Abstract
Background:
Molecular markers of WHO grade II/III glioma are known to have important prognostic and predictive implications and may be associated with unique imaging phenotypes. The purpose of this study is to determine whether three clinically relevant molecular markers identified in gliomas—IDH, 1p/19q, and MGMT status—show distinct quantitative MRI characteristics on FLAIR imaging.
Methods:
Sixty-one patients with grade II/III gliomas who had molecular data and MRI available prior to radiation were included. Quantitative MRI features were extracted that measured tissue heterogeneity (homogeneity and pixel correlation) and FLAIR border distinctiveness (edge contrast; EC). T-tests were conducted to determine whether patients with different genotypes differ across the features. Logistic regression with LASSO regularization was used to determine the optimal combination of MRI and clinical features for predicting molecular subtypes.
Results:
Patients with IDH wildtype tumors showed greater signal heterogeneity (p = 0.001) and lower EC (p = 0.008) within the FLAIR region compared to IDH mutant tumors. Among patients with IDH mutant tumors, 1p/19q co-deleted tumors had greater signal heterogeneity (p = 0.002) and lower EC (p = 0.005) compared to 1p/19q intact tumors. MGMT methylated tumors showed lower EC (p = 0.03) compared to the unmethylated group. The combination of FLAIR border distinctness, heterogeneity, and pixel correlation optimally classified tumors by IDH status.
Conclusion:
Quantitative imaging characteristics of FLAIR heterogeneity and border pattern in grade II/III gliomas may provide unique information for determining molecular status at time of initial diagnostic imaging, which may then guide subsequent surgical and medical management.
Keywords: grade II/III gliomas, texture analysis, IDH, 1p/19q, Magnetic Resonance Imaging
Introduction
WHO grade II and III gliomas include a heterogeneous group of infiltrative neoplasms with astrocytic and oligodendroglial morphology. These tumors have a wide range of both progression-free survival (PFS) and overall survival (OS); some respond to therapy with OS greater than 13 years, while others precipitously progress to glioblastoma (GBM) [1,2]. Although the prognosis for grade II/III gliomas was previously thought to depend mostly on histopathological grade, it is now recognized that OS is highly influenced by specific molecular markers.
Over the past several decades, tremendous progress has been made in revealing the underlying molecular alterations that influence prognosis in grade II/III gliomas [3]. Recent studies have shown that the molecular characterization of glioma [i.e., isocitrate dehydrogenase (IDH), codeletion of chromosome arms 1p and 19q (1p/19q), and methylguanine methyltransferase promoter methylation (MGMT status)] is more robust for the prediction of clinical outcomes in comparison to histological classification [1,3–7]. Codeletion of 1p/19q was initially determined to be a prognostic marker associated with oligodendroglial morphology and shown to predict response to chemoradiation [8–11]. IDH gene mutations are thought to be an early step in gliomagenesis, and are estimated to occur in 79% to 94% of grade II/III gliomas [12]. The status of IDH mutation [i.e., IDH mutant (mt) vs IDH wild-type (wt) tumors] has also been well-validated as both a prognostic and predictive marker in glioma[5,13–16]. In fact, it has been shown that most adult grade II/III glioma patients who are IDH-wt resemble GBM, molecularly and clinically [1,17] and all are presumed to have 1p19q intact [17]. In addition, MGMT promoter methylation status has known prognostic and predictive value. In patients with grade III gliomas, response to alkylating chemotherapy is better with MGMT promoter methylation; however, this may be due to the common co-occurrence of IDH mutations and MGMT promoter methylation [7,18].
Because both molecular and histological classification require invasive measures (i.e., biopsy/resection), there is increased interest in identifying non-invasive surrogates for tumor genotypes [19–25]. In recent years, distinct imaging features have been identified on MRI and other imaging modalities (i.e., MR spectroscopy) that have shown initial promise for classifying tumors by specific molecular markers [26,27]. Qi et al. showed that IDH-mt tumors were more likely to have less contrast enhancement (CE) in patients with astrocytic tumors than IDH-wt tumors [26]. Also, Tietze et al. showed in vivo MRS for 2-hydroxyglutarate identified IDH status in almost 88% of the gliomas [27]. In previous studies, it was also noted that IDH-mt tumors, regardless of the tumor grade, were more likely to have sharp tumor margins and homogeneous signal intensity relative to IDH-wt tumors [26,28] On the contrary, “indistinct” tumor margins have been described in patients with 1p/19q codeleted tumors [29–31]. Similarly, 1p/19q codeleted tumors are reported to have more heterogeneity of T2 signal compared to intact tumors [32,33]. It has been proposed that the increased heterogeneity in 1p/19q codeleted tumors is associated with calcification and paramagnetic susceptibility, which increases heterogeneity of the T2 signal [32,33]. In addition to IDH and 1p/19q codeletion status, MGMT status has been associated with specific T2 features [34]. Noushmehr et al. found that in GBM, methylated tumors are more likely to show a higher level of tumor infiltration and more indistinct borders relative to unmethylated tumors [16]. However, T2/FLAIR features associated with MGMT status have not been studied in patients with grade II/III gliomas.
While previous studies have reported that IDH, 1p/19q, and MGMT status appear phenotypically different on conventional imaging in subsets of glioma patients [35,36], few studies have evaluated all three markers simultaneously in patients with grade II or III glioma. Importantly, most of the existing studies have been based on qualitative descriptions, which may vary greatly across readers and sites, explaining some of the contradictory findings described in previous reports [26]. Thus, there is a need for reliable, quantitative imaging features that can differentiate tumor genotypes without clinician and/or researcher bias.
