Abstract
Serine/arginine-rich splicing factor 2 (SRSF2) is a splicing factor that is widely expressed in a variety of mammalian cell types. Increasing evidence has confirmed that SRSF2 plays vital roles in a number of biological and pathological processes. Therefore, it is important to understand how its expression is regulated, and how it regulates the expression of its target genes. Recently, we found that SRSF2 expression could be upregulated by herpes simplex virus-1 (HSV-1) infection, and that altered SRSF2 expression, in turn, epigenetically regulates the transcription of HSV-1 genes. Further studies on T cell exhaustion demonstrated that upregulated SRSF2 in exhausted T cells elevated the levels of multiple immune checkpoint molecules by associating with the acyl-transferases, P300 and CBP, and by altering histone modification near the transcription start sites of these genes, thereby influencing signal transducer and activator of transcription 3 binding to these gene promoters. These findings suggest that SRSF2 acts as an important sensor and effector during disease progression. Here, we discuss the molecules that regulate SRSF2 gene expression and their associated mechanisms, and the mechanisms via which SRSF2 regulates the expression of target genes, thus providing novel insights into the central role of SRSF2 in gene regulation.
Keywords: SRSF2, SC35, gene regulation, transcription, splicing, mRNA stability.
Introduction
Serine/arginine-rich splicing factor 2 (SRSF2), previously known as SC35, is a member of a well-known serine/arginine-rich (SR) protein family. SRSF2 is an important component of the nuclear structure, speckle. It consists of an RNA recognition motif (RRM) and a domain rich in arginine and serine residues (RS domain). SRSF2 has been shown to control the splicing of pre-mRNAs by recognizing and binding to SRSF2-binding sites on pre-mRNA via the RRM and by interacting with other SR splicing factors via the RS domain 1,2. Unlike other SR splicing factors, SRSF2 has a longer L3 loop region and prefers to recognize highly degenerated RNA sequences of exonic splicing enhancers (ESEs) 3,4. However, recent studies have found that SRSF2 is also involved in regulating genomic stability, gene transcription, mRNA stability, and translation 5-8.
Increasing evidence has demonstrated that SRSF2 and other SR proteins are associated with the progression of a variety of diseases, including viral infection and tumorigenesis. Majerciak et al. reported that serine/arginine-rich splicing factor 3 (SRSF3) interacted with open reading frame 57 (ORF57) to mediate splicing of Kaposi sarcoma-associated herpesvirus (KSHV) K8β RNA via RNA binding motif protein 15 (RBM15) and OTT3 (RBM15B), a close member to RBM15 9. Our previous study of the interaction between host cellular factors and herpes simplex virus-1 (HSV-1) found that SRSF2 enhances HSV-1 replication and viral gene expression by epigenetically regulating the transcription of viral genes 5. Jacquenet et al. reported that SRSF2 modulates human immunodeficiency virus type 1 (HIV-1) replication by downregulating the levels of HIV-1 structural proteins and genomic RNA 10, and by mediating the splicing of Tat 11-14 and Rev 15, which are required for HIV-1 replication. In another study, we investigated the role of SRSF2 in T cell exhaustion during tumorigenesis and demonstrated that SRSF2 expression is significantly elevated in exhausted T cells, and that SRSF2 knockdown reverses the exhaustion of T cells by epigenetically regulating the expression of multiple immune checkpoint molecules 6. In addition, an increasing number of studies have recently shown that SRSF2 gene mutation and dysregulated SRSF2 expression are significantly associated with acute myeloid leukemia (AML) 16, 17, myelodysplastic syndromes 18-20, systemic mastocytosis 21, 22, chronic myelomonocytic leukemia 23-25, lung carcinoma 26, 27, hepatocellular carcinoma 28, and sinonasal squamous cell carcinoma 29. These findings indicate that SRSF2 functions as a promising therapeutic target for various diseases.
In this review, we discuss the molecules that regulate SRSF2 expression, and their associated regulatory mechanisms. These molecules are mainly transcription factors, miRNAs, protein kinases, and acetyltransferases that can influence SRSF2-mediated gene regulation by altering the transcriptional and post-transcriptional levels and the modification of SRSF2 protein. We also discuss the molecular mechanisms via which SRSF2 regulates the expression of its target genes.
Factors and regulatory patterns of SRSF2 expression and protein modification
To date, many factors have been found to regulate SRSF2 gene expression and modification of SRSF2 protein. Some factors influence SRSF2 gene transcription, pre-mRNA splicing, and mRNA translation. Others have been shown to phosphorylate, acetylate, or deacetylate SRSF2 protein. These factors, which are summarized in Table 1, are generally transcriptional factors, miRNAs, protein kinases, and acetyltransferases.
Table 1.
Factors regulating SRSF2 expression and protein modification.
