Cancer Treatment. Precis. Clin. Med. 1, 29–48. 10.1093/pcmedi/pby007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jeppesen D. K., Higginbotham J. N., Demory Beckler M., Poulin E. J., Walsh A. J., et al. (2018). Mutant KRAS Exosomes Alter the Metabolic State of Recipient Colonic Epithelial Cells. Cell Mol. Gastroenterol. Hepatol. 5, 627–629.e626. 10.1016/j.jcmgh.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu R.-X., Chan K.-W., Hu J., Zhang J., Wei L., et al. (2019). Exosomal Transfer of P-STAT3 Promotes Acquired 5-FU Resistance in Colorectal Cancer Cells. J. Exp. Clin. Cancer Res. 38, 320. 10.1186/s13046-019-1314-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Cai X., Yu J., Lu X., Qian Q., Qian W. (2018). Exosome-mediated Transfer of lncRNA RP11838N2.4 Promotes Erlotinib Resistance in Non-small Cell Lung Cancer. Int. J. Oncol. 53, 527–538. 10.3892/ijo.2018.4412 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu X., You Y., Li Q., Zeng C., Fu F., Guo A., et al. (2014). BCR-ABL1-positive Microvesicles Transform Normal Hematopoietic Transplants through Genomic Instability: Implications for Donor Cell Leukemia. Leukemia 28, 1666–1675. 10.1038/leu.2014.51 [DOI] [PubMed] [Google Scholar]
- Zocco D., Ferruzzi P., Cappello F., Kuo W. P., Fais S. (2014). Extracellular Vesicles as Shuttles of Tumor Biomarkers and Anti-tumor Drugs. Front. Oncol. 4, 267. 10.3389/fonc.2014.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]