In this study, we investigate the ability of quantitative FLAIR and T1-post-contrast images to aid in differentiating molecular subtypes of patients with grade II/III gliomas. In particular, given recent evidence that features of the FLAIR signal may be especially informative, we introduce several novel imaging parameters obtained from FLAIR texture analysis that measure the heterogeneity (i.e., signal homogeneity/heterogeneity and pixel correlation) and border patterns (Edge Contrast; EC) within the FLAIR signal. We test the ability of our quantitative texture features to differentiate molecular subtypes compared to FLAIR and CE volumes. We accomplish this by comparing all patients who are IDH-wt to those who are IDH-mt. Next, we test the contribution of 1p19q status by comparing patients who are IDH-mt and 1p19q codeleted to those who are IDH-mt and 1p19q intact. Patients who are IDH-wt are excluded from this analysis due to evidence that all are 1p19q intact [1]. Finally, in an exploratory analysis, we examine our quantitative imaging features in patients who are MGMT methylated versus unmethylated. Based on our previous work [36] and other published studies [37], we hypothesize that: 1) measures of FLAIR heterogeneity will be greater in patients with IDH-wt compared to IDH-mt tumors. In those with IDH-mt tumor, we hypothesize that heterogeneity will be greater in 1p/19q-codeleted tumors compared to those with 1p/19q-intact tumors; 2) FLAIR borders will be less distinct in patients with IDH-wt compared to IDH-mt tumors. In those with IDH-mt, FLAIR borders will be less distinct in 1p/19q-codeleted compared to 1p/19q-intact tumors. Finally, we hypothesize that a combination of quantitative features derived from FLAIR and CE images will provide better classification of patient tumors according to molecular status compared to any single imaging feature.
Methods and Materials
Patients
This retrospective study was approved by the institutional review board. From 2010 to 2017, 115 patients with grade II/III gliomas were identified at our institution who had MR imaging, including 3D IR-SPGR pre- and post- contrast and 3D FLAIR sequences. Patients were excluded if they did not have the specific MR sequences performed at our institution prior to radiation (n = 42), there was significant artifact on imaging (n=3), or molecular information was not available (n= 9). Sixty-one patients with grade II/III gliomas met inclusion criteria to form the final study cohort (see figure 1). The final cohort included 32 males and 29 females, and the average age was 46.18 years with a range of 23 to 71 years (see table 1). All patients had undergone either biopsy (N = 20) or resection (N = 41) prior to MRI acquisition. However, all imaging was performed before start of radiation or chemotherapy.
Figure 1.
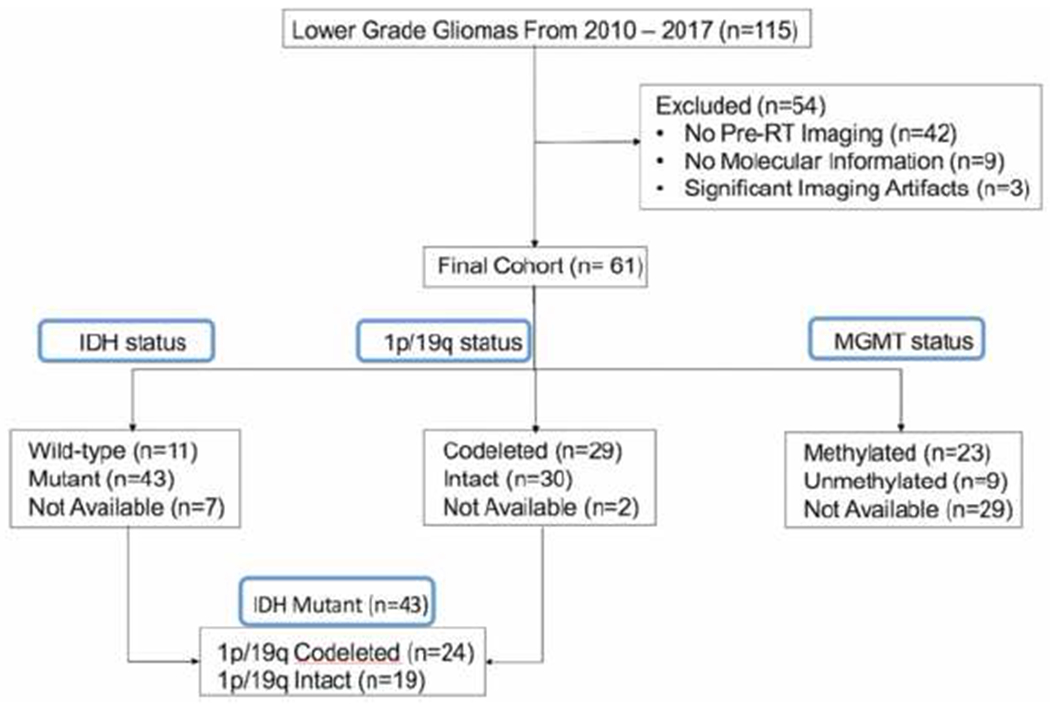
Consort diagram illustrates the exclusion and inclusion criteria and the final cohort.
Table 1.
Demographic and clinical characteristics of the patient sample.
| Characteristic | Total (N=61) |
|---|---|
| Sex – no. (%) | |
| Male | 32 (52.5) |
| Female | 29 (47.5) |
| Age – year | |
| Median | 45.0 |
| Range | 23.0–71.0 |
| Histopathology – no. (%) | |
| Oligodendroglioma | 19 (31.1) |
| Astrocytoma | 13 (21.3) |
| Anaplastic Astrocytoma | 12 (19.7) |
| Anaplastic Oligodendroglioma | 10 (16.4) |
| Oligoastrocytoma | 5 (8.2) |
| Anaplastic Oligoastrocytoma | 2 (3.3) |
| Tumor Grade – no. (%) | |
| Grade II | 35 (57.4) |
| Grade III | 26 (42.6) |
| Location – no. (%) | |
| Left hemisphere | 36 (59.0) |
| Right hemisphere | 25 (41.0) |
| 1p/19q status – no. (%) | |
| Codeleted | 29 (47.5) |
| Intact | 30 (49.2) |
| Not available | 2 (3.3) |
| IDH status – no. (%) | |
| Mutant | 43 (70.4) |
| Wild-type | 11 (18.0) |
| Not available | 7 (11.4) |
| MGMT status – no. (%) | |
| Methylated | 23 (37.7) |
| Unmethylated | 9 (14.8) |
| Not available | 29 (47.5) |
| Procedure – no (%) | |
| Resection | 41 (67.2) |
| Biopsy | 20 (32.8) |
Molecular Analysis
OncoScan microarray analysis (Affymetrix, Santa Clara, CA) or fluorescence in situ hybridization (FISH) was performed on formalin fixed paraffin embedded tissue to determine the 1p/19q status. For most patients, IDH status was determined by whole exome next generation sequencing on a panel of known cancer genes that included IDH1 and IDH2. Two patients had IDH status determined by immunohistochemistry for the R132H mutation. MGMT promoter methylation status was determined by Methylation-specific PCR. Two of the patients that were found to have the 1p/19q codeletion by FISH, prior to routine testing for IDH, were included in the 1p/19q codeletion and IDH-mt group. As shown in figure 1, at least one molecular marker was missing for a subset of patients (IDH status; n = 7, 1p/19q status; n = 2, MGMT status, n = 29).