| Cell Type | Factor | Regulatory Pattern | Function | Reference |
|---|---|---|---|---|
| H358 and H1299 cells | E2F1 | Transcription | Up (alternative splicing) | 30 |
| HeLa Tet-On cells | SRSF2 | Alternative splicing | / | 32 |
| HeLa cells | SRSF2 | / | 33 | |
| SMMC-7721 and QGY-7703 cells | miR-193a-3p | mRNA stability and translation | Down (alternative splicing) | 34 |
| 5637 cells and H-bc cells | miR-193a-3p | Down (alternative splicing) | 35 | |
| CNE-1 and CNE-2 cells | miR-193a-3p | Down (alternative splicing) | 36 | |
| HeLa cells | miR-183-5p and miR-33a-5p | / | 37 | |
| HeLa and MG63 cells | SRPK1 | Phosphorylation and acetylation | Down (alternative splicing) | 38 |
| DT40 cells | PI3K/Akt | Up (alternative splicing) | 39 | |
| HEK293T cells | PI3K/Akt | Up (alternative splicing) | 40 | |
| MEFs | PHLPP1 and 2 | / | 41 | |
| HeLa cells | GSK-3β | Down (alternative splicing) | 46 | |
| Cortical neurons | GSK-3β | Down (alternative splicing) | 47 | |
| HEK-293T cells | Dyrk1A | Down (alternative splicing) | 48 | |
| HEK-293FT and HeLa cells | PKA | Up (alternative splicing) | 49 | |
| Embryonic neurons | HIV-1 Tat | Down (alternative splicing) | 50 | |
| HeLa and HEK-293FT cells | SIRT1 | Down (alternative splicing) | 51 | |
| H1299 and H358 cells | TIP60 | Up (alternative splicing) | 52 |
Transcription
E2F transcription factor 1 (E2F1) is a transcriptional regulator of SRSF2. As an important trans-activator of pro-apoptotic target genes, E2F1 has been shown to regulate the alternative splicing of various apoptotic genes, including FADD-like apoptosis regulator (c-Flip), caspases-8 (Casp8) and -9 (Casp9), and Bcl-x, by increasing SRSF2 gene transcription. Molecular mechanism investigation showed E2F1 directly targeted the promoter region 296-79 bp upstream of SRSF2 to initiate its transcription, which is required for apoptosis in response to drugs that induce DNA damage 30.
Alternative splicing
SRSF2 mRNAs consist of different 3' untranslated regions (UTRs) and exhibit significantly different stabilities 31 and the splicing of SRSF2 pre-mRNA has been shown to be autoregulated by SRSF2. SRSF2 enhances both exon inclusion and intron excision in the 3' UTR of the SRSF2 pre-mRNA, resulting in a significant decrease in endogenous SRSF2 mRNA levels and an increase in the levels of alternatively spliced SRSF2 mRNAs, both of which have a short half-life 32. Further investigation showed that there are multiple low-affinity SRSF2-binding sites located within a highly conserved stem-loop region of the terminal exon 33.
mRNA stability and translation
Several miRNAs have been shown to target SRSF2 to regulate SRSF2 mRNA stability and translation. In a study aimed at understanding the resistance of hepatocellular carcinoma to 5-fluorouracil (5-FU), the stability of SRSF2 mRNA, the splicing regulator of caspase 2, which contributes to tumor apoptosis in response to 5-FU, was found to be decreased by DNA methylation-regulated miR-193a-3p 34. Another investigation of the resistance of bladder cancer to pirarubicin, paclitaxel, adriamycin, and epirubicin hydrochloride found that miR-193a-3p targeted SRSF2 mRNA to repress its expression levels, which then activated Notch and oxidative stress, two chemoresistance-associated signaling pathways 35. In addition to chemoresistance, miR-193a-3p-regulated SRSF2 has been shown to be involved in radio-resistance. miR-193a-3p was found to increase the resistance of nasopharyngeal cancer cells to radiation by targeting SRSF2 and thus, activating the hypoxia signaling pathway 36. Moreover, two miRNAs, miR-183-5p and miR-33a-5p, were found to contribute to the upregulation of SRSF2 expression by methylxanthine caffeine by targeting SRSF2 and repressing the translation of SRSF2 37.
Protein modification
Growing evidence has shown that the splicing activity of SRSF2 and other SR proteins are affected by its phosphorylation. SRSF protein kinase 1 (SRPK1) 38, phosphatidylinositol 3-kinase (PI3K)/Akt pathway 39, 40, and PH domain and leucine rich repeat protein phosphatase 1/2 (PHLPP1/2) 41 were found to regulate splicing by mediating the phosphorylation and dephosphorylation of SR proteins, respectively. SRSF2 is an important splicing regulator of tau pre-mRNA 8, and the dysregulation of its splicing often results in neurodegenerative disorders 42, 43. To date, many factors have been reported to regulate tau pre-mRNA splicing by mediating the phosphorylation and acetylation of SRSF2. Glycogen synthase kinase-3β, a serine-threonine kinase that functions as a regulator of pre-mRNA processing through the phosphorylation of splicing factors 44,45, has been shown to interact with and phosphorylate SRSF2, resulting in the enrichment of SRSF2 in nuclear speckles and the loss of its ability to mediate splicing events 46. This contributes to aberrant tau splicing 47. Moreover, another protein kinase, dual-specificity tyrosine-phosphorylated and regulated kinase 1A (Dyrk1A), also regulates SRSF2-mediated splicing of tau pre-mRNA. Dyrk1A suppresses the ability of SRSF2 to promote tau exon 10 inclusion, by interacting with the N-terminus of SRSF2 and phosphorylating SRSF2 48. However, the phosphorylation of SRSF2, regulated by protein kinase A (PKA) 49 and HIV-1 Tat 50, enhances SRSF2-mediated tau exon 10 inclusion. In addition, the deacetylase, sirtuin-1, inhibits the SRSF2-promoted tau exon 10 inclusion by interacting with and deacetylating SRSF2 51.
In addition to the splicing ability of SRSF2, modifications of SRSF2 protein influence its turnover. The acetyltransferase, Tip60, has been found to acetylate SRSF2 on lysine 52 in the RRM, resulting in the proteasomal degradation of SRSF2, whereas histone deacetylase 6 counters this acetylation to function as a positive regulator of SRSF2 protein levels. Moreover, Tip60 inhibits SRSF2 phosphorylation by downregulating the nuclear levels of SRPK1 and SRPK2 52, two serine/arginine protein kinases specific for the SR domain family 38, 53.
Regulation of gene expression by SRSF2
SRSF2 has been reported to be involved in the regulation of gene transcription, pre-mRNA splicing, mRNA transport, and mRNA stability. In this section, we will discuss the mechanisms by which SRSF2 regulates its target genes (Table 2).
Table 2.
SRSF2 regulate genes expression.
| Cell type | Target | Regulatory pattern | References |
|---|---|---|---|
| HeLa cells | HSV-1 ICP0, ICP27, and TK | Transcription | 5 |
| JurkateE6.1 cells | PD-L1, BTLA, CTLA4, LAG3, and CD160 | 6 | |
| H1299 and HeLa cells | Skp2 and Cyclin E | 55 | |
| HeLa cells | HSV-1 ICP0 | Alternative splicing | 5 |
| HEK-293T and HeLa cells | Adenovirus E1a | 56 | |
| HEK-293T and HeLa cells | SV40 early pre-mRNA | 56 | |
| HEK-293T and HeLa cells | HIV-1 Tat | 10,12,14 | |
| MCF-7 cells | MDM2 | 60 | |
| KG-1, HL-60 and promyelocytic cells | CSF3R | 16 | |
| MGH7 cells | VEGFR1 | 64 | |
| H358 cells | VEGF-A | 68 | |
| MDA MB 231 and HeLa cells | Ron | 71 | |
| MCF-7 cells | CD44 | 77 | |
| H358 cells | c-Flip, Caspase -8, -9, and Bcl-x | 30 | |
| SMMC-7721 and QGY-7703 cells | Caspase 2 | 34 | |
| NT2/D1 cells | PKCδ | 79 | |
| HeLa cells | KLF6 | 82 | |
| HEK293 cells | APP | 86 | |
| HEK-293T cells | TAU | 8 | |
| HEK-293T and HeLa cells | SMN2 | 89-91 | |
| HEK-293T cells | TAU | RNA stability and transport | 8 |
| W12ti tumor cells | HPV16 E6E7 | 93 | |
| HEK-293T and HeLa cells | SV40 early pre-mRNA | 56 |
Transcription
Our previous study investigating the role of SRSF2 in HSV-1 infection found that SRSF2 facilitated HSV-1 viral replication and gene expression by binding to viral gene promoters and associating with RNA polymerase II, infected cell protein 27 (ICP27), and infected cell protein 8 (ICP8), to promote gene transcription 5. In another study investigating the role of SRSF2 in T cell exhaustion, we found that SRSF2 expression is upregulated in exhausted T cells and the inhibition of SRSF2 downregulates the transcription of many immune checkpoint genes, which contributes to dysfunctional cytokine secretion by T cells, cells proliferation, tumor cell cytotoxicity, and effective memory cell generation 54, through associating with the acetyltransferase CBP/P300 complex, altering histone modifications, and thereby recruiting the transcriptional factor signal transducer and activator of transcription 3 to the promoter of immune checkpoint genes to initial gene transcription 6.
In addition, SRSF2 acts as a transcriptional regulator during cell cycle progression. SRSF2 has been shown to be a cell cycle-related protein that is involved in regulating entry and progression into S phase, by associating with the transcription factor, E2F1, and recruiting it to the promoters of Skp2 or cyclin E, to upregulate the transcription of these cell cycle-related genes 55.
Alternative splicing
Alternative splicing of pre-mRNAs contributes significantly to human proteomic complexity, and aberrant alternative splicing plays a key role in the progression of various diseases. SRSF2 has been shown to be involved in the alternative splicing of many target pre-mRNAs that are associated with human diseases, including viral infection, tumors, and neurodegenerative diseases.
In our previous study, we found that SRSF2 mediated the alternative splicing of HSV-1 infected cell protein (ICP0) pre-mRNA by associating with ICP0 exon 3, which contains the SRSF2-binding motif 5. In HIV-infected cells, SRSF2 profoundly changes the HIV-1 splicing pattern, in which SRSF2 mainly promotes Tat1 production by enhancing splicing at site A3, which is necessary for Tat mRNA synthesis 10,13. Further investigation identified a complex cis-acting element in tat exon 2 that is required for splicing regulation. An ESE was found to be located within the regulatory element and SRSF2 was reported to activate tat exon 2 splicing by binding to the ESE 14. For adenovirus E1a pre-mRNAs, SRSF2 promotes 13S production via an increase in the utilization of most downstream 5′ splicing sites 56. However, for simian vacuolating virus 40 (SV40) early pre-mRNA, SRSF2 overexpression resulted in a significant inhibition of splicing 56.
Many tumor-associated genes are also regulated by SRSF2. MDM2 is a nuclear-localized E3 ubiquitin ligase that accelerates tumor formation by targeting p53, a tumor suppressor, and mediating its proteasomal degradation 57-59. A recent study investigating genotoxic stress-induced MDM2 splicing found that SRSF2 promotes the inclusion of MDM2 exon 11 by binding to two conserved ESEs located at exon 11 60.
Colony-stimulating factor 3 receptor (CSF3R), a member of the family of cytokine receptors that function to regulate the production, differentiation, and function of granulocytes 61, contains three CSF3R mRNA splice variants, namely V1, V3, and V4, with different expression levels. A study to examine the expression levels of these CSF3R mRNA variants in patients with AML found that those harboring SRSF2 mutations and SRSF2 knock-out cells both exhibit a significant alteration in the V3/V1 ratio, suggesting SRSF2-mediated CSF3R splicing 16.