Magnetic Resonance Imaging
All MRI scans were acquired on a 3.0 T GE Signa Excite HDx scanner using an 8-channel head coil. The imaging protocol included pre- and post-gadolinium 3D T1-weighted inversion recovery-spoiled gradient recalled echo (IR-SPGR) with TE/TR = 2.8/6.5 ms, TI = 450 ms, flip angle = 8 degrees. FOV = 24 cm, voxel size = 0.93×0.93×1.2 mm, and a 3D T2-weighted FLAIR sequence with TE/TR = 126/6000 ms, TI = 863, FOV = 24 cm, voxel size = 0.93×0.93×1.2 mm. Prior to analysis, raw data were corrected for bias field and distortion [38]. Subsequently, correction for patient motion was carried out using in-house software. The pre- and post-contrast 3D IR-SPGR and FLAIR images were registered to each other using rigid body registration.
Image Pre- and Post- Processing
Contrast-enhanced volumes (CEVOL) and FLAIR hyperintense volumes (FLAIRVOL) were segmented semi-automatically (Amira software package, Visage Imaging) on the co-registered post-contrast 3D IR-SPGR and FLAIR images, while regions of necrosis and the resection cavity were excluded. Volumes of interest (VOIs) were drawn for the entire tumor volume by 2 trained image analysts (R.D. and S.H.) and approved by a board-certified neuroradiologist with expertise in neuro-oncology (N.F.). Quantitative texture analyses were conducted to measure the signal heterogeneity and FLAIR borders using three-dimensional co-occurrence matrix (3D-COM) and EC, respectively.
Signal heterogeneity:
3D-COM was applied to extract the heterogeneity features from the FLAIR hyperintense ROI. Histogram normalization was applied to the MR images when generating the gray-level co-occurrence matrix (GLCM) prior to the texture analysis [39]. This method provides statistics on the probability density function Pd,θ (i, j), that is the probability of finding a joint relationship between a pair of pixels composed of a central pixel of gray level i and a neighboring one of gray level j. These two pixels are separated by a distance d (pixel distance) and angle θ of one of the four values (0°, 45°, 90° and 135°) [40]. Texture features extracted by the 3D-COM method are represented by homogeneity and pixel correlation. Homogeneity is a metric that indicates the level of homogeneity/heterogeneity of the FLAIR signal on a global level (i.e., within the entire VOI), whereas pixel correlation indicates the level of homogeneity/heterogeneity on a “local” level (i.e., across adjacent pixels). Higher homogeneity indicates more uniformity of the entire signal, and higher pixel correlation indicates more homogeneity across neighboring voxels.
Border distinctness:
EC is defined as the gradient magnitude of the lesion edges, where higher EC indicates a sharper and more distinct border compared to lower EC. A 3D analysis was applied to the FLAIR VOI to enhance the local precision and decrease the partial volume effect [41–44]. The technique of EC extraction has been described in our previous study [45], where the morphological operations of erosion and dilation were applied to the FLAIRVOL binary mask using a spherical 3D mask. Then, the contour of the FLAIRVOL binary mask was drawn in 3D indexing the surface of the FLAIR VOL lesion. The gradients of the FLAIR image were calculated and were overlaid on the surface of the 3D binary mask to create the hyperintense surface representing EC.
Quantitative MRI measurements including FLAIRVOL, CEVOL, ratio of FLAIRVOL/CEVOL, homogeneity, pixel correlation, and EC were calculated for each patient. Despite the complexity of the model and processing pipeline, the parameters are generated in a very time efficient manner within the seconds.
Statistical Analysis
Independent t-tests were used to test for differences in continuous variables as functions of IDH, 1p19q, and MGMT status. A chi square test was used to test for differences in sex distribution. A generalized linear model (GLM) was used to test the association between imaging paramaters and molecular status of the patient using a logit linear function. The Least Absolute Shrinkage and Selection Operator (LASSO) was used to select imaging predictors for use in a logistic regression model in order to predict each genotype status. Model selection was based on the Akaike Information Criteria (AIC). A second model was run that included age and gender to determine the additional value of adding these variables to the prediction of each marker.
Results
Patients with IDH-wt tumors were significantly older than the patients with IDH-mt tumors (p = 0.006). Within the IDH-mt group, patients with 1p/19q co-deleted tumors were also older than patients with 1p/19q intact tumors (p = 0.011). There were no significant age differences between the two MGMT groups (p = 0.42). There were no significant differences in sex distribution across IDH and 1p/19q groups (p > 0.05), whereas there was a higher proportion of females in the methylated (1 male vs 8 female) compared to the unmethylated (10 male vs 13 female) group (p = 0.02) (see table 2).
Table 2.
Imaging parameters for the three molecular subgroups; 1) IDH wildtype vs mutant, 2) IHD mutant/1p/19q codeleted vs IDH mutant/1p/19q intact, and 3) MGMT methylated vs MGMT unmethylated.
| IDH wildtype (n=11) Mean(range) | IDH mutant (n = 43) Mean(range) | p-value | Test Statistic | HDH-mt 1p/19q codeleted (n = 24) Mean(range) | IDH-mt 1p/19q intact (n = 19) Mean(range) | p-value | Test Statistic | MGMT methylated Mean( range) (n = 23) | MGMT unmethylated Mean(range) (n = 9) | p-value | Test Statistic | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 57.09(27–71) | 43.23(23–71) | 0.006** | t=320 | 44.2(30–71) | 38.7(23–67) | 0.011* | t=2.81 | 48.6(32–65) | 48.6(23–67) | 0.44 | t=−0.83 |
| Sex | M, F = 3, 8 | M, F = 25, 18 | 0.181 | r = 3.14 | M, F = 13, 11 | M, F = 13, 6 | 0.34 | χ2 = 2.31 | M, F = 1, 8 | M, F = 13, 10 | 0.02* | χ2 = 7.39 |
| FLAIRVOL | 29052(1445–75116) | 34192(0–218140) | 0.63 | t=−0.47 | 20774(0–218140) | 27783(146–146030) | 0.45 | t=0.88 | 26997(1445–83239) | 33727(3461–218140) | 0.31 | t=−0.48 |
| CEVOL | 2203(0–7077) | 3090(0–27664) | 0.51 | t=−0.66 | 3420(0–27664) | 2171(0–24497) | 0.35 | t=−1.11 | 2212(0–7077) | 4033(0–27664) | 0.17 | t=−0.95 |
| CEVOL / FLAIRVOL | 0.17(0–1.15) | 0.14(0–1.82) | 0.79 | t=0.26 | 0.22(0–2.05) | 0.11(0–1.15) | 0.38 | t=0.98 | 0.19(0–1.15) | 0.12(0–052) | 0.31 | t=0.49 |
| Edge Contrast | 544.7(72–1561) | 1058.5(46–2741) | 0.008** | t=−2.97 | 632(46–2741) | 1369(72–1967) | 0.005** | t=−328 | 643(72–1368) | 1055(142–2741) | 0.03* | t=−1.88 |
| Homogeneity | 1.9(02–4.5) | 3.1(0.3–6.3) | 0.013* | t=−2.75 | 2.4(0.2–5.1) | 3.8(0.2–6.3) | 0.002** | t=−3.46 | 2.5(0.8–4.5) | 2.6(0.2–5.2) | 0.44 | t=−0.14 |
| Pixel Correlation | 6.8(0.9–11.3) | 11.8(2.2–33.3) | 0.001** | t=−3.65 | 12.3(2.2–28.5) | 13.2(0.9–33.3) | 0.17 | t=−1.26 | 8.1(4.8–14.8) | 9.8(0.9–18.3) | 0.10 | t=−1.26 |
p-value < 0.01
p-value < 0.05
IDH status:
IDH-wt status was detected in 11/54 (21%) of tumors. FLAIR borders in the IDH-wt group showed lower EC compared to the IDH-mt group (p = 0.008). Homogeneity of the FLAIRVOL was higher in patients with IDH-mt tumors compared to patients with IDH-wt tumors (p = 0.013). Pixel correlations in the hyperintense FLAIRVOL were also higher in the IDH-mt tumors compared to the IDH-wt (p = 0.001) group. There were no significant differences in the FLAIRVOL, CEVOL, or CEVOL/FLAIRVOL between these two groups.
1p/19q status:
The ratio of patients with IDH-mt-1p/19q-codeleted to IDH-mt-1p/19q-intact tumors was 24:19 (55%). Homogeneity and EC were lower in patients with IDH-mt-1p/19q-codeleted tumors compared to those with IDH-mt-1p/19q-intact tumors (p = 0.002 and p = 0.005, respectively).
MGMT status:
There were data on MGMT promoter methylation for 32/61 patients (52%). Twenty-three out of 32 (71%) patients with avaialbe MGMT promoter status were in the MGMT methylated group (see table 1). MGMT methylated tumors showed lower EC compared to MGMT unmethylated tumors (p = 0.03).
A pictorial summary of the univariate results for each texture feature by molecular subgroup is provided in table 3.
Table 3.
Relationship between imaging parameters and molecular information based on the univariate t-tests.
| Molecular subgroups | IDH | 1p/19q | MGMT | |||
|---|---|---|---|---|---|---|
| Imaging parameters | wildtype | mutant | Codeleted | intact | methylated | unmethylated |
| Edge Contrast | ↓↓ | ↑↑ | ↓↓ | ↑↑ | ↓↓ | ↑↑ |
| Homogeneity | ↓↓ | ↑↑ | ↓↓ | ↑↑ | - | - |
| Pixel Correlation | ↓↓ | ↑↑ | - | - | - | - |
↓↓ significantly lower (p-value < 0.05), ↑↑ significantly higher (p-value < 0.05), - no significant changes (p-value > 0.1)
Logistic Regression Model
EC, homogeneity, and pixel correlation survived as predictors of IDH status among all the quantitative imaging features extracted using logistic regression with LASSO regularization (ECcoefficient = −6.36e-4, homogeneitycoefficient = −8.98e-05, pixel correlationcoefficient = −6.76e-02) (see figure 3). The stepwise AIC of the model for IDH status prediction was 51.7 using these three imaging parameters. When clinical/demographic data were also included in the model (i.e., age, sex), five features showed non-zero coefficients for predicting IDH status (agecoefficient = 4.62e-02, sexcoefficient = −1.02e-1, ECcoefficient = −3.74e-4, homogeneitycoefficient = −7.69e-05, and pixel correlationcoefficient = −4.14e-02). The stepwise AIC of this model for IDH status prediction was 44.0. The lower AIC of the second model indicates that the quality of the multivariate model for prediction of IDH status was slightly better when including age and sex with the imaging parameters compared to using only the imaging parameters. There were no non-zero coefficients in the logistic regression model using the imaging and clinical features to predict the status of 1p/19q and MGMT tumors.
Figure 3.