Soluble isoforms of vascular endothelial growth factor receptors 1 (sVEGFR1) have been implicated in several physical and pathological processes, including tumorigenesis 62, 63. In lung cancer cells, SRSF2 has been found to positively regulate the expression level of sVEGFR1-i13, one of the sVEGFR1 mRNA splice variants, with no correlation between SRSF2 and VEGFR1 mRNA levels, indicating that SRSF2 controls the splicing of sVEGFR1-i13 64.
Vascular endothelial growth factor A (VEGF-A), a growth factor active in angiogenesis and endothelial cell growth, is correlated with tumor progression via the formation of blood vessels 65. VEGF-A mRNAs mainly consists of three isoforms, namely VEGF121, VEGF165, and VEGF189 66. A study aimed at determining the roles of E2F1 and SRSF2 in VEGF-A expression and pre-mRNA splicing found that SRSF2 overexpression increased the production of the VEGF165b splice variant, an inhibitor of the growth of several types of tumors, by inhibiting the migration and proliferation of endothelial cells 67, resulting in a decrease in tumor neovascularization and tumor formation 68.
Ron, also known as MST1R, PTK8, or SEA, is a member of the MET proto-oncogene family 69, whose members function as inducers of tumor progression by binding to macrophage-stimulating protein, and thereby stimulating the phosphorylation on C-terminal docking sites of multiple transducer and adaptor proteins 70. By physically interacting with the CGAG sequence in exon 11, SRSF2 enhances the inclusion of exon 11 in Ron pre-mRNA, resulting in an increase in the production of Ron△165 71, which promotes invasive growth and metastasis 72.
CD44 is a cell membrane glycoprotein that mediates cell growth, differentiation, and motility 73. The V6 exon-containing isoforms of CD44 mRNA (CD44 V6) have been implicated in tumorigenesis by promoting tumor cell invasion and metastasis 74-76. SRSF2 has been shown to positively regulate the expression level of CD44 V6 and the V6 exon and its flanking introns contain SRSF2 response elements, indicating that SRSF2 is involved in the inclusion or exclusion of the V6 exon 77.
Protein kinase C delta (PKCδ), a serine/threonine kinase, plays a key role in cell proliferation, differentiation, and apoptosis 78. A study investigating the role of retinoic acid in the splicing of PKCδ pre-mRNA identified that SRSF2 upregulates the expression level of PKCδVIII, a pro-survival splice variant of PKCδ, by promoting the selection of 5' splice site II and binding to an ESE in PKCδ exon 10 79.
Kruppel-like factor 6 (KLF6), a member of the Kruppel-like family of transcription factors, functions as a tumor suppressor and is implicated in tumorigenesis 80, 81. A study of the regulation of the alternative splicing of KLF6 pre-mRNA by caffeine found that caffeine induced the expression of SRSF2, and upregulated SRSF2 levels promoted exon 1a inclusion in KLF6 pre-mRNA 82.
Recent studies have also shown that SRSF2 promotes the pre-mRNA splicing of several apoptosis-related genes, including Casp8, Casp9, c-Flip, and Bcl-x, in response to DNA-damaging agents, resulting in an increase in the expression levels of proapoptotic splice forms of these apoptotic genes, and subsequently, inducing apoptosis 30,34.
In addition to genes related to virus infection and tumor progression, SRSF2 has been shown to mediate the pre-mRNA splicing of multiple neurodegenerative disease-related genes. During the pathogenesis of Alzheimer's disease (AD), disrupted alternative splicing of β-amyloid precursor protein (APP) and tau pre-mRNA has been found to contribute to the accumulation of β-amyloid peptide (Aβ) and tau, the characteristics of AD 83-85. A study of the regulation of APP splicing showed that SRSF2 is involved in the alternative splicing of APP exons 7 and 8 by binding to Alu elements on either side of exon 86. Another study of the role of SRSF2 in regulating tau expression found that SRSF2 positively regulated the expression of tau isoforms containing exon 10 by binding to this exon, suggesting that the regulation of tau pre-mRNA splicing by SRSF2 is dependent on exon 10 8.
In addition to the promotion of pre-mRNA splicing, SRSF2 has also been reported to directly repress exon inclusion. Studies of the splicing regulator of survival of motor neuron 2 (SMN2) gene, whose aberrant splicing contributes to disease severity in spinal muscular atrophy patients, by the deletion or mutation of the SMN1 gene 87, 88, found that overexpression of SRSF2 reduces SMN2 exon 7 inclusion by binding to a 10 nt RNA sequence surrounding the branch-point, located upstream of the 3′ splice site (3′ SS), thus promoting the selection of the 3′ SS 89-91.
RNA stability and transport
In addition to the regulation of gene transcription and alternative splicing, SRSF2 has been reported to be involved in regulating RNA stability and transport. A study investigating the molecular mechanism by which SRSF2 promotes tau40 expression found that SRSF2 overexpression leads to the accumulation of tau40 mRNA and prevents its degradation by actinomycin D, an inhibitor of DNA transcription and replication 92, via association with the RRM domain of tau exon 10 8. This indicates that SRSF2 stabilizes tau40 mRNA.