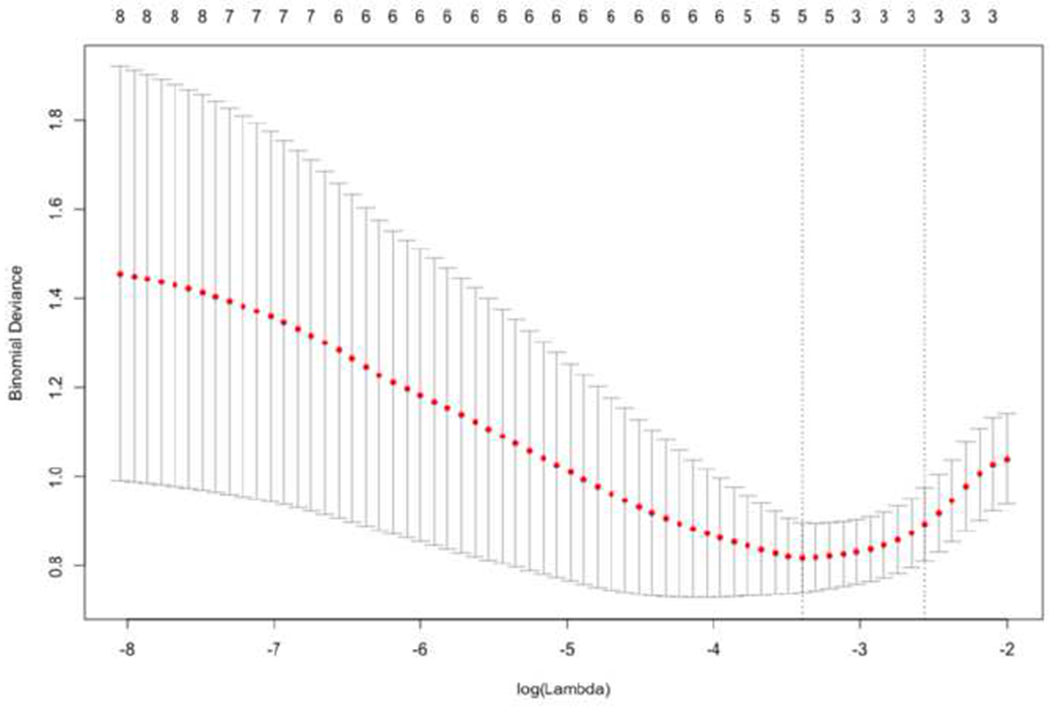
Depicts the features selection and regularization using LASSO on multiple logistic regression for predicting the IDH-status. The tuning parameter (lambda) has the most parsimonious model where its error is within one standard error of the minimum bionomial deviance. The optimum lambda occurs using three selected features (homogeneity, EC, and pixel correlation).
Discussion
In this study, we investigated whether three clinically validated molecular markers in patients with grade II/III gliomas—IDH, 1p19q, and MGMT status—have unique imaging phenotypes. Using novel quantitative imaging parameters of FLAIR homogeneity/heterogeneity and border patterns, we demonstrated that whereas these three groups do not differ in FLAIR or CE volumes, they differ across a range of FLAIR texture features that could help to determine genotype and facilitate treatment planning prior to surgery.
Higher signal homogeneity indicates a more uniform signal throughout the FLAIR region, whereas higher pixel correlation indicates more uniform texture at a local level (i.e., within neighboring regions of the tumor). Higher EC indicates more distinct and sharp borders surrounding the FLAIR region. Heterogeneous FLAIR signal may result from hemorrhage, necrosis, or calcification, whereas poorly defined FLAIR borders may reflect tumor infiltration from the tumor bed to adjacent tissue [31]. In this study, we apply these texture features to patients with grade II/III gliomas and demonstrate their utility in delineating the molecular subtypes described below.
IDH status:
Our results demonstrate that patients with IDH-wt tumors show greater global and local heterogeneity within the FLAIR hyperintense region in comparison to IDH-mt tumors. These findings are concordant with a previous study from Qi et al., which reported that IDH-mt tumors were more likely to show more homogeneous signal intensity compared to IDH-wt tumors [26]. In addition, we found that patients with IDH-wt tumors showed less distinct FLAIR borders compared with patients with IDH-wt tumors. These findings are supported by several other studies that have shown less distinct borders in IDH-wt tumors based on visual analysis [26,28]. Previous studies have shown the association between an “invasive” FLAIR border and tumor growth [46] in IDH-wt tumors. Interestingly, we found that a combination of imaging features including global and local FLAIR heterogeneity combined with FLAIR border patterns provides the best classification of tumors based on IDH status. As in previous studies, patients with IDH-wt tumors were older than IDH-mt group. Therefore, age also contributed to the model. These findings indicate that multiple feature of the FLAIR signal should be considered simultaneously when determining tumor genotypes—which may be easier to achieve with quantitative measures.
1p/19q codeletion:
We demonstrate that among patients with IDH-mt tumors, those with 1p/19q codeleted tumors show a more heterogeneous FLAIR signal and less distinct borders compared to the intact group. Consistent with our findings, previous studies have shown that a more heterogeneous intratumoural signal is present in patients with 1p/19q codeleted anaplastic tumors [31][32,33]. Similar findings were also reported when determining heterogeneity using a qualitative, visual analysis of tumor characteristics [32,33]. Our data show that patients with IDH-mt-1p/19q-intact tumors showed less heterogeneity in the FLAIR hyperintense region compared with patients who had IDH-mt-1p/19-codeleted tumors (see figure 2). Although the reason for the lower heterogeneity in the IDH-mt-1p/19q-intact tumors compared to the co-deleted tumors is unclear, calcification and paramagnetic susceptibility present in co-deleted tumors may increase heterogeneity in this subset of otherwise IDH-mt patients [33]. Although the goal of this paper was to characterize the imaging phenotypes associated with each individual marker, these data also speak to the value of stratifying patients by multiple molecular markers in larger patient cohorts.
Figure 2.
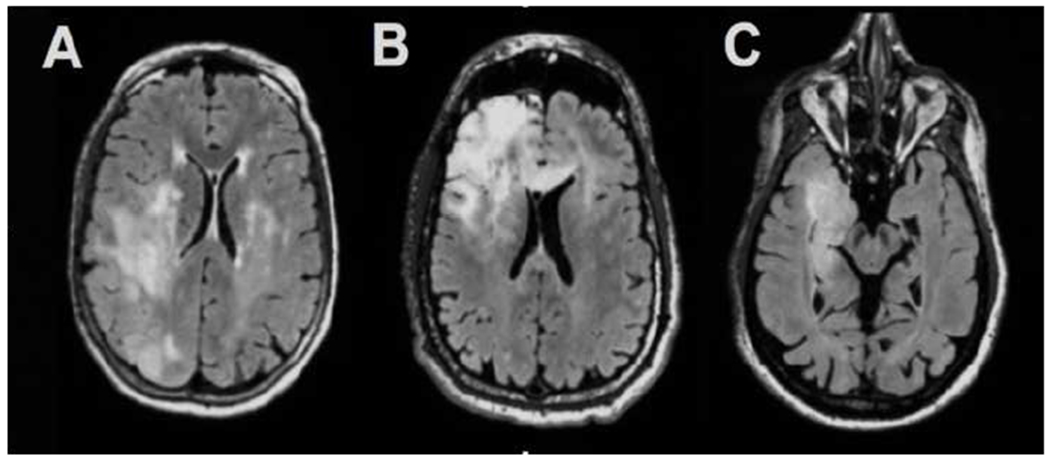
A) IDH-wildtype tumor demonstrating very indistinct borders and heterogeneous signal on FLAIR imaging, B) IDH-mutant-1p/19q codeleted tumor demonstrating heterogeneous signal on FLAIR imaging and C) IDH-mutant-1p/19q-intact tumor demonstrating homogeneous signal on FLAIR imaging.
MGMT prediction:
In this study, patients with MGMT methylated tumors showed less distinct borders compared to patients with MGMT unmethylated tumors. MGMT promoter methylation status predicts treatment response to alkylating agent chemotherapy, and may be used in conjunction with IDH status for guidance in clinical decision-making [18,34,47–49]. In terms of imaging characteristics, Drabycz et al. found associations between texture on FLAIR images and MGMT methylation status in patients with GBM [50]. We extend the literature by demonstrating differences in texture features in patients with grade II/III gliomas. Although it is unclear what advantage MGMT methylation status holds in patients with grade II/III gliomas, our findings of differences in EC between the methylated and unmethylated patients is of interest and could help to guide further hypotheses about the relevance of this marker in this group of patients.
With the rapid expansion of radiogenomics, there is increased interest in identifying non-invasive means of determining a patient’s tumor genotype. However, previous studies have primarily classified brain tumors based on visual analysis of imaging features, which may not provide a reliable, systematic characterization of texture features. The current work introduces quantitative imaging metrics to capture FLAIR signal homogeneity/heterogeneity and border distinctness. These measures may serve as adjuncts to standard clinical analysis and interpretation and provide information regarding the patient’s molecular status prior to biopsy/resection. These quantitative features may be particularly useful in large-scale clinical trials where measures must be quantifiable, reproducible across sites, and free of reader bias.
This study has some limitations that should be noted. First, our sample size is modest compared to many large-scale studies. Nevertheless, the fact that we demonstrate group differences as a function of genotype in a relatively small sample may speak to the robustness of our measures. Second, we did not have MGMT promoter methylation status for many of the patients, limiting the conculsions that we can draw about these data. However, we hope that are preliminary results with MGMT status will motivate future studies with robust sample sizes. As expected in patients with grade II/III gliomas, our groups were also unbalanced with respect to IDH status, with a lower number of IDH-wt tumors. However, our numbers are commensurate with those reported in the literature (5–15%) [51,52]. Third, our patients were not treatment-naïve, as many had undergone biopsy or subtotal resection. Therefore, our findings will need to be further validated in the pre-operative setting. Fourth, although several studies, including ours, have proposed novel imaging markers to classify molecular subtypes, an important extension of this work will be to derive reliable cut-offs for each imaging parameter that would be invariant to sample. However, a very large multi-site study would be needed to systematically address this. Finally, we did not include survival data in our study as most patients with grade II/III gliomas do not reach meaningful progression-free or overall survival endpoints over a short time frame (i.e., 7 years in our study). However, we intend to follow this patient cohort and report on the prognostic value of our imaging features overall and within subtypes to address specific questions related to risk statification (e.g., do patients with IDH-mt tumors with less distinct borders have poorer outcomes than patients with IDH-mt tumors with sharper borders?) in a longitudinal study. Although our current sample size was not large enough to subdivide patients within each molecular subtype by tumor grade, we plan to investigate the differences in imaging characteristics of patients with grade II versus III gliomas separately in a larger prospective study.
In conclusion, our study demonstrates the utility of novel, quantitative FLAIR texture features, specifically signal heterogeneity and border sharpness, that may serve as powerful imaging biomarkers for determining tumor molecular status in patients with grade II/III gliomas.
Funding and Acknowledgement
We would like to thank patients at the UCSD Moores Cancer Center Neuro-Oncology Program for their generous participation. We also acknowledge the funding from National Institutes of Health grants R01NS065838 (C.R.M.); National Institutes of Health UL1TR000100 (J.A.H.) and KL2TR00144 (J.A.H.); American Cancer Society Award ACS-IRG 70–002 (J.A.H.) and American Cancer Society RSG-15–229-01-CCE (C.R.M).
Funding:
National Institutes of Health grants R01NS065838 (C.R.M.); National Institutes of Health UL1TR000100 (J.A.H.) and KL2TR00144 (J.A.H.); American Cancer Society Award ACS-IRG 70–002 (J.A.H.) and American Cancer Society RSG-15–229-01-CCE (C.R.M).
Footnotes
Conflict of Interest:
None of the authors have any personal or financial interest in drugs, materials, or devices described in this submission.