Overexpression of SRSF2 also results in an accumulation of the unspliced SV40 pre-mRNA, and the exporting of most of the unspliced SV40 pre-mRNA to the cytoplasm, suggesting a role of SRSF2 in regulating RNA stability and transport 56. In addition, a study investigating the role of SRSF2 in human papillomavirus 16 gene expression found that SRSF2 knock-down decreases the expression levels of E6E7 RNAs. A mechanistic study showed that SRSF2 maintains the stability of E6E7 mRNAs 93.
Conclusion
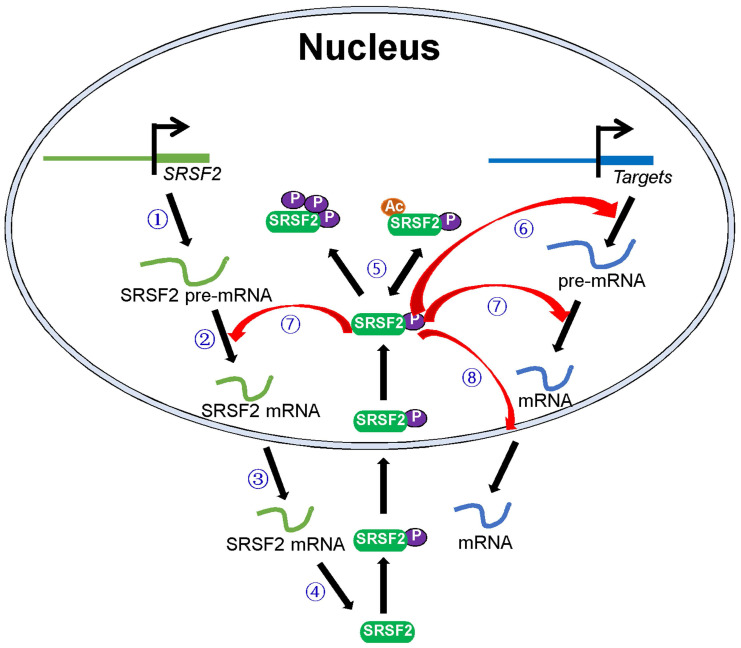
SRSF2 mutations and dysregulated SRSF2 expression have been shown to be associated with various diseases. In this review, we focused on how these pathological processes regulate SRSF2 expression and how SRSF2 regulates these processes. In particular, we discussed the molecules that regulate SRSF2 expression and their associated mechanisms. Most of these regulatory molecules are transcription factors, miRNAs, protein kinases, and acetyltransferases. They play important roles in SRSF2 gene transcription, pre-mRNA splicing, mRNA stability, and translation and SRSF2 protein modification. In addition, SRSF2, in turn, was found to mediate the processes via which it regulates the expression of target genes (Figure 1). Overall, this review discusses SRSF2-centric gene regulation and provides insights into its potential clinical utilities.
Figure 1.
Schematic model of the central roles of SRSF2 in genes expression. Numbers of regulators are found to modulate SRSF2 gene transcription ①, pre-mRNA splicing ②, mRNA stability ③, mRNA translation ④, and SRSF2 modification (phosphorylation and acetylation) ⑤. In turn, SRSF2 regulates target gene transcription ⑥, pre-mRNA splicing ⑦, and mRNA transport and stability ⑧.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32000878), and the Natural Science Foundation of Shandong Province (ZR2020LZL008).
Author Contributions
K.L. and Z.W. prepared the manuscript. Z.W. reviewed and edited the manuscript. Both authors read and approved the final manuscript.
Abbreviations
- SRSF2
Serine/arginine-rich splicing factor 2
- SR
Serine/arginine-rich
- RRM
RNA recognition motif
- RS domain
Arginine and serine residues
- ESE
Exonic splicing enhancer
- ORF57
Open reading frame 57
- KSHV
Kaposi sarcoma-associated herpesvirus
- RBM15
RNA binding motif protein 15
- HSV-1
Herpes simplex virus-1
- HIV-1
Human immunodeficiency virus type 1
- AML
Acute myeloid leukemia
- E2F1
E2F transcription factor 1
- c-FLIP
Cellular FADD-like IL-1β-converting enzyme-inhibitory protein
- Casp8
Caspases-8
- Casp9
Caspases-9
- UTRs
Untranslated regions
- 5-FU
5-Fluorouracil
- SRPK1
SRSF protein kinase 1
- PI3K
Phosphatidylinositol 3-kinase
- Dyrk1A
Dual-specificity tyrosine-phosphorylated and regulated kinase 1A
- PKA
Protein kinase A
- ICP27
Infected cell protein 27
- ICP8
Infected cell protein 8
- ICP0
Infected cell protein 0
- SV40
Imian vacuolating virus 40
- CSF3R
Colony-stimulating factor 3 receptor
- sVEGFR1
Soluble isoforms of vascular endothelial growth factor receptors 1
- VEGF-A
Vascular endothelial growth factor A
- CD44 V6
V6 exon-containing isoforms of CD44 mRNA
- PKCδ
Protein kinase C delta
- KLF6
Kruppel-like factor 6
- AD
Alzheimer's disease
- APP
Amyloid precursor protein
- Aβ
β-Amyloid peptide
- SMN2
Survival of motor neuron 2
- 3′ SS
3′ splice site
References
- 1.Fu XD, Mayeda A, Maniatis T. et al. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5' and 3' splice site selection. Proc Natl Acad Sci U S A. 1992;89:11224–8. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3' splice site. Proc Natl Acad Sci U S A. 1992;89:1725–9. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phelan MM, Goult BT, Clayton JC. et al. The structure and selectivity of the SR protein SRSF2 RRM domain with RNA. Nucleic Acids Res. 2012;40:3232–44. doi: 10.1093/nar/gkr1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daubner GM, Cléry A, Jayne S. et al. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–74. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Liu Q, Lu J. et al. Serine/Arginine-rich Splicing Factor 2 Modulates Herpes Simplex Virus Type 1 Replication via Regulating Viral Gene Transcriptional Activity and Pre-mRNA Splicing. J Biol Chem. 2016;291:26377–26387. doi: 10.1074/jbc.M116.753046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Li K, Chen W. et al. Modulation of SRSF2 expression reverses the exhaustion of TILs via the epigenetic regulation of immune checkpoint molecules. Cell Mol Life Sci. 2020;77:3441–3452. doi: 10.1007/s00018-019-03362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao R, Sun Y, Ding JH. et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian W, Iqbal K, Grundke-Iqbal I. et al. Splicing factor SC35 promotes tau expression through stabilization of its mRNA. FEBS Lett. 2011;585:875–80. doi: 10.1016/j.febslet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majerciak V, Lu M, Li X. et al. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20:1747–58. doi: 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquenet S, Decimo D, Muriaux D. et al. Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology. 2005;2:33. doi: 10.1186/1742-4690-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erkelenz S, Hillebrand F, Widera M. et al. Balanced splicing at the Tat-specific HIV-1 3' ss A3 is critical for HIV-1 replication. Retrovirology. 2015;12:29. doi: 10.1186/s12977-015-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallay H, Locker N, Ayadi L. et al. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J Biol Chem. 2006;281:37159–74. doi: 10.1074/jbc.M603864200. [DOI] [PubMed] [Google Scholar]