References
- [1].Cancer Genome Atlas Research Network, Brat DJ, Verhaak, Aldape KD, Yung WKA, Salama SR et al. (2015) Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England Journal of Medicine, 372, 2481–98. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Buckner J, Giannini C, Eckel-Passow J, Lachance D, Parney I, Laack N et al. (2017) Management of diffuse low-grade gliomas in adults - use of molecular diagnostics. Nature Reviews Neurology, 13, 340–51. 10.1038/nrneurol.2017.54 [DOI] [PubMed] [Google Scholar]
- [3].Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H et al. (2015) Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. New England Journal of Medicine, 372, 2499–508. 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Y, Zhang T, Li S, Fan X, Ma J, Wang L et al. (2015) Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. European Journal of Neurology, 22, 348–54. 10.1111/ene.12578 [DOI] [PubMed] [Google Scholar]
- [5].Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F et al. (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 27, 4150–4. 10.1200/JCO.2009.21.9832 [DOI] [PubMed] [Google Scholar]
- [6].Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al. (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Research, 66, 9852–61. 10.1158/0008-5472.CAN-06-1796 [DOI] [PubMed] [Google Scholar]
- [7].van den Bent MJ, Erdem-Eraslan L, Idbaih A, de Rooi J, Eilers PHC, Spliet WGM et al. (2013) MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic Oligodendrogliomas and Oligoastrocytomas. A report from EORTC study 26951. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 19, 5513–22. 10.1158/1078-0432.CCR-13-1157 [DOI] [PubMed] [Google Scholar]
- [8].Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W and Collins VP (1994) Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. The American Journal of Pathology, 145, 1175–90. [PMC free article] [PubMed] [Google Scholar]
- [9].Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al. (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Research, 66, 9852–61. 10.1158/0008-5472.CAN-06-1796 [DOI] [PubMed] [Google Scholar]
- [10].Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J et al. (2013) Phase III Trial of Chemoradiotherapy for Anaplastic Oligodendroglioma: Long-Term Results of RTOG 9402. Journal of Clinical Oncology, 31, 337–43. 10.1200/JCO.2012.43.2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van den Bent MJ, Brandes AA, Taphoorn MJB, Kros JM, Kouwenhoven MCM, Delattre J-Y et al. (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 31, 344–50. 10.1200/JCO.2012.43.2229 [DOI] [PubMed] [Google Scholar]
- [12].Watanabe T, Nobusawa S, Kleihues P and Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. The American Journal of Pathology, 174, 1149–53. 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al. (2009) IDH1 and IDH2 Mutations in Gliomas. New England Journal of Medicine, 360, 765–73. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A et al. (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathologica, 118, 469–74. 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- [15].Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, Powell DR et al. (2012) A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Research, 22, 2339–55. 10.1101/gr.132738.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell, 17, 510–22. 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica, 131, 803–20. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- [18].Wick W, Meisner C, Hentschel B, Platten M, Schilling A, Wiestler B et al. (2013) Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology, 81, 1515–22. 10.1212/WNL.0b013e3182a95680 [DOI] [PubMed] [Google Scholar]
- [19].Dean BL, Drayer BP, Bird CR, Flom RA, Hodak JA, Coons SW et al. (1990) Gliomas: classification with MR imaging. Radiology, 174, 411–5. 10.1148/radiology.174.2.2153310 [DOI] [PubMed] [Google Scholar]
- [20].Hajnal JV, Bryant DJ, Kasuboski L, Pattany PM, De Coene B, Lewis PD et al. (1992) Use of fluid attenuated inversion recovery (FLAIR) pulse sequences in MRI of the brain. Journal of Computer Assisted Tomography, 16, 841–4. [DOI] [PubMed] [Google Scholar]
- [21].Law M, Oh S, Babb JS, Wang E, Inglese M, Zagzag D et al. (2006) Low-Grade Gliomas: Dynamic Susceptibility-weighted Contrast-enhanced Perfusion MR Imaging—Prediction of Patient Clinical Response. Radiology, 238, 658–67. 10.1148/radiol.2382042180 [DOI] [PubMed] [Google Scholar]
- [22].Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T et al. (1999) Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. Journal of Magnetic Resonance Imaging, 9, 53–60. [DOI] [PubMed] [Google Scholar]
- [23].Forst DA, Nahed BV, Loeffler JS and Batchelor TT (2014) Low-Grade Gliomas. The Oncologist, 19, 403–13. 10.1634/theoncologist.2013-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Narang AK, Chaichana KL, Weingart JD, Redmond KJ, Lim M, Olivi A et al. (2017) Progressive Low-Grade Glioma: Assessment of Prognostic Importance of Histologic Reassessment and MRI Findings. World Neurosurgery, 99, 751–7. 10.1016/j.wneu.2016.04.030 [DOI] [PubMed] [Google Scholar]
- [25].Cuccarini V, Erbetta A, Farinotti M, Cuppini L, Ghielmetti F, Pollo B et al. (2016) Advanced MRI may complement histological diagnosis of lower grade gliomas and help in predicting survival. Journal of Neuro-Oncology, 126, 279–88. 10.1007/s11060-015-1960-5 [DOI] [PubMed] [Google Scholar]
- [26].QI S, YU L, LI H, OU Y, QIU X, DING Y et al. (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncology Letters, 7, 1895–902. 10.3892/ol.2014.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tietze A, Choi C, Mickey B, Maher EA, Parm Ulhøi B, Sangill R et al. (2018) Noninvasive assessment of isocitrate dehydrogenase mutation status in cerebral gliomas by magnetic resonance spectroscopy in a clinical setting. Journal of Neurosurgery, 128, 391–8. 10.3171/2016.10.JNS161793 [DOI] [PubMed] [Google Scholar]
- [28].Metellus P, Coulibaly B, Colin C, de Paula AM, Vasiljevic A, Taieb D et al. (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathologica, 120, 719–29. 10.1007/s00401-010-0777-8 [DOI] [PubMed] [Google Scholar]