- 13.Ropers D, Ayadi L, Gattoni R. et al. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J Biol Chem. 2004;279:29963–73. doi: 10.1074/jbc.M404452200. [DOI] [PubMed] [Google Scholar]
- 14.Zahler AM, Damgaard CK, Kjems J. et al. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279:10077–84. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- 15.Boe SO, Bjorndal B, Rosok B. et al. Subcellular localization of human immunodeficiency virus type 1 RNAs, Rev, and the splicing factor SC-35. Virology. 1998;244:473–82. doi: 10.1006/viro.1998.9110. [DOI] [PubMed] [Google Scholar]
- 16.Lance A, Druhan LJ, Vestal CG. et al. Altered expression of CSF3R splice variants impacts signal response and is associated with SRSF2 mutations. Leukemia. 2020;34:369–379. doi: 10.1038/s41375-019-0567-9. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Yao DM, Ma JC. et al. The prognostic implication of SRSF2 mutations in Chinese patients with acute myeloid leukemia. Tumour Biol. 2016;37:10107–14. doi: 10.1007/s13277-015-4716-0. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Song L, Xu L. et al. Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes. Tumour Biol. 2016;37:4633–40. doi: 10.1007/s13277-015-4305-2. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Yang J, Wen XM. et al. Detection of SRSF2-P95 mutation by high-resolution melting curve analysis and its effect on prognosis in myelodysplastic syndrome. PLoS One. 2014;9:e115693. doi: 10.1371/journal.pone.0115693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SJ, Kuo YY, Hou HA. et al. The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012;120:3106–11. doi: 10.1182/blood-2012-02-412296. [DOI] [PubMed] [Google Scholar]
- 21.Jawhar M, Schwaab J, Schnittger S. et al. Additional mutations in SRSF2, ASXL1 and/or RUNX1 identify a high-risk group of patients with KIT D816V(+) advanced systemic mastocytosis. Leukemia. 2016;30:136–43. doi: 10.1038/leu.2015.284. [DOI] [PubMed] [Google Scholar]
- 22.Hanssens K, Brenet F, Agopian J. et al. SRSF2-p95 hotspot mutation is highly associated with advanced forms of mastocytosis and mutations in epigenetic regulator genes. Haematologica. 2014;99:830–5. doi: 10.3324/haematol.2013.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Federmann B, Abele M, Rosero Cuesta DS. et al. The detection of SRSF2 mutations in routinely processed bone marrow biopsies is useful in the diagnosis of chronic myelomonocytic leukemia. Hum Pathol. 2014;45:2471–9. doi: 10.1016/j.humpath.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Patnaik MM, Lasho TL, Finke CM. et al. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: prevalence, clinical correlates, and prognostic relevance. Am J Hematol. 2013;88:201–6. doi: 10.1002/ajh.23373. [DOI] [PubMed] [Google Scholar]
- 25.Meggendorfer M, Roller A, Haferlach T. et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–8. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherine Abou Faycal, Sylvie Gazzeri, Beatrice Eymin. A VEGF-A/SOX2/SRSF2 network controls VEGFR1 pre-mRNA alternative splicing in lung carcinoma cells. Sci Rep. 2019;9:336. doi: 10.1038/s41598-018-36728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gout S, Brambilla E, Boudria A. et al. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non small cell lung carcinoma. PLoS One. 2012;7:e46539. doi: 10.1371/journal.pone.0046539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo C, Cheng Y, Liu Y. et al. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res. 2017;77:1168–1178. doi: 10.1158/0008-5472.CAN-16-1919. [DOI] [PubMed] [Google Scholar]
- 29.Yu B, Qu L, Wu T. et al. A Novel LncRNA, AC091729.7 Promotes Sinonasal Squamous Cell Carcinomas Proliferation and Invasion Through Binding SRSF2. Front Oncol. 2020;9:1575. doi: 10.3389/fonc.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merdzhanova G, Edmond V, De Seranno S. et al. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ. 2008;15:1815–23. doi: 10.1038/cdd.2008.135. [DOI] [PubMed] [Google Scholar]
- 31.Sureau A, Perbal B. Several mRNAs with variable 3' untranslated regions and different stability encode the human PR264/SC35 splicing factor. Proc Natl Acad Sci U S A. 1994;91:932–6. doi: 10.1073/pnas.91.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sureau A, Gattoni R, Dooghe Y. et al. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–96. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreumont N, Hardy S, Behm-Ansmant I. et al. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2010;38:1353–66. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma K, He Y, Zhang H. et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287:5639–49. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv L, Deng H, Li Y. et al. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis. 2014;5:e1402. doi: 10.1038/cddis.2014.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong L, Wei Q, Hu X. et al. miR-193a-3p Promotes Radio-Resistance of Nasopharyngeal Cancer Cells by Targeting SRSF2 Gene and Hypoxia Signaling Pathway. Med Sci Monit Basic Res. 2019;25:53–62. doi: 10.12659/MSMBR.914572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Pabon K, Scotto KW. Methylxanthines Increase Expression of the Splicing Factor SRSF2 by Regulating Multiple Post-transcriptional Mechanisms. J Biol Chem. 2015;290:14986–5003. doi: 10.1074/jbc.M114.624254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–82. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Mayeda A, Tao M. et al. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3' splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J Virol. 2003;77:2105–15. doi: 10.1128/JVI.77.3.2105-2115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z, Qiu J, Liu W. et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–33. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Zhou Z, Hu A. et al. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol Cell. 2014;54:378–91. doi: 10.1016/j.molcel.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu J, Chen F, Iqbal K. et al. Transactive response DNA-binding protein 43 (TDP-43) regulates alternative splicing of tau exon 10: Implications for the pathogenesis of tauopathies. J Biol Chem. 2017;292:10600–10612. doi: 10.1074/jbc.M117.783498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinde MY, Sidoli S, Kulej K. et al. Phosphoproteomics reveals that glycogen synthase kinase-3 phosphorylates multiple splicing factors and is associated with alternative splicing. J Biol Chem. 2017;292:18240–18255. doi: 10.1074/jbc.M117.813527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goncalves V, Henriques AF, Pereira JF. et al. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA. 2014;20:474–482. doi: 10.1261/rna.041376.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An Y, Zou Y, Cao Y. et al. The nuclear GSK-3β regulated post-transcriptional processing of mRNA through phosphorylation of SC35. Mol Cell Biochem. 2019;451:55–67. doi: 10.1007/s11010-018-3393-x. [DOI] [PubMed] [Google Scholar]
- 47.Hernández F, Pérez M, Lucas JJ. et al. Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35. Implications for Alzheimer's disease. J Biol Chem. 2004;279:3801–6. doi: 10.1074/jbc.M311512200. [DOI] [PubMed] [Google Scholar]
- 48.Qian W, Liang H, Shi J. et al. Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic Acids Res. 2011;39:6161–71. doi: 10.1093/nar/gkr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Jin N, Qian W. et al. Cyclic AMP-dependent protein kinase enhances SC35-promoted Tau exon 10 inclusion. Mol Neurobiol. 2014;49:615–24. doi: 10.1007/s12035-013-8542-3. [DOI] [PubMed] [Google Scholar]
- 50.Kadri F, Pacifici M, Wilk A. et al. HIV-1-Tat Protein Inhibits SC35-mediated Tau Exon 10 Inclusion through Up-regulation of DYRK1A Kinase. J Biol Chem. 2015;290:30931–46. doi: 10.1074/jbc.M115.675751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin X, Jiang X, Wang J. et al. SIRT1 Deacetylates SC35 and Suppresses Its Function in Tau Exon 10 Inclusion. J Alzheimers Dis. 2018;61:561–570. doi: 10.3233/JAD-170418. [DOI] [PubMed] [Google Scholar]
- 52.Edmond V, Moysan E, Khochbin S. et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011;30:510–23. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang HY, Lin W, Dyck JA. et al. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–50. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okoye IS, Houghton M, Tyrrell L. et al. Coinhibitory Receptor Expression and Immune Checkpoint Blockade: Maintaining a Balance in CD8+ T Cell Responses to Chronic Viral Infections and Cancer. Front Immunol. 2017;8:1215. doi: 10.3389/fimmu.2017.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edmond V, Merdzhanova G, Gout S. et al. A new function of the splicing factor SRSF2 in the control of E2F1-mediated cell cycle progression in neuroendocrine lung tumors. Cell Cycle. 2013;12:1267–78. doi: 10.4161/cc.24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–46. [PMC free article] [PubMed] [Google Scholar]
- 57.Haupt Y, Maya R, Kazaz A. et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 58.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 59.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 60.Comiskey DF Jr, Montes M, Khurshid S. et al. SRSF2 Regulation of MDM2 Reveals Splicing as a Therapeutic Vulnerability of the p53 Pathway. Mol Cancer Res. 2020;18:194–203. doi: 10.1158/1541-7786.MCR-19-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong F, Hoefsloot LH, Schelen AM. et al. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci U S A. 1994;91:4480–4. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abou-Fayçal C, Hatat AS, Gazzeri S. et al. Splice Variants of the RTK Family: Their Role in Tumour Progression and Response to Targeted Therapy. Int J Mol Sci. 2017;18:383. doi: 10.3390/ijms18020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abou Faycal C, Brambilla E, Agorreta J. et al. The sVEGFR1-i13 splice variant regulates a β1 integrin/VEGFR autocrine loop involved in the progression and the response to anti-angiogenic therapies of squamous cell lung carcinoma. Br J Cancer. 2018;118:1596–1608. doi: 10.1038/s41416-018-0128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abou Faycal C, Gazzeri S, Eymin B. A VEGF-A/SOX2/SRSF2 network controls VEGFR1 pre-mRNA alternative splicing in lung carcinoma cells. Sci Rep. 2019;9:336. doi: 10.1038/s41598-018-36728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–76. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 66.Bates DO, Cui TG, Doughty JM. et al. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–31. [PubMed] [Google Scholar]
- 67.Rennel E, Waine E, Guan H. et al. The endogenous anti-angiogenic VEGF isoform, VEGF165b inhibits human tumour growth in mice. Br J Cancer. 2008;98:1250–7. doi: 10.1038/sj.bjc.6604309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merdzhanova G, Gout S, Keramidas M. et al. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene. 2010;29:5392–403. doi: 10.1038/onc.2010.281. [DOI] [PubMed] [Google Scholar]
- 69.Ronsin C, Muscatelli F, Mattei MG. et al. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–202. [PubMed] [Google Scholar]
- 70.Iwama A, Yamaguchi N, Suda T. STK/RON receptor tyrosine kinase mediates both apoptotic and growth signals via the multifunctional docking site conserved among the HGF receptor family. EMBO J. 1996;15:5866–75. [PMC free article] [PubMed] [Google Scholar]
- 71.Moon H, Cho S, Loh TJ. et al. SRSF2 promotes splicing and transcription of exon 11 included isoform in Ron proto-oncogene. Biochim Biophys Acta. 2014;1839:1132–40. doi: 10.1016/j.bbagrm.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collesi C, Santoro MM, Gaudino G. et al. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16:5518–26. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–67. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 74.Wang HY, Kim H, Park S. et al. Analytical performance evaluation of the HPV OncoCheck assay for detection of high-risk HPV infection in liquid-based cervical samples. Exp Mol Pathol. 2019;106:149–156. doi: 10.1016/j.yexmp.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 75.Günthert U, Hofmann M, Rudy W. et al. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-l. [DOI] [PubMed] [Google Scholar]
- 76.Rudy W, Hofmann M, Schwartz-Albiez R. et al. The two major CD44 proteins expressed on a metastatic rat tumor cell line are derived from different splice variants: each one individually suffices to confer metastatic behavior. Cancer Res. 1993;53:1262–8. [PubMed] [Google Scholar]
- 77.Loh TJ, Moon H, Cho S. et al. SC35 promotes splicing of the C5-V6-C6 isoform of CD44 pre-mRNA. Oncol Rep. 2014;31:273–9. doi: 10.3892/or.2013.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–12. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- 79.Apostolatos H, Apostolatos A, Vickers T. et al. Vitamin A metabolite, all-trans-retinoic acid, mediates alternative splicing of protein kinase C deltaVIII (PKCdeltaVIII) isoform via splicing factor SC35. J Biol Chem. 2010;285:25987–95. doi: 10.1074/jbc.M110.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Syafruddin SE, Rodrigues P, Vojtasova E. et al. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma. Nat Commun. 2019;10:1152. doi: 10.1038/s41467-019-09116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang B, Guo DD, Zheng JY. et al. Expression of KLF6-SV2 in colorectal cancer and its impact on proliferation and apoptosis. Eur J Cancer Prev. 2018;27:20–26. doi: 10.1097/CEJ.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J, Hu Z, Pabon K. et al. Caffeine regulates alternative splicing in a subset of cancer-associated genes: a role for SC35. Mol Cell Biol. 2008;28:883–95. doi: 10.1128/MCB.01345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masters CL, Simms G, Weinman NA. et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;2:4245–9. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D'Souza I, Schellenberg GD. Regulation of tau isoform expression and dementia. Biochim Biophys Acta. 2005;1739:104–15. doi: 10.1016/j.bbadis.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 85.Goedert M, Jakes R. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta. 2005;1739:240–50. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 86.Donev R, Newall A, Thome J. et al. A role for SC35 and hnRNPA1 in the determination of amyloid precursor protein isoforms. Mol Psychiatry. 2007;12:681–90. doi: 10.1038/sj.mp.4001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lefebvre S, Burlet P, Liu Q. et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–9. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 88.Feldkötter M, Schwarzer V, Wirth R. et al. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–68. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moon H, Jang HN, Liu Y. et al. Activation of Cryptic 3' Splice-Sites by SRSF2 Contributes to Cassette Exon Skipping. Cells. 2019;8:696. doi: 10.3390/cells8070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moon H, Cho S, Loh TJ. et al. SRSF2 directly inhibits intron splicing to suppresses cassette exon inclusion. BMB Rep. 2017;50:423–428. doi: 10.5483/BMBRep.2017.50.8.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wee CD, Havens MA, Jodelka FM. et al. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS One. 2014;9:e115205. doi: 10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barret JM, Salles B, Provot C. et al. Evaluation of DNA repair inhibition by antitumor or antibiotic drugs using a chemiluminescence microplate assay. Carcinogenesis. 1997;18:2441–5. doi: 10.1093/carcin/18.12.2441. [DOI] [PubMed] [Google Scholar]
- 93.McFarlane M, MacDonald AI, Stevenson A. et al. Human Papillomavirus 16 Oncoprotein Expression Is Controlled by the Cellular Splicing Factor SRSF2 (SC35) J Virol. 2015;89:5276–87. doi: 10.1128/JVI.03434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]