- [29].Chawla S, Krejza J, Vossough A, Zhang Y, Kapoor GS, Wang S et al. (2013) Differentiation between Oligodendroglioma Genotypes Using Dynamic Susceptibility Contrast Perfusion-Weighted Imaging and Proton MR Spectroscopy. American Journal of Neuroradiology, 34, 1542–9. 10.3174/ajnr.A3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Weller M, Stupp R, Hegi ME, van den Bent M, Tonn JC, Sanson M et al. (2012) Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro-Oncology, 14 Suppl 4, iv100–108. 10.1093/neuonc/nos206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim JW, Park C-K, Park S-H, Kim YH, Han JH, Kim C-Y et al. (2011) Relationship between radiological characteristics and combined 1p and 19q deletion in World Health Organization grade III oligodendroglial tumours. Journal of Neurology, Neurosurgery & Psychiatry, 82, 224–7. 10.1136/jnnp.2009.178806 [DOI] [PubMed] [Google Scholar]
- [32].Megyesi JF, Kachur E, Lee DH, Zlatescu MC, Betensky RA, Forsyth PA et al. (2004) Imaging correlates of molecular signatures in oligodendrogliomas. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 10, 4303–6. 10.1158/1078-0432.CCR-04-0209 [DOI] [PubMed] [Google Scholar]
- [33].Jenkinson MD, du Plessis DG, Smith TS, Joyce KA, Warnke PC and Walker C (2006) Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain: A Journal of Neurology, 129, 1884–91. 10.1093/brain/awl108 [DOI] [PubMed] [Google Scholar]
- [34].Eoli M, Menghi F, Bruzzone MG, De Simone T, Valletta L, Pollo B et al. (2007) Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 13, 2606–13. 10.1158/1078-0432.CCR-06-2184 [DOI] [PubMed] [Google Scholar]
- [35].Darlix A, Deverdun J, Menjot de Champfleur N, Castan F, Zouaoui S, Rigau V et al. (2017) IDH mutation and 1p19q codeletion distinguish two radiological patterns of diffuse low-grade gliomas. Journal of Neuro-Oncology, 133, 37–45. 10.1007/s11060-017-2421-0 [DOI] [PubMed] [Google Scholar]
- [36].Delfanti R, Piccioni D, Bahrami N, Dale A, McDonald CR and Farid N (2017) Imaging Correlates for the 2016 Update on WHO Classification of Grade II/III Gliomas: Implications for IDH, 1p/19q and ATRX Status. Journal of Neuro-Oncology,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smits M and van den Bent MJ (2017) Imaging Correlates of Adult Glioma Genotypes. Radiology, 284, 316–31. 10.1148/radiol.2017151930 [DOI] [PubMed] [Google Scholar]
- [38].Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R et al. (2006) Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage, 30, 436–43. 10.1016/j.neuroimage.2005.09.046 [DOI] [PubMed] [Google Scholar]
- [39].Matlab - graycoprops [Internet]. [Google Scholar]
- [40].Mahmoud-Ghoneim D, Toussaint G, Constans JM and de Certaines JD (2003) Three dimensional texture analysis in MRI: a preliminary evaluation in gliomas. Magnetic Resonance Imaging, 21, 983–7. [DOI] [PubMed] [Google Scholar]
- [41].Nachimuthu DS and Baladhandapani A (2014) Multidimensional Texture Characterization: On Analysis for Brain Tumor Tissues Using MRS and MRI. Journal of Digital Imaging, 27, 496–506. 10.1007/s10278-013-9669-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li X, Xia H, Zhou Z and Tong L (2010) 3D texture analysis of hippocampus based on MR images in patients with alzheimer disease and mild cognitive impairment. 2010 3rd International Conference on Biomedical Engineering and Informatics, p. 1–4. 10.1109/BMEI.2010.5639520 [DOI] [Google Scholar]
- [43].Suoranta S, Holli-Helenius K, Koskenkorva P, Niskanen E, Könönen M, Äikiä M et al. (2013) 3D Texture Analysis Reveals Imperceptible MRI Textural Alterations in the Thalamus and Putamen in Progressive Myoclonic Epilepsy Type 1, EPM1. PLOS ONE, 8, e69905. 10.1371/journal.pone.0069905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang J, Yu C, Jiang G, Liu W and Tong L (2012) 3D texture analysis on MRI images of Alzheimer’s disease. Brain Imaging and Behavior, 6, 61–9. 10.1007/s11682-011-9142-3 [DOI] [PubMed] [Google Scholar]
- [45].Bahrami N, Dale A, White NS, Hattangadi-Gluth JA, Farid N, Piccioni DE et al. (2017) Edge Contrast of the FLAIR Hyperintense Region Predicts Survival in Patients with High Grade Gliomas Following Treatment with Bevacizumab. American Journal of Neuroradiology,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Claes A, Idema AJ and Wesseling P (2007) Diffuse glioma growth: a guerilla war. Acta Neuropathologica, 114, 443–58. 10.1007/s00401-007-0293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang Y, Fan X, Zhang C, Zhang T, Peng X, Li S et al. (2014) Anatomical specificity of O6-methylguanine DNA methyltransferase protein expression in glioblastomas. Journal of Neuro-Oncology, 120, 331–7. 10.1007/s11060-014-1555-6 [DOI] [PubMed] [Google Scholar]
- [48].Carrillo JA, Lai A, Nghiemphu PL, Kim HJ, Phillips HS, Kharbanda S et al. (2012) Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR American Journal of Neuroradiology, 33, 1349–55. 10.3174/ajnr.A2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bahrami N, Seibert TM, Karunamuni R, Bartsch H, Krishnan A, Farid N et al. (2017) Altered Network Topology in Patients with Primary Brain Tumors After Fractionated Radiotherapy. Brain Connectivity, 7, 299–308. 10.1089/brain.2017.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Drabycz S, Roldán G, de Robles P, Adler D, McIntyre JB, Magliocco AM et al. (2010) An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage, 49, 1398–405. 10.1016/j.neuroimage.2009.09.049 [DOI] [PubMed] [Google Scholar]
- [51].Cohen A, Holmen S and Colman H (2013) IDH1 and IDH2 Mutations in Gliomas. Current Neurology and Neuroscience Reports, 13, 345. 10.1007/s11910-013-0345-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Claus EB, Walsh KM, Wiencke J, Molinaro AM, Wiemels JL, Schildkraut JM et al. (2015) Survival and low grade glioma: the emergence of genetic information. Neurosurgical Focus, 38, E6. 10.3171/2014.10.FOCUS12367 [DOI] [PMC free article] [PubMed] [Google Scholar